Abstract
The family of Mps One binder (MOB) co-activator proteins is highly conserved from yeast to man. At least two different MOB proteins have been identified in every eukaryote analysed to date. Initially, yeast genetics revealed essential roles for Mob1p and Mob2p in the regulation of mitotic exit and cell morphogenesis. Studies in flies then showed that dMOB1/MATS is a core component of Hippo signalling. Loss of dMOB1 resulted in increased cell proliferation and decreased cell death, suggesting that MOB1 acts as tumour suppressor protein. Recent work focused primarily on mammalian cells has shown how hMOB1 can regulate NDR/LATS kinases, a function that can to be counteracted by hMOB2. Here we summarise and discuss our current knowledge of this emerging protein family, with emphasis on subcellular localisation, protein-protein interactions and biological functions in apoptosis, mitosis, morphogenesis, cell proliferation and centrosome duplication.
Keywords: Mps one binder; Protein kinase signal transduction; NDR/STK38 and LATS kinases; Protein-protein interactions; Phosphorylation; MEN, SIN and Hippo pathways
1. Introduction
Signal transduction cascades control essential biological processes, such as cell division, morphogenesis, cell growth, and controlled cell death/apoptosis. The vast majority of signalling machineries transmit extra- and intracellular inputs through protein kinases, hence it is not surprising that protein kinases are one of the largest superfamilies found in the human genome [1]. Research has shown that members of the AGC kinase subfamily of protein kinases play diverse and crucial cellular functions [2]. In particular, members of a subgroup of the AGC group of protein kinases termed the LATS (large tumour suppressor) / NDR (nuclear Dbf2-related) family of kinases have attracted much attention over the past few years. This family of kinases is highly conserved from yeast to man [3]. Human cells express four related NDR kinases: NDR1 (also known as serine/threonine kinase 38 or STK38), NDR2 (or STK38L), LATS1 (large tumour suppressor-1) and LATS2. In Drosophila melanogaster, Trc (tricornered) and Warts kinases correspond to human NDR1/2 and LATS1/2, respectively. In Saccharomyces cerevisiae, Dbf2p/Dbf20p and Cbk1p have been classified as NDR/LATS kinases, and in Schizosaccharomyces pombe the kinases Sid2p and Orb6p are the counterparts of mammalian NDR/LATS.
A decade ago, members of the NDR/LATS family were of central interest to several yeast geneticists based on the essential roles of Dbf2p and Sid2p in mitotic exit of yeast cells [4]. Over the past years, fly geneticists grew more and more interested in Trc and Warts due to the involvement of NDR/LATS kinases in Hippo signalling, a novel tumour suppressor cascade that controls the proto-oncogene Yorkie (Yki). Yki has attracted a lot of attention in its own right since the proto-oncogenes YAP and TAZ are the human orthologues of Yki [2, 5-9]. Significantly, NDR/LATS kinases do not function independently. On the one hand, members of the Ste20 kinase family are required to phosphorylate and thereby activate NDR/LATS kinases [3]. On the other hand, binding of members of the MOB (Mps one binder) protein family is essential for NDR/LATS kinases to fully function (see section 5 for a detailed discussion).
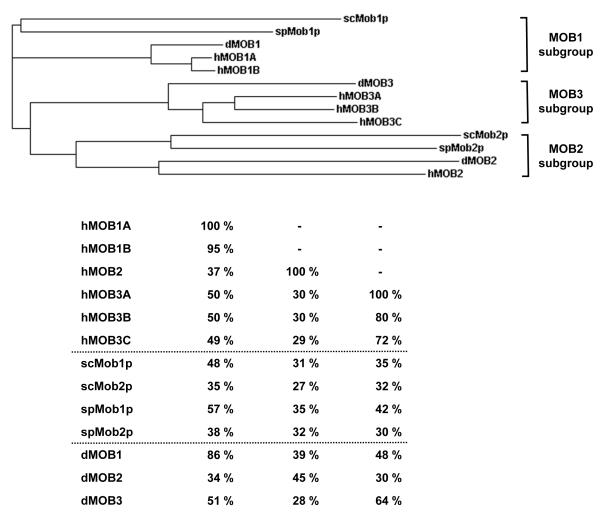
In budding and fission yeast, MOB proteins are expressed by two independent genes. Mob1p associates with Dbf2p and Sid2p, and Mob2p binds to Cbk1p and Orb6p, respectively. In Drosophila, the picture is more complicated, as the fly genome encodes three different MOBs, termed dMOBs (Drosophila MOB proteins). In mammals, the complexity is even higher, because at least six different MOB proteins are encoded by independent genes. As if the biological complexity is not challenging enough, the research community has given each human MOB protein several names. Therefore, the nomenclature used in this review has been simplified as follows: hMOB1A (also termed MOBKL1B, MATS1, Mob1A, hMOB1B and Mob4a), hMOB1B (MOBKL1A, Mob1b, hMOB1A, and Mob4b), hMOB2 (HCCA2, and hMOB3), hMOB3A (MOBKL2A, Mob3A, MOB-LAK, hMOB2A, Mob1C), hMOB3B (MOBKL2B, Mob3B, hMOB2B, Mob1D), and hMOB3C (MOBKL2C, Mob3C, hMOB2C, Mob1E). The phylogenetic tree and sequence identities of MOB proteins expressed in yeast, fly and human cells are displayed in Fig. 1 using this proposed nomenclature.
Figure 1. Phylogenetic relationships within the MOB protein family.
Top: Phylogenetic tree using Clustal W phylogenetic calculation based on the neighbour-joining method. Budding and fission yeast scMob1p and spMob1p, respectively, group together with dMOB1 and hMOB1A/B (MOB1 subgroup), while scMob2p and spMob2p fall into a group together with dMOB2 and hMOB2 (MOB2 subgroup). dMOB3 together with hMOB3A/B/C forms a third group (MOB3 subgroup). Bottom: Display of primary sequence identities within the MOB protein family. hMOB1 is closest related to scMob1p, spMob1p, and dMOB1 in the respective species. hMOB2 is not as well conserved as hMOB1, since it aligns similarly with several MOB proteins in yeast and flies. The MOB3 subgroup displays close identity between dMOB3 and hMOB3A/B/C.
2. MOB proteins in budding and fission yeast
In 1998, Luca and Winey described the first MOB protein [10]. By screening for novel interactors with the Mps1p kinase, they discovered Mob1p, standing for Mps one binder 1p. Their initial data suggested that Mob1p is essential for the survival of cells, and plays a role in spindle pole body duplication of Saccharomyces cerevisiae (the yeast equivalent of centrosome duplication in multicellular organisms). However, follow up studies by several laboratories revealed that Mob1p also plays a central role in the mitotic exit network (MEN) of budding yeast [11-16]. In summary these studies showed that after the initial activation of Tem1p, the Cdc15p protein kinase is stimulated, followed by increased activity of the Mob1p/Dbf2p complex. This results in the release of protein phosphatase Cdc14p from the nucleolus into the cytoplasm, thereby leading to a dephosphorylation wave through which cyclin B/cdc34p is inactivated, finally allowing the exit from mitosis (Fig. 2, see also [4]). For some time it was unclear how Mob1p in complex with Dbf2p relayed the signal to Cdc14p [4], however, Mohl et al. (2009) recently showed that the Mob1p/Dbf2p complex can directly phosphorylate Cdc14p, thereby triggering the required relocalisation of Cdc14p at the end of mitosis [17].
Figure 2. MOB signalling in mitosis and morphogenesis in Saccharomyces cerevisiae.
