Abstract
High accuracy (fidelity) of DNA replication is important for cells to preserve genetic identity and to prevent accumulation of deleterious mutations. The error rate during DNA replication is as low as 10−9 to 10−11 errors per base pair. How this low level is achieved is an issue of major interest. This review is concerned with the mechanisms underlying the fidelity of the chromosomal replication in the model system Escherichia coli by DNA polymerase III holoenzyme (HE), with further emphasis on participation of the other, accessory DNA polymerases, of which E. coli contains four (Pols I, II, IV, and V). Detailed genetic analysis of mutation rates revealed that (i) Pol II has an important role as a back-up proofreader for Pol III, (ii) Pols IV and V do not normally contribute significantly to replication fidelity, but can readily do so under conditions of elevated expression, (iii) participation of Pols IV and V, in contrast to that of Pol II, is specific to the lagging strand, and (iv) Pol I also makes a lagging-strand specific fidelity contribution, limited however to the faithful filling of the Okazaki fragment gaps. The fidelity role of the Pol III τ subunit is also reviewed.
Keywords: DNA replication fidelity, accessory DNA polymerases, polymerase switching, untargeted mutagenesis, leading and lagging DNA replication, exonucleolytic proofreading
Introduction
The mechanisms by which organisms are able to efficiently and faithfully replicate their DNA are of critical importance in genetics, and biology, and medical science, and research into these mechanisms has received significant attention. Overall, DNA replication is a high-accuracy process that enables cells and organisms to maintain their genetic identity. On the other hand, the replication process is not without error and mutations will occur with defined frequencies. While mutations are essential factors in mediating long-term processes of adaptation and evolution or in generating antibody diversity, in the short run mutations are generally deleterious and may be a cause of human disease, like birth defects or cancer (Jackson & Loeb, 1998; Preston et al., 2010; Tuppen et al., 2010). Studies of spontaneous mutation rates in various systems have provided a base line value for the in vivo replication error rate. When expressed per base pair (per round of replication), the rates can be 5 × 10−10 for bacterial systems (E. coli) or 5 × 10−11 for mammalian systems (Drake et al., 1998). It is clear that such low error rates must be the outcome of multiple fidelity pathways acting in parallel and/or consecutively.
In broad outline, high fidelity must be based on two major components. First, the DNA must be kept intact and, to the extent possible, free of any damage before DNA replication is to proceed. Obviously, damaged DNA is unlikely to be copied accurately. The multitude of DNA surveying and repair systems used by cells to monitor their DNA attest to the critical importance of this issue (Friedberg et al., 2006). Second, the intrinsic accuracy of DNA replication, when performed on essentially damage-free DNA, has its limits. Therefore, unavoidably, DNA polymerases will commit errors, leading to mutations. The present review focuses on this particular aspect of DNA replication fidelity, and will cover insights obtained regarding this process from the model system Escherichia coli. The data used are derived from mutation frequencies in actively growing E. coli cultures under aerobic conditions in the absence of external stresses. Nevertheless, spontaneous DNA damage (e.g., hydrolytic DNA damage leading to deaminated bases or abasic sites) does occur under these conditions, which, if unrepaired, may contribute to observed mutations. Thus, an absolute distinction between the two types of mutation production may not be possible. In our studies, we have minimized the possible contribution of spontaneous DNA damage by performing most experiments in DNA mismatch-repair defective strains, in which uncorrected replication errors are assumed to predominate (see below).
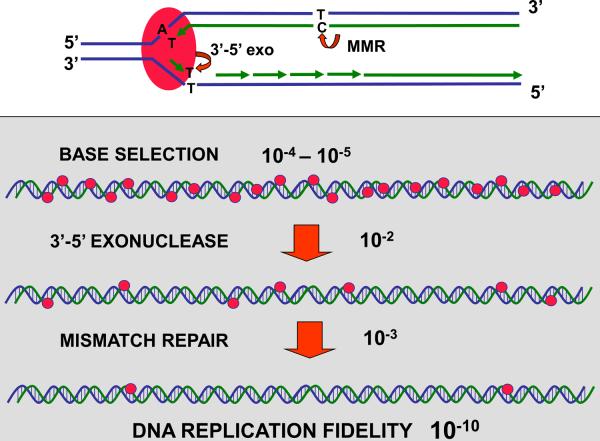
As a general outline, we present in Fig. 1 a cartoon showing how in most organisms the overall accuracy of DNA replication can be considered ax the outcome of three (highly conserved) components acting sequentially: (i) selection of the correct DNA nucleotide by the replicative DNA polymerase, (ii) removal of any misinserted nucleotides by the 3'→5' exonuclease proofreading activity associated with the polymerase, and (iii) postreplicative correction of polymerase errors that have escaped proofreading by the DNA mismatch repair system (MMR) (Schaaper, 1993a; Kunkel & Bebenek, 2000; Kunkel, 2004). In E. coli, the mismatch repair step is performed by the mutHLS system, which is capable of distinguishing the (incorrect) newly-synthesized strand from the (correct) parental strand based on the transient undermethylation (at 5'-GATC-3' sequences) of the newly-made strand (reviewed in Modrich, 1987; Kunkel & Erie, 2005). Roughly, the contribution of each of these components to the error rate can be estimated at 10−5 (insertion), 10−2 (proofreading), and 10−3 (mismatch repair), accounting for the 10−10 overall rate, although each contribution is highly dependent on the precise type of error under question (Schaaper, 1993a; Kunkel & Bebenek, 2000; Kunkel, 2004).
Fig. 1.
General scheme indicating how three serial fidelity steps during chromosomal replication can produce the low error rate of ~10−10 errors per base per round of replication. The steps are: (a) discrimination by the polymerase against inserting an incorrect base, (error rate ~10−5); (b) proofreading (editing) of misinserted bases (T·T, example) by the 3'→5' exonuclease associated with the polymerase (escape rate ~10−2); and (c) removal of remaining mismatches (G·G, example) by postreplicative DNA Mismatch Repair (MMR) (escape rate ~10−3).
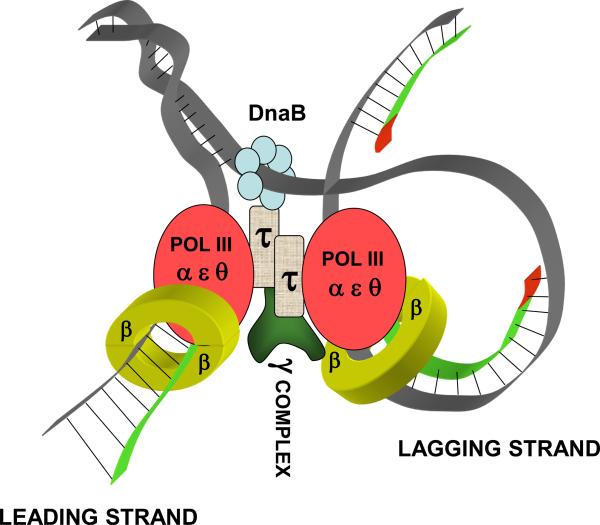
Chromosomes are replicated generally by large, multi-subunit, dimeric complexes - termed replicases, which are capable of copying the two strands, leading and lagging, at a replication fork in a coordinated fashion. In E. coli the replicase is DNA polymerase III holoenzyme (HE), a dimeric enzyme that contains two copies of DNA polymerase III, one for each strand. A schematic depiction of HE containing its various associated subunits at a replication fork (replisome) is shown in Fig. 2. Replicases and their functioning at the replication fork are subjects of intense current interest (e.g., McHenry, 2012).
Fig. 2.
A model of the E. coli DNA polymerase III holoenzyme (HE) at the chromosomal replication fork, synthesizing, simultaneously, leading and lagging strands in, respectively, continous (leading strand) and discontinuous (lagging strand) fashion. The αεθ complex represents the Pol III core, in which α is the polymerase, ε is the exonuclease (proofreading) subunit, and θ is a stabilizing subunit. See text for details. Not shown is the DnaG primase, which, in association with the DnaB helicase, produces lagging-strand RNA primers supporting the discontinous synthesis in this strand. The γ complex (γδδ'χψ) conducts the cycling (loading and unloading) of the β2 processivity clamps, which is particularly important for the cycling of the polymerase in the lagging strand. Recent studies have suggested that the relevant form of the DnaX assembly (τ2γδδ'χψ as shown here) may be τ3δδ'χψ (noting that γ and τ are both products of the dnaX gene and differ only in their C-termini). As τ contains the extra C-terminal extension that mediates the τ-α interaction, the τ3δδ'χψ-containing HE is capable of binding a third Pol III core (McInerney et al., 2007; Reyes-Lamothe et al., 2010; Georgescu et al., 2012). This third Pol III core (not shown) may participate in polymerase switching and hence contribute to the chromosomal replication process (see text).
Specific questions regarding the functioning and fidelity of chromosomal replisomes center around: (i) what is the precise composition of the complex, (ii) which DNA polymerase copies which strand, (iii) is there differential fidelity between leading and lagging strands, (iv) what is the contribution to fidelity of the non-catalytic subunits within the replicase, and (v) what is the fidelity contribution, if any, of the additional DNA polymerases that cells possess? The present review will focus, in part, on the latter aspect, namely the possible role of the additional polymerases, often referred to as 'polymerase switching'.
Multiple DNA polymerases in the cell
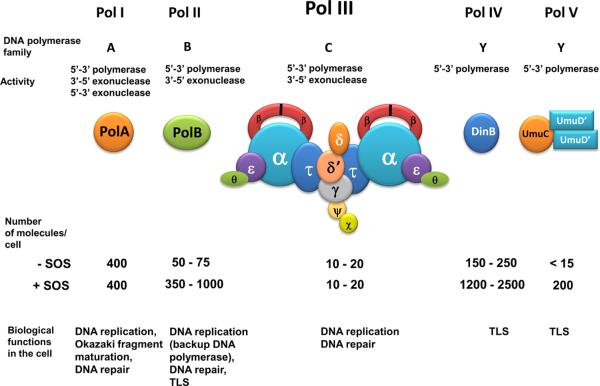
E. coli encodes five distinct DNA polymerases: Pol I, Pol II, Pol III, Pol IV, and Pol V. These polymerases, along with some of their relevant properties, are listed in Fig. 3. The presence of multiple DNA polymerases in the cell is a general phenomenon: for example, the yeast Saccharomyces cerevisiae has eight, while human cells possess at least sixteen DNA-template-dependent DNA polymerases (Friedberg et al., 2006; Shcherbakova & Fijalkowska, 2006, Kunkel, 2009). Some of these, functioning usually as replicative DNA polymerases, are high-fidelity enzymes possessing a 3'→5' exonuclease proofreading activity. But other polymerases are error-prone enzymes, lacking a proofreading activity and having an adjusted (or enlarged) active site permitting higher frequencies of base misinsertions (reviewed in Friedberg et al., 2002; Goodman, 2002; Nohmi, 2006; Shcherbakova & Fijalkowska, 2006; Jarosz et al., 2007; Lehmann et al., 2007; McCulloch & Kunkel, 2008). Most of the error-prone polymerases, referred to as TLS polymerases (translesion synthesis polymerases), may have evolved to bypass replication-blocking template lesions. TLS allows strand extension when the replicative DNA polymerase is stalled at a DNA lesion, a subject that has been extensively covered (reviewed in Friedberg et al., 2002; Goodman, 2002; Nohmi, 2006; Shcherbakova & Fijalkowska, 2006; Jarosz et al., 2007; Fuchs & Fujii, 2007; Lehmann et al., 2007; Guo et al., 2009; Takata & Wood, 2009; Sutton, 2010; Andersson et al., 2010; Livneh et al., 2010; Ho & Scharer, 2010).
