SUMMARY
Trans-acyltransferase polyketide synthases (trans-AT PKSs) are an important group of bacterial enzymes producing bioactive polyketides. One difference from textbook PKSs is the presence of one or more free-standing AT-like enzymes. While one homolog loads the PKS with malonyl units, the function of the second copy (AT2) was unknown. We studied the two ATs PedC and PedD involved in pederin biosynthesis in an uncultivated symbiont. PedD displayed malonyl- but not acetyltransferase activity toward various acyl carrier proteins (ACPs). In contrast, the AT2 PedC efficiently hydrolyzed acyl units bound to N-acetylcysteamine or ACP. It accepted substrates with various chain lengths and functionalizations but did not cleave malonyl-ACP. These data are consistent with the role of PedC in PKS proofreading, suggesting a similar function for other AT2 homologs and providing strategies for polyketide titer improvement and biosynthetic investigations.
INTRODUCTION
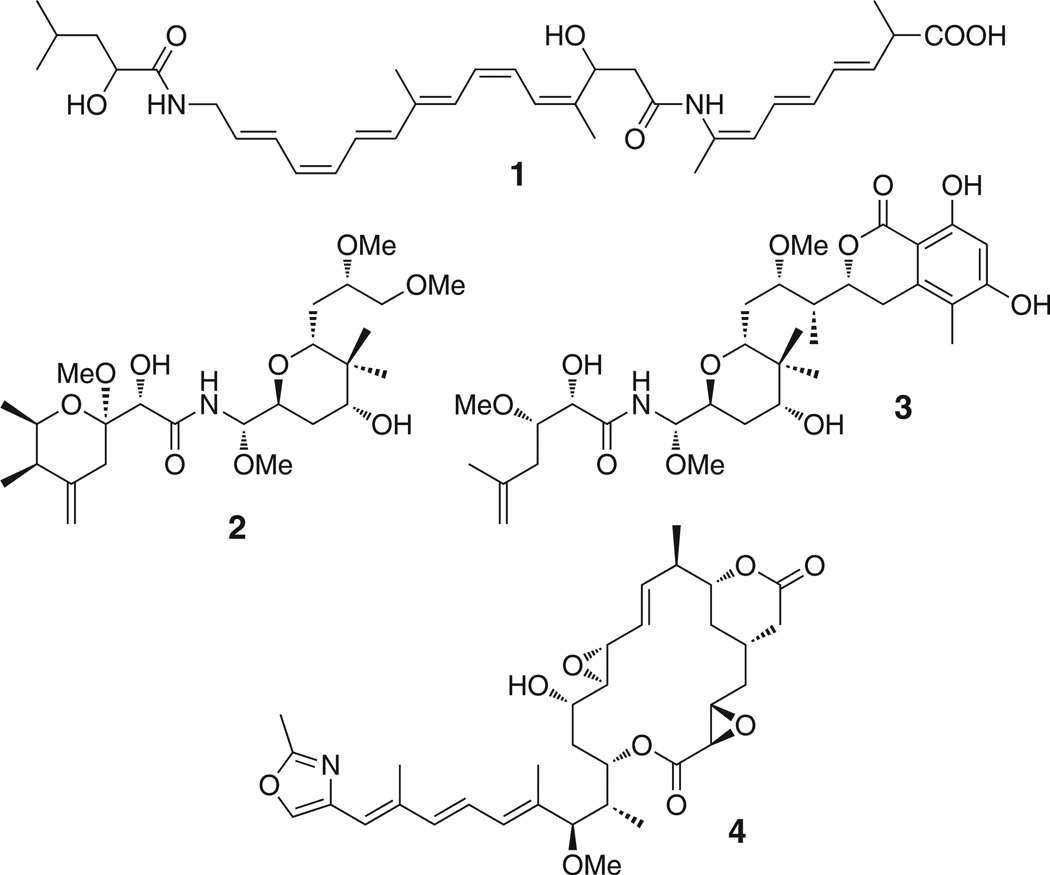
Bacterial complex polyketides are biosynthesized on giant multifunctional enzymatic assembly lines called modular polyketide synthases (PKSs) (Hertweck, 2009; Piel, 2010; Staunton and Weissman, 2001). These enzymes contain numerous catalytic domains, the order of which typically determines the structure of the polyketide chain. In addition, multiple acyl carrier protein (ACP) domains serve as anchors that covalently bind intermediates of increasing chain length via thioester linkages. After full elongation, a C-terminal thioesterase (TE) domain usually cleaves the thioester bond by hydrolysis or lactonization to release the polyketide (Du and Lou, 2010). In textbook PKSs, all biosynthetic components needed to generate the polyketide core structure are domains incorporated in the megaenzyme. In contrast, the recently discovered group of trans-acyltransferase (trans-AT) PKSs diverges from this architecture in using multiple covalently and noncovalently joined components (Piel, 2010). Trans-AT PKSs produce diverse bioactive polyketides, including the antibiotic bacillaene (1) from bacilli (Butcher et al., 2007; Chen et al., 2006b; Moldenhauer et al., 2007) and the cytotoxins pederin (2) (Piel, 2002), psymberin/irciniastatin A (3) (Cichewicz et al., 2004; Fisch et al., 2009; Pettit et al., 2004), and rhizoxin (4) (Iwasaki et al., 1986; Partida-Martinez and Hertweck, 2005) from various endosymbiotic bacteria (Figure 1). Trans-acting components identified in these poorly understood systems are ATs needed for loading the PKS with biosynthetic building blocks (Cheng et al., 2003; Wong et al., 2011), enoylreductases (ER) for reduction of α,β-unsaturated intermediates (Bumpus et al., 2008), and enzymes to convert β-keto groups to carbon branches (Calderone, 2008). The polyketide products are almost (Musiol et al., 2011) exclusively constructed from malonyl units, and a single, free-standing AT resembling those from bacterial fatty acid synthases (here termed AT1) has been shown to be sufficient for loading multiple trans-AT PKS modules with these units (Cheng et al., 2003; Lopanik et al., 2008). It is therefore peculiar that more than half of all characterized trans-AT PKS systems, including those of 1, 2, and 4, contain another AT homolog (AT2). These enzymes are either free standing or fused with the malonyl-loading AT and/or the trans-ER. Except for the fact that their inactivation can result in reduced polyketide titers (Lopanik et al., 2008), nothing is known about the function of these ubiquitous enzymes.
Figure 1. Selected Products of Trans-AT PKS Pathways.
Compounds are bacillaene (1), pederin (2), psymberin (3), and rhizoxin (4).
Much of the current knowledge about trans-AT PKS functionality stems from studies on the polyketide antibiotic bacillaene (1) in Bacillus amyloliquefaciens and Bacillus subtilis (Bumpus et al., 2008; Calderone et al., 2008, 2006; Chen et al., 2009; Dorrestein et al., 2006; Moldenhauer et al., 2007, 2010). Functional dissection of the bacillaene (bae) PKS was greatly facilitated by the discovery that mutants with an inactivated TE domain produce polyketides of virtually all elongation stages (Moldenhauer et al., 2007, 2010). This phenomenon—which has, with one exception (Yu et al., 1999), not been reported for textbook (cis-AT) PKSs—suggests the existence of an as-yet uncharacterized proofreading mechanism that releases stalled intermediates from the enzyme. Such truncated products were also observed for the rhizoxin (4) (Kusebauch et al., 2009) and, for some elongation stages, the mupirocin (Wu et al., 2008) pathways, indicating that intermediate hydrolysis is a more general phenomenon for trans-AT PKSs. Here, we report genetic, biochemical, and chemical studies of a member of the AT2 group, identifying it as a thioester hydrolase for structurally diverse N-acetylcysteamine- or ACP-bound acyl units. Our data suggest that members of the AT2 group are not involved in acylation of PKS modules but act as proofreading factors that reactivate PKS modules by releasing stalled acyl units.
RESULTS
Study of BaeB, a Hydrolase Candidate from Bacillaene Biosynthesis