(A) Mob1p together with the Ste20-like kinase Cdc15p activates the Dbf2p NDR/LATS kinase. The Mob1p/Dbf2p complex phosphorylates the phosphatase Cdc14p, thereby triggering mitotic exit of yeast cells. (B) Mob2p cooperates with the Ste20-like kinase Kic1p to stimulate the Cbk1p NDR/LATS kinase. The Mob2p/Cbk1p complex targets Ace2p and other factors to regulate Ace2p activity and cellular morphogenesis (RAM). Of note, Mob1p functions in complex with Dbf2p and not Cbk1p, and Mob2p functions together with Cbk1p and not Dbf2p. Signalling components are not interchangeable. Ste20-like kinases are in yellow. NDR/LATS kinases are green, and MOB proteins are shown in red. Phosphorylations are indicated by “P” in blue.
In Schizosaccharomyces pombe, Mob1p is also essential for controlled exit of mitosis [4]. Given the differences between budding and fission yeast in mitosis, the budding yeast MEN was termed septation initiation network (SIN) in S. pombe. Using a similar signalling cascade as reported for MEN, Mob1p in complex with the kinase Sid2p (the fission yeast counterpart of Dbf2) controls the Clp1p phosphatase (the counterpart of Cdc14p) [18-20]. Significantly, the Mob1p/Sid2p complex also phosphorylates Clp1p, thereby affecting the subcellular localisation of Clp1p [21]. In summary, in budding and fission yeast, Mob1p in complex with the NDR/LATS kinases Dbf2p or Sid2p is essential for the functionality of MEN and SIN, respectively [4].
Significantly, Luca and Winey also laid the foundation for research on the second MOB protein in S. cerevisiae [10]. In addition to defining the first function of Mob1p, they showed in the same report that Mob2p is a non-essential protein in yeast [10]. Nevertheless, research revealed an important role for Mob2p in the regulation of asymmetry in budding yeast by inducing Ace2p daughter-specific genetic programs [22, 23]. Additional studies showed that Mob2p in complex with Cbk1p (the second NDR/LATS kinase in S. cerevisiae) controls polarized growth, hence the signalling network centering around Mob2p was termed RAM (regulation of Ace2p activity and cellular morphogenesis) [24, 25]. In S. pombe, Mob2p was also found in complex with the second NDR/LATS kinase Orb6p [26]. In full support of the findings in budding yeast, studies showed that the Mob2p/Orb6p complex coordinates a morphogenesis network in S. pombe [26, 27].
In summary, budding and fission yeast express two different MOB proteins. Mob1p binds to Dbf2p and Sid2p, respectively, thereby having a central function in the control of exit from mitosis. Mob2p, on the other hand, associates with Cbk1p and Orb6p, respectively, playing a key part in the regulation of cell morphogenesis networks. Significantly, Mob1p does not interact with Orb6p, and Mob2p does not associate with Sid2p [18, 26]. Therefore, in yeast Mob1p and Mob2p function in independent, non-interchangeable complexes, thereby regulating two different biological processes by their binding to the corresponding NDR/LATS kinases (Fig. 2).
3. MOB proteins in Drosophila melanogaster
In 2005, fly geneticists started the hunt for understanding physiological functions of MOB proteins in multicellular organisms. Lai et al. (2005) described that dMOB1 (also termed MATS for MOB as tumour suppressor) is essential for the control of cell proliferation and death in D. melanogaster [28]. Significantly, they showed that dMOB1 is the counterpart of hMOB1A, since the loss of dMOB1 could be rescued by hMOB1A expression. Furthermore, they reported that complete loss of dMOB1 is lethal for flies [28]. Lai and colleagues then studied flies lacking dMOB1 in more detail, revealing that dMOB1 is essential for early development, most likely because dMOB1 is required for proper chromosomal segregation in developing embryos [29]. However, since the mitotic spindle checkpoint does not seem to be compromised upon loss of dMOB1 [29], it is yet unclear how loss of dMOB1 can lead to aberrant mitoses. One possibility is that dMOB1 functions in mitosis together with Warts (the LATS1/2 kinases counterpart in flies) as already shown by Lai and colleagues for the control of cell proliferation and death [28, 30]. Nevertheless, future research has yet to determine whether dMOB1 signals in complex with Warts in mitosis.
Since hMOB1 can activate LATS1 when targeted to the plasma membrane [31], the Lai laboratory also analysed the role of dMOB1 at the plasma membrane [32]. Significantly, they observed that endogenous dMOB1 is located at the plasma membrane in developing tissues. By constitutively targeting dMOB1 to the plasma membrane, they further showed that membrane-targeted dMOB1 can cause reduced tissue growth and organ size by regulating Warts kinase [32]. Therefore, it is very likely that at least in flies the plasma membrane is an important site of action for dMOB1 with respect to its role in tumour suppression. Taken together, dMOB1 is now an established core component of Hippo tumour suppressor signalling by functioning as an activator of Warts, one of two NDR/LATS kinases in D. melanogaster [8, 9]. Nevertheless, the current picture is more complicated, as dMOB1 also interacts genetically with Trc, the second NDR/LATS kinase in flies [33]. The Trc dominant negative phenotype is enhanced by a mutation in dMOB1, and larval phenotypes of flies lacking dMOB1 are similar to that of flies without functional Trc [33]. These findings indicate that dMOB1 does not specifically bind to a single NDR/LATS kinase (Fig. 3), as observed in budding and fission yeast (Fig. 2). Perhaps future research on dMOB1 will unveil how dMOB1 can regulate Warts and Trc kinases in their respective biological processes, as already reported for their upstream kinase Hippo in dendritic tiling and maintenance [34].
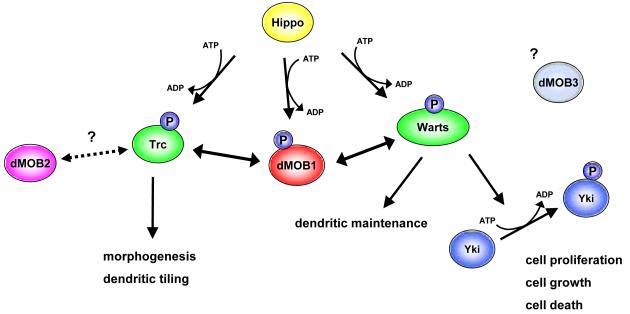
Figure 3. MOB signalling in Drosophila melanogaster.
dMOB1/MATS together with the Ste20-like kinase Hippo activates both NDR/LATS kinases, namely Trc and Warts. While the Trc branch is important for morphological changes such as outgrowth of epidermal hair and dendritic tiling in sensory neurons, the Warts branch is required for dendritic maintenance in sensory neurons, in addition to controlling the balance of cell proliferation and apoptosis as part of Hippo signalling. In Hippo signalling, the dMOB1/Warts complex phosphorylates the proto-oncogene Yki, which in turn inactivates the transcriptional co-activator Yki. Substrates of the dMOB1/Trc complex are unknown. dMOB2 might function together with Trc in controlling morphogenesis. The role of dMOB3 is not yet understood. Ste20-like kinases are in yellow. NDR/LATS kinases are green, and MOB proteins are shown in red, purple and light blue. Phosphorylations are indicated by “P” in blue.