Fig. 3.
The five DNA polymerases of E. coli and some of their relevant properties.
In addition to lesion bypass performed by specialized TLS polymerases, evidence has accumulated, from both in vitro and in vivo experiments, that the replicase is a dynamic entity, in which DNA polymerase exchanges can occur, thus providing access of accessory polymerases to the replication fork also during synthesis of undamaged DNA (Banach-Orlowska et al., 2005; Gawel et al., 2008; Furukohri et al., 2008; Uchida et al., 2008; Curti et al., 2009; Makiela-Dzbenska et al., 2009; Heltzel et al., 2009a). These observations raise important fidelity questions. In some cases this participation of accessory polymerases may lead to improved fidelity, when the accessory DNA polymerase is accurate (e.g., a proofreading-proficient polymerase), while in other cases this may lead to lower fidelity when the polymerase is error-prone.
In this review, we will summarize the results from work done in the bacterium E. coli, showing that during normal DNA replication both replicative and accessory DNA polymerases contribute to the overall fidelity of DNA replication. Furthermore, we will describe studies suggesting that certain non-catalytic subunits of HE, like the τ subunit, have a role in controlling the access of these polymerases to the fork, affecting, in this manner, the fidelity of replication. We will first discuss the major replicase, DNA Polymerase III holoenzyme, then accessory polymerases II, IV, and V, which may have occasional access to the replication fork, and finally Pol I, whose fidelity role may be limited to the processing of Okazaki fragments.
DNA Polymerase III holoenzyme (HE)
The major replicative DNA polymerase in E. coli cells is DNA polymerase III holoenzyme (Pol III HE) able to synthesize DNA with high processivity (over 50 kb/binding event) (Yao et al., 2009) and with high speed (up to 1000 nucleotides per second). As shown in Fig. 2, Pol III HE in E. coli is an asymmetric, dimeric enzyme, able to perform coupled, high-speed replication of the two, leading and lagging, DNA strands at the replication fork (for reviews, see: McHenry, 2003; O'Donnell, 2006; Pomerantz & O'Donnell, 2007; Yao & O'Donnell, 2008; Langston et al., 2009; McHenry 2012). Pol III HE is a complex of 17 subunits (10 distinct proteins) with an overall composition (αεθ)2(β□)2τ2γδδ'χψ. It contains two Pol III core subassemblies, one for each strand, each consisting of three tightly bound subunits: α (dnaE gene product, the DNA polymerase), ε (dnaQ gene product, the 3'→5' proofreading exonuclease) and θ (holE gene product, stabilizer for ε subunit) (Gefter et al., 1971; Takano et al., 1986; Taft-Benz & Schaaper, 2004). Each core is tethered to the DNA by a ring-shaped sliding-clamp processivity factor (β2 homodimer) (Kong et al., 1992). Additionally, Pol III HE contains the seven-subunit (τ2γδδ'χψ) DnaX complex (Pritchard et al., 2000). This complex contains the τ2 dimer, which connects the two Pol III α subunits, creating the dimeric polymerase unit capable of coupled synthesis of leading and lagging strands. The DnaX complex is also responsible, due to the activity of the δ and δ' subunits, for the loading and unloading of the β2 sliding-clamp (Jeruzalmi et al., 2001). This important activity is especially critical for the lagging-strand polymerase, which needs to cycle on and off during Okazaki fragment synthesis (McHenry 2012).
The form of Pol III HE depicted here, with the DnaX complex containing two τ subunits and one γ subunit, is the form of HE historically found when isolated from cells (McHenry 2012). More recently, a form of Pol III has been described, upon reconstitution of HE in vitro, in which the DnaX complex contains three τ subunits and no γ subunit (τ3δδ'χψ) leading to incorporation of three pol III core units (McInerney et al., 2007; Reyes-Lamothe et al., 2010; Georgescu et al., 2012). As τ and γ are products of the same (dnaX) gene and are identical for the first 430 amino acids, they share several functions and can partially substitute for each other (Flower and McHenry, 1986; Blinkowa and Walker, 1990; McHenry, 2012). However, τ, as the longer, full-length dnaX gene product (630 residues) has additional capabilities not present in γ, including its interaction with the α subunit, which is responsible for the dimerization of the polymerase. The alternative τ3δδ'χψ DnaX complex is, however, capable of binding three pol III cores, and this trimeric form of HE has been demonstrated to operate efficiently in vitro (McInerney et al., 2007; Reyes-Lamothe et al., 2010; Georgescu et al., 2012). While a trimeric form of HE may have certain advantages over the dimeric form (Georgescu et al., 2012), including possibly its fidelity, the significance of the trimeric form for HE operating in vivo remains to be established (McHenry 2012).
The fidelity properties of Pol III have been investigated in vivo and in vitro. The in vivo experiments have suggested that indeed HE is highly accurate, as expected for the major replicating enzyme (Schaaper 1993a). The importance of Pol III for controlling the mutation rate is also evidenced by the properties of several E. coli mutator mutants (which have enhanced mutation rates) carrying a defect in the dnaE gene encoding the Pol III α subunit (Sevastopoulos and Glaser, 1977; Fijalkowska et al., 1993; Oller et al., 1993; Vandewiele et al., 2002). Strong mutators have also been found residing in the dnaQ gene, encoding the ε proofreading subunit (Fijalkowska and Schaaper 1996; Taft-Benz and Schaaper, 1998), establishing the importance of the proofreading process for the chromosomal replication process. A modest mutator effect was also detected for strain lacking the holE gene, encoding the θ subunit (Taft-Benz and Schaaper, 2004). This mutator effect likely reflects the stabilizing role that θ subunit plays for ε subunit (the three subunits are arranged in the linear order α–ε–θ) (Taft-Benz and Schaaper, 2004; Chikova and Schaaper 2005).
While the studies on E. coli mutator mutants described above bear evidence to the important role that (wild-type) Pol III HE plays in preventing replication errors, they do not necessarily prove that errors made by the wild-type enzyme contribute to the observed replication error rate or bacterial mutation rate. That this is indeed the case was evidenced by the isolation of a series of dnaE antimutator mutants (Fijalkowska et al., 1993; Fijalkowska and Schaaper, 1993; Oller and Schaaper, 1994; Schaaper 1996, 1998). In these strains, replication has become more accurate due to the altered α subunit of HE, resulting in a detectably lower mutation rate. The mechanism underlying the antimutator effects could be enhanced fidelity at the insertion step or, more likely, an effect on the interplay between polymerase and associated proofreading exonuclease. Following a nucleotide misinsertion, competition arises between (i) the forward polymerization rate, extending the mismatch and hence fixing the error as a potential mutation, and (ii) the reverse reaction by movement of the incorrect base from the polymerase active site to the exonuclease site leading to removal of the base (proofreading). Obviously, impairment in the polymerase forward rate will lead to a different partitioning between the two reactions, leading to enhanced proofreading and a reduction in the error rate. This enhanced-proofreading mechanism for antimutators was first demonstrated in studies with the bacteriophage T4 DNA polymerase (Drake & Allen, 1968; Gillin & Nossal, 1976; Drake, 1993; Reha-Krantz & Nonay 1994; Reha-Krantz, 1995).
Differential fidelity of Pol III HE in leading and lagging strands
Based on the differential enzymology of leading and lagging strand polymerization (continuous synthesis in the leading strand, discontinuous in the lagging strand) it is a relevant question to ask whether replication fidelity is identical in the two strands or whether they might differ. In the latter case, most of the replication errors may result predominantly from one of the two strands. This question has been addressed in E. coli by investigating the reversion of certain lacZ alleles when placed in the two possible orientations on the E. coli chromosome. The logic for this type of experiment is that when a given base-substitution is mediated by a known mispair (for example, Ttemplate·G errors causing A·T→G·C base substitutions) inversion of the gene orientation will place the causative error from, for example, the leading strand to lagging strand or vice versa. Thus, any difference observed in the mutation rate between the two orientations may be indicative of strand-dependent replication fidelity. These experiments have been performed with several lacZ alleles under a large series of circumstances (Fijalkowska et al., 1998; Maliszewska-Tkaczyk et al., 2000; Gawel et al., 2002; Banach-Orlowska et al., 2005; Kuban et al., 2005; Makiela-Dzbenska et al., 2009; Gawel et al., 2011). The conclusion reached from these experiments is that indeed there is a significant difference in the error rate depending on the gene orientation. This difference is generally 2- to 6-fold, with the lagging-strand replication being more accurate. Therefore, the majority of replication errors result from leading-strand replication (Fijalkowska et al., 1998). Note that the critical experiments were all performed in a mismatch-repair-defective (mutL) background (see Fig. 1), so that the outcomes reflect the direct production of the errors without interference of the subsequent mismatch repair step, which may have its own specificity.
Differential leading and lagging strand replication have also been described in eukaryotic systems, like S. cerevisiae (Pavlov et al., 2002; Pursell et al., 2007; Nick McElhinny et al., 2008). In this case, the fidelity differences are ascribed to replication being performed by different enzymes in the two strands (for example, Pol ε and Pol δ in leading and lagging strands, respectively). However, in the E. coli case both strands are copied by the same polymerase (Pol III) and the differential outcome must reflect a different behavior of the same enzyme in its two configurations. The reason why in E. coli leading and lagging strands are copied with differential fidelity is still open at this time, but some clues may be found in the experiments focusing on the role of the accessory DNA polymerases, as described in the following sections.
DNA polymerase switching
The question whether the E. coli accessory DNA polymerases have access to the DNA replication process and hence may contribute to the observed error rate may be raised for several reasons, but most directly based on the relatively high intracellular concentrations of Pol I (400 molecules/cell), Pol II (50 molecules/cell) or Pol IV (200–250 molecules/cell), compared to those estimated for Pol III HE (10–20 molecules/cell) (Kornberg & Baker, 1992; Qui & Goodman, 1997; Kim et al., 2001; McHenry, 2003). The term DNA polymerase switching is used to describe a process by which one DNA polymerase (e.g., the major replicative DNA polymerase) is replaced, temporarily, by another DNA polymerase at the 3'-OH end of growing chain, possibly in a coordinated manner. This type of DNA polymerase exchange occurs normally during DNA replication of lagging-strand DNA synthesis. Lagging-strand replication starts with synthesis of a short RNA primer by DNA primase (DnaG in E. coli) followed by a switch replacing the primase by the main replicating polymerase for elongation of the strand. When the polymerase reaches the end of the Okazaki fragment, a second switch occurs, replacing the polymerase by an enzyme (Pol I in E. coli) capable of removing the RNA primer and filling the remaining gap.
Do other, similar types of DNA polymerase exchanges occur at the replication fork during ongoing DNA replication, allowing alternative polymerases to synthesize parts of the chromosome and contribute to the overall replication fidelity? The possible mechanisms that promote or enable polymerase switches have been the subject of several investigations. In vivo data clearly indicate that in prokaryotic cells the β sliding-clamp, like PCNA in eukaryotes, is the important platform that coordinates the participation of different polymerases (Becherel et al., 2002; Lenne-Samuel et al., 2002; Heltzel et al., 2009a; reviewed in Sutton, 2010). In vitro data also demonstrate the possibility of polymerase exchanges during ongoing DNA synthesis showing that DNA polymerases compete depending on polymerase concentrations and their affinity to the β-clamp (Indiani et al., 2005; Furukohri et al., 2008; Indiani et al., 2009; Heltzel et al., 2009b).
Role of DNA polymerase II (Pol II)
DNA polymerase II, encoded by the polB (dinA) gene, was discovered in 1970 (Knippers, 1970). Some of its properties are presented in Fig. 3. Pol II is a prototype for the B-family polymerases, which mostly contain replicative DNA polymerases, including the major eukaryotic DNA polymerases α, δ, ε, as well as DNA polymerase ζ (Bonner et al., 1990). Purified Pol II protein is a single 90 kDa polypeptide possessing both a DNA polymerase and a 3'→5' exonuclease (Cai et al., 1995). There are 30 to 50 molecules of Pol II per cell, which is several times more than the number of Pol III HE molecules (Qui & Goodman, 1997). The level of Pol II increases about 7-fold following SOS induction (Bonner et al., 1990; Iwasaki et al., 1990). Genetic studies indicate that Pol II may be involved in a variety of cellular activities including repair of intrastrand cross-links (Berardini et al., 1999), repair of DNA damaged by UV irradiation and replication restart after UV irradiation (Masker et al., 1973; Rangarajan et al., 1999), repair of DNA damaged by oxidation (Escarceller et al., 1994), stress-induced mutagenesis (Foster et al., 1995; Hastings et al., 2010), and long-term survival (Yeiser et al., 2002). It has been also shown that, although Pol II is a proofreading-proficient enzyme, it can carry out error-prone translesion synthesis at certain lesions (Becherel & Fuchs, 2001; Wang & Yang, 2009). In vitro studies have shown that Pol II interacts with the β-clamp (β□) and the clamp-loading complex (γδδ'χψ), both major Pol III accessory proteins, to become competent for synthesizing DNA with high fidelity and processivity (Wickner & Hurwitz, 1974; Hughes et al., 1991; Bonner et al., 1992; Heltzel et al., 2009a). Thus, E. coli, besides Pol III, possesses another DNA polymerase capable of participating, at least in principle, in chromosomal DNA synthesis.
The above properties of Pol II have given rise to the concept that this enzyme may function as a back-up polymerase for Pol III HE (Banach-Orlowska et al., 2005). In this model, Pol II may occasionally take the place of Pol III through polymerase switching. Initially, this concept was rather difficult to accept, as experiments with E. coli strains deleted for Pol II did not show significant effects on the chromosomal error rate (Rangarajan et al., 1997, Banach-Orlowska et al., 2005). However, a role for Pol II in chromosomal DNA synthesis was revealed through experiments using mutants carrying a proofreading-deficient form of Pol II (PolBex1). The underlying rationale for the experiments replacing the accurate, proofreading-proficient form of Pol II by its proofreading-deficient and error-prone form (PolBex1), is that in a background of high-fidelity DNA synthesis (by Pol III HE) even a small amount of Pol II-mediated synthesis may be detectable through an increase in the mutation frequency (mutator phenotype). Initially, this strategy was used to demonstrate a polBex1-mediated increase in chromosomal rpoB mutations in the dnaE915 antimutator background (Rangarajan et al., 1997). The dnaE915 allele encodes a mutant Pol III α-subunit that promotes higher accuracy of chromosomal replication (Fijalkowska et al., 1993; Fijalkowska and Schaaper, 1993), and it has been assumed that the more prominent apparent role of the polBex allele in the dnaE915 background is a consequence of either the reduced background of HE-mediated mutations, making it easier to see the Pol II effect, or of a possibly enhanced tendency of the DnaE915-encoded polymerase to dissociate from the template and, hence, to exchange places with Pol II (Fijalkowska et al., 1993; Fijalkowska & Schaaper, 1995; Rangarajan et al., 1997). Regardless of the precise mechanisms, these experiments highlight the potential of Pol II to participate in chromosomal DNA synthesis.
Using the lacZ leading/lagging system described earlier, Banach-Orlowska et al. (2005) also demonstrated that the presence of the proofreading-deficient Pol II (PolBex1) in the cell significantly increased the mutant frequencies, however not only in cells carrying dnaE915 but also in dnaE+ cells. Even more interestingly, the polBex1 mutator effect was more pronounced for the lagging strand. These results suggested that Pol II has access to the chromosomal replication fork and participates in the replication process, possibly in a strand-preferential way. Interestingly, no significant changes in mutability were observed in cells lacking Pol II (ΔpolB strains), suggesting that any DNA synthesis performed by wild-type (proofreading-proficient) Pol II is generally error-free.
We have proposed that the role of Pol II in DNA replication is that of a back-up polymerase to Pol III HE. There are likely to be situations where HE may temporarily dissociate from the primer terminus, possibly randomly, but more plausibly only in special situations where HE could temporarily stall, such as at DNA lesions or secondary structures. We have proposed that the fidelity role of Pol II is most easily explained by a switch-over from HE to Pol II at terminal mismatches created by a HE misinsertion error. Extension of terminal mismatches is generally difficult for DNA polymerases (Perrino & Loeb, 1989; Mendelman et al., 1990; Joyce et al., 1992) and in vitro data on α subunit and HE have confirmed this to be the case also for Pol III (Mo & Schaaper, 1996; Kim & McHenry, 1996; Pham et al., 1998, 1999). While a majority of terminal mismatches is expected to removed by the proofreading activity of Pol III (ε subunit), a certain fraction of errors will escape the exonuclease, presenting a critical decision point for the polymerase. Extension of the mismatch would fix the error into a potential mutation, while Pol III dissociation from the mismatch would allow allow for additional scenarios. Take-over of the terminal mismatch by Pol II is expected to lead to removal of the mismatch by the Pol II 3'-exonuclease, a clear fidelity function. On the other hand, a take-over by the exonuclease-deficient Pol II would likely lead to extension of the mismatch - fixing the error - consistent with the observed polBex mutator effect. This model has the added advantage that extensive chromosomal synthesis by Pol II need not be invoked to explain the polBex mutator effect (Banach-Orlowska et al., 2005). The possibility that the proofreading activity of one polymerase can correct errors made by another polymerase has been shown in Saccharomyces cerevisiae cells (Pavlov et al., 2004).
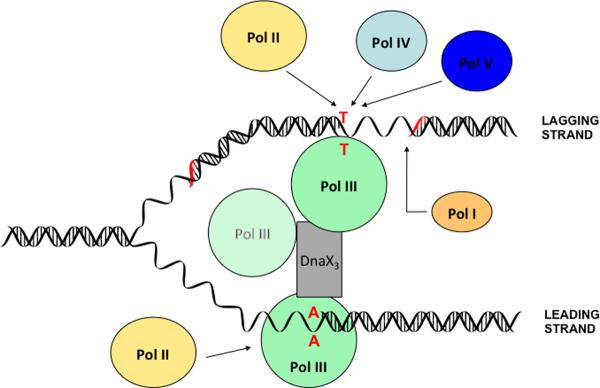
Further support for involvement of Pol II in DNA replication and the proposed sequential action of Pol III and Pol II came from experiments showing that the polBex allele synergistically enhanced a Pol III proofreading defect (dnaQ mutant) or Pol III mutator effect (dnaE486 or dnaE511 mutants) (Banach-Orlowska et al., 2005). Again, the simplest interpretation of these results is that when Pol III HE has problems (e.g. stalling due to increased mismatch production or/and defective polymerase) and the frequency of its dissociation increases, the access of Pol II to the terminal mismatch also increases. This model for the participation of Pol II is presented in Fig. 4.
Fig. 4.
Proposed scheme for the involvement of E. coli DNA polymerases in error production and prevention at the replication fork, as based on the work described in this review. Events start when Pol III commits a misinsertion error (indicated as T:T or A:A) in either DNA strand. While Pol III may handle the mismatch by itself by either extending the mismatch, yielding a potential mutation, or by removing it using its proofreading activity (not shown), in a fraction of cases Pol III may dissociate from the mismatch, leading to a possible polymerase exchange (arrows). The data suggest that, in both strands, Pol II is the primary enzyme for such a polymerase exchange, leading to removal of the terminal mismatch by the 3' exonuclease of Pol II (back-up proofreading function). When dissociation occurs in the lagging strand, Pol IV and Pol V may also compete for the primer terminus, leading to extension of the mismatch and fixing the error as a potential mutation. In contrast, Pol I does not compete for the primer terminus in either strand. Instead, its fidelity function is limited to the error-free filling of the Okazaki fragment gaps. We have also indicated in the scheme the third Pol III core (shaded green) that may be present in HE if the indicated DnaX3 assembly were composed of three τ subunits (McInerney et al., 2007; Reyes-Lamothe et al., 2010; Georgescu et al., 2012). If present, this third core might be expected to behave similarly to Pol II, leading, upon binding to the terminus, to mismatch removal (preferential binding with exonuclease site). See text for further details and justification.
Role of Pol IV at the replication fork
DNA Polymerase IV (Pol IV) is one of the two main TLS polymerases of E. coli, the other being Pol V (see section below), both members of Y family of polymerases (Wagner et al., 1999; Reuven et al., 1999; Tang et al., 1999). Pol IV, encoded by the damage-inducible dinB gene, is a single polypeptide enzyme lacking intrinsic proofreading activity, and its fidelity is considered to be relatively low (Tang et al., 2000; Goodman 2002). The constitutive intracellular concentration of Pol IV is fairly high (250 molecules/ cell) and increases by about 10-fold upon SOS induction (Kim et al., 2001). The high constitutive presence of this polymerase in the cell raises the question what its role might be and whether it might have occasional access to the replication fork and affect overall chromosomal fidelity. Due to the lack of exonucleolytic proofreading activity, its involvement in replication, if any, is expected to be error prone.
Goodman (2002) proposed that in certain situations, for example at blocked replication forks, Pol IV might gain access to the nascent 3'-end and perform remedial DNA synthesis. This possibility again raised the question of any possible Pol IV contribution to spontaneous mutagenesis. However, three independent studies demonstrated that in growing cells the lack of Pol IV did not significantly reduce the level or types of spontaneous chromosomal mutations (McKenzie et al., 2003; Kuban et al., 2004; Wolf et al., 2004). This indicated that access of Pol IV to the replication fork must be limited under normal growth conditions, at least as far as any error-prone role is involved (an error-free role would not be readily detectable in these assays). On the other hand, overproduction of Pol IV, from low- or medium-copy number plasmids, clearly resulted in a mutator phenotype (Kim et al., 1997; Wagner & Nohmi, 2000; Kuban et al., 2005). Interestingly, our group also showed that this Pol IV mutator effect occurs preferentially during lagging-strand synthesis, similarly to the mutator effect of polBex1 strains (Banach-Orlowska et al., 2005; Kuban et al., 2005). Furthermore, expression of dinB from a medium-copy plasmid led to a 3-fold decrease in viability, indicative of some deleterious effects of Pol IV overproduction (Kuban et al., 2005), consistent with experiments in which Pol IV was overexpressed from an arabinose-inducible promoter (Uchida et al., 2008), which showed inhibition of DNA synthesis and a dramatic loss of viability when Pol IV levels were increased up to 15-fold higher than observed in SOS-induced cells (Uchida et al., 2008). Clearly, Pol IV can compete with HE when its concentration is sufficiently elevated.
