Summary
Bacterial attachment to host surfaces is a pivotal event in the biological and infectious processes of both commensal and pathogenic bacteria, respectively. Serine-rich Repeat Proteins (SRRPs) are a family of adhesins in Gram-Positive bacteria that mediate attachment to a variety of host and bacterial surfaces. As such, they contribute towards a wide-range of diseases including sub-acute bacterial endocarditis, community-acquired pneumonia, and meningitis. SRRPs are unique in that they are glycosylated, require a non-canonical Sec-translocase for transport, and are largely composed of a domain containing hundreds of alternating serine residues. These serine-rich repeats are thought to extend a unique non-repeat (NR) domain outward away from the bacterial surface to mediate adhesion. Thus far, NR domains have been determined to bind to sialic acid moieties, keratins, or other NR domains of a similar SRRP. This review summarizes how this important family of bacterial adhesins mediates bacterial attachment to host and bacterial cells, contributes to disease pathogenesis, and might be targeted for pharmacological intervention or used as novel protective vaccine antigens. This review also highlights recent structural findings on the NR domains of these proteins.
Keywords: Gram-positive bacteria, Serine-rich repeat proteins, pathogenesis, adhesion, biofilm, review
Introduction
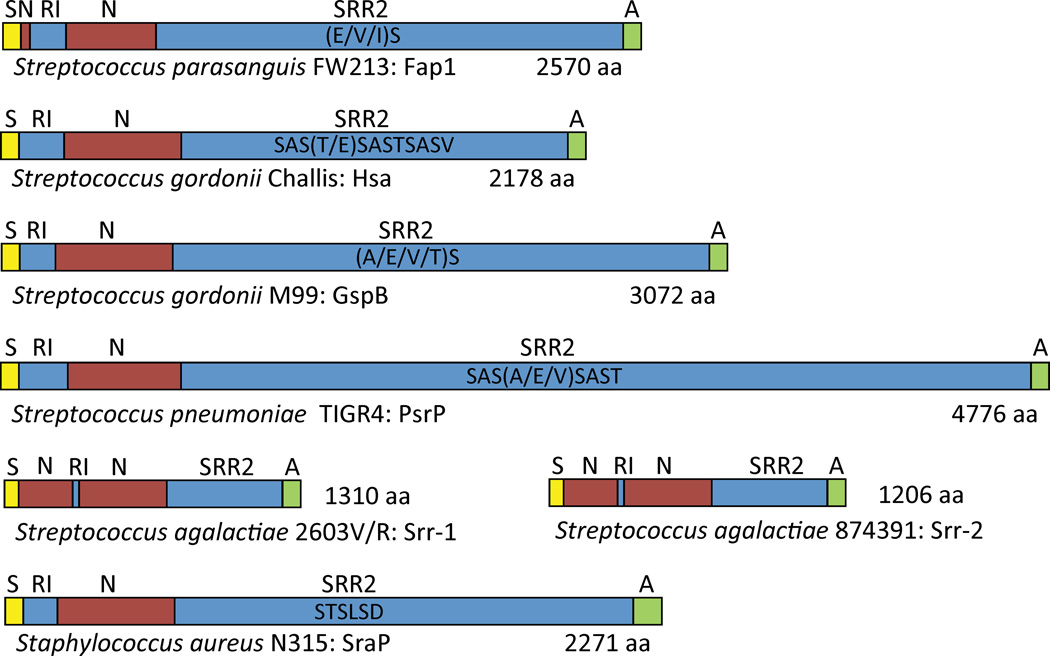
Serine-rich repeat proteins (SRRPs) are a family of large, surface-exposed proteins exclusive to Gram-positive bacteria and are predominately found within the oral streptococci(Stephenson et al., 2002, Wu & Fives-Taylor, 1999, Zhou et al., 2008, Wu et al., 1998, Froeliger & Fives-Taylor, 2001, Rigel & Braunstein, 2008, Takahashi et al., 2002, Handley et al., 2005, Bensing & Sullam, 2002, Lauer et al., 2005, Zhou & Wu, 2009, Vickerman et al., 2007, Xu et al., 2007, Glaser et al., 2002, Tettelin et al., 2001, Baba et al., 2002, Zhang et al., 2003, Takeuchi et al., 2005).Scanning electron microscopy suggests that SRRPs form large stalks or fimbriae-like structures, similar to the recently characterized Gram-positive pilus (Lauer et al., 2005), that extend outward from the bacterial surface to mediate adhesion (Wu et al., 1998, Wu & Fives-Taylor, 1999). Defining features of SRRPs includes two serine-repeat regions (SRR), one of which comprises the majority of the protein, one or two species unique non-repeat regions (NR domain) near the N-terminal domain that mediate adhesion, and aC-terminal cell wall anchor domain (Figure 1). SRRs are typically composed of alternating serine residues, most frequently separated by an alanine, valineor threonine. Importantly, there are O-linked glycoconjugates attached to the SRRS, this modification occurs in the cytoplasm and is required for protein stability(Bensing et al., 2004, Takamatsu et al., 2004a). Export of SRRPs onto the bacterial surface occurs through a dedicated non-canonical Sec translocase, Sec-Y2A2, following recognition of an extended atypical signal sequence peptide at the N-terminus(Bensing & Sullam, 2002, Bensing et al., 2005, Bensing et al., 2007, Bensing & Sullam, 2009, Bensing & Sullam, 2010, Takamatsu et al., 2004a, Takamatsu et al., 2004b, Takamatsu et al., 2005b, Yen et al., 2011, Mistou et al., 2009, Bensing et al., 2004, Siboo et al., 2008, Rigel & Braunstein, 2008).
Figure 1. Structural domain organization of select Serine-rich repeat proteins (SRRPs).
SRRPs are composed of an N-terminal signal peptide (S); most often a single, but sometimes two, unique non-repeat domains (N) that mediate adhesion; two serine-rich repeat domains (SRR) flanking the single or second N domain (SRR1 and SRR2); and a C-terminal cell wall anchor domain (A). The SRRs are composed of serine residues alternating, most frequently, either an alanine, valine, or threonine residue. The large number of repeats in the SRR2 domain is responsible for the large size of most SRRPs. Domains are not drawn to scale.
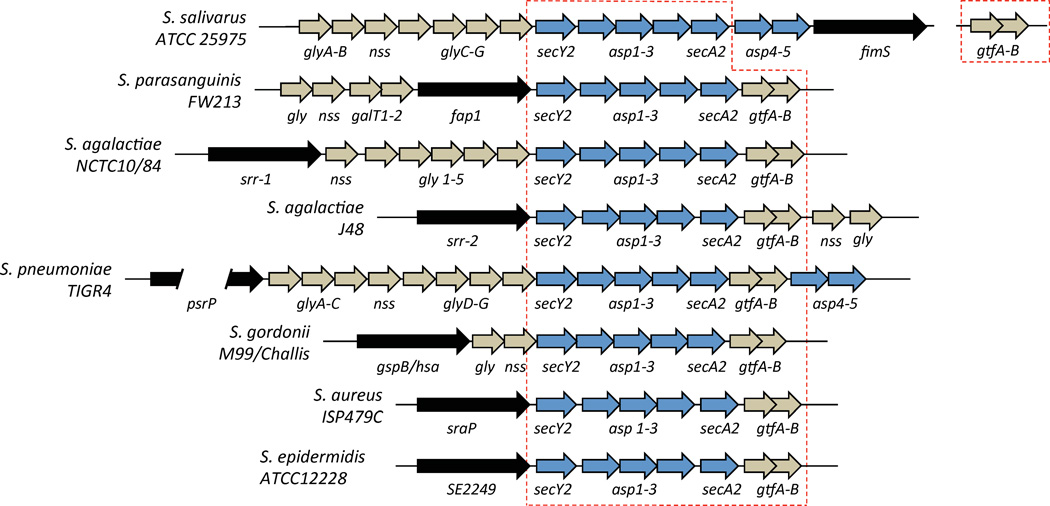
Genes encoding each SRRP, its specific glycosyltransferases, and the required accessory components of an alternate secY2A2 translocase are found on genomic islands present in the chromosome. These islands vary considerably in size, with the largest being psrP-secY2A2 in Streptococcus pneumoniae, at 37-kb (Obert et al., 2006, Wu et al., 2007a, Bensing & Sullam, 2002, Siboo et al., 2005, Seifert et al., 2006, Chen et al., 2004, Chen et al., 2006, Takahashi et al., 2002, Takahashi et al., 2004, Handley et al., 2005, van Sorge et al., 2009); in at least one case, the SRRP locus is considered to be a pathogenicity island(Obert et al., 2006). The core genetic component of these loci, found in all strains that carry an SRRP, includes two genes (most commonly called GtfA and GtfB) encoding glycosyltransferases involved in the first step of SRRP glycosylation that is required for protein stability, and five genes encoding components of the alternate secY2A2 complex necessary for SRRP transport [secA2, secY2, Asp 1-3 (also known as gap 1-3); the latter may also be involved in glycosylation(Peng et al., 2008a, Peng et al., 2008b)].(Figure 2) (Bensing et al., 2004, Bu et al., 2008, Li et al., 2008, Wu et al., 2007a, Seepersaud et al., 2010, Siboo et al., 2008, Zhou & Wu, 2009, Zhou et al., 2011, Zhou et al., 2010, Zhou et al., 2012, Zhu et al., 2011). Species and strain-specific variation does occur and can include the presence of additional glycosyltransferases, as many as five in the psrP-secY2A2 island in S. pneumoniae, or additional secY2A2 accessory components, as in the Fap-1 locus in S. parasanguinis. Consequently, differences in the complexity of SRRP glycosylation are presumed to occur and most likely impact the individual activities of these proteins. SRRP biogenesis and glycosylation will not be expanded upon here because an excellent and extensive review on these topics was written recently by Zhou et al.(Zhou & Wu, 2009).
Figure 2. Genetic organization of loci encoding SRRPs in Streptococcal and Staphylococcal species.
SRRPs are encoded on genomic islands present in the chromosome. In addition to the SRRP (black arrow), these loci also contain numerous genes encoding enzymes responsible for the O-linked glycosylation of the protein (tan arrows) and for transport of the protein (blue arrows). No obvious regulatory elements are present. All SRRP loci have a core set of eight genes consisting of an SRRP and secY2, secA2, asp1-3 and gtfA-B (red dashed line).
After the initial description of Fap1 (fimbriae-associated protein) by Wu et al. in Streptococcus parasanguis (Wu et al., 1998), SRRPs were identified in numerous medically relevant Streptococci. This includes assorted oral streptococci, the neonatal pathogen S. agalactiae, and the respiratory tract pathogen S.pneumoniae (Plummer et al., 2005, Xiong et al., 2008, Nobbs et al., 2009, Mistou et al., 2009, Seifert et al., 2006, Shivshankar et al., 2009).SRRPs have also been identified in Staphylococcus aureus, S. saprophyticus and Staphylococcus epidermidis (Takeuchi et al., 2005, Zhang et al., 2003), as well as in other less virulent bacteria including Lactococcusjohnsonii (Zhou & Wu, 2009). Importantly, it is now understood that SRRPs function both as intra- and inter-species adhesins, facilitating colonization of host tissue through the formation of biofilms, and that they are involved in the development of tissue infections such as oral infectious diseases, infective endocarditis, pneumonia, and meningitis (Sanchez et al., 2010, Handley et al., 2005, van Sorge et al., 2009). The focus of this review will be to summarize how this important family of bacterial adhesins mediates bacterial attachment to host and bacterial cells, contributes to disease pathogenesis, and might be targeted for pharmacological intervention or used as novel protective vaccine antigens.
Model for SRRP function
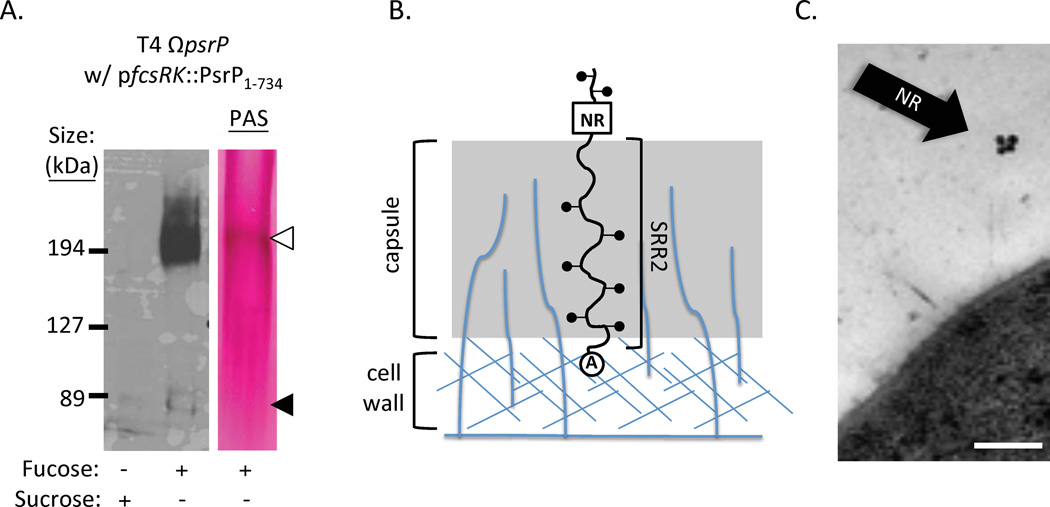
The number and amino acid composition of serine repeats in the SRR2 region is highly variable and is the primary reason why these proteins are tremendously diverse in size. For example, in S. pneumoniae strain TIGR4, PsrP is 4,776 amino acids long with 539 repeats of the amino acid sequence SAS[A/E/V]SAS[E/T] in its SRR2 domain; it has a predicted non-glycosylated molecular weight of 512 kDa. In contrast, Srr-1 of S. agalactiae strain A909 is 1,259 amino acids in size with an SRR2 domain containing only103 SASAS [T/M] repeats. Because these proteins are glycosylated they migrate at sizes much larger than their predicted molecular weight when separated by SDS-PAGE (Figure 3A). Based on the conserved C-terminal location of the CWAD for SRRPs and images of Fap1 taken by transmission electron microscopy (Wu & Fives-Taylor, 1999, Wu et al., 1998), it has been proposed that the primary function of the SRR2 domain is to extend the NR domain, which has adhesive properties (discussed below), beyond the cell wall and other surface components in order to be able to mediate attachment (Figure 3B)(Pyburn et al., 2011, Ramboarina et al., 2010).The best experimental evidence supporting this hypothesis is with PsrP of S. pneumoniae, where complementation of a psrP deletion mutant with a truncated version having only 33 of its normal 539 SRRs in its SRR2 domain was unable to restore bacterial adhesion to lung cells. However, the same truncated construct restored adhesion if transformed into an unencapsulated psrP deficient mutant. This indicates that the length of the SRR2 domain affects its ability to avoid obstruction by the capsular polysaccharide (Shivshankar et al., 2009). Likewise, immunogold electron microscopy of the NR domain of PsrP found the presence of gold particles located distally from the bacterial cell wall surface (Figure 3C).
Figure 3. SRRPs are glycosylated and extend the NR domains outward from the bacterial surface.
A) Western blot and carbohydrate staining of whole cell lysates of a S. pneumoniae mutant deleted of its chromosomal PsrP but complemented with a plasmid (pfcsRK::PsrP1–734) that expresses a truncated version of the protein with only 33 SRR repeats when grown in THY media supplemented with 1% fucose. Carbohydrate staining was done using a Periodic Acid-Schiff (PAS) stain. Note the presence of a faint fucose-inducible band at 80 kDa that is the predicted unglycosylated size of the recombinant protein (black triangle), and a stronger band at 210 kDa (white triangle). Only the 210 kDa band tested positive for carbohydrates by PAS, suggesting PsrP is glycosylated. Taken from (Shivshankar et al., 2009) B) Hypothetical model showing how SRRPs might employ the SRR2 domain to extend the NR domain outward away from the cell through surface components such that it can mediate adhesion. C) Immunogold electron microscopy using antibodies against the NR domain of PsrP. Note that the gold particles are found distal to the bacterial surface, supporting the model shown in panel B. Findings first described in (Shivshankar et al., 2009).
Such a model suggests that the number of repeats in the SRR2 domains my have evolved in a manner that optimally matches the span of extracellular components such as capsule for each species and strain. This model also suggests that the exposed SRR1 domain may have an unknown functional role mediated by its attached glycoconjugates. Finally, SRRPs might serve as “tent-poles” and stabilize/interact with capsule or other loosely attached surface components on the bacterial surface. The last two proposed functions may explain why loci encoding SRRPs such as Fap1 encode additional glycosyltranferases and SRRPs can be glycosylated in a manner that exceeds the minimum required for protein stability. Although these concepts are merely speculative, they are areas of extreme interest because of the possible contribution of SRRPs to Gram-positive biology and pathogenesis.
The NR domains of SRRPs are highly variable but mediate attachment to either conjugated sialic acid or keratin
While the overall domain structures of SRRPs are conserved between species, the individual NR domains are highly diverse with limited to no homology when compared by BLAST analysis (Zhou & Wu, 2009). The NR domains also vary considerably in size with the largest, 1231 amino acids, and smallest, 129 amino acids, known versions present in the same strain of L. johnsonii strain. In general, most NR domains are 250–500 amino acids in length with a pI that is either strongly acidic or basic(Zhou & Wu, 2009). Currently, it is not known if the pI value of the NR domain directs the binding properties of the protein.
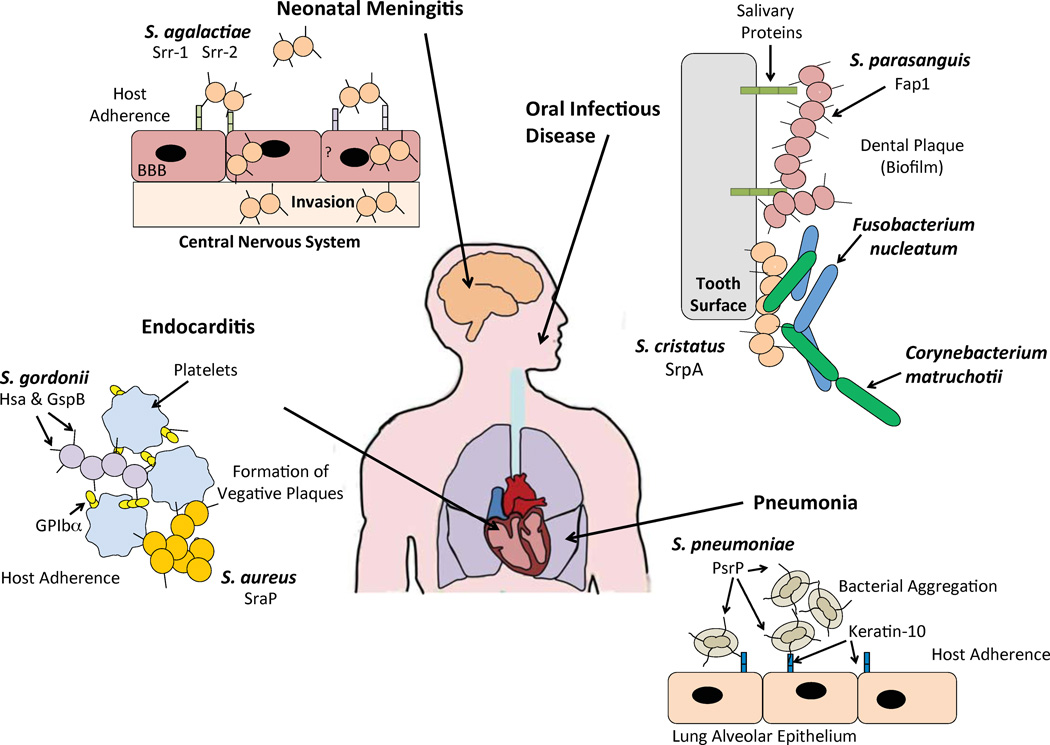
The NR domain in GspB of S. gordonii strain M99 has been shown to bind only to sialyl-T antigen [NeuAcα(2–3)Galβ(1–3)GalNAc]. In contrast, the NR in Hsa of S. gordonii strain Challis binds both α(2–3) sialyllactosamine [NeuAcα(2–3)Galβ(1–4)GlcNAc] and sialyl-T antigen (Takamatsu et al., 2005a).Importantly, the ability of these NR domains to attach to glycoconjugates enables them to mediate bacterial attachment to cell surfaces, primarily platelet membranes and dental surfaces that carry sialyllactosamine and sialyl-T antigen respectively. This binding, in some instances, has been clearly shown to contribute to disease. For example, GspB and Hsa have been shown to bind to salivary mucin MG2 and salivary agglutinin (Takamatsu et al., 2006), and an S. parasanguis mutant deficient in Fap1 fails to attach to saliva-coated hydroxyapatite, an in vitro tooth surface model (Fachon-Kalweit et al., 1985, Stephenson et al., 2002, Wu & Fives-Taylor, 1999). In addition, attachment of GspB and Hsa to sialyl-T antigen on platelet membrane GPIbα is thought to play a major role in the ability of these oral bacteria to colonize heart valves and cause endocarditis. Indeed when wild type S. gordonii and S. aureus are injected intravenously through a catheter into rats, they are both able to form aortic vegetations with platelets in the heart; whereas mutants deficient in GspB and SraP, respectively, had an absence of aortic vegetations (Xiong et al., 2008, Siboo et al., 2005).
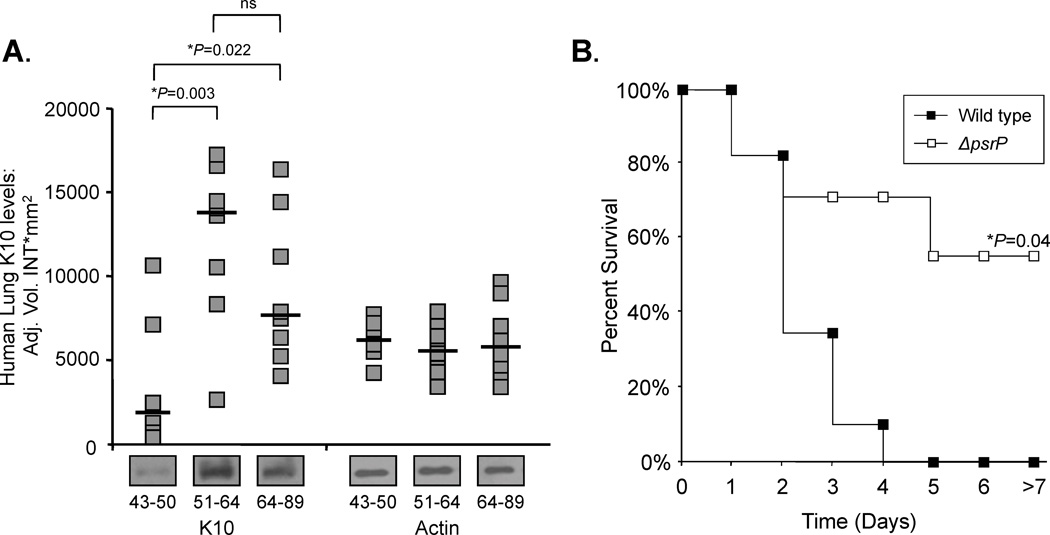
For S. pneumoniae, the pathogenicity island encoding PsrP, psrP-secY2A2, has been shown to be positivity correlated with the propensity of a strain to cause pneumonia (Obert et al., 2006). One reason for this is that the NR domain of PsrP binds to surface exposed keratin 10, which is present on lung epithelial cells(Shivshankar et al., 2009). Interestingly, PsrP is distinct from GspB or Hsa in that binding of its NR to keratin 10 is independent of any lectin properties its NR domain might have. In fact, treatment of lung cells with glycosidases enhanced bacterial adhesion to lung cells, and recombinant NR was able to bind unglycosylated recombinant keratin 10 (Shivshankar et al., 2009). More recently, we have demonstrated that keratin 10 is elevated on lung cells that have experienced DNA damage and are senescent, such as in older individuals (Figure 4A). Such non-replicating cells are commonly found in the lung of elderly individuals, smokers, or those who have developed chronic obstructive pulmonary disease(Tsuji et al., 2010). We have also demonstrated that the increased susceptibility of aged mice to S. pneumoniae is in part PsrP-dependent (Shivshankar et al., 2011)(Figure 4B). Thus, enhanced susceptibility to pneumococcal pneumonia for those who smoke and are >65 year of age (two recognized at risk populations), is due in part, to increase PsrP/keratin 10 interactions.
Figure 4. Aging enhances susceptibility to pneumococcal pneumonia in a K10-dependent Manner.
A) Densitometric analyses and representative Western blot bands for K10 in human lung biopsies obtained from young (43–50 years; n = 6), mature (51–64 years; n = 8), and aged (64–89 years; n = 8) humans. Asterisks denote a statistical significant difference when using one-way ANOVA (Duncan’s Method; P < 0.05). B) Kaplan–Meier plot showing percent survival of aged Balb/c mice intranasally challenged with 105 CFU of wild type S. pneumoniae (Wild type: n=8) or an isogenic mutant deficient in the SRRP PsrP (ΔpsrP: n=11). Statistical analysis was performed using a Kaplan–Meier log-rank Test. These findings where first described in (Shivshankar et al., 2011).
Streptococcus agalactiae, which colonizes the genital tract of women, is the leading cause of neonatal sepsis (Glaser et al., 2002); neonates become infected during passage through the birth canal. Consistent with colonization pattern, Srr-1 from S. agalactiae binds tokeratin4, which is present on the stratified epithelia lining the vaginal tract (Leube et al., 1988). In addition, Incubation of HEp-2 epithelial cells with the NR domain of Srr-1 blocked S. agalactiae adherence to cells in a dose-dependent fashion (Samen et al., 2007). However, it is not clear if Srr-1 binding to keratin 4 is glycoconjugate dependent or independent. Separately, studies with an Srr-1 deletion mutant in a mouse model implicated this protein in S. agalactiae’s ability to penetrate the blood-brain barrier (van Sorge et al., 2009). As keratin is not expressed by endothelial cells that compose the vasculature(Gu & Coulombe, 2007), one must speculate that this host-cell interaction might be limited to the cuboidal epithelial cells of the choroid plexus or that another host ligand is involved. Interestingly, Serotype III multi-locus sequence type clone ST-17 of S. agalactiae, which is associated with a propensity for invasive disease and meningitis, has been found to carry an SRRP named Srr-2; although the ligands for Srr-2 also remain unknown (van Sorge et al., 2009, Seifert et al., 2006). Interestingly, S. pneumoniae is also thought to penetrate the blood brain barrier through the choroid plexus, although a role for PsrP in pneumococcal meningitis has not been described. Figure 5 summarizes the functions of SRRPs that have thus far been demonstrated to play a role in human disease.
Figure 5. SRRPs expressed in medically relevant bacteria play a role in pathogenesis.
PsrP in S. pneumoniae is an intra-species adhesin that initiates attachment to the host by binding to keratin 10 expressed on lung alveolar epithelium. Likewise, it can also mediate attachment to another PsrP molecule expressed on another pneumococci, facilitating bacterial aggregation, leading to pneumonia. GspB and Hsa are present on S. gordonii M99 and challis, respectively. These SRRPs bind to sialic acid moieties present on the platelet receptor GPIbα forming vegetative plaques causing infective endocarditis. SraP present in S. aureus mediates direct binding to platelets also forming vegetative plaques, and leading to infective endocarditis. Neonatal sepsis and meningitis is highly associated with the presence of Srr-2 in S. agalactiae. The receptor or ligand of Srr-2 has not been identified, but it is suggested that expression of Srr-2 accelerates disease onset in neonates by facilitating transmigration of the BBB. Periodontal disease is associated with the expression of Fap1 from S. parasanguinis. Fap1 binds to glycosylated salivary proteins forming biofilms to cause caries. SrpA in S. cristatus is required for the formation of corn-cob structures with Corynbacteriummatruchoiti and Fusobacteriumnulceatum, mediating intra-species adhesion seen in dental plaque.
SRRPs mediate intra-species aggregation
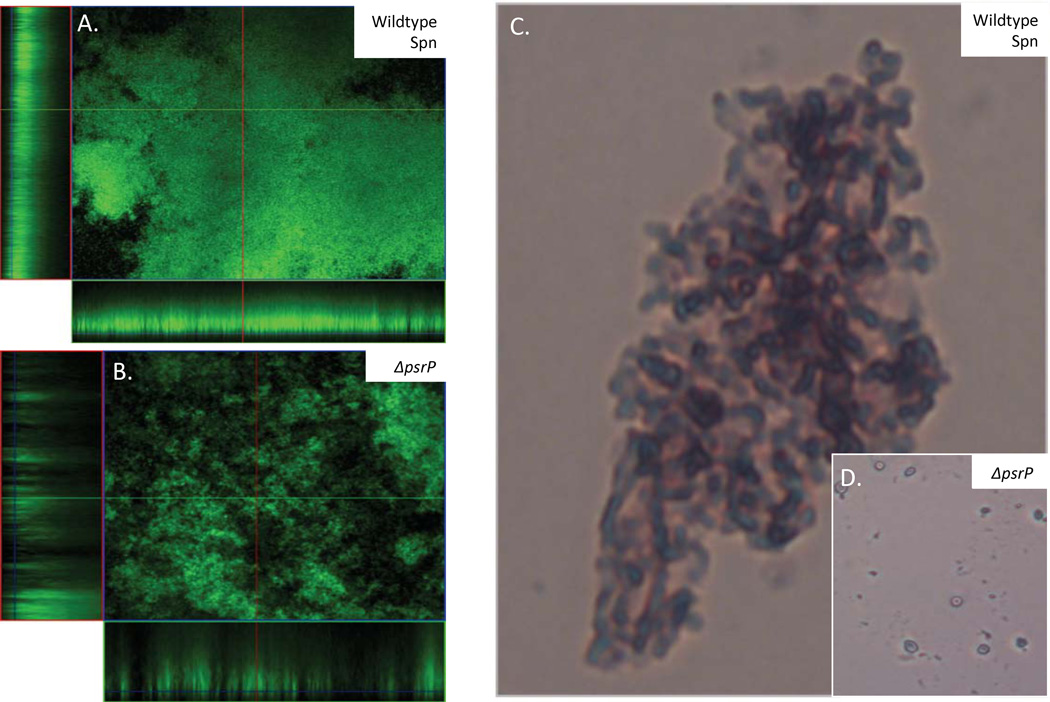
Whereas the initial studies with Fap1 demonstrated that the protein was required for S. parasanguis attachment to and robust biofilm formation on glass slides or saliva-coated hydroxyapatite (Wu et al., 1998, Wu et al., 2007b), and studies with SrpA of S. cristatus determined that the protein was required for the formation of corn-cob structures with Corynbacteriummatruchoiti and Fusobacteriumnulceatum (Handley et al., 2005) (Figure 5), only recently has it been fully appreciated that SRRPs might serve as intra- or inter-species adhesins for non-dental plaque Gram-positive bacteria. For example, we have demonstrated that amino acids 122–166 of PsrP in the NR domain mediate a bacterial self-interaction that contributes to robust biofilm formation in vitro and bacterial aggregation in vivo (Figure 6) (Sanchez et al., 2010). The affinity of recombinant PsrP NR for other pneumococci is dependent on the presence of the NR domain of PsrP on the bacterial surface, suggesting dimerization of the NR. Similar to keratin 10 binding, which is mediated through a different stretch of amino acids in NR, the intra-bacterial interaction was found to be glycoconjugate independent (Shivshankar et al., 2009).
Figure 6. PsrP in S. pneumoniae mediates robust biofilms in vitro and pneumococcal aggregates in vivo.
Micrographs of A) TIGR4 and B) T4 ΔpsrP biofilms grown in a flow cell under once-through flow conditions for 3 days. Bacteria were visualized with Live/Dead BacLight stain using an inverted confocal laser scanning microscope at 400× magnification. Representative micrographs of C) TIGR4 and D) T4 ΔpsrP Gram-stained bacteria from bronchial alveolar lavage samples from mice. These findings were first described in (Sanchez et al., 2010).
We subsequently determined that SraP (S. aureus) and GspB (S. gordonii) also promote bacterial interactions during in vitro biofilm growth. Unlike the NR domain of PsrP, the NRs of these SRRPs have an affinity not only for bacteria carrying the SRRP, but also for cell lysates from other bacteria, whether they carry an SRRP or not (Sanchez et al., 2010). Thus, as was originally demonstrated with S. cristatus SrpA, SRRPs most likely have the ability to serve as inter-species adhesins. Although untested, one possible explanation for the limited interactions of PsrP versus the other SRRPs lies in its cognate ligand that is a specific amino acid motif, whereas the ligand for GspB is a sialic acid moiety. Thus, the NR domains of GspB and SraP may mediate interactions with any surface components that carry the target glycoconjugate. Furthermore, the possibility exists that these NR domains may bind to glycoconjugates that are found on the SRRP itself, for example on the extended SRR1 domains of another SRRP molecule.
Importantly, the fact that SRRPs can mediate host surface binding and bacterial aggregation clarifies the role these proteins play during pathogenesis. SRRP-mediated interactions facilitate formation of dental plaque, which is recognized to be a multi-species biofilm. One can also imagine that SRRPs on S. aureus or the oral streptococci facilitate attachment to platelets and cells in an endocarditic lesion, while at the same time mediating adhesion to other bacteria. Similarly, one could envision a micro colony of the pneumococcus in the lungs or middle ear with some bacteria attached to host cells via PsrPNR::K10 interactions and other bacteria attached to these bacteria through PsrPNR::PsrPNR interactions. In all, since SRRPs seem to be involved in bacterial aggregation and biofilm formation, they may play a role in any aspect of disease that involves biofilms. Consistent with this idea, microarray studies and proteomic studies show that PsrP is one of the few pneumococcal virulence determinants that is up regulated during pneumococcal biofilm formation(Sanchez et al., 2011b).
Transcriptional regulation of SRRPs
Other than the finding that PsrP is upregulated during biofilm growth (Sanchez et al., 2011b), nothing else is known about the regulation of SRRPs or their accessory genes. Since no obvious transcription factors are encoded in any of the SRRP loci, this suggests that the genes may be regulated by trans acting elements or non-coding RNAs embedded within the loci. Future studies are warranted to determine how these large, and most likely highly metabolically expensive, determinants are transcriptionally regulated. Importantly, it is also unknown if SRRPs are also post-translationally modified on the bacterial surface.
The NR domains of Fap1 and GspB have been crystallized and contain distinct domains
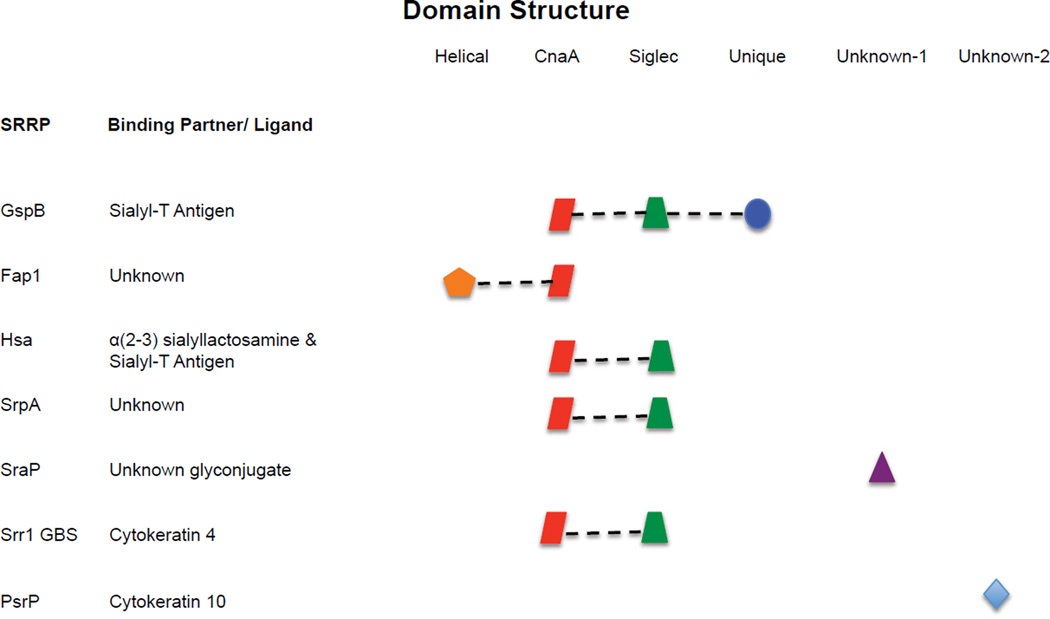
In order to gain insight into the mechanism of NR mediated adhesion in the SRRPs, high-resolution X-ray crystal structures of two NRs, Fap1 and GspB, have been determined. In addition, the Srr-1 keratin 4-binding domain from S. agalactiae has been expressed, purified, and crystalized. However, further crystallization trials are needed in order to obtain diffraction-quality crystals (Sundaresan et al., 2011). Analysis of the NR domains of Fap1 from S.parasanguinis and GspB from S. gordonii revealed that the predicted binding regions of these adhesins contain different modular organizations (Ramboarina et al., 2010, Garnett et al., 2011, Garnett et al., 2012). The GspBNR structure folds into a linear elongated rod, containing 3 independent subdomains. Interestingly, all three subdomains are predominated by β-strands as a secondary structure. The first subdomain has a folding topology similar to the S. aureus Microbial Surface Components Recognizing Adhesive Matrix Molecule (MSCRAMM) surface adhesin protein CNA; therefore, it is has been referred to as the CnaA subdomain. The second subdomain identified in GspBNR is very similar in topology to the sialic acid binding immunoglobulin-like lectin 5 (Siglec-5) structure, mostly present on leukocytes. Siglecs are a family of structurally related transmembrane proteins shown to be responsible for recognizing sialylated glycoconjugates (Pyburn et al., 2011). Accordingly, it is this domain that mediates GspB NR binding to sialyl-T antigen (Takamatsu et al., 2005a, Pyburn et al., 2011). The third domain located on the C-terminal part of GspB NR did not have identity with any know protein structures. Therefore, this subdomain has been termed the unique subdomain (Pyburn et al., 2011).
After a similar investigation, the structure of Fap1NR has been shown to contain two distinct subdomains, NRα and NRβ, with different secondary structures (Ramboarina et al., 2010, Garnett et al., 2011, Garnett et al., 2012). NRαis composed of a three-helix bundles packed against each other in an up-down-up arrangement. This subdomain is very similar to the domain of the giant extracellular matrix-protein Ebh (ECM-binding protein homolog) from staphylococci (Ramboarina et al., 2010, Tanaka et al., 2008). Interestingly, Ebh also forms a bridge between the cytoplasm and cell wall and is involved in tolerance to transient hyperosmotic pressure (Kuroda et al., 2008). Whether SRRP function in a similar role remains unexplored. The structure of the NRβ contains folding topology similar to the CnaA subdomain of GspB and MSCRAMMs with a similar secondary structure predominated by β-strands.
Although the GspB-CnaA and Fap1-NRβ subdomains have similar structures, they have distinct binding affinities. Only the Siglec domain in GspB NR binds to sialyl-T antigen, whereas in the Fap1 NR both subdomains have been shown to be important for binding saliva-coated hydroxyapatite; although the specific ligand for Fap1 remains unknown. Importantly, the Fap1NRα structure is unique among the other NRs based on the sequence alignments. Alternatively, the CnaA subdomain in GspB and Fap-1 is likely to be found in other SRRPs (Figure 7) (Pyburn et al., 2011). Thus, it seems possible that neutralizing antibodies could individually target distinct functional aspects of these NR domains, or alternately small molecules might be designed to target the CnaA domain.
Figure 7. Modular organization of the NR regions within the SRRP family.
Subdomains of the NR region organized according to structural similarities. Similarities are based on crystal structure and predicted sequence identity after BLAST or Clustal W sequence alignments(Garnett et al., 2012, Pyburn et al., 2011, Chenna et al., 2003, Altschul et al., 1990). The middle column lists the known ligands for these NR. Figure adapted from Pyburn et al.(Pyburn et al., 2011).
SRRPs as novel vaccine candidates
The pivotal role of SRRPs during disease opens the possibility that their neutralization with antibody might confer protection. In vitro studies have demonstrated that monoclonal and polyclonal antibodies against Fap1 block the ability of the bacteria to form biofilms on glass slides. Interestingly, these monoclonal antibodies are specific to the glycoconjugates present on Fap1, suggesting that in addition to the adhesive property of the NR domain, the glycosylated SRR2 might also function as an adhesin(Stephenson et al., 2002). In such an instance the SRRP might be functioning similar to mucin. For S. pneumoniae polyclonal antibodies against the NR domain have been shown to block bacterial adhesion to A549 cells, inhibit biofilm formation, and protect mice following both passive and active immunization (Rose et al., 2008, Sanchez et al., 2010, Shivshankar et al., 2009). Notably, individuals who have recovered from invasive pneumococcal disease have detectable antibody against PsrP, thus providing direct evidence that this protein is produced during human infection (Sanchez et al., 2011a). It remains unclear as to which part of PsrP elicits a protective humoral response.
Due to the modular domain structure of the NR domains, it seems possible that individual epitopes of the protein could be targeted. For example antibodies against the MSCRAMM component or the Siglec domain would presumably neutralize these individual activities. Generation of monoclonal antibodies against these subdomains could be used to confirm the individual activity of these proteins and be tested as therapeutic agents. Unfortunately, SRRPs are not universally present in the Gram-positive bacteria that cause infectious disease and can have distinct forms even within the same species. This suggests that immunization with SRRP fragments would not stand alone as a vaccine antigen and would most likely need to be part of a multi-valent protein vaccine.
Summary
SRRPs are an important family of conserved surface proteins involved in Gram-positive pathogenesis. While evidence now strongly supports their role as intra- and inter-species adhesins numerous questions remain unanswered. The most pressing of these include the role of the glycoconjugates forms of these proteins, how they are transcriptionally regulated, and full characterization of their cognate ligands. Given their important role in pathogenesis, SRRPs might be targeted for intervention. If so information collected from structural analyses will be critical for the design of either antigens that elicit neutralizing antibodies or small molecules that bind specific subdomains.
Acknowledgments
AL and CJS received support from NIDCR TE-32 DE 16318. CJO is supported by NIH AI078972. We thank Dr. David Kolodrubetz for his helpful review of the manuscript.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- Bensing BA, Gibson BW, Sullam PM. The Streptococcus gordonii platelet binding protein GspB undergoes glycosylation independently of export. J Bacteriol. 2004;186:638–645. doi: 10.1128/JB.186.3.638-645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Siboo IR, Sullam PM. Glycine residues in the hydrophobic core of the GspB signal sequence route export toward the accessory Sec pathway. J Bacteriol. 2007;189:3846–3854. doi: 10.1128/JB.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Sullam PM. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol Microbiol. 2002;44:1081–1094. doi: 10.1046/j.1365-2958.2002.02949.x. [DOI] [PubMed] [Google Scholar]
- Bensing BA, Sullam PM. Characterization of Streptococcus gordonii SecA2 as a paralogue of SecA. J Bacteriol. 2009;191:3482–3491. doi: 10.1128/JB.00365-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Sullam PM. Transport of preproteins by the accessory Sec system requires a specific domain adjacent to the signal peptide. J Bacteriol. 2010;192:4223–4232. doi: 10.1128/JB.00373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Takamatsu D, Sullam PM. Determinants of the streptococcal surface glycoprotein GspB that facilitate export by the accessory Sec system. Mol Microbiol. 2005;58:1468–1481. doi: 10.1111/j.1365-2958.2005.04919.x. [DOI] [PubMed] [Google Scholar]
- Bu S, Li Y, Zhou M, Azadin P, Zeng M, Fives-Taylor P, Wu H. Interaction between two putative glycosyltransferases is required for glycosylation of a serine-rich streptococcal adhesin. J Bacteriol. 2008;190:1256–1266. doi: 10.1128/JB.01078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Wu H, Fives-Taylor PM. Investigating the role of secA2 in secretion and glycosylation of a fimbrial adhesin in Streptococcus parasanguis FW213. Mol Microbiol. 2004;53:843–856. doi: 10.1111/j.1365-2958.2004.04116.x. [DOI] [PubMed] [Google Scholar]
- Chen Q, Wu H, Kumar R, Peng Z, Fives-Taylor PM. SecA2 is distinct from SecA in immunogenic specificity, subcellular distribution and requirement for membrane anchoring in Streptococcus parasanguis . FEMS Microbiol Lett. 2006;264:174–181. doi: 10.1111/j.1574-6968.2006.00455.x. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachon-Kalweit S, Elder BL, Fives-Taylor P. Antibodies that bind to fimbriae block adhesion of Streptococcus sanguis to saliva-coated hydroxyapatite. Infect Immun. 1985;48:617–624. doi: 10.1128/iai.48.3.617-624.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froeliger EH, Fives-Taylor P. Streptococcus parasanguis fimbria-associated adhesin fap1 is required for biofilm formation. Infect Immun. 2001;69:2512–2519. doi: 10.1128/IAI.69.4.2512-2519.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett JA, Ramboarina S, Lee WC, Tagliaferri C, Wu W, Matthews S. Crystallization and initial crystallographic analysis of the Streptococcus parasanguinis FW213 Fap1-NRalpha adhesive domain at pH 5.0. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:274–276. doi: 10.1107/S1744309110052772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett JA, Simpson PJ, Taylor J, Benjamin SV, Tagliaferri C, Cota E, Chen YY, Wu H, Matthews S. Structural insight into the role of Streptococcus parasanguinis Fap1 within oral biofilm formation. Biochem Biophys Res Commun. 2012;417:421–426. doi: 10.1016/j.bbrc.2011.11.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser P, Rusniok C, Buchrieser C, Chevalier F, Frangeul L, Msadek T, Zouine M, Couve E, Lalioui L, Poyart C, Trieu-Cuot P, Kunst F. Genome sequence of Streptococcus agalactiae a pathogen causing invasive neonatal disease. Mol Microbiol. 2002;45:1499–1513. doi: 10.1046/j.1365-2958.2002.03126.x. [DOI] [PubMed] [Google Scholar]
- Gu LH, Coulombe PA. Keratin function in skin epithelia: a broadening palette with surprising shades. Curr Opin Cell Biol. 2007;19:13–23. doi: 10.1016/j.ceb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Handley PS, Correia FF, Russell K, Rosan B, DiRienzo JM. Association of a novel high molecular weight, serine-rich protein (SrpA) with fibril-mediated adhesion of the oral biofilm bacterium Streptococcus cristatus . Oral Microbiol Immunol. 2005;20:131–140. doi: 10.1111/j.1399-302X.2004.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M, Tanaka Y, Aoki R, Shu D, Tsumoto K, Ohta T. Staphylococcus aureus giant protein Ebh is involved in tolerance to transient hyperosmotic pressure. Biochem Biophys Res Commun. 2008;374:237–241. doi: 10.1016/j.bbrc.2008.07.037. [DOI] [PubMed] [Google Scholar]
- Lauer P, Rinaudo CD, Soriani M, Margarit I, Maione D, Rosini R, Taddei AR, Mora M, Rappuoli R, Grandi G, Telford JL. Genome analysis reveals pili in Group B Streptococcus. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- Leube RE, Bader BL, Bosch FX, Zimbelmann R, Achtstaetter T, Franke WW. Molecular characterization and expression of the stratification-related cytokeratins 4 and 15. J Cell Biol. 1988;106:1249–1261. doi: 10.1083/jcb.106.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chen Y, Huang X, Zhou M, Wu R, Dong S, Pritchard DG, Fives-Taylor P, Wu H. A conserved domain of previously unknown function in Gap1 mediates protein-protein interaction and is required for biogenesis of a serine-rich streptococcal adhesin. Mol Microbiol. 2008;70:1094–1104. doi: 10.1111/j.1365-2958.2008.06456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistou MY, Dramsi S, Brega S, Poyart C, Trieu-Cuot P. Molecular dissection of the secA2 locus of Group B Streptococcus reveals that glycosylation of the Srr1 LPXTG protein is required for full virulence. J Bacteriol. 2009;191:4195–4206. doi: 10.1128/JB.01673-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus adherence and colonization. Microbiol Mol Biol Rev. 2009;73:407–450. doi: 10.1128/MMBR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obert C, Sublett J, Kaushal D, Hinojosa E, Barton T, Tuomanen EI, Orihuela CJ. Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect Immun. 2006;74:4766–4777. doi: 10.1128/IAI.00316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Fives-Taylor P, Ruiz T, Zhou M, Sun B, Chen Q, Wu H. Identification of critical residues in Gap3 of Streptococcus parasanguinis involved in Fap1 glycosylation, fimbrial formation and in vitro adhesion. BMC Microbiol. 2008a;8:52. doi: 10.1186/1471-2180-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Wu H, Ruiz T, Chen Q, Zhou M, Sun B, Fives-Taylor P. Role of gap3 in Fap1 glycosylation, stability, in vitro adhesion, and fimbrial and biofilm formation of Streptococcus parasanguinis . Oral Microbiol Immunol. 2008b;23:70–78. doi: 10.1111/j.1399-302X.2007.00401.x. [DOI] [PubMed] [Google Scholar]
- Plummer C, Wu H, Kerrigan SW, Meade G, Cox D, Ian Douglas CW. A serine-rich glycoprotein of Streptococcus sanguis mediates adhesion to platelets via GPIb. Br J Haematol. 2005;129:101–109. doi: 10.1111/j.1365-2141.2005.05421.x. [DOI] [PubMed] [Google Scholar]
- Pyburn TM, Bensing BA, Xiong YQ, Melancon BJ, Tomasiak TM, Ward NJ, Yankovskaya V, Oliver KM, Cecchini G, Sulikowski GA, Tyska MJ, Sullam PM, Iverson TM. A structural model for binding of the serine-rich repeat adhesin GspB to host carbohydrate receptors. PLoS Pathog. 2011;7:e1002112. doi: 10.1371/journal.ppat.1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramboarina S, Garnett JA, Zhou M, Li Y, Peng Z, Taylor JD, Lee WC, Bodey A, Murray JW, Alguel Y, Bergeron J, Bardiaux B, Sawyer E, Isaacson R, Tagliaferri C, Cota E, Nilges M, Simpson P, Ruiz T, Wu H, Matthews S. Structural insights into serine-rich fimbriae from Gram-positive bacteria. J Biol Chem. 2010;285:32446–32457. doi: 10.1074/jbc.M110.128165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigel NW, Braunstein M. A new twist on an old pathway - accessory Sec systems. Mol Microbiol. 2008;69:291–302. doi: 10.1111/j.1365-2958.2008.06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose L, Shivshankar P, Hinojosa E, Rodriguez A, Sanchez CJ, Orihuela CJ. Antibodies against PsrP, a novel Streptococcus pneumoniae adhesin, block adhesion and protect mice against pneumococcal challenge. J Infect Dis. 2008;198:375–383. doi: 10.1086/589775. [DOI] [PubMed] [Google Scholar]
- Samen U, Eikmanns BJ, Reinscheid DJ, Borges F. The surface protein Srr-1 of Streptococcus agalactiae binds human keratin 4 and promotes adherence to epithelial HEp-2 cells. Infect Immun. 2007;75:5405–5414. doi: 10.1128/IAI.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez CJ, Hurtgen BJ, Lizcano A, Shivshankar P, Cole GT, Orihuela CJ. Biofilm and planktonic pneumococci demonstrate disparate immunoreactivity to human convalescent sera. BMC Microbiol. 2011a;11:245. doi: 10.1186/1471-2180-11-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez CJ, Kumar N, Lizcano A, Shivshankar P, Dunning Hotopp JC, Jorgensen JH, Tettelin H, Orihuela CJ. Streptococcus pneumoniae in biofilms are unable to cause invasive disease due to altered virulence determinant production. PLoS One. 2011b;6:e28738. doi: 10.1371/journal.pone.0028738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez CJ, Shivshankar P, Stol K, Trakhtenbroit S, Sullam PM, Sauer K, Hermans PW, Orihuela CJ. The pneumococcal serine-rich repeat protein is an intra-species bacterial adhesin that promotes bacterial aggregation in vivo and in biofilms. PLoS Pathog. 2010;6:e1001044. doi: 10.1371/journal.ppat.1001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seepersaud R, Bensing BA, Yen YT, Sullam PM. Asp3 mediates multiple protein-protein interactions within the accessory Sec system of Streptococcus gordonii . Mol Microbiol. 2010;78:490–505. doi: 10.1111/j.1365-2958.2010.07346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert KN, Adderson EE, Whiting AA, Bohnsack JF, Crowley PJ, Brady LJ. A unique serine-rich repeat protein (Srr-2) and novel surface antigen (epsilon) associated with a virulent lineage of serotype III Streptococcus agalactiae . Microbiol. 2006;152:1029–1040. doi: 10.1099/mic.0.28516-0. [DOI] [PubMed] [Google Scholar]
- Shivshankar P, Boyd AR, Le Saux CJ, Yeh IT, Orihuela CJ. Cellular senescence increases expression of bacterial ligands in the lungs and is positively correlated with increased susceptibility to pneumococcal pneumonia. Aging Cell. 2011;10:798–806. doi: 10.1111/j.1474-9726.2011.00720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivshankar P, Sanchez C, Rose LF, Orihuela CJ. The Streptococcus pneumoniae adhesin PsrP binds to Keratin 10 on lung cells. Mol Microbiol. 2009;73:663–679. doi: 10.1111/j.1365-2958.2009.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siboo IR, Chaffin DO, Rubens CE, Sullam PM. Characterization of the accessory Sec system of Staphylococcus aureus . J Bacteriol. 2008;190:6188–6196. doi: 10.1128/JB.00300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siboo IR, Chambers HF, Sullam PM. Role of SraP, a Serine-Rich Surface Protein of Staphylococcus aureus in binding to human platelets. Infect Immun. 2005;73:2273–2280. doi: 10.1128/IAI.73.4.2273-2280.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson AE, Wu H, Novak J, Tomana M, Mintz K, Fives-Taylor P. The Fap1 fimbrial adhesin is a glycoprotein: antibodies specific for the glycan moiety block the adhesion of Streptococcus parasanguis in an in vitro tooth model. Mol Microbiol. 2002;43:147–157. doi: 10.1046/j.1365-2958.2002.02725.x. [DOI] [PubMed] [Google Scholar]
- Sundaresan R, Samen U, Ponnuraj K. Expression, purification, crystallization and preliminary X-ray diffraction studies of the human keratin 4-binding domain of serine-rich repeat protein 1 from Streptococcus agalactiae . Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:1582–1585. doi: 10.1107/S1744309111040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Konishi K, Cisar JO, Yoshikawa M. Identification and characterization of has, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect Immun. 2002;70:1209–1218. doi: 10.1128/IAI.70.3.1209-1218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Yajima A, Cisar JO, Konishi K. Functional analysis of the Streptococcus gordonii DL1 sialic acid-binding adhesin and its essential role in bacterial binding to platelets. Infect Immun. 2004;72:3876–3882. doi: 10.1128/IAI.72.7.3876-3882.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu D, Bensing BA, Cheng H, Jarvis GA, Siboo IR, Lopez JA, Griffiss JM, Sullam PM. Binding of the Streptococcus gordonii surface glycoproteins GspB and Hsa to specific carbohydrate structures on platelet membrane glycoprotein Ibalpha. Mol Microbiol. 2005a;58:380–392. doi: 10.1111/j.1365-2958.2005.04830.x. [DOI] [PubMed] [Google Scholar]
- Takamatsu D, Bensing BA, Prakobphol A, Fisher SJ, Sullam PM. Binding of the streptococcal surface glycoproteins GspB and Hsa to human salivary proteins. Infect Immun. 2006;74:1933–1940. doi: 10.1128/IAI.74.3.1933-1940.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu D, Bensing BA, Sullam PM. Four proteins encoded in the gspB-secY2A2 operon of Streptococcus gordonii mediate the intracellular glycosylation of the platelet-binding protein GspB. J Bacteriol. 2004a;186:7100–7111. doi: 10.1128/JB.186.21.7100-7111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu D, Bensing BA, Sullam PM. Genes in the accessory sec locus of Streptococcus gordonii have three functionally distinct effects on the expression of the platelet-binding protein GspB. Mol Microbiol. 2004b;52:189–203. doi: 10.1111/j.1365-2958.2004.03978.x. [DOI] [PubMed] [Google Scholar]
- Takamatsu D, Bensing BA, Sullam PM. Two additional components of the accessory sec system mediating export of the Streptococcus gordonii platelet-binding protein GspB. J Bacteriol. 2005b;187:3878–3883. doi: 10.1128/JB.187.11.3878-3883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F, Watanabe S, Baba T, Yuzawa H, Ito T, Morimoto Y, Kuroda M, Cui L, Takahashi M, Ankai A, Baba S, Fukui S, Lee JC, Hiramatsu K. Whole-genome sequencing of staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J Bacteriol. 2005;187:7292–7308. doi: 10.1128/JB.187.21.7292-7308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sakamoto S, Kuroda M, Goda S, Gao YG, Tsumoto K, Hiragi Y, Yao M, Watanabe N, Ohta T, Tanaka I. A helical string of alternately connected three-helix bundles for the cell wall-associated adhesion protein Ebh from Staphylococcus aureus . Structure. 2008;16:488–496. doi: 10.1016/j.str.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae . Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- Tsuji T, Aoshiba K, Nagai A. Alveolar cell senescence exacerbates pulmonary inflammation in patients with chronic obstructive pulmonary disease. Respiration. 2010;80:59–70. doi: 10.1159/000268287. [DOI] [PubMed] [Google Scholar]
- van Sorge NM, Quach D, Gurney MA, Sullam PM, Nizet V, Doran KS. The Group B Streptococcal serine-rich repeat 1 glycoprotein mediates penetration of the blood-brain barrier. J Infect Dis. 2009;199:1479–1487. doi: 10.1086/598217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman MM, Iobst S, Jesionowski AM, Gill SR. Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J Bacteriol. 2007;189:7799–7807. doi: 10.1128/JB.01023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Bu S, Newell P, Chen Q, Fives-Taylor P. Two gene determinants are differentially involved in the biogenesis of Fap1 precursors in Streptococcus parasanguis . J Bacteriol. 2007a;189:1390–1398. doi: 10.1128/JB.00836-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Fives-Taylor PM. Identification of dipeptide repeats and a cell wall sorting signal in the fimbriae-associated adhesin, Fap1, of Streptococcus parasanguis . Mol Microbiol. 1999;34:1070–1081. doi: 10.1046/j.1365-2958.1999.01670.x. [DOI] [PubMed] [Google Scholar]
- Wu H, Mintz KP, Ladha M, Fives-Taylor PM. Isolation and characterization of Fap1, a fimbriae-associated adhesin of Streptococcus parasanguis FW213. Mol Microbiol. 1998;28:487–500. doi: 10.1046/j.1365-2958.1998.00805.x. [DOI] [PubMed] [Google Scholar]
- Wu H, Zeng M, Fives-Taylor P. The glycan moieties and the N-terminal polypeptide backbone of a fimbria-associated adhesin, Fap1, play distinct roles in the biofilm development of Streptococcus parasanguinis . Infect Immun. 2007b;75:2181–2188. doi: 10.1128/IAI.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong YQ, Bensing BA, Bayer AS, Chambers HF, Sullam PM. Role of the serine-rich surface glycoprotein GspB of Streptococcus gordonii in the pathogenesis of infective endocarditis. Microb Pathog. 2008;45:297–301. doi: 10.1016/j.micpath.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Alves JM, Kitten T, Brown A, Chen Z, Ozaki LS, Manque P, Ge X, Serrano MG, Puiu D, Hendricks S, Wang Y, Chaplin MD, Akan D, Paik S, Peterson DL, Macrina FL, Buck GA. Genome of the opportunistic pathogen Streptococcus sanguinis . J Bacteriol. 2007;189:3166–3175. doi: 10.1128/JB.01808-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen YT, Seepersaud R, Bensing BA, Sullam PM. Asp2 and Asp3 interact directly with GspB, the export substrate of the Streptococcus gordonii accessory Sec System. J Bacteriol. 2011;193:3165–3174. doi: 10.1128/JB.00057-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, Ren SX, Li HL, Wang YX, Fu G, Yang J, Qin ZQ, Miao YG, Wang WY, Chen RS, Shen Y, Chen Z, Yuan ZH, Zhao GP, Qu D, Danchin A, Wen YM. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228) Mol Microbiol. 2003;49:1577–1593. doi: 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- Zhou M, Peng Z, Fives-Taylor P, Wu H. A conserved C-terminal 13-amino-acid motif of Gap1 is required for Gap1 function and necessary for the biogenesis of a serine-rich glycoprotein of Streptococcus parasanguinis . Infect Immun. 2008;76:5624–5631. doi: 10.1128/IAI.00534-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Wu H. Glycosylation and biogenesis of a family of serine-rich bacterial adhesins. Microbiol. 2009;155:317–327. doi: 10.1099/mic.0.025221-0. [DOI] [PubMed] [Google Scholar]
- Zhou M, Zhang H, Zhu F, Wu H. Canonical SecA associates with an accessory secretory protein complex involved in biogenesis of a streptococcal serine-rich repeat glycoprotein. J Bacteriol. 2011;193:6560–6566. doi: 10.1128/JB.05668-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Zhu F, Dong S, Pritchard DG, Wu H. A novel glucosyltransferase is required for glycosylation of a serine-rich adhesin and biofilm formation by Streptococcus parasanguinis . J Biol Chem. 2010;285:12140–12148. doi: 10.1074/jbc.M109.066928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Zhu F, Li Y, Zhang H, Wu H. Gap1 functions as a molecular chaperone to stabilize its interactive partner Gap3 during biogenesis of serine-rich repeat bacterial adhesin. Mol Microbiol. 2012;83:866–878. doi: 10.1111/j.1365-2958.2012.07970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Erlandsen H, Ding L, Li J, Huang Y, Zhou M, Liang X, Ma J, Wu H. Structural and functional analysis of a new subfamily of glycosyltransferases required for glycosylation of serine-rich streptococcal adhesins. J Biol Chem. 2011;286:27048–27057. doi: 10.1074/jbc.M110.208629. [DOI] [PMC free article] [PubMed] [Google Scholar]