In contrast to yeast, flies express not two but three different MOB proteins. In spite of growing interest in dMOB1, the biological functions of dMOB2 and dMOB3 are less understood. dMOB2 seems to play a role in wing hair morphogenesis [33]. The Adler laboratory showed further by co-immunoprecipitation and yeast two hybrid experiments that dMOB2 can form a complex with Trc [33]. This suggests that dMOB2 might function in complex with Trc. Notably, the role of dMOB2 in flies seems to differ from that of dMOB1, given that mutations in the dMOB2 gene do not significantly synergise with loss or gain of Trc kinase in enhancing morphological phenotypes [33]. Nevertheless, overexpression of a truncated form of dMOB2 leads to fly wing phenotype similar to the Trc mutant [33], suggesting a potential dominant-negative role of dMOB2 in flies. In this context, it is very intriguing that the truncated dMOB2 variant displays high sequence similarity with full-length hMOB2 [35], recently been shown to function as an inhibitor of NDR1/2 kinase signalling by competing with hMOB1 in mammalian cells [35]. Therefore, it is very tempting to speculate that dMOB2 can inhibit Trc by a similar mechanism as reported for the human counterpart [35]. Research is now required to test whether dMOB2 can negatively regulate the Trc kinase by competing with dMOB1. Recently, dMOB2 was also reported to play a role in the development of photoreceptor cells [36], but the mechanism of action is currently unknown. Any clear function or binding partner of dMOB3 also remains elusive, although dMOB3 was initially reported to genetically interact with Trc [33].
In summary, one can conclude that, unlike in yeast, D. melanogaster individual Mob proteins interact with both NDR/LATS kinases (Fig. 3). Furthermore, individual NDR/LATS kinases are able to interact with different Mob proteins (Fig. 3). dMOB1 plays a role in the control of Trc and Warts kinases. The interaction of dMOB1 with Warts is much better understood, since the dMOB1/Warts complex is a core component of Hippo signalling, which is an essential tumour suppressor cascade required for the correct coordination of cell proliferation and apoptosis. Consequently, loss of dMOB1 results in tissue overgrowth that is associated with increased cell proliferation and decreased cell death. In addition to dMOB1, dMOB2 seems to control Trc, but we currently cannot exclude the possibility that dMOB2 might also play a role in the regulation of Warts. dMOB3 might also play a part in Trc and Warts signalling, however, this research aspect is yet to be addressed. Taken together, the current evidence supports the notion that in multicellular organisms, the binding of MOB proteins is not restricted to a unique NDR/LATS kinase.
4. MOB proteins in mammals
4.1 Subcellular localisation and binding partners of mammalian MOB proteins
hMOB1A and hMOB1B (also termed hMOB1) are mainly cytoplasmic proteins [37, 38], but can hyperactivate NDR/LATS kinases when targeted to the plasma membrane of mammalian cells [31, 37]. hMOB1 also plays a role in centrosome duplication [38], but although hMOB1 has been reported to localise on centrosomes [39, 40], it is yet unclear whether the centrosomal pool of hMOB1 is the key player in centrosome duplication. Recently hMOB1 was also found on kinetochore structures of mitotic cells [39], but the biological importance of this kinetochore association is yet to be established.
hMOB1 can bind to all four human NDR/LATS kinases [31, 35, 37, 38, 40-48]. Intriguingly, these interactions are mediated through a domain in NDR/LATS kinases which is conserved from yeast to man [3]. The importance of these interactions is discussed in section 5 below. Furthermore, hMOB1A has been reported to associate with various proteins, such as NDR2/STK38L, S100A8, TRIP6, TSSC1, Nup98, HDAC3, DEGS, SGK, and TRAF6 [49]. However, except for the interaction of hMOB1A with NDR kinases [37, 38, 41, 44], the binding of these factors to hMOB1A has not been confirmed by conventional interaction assays. A second large-scale screen revealed that hMOB1A appears to associate with DIPA [50], which may be of interest, since this coiled-coil protein has been detected on centrosomes [51]. However, the importance of this interaction has as yet not been studied. hMOB1B has not been found in any screens addressing protein-protein interactions, most likely because hMOB1B is 95% identical to hMOB1A ([44, 52] see also Fig. 1) and hence, researchers did not discriminate between the two proteins when analysing their samples.
In contrast to all other MOB proteins, a substantial fraction of hMOB2 is detected in the nucleus [37, 38]. hMOB2 binds to NDR1/2 kinases, but does not associate with LATS1/2 kinases [31, 35, 37, 38, 40, 44]. Significantly, hMOB2 binds to the same domain on NDR1/2 as hMOB1 [35], and this binding opposes the activation of NDR1/2 by hMOB1 (please see section 5 for a more detailed discussion of this point).
Similar to hMOB1, hMOB3A/B/C are mainly cytoplasmic proteins [38, 53], which neither bind to NDR1/2 nor to LATS1/2 kinases [35]. hMOB3A/B/C do not activate NDR1/2 and LATS1/2 kinases [35], in spite of high sequence similarity between hMOB3A/B/C and hMOB1 (Fig. 1). hMOB3B seems to interact with Mitogen activated protein kinase p38 alpha (also termed MAPK14) [54], CNK2/KSR2 [49], and NT5C2 [50]. However, these interactions have as yet not been confirmed by standard co-immunoprecipitation and co-fractionation experiments. No binding partners for hMOB3A or hMOB3C have been reported to date.
4.2 Mammalian MOB proteins in disease
Human cells express two hMOB1 proteins (hMOB1A and hMOB1B) that are 95% identical at the protein level ([37, 44, 52] see also Fig. 1). Mutations in hMOB1A have been found in human cancer cells [28]. hMOB1A mRNA levels are low in human colorectal and lung cancer samples [55, 56]. In support of this putative tumour suppressor role, expression of hMOB1A in flies lacking Mats/dMOB1 (the fly counterpart of hMOB1) is sufficient to restore normal tissue growth [28], suggesting that hMOB1A is indeed a tumour suppressor protein. In this context it is noteworthy to mention that inactivation of the murine MOB1B locus seems to correlate with cancer development [57]. In further support of a tumour suppressor role for hMOB1, hMOB1 phosphorylation of Thr12 was significantly decreased in human liver cancer samples, displaying a strong correlation with reduction of inactivating phosphorylation of the proto-oncogene YAP [58]. However, a clear tumour suppressive function of hMOB1 has not yet been validated in a mammalian system. Most likely, given that MOB1B phosphorylation on Thr35 was observed in the spleen, brown fat, liver, pancreas and testis [59], the inactivation of MOB1 in various tissues will have to be analysed in the context of tumour suppression.
Elevated expression of the mRNA encoding for hMOB2 is associated with hepatocellular carcinoma development and progression [60]. However, the hMOB2 mRNA also encodes an overlapping (but entirely distinct) ORF [60]. Therefore, it is currently unclear whether the putative 50 kDa HCCA2 (HepatoCellular Carcinoma-associated gene 2) protein or hMOB2 is driving cancer development. Unfortunately, in spite of the clear differences between hMOB2 and HCCA2, the vast majority of databases still use the synonyms hMOB2 and HCCA2 interchangeably.
As mentioned in section 4.1, hMOB3B seems to interact with MAPK14, CNK2/KSR2, and NT5C2. Given that NT5C2 expression levels have been used as a prognostic marker in lung cancer and haematological malignancies [61-63], it is tempting to speculate that the interaction with NT5C2 might be the most relevant in human disease. In this context, one should note that altered expression of hMOB3A and hMOB3B has been reported in mantle cell lymphoma [64, 65]. However, studies addressing the biological significance of changes in hMOB3A/B expression are missing to date. Any involvement of hMOB3C in human diseases is yet to be reported.
Taken together, hMOB1 seems to represent a tumour suppressor protein. Potentially, loss of MOB1 could result in similar loss of function phenotypes as reported for its binding partners NDR1 [66] and LATS1/2 [5]. However, MOB1 knock-out animals have not been reported to date. Transgenic animals allowing researchers to address the physiological functions of MOB2 and MOB3A/B/C are also not available. We are therefore likely to see exciting new discoveries related to MOB proteins and their role in cancer biology in the near future.
5. Regulation and functions of mammalian MOB proteins
5.1. Regulation of and by MOB proteins
In 1998, besides describing the MOB proteins Mob1p and Mob2p, Luca and Winey also provided the first lead in the regulation of Mob1p by observing that Mob1p is a phospho-protein [10]. Mah et al. (2001) then reported that Cdc15p (the yeast counterpart of the Hippo kinase in flies) can phosphorylate Mob1p [15]. Next, Wei and colleagues published that the phosphorylation of dMOB1 by Hippo could play a role in Hippo signalling [30]. Thus, genetic and biochemical studies of yeast and flies not only revealed the involvement of MOBs in MEN, SIN, Hippo and morphogenesis networks, but also laid the foundation for studies addressing how MOB proteins are regulated. Nevertheless, the precise mechanisms of MOB regulation have been established in mammalian systems. The Avruch laboratory studied in detail the phosphorylation of hMOB1 by MST1/2 kinases (the two human counterparts of Cdc15p and Hippo kinases in yeast and flies, respectively) [67]. Praskova et al. observed that hMOB1 is specifically phosphorylated on Thr12 and Thr35 by MST1 and MST2, where active MST2 bound to hMOB1, while kinase-dead MST2 and wild-type MST1 did not [67]. The significance of these differences in protein-protein interactions is unknown, but does not seem to affect the phosphorylation of hMOB1 by MST1 or MST2. Therefore, Praskova et al. focused on understanding the importance of the phosphorylation of hMOB1, and not the differences in complex formations. Their efforts revealed that the phosphorylation of hMOB1 on Thr12/Thr35 is required for the interaction of hMOB1 with NDR/LATS kinases [67], which was confirmed later by another laboratory [68]. Most likely phospho-hMOB1 has an increased affinity for NDR/LATS kinases due to conformational changes triggered by the phosphorylation of Thr12/Thr35. Unfortunately, the published crystal structure of hMOB1A does not contain the N-terminal 32 residues [52], therefore limiting our current understanding on how phosphorylation on Thr12/Thr35 of hMOB1A could affect the three-dimensional structure of this globular co-activator protein. Insight into this aspect will be crucial, since the phosphorylation of hMOB1 on Thr12/Th35 appears to be important in cell proliferation [67], apoptosis signalling [58], and the control of the proto-oncogene YAP [58]. As mentioned earlier, it is also striking that phosphorylation on Thr35 appears to be tissue specific [59], suggesting tissue specific roles for the phosphorylation of hMOB1 have to be addressed by future research projects. Furthermore, in yeast, Mob1p is phosphorylated and regulated by the highly conserved Cdk1 kinase [69]. However, the phosphorylation sites are not conserved between yeast Mob1p and human MOB1 [69], suggesting that this crosstalk might be specific to yeast cells. Nevertheless, it could well be that hMOB1 is regulated by additional post-translational modifications other than Thr12/Thr35 phosphorylation. Regulatory modifications of hMOB2 or hMOB3A/B/C have not been reported.
Having somewhat understood how hMOB1 can be regulated, another question was (and to some degrees still is) how hMOB1 could regulate other proteins. As already discussed in section 4.2, hMOB1 appears to bind to a variety of components, however, the complex formations with NDR/LATS kinases are the only protein-protein interactions studied in detail. Combined structural and biochemical studies have shown that hMOB1 binds to NDR/LATS kinases through conserved residues on hMOB1 and NDR/LATS kinases [3, 52, 70, 71]. A stretch of conserved hydrophobic and positively charged residues N-terminal of the catalytic domains of NDR/LATS kinases is required for the association of hMOB1 with NDR/LATS kinases [31, 35, 37, 38, 40, 41]. On the other hand, a conserved cluster of negatively charged residues on hMOB1 is needed for the interaction of hMOB1 with NDR/LATS kinases [38, 52, 71].
The binding of hMOB1 to NDR1/2 kinases through these conserved domains triggers auto-phosphorylation of NDR1/2 on the activation segment (also known as T-loop phosphorylation), and also facilitates the phosphorylation of NDR1/2 on the hydrophobic motif (HM phosphorylation) by MST kinases [35, 37, 38, 41, 46, 47]. In contrast, binding of hMOB1 to LATS1/2 kinases seems to be only required for T-loop phosphorylation, while HM phosphorylation is independent of hMOB1/LATS complex formation [67]. Somewhat surprisingly T-loop and HM phosphorylation of LATS1/2 kinases can also occur without increased hMOB1 phosphorylation on Thr12/Th35 [58], suggesting that hMOB1/LATS complex formation is not required at all for efficient phosphorylation of LATS1/2 (at least in hydrogen peroxide trigger signalling). Taken together, these findings illustrate that hMOB1/NDR and hMOB1/LATS complexes are not equal, thereby allowing cells to trigger different biological outputs through hMOB1 signalling (see section 5.2 below for a summary).
Another difference between hMOB1/NDR and hMOB1/LATS complexes is how they can be influenced by hMOB2. Surprisingly, in spite of the highly conserved hMOB1 binding domain shared between NDR and LATS kinases [3, 31], hMOB2 interacts with NDR1/2 kinases through the hMOB1 binding domain, but does not associate with LATS1/2 kinases [35]. Significantly, binding of hMOB2 to NDR1/2 kinases inhibits the phosphorylation of NDR on the T-loop and HM motifs, thereby blocking kinase activation [35]. This inhibition is most likely caused by competition between hMOB1 and hMOB2 for direct binding to the same domain within NDR1/2 kinases, since a hMOB2 mutant deficient in NDR1/2 binding could not interfere with the activation of NDR by hMOB1 [35].
In summary (see also [5]), the efficacy of hMOB1 binding to NDR/LATS kinases is dramatically increased upon Thr12/Thr35 phosphorylation of hMOB1 by MST1/2 kinases. Increased formation of hMOB1/NDR and hMOB1/LATS complexes results in increased T-loop auto-phosphorylation of NDR/LATS kinases. The interaction of hMOB1 with NDR is also important for HM phosphorylation of NDR1/2 by MST kinases, while the HM phosphorylation of LATS1/2 by MST kinases can be uncoupled from hMOB1/LATS complex formation. A second human MOB protein, hMOB2, also plays a role in the regulation of NDR/LATS kinase signalling, more specifically in the control of NDR signalling. hMOB2 competes with hMOB1 for the binding to a conserved domain of NDR1/2 kinases, thereby interfering with the activation of NDR kinases by hMOB1. Therefore, hMOB2 has been classified as an inhibitor of NDR signalling, while hMOB1 has been classified as a co-activator of NDR/LATS signalling cascades.
5.2. Biological functions of MOB proteins in mammals
As mentioned before (see section 4), human cells express at least six different MOB proteins. As outlined above (see section 5.1), reports have revealed hMOB1 and hMOB2 influence NDR/LATS signalling, while hMOB3A/B/C are currently rather uncharacterised proteins without any known binding partner (Fig. 4). Not surprisingly, given that hMOB1 can bind to NDR1/2 as well as LATS1/2 kinases, researchers have found that hMOB1 plays a role in different biological processes (Fig. 4). Considering that Mob1p has a key role in the yeast MEN/SIN pathways [4], it was not surprising that loss of hMOB1 was associated with altered spindle checkpoint signalling and prolonged telophase progression in human cells [40]. In this report [40] hMOB1 was published as a centrosomal protein, a subcellular localisation also reported recently by Wilmeth et al. [39]. However, the Schuster laboratory also observed endogenous and exogenous hMOB1 on kinetochore structures in early mitosis, and the spindle midzone/midbody in later mitotic stages [39], suggesting that hMOB1 might have several roles during mitotic progression. This conclusion is further supported by the notion that in budding yeast the Luca laboratory found Mob1p associated with centromeres, thereby regulating the recruitment of factors to kinetochore structures [72]. Similarly, depletion of hMOB1 also alters the subcellular localisation of chromosomal passenger complex proteins [39]. Nevertheless, although these observations make sense when considering potential functional conservation from yeast to man, the biological significance has not been reported yet. Therefore, more detailed analyses remain to be undertaken to decipher the involvement of hMOB1 in mammalian MEN/SIN pathways. This research avenue could be very interesting, since loss of dMOB1 results in aberrant mitosis in fly embryos [29], indicating that researchers will be able to study mitotic functions of MOB1 in yeast, fly and human cells.
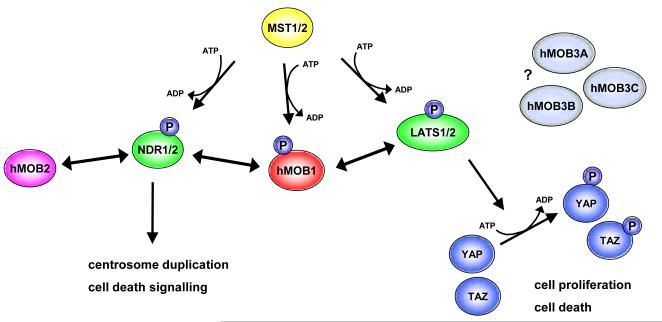
Figure 4. MOB signalling in human cells.
hMOB1 together with the Ste20-like kinases MST1/2 activates all four NDR/LATS kinases, namely NDR1/2 and LATS1/2. While the NDR branch is important for centrosome and apoptosis biology, the LATS branch is required for controlling cell proliferation and apoptosis as part of Hippo signalling. In mammalian Hippo signalling, the hMOB1/LATS complex phosphorylates the proto-oncogenes YAP and TAZ, which in turn inactivates these transcriptional co-activators. Substrates of the hMOB1/NDR complex in centrosome duplication and apoptosis signalling are unknown. hMOB2 counteracts hMOB1/NDR signalling by competing with hMOB1 for binding to NDR. Functions and binding partners of hMOB3A/B/C are currently unknown. Ste20-like kinases are in yellow. NDR/LATS kinases are green, and MOB proteins are shown in red, purple and light blue. Phosphorylations are indicated by “P” in blue.
The involvement of MOB1 in centrosome duplication (the equivalent of spindle pole body duplication in yeast) could potentially also be studied in different cellular systems. More than a decade ago, it was reported that loss of Mob1p results in defective spindle pole body duplication [10]. Recently, our research has shown that hMOB1 is required for efficient centrosome duplication in human cells [38], suggesting perhaps centrosome-associated hMOB1 plays a role in centrosome duplication in S-phase. However, hMOB1 localisation on centrosomes [39, 40] does not necessarily imply the centrosomal pool of hMOB1 is the key player in centrosome duplication. Nevertheless, we know that hMOB1/NDR complex formation is required for centrosome duplication in human cells [38], a function counteracted by overexpression of hMOB2 [35]. Most likely centrosome-associated hMOB1 has different functions depending on the cell cycle stage of a cell. Likely, in S-phase centrosome duplication is supported by centrosome-bound hMOB1, and in mitosis chromosomal passenger proteins are controlled by hMOB1 using centrosomes as a signalling platform. Significantly, centrosomal hMOB1 may also function in centrosome disjunction (the detachment of mature centrioles in G2/M-phase). The Schiebler and Fry laboratories reported that depletion of hMOB1 had some effect on Nek2 centrosome localisation, and thereby could potentially play a role in centrosome disjunction [73]. Taken together, these publications suggest that we have just started to understand the roles of different subcellular pools of hMOB1, and many interesting discoveries are still to come.
Significantly, hMOB1 does not only function in mitosis and centrosome biology. Depletion of hMOB1 also negatively affects apoptotic signalling [47], and might have some effect on p16/INK4A controlled biological processes [74]. While the involvement of hMOB1 in apoptosis has been addressed in some detail [47], the crosstalk between hMOB1 and p16/INK4A signalling remains to be defined clearly. Furthermore, loss of Thr12/Thr35 phosphorylation on hMOB1 has been associated with decreased inhibitory phosphorylation of the proto-oncogene YAP [58]. Given that Thr12/Thr35 phosphorylation on hMOB1 is required for efficient binding to NDR/LATS kinases [67], this finding suggests that YAP phosphorylation is decreased due to reduced hMOB1/NDR and/or hMOB1/LATS complex formation. Puzzlingly, in spite of decreased YAP phosphorylation, LATS phosphorylation was not affected upon loss of hMOB1 phosphorylation [58], suggesting that another kinase than LATS is controlling YAP in liver cells. Perhaps NDR kinase in complex with hMOB1 does the job in this setting. Whatever the case, this surprising finding by Zhou et al. [58] warrants novel research avenues on mechanisms controlling YAP activity. Furthermore, the findings by Zhou et al. [58] highlight that mammalian Hippo signalling appears to be more complex than Hippo signalling in flies [8, 9]. Last, but not least, it is important to note when summarising biological roles of hMOB1 that Praskova et al. (2008) reported that loss of Thr12/Thr35 phosphorylation on hMOB1 resulted in increased cell proliferation of human cells [67]. This suggests that phospho-hMOB1 can inhibit cell cycle progression. Therefore, considering that Thr12/Thr35 phosphorylation on hMOB1 is required for efficient binding to NDR/LATS kinases [67, 68], it might be that uncontrolled increase of hMOB1/NDR and hMOB1/LATS complex formation has a negative effect on the cell cycle. Of course, another possibility could be that an as yet unidentified hMOB1 binding partner is the main player in hMOB1 controlled cell proliferation. Whatever the case, various biological processes are to be (re-)addressed in order to decipher the function of endogenous hMOB1, with focus on physiologically relevant interactions of hMOB1 with kinases and other signalling components.
As discussed in sections 4 and 5.1, hMOB1 is not the only MOB protein that plays a role in centrosome duplication and apoptosis signalling (Fig. 4). Significantly, overexpression of wild-type hMOB2 was sufficient to block centrosome overduplication and strongly delay induction of apoptosis upon activation of death receptor signalling [35]. Importantly, we defined how hMOB2 functions in these settings. By using a hMOB2 mutant deficient in NDR1/2 binding, we showed that hMOB2 directly competes with hMOB1 for binding to NDR1/2 [35], thereby negatively regulating two crucial biological processes, namely centrosome and apoptosis biology. In other words, while hMOB1 can stimulate T-loop autophosphorylation and facilitate HM phosphorylation of NDR/LATS [3, 38, 47, 67], binding of hMOB2 to NDR1/2 kinases inhibits NDR activation [35]. Therefore, hMOB1 and hMOB2 appear to have opposing functions, with hMOB1 as co-activator and hMOB2 as inhibitor of NDR1/2 kinases.
Besides functioning in centrosome and apoptosis biology [35], hMOB2 also has a role to play in controlling cell morphology. Lin et al. (2011) reported recently that manipulation of MOB2 expression resulted in altered neurite formation in a mouse neuroblastoma cell line [75]. Depletion of MOB2 caused decreased neurite outgrowth, while overexpression of MOB2 dramatically elevated neurite formation [75]. The authors also provide evidence suggesting that MOB2 co-operates with NDR2 kinase in neuritogenesis [75]. However, since they did not address the interaction of MOB2 with NDR2 by mutants deficient in binding, it is currently unclear how direct this synergetic effect actually is. Furthermore, considering that hMOB2 has been defined by different experimental approaches as an inhibitor of NDR signalling [35], the positive co-operation between MOB2 and NDR2 in neuritogenesis as reported by Lin et al. [75] is difficult to explain by a direct protein-protein interaction between MOB2 and NDR2. Nevertheless, in the context of describing a function conserved from yeast to mammals, it makes sense that MOB2 plays a role in the control of cell morphological changes. In budding and fission yeast, Mob2p (the putative counterpart of dMOB2 and hMOB2 in flies and humans, see Fig. 1) is a central component of the RAM network regulating cell morphogenesis (see section 2, and Fig. 2). Mob2p in complex with Cbk1p or Orb6p (the second NDR/LATS kinase in S. cerevisiae and S. pombe, respectively) controls polarized growth (see section 2). Furthermore, T-loop and HM phosphorylation of Cbk1p is fully dependent on Mob2p in budding yeast [24]. Therefore, when translating the available data from yeast studies into a specific mammalian system, it might well be that MOB2 can act as a co-activator of NDR kinases during the regulation of highly specific morphological changes. Nevertheless, although studies in yeast showed that Mob2p has important roles in the control of morphological changes, studies addressing the involvement of MOB2 in morphogenesis networks have just started to emerge using cell systems from multicellular eukaryotes.
6. Conclusions
In summary, members of the MOB protein family have been shown to regulate mitosis, cell proliferation, apoptosis, centrosome biology and morphological changes. In budding and fission yeast, Mob1p and Mob2p control MEN/SIN and RAM/morphogenesis pathways by their association with the Dbf2p/Sid2p and Cbk1p/Orb6p kinases, respectively (Fig. 2). In D. melanogaster, dMOB1 plays a central role in Hippo signalling by forming a complex with Warts kinase, thereby controlling the balance between cell proliferation and death (Fig. 3). dMOB2 and dMOB3 might function together with the Trc kinase in controlling morphological changes, but specific studies addressing the biological roles of dMOB2 and dMOB3 are yet to come. In human cells, hMOB1A and hMOB1B (also referred to as hMOB1) function in cell proliferation, apoptosis and centrosome duplication by binding to NDR/LATS kinases (Fig. 4). hMOB2 opposes the function of the hMOB1/NDR complex in apoptosis and centrosome biology, but possibly co-operates positively with NDR2 in the formation of neurites. The biological roles of hMOB3A, hMOB3B and hMOB3C are unknown at the moment.
As discussed in this review, budding and fission yeast express two different MOB proteins. Intriguingly, in yeast Mob1p and Mob2p function in independent and non-interchangeable complexes, thereby regulating different biological processes by their binding to the corresponding NDR/LATS kinases (Fig. 2). In contrast, in multicellular organisms, the binding of MOB proteins is not restricted to a unique NDR kinase (Figs. 3 and 4). For example, flies express three MOB proteins: dMOB1 interacts with Trc and Warts, the two NDR/LATS kinases in D. melanogaster. dMOB2 can bind to Trc, and dMOB3 maybe also associates with Trc. In mammals, the situation is even more complex: hMOB1 forms a complex with all four NDR/LATS kinases, namely NDR1/2 and LATS1/2. hMOB2 binds selectively to NDR1/2, and does not interact with LATS1/2. hMOB3A/B/C do not associate with any NDR/LATS kinase, although hMOB3A/B/C are more similar to hMOB1 than hMOB2 is to hMOB1 (Fig. 1). Given that the binding partners of hMOB3A/B/C are currently unknown, it is not surprising that functions of hMOB3A/B/C are also yet to be defined. Since potential binding partners have been identified, future progress is warranted in this direction.
In the future, the biological roles of hMOB1 and hMOB2 also need to be addressed in more detail. One pressing question is whether NDR/LATS kinases are the only binding partners of hMOB1/2 in human cells. Potential additional interactors have been found in large scale interaction screens, but it is not yet clear whether any of these novel protein-protein interactions is relevant for hMOB1 signalling. Even for some of the already defined functions of hMOB1 and hMOB2 more research is needed to determine whether hMOB1/NDR, hMOB2/NDR and hMOB1/LATS protein complex formations are important. Another essential point for future research is the development of animal models that allow researchers to study loss and gain of function phenotypes upon manipulation of MOB1 and MOB2 expression. Many different research avenues are now needed to address the complexity of MOB signalling in mammalian systems. More detailed studies of dMOB2 and dMOB3 in D. melanogaster could also be of help in this respect. Taken together, several features of MOB proteins are conserved from yeast to man, but research in multicellular organisms suggests that MOB signalling is more complex in flies when compared to yeast, and even more complex in human cells when compared to flies and yeast. Thus, one future challenge is to understand this complexity.
Given that MOB proteins are involved in cellular processes that are deregulated in human disease, research on MOB proteins is timely and relevant. Importantly, by understanding how MOB proteins function in cells, we might also enable studies pursuing novel research avenues. For example, MOB1 is essential for cytokinesis in blood stream forms of Trypanosoma brucei [76], the protozoan parasite that is the causative agent of African trypanosomiasis. Initially, it was believed that MOB1 functions in complex with TBPK50 [76], one of two NDR/LATS kinases in T. brucei [77]. However, a recent report by the Hammarton laboratory suggests that both NDR/LATS kinases in T. brucei, TBPK50 and TBPK53, can function independent of MOB1 [78]. This suggests that MOB1 could be targeted independently to block proliferation of T. brucei, thereby making MOB1 a potential drug target candidate, in addition to TBPK50 kinase, which is already a promising drug target candidate in trypanosomes. Therefore, further research on various MOB proteins from different species could provide important leads in the discovery of anti-parasitic therapeutics, in addition to establishing how members of the MOB protein family fulfil their essential cellular functions.
Acknowledgements
We thank D. Cook, C. Ender, and J. Lisztwan for their critical input on this manuscript. This work was supported by the Wellcome Trust grant 090090/Z/09/Z. A.H. is a Wellcome Trust Research Career Development fellow at the UCL Cancer Institute.
References
- [1].Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- [2].Pearce LR, Komander D, Alessi DR. Nat Rev Mol Cell Biol. 2010;11(1):9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- [3].Hergovich A, Stegert MR, Schmitz D, Hemmings BA. Nat Rev Mol Cell Biol. 2006;7(4):253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- [4].Bardin AJ, Amon A. Nat Rev Mol Cell Biol. 2001;2(11):815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- [5].Hergovich A, Hemmings BA. Biofactors. 2009;35(4):338–345. doi: 10.1002/biof.47. [DOI] [PubMed] [Google Scholar]
- [6].Pan D. Dev Cell. 2010;19(4):491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sudol M, Harvey KF. Trends Biochem Sci. 2010;35(11):627–633. doi: 10.1016/j.tibs.2010.05.010. [DOI] [PubMed] [Google Scholar]
- [8].Zhao B, Li L, Guan KL. J Cell Sci. 2010;123(Pt 23):4001–4006. doi: 10.1242/jcs.069070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhao B, Li L, Lei Q, Guan KL. Genes Dev. 2010;24(9):862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Luca FC, Winey M. Mol Biol Cell. 1998;9(1):29–46. doi: 10.1091/mbc.9.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Frenz LM, Lee SE, Fesquet D, Johnston LH. J Cell Sci. 2000;113(Pt 19):3399–3408. doi: 10.1242/jcs.113.19.3399. [DOI] [PubMed] [Google Scholar]
- [12].Komarnitsky SI, Chiang YC, Luca FC, Chen J, Toyn JH, Winey M, Johnston LH, Denis CL. Mol Cell Biol. 1998;18(4):2100–2107. doi: 10.1128/mcb.18.4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lee SE, Frenz LM, Wells NJ, Johnson AL, Johnston LH. Curr Biol. 2001;11(10):784–788. doi: 10.1016/s0960-9822(01)00228-7. [DOI] [PubMed] [Google Scholar]
- [14].Luca FC, Mody M, Kurischko C, Roof DM, Giddings TH, Winey M. Mol Cell Biol. 2001;21(20):6972–6983. doi: 10.1128/MCB.21.20.6972-6983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mah AS, Jang J, Deshaies RJ. Proc Natl Acad Sci U S A. 2001;98(13):7325–7330. doi: 10.1073/pnas.141098998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yoshida S, Toh-e A. Genes Genet Syst. 2001;76(2):141–147. doi: 10.1266/ggs.76.141. [DOI] [PubMed] [Google Scholar]
- [17].Mohl DA, Huddleston MJ, Collingwood TS, Annan RS, Deshaies RJ. J Cell Biol. 2009;184(4):527–539. doi: 10.1083/jcb.200812022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hou MC, Guertin DA, McCollum D. Mol Cell Biol. 2004;24(8):3262–3276. doi: 10.1128/MCB.24.8.3262-3276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hou MC, Salek J, McCollum D. Curr Biol. 2000;10(10):619–622. doi: 10.1016/s0960-9822(00)00492-9. [DOI] [PubMed] [Google Scholar]
- [20].Salimova E, Sohrmann M, Fournier N, Simanis V. J Cell Sci. 2000;113(Pt 10):1695–1704. doi: 10.1242/jcs.113.10.1695. [DOI] [PubMed] [Google Scholar]
- [21].Chen CT, Feoktistova A, Chen JS, Shim YS, Clifford DM, Gould KL, McCollum D. Curr Biol. 2008;18(20):1594–1599. doi: 10.1016/j.cub.2008.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Colman-Lerner A, Chin TE, Brent R. Cell. 2001;107(6):739–750. doi: 10.1016/s0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- [23].Weiss EL, Kurischko C, Zhang C, Shokat K, Drubin DG, Luca FC. J Cell Biol. 2002;158(5):885–900. doi: 10.1083/jcb.200203094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jansen JM, Barry MF, Yoo CK, Weiss EL. J Cell Biol. 2006;175(5):755–766. doi: 10.1083/jcb.200604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nelson B, Kurischko C, Horecka J, Mody M, Nair P, Pratt L, Zougman A, McBroom LD, Hughes TR, Boone C, Luca FC. Mol Biol Cell. 2003;14(9):3782–3803. doi: 10.1091/mbc.E03-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hou MC, Wiley DJ, Verde F, McCollum D. J Cell Sci. 2003;116(Pt 1):125–135. doi: 10.1242/jcs.00206. [DOI] [PubMed] [Google Scholar]
- [27].Kanai M, Kume K, Miyahara K, Sakai K, Nakamura K, Leonhard K, Wiley DJ, Verde F, Toda T, Hirata D. EMBO J. 2005;24(17):3012–3025. doi: 10.1038/sj.emboj.7600782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Cell. 2005;120(5):675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- [29].Shimizu T, Ho LL, Lai ZC. Genetics. 2008;178(2):957–965. doi: 10.1534/genetics.107.081570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wei X, Shimizu T, Lai ZC. EMBO J. 2007;26(7):1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hergovich A, Schmitz D, Hemmings BA. Biochem Biophys Res Commun. 2006;345(1):50–58. doi: 10.1016/j.bbrc.2006.03.244. [DOI] [PubMed] [Google Scholar]
- [32].Ho LL, Wei X, Shimizu T, Lai ZC. Dev Biol. 2010;337(2):274–283. doi: 10.1016/j.ydbio.2009.10.042. [DOI] [PubMed] [Google Scholar]
- [33].He Y, Emoto K, Fang X, Ren N, Tian X, Jan YN, Adler PN. Mol Biol Cell. 2005;16(9):4139–4152. doi: 10.1091/mbc.E05-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Emoto K, Parrish JZ, Jan LY, Jan YN. Nature. 2006;443(7108):210–213. doi: 10.1038/nature05090. [DOI] [PubMed] [Google Scholar]
- [35].Kohler RS, Schmitz D, Cornils H, Hemmings BA, Hergovich A. Mol Cell Biol. 2010;30(18):4507–4520. doi: 10.1128/MCB.00150-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liu LY, Lin CH, Fan SS. Cell Tissue Res. 2009;338(3):377–389. doi: 10.1007/s00441-009-0878-7. [DOI] [PubMed] [Google Scholar]
- [37].Hergovich A, Bichsel SJ, Hemmings BA. Mol Cell Biol. 2005;25(18):8259–8272. doi: 10.1128/MCB.25.18.8259-8272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hergovich A, Kohler RS, Schmitz D, Vichalkovski A, Cornils H, Hemmings BA. Curr Biol. 2009;19(20):1692–1702. doi: 10.1016/j.cub.2009.09.020. [DOI] [PubMed] [Google Scholar]
- [39].Wilmeth LJ, Shrestha S, Montano G, Rashe J, Shuster CB. Mol Biol Cell. 2010;21(3):380–392. doi: 10.1091/mbc.E09-06-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bothos J, Tuttle RL, Ottey M, Luca FC, Halazonetis TD. Cancer Res. 2005;65(15):6568–6575. doi: 10.1158/0008-5472.CAN-05-0862. [DOI] [PubMed] [Google Scholar]
- [41].Bichsel SJ, Tamaskovic R, Stegert MR, Hemmings BA. J Biol Chem. 2004;279(34):35228–35235. doi: 10.1074/jbc.M404542200. [DOI] [PubMed] [Google Scholar]
- [42].Chiba S, Ikeda M, Katsunuma K, Ohashi K, Mizuno K. Curr Biol. 2009;19(8):675–681. doi: 10.1016/j.cub.2009.02.054. [DOI] [PubMed] [Google Scholar]
- [43].Chow A, Hao Y, Yang X. Int J Cancer. 2010;126(9):2079–2089. doi: 10.1002/ijc.24878. [DOI] [PubMed] [Google Scholar]
- [44].Devroe E, Erdjument-Bromage H, Tempst P, Silver PA. J Biol Chem. 2004;279(23):24444–24451. doi: 10.1074/jbc.M401999200. [DOI] [PubMed] [Google Scholar]
- [45].Hirabayashi S, Nakagawa K, Sumita K, Hidaka S, Kawai T, Ikeda M, Kawata A, Ohno K, Hata Y. Oncogene. 2008;27(31):4281–4292. doi: 10.1038/onc.2008.66. [DOI] [PubMed] [Google Scholar]
- [46].Stegert MR, Hergovich A, Tamaskovic R, Bichsel SJ, Hemmings BA. Mol Cell Biol. 2005;25(24):11019–11029. doi: 10.1128/MCB.25.24.11019-11029.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Vichalkovski A, Gresko E, Cornils H, Hergovich A, Schmitz D, Hemmings BA. Curr Biol. 2008;18(23):1889–1895. doi: 10.1016/j.cub.2008.10.060. [DOI] [PubMed] [Google Scholar]
- [48].Yabuta N, Okada N, Ito A, Hosomi T, Nishihara S, Sasayama Y, Fujimori A, Okuzaki D, Zhao H, Ikawa M, Okabe M, Nojima H. J Biol Chem. 2007;282(26):19259–19271. doi: 10.1074/jbc.M608562200. [DOI] [PubMed] [Google Scholar]
- [49].Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O’Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y, Vasilescu J, Abu-Farha M, Lambert JP, Duewel HS, Stewart II, Kuehl B, Hogue K, Colwill K, Gladwish K, Muskat B, Kinach R, Adams SL, Moran MF, Morin GB, Topaloglou T, Figeys D. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Nature. 2005;437(7062):1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- [51].Du X, Wang Q, Hirohashi Y, Greene MI. Exp Mol Pathol. 2006;81(3):184–190. doi: 10.1016/j.yexmp.2006.07.008. [DOI] [PubMed] [Google Scholar]
- [52].Stavridi ES, Harris KG, Huyen Y, Bothos J, Verwoerd PM, Stayrook SE, Pavletich NP, Jeffrey PD, Luca FC. Structure. 2003;11(9):1163–1170. doi: 10.1016/s0969-2126(03)00182-5. [DOI] [PubMed] [Google Scholar]
- [53].Simpson JC, Wellenreuther R, Poustka A, Pepperkok R, Wiemann S. EMBO Rep. 2000;1(3):287–292. doi: 10.1093/embo-reports/kvd058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Bandyopadhyay S, Chiang CY, Srivastava J, Gersten M, White S, Bell R, Kurschner C, Martin CH, Smoot M, Sahasrabudhe S, Barber DL, Chanda SK, Ideker T. Nat Methods. 2010;7(10):801–805. doi: 10.1038/nmeth.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kosaka Y, Mimori K, Tanaka F, Inoue H, Watanabe M, Mori M. Int J Oncol. 2007;31(2):333–338. [PubMed] [Google Scholar]
- [56].Sasaki H, Kawano O, Endo K, Suzuki E, Yukiue H, Kobayashi Y, Yano M, Fujii Y. Clin Lung Cancer. 2007;8(4):273–276. doi: 10.3816/CLC.2007.n.006. [DOI] [PubMed] [Google Scholar]
- [57].Uren AG, Kool J, Matentzoglu K, de Ridder J, Mattison J, van Uitert M, Lagcher W, Sie D, Tanger E, Cox T, Reinders M, Hubbard TJ, Rogers J, Jonkers J, Wessels L, Adams DJ, van Lohuizen M, Berns A. Cell. 2008;133(4):727–741. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, Bardeesy N. Cancer Cell. 2009;16(5):425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. Cell. 2010;143(7):1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Wang ZX, Wang HY, Wu MC. Br J Cancer. 2001;85(8):1162–1167. doi: 10.1054/bjoc.2001.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Galmarini CM, Cros E, Thomas X, Jordheim L, Dumontet C. Haematologica. 2005;90(12):1699–1701. [PubMed] [Google Scholar]
- [62].Seve P, Mackey JR, Isaac S, Tredan O, Souquet PJ, Perol M, Cass C, Dumontet C. Lung Cancer. 2005;49(3):363–370. doi: 10.1016/j.lungcan.2005.04.008. [DOI] [PubMed] [Google Scholar]
- [63].Suzuki K, Sugawara T, Oyake T, Uchiyama T, Aoki Y, Tsukushi Y, Onodera S, Ito S, Murai K, Ishida Y. Leuk Res. 2007;31(10):1343–1349. doi: 10.1016/j.leukres.2007.01.018. [DOI] [PubMed] [Google Scholar]
- [64].Bea S, Salaverria I, Armengol L, Pinyol M, Fernandez V, Hartmann EM, Jares P, Amador V, Hernandez L, Navarro A, Ott G, Rosenwald A, Estivill X, Campo E. Blood. 2009;113(13):3059–3069. doi: 10.1182/blood-2008-07-170183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hartmann EM, Campo E, Wright G, Lenz G, Salaverria I, Jares P, Xiao W, Braziel RM, Rimsza LM, Chan WC, Weisenburger DD, Delabie J, Jaffe ES, Gascoyne RD, Dave SS, Mueller-Hermelink HK, Staudt LM, Ott G, Bea S, Rosenwald A. Blood. 2010;116(6):953–961. doi: 10.1182/blood-2010-01-263806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Cornils H, Stegert MR, Hergovich A, Hynx D, Schmitz D, Dirnhofer S, Hemmings BA. Sci Signal. 2010;3(126):ra47. doi: 10.1126/scisignal.2000681. [DOI] [PubMed] [Google Scholar]
- [67].Praskova M, Xia F, Avruch J. Curr Biol. 2008;18(5):311–321. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Bao Y, Sumita K, Kudo T, Withanage K, Nakagawa K, Ikeda M, Ohno K, Wang Y, Hata Y. Genes Cells. 2009;14(12):1369–1381. doi: 10.1111/j.1365-2443.2009.01354.x. [DOI] [PubMed] [Google Scholar]
- [69].Konig C, Maekawa H, Schiebel E. J Cell Biol. 2010;188(3):351–368. doi: 10.1083/jcb.200911128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Mrkobrada S, Boucher L, Ceccarelli DF, Tyers M, Sicheri F. J Mol Biol. 2006;362(3):430–440. doi: 10.1016/j.jmb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- [71].Ponchon L, Dumas C, Kajava AV, Fesquet D, Padilla A. J Mol Biol. 2004;337(1):167–182. doi: 10.1016/j.jmb.2004.01.010. [DOI] [PubMed] [Google Scholar]
- [72].Stoepel J, Ottey MA, Kurischko C, Hieter P, Luca FC. Mol Biol Cell. 2005;16(12):5465–5479. doi: 10.1091/mbc.E05-04-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Mardin BR, Lange C, Baxter JE, Hardy T, Scholz SR, Fry AM, Schiebel E. Nat Cell Biol. 2010;12(12):1166–1176. doi: 10.1038/ncb2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Bishop CL, Bergin AM, Fessart D, Borgdorff V, Hatzimasoura E, Garbe JC, Stampfer MR, Koh J, Beach DH. Mol Cell. 2010;40(4):533–547. doi: 10.1016/j.molcel.2010.10.027. [DOI] [PubMed] [Google Scholar]
- [75].Lin CH, Hsieh M, Fan SS. FEBS Lett. 2011;585(3):523–530. doi: 10.1016/j.febslet.2011.01.003. [DOI] [PubMed] [Google Scholar]
- [76].Hammarton TC, Lillico SG, Welburn SC, Mottram JC. Mol Microbiol. 2005;56(1):104–116. doi: 10.1111/j.1365-2958.2005.04542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Naula C, Parsons M, Mottram JC. Biochim Biophys Acta. 2005;1754(1-2):151–159. doi: 10.1016/j.bbapap.2005.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Ma J, Benz C, Grimaldi R, Stockdale C, Wyatt P, Frearson J, Hammarton TC. J Biol Chem. 2010;285(20):15356–15368. doi: 10.1074/jbc.M109.074591. [DOI] [PMC free article] [PubMed] [Google Scholar]