The effort to solve Alzheimer's disease (AD) has led to a salutary intersection between the desire to prevent a major health problem and the fundamental study of membrane protein biology. It has become apparent in the last few years that a compelling strategy to treat and prevent AD may involve understanding how certain integral membrane proteins undergo unusual proteolytic cleavages within their transmembrane domains, and then inhibiting this process pharmacologically. The substrate of interest in AD is the βamyloid precursor protein (APP), but several other proteins apparently are cleaved by a highly similar or identical proteolytic activity within their respective transmembrane domains, including the Notch family of cell-surface receptors required for cell fate determination (1) and the Ire1 proteins that initiate signaling in the unfolded protein response pathway (2). The unknown protease that affects the intramembranous cleavage of APP (and perhaps Notch and Ire1) is referred to as γ-secretase (reviewed in ref. 3). Its identity is being actively sought because it is the second of two proteases that sequentially cleave APP to release a small fragment called the amyloid β-protein (Aβ). [The other protease, β-secretase, recently has been cloned (4–8).] Progressive accumulation of Aβ in brain regions subserving memory and cognition is an early, invariant, and necessary step in the pathogenesis of AD. For these reasons, β- and γ-secretases are considered important targets for the development of effective therapeutics to treat AD.
In this issue of PNAS, Li and colleagues (9) report the solubilization and enrichment of γ-secretase activity from cultured human cells and examine its relationship to the protein presenilin 1 (PS1). To understand the rationale for their experiments, we should review briefly certain current tenets of AD pathobiology. The brains of patients dying with AD contain abundant deposits of fibrillar Aβ (amyloid) surrounded by clusters of damaged axons and dendrites (the neuritic plaques), as well as many neuronal cell bodies containing abnormal filamentous assemblies of the microtubule-associated protein tau (the neurofibrillary tangles). Genetic research so far has confirmed three distinct genes, mutations that cause severe, autosomal dominant forms of AD (reviewed in ref. 10). Missense mutations in APP itself were the first genetic cause of AD to be identified, and these mutations are principally located at or near either the β- or γ-secretase cleavage sites. The mutations enhance these respective cleavages, resulting in overproduction of the highly amyloidogenic 42-residue form of Aβ (Aβ42). The other two causative genes encode homologous eight-transmembrane proteins, PS1 and PS2, and missense mutations again lead to excessive cellular production of Aβ42 by somehow altering γ-secretase activity. Deletion of PS1 in mice markedly reduces γ-secretase activity, i.e., it lowers Aβ production and increases levels of the C-terminal fragments of APP that are substrates for γ-secretase (11). Thus, PS1 mediates most γ-secretase activity, with the residual activity likely caused by the presence of PS2.
Other findings increasingly suggested that γ-secretase activity was intimately associated with presenilins. APP could be coimmunoprecipitated with PS1 and PS2 (12), and this interaction with PS recently was shown to occur for the APP C-terminal fragments that are the immediate substrates for γ-secretase (13). Meanwhile, site-directed mutagenesis (14) and molecular modeling (15) supported a helical conformation of the γ-secretase cleavage site within APP, typical of transmembrane regions and consistent with an intramembranous proteolysis. The development of transition state analogs that mimic the APP cleavage region and inhibit γ-secretase suggested that this enzyme is an aspartyl protease (15). Together, these findings led to the observation of two conserved intramembranous aspartate residues in TM6 and TM7 of PS1, and their subsequent mutation to alanine or glutamate markedly decreased γ-secretase cleavage of APP (16). As seen with the analogous γ-secretase processing of APP, the putative intramembranous cleavage of Notch to release its cytoplasmic signaling domain to the nucleus was similarly inhibited by knockout of the PS1 gene (17), peptidomimetic γ-secretase inhibitors (17), and PS Asp-to-Ala mutations (18, 19) (see Fig. 1). Moreover, mutation of the aspartates also abrogated the normal endoproteolysis of PS that creates its heterodimeric complexes, which are believed to be the biologically active form of the protein (16, 19, 20). These observations led to the hypothesis that the presenilins are either unique diaspartyl cofactors for γ-secretase or are themselves the long-sought protease (16).
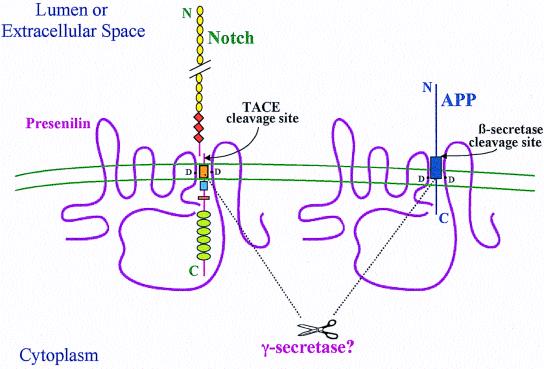
Figure 1.
Hypothetical model of the role of PS in Notch and APP processing based on current information. The diagram shows the predicted eight-transmembrane (TM) domain topology of PS, which occurs principally as a cleaved heterodimer. Some Notch and APP molecules form complexes with PS. Two aspartates (D), one in TM6 and one in TM7 of PS, are required for the cleavages of Notch and APP within their TM domains, and these aspartates may align with each other and with the respective sites of cleavage in the two substrates. It is unknown whether PS directly effects these cleavages or whether a still unidentified aspartyl protease (γ-secretase) present in the complexes does so. PS-mediated proteolysis is preceded by ectodomain shedding caused by tumor necrosis factor α-converting enzyme (TACE) for Notch and β-secretase for APP. Several motifs are depicted in Notch: epidermal growth factor-like repeats (yellow circles), LNG repeats (orange diamonds), a single TM (orange box), the RAM23 domain (blue square), a nuclear localization sequence (red rectangle), and six cdc10/ankyrin repeats (green ovals). After the putative intramembranous cleavage mediated by PS, the Notch intracellular domain is released to the nucleus to activate transcription of target genes. APP contains the Aβ region (dark blue box), which is released into the lumen after sequential cleavages of APP by β-secretase and then γ-secretase/PS. The fate of the APP intracellular domain is unknown.
Although γ-secretase activity had been studied in intact cells and isolated microsomes, the solubilization of the enzyme and its biochemical enrichment had not been published. Now, Li et al. (9) have found that recovery of catalytically competent, soluble γ-secretase activity from HeLa cells can be accomplished by using low concentrations of the detergents CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) or CHAPSO (3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate) but not Triton X-100. Remarkably, the solubilized γ-secretase activity faithfully reproduces the ratio of Aβ42 to Aβ40 peptides produced in intact cells (90:10), suggesting that Aβ42 formation is a normal, intrinsic property of the protease and not primarily dependent on membrane composition or thickness. The solubilized activity is inhibited by pepstatin, a classic aspartyl protease inhibitor. Moreover, the protease activity is inhibited far more potently by a transition state analog closely related to known inhibitors of aspartyl proteases such as renin (21) and HIV protease (22). This pharmacological profile provides further strong evidence that γ-secretase is an aspartyl protease. Li et al. further show that solubilized γ-secretase coelutes with PS1 during size-exclusion chromatography. Most importantly, they report that the coimmunoprecipitation of PS1 heterodimeric complexes from the solubilized preparation recovers γ-secretase activity, demonstrating an intimate association of the solubilized protease with PS subunits. Although these results are consistent with the hypothesis that PS1 is γ-secretase, Li et al. appropriately emphasize that their recovery of PS1 and Aβ-generating activity in a high molecular weight complex, roughly estimated at greater than 106 Da, indicates that there are other components present. Indeed, their method provides a starting point for the identification of additional proteins in the catalytic complex, presumably including the postulated “limiting cellular factor(s)” that are believed to bind to and stabilize presenilins and tightly regulate their conversion to steady-state levels of heterodimers (23, 24). Parenthetically, it is this strict regulation of the amount of bioactive PS in the cell that prevents one from simply overexpressing the protein to prove that it can generate more Aβ and is therefore the catalytic entity.
Although the new data of Li et al., coupled with earlier findings, appear to bring us tantalizingly close to identifying γ-secretase, the final answer must await further experimentation. It had been assumed that reconstitution of Aβ-generating activity from purified protein components would be the best way to prove the identity of the protease. However, the likelihood of a substantial number of associated proteins that emerges from the crude size estimates of Li et al. signifies that: (i) it will be no mean feat to identify, purify, and correctly reassemble the components into an active complex, and (ii) even stepwise addition of the components to achieve proteolysis may not definitively identify which one actually contains the catalytic site. A more tractable approach would be the identification of the immediate protein target of transition-state analog inhibitors directed specifically at the active site.
Other important questions remain. How is specificity conferred on γ-secretase for cleaving the Aβ40–41 versus the Aβ42–43 peptide bonds within APP? If one could understand the conformational basis for this choice of scissile bonds, one might be able to design an inhibitor that would act selectively on the 42–43 cleavage, thereby decreasing the levels of the less abundant but far more amyloidogenic Aβ42 species and leaving more than 90% of Aβ production untouched. Do the several potential substrates of γ-secretase (APP, APP-like proteins APLP1 and APLP2, Notch, Ire1, and perhaps PS itself) all share an identical active site, or are there subtle conformational distinctions in the substrate pockets that might allow selective inhibition of only the APP scission? This question relates to the concern that therapeutically inhibiting Aβ production may interfere with Notch signaling, resulting in toxic effects. However, the realization that miniscule amounts of the Notch cytoplasmic domain may be sufficient to maintain signaling (1) and the possibility that there are additional pathways to activate downstream targets that do not require Notch intramembranous cleavage (25) suggest that chronic γ-secretase inhibition may be tolerated. And precisely where in the cell do these various proteolytic events occur? Is Aβ42 generated earlier in the secretory trafficking of APP and PS than is Aβ40, e.g., in the endoplasmic reticulum or intermediate compartment (26–28), or are both Aβ peptides principally produced at or near the surface, e.g., in recycling endosomes (29, 30). Because Notch intramembranous cleavage requires an earlier cleavage in its proximal ectodomain triggered by binding of extracellular ligand (e.g., Delta) (31, 32), is the γ-secretase cleavage of APP, which is likewise preceded by the α- or β-secretase cleavages in the proximal ectodomain, also ligand dependent, and if so, what is the ligand?
Regardless of the outcome of these interesting issues, the original reason for this focus on the intramembranous proteolysis of APP—the treatment and prevention of AD—continues to receive the attention it deserves. Indeed, before anything was known about the nature of the β- and γ-secretases, the discovery that Aβ was constitutively secreted by cells throughout life (33–35) led to widespread screening of compound libraries on whole cells to identify “hits” that lowered Aβ production without causing cytoxicity. The preclinical and clinical development of such compounds has brought us to the verge of human trials of Aβ-lowering agents. Emerging information about how to solubilize and purify γ-secretase should lead shortly to additional screens capable of identifying and characterizing therapeutic inhibitors. Such progress should not only enable the identification of the elusive γ-secretase but also provide us with more potent and specific drugs to block it, thereby enabling the ultimate test of the amyloid hypothesis of AD.
Footnotes
See companion article on page 6138.
References
- 1.Schroeter E H, Kisslinger J A, Kopan R. Nature (London) 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 2.Niwa M, Sidrauski C, Kaufman R J, Walter P. Cell. 1999;99:691–702. doi: 10.1016/s0092-8674(00)81667-0. [DOI] [PubMed] [Google Scholar]
- 3.Brown M S, Ye J, Rawson R B, Goldstein J L. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 4.Vassar R, Bennett B D, Babu-Khan S, Kahn S, Mendiaz E A, Denis P, Teplow D B, Ross S, Amarante P, Loeloff R, et al. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 5.Yan R, Bienkowski M J, Shuck M E, Miao H, Tory M C, Pauley A M, Brashier J R, Stratman N C, Mathews W R, Buhl A E, et al. Nature (London) 1999;402:533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- 6.Sinha S, Anderson J P, Barbour R, Basi G S, Caccavello R, Davis D, Doan M, Dovey H F, Frigon N, Hong J, et al. Nature (London) 1999;402:537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- 7.Hussain I, Powell D, Howlett D R, Tew D G, Meek T D, Chapman C, Gloger I S, Murphy K E, Southan C D, Ryan D M, et al. Mol Cell Neurosci. 1999;14:419–427. doi: 10.1006/mcne.1999.0811. [DOI] [PubMed] [Google Scholar]
- 8.Lin X, Koelsch G, Wu S, Downs D, Dashti A, Tang J. Proc Natl Acad Sci USA. 2000;97:1456–1460. doi: 10.1073/pnas.97.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y-M, Lai M-T, Xu M, Huang Q, DiMuzio-Mower J, Sardana M K, Shi X-P, Yin K-C, Shafer J A, Gardell S J. Proc Natl Acad Sci USA. 2000;97:6138–6143. doi: 10.1073/pnas.110126897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardy J. Proc Natl Acad Sci USA. 1997;94:2095–2097. doi: 10.1073/pnas.94.6.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Nature (London) 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 12.Xia W, Zhang J, Perez R, Koo E H, Selkoe D J. Proc Natl Acad Sci USA. 1997;94:8208–8213. doi: 10.1073/pnas.94.15.8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia W, Wolfe M S, Ostaszewski B L, Rahmati T, Zhang J, Selkoe D J. Soc Neurosci Abstr. 1999;25:219.7. [Google Scholar]
- 14.Lichtenthaler S F, Wang R, Grimm H, Uljon S N, Masters C L, Beyreuther K. Proc Natl Acad Sci USA. 1999;96:3053–3058. doi: 10.1073/pnas.96.6.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolfe M S, Xia W, Moore C L, Leatherwood D D, Ostaszewski B, Donkor I O, Selkoe D J. Biochemistry. 1999;38:4720–4727. doi: 10.1021/bi982562p. [DOI] [PubMed] [Google Scholar]
- 16.Wolfe M S, Xia W, Ostaszewski B L, Diehl T S, Kimberly W T, Selkoe D J. Nature (London) 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 17.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm J S, Schroeter E H, Schrijvers V, Wolfe M S, Ray W J, et al. Nature (London) 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 18.Ray W J, Yao M, Mumm J, Schroeter E H, Saftig P, Wolfe M, Selkoe D J, Kopan R, Goate A M. J Biol Chem. 1999;274:36801–36807. doi: 10.1074/jbc.274.51.36801. [DOI] [PubMed] [Google Scholar]
- 19.Steiner H, Duff K, Capell A, Romig H, Grim M G, Lincoln S, Hardy J, Yu X, Picciano M, Fechteler K, et al. J Biol Chem. 1999;274:28669–28673. doi: 10.1074/jbc.274.40.28669. [DOI] [PubMed] [Google Scholar]
- 20.Leimer U, Lun K, Romig H, Walter J, Grunberg J, Brand M, Haass C. Biochemistry. 1999;38:13602–13609. doi: 10.1021/bi991453n. [DOI] [PubMed] [Google Scholar]
- 21.Kati W M, Pals D T, Thaisrivongs S. Biochemistry. 1987;26:7621–7626. doi: 10.1021/bi00398a014. [DOI] [PubMed] [Google Scholar]
- 22.Huff J R. J Med Chem. 1991;34:2305–2314. doi: 10.1021/jm00112a001. [DOI] [PubMed] [Google Scholar]
- 23.Ratovitski T, Slunt H H, Thinakaran G, Price D L, Sisodia S S, Borchelt D R. J Biol Chem. 1997;272:24536–24541. doi: 10.1074/jbc.272.39.24536. [DOI] [PubMed] [Google Scholar]
- 24.Thinakaran G, Harris C L, Ratovitski T, Davenport F, Slunt H H, Price D L, Borchelt D R, Sisodia S S. J Biol Chem. 1997;272:28415–28422. doi: 10.1074/jbc.272.45.28415. [DOI] [PubMed] [Google Scholar]
- 25.Artavanis-Tsakonas S, Rand M D, Lake R J. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 26.Wild-Bode C, Yamazaki T, Capell A, Leimer U, Steiner H, Ihara Y, Haass C. J Biol Chem. 1997;272:16085–16088. doi: 10.1074/jbc.272.26.16085. [DOI] [PubMed] [Google Scholar]
- 27.Cook D G, Forman M S, Sung J C, Leight S, Kolson D L, Iwatsubo T, Lee V M, Doms R W. Nat Med. 1997;3:1021–1023. doi: 10.1038/nm0997-1021. [DOI] [PubMed] [Google Scholar]
- 28.Hartmann T, Bieger S C, Bruhl B, Tienari P J, Ida N, Allsop D, Roberts G W, Masters C L, Dotti C G, Unsicker K, Beyreuther K. Nat Med. 1997;3:1016–1020. doi: 10.1038/nm0997-1016. [DOI] [PubMed] [Google Scholar]
- 29.Koo E H, Squazzo S L. J Biol Chem. 1994;269:17386–17389. [PubMed] [Google Scholar]
- 30.Perez R G, Soriano S, Hayes J D, Ostaszewski B, Xia W, Selkoe D J, Chen X, Stokin G B, Koo E H. J Biol Chem. 1999;274:18851–18856. doi: 10.1074/jbc.274.27.18851. [DOI] [PubMed] [Google Scholar]
- 31.Mumm J S, Schroeter E H, Saxena M T, Griesemer A, Tian X, Pan D J, Ray W J, Kopan R. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 32.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens J R, Cumano A, Roux P, Black R A, Israël A. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 33.Haass C, Schlossmacher M G, Hung A Y, Vigo-Pelfrey C, Mellon A, Ostaszewski B L, Lieberburg I, Koo E H, Schenk D, Teplow D B, et al. Nature (London) 1992;359:322–325. doi: 10.1038/359322a0. [DOI] [PubMed] [Google Scholar]
- 34.Seubert P, Vigo-Pelfrey C, Esch F, Lee M, Dovey H, Davis D, Sinha S, Schlossmacher M, Whaley J, Swindlehurst C, et al. Nature (London) 1992;359:325–327. doi: 10.1038/359325a0. [DOI] [PubMed] [Google Scholar]
- 35.Shoji M, Golde T E, Ghiso J, Cheung T T, Estus S, Shaffer L M, Cai X D, McKay D M, Tintner R, Frangione B, et al. Science. 1992;258:126–129. doi: 10.1126/science.1439760. [DOI] [PubMed] [Google Scholar]