Abstract
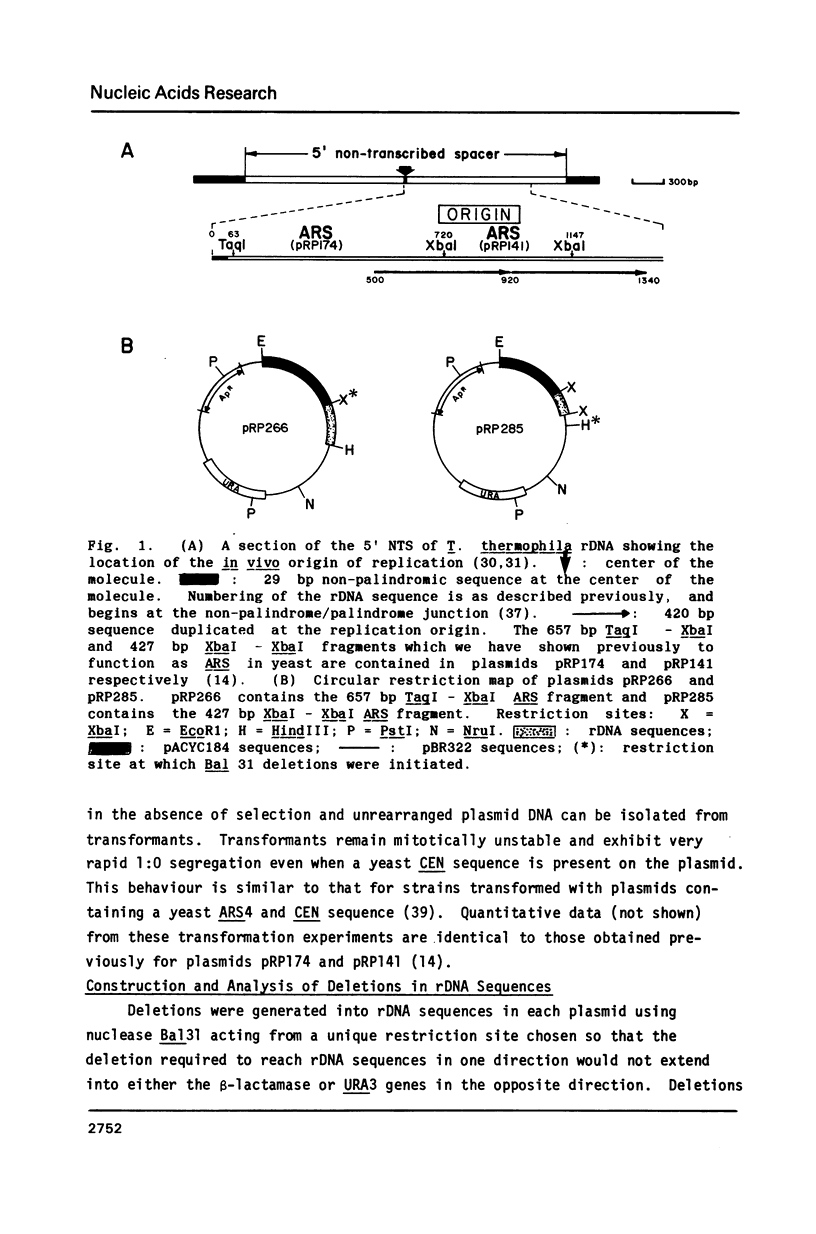
Two adjacent but non-overlapping restriction fragments that encompass the replication origin of the macronuclear copy of rDNA from Tetrahymena thermophila allow autonomous replication of plasmids in the yeast Saccharomyces cerevisiae; i.e. they function as autonomously replicating segments (ARS). Deletions generated in vitro into these fragments yield an 82 bp segment from each as the smallest sequence specifying ARS function. These 82 bp segments are at the 5' end of a 220 bp region of homology between the two original ARS restriction fragments. A 39 bp region of almost complete sequence identity between the two 82 bp fragments is suggested to be a core sequence element necessary for ARS function. This 39 bp sequence contains a region identical or nearly identical to the 11 bp yeast ARS consensus sequence (T/ATTTATPuTTTA/T) which is suggested to be essential for ARS function. Detailed comparisons of the 82 bp segments and of the 39 bp core with other ARS sequences reveal no extensive homologies aside from the consensus.
Full text
PDF











Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amin A. A., Pearlman R. E. Autonomously replicating sequences from the non transcribed spacers of Tetrahymena thermophila ribosomal DNA. Nucleic Acids Res. 1985 Apr 11;13(7):2647–2659. doi: 10.1093/nar/13.7.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., Li Y. Y., Feldman J., Jayaram M., Abraham J., Nasmyth K. A., Hicks J. B. Localization and sequence analysis of yeast origins of DNA replication. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1165–1173. doi: 10.1101/sqb.1983.047.01.132. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Brehm S. L. Replication of the extrachromosomal ribosomal RNA genes of Tetrahymena thermophilia. Nucleic Acids Res. 1981 Jul 24;9(14):3531–3543. doi: 10.1093/nar/9.14.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S. E., Campbell J. L. Yeast DNA replication in vitro: initiation and elongation events mimic in vivo processes. Cell. 1982 Nov;31(1):201–213. doi: 10.1016/0092-8674(82)90420-2. [DOI] [PubMed] [Google Scholar]
- Celniker S. E., Sweder K., Srienc F., Bailey J. E., Campbell J. L. Deletion mutations affecting autonomously replicating sequence ARS1 of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2455–2466. doi: 10.1128/mcb.4.11.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challoner P. B., Amin A. A., Pearlman R. E., Blackburn E. H. Conserved arrangements of repeated DNA sequences in nontranscribed spacers of ciliate ribosomal RNA genes: evidence for molecular coevolution. Nucleic Acids Res. 1985 Apr 11;13(7):2661–2680. doi: 10.1093/nar/13.7.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. S., Tye B. K. Autonomously replicating sequences in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6329–6333. doi: 10.1073/pnas.77.11.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dron M., Rahire M., Rochaix J. D., Mets L. First DNA sequence of a chloroplast mutation: a missense alteration in the ribulosebisphosphate carboxylase large subunit gene. Plasmid. 1983 May;9(3):321–324. doi: 10.1016/0147-619x(83)90009-4. [DOI] [PubMed] [Google Scholar]
- Fangman W. L., Hice R. H., Chlebowicz-Sledziewska E. ARS replication during the yeast S phase. Cell. 1983 Mar;32(3):831–838. doi: 10.1016/0092-8674(83)90069-7. [DOI] [PubMed] [Google Scholar]
- Gorman J. A., Dove W. F., Warren N. Isolation of Physarum DNA segments that support autonomous replication in yeast. Mol Gen Genet. 1981;183(2):306–313. doi: 10.1007/BF00270633. [DOI] [PubMed] [Google Scholar]
- Hartley J. L., Donelson J. E. Nucleotide sequence of the yeast plasmid. Nature. 1980 Aug 28;286(5776):860–865. doi: 10.1038/286860a0. [DOI] [PubMed] [Google Scholar]
- Hieter P., Mann C., Snyder M., Davis R. W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985 Feb;40(2):381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Higashinakagawa T., Saiga H., Shintani N., Narushima-Iio M., Mita T. Localization of putative transcription initiation site on the cloned rDNA fragment of Tetrahymena pyriformis. Nucleic Acids Res. 1981 Nov 25;9(22):5905–5916. doi: 10.1093/nar/9.22.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski S. M., Edelman G. M. Evidence for participation of a multiprotein complex in yeast DNA replication in vitro. J Biol Chem. 1984 Jun 10;259(11):6852–6857. [PubMed] [Google Scholar]
- Jazwinski S. M., Niedzwiecka A., Edelman G. M. In vitro association of a replication complex with a yeast chromosomal replicator. J Biol Chem. 1983 Mar 10;258(5):2754–2757. [PubMed] [Google Scholar]
- Kearsey S. Structural requirements for the function of a yeast chromosomal replicator. Cell. 1984 May;37(1):299–307. doi: 10.1016/0092-8674(84)90326-x. [DOI] [PubMed] [Google Scholar]
- Kiss G. B., Amin A. A., Pearlman R. E. Two separate regions of the extrachromosomal ribosomal deoxyribonucleic acid of Tetrahymena thermophila enable autonomous replication of plasmids in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Jun;1(6):535–543. doi: 10.1128/mcb.1.6.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D., Kent J. C., Hartwell L. H. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 1985 Feb;40(2):393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K., Wright A. P., Piper M., Shall S. Evaluation of heterologous ARS activity in S. cerevisiae using cloned DNA from S. pombe. Nucleic Acids Res. 1985 May 24;13(10):3711–3722. doi: 10.1093/nar/13.10.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Montiel J. F., Norbury C. J., Tuite M. F., Dobson M. J., Mills J. S., Kingsman A. J., Kingsman S. M. Characterization of human chromosomal DNA sequences which replicate autonomously in Saccharomyces cerevisiae. Nucleic Acids Res. 1984 Jan 25;12(2):1049–1068. doi: 10.1093/nar/12.2.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méchali M., Kearsey S. Lack of specific sequence requirement for DNA replication in Xenopus eggs compared with high sequence specificity in yeast. Cell. 1984 Aug;38(1):55–64. doi: 10.1016/0092-8674(84)90526-9. [DOI] [PubMed] [Google Scholar]
- Niles E. G., Cunningham K., Jain R. Structure of the Tetrahymena pyriformis rRNA gene. Nucleotide sequence of the transcription termination region. J Biol Chem. 1981 Dec 25;256(24):12857–12860. [PubMed] [Google Scholar]
- Palen T. E., Cech T. R. Chromatin structure at the replication origins and transcription-initiation regions of the ribosomal RNA genes of Tetrahymena. Cell. 1984 Apr;36(4):933–942. doi: 10.1016/0092-8674(84)90043-6. [DOI] [PubMed] [Google Scholar]
- Roth G. E., Blanton H. M., Hager L. J., Zakian V. A. Isolation and characterization of sequences from mouse chromosomal DNA with ARS function in yeasts. Mol Cell Biol. 1983 Nov;3(11):1898–1908. doi: 10.1128/mcb.3.11.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Skryabin K. G., Eldarov M. A., Larionov V. L., Bayev A. A., Klootwijk J., de Regt V. C., Veldman G. M., Planta R. J., Georgiev O. I., Hadjiolov A. A. Structure and function of the nontranscribed spacer regions of yeast rDNA. Nucleic Acids Res. 1984 Mar 26;12(6):2955–2968. doi: 10.1093/nar/12.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srienc F., Bailey J. E., Campbell J. L. Effect of ARS1 mutations on chromosome stability in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Jul;5(7):1676–1684. doi: 10.1128/mcb.5.7.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb D. T., Struhl K., Davis R. W. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979 Nov 1;282(5734):39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Delta sequences and double symmetry in a yeast chromosomal replicator region. J Mol Biol. 1982 Apr 5;156(2):293–307. doi: 10.1016/0022-2836(82)90330-8. [DOI] [PubMed] [Google Scholar]
- Vallet J. M., Rahire M., Rochaix J. D. Localization and sequence analysis of chloroplast DNA sequences of Chlamydomonas reinhardii that promote autonomous replication in yeast. EMBO J. 1984 Feb;3(2):415–421. doi: 10.1002/j.1460-2075.1984.tb01822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. B., Johnson M. J., Suggs S. V., Miyoshi K., Bhatt R., Itakura K. A set of synthetic oligodeoxyribonucleotide primers for DNA sequencing in the plasmid vector pBR322. Gene. 1981 Dec;16(1-3):21–26. doi: 10.1016/0378-1119(81)90057-3. [DOI] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Sequence-induced DNA curvature at the bacteriophage lambda origin of replication. Nature. 1985 Oct 3;317(6036):451–453. doi: 10.1038/317451a0. [DOI] [PubMed] [Google Scholar]
- Zakian V. A., Scott J. F. Construction, replication, and chromatin structure of TRP1 RI circle, a multiple-copy synthetic plasmid derived from Saccharomyces cerevisiae chromosomal DNA. Mol Cell Biol. 1982 Mar;2(3):221–232. doi: 10.1128/mcb.2.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]