To account for the mutator effect resulting from Pol IV overproduction, two simple models can be proposed. In the first model, the increased number of mutations results from errors made by Pol IV during ongoing synthesis. However, this would require extensive synthesis by Pol IV, especially in view of the low processivity of Pol IV (even when supported by the β-clamp) (Wagner et al. 2000; Kobayashi et al. 2002). For example, assuming that Pol IV is 100-fold less accurate than Pol III, the 10-fold mutator effect would require 10% of the chromosome to be replicated by Pol IV. In the second, probably more plausible model, the Pol IV mutator activity results from the error-prone extension of mismatches made by Pol III HE, similar to the proposed model for the PolBex mutator effect (see above). Again, terminal mismatches present a logical step-up point for accessory polymerases, and their effect could either be mutagenic or antimutagenic depending on the nature of the accessory polymerase. Support for this view of Pol IV-mediated mismatch extension is found in experiments where greater than additive effects were observed when Pol IV overproduction is performed in dnaQ928, dnaE486, or dnaE511 mutator strains, which contain functionally impaired ε (dnaQ) or α (dnaE) subunits (Kuban et al., 2005). As the dnaQ or dnaE mutator alleles are expected to increase the number of replication errors (i.e., terminal mismatches), a multiplicative or at least a greater than additive effect is expected.
The experiments with the dnaQ or dnaE mutator alleles, where replication produces increased number of terminal mismatches, provide a useful testing ground for probing the competition between various polymerases, in particular competition between accessory polymerases Pol II and Pol IV. These experiments were performed in dnaE or dnaQ mutator strains lacking Pol II, Pol IV, or both (Banach-Orlowska et al., 2005; Makiela-Dzbenska et al., 2009). In contrast to what is observed in dnaE+ strains, the lack of Pol II (ΔpolB) in dnaE486 or dnaE511 strains led to a 10- or 7-fold increase of the mutator effect, respectively. The lack of Pol IV (ΔdinB) reduced the mutator activity of dnaE486 or dnaE511 by 2-fold. Both results are consistent with an important role of the two accessory polymerases in establishing the replication error rate, Pol II in a mutation-preventive mode, Pol IV in a mutation-enhancing mode. When both accessory polymerases were deleted, the mutability of dnaE strains was similar to that observed when only Pol IV was lacking. The results are fully consistent with a model in which access of Pol IV leads to mutagenesis, and this is a major component of the dnaE or dnaQ mutator effects, whereas Pol II is strongly antimutagenic, most simply envisaged by competing out Pol IV for access to the mismatch.
Interestingly, the (dinB-dependent) 10- or 7-fold increase of mutagenesis of dnaE486 ΔpolB or dnaE511 ΔpolB strains is much lower than the increase caused by the presence of polBex1 allele (55- to 280-fold) (polBex dnaE486 or polBex dnaE511). This large 55- to 280-fold effect indicates that Pol II's role in editing mismatches in dnaE486 or dnaE511 cells is much larger than anticipated from the ΔpolB or ΔpolB ΔdinB experiments. Presumably, in the absence of Pol II, Pol IV still has limited access and will need to compete with other factors, most likely Pol III HE. Taken together, these in vivo experiments indicate that Pol II along with its proofreading activity serves as an important backup system for error removal and at the same time protects primer termini against the activity of error-prone Pol IV. These functions become particularly important under conditions of a malfunctioning of Pol III HE. The role of Pol IV in lagging-strand replication and its competition with Pol II is schematically presented in Fig. 4.
Role of DNA Polymerase V (Pol V)
Pol V, encoded by the umuDC operon, is, like Pol IV, a member of the Y family of polymerases (Wagner et al., 1999; Reuven et al., 1999; Tang et al., 1999). See Fig. 2 for some of its properties. Pol V is a heterotrimeric complex (UmuD'2C), containing the umuC gene product representing the catalytic subunit, and two copies of UmuD', a product of RecA-facilitated autodigestion of UmuD protein (Shinagawa et al., 1988; Nohmi et al., 1988; McDonald et al., 1998). Pol V is clearly an error-prone enzyme, as it lacks proofreading activity and has a promiscuous base insertion fidelity that allows it to copy across a variety of DNA lesions (TLS polymerase) at the expense of high levels of mutagenesis (Reuven et al., 1999; Tang et al., 1999, Tang et al., 2000). The level of Pol V in normal (non-induced) cells is low and largely undetectable (Woodgate & Ennis 1991), although certain genetic experiments have suggested that it is not entirely absent (e.g., Bhamre et al., 2001). However, importantly, upon SOS induction the level of Pol V increases dramatically, producing 2400 molecules of UmuD and 200 molecules of UmuC (Woodgate & Ennis, 1991; Kim et al., 2001).
As normally Pol V is rarely expressed in the cell, it does not play a significant role in determining the chromosomal replication fidelity under normal (non-SOS-induced) growth conditions (Nowosielska et al., 2004). On the other hand, its potential of interfering with ongoing chromosomal replication can be seen in strains in which the SOS system (and hence Pol V) is induced constitutively in the absence of exogenous DNA damage. This condition of `spontaneous SOS induction' is achieved in certain recA strains (recA441, recA730) in which the RecA regulatory protein is constitutively active, leading to constitutive expression of the SOS system, including overproduction of Pol V as well as Pol IV (Witkin, 1974; Witkin et al., 1982; Tessman & Peterson, 1985). In these strains, increased mutagenesis is observed in the absence of any DNA damaging treatment (Castellazzi et al., 1972; Ciesla, 1982; Witkin & Kogoma, 1984; Sweasy et al., 1990). This phenomenon of increased spontaneous mutagenesis is named the `SOS mutator phenotype' or `untargeted SOS mutagenesis'. Genetic studies on the recA730-induced SOS mutator phenotype led to a postulated model in which the Pol V-mediated increase in mutagenesis results from Pol V displacing HE at HE-generated terminal mismatches (insertion errors) leading to error prone extension of these mispairs (Fijalkowska et al., 1997). This type of mispair extension is likely further aided by the observed increases in cellular dNTP levels in SOS-induced cells (Gon et al., 2011). As noted above for Pol II and Pol IV, error-prone extension of preexisting mispairs is an economical way of accounting for the increased mutation rates, as opposed to postulating extensive synthesis by Pol V and accounting for the high error rate by Pol V-mediated synthesis errors (Maliszewska-Tkaczyk et al., 2000).
Among the base substitution mutations that are enhanced in recA730 strains transversions predominate (Fijalkowska et al., 1997; Maliszewska-Tkaczyk et al., 2000; Kuban et al., 2006). This finding is consistent with in vitro data showing that transversion mismatches are most difficult to extend by HE (and more difficult to proofread) and result in DNA polymerase stalling (Kim & McHenry, 1996). It was suggested that the transient stalling of Pol III HE at terminal mismatches may stimulate dissociation of HE, allowing extension of mismatches by Pol V (Fijalkowska et al., 1997). Overall, the data strongly support the idea that in recA730 strains, and in SOS-induced cells in general, there is a competition between the major replicase (HE) and error-prone Pol V (Fijalkowska et al., 1997; Timms & Bridges, 2002; Vandewiele et al., 2002).
As SOS induction leads to enhanced levels of both Pol V and Pol IV, the participation of the latter in the SOS mutator activity also requires consideration. To address this question, our group showed that deletion of Pol IV from recA730 cells (recA730 ΔdinB::kan) reduced mutagenesis relative to the dinB+ recA730 strain by at least two-fold (Kuban et al., 2006). As loss of Pol V (ΔumuDC recA730 strain) completely abolishes the recA730 mutator effect (Witkin et al., 1984; Sweasy et al., 1990) it was concluded that both Pol V and Pol IV are required for full expression of the mutator effect, with Pol V playing the critical role, presumably in extending the primary mismatch. Pol IV may participate in further extension of the Pol V-generated product, thereby improving the likelihood that the mismatch-containing DNA is ultimately converted into a mutation (Kuban et al., 2006).
Studies of the strandedness of the Pol V-mediated SOS mutator effect clearly indicated that the effect results primarily, if not exclusively, from events during lagging-strand replication (Maliszewska-Tkaczyk et al., 2000; Kuban et al., 2005). This preference mimics that of the Pol IV mutator effect, and to some extent the PolBex mutator effect (see above). Clearly, the lagging strand is more accessible for accessory polymerases. This is consistent with the greater dissociativity of HE in the lagging strand. See also Fig. 4.
Role of DNA polymerase I (Pol I)
DNA polymerase I, encoded by the polA gene, was the first polymerase ever identified (Bessman et al., 1956; De Lucia & Cairns, 1969). Pol I is the most abundant DNA polymerase in E. coli (~400 molecules per cell) (Kornberg & Baker, 1992). Pol I, a 103 kDa protein, possesses three enzymatic activities: the 5'→3' polymerase, a 5'→3' exonuclease, and a 3'→5' proofreading exonuclease (Joyce & Grindley, 1984). The polymerase and 5'→3' exonuclease activities enable Pol I to fulfill its primary role in DNA replication: to remove RNA primers and fill the resulting gap during maturation of Okazaki fragments in the lagging strand (Okazaki et al., 1971). Pol I also fills gaps generated during DNA repair and recombination (Cooper & Hanawalt, 1972; Kornberg & Baker, 1992; Friedberg et al., 2006). Pol I, which is presumably capable of interacting with the β-processivity clamp, is a competent high-fidelity enzyme, and should be capable, in principle, of competing with HE, similarly to Pol II and Pol IV (Lopez de Saro and O'Donnell, 2001).
The role of Pol I in chromosomal replication fidelity has been investigated (Curti et al., 2008; Makiela-Dzbenska et al., 2009, 2011), most productively using a proofreading-deficient variant of Pol I (polAexo mutant), analogously to the polBex mutant described above. The logic behind these experiments is again that small, normally error-free, contributions of Pol I would be observable as an increase in the mutation frequency when substituted by its error-prone proofreading-deficient counterpart. A 1.5- to 4-fold increase in mutant frequencies was observed in polAexo cells, indicating that Pol I indeed plays a role in fidelity. Three separate observations in these studies led us to further define this role: (i) the polAexo mutator effect has a lagging-strand preference, (ii) there is no synergism between the polAexo mutator effect and the mutator effects exerted by other DNA polymerases or replication defects, and (iii) no competition could be demonstrated between Pol I, on the one hand, and Pol II and Pol IV on the other. Based on these results, we concluded that Pol I does not compete directly at the growing point in the replication fork. Instead, the fidelity role of Pol I is concerned primarily with the error-free processing of the Okazaki fragment gaps, consistent with the established role of Pol I in maturation of Okazaki fragments. In E. coli, Okazaki fragments average between 1000 to 2000 nucleotides, while the length of RNA primers is 10 to 20 nucleotides (Zechner et al., 1992a; Zechner et al., 1992b). Thus, Pol I is likely responsible for 1 to 2% of DNA synthesis in the lagging strand. However, if even such a relatively small fraction of chromosomal DNA is synthesized by an enzyme that is 50- to 100-fold less accurate (polAexo) than Pol III, then a 2% contribution in DNA replication may produce a 2-fold increase of the chromosomal error rate. See Fig. 4 for inclusion of Pol I in the overall chromosomal fidelity scheme.
Role of the Pol III HE τ subunit in fidelity and polymerase switching
The dnaX gene encodes both the τ and γ subunits of HE (Fig.2). τ is the gene's full length (643 amino acid) product, while γ is a truncated product lacking the C-terminal residues due to a translational frameshifting event at residues 428–430 (Flower & McHenry 1986; Blinkowa & Walker 1990). While γ, as part of the γδδ' complex, is responsible for loading and unloading the β-clamps from DNA (reviewed in Bowman et al., 2005), τ fulfills several additional critical functions, both structurally and mechanistically, due its unique C-terminus (for a review, see McHenry, 2012). Foremost, τ dimerizes the two pol III cores within HE, creating the dimeric polymerase that is capable of copying the two DNA strands simultaneously. τ interacts with the DnaB helicase (Gao & McHenry 2001) and regulates in this manner the speed of the replication fork (Dallmann et al., 2000). τ also controls the cycling of the lagging strand polymerase, as it mediates the release of the Pol III core from its β-processivity clamp upon completion of Okazaki fragments (Leu et al., 2003; Lopez de Saro et al., 2003). Based on the central role of τ within HE, including its direct interaction with both the leading and lagging strand polymerase, a role of τ in determining the fidelity of HE, either directly or via an effect on polymerase switching, might be expected. Indeed, a survey of several dnaX(Ts) mutants revealed a mutator phenotype, at permissive temperatures, for the dnaX36 and dnaX2016 alleles (Pham et al., 2006). The dnaX36 mutator was considered particularly interesting because its underlying mutation (E601K) is located in the C-terminus of DnaX and is, hence, unique to τ (Pham et al., 2006). Analysis of the specificity of mutagenesis of the dnaX36 mutator revealed a unique pattern: transversion base substitutions as well as (−1) frameshifts in non-run sequences are specifically enhanced (Pham et al., 2006). Based on these and other data it was concluded that τ subunit is an important factor in controlling the accuracy of chromosomal replication. Further, its effect is likely mediated at the processing stage of HE-made replication errors - rather than the initial error production by HE.
Studies on the strandedness of the dnaX36 mutator effect revealed that the effect applied to both strands, although with a slight preference for the lagging strand (Gawel et al., 2011). It was also found that the dnaX36 mutator effect was largely independent of dinB and that, hence, it is not mediated by an increased switch over of HE to error-prone Pol IV. On the other hand, an enhancement of the dnaX36 mutator effect was observed in the ΔpolB background, most pronounced for the lagging strand (Gawel et al., 2011). These results indicate that Pol II plays a role in preventing mutations in dnaX36 strains and that this role may be more important in lagging-strand replication. This is in contrast to the situation in dnaX+ strains, where the ΔpolB has no effect (Rangarajan et al., 1997; Banach-Orlowska et al., 2005). This suggests that in the dnaX36 mutant Pol II has increased access to the replication fork and that in this case Pol II acts to prevent mutations (Gawel et al., 2008, 2011). Part of this role of Pol II is to outcompete access by Pol IV, because the dnaX36 ΔpolB ΔdinB triple mutant has reduced mutability relative to the dnaX36 ΔpolB strain (Gawel et al., 2008, 2011).
The important role of Pol II in preventing replication errors was demonstrated most dramatically when dnaX36 was combined with the polBex1 proofreading deficiency. In this case, the polBex1 deficiency increased the mutant frequencies dramatically (20- to 50-fold) above the dnaX36 level, and this was true for both leading and lagging strands. The strong polBex1 mutator effect resulting from the Pol II proofreading deficiency is not dependent on the action of Pol IV, since the dnaX36 polBex1 ΔdinB triple mutant behaved essentially as the dnaX36 polBex1 strain (Gawel et al., 2008, 2011). This again indicates that Pol II is very effective in preventing access by Pol IV. Overall, these results highlight the important role that Pol II can play in controlling the fidelity. Note that within the favored model of Pol II as a back-up proofreader for HE-made insertion errors, a 50-fold mutant frequency increase by the polBex1 allele could indicate that some 98% of Pol III errors become a substrate for Pol II. When the normal, wild-type Pol II is present, these errors are efficiently removed by Pol II's proofreading activity, while in case of the polBex1 allele, they have a high chance of being extended by Pol II to fix the error into a potential mutation (Gawel et al., 2018, 2011).
What do these results tell us about the fidelity function of τ subunit? The defect in dnaX36 is in Domain V of DnaX, which is responsible for the α-τ interaction (Gao & McHenry 2001; Pham et al., 2006; Su et al., 2007). The relevance of this location to the mutator effect was further evidenced by the isolation of an additional set of dnaX mutators with properties similar to dnaX36 and all located in Domain V (Pham et al., 2006). Thus, an altered or impaired α-τ interaction underlies the observed mutator effect. It may be that the impaired α-τ interaction leads to more frequent Pol III dissociations. If so, the sole fidelity function of τ would be to keep HE together in its dimeric state and on the primer-templates; upon dissociation less accurate polymerases may take a turn, accounting for the mutator phenotype. While this is a possibility, the model does not readily account for (i) the strength of the dnaX36 mutator effect (too much synthesis by error-prone enzymes required), (ii) the independence of the mutator effect of Pol IV, the major candidate for error-prone interference, (iii) the substantial mutator effect of dnaX36 ΔpolB ΔdinB strains (i.e., even in the absence of Pol II and Pol IV), as well as (iv) the unusual specificity of the mutator effect: preferential enhancement of transversion base substitutions as well as −1 frameshifts in non-run sequences (Pham et al., 2006; Gawel et al., 2008, 2011).
As a preferred model, we have suggested that the fidelity function of τ results from its role in assisting HE when it is faced with terminal mismatches created by the α subunit (Maliszewska-Tkaczyk et al., 2000; Banach-Orlowska et al., 2005; Kuban et al., 2005; Gawel et al., 2008; Makiela-Dzbenska et al., 2009). Terminal mismatches are more difficult to extend (Perrino & Loeb 1989; Mendelman et al., 1990; Joyce et al., 1992; Kim & McHenry 1996) and temporary stalling will ensue. In most cases this stalling will lead to a conformational switch moving the primer terminus to the exonuclease (proofreading) subunit leading to removal of the incorrect nucleotide. However, a subset of errors may be refractory to this mechanism (Kim & McHenry 1996; Pham et al., 2006), leading to longer-lived type of stalling, which may impede the progress of the replication fork. Evidence for this type of HE stalling has been obtained in in vitro fidelity experiments (Pham et al., 2006; RMS, unpublished data). We propose that, in vivo, this type of stalling is sensed and remedied by τ subunit, leading to removal of the error and continuation of DNA synthesis. This role of τ as sensor for a stalled polymerase is similar to, and may share features with the proposed sensing role of τ in the lagging strand that leads to release of the polymerase in this strand when it reaches the end of the Okazaki fragment (Leu et al., 2003; López de Saro et al., 2003; McInerney et al., 2007).
This τ-mediated fidelity mechanism operates on both leading and lagging strands (Gawel et al., 2011). How τ promotes error removal is not yet known, but in the simplest case it may involve enforcing the required conformational switch in α that places the erroneous terminal nucleotide in the ε exonuclease site. Alternatively, τ may dislodge Pol III and channel the mismatch toward the exonuclease of Pol II or, possibly, to the third Pol III core if present at the replication fork (Georgescu et al., 2012). Obviously, in the absence of this τ function (as in the dnaX36 mutant), this mechanism is inoperative, and Pol III may eventually extend the mismatch, accounting for the dnaX mutator effect. This model is supported by the specificity of the dnaX mutator effect: transversions and non-run (−1) frameshifts are specifically enhanced. Polymerase stalling is most pronounced at transversion mismatches (purine·purine or pyrimidine·pyrimidine mispairs) (Perrino & Loeb 1989; Mendelman et al., 1990; Joyce et al., 1992; Kim & McHenry 1996), and their ability to be proofread may also be limited (Kim & McHenry 1996; Pham et al., 1998, 1999). Forced extension of such mismatches will naturally lead to transversion base-pair substitutions or, a preferred option in permissible sequence contexts, (−1) frameshifts by misalignment of the incorrect base on the next complementary template base (Mo and Schaaper, 1996; Pham et al., 1998, 1999).
Summary and concluding remarks
The results of the studies clearly indicate that the accessory DNA polymerases of E. coli can, at least occasionally, obtain access to the replication fork and participate in chromosomal replication, affecting in this manner the overall replication fidelity and the resulting bacterial mutation rate. Depending on the polymerase, the effect may be to improve fidelity (for the proofreading-proficient enzymes, such as Pol I or Pol II) or to produce an increased number of mutations (for the error-prone polymerases, Pol IV or V). Our data and interpretations are presented most straightforwardly using the cartoon of Fig. 4.
In a most parsimonious model, DNA Polymerase III HE, as an efficient and high-speed polymerase machine, is not expected to give way to accessory polymerases unless provoked by conditions impeding its progress. One such case, most relevant to the fidelity issue, is the expected stalling on terminal mismatches, from which it is difficult to continue synthesis especially for high-efficiency enzymes with `tight' catalytic sites (Beard & Wilson 2003). We have indicated this in Fig. 4, where HE is temporarily stalled on a (e.g. T:T or A:A) mismatch. If the mismatch cannot be rapidly proofread by HE, the enzyme may temporarily release the 3' terminus, providing access opportunities for accessory polymerases. Entry by an exonuclease-proficient enzyme like Pol II would lead to removal of the terminal mismatch, especially since DNA polymerases - when first binding to a primer terminus - do so with their exonuclease site rather than the polymerase site (Johnson 1993). This represents the basis of the postulated role of Pol II as a backup proofreader for Pol III-mediated replication errors. Importantly, our data indicate that Pol II performs this role in both the leading and lagging strands.
Under conditions where Pol IV or Pol V are present at elevated levels, they too can gain access to the abandoned primer termini. However, they have to compete with Pol II. Our results clearly indicate that, for example, the mutagenic potential of Pol IV is strongly limited by the presence of Pol II (Banach-Orlowska et al., 2005; Gawel et al., 2008, 2011). However, the issues are clearly more complicated than the simple model might suggest, as Pol IV and Pol V, in contrast to Pol II, appear restricted in their actions primarily to the lagging strand. Thus, the dissociative events at terminal mismatches in the two strands are likely to be different mechanistically, perhaps reflective of the normally different behavior of the Pol III core in the two strands (highly processive vs. discontinuous in leading vs. lagging strands, respectively). It may be that the polymerase switch in the leading strand follows a prescribed and controlled pattern that favors recruitment of Pol II or a return of Pol III itself, while the switch in the lagging strand may resemble more a “free-for-all” type of switch.
The precise nature of these events remains to be determined, although, as discussed above, the action of τ subunit is likely to be a critical factor, acting as a sensor for stalled polymerases and/or as a switch that remedies the stalled situation. One particular class of dnaX mutants, exemplified by dnaX36, displays a unique mutator effect that appears fully consistent with a malfunctioning of this τ-mediated sensing and/or switching mechanism, and in these strains Pol III might be forced to extend errors that would normally be resolved. Additional genetic experiments as well as biochemical approaches will be needed to provide further insight into these intriguing mechanisms.
The other important issue that needs to be further addressed is why in normal, wild-type cells the fidelity of lagging-strand replication is several-fold higher than that of the leading strand. It was suggested that the lagging-strand replication has at least one additional mechanism available for error correction, possibly related to more facile polymerase dissociation in this strand (Fijalkowska et al., 1998). If enhanced dissociation leads to more frequent engagement of Pol II, the greater fidelity of lagging-strand replication could be readily explained. Consistent with this idea is the stronger lagging-strand mutator effect associated with the exonuclease-deficient polBex1 allele (Banach-Orlowska et al., 2005). However, the situation is not so simple, as in the polBex1 strain an inversion of strand bias is actually observed for some of the tested lacZ markers (i.e., leading-strand replication is now more accurate). Thus, this question will also require further investigation.
Acknowledgements
The authors thank Drs. K. Bebenek and A. Clausen for their careful reading of the manuscript for this paper. The work described in this review was supported by grant 2 PO4A 061 30 (to IJF and PJ) from the Polish Ministry of Science and Higher Education, and project number Z01 ES065086 of the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (to RMS), and by the Foundation for Polish Science TEAM Program (to IF).
References
- Andersson DI, Koskiniemi S, Hughes D. Biological roles of translesion synthesis DNA polymerases in eubacteria. Microbiol. 2010;77:540–548. doi: 10.1111/j.1365-2958.2010.07260.x. [DOI] [PubMed] [Google Scholar]
- Banach-Orlowska M, Fijalkowska IJ, Schaaper RM, Jonczyk P. DNA polymerase II as a fidelity factor in chromosomal DNA synthesis in Escherichia coli. Mol Microbiol. 2005;58:61–70. doi: 10.1111/j.1365-2958.2005.04805.x. [DOI] [PubMed] [Google Scholar]
- Beard WA, Shock DD, VandeBerg BJ, Wilson SH. Efficiency of correct nucleotide insertion governs DNA polymerase fidelity. J Biol Chem. 2002;277:47393–47398. doi: 10.1074/jbc.M210036200. [DOI] [PubMed] [Google Scholar]
- Becherel OJ, Fuchs RP. Mechanism of DNA polymerase II-mediated frameshift mutagenesis. Proc Natl Acad Sci USA. 2001;98:8566–8571. doi: 10.1073/pnas.141113398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherel OJ, Fuchs RP, Wagner J. Pivotal role of the beta-clamp in translesion DNA synthesis and mutagenesis in E. coli cells. DNA Repair (Amst) 2002;1:703–708. doi: 10.1016/s1568-7864(02)00106-4. [DOI] [PubMed] [Google Scholar]
- Berardini M, Foster PL, Loechler EL. DNA polymerase II (polB) is involved in a new DNA repair pathway for DNA interstrand cross-links in Escherichia coli. J Bacteriol. 1999;181:2878–2882. doi: 10.1093/gao/9781884446054.article.t031385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessman MJ, Kornberg A, Lehman IR, Simms ES. Enzymatic synthesis of deoxyribonucleic acid. Biochim Biophys Acta. 1956;21:197–198. doi: 10.1016/0006-3002(56)90127-5. [DOI] [PubMed] [Google Scholar]
- Bhamre S, Gadea BB, Koyama CA, White SJ, Fowler RG. An aerobic recA-, umuC- dependent pathway of spontaneous base-pair substitution mutagenesis in Escherichia coli. Mutat Res. 2001;473:229–247. doi: 10.1016/s0027-5107(00)00155-x. [DOI] [PubMed] [Google Scholar]
- Blinkowa AL, Walker JR. Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III γ subunit from within the τ subunit reading frame. Nucleic Acids Res. 1990;18:1725–1729. doi: 10.1093/nar/18.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner CA, Hays S, McEntee K, Goodman MF. DNA polymerase II is encoded by the DNA damage inducible dinA gene of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:7663–7667. doi: 10.1073/pnas.87.19.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner CA, Stukenberg PT, Rajagopalan M, Eritja R, O'Donnell M, McEntee K, Echols H, Goodman MF. Processive DNA synthesis by DNA polymerase II mediated by DNA polymerase III accessory proteins. J Biol Chem. 1992;267:11431–11438. [PubMed] [Google Scholar]
- Bowman GD, Goedken ER, Kazmirski SL, O'Donnell M, Kuryan J. FEBS Lett. 2005;579:863–867. doi: 10.1016/j.febslet.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Cai H, Yu H, McEntee K, Kunkel TA, Goodman MF. Purification and properties of wild-type and exonuclease-deficient DNA polymerase II from Escherichia coli. J Biol Chem. 1995;270:15327–15335. doi: 10.1074/jbc.270.25.15327. [DOI] [PubMed] [Google Scholar]
- Caillet-Fauquet P, Maenhaut-Michel G. Nature of the SOS mutator activity: genetic characterization of untargeted mutagenesis in Escherichia coli. Mol Gen Genet. 1988;213:491–498. doi: 10.1007/BF00339621. [DOI] [PubMed] [Google Scholar]
- Castellazzi M, George J, Buttin G. Prophage induction and cell division in E. coli. I. Further characterization of the thermosensitive mutation tif-1 whose expression mimics the effect of UV irradiation. Mol Gen Genet. 1972;119:139–152. doi: 10.1007/BF00269133. [DOI] [PubMed] [Google Scholar]
- Chikova AK, Schaaper RM. The bacteriophage P1 hot gene can substitute for the Escherichia coli DNA polymerase III θ subunit. J Bacteriol. 2005;187:5528–5536. doi: 10.1128/JB.187.16.5528-5536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesla Z. Plasmid pKM101-mediated mutagenesis in Escherichia coli is inducible. Mol Gen Genet. 1982;186:298–300. doi: 10.1007/BF00331866. [DOI] [PubMed] [Google Scholar]
- Cooper PK, Hanawalt PC. Role of DNA polymerase I and the rec system in excision-repair in Escherichia coli. Proc Natl Acad Sci USA. 1972;69:1156–1160. doi: 10.1073/pnas.69.5.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curti E, McDonald JP, Mead S, Woodgate R. DNA polymerase switching: effects on spontaneous mutagenesis in Escherichia coli. Mol Microbiol. 2009;71:315–331. doi: 10.1111/j.1365-2958.2008.06526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- De Lucia P, Cairns J. Isolation of an E. coli strain with a mutation affecting DNA polymerase. Nature. 1969;224:1164–1166. doi: 10.1038/2241164a0. [DOI] [PubMed] [Google Scholar]
- Drake JW, Allen EF. Antimutagenic DNA polymerase of bacteriophage T4. Cold Spring Harbor Symp Quant Biol. 1968;35:339–344. doi: 10.1101/sqb.1968.033.01.039. [DOI] [PubMed] [Google Scholar]
- Drake JW. Spontaneous mutation. Annu Rev Genet. 1991;25:125–146. doi: 10.1146/annurev.ge.25.120191.001013. [DOI] [PubMed] [Google Scholar]
- Drake JW. General antimutators are improbable. J Mol Biol. 1993;229:8–13. doi: 10.1006/jmbi.1993.1002. [DOI] [PubMed] [Google Scholar]
- Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escarceller M, Hicks J, Gudmundsson G, Trump G, Touati D, Lovett S, Foster PL, McEntee K, Goodman MF. Involvement of Escherichia coli DNA polymerase II in response to oxidative damage and adaptive mutation. J Bacteriol. 1994;176:6221–6228. doi: 10.1128/jb.176.20.6221-6228.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijalkowska IJ, Dunn RL, Schaaper RM. Mutants of Escherichia coli with increased fidelity of DNA replication. Genetics. 1993;134:1023–1030. doi: 10.1093/genetics/134.4.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijalkowska IJ, Dunn RL, Schaaper RM. Genetic requirements and mutational specificity of the Escherichia coli SOS mutator activity. J Bacteriol. 1997;179:7435–7445. doi: 10.1128/jb.179.23.7435-7445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijalkowska IJ, Jonczyk P, Tkaczyk MM, Bialoskorska M, Schaaper RM. Unequal fidelity of leading and lagging strand DNA replication on the Escherichia coli chromosome. Proc Natl Acad Sci USA. 1998;95:10020–10025. doi: 10.1073/pnas.95.17.10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijalkowska IJ, Schaaper RM. Antimutator mutations in the α subunit of Escherichia coli DNA polymerase III: Identification of the responsible mutations and alignment with other DNA polymerases. Genetics. 1993;135:1039–1044. doi: 10.1093/genetics/134.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijalkowska IJ, Schaaper RM. Effects of Escherichia coli DNA antimutator alleles in proofreading deficient mutD5 strain. J Bacteriol. 1995;177:5979–5986. doi: 10.1128/jb.177.20.5979-5986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijalkowska IJ, Schaaper RM. Mutants in the Exo I motif of E. coli dnaQ: defective proofreading and inviability due to error catastrophe. Proc Natl Acad Sci USA. 1996;93:2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower AM, McHenry CS. The γ subunit of DNA polymerase III of Escherichia coli is produced by ribosomal frameshifting. Proc Natl Acad Sci USA. 1986;87:3713–3717. doi: 10.1073/pnas.87.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL, Gudmundsson G, Trimarchi JM, Cai H, Goodman MF. Proofreading-defective DNA polymerase II increases adaptive mutation in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:7951–7955. doi: 10.1073/pnas.92.17.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Wagner R, Radman M. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science. 2002;296:1627–1630. doi: 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schult RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd ed ASM Press; Washington D.C.: 2006. [Google Scholar]
- Fuchs RP, Fujii S. Translesion synthesis in Escherichia coli; lessons from the NarI mutation hot spot. DNA Repair (Amst) 2007;6:1032–1041. doi: 10.1016/j.dnarep.2007.02.021. [DOI] [PubMed] [Google Scholar]
- Furukohri A, Goodman MF, Maki H. A dynamic polymerase exchange with Escherichia coli DNA polymerase IV replacing DNA polymerase III on the sliding clamp. J Biol Chem. 2008;283:11260–11269. doi: 10.1074/jbc.M709689200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawel D, Bialoskorska M, Jonczyk P, Schaaper RM, Fijalkowska IJ. Asymmetry of frameshift mutagenesis during leading and lagging strand replication in Escherichia coli. Mutat Res. 2002;501:129–136. doi: 10.1016/s0027-5107(02)00020-9. [DOI] [PubMed] [Google Scholar]
- Gawel D, Maliszewska-Tkaczyk M, Jonczyk P, Schaaper RM, Fijalkowska IJ. Lack of strand bias in UV-induced mutagenesis in Escherichia coli. J Bacteriol. 2002;184:4449–4454. doi: 10.1128/JB.184.16.4449-4454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawel D, Pham PT, Fijalkowska IJ, Jonczyk P, Schaaper RM. Role of accessory DNA polymerases in DNA replication in Escherichia coli: analysis of the dnaX36 mutator mutant. J Bacteriol. 2008;190:1730–1742. doi: 10.1128/JB.01463-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefter ML, Hirota Y, Kornberg T, Wechsler JA, Barnoux C. Analysis of DNA polymerases II and III in mutants of Escherichia coli thermosensitive for DNA synthesis. Proc Natl Acad Sci USA. 1971;68:3150–3153. doi: 10.1073/pnas.68.12.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu RE, Kurth I, O'Donnell ME. Single-molecule studies reveal the function of a third polymerase in the replisome. Nature Struct Mol Biol. 2012;19:113–116. doi: 10.1038/nsmb.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillin FD, Nosal NG. Control of mutation frequency by bacteriophage T4 DNA polymerase. I. The CB120 antimutator DNA polymerase is defective in strand displacement. J Biol Chem. 1976;251:5219–5224. [PubMed] [Google Scholar]
- Gon S, Napolitano R, Rocha W, Coulon S, Fuchs RP. Increase in dNTP pool size during the DNA damage response plays a key role in spontaneous and induced-mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 2011;108:19311–19316. doi: 10.1073/pnas.1113664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- Guo C, Kosarek-Stancel JN, Tang TS, Friedberg EC. Y-family DNA polymerases in mammalian cells. Cell Mol Life Sci. 2009;66:2363–2381. doi: 10.1007/s00018-009-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon D. The aging process. Proc Natl Acad Sci USA. 1981;78:7124–7128. doi: 10.1073/pnas.78.11.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Hersh MN, Thornton PC, Fonville NC, Slack A, Frish RL, Ray MP, Harris RS, Leal SM, Rosenberg SM. Competition of Escherichia coli DNA polymerases I, II and III with DNA Pol IV in stressed cells. PLoS One. 2010;5:e10862. doi: 10.1371/journal.pone.0010862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heltzel JMH, Scouten-Ponticelli SK, Sanders LH, Duzen JM, Cody V, Pace J, Snell EH, Sutton MD. Sliding clamp-DNA interactions are required for viability and contribute to DNA polymerase management in Escherichia coli. J Mol Biol. 2009a;387:74–92. doi: 10.1016/j.jmb.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heltzel JMH, Maul RW, Scouten-Ponticelli SK, Sutton MD. A model for DNA polymerase switching involving a single cleft and the rim of the sliding clamp. Proc Natl Acad Sci USA. 2009b;106:12664–12669. doi: 10.1073/pnas.0903460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TV, Scharer Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ Mol Mutagen. 2010;51:552–566. doi: 10.1002/em.20573. [DOI] [PubMed] [Google Scholar]
- Hughes AJ, Bryan SK, Chen H, Moses RE, McHenry CS. Escherichia coli DNA polymerase II is stimulated by DNA polymerase III holoenzyme auxiliary subunits. J Biol Chem. 1991;266:4568–4573. [PubMed] [Google Scholar]
- Indiani C, McInerney P, Georgescu R, Goodman MF, O'Donnell M. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol Cell. 2005;19:805–815. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Indiani C, Langston LD, Yurieva O, Goodman MF, O'Donnell M. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc Natl Acad Sci USA. 2009;106:6031–6038. doi: 10.1073/pnas.0901403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Nakata A, Walker GC, Shinagawa H. The Escherichia coli polB gene, which encodes DNA polymerase II, is regulated by the SOS system. J Bacteriol. 1990;172:6268–6273. doi: 10.1128/jb.172.11.6268-6273.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Loeb LA. On the origin of multiple mutations in human cancers. Semin Cancer Biol. 1998;8:421–429. doi: 10.1006/scbi.1998.0113. [DOI] [PubMed] [Google Scholar]
- Jarosz DF, Beuning PJ, Cohen SA, Walker CS. Y-family DNA polymerases in Escherichia coli. Trends Microbiol. 2007;15:70–77. doi: 10.1016/j.tim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D, Yurieva O, Zhao Y, Young M, Stewart J, Hingorani M, O'Donnell M, Kuriyan J. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell. 2001;106:417–428. [PubMed] [Google Scholar]
- Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- Joyce CM, Grindley ND. Method for determining whether the gene of Escherichia coli is essential – application to the polA gene. J Bacteriol. 1984;158:636–643. doi: 10.1128/jb.158.2.636-643.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce CM, Sun XC, Grindley NDF. Reactions at the polymerase active site that contribute to the fidelity of Escherichia coli DNA polymerase I (Klenow fragment) J Biol Chem. 1992;267:24485–24500. [PubMed] [Google Scholar]
- Kim DR, McHenry CS. In vivo assembly of overproduced DNA polymerase III. Overproduction, purification and characterization of the α, α-ε and α-ε-θ subunits. J Biol Chem. 1996;271:20681–20689. doi: 10.1074/jbc.271.34.20681. [DOI] [PubMed] [Google Scholar]
- Kim SR, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc Natl Acad Sci USA. 1997;94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SR, Matsui K, Yamada M, Gruz P, Nohmi T. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol Genet Genomics. 2001;266:207–215. doi: 10.1007/s004380100541. [DOI] [PubMed] [Google Scholar]
- Knippers R. DNA polymerase II. Nature (London) 1970;228:1050–1053. doi: 10.1038/2281050a0. [DOI] [PubMed] [Google Scholar]
- Kong XP, Onrust R, O'Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Valentine MR, Pham P, O'Donnell M, Goodman MF. Fidelity of Escherichia coli DNA polymerase IV. Preferential generation of small deletion mutations by dNTP-stabilized misalignments. J Biol Chem. 2002;277:34198–34207. doi: 10.1074/jbc.M204826200. [DOI] [PubMed] [Google Scholar]
- Kornberg A, Baker TA. DNA Replication. W.H. Freeman & Co; New York, NY: 1992. [Google Scholar]
- Kuban W, Jonczyk P, Gawel D, Malanowska K, Schaaper RM, Fijalkowska IJ. Role of Escherichia coli DNA polymerase IV in in vivo replication fidelity. J Bacteriol. 2004;186:4802–4807. doi: 10.1128/JB.186.14.4802-4807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuban W, Banach-Orlowska M, Bialoskorska M, Lipowska A, Schaaper RM, Jonczyk P, Fijalkowska IJ. Mutator phenotype resulting from DNA polymerase IV overproduction in Escherichia coli: preferential mutagenesis on the lagging strand. J Bacteriol. 2005;187:6862–6866. doi: 10.1128/JB.187.19.6862-6866.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuban W, Banach-Orlowska M, Schaaper RM, Jonczyk P, Fijalkowska IJ. Role of DNA polymerase IV in Escherichia coli SOS mutator activity. J Bacteriol. 2006;188:7977–7980. doi: 10.1128/JB.01088-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2004;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Kunkel TA. Evolving views of DNA replication (in)fidelity. Cold Spring Harb Symp Quant Biol. 2009;74:91–101. doi: 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston LD, Indiani C, O'Donnell M. Whither the replisome: emerging perspectives on the dynamic nature of the DNA replication machinery. Cell Cycle. 2009;8:2686–2691. doi: 10.4161/cc.8.17.9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerase and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Lenne-Samuel N, Wagner J, Etienne H, Fuchs RP. The processivity factor beta controls DNA polymerase IV traffic during spontaneous mutagenesis and translesion synthesis in vivo. EMBO Rep. 2002;3:45–49. doi: 10.1093/embo-reports/kvf007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livneh Z, Ziv O, Shachar S. Multiple two-polymerase mechanisms in mammalian translesion synthesis. Cell Cycle. 2010;9:729–735. doi: 10.4161/cc.9.4.10727. [DOI] [PubMed] [Google Scholar]
- Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- López de Saro FJ, O'Donnell M. Interaction of β sliding clamp with MutS, ligase and DNA polymerase I. Proc Natl Acad Sci USA. 2001;98:8376–8380. doi: 10.1073/pnas.121009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López de Saro FJ, Georgescu RE, O'Donnell M. A peptide switch regulates DNA polymerase processivity. Proc Natl Acad Sci USA. 2003;100:14689–14694. doi: 10.1073/pnas.2435454100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makiela-Dzbenska K, Jaszczur M, Banach-Orlowska M, Jonczyk P, Schaaper RM, Fijalkowska IJ. Role of Escherichia coli DNA polymerase I in chromosomal DNA replication fidelity. Mol Microbiol. 2009;74:1114–1127. doi: 10.1111/j.1365-2958.2009.06921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makiela-Dzbenska K, Jonczyk P, Schaaper RM, Fijalkowska IJ. Proofreading deficiency of Pol I increases the levels of spontaneous rpoB mutations in E. coli. Mutat Res. 2011;712:28–32. doi: 10.1016/j.mrfmmm.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliszewska-Tkaczyk M, Jonczyk P, Bialoskorska M, Schaaper RM, Fijalkowska IJ. SOS mutator activity: unequal mutagenesis on leading and lagging strands. Proc Natl Acad Sci USA. 2000;97:12678–12683. doi: 10.1073/pnas.220424697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masker W, Hanawalt P, Shizuya H. Role of DNA polymerase II in repair replication in Escherichia coli. Nat New Biol. 1973;244:242–243. doi: 10.1038/newbio244242a0. [DOI] [PubMed] [Google Scholar]
- McCullock SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JP, Frank EG, Levine AS, Woodgate R. Intramolecular cleavage of the UmuD-like mutagenesis proteins. Proc Natl Acad Sci USA. 1998;95:1478–1483. doi: 10.1073/pnas.95.4.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenry CS. Chromosomal replicases as asymmetric dimers: studies of subunit arrangements and functional consequences. Mol Microbiol. 2003;49:1157–1165. doi: 10.1046/j.1365-2958.2003.03645.x. [DOI] [PubMed] [Google Scholar]
- McHenry CS. Breaking the rules: bacteria that use several DNA polymerase IIIs. EMBO Reports. 2011;12:408–414. doi: 10.1038/embor.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenry CS. DNA replicases from a bacterial perspective. Annu. Rev. Biochem. 2011;80:403–436. doi: 10.1146/annurev-biochem-061208-091655. [DOI] [PubMed] [Google Scholar]
- McInerney P, Johnson A, Katz F, O'Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27:527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- McKenzie GJ, Magner DB, Lee PL, Rosenberg SM. The dinB operon and spontaneous mutation in Escherichia coli. J Bacteriol. 2003;185:3972–3977. doi: 10.1128/JB.185.13.3972-3977.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelman LV, Petrusca J, Goodman MF. Base mispair extension kinetics. Comparison of DNA polymerase alpha and reverse transcriptase. J Biol Chem. 1990;265:2338–2346. [PubMed] [Google Scholar]
- Mo JY, Schaaper RM. Fidelity and error specificity of the alpha catalytic subunit of Escherichia coli DNA polymerase III. J Biol Chem. 1996;271:18947–18953. doi: 10.1074/jbc.271.31.18947. [DOI] [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Ann Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–44. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohmi T. Environmental stress and lesion-bypass DNA polymerases. Ann Rev Microbiol. 2006;60:231–253. doi: 10.1146/annurev.micro.60.080805.142238. [DOI] [PubMed] [Google Scholar]
- Nohmi T, Battista JR, Dodson LA, Walker GC. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and the posttranstional activation. Proc Natl Acad Sci USA. 1988;85:4331–4335. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowosielska A, Janion C, Grzesiuk E. Effect of deletion of SOS-induced polymerase II, IV and V, on spontaneous mutagenesis in Escherichia coli mutD5. Environ Mol Mutagen. 2004;43:226–234. doi: 10.1002/em.20019. [DOI] [PubMed] [Google Scholar]
- O'Donnell M. Replisome architecture and dynamics in Escherichia coli. J Biol Chem. 2006;281:10653–10656. doi: 10.1074/jbc.R500028200. [DOI] [PubMed] [Google Scholar]
- Okazaki R, Arisawa M, Sugino A. Slow joining of newly replicated DNA chains in DNA polymerase I-deficient Escherichia coli mutants. Proc Natl Acad Sci USA. 1971;68:2954–2957. doi: 10.1073/pnas.68.12.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oller AR, Fijalkowska IJ, Schaaper RM. The Escherichia coli galK2 papillation assay: its specificity and application to seven newly isolated mutator strains. Mutat Res. 1993;292:175–185. doi: 10.1016/0165-1161(93)90145-p. [DOI] [PubMed] [Google Scholar]
- Oller AR, Schaaper RM. Spontaneous mutation in Escherichia coli containing the dnaE911 DNA polymerase antimutator allele. Genetics. 1994;138:263–270. doi: 10.1093/genetics/138.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov YI, Newlon CS, Kunkel CA. Yeast origins establish a strand bias for replicational mutagenesis. Mol Cell. 2002;10:207–213. doi: 10.1016/s1097-2765(02)00567-1. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Maki S, Maki H, Kunkel TA. Evidence for interplay among yeast replicative polymerase alpha, delta and epsilon from studies of exonuclease and polymerase active site mutations. BMC Biol. 2004;2:11. doi: 10.1186/1741-7007-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrino FW, Loeb LA. Differential extension of 3' mispairs is a major contribution to the high fidelity of calf thymus DNA polymerase-alpha. J Biol Chem. 1989;264:2898–2905. [PubMed] [Google Scholar]
- Pham PT, Olson MW, McHenry CS, Schaaper RM. The base substitution and frameshift fidelity of Escherichia coli DNA polymerase III holoenzyme in vitro. J Biol Chem. 1998;273:23575–23584. doi: 10.1074/jbc.273.36.23575. [DOI] [PubMed] [Google Scholar]
- Pham PT, Olson MW, McHenry CS, Schaaper RM. Mismatch extension by Escherichia coli DNA polymerase III holoenzyme. J Biol Chem. 1999;274:3705–3710. doi: 10.1074/jbc.274.6.3705. [DOI] [PubMed] [Google Scholar]
- Pomerantz RT, O'Donnell M. Replisome mechanics: insights into twin DNA polymerase machine. Trends Microbiol. 2007;15:156–164. doi: 10.1016/j.tim.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Preston BD, Albertson TM, Herr AJ. DNA replication fidelity and cancer. Semin Cancer Biol. 2010;20:281–293. doi: 10.1016/j.semcancer.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard AE, Dallmann HG, Glover BP, McHenry CS. A novel assembly mechanism for the DNA polymerase III holoenzyme DnaX complex: association of δδ' with DnaX4 forms DnaX3δδ'. EMBO J. 2000;19:6536–6545. doi: 10.1093/emboj/19.23.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell ZF, Isoz I, Lundström EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qui Z, Goodman MF. The Escherichia coli polB locus is identical to dinA, the structural gene for DNA polymerase II. Characterization of Pol II purified from a polB mutant. J Biol Chem. 1997;272:8611–8617. doi: 10.1074/jbc.272.13.8611. [DOI] [PubMed] [Google Scholar]
- Rangarajan S, Gudmundsson G, Qui Z, Foster PL, Goodman MF. Escherichia coli DNA polymerase II catalyzes chromosomal and episomal DNA synthesis in vivo. Proc Natl Acad Sci USA. 1997;96:946–951. doi: 10.1073/pnas.94.3.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan S, Woodgate R, Goodman MF. A phenotype for engmatic DNA polymerase II: a pivotal role for pol II in replication restart in UV-irradiated Escherichia coli. Proc Natl Acad Sci USA. 1999;94:9224–9229. doi: 10.1073/pnas.96.16.9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reha-Krantz LJ. Learning about DNA polymerase function by studying antimutator DNA polymerases. Trends Biochem Sci. 1995;20:136–140. doi: 10.1016/s0968-0004(00)88987-2. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz LJ, Nonay RL. Genetic and biochemical studies of bacteriophage T4 DNA polymerase 3'→' exonuclease activity. J Biol Chem. 1993;268:27100–27108. [PubMed] [Google Scholar]
- Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD', RecA and SSB and specialized for translesion synthesis. J Biol Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Sherratt DJ, Leake MC. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper RM. Base selection, proofreading and mismatch repair during DNA replication in Escherichia coli. J Biol Chem. 1993a;273:23762–23775. [PubMed] [Google Scholar]
- Schaaper RM. The mutational specificity of two Escherichia coli dnaE antimutator alleles as determined from lacI mutation spectra. Genetics. 1993b;134:1031–1038. doi: 10.1093/genetics/134.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper RM. Suppressors of Escherichia coli mutT: antimutators for DNA replication errors. Mutat Res. 1996;350:17–23. doi: 10.1016/0027-5107(95)00086-0. [DOI] [PubMed] [Google Scholar]
- Schaaper RM. Antimutator mutants in bacteriophage T4 and Escherichia coli. Genetics. 1998;148:1579–1585. doi: 10.1093/genetics/148.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper RM, Danforth BN, Glickman BW. Mechanisms of spontaneous mutagenesis: an analysis of the spectrum of spontaneous mutation in the Escherichia coli lacI gene. J Mol Biol. 1986;189:273–284. doi: 10.1016/0022-2836(86)90509-7. [DOI] [PubMed] [Google Scholar]
- Schaaper RM, Dunn RL. Spontaneous mutation in the Escherichia coli lacI gene. Genetics. 1991;129:317–326. doi: 10.1093/genetics/129.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevastopoulos CG, Glaser DA. Mutator action by Escherichia coli strains carrying dnaE mutations. Proc Natl Acad Sci USA. 1977;74:3947–3950. doi: 10.1073/pnas.74.9.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova PV, Fijalkowska IJ. Translesion synthesis DNA polymerases and control of genome stability. Front Biosci. 2006;11:2496–2517. doi: 10.2741/1985. [DOI] [PubMed] [Google Scholar]
- Shinagawa H, Iwasaki H, Kato T, Nakata A. RecA-dependent cleavage of UmuD protein and SOS mutagenesis. Proc Natl Acad Sci USA. 1988;85:1806–1810. doi: 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweasy JB, Witkin EM, Sinha N, Roegner-Maniscalco V. RecA protein of Escherichia coli has a third essential role in SOS mutator activity. J Bacteriol. 1990;172:3030–3036. doi: 10.1128/jb.172.6.3030-3036.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MD. Coordinating DNA polymerase traffic during high and low fidelity synthesis. Biochim Biophys Acta. 2010;1804:1167–1179. doi: 10.1016/j.bbapap.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft-Benz SA, Schaaper RM. Mutational analysis of the 3'→5' proofreading exonuclease of Escherichia coli DNA polymerase III. Nucleic Acids Res. 1998;26:4005–4011. doi: 10.1093/nar/26.17.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft-Benz SA, Schaaper RM. The theta subunit of Escherichia coli DNA polymerase III; a role in stabilizing the epsilon proofreading subunit. J Bacteriol. 2004;186:2774–2780. doi: 10.1128/JB.186.9.2774-2780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano K, Nakabeppu Y, Maki H, Horiuchi T, Sekiguchi M. Structure and function of dnaQ and mutD5 mutators of Escherichia coli. Molec Gen Genet. 1986;205:9–13. doi: 10.1007/BF02428026. [DOI] [PubMed] [Google Scholar]
- Takata K, Wood RD. Bypass specialists operate together. EMBO J. 2009;28:313–314. doi: 10.1038/emboj.2008.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Shen X, Frank EG, O'Donnell M, Woodgate R, Goodman MF. UmuD'2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Pham P, Shen X, Taylor JS, O'Donnell M, Woodgate R, Goodman MF. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- Tessman ES, Peterson P. Plaque color method for rapid isolation of novel recA mutants of Escherichia coli K-12: new classes of protease-constitutive recA mutants. J Bacteriol. 1985;163:677–687. doi: 10.1128/jb.163.2.677-687.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timms AR, Bridges BA. DNA Pol V-dependent mutator activity in an SOS-induced Escherichia coli strains with a temperature-sensitive DNA polymerase III. Mutat Res. 2002;499:97–101. doi: 10.1016/s0027-5107(01)00267-6. [DOI] [PubMed] [Google Scholar]
- Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim Biophys Acta. 2010;1797:113–128. doi: 10.1016/j.bbabio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Uchida K, Furukohri A, Shinozaki Y, Mori T, Ogawara D, Kanaya S, Nohmi T, Maki H, Akiyama M. Overproduction of Escherichia coli DNA polymerase DinB, (Pol IV) inhibits replication fork progression and is lethal. Mol Microbiol. 2008;70:608–622. doi: 10.1111/j.1365-2958.2008.06423.x. [DOI] [PubMed] [Google Scholar]
- Vandewiele D, Fernan dez deHenestrosa AR, Timms AR, Bridges BA, Woodgate R. Sequence analysis and phenotypes of five temperature sensitive mutator alleles of dnaE, encoding modified alpha-catalytic subunits of Escherichia coli DNA polymerase III holoenzyme. Mutat Res. 2002;499:85–95. doi: 10.1016/s0027-5107(01)00268-8. [DOI] [PubMed] [Google Scholar]
- Wagner J, Gruz P, Kim SR, Yamada M, Matsui K, Fuchs RPP, Nohmi T. The dinB encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol Cell. 1999;4:281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- Wagner J, Nohmi T. Escherichia coli DNA polymerase IV mutator activity: genetic requirements and mutational specificity. J Bacteriol. 2000;182:4587–4595. doi: 10.1128/jb.182.16.4587-4595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J, Fujii S, Gruz P, Nohmi T, Fuchs RPP. The β clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Reports. 2000;1:484–488. doi: 10.1093/embo-reports/kvd109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Yang W. Structural insight into translesion synthesis by DNA Pol II. Cell. 2009;139:1279–1289. doi: 10.1016/j.cell.2009.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S, Hurwitz J. Conversion of oX174 viral DNA to double-stranded form by purified Escherichia coli proteins. Proc Natl Acad Sci USA. 1974;71:4120–4124. doi: 10.1073/pnas.71.10.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintersberger U. On the origins of genetic variants. FEBS Lett. 1991;285:160–164. doi: 10.1016/0014-5793(91)80796-6. [DOI] [PubMed] [Google Scholar]
- Witkin EM. Thermal enhancement of ultraviolet mutability in a tif-1 uvrA derivative of Escherichia coli B-r: evidence that ultraviolet mutagenesis depends upon the inducible function. Proc Natl Acad Sci USA. 1974;71:1930–1934. doi: 10.1073/pnas.71.5.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin EM, Kogoma T. Involvement of the activated form of RecA protein in SOS mutagenesis and stable DNA replication in Escherichia coli. Proc Natl Acad Sci USA. 1984;81:7539–7543. doi: 10.1073/pnas.81.23.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin EM, McCall JO, Volkert MR, Wermundsen IE. Constitutive expression of SOS functions and modulation of mutagenesis resulting from resolution of genetic instability at or near the recA locus of Escherichia coli. Mol Gen Genet. 1982;185:43–50. doi: 10.1007/BF00333788. [DOI] [PubMed] [Google Scholar]
- Wolff E, Kim M, Hu K, Yang H, Miller JH. Polymerase leave fingerprints: analysis of the mutational spectrum in Escherichia coli rpoB to assess the role of polymerase IV in spontaneous mutation. J Bacteriol. 2004;186:2900–2905. doi: 10.1128/JB.186.9.2900-2905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgate R, Ennis DG. Levels of chromosomally encoded Umu proteins and requirements for in vitro UmuD cleavage. Mol Gen Genet. 1991;229:10–16. doi: 10.1007/BF00264207. [DOI] [PubMed] [Google Scholar]
- Yao NY, O'Donnell M. Replisome dynamics and use of DNA trombone loops to bypass replication blocks. Mol Biosyst. 2008;4:1075–1084. doi: 10.1039/b811097b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao NY, Georgescu RE, Finkelstein J, O'Donnell ME. Single-molecule analysis reveals that the lagging strand increases replisome processivity but slows replication fork progression. Proc Natl Acad Sci USA. 2009;106:13236–13241. doi: 10.1073/pnas.0906157106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeiser B, Pepper ED, Goodman MF, Finkel SE. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc Natl Acad Sci USA. 2002;99:8737–8741. doi: 10.1073/pnas.092269199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner EL, Wu C, Marians KJ. Coordinated leading and lagging strand synthesis at the Escherichia coli DNA replication fork: II. Frequency of primer synthesis and efficiency of primer utilization controls Okazaki fragment size. J Biol Chem. 1992a;267:4045–4053. [PubMed] [Google Scholar]
- Zechner EL, Wu C, Marians KJ. Coordinated leading and lagging strand synthesis at the Escherichia coli DNA replication fork: III. Polymerase-primer interaction governs primer size. J Biol Chem. 1992b;267:4054–4063. [PubMed] [Google Scholar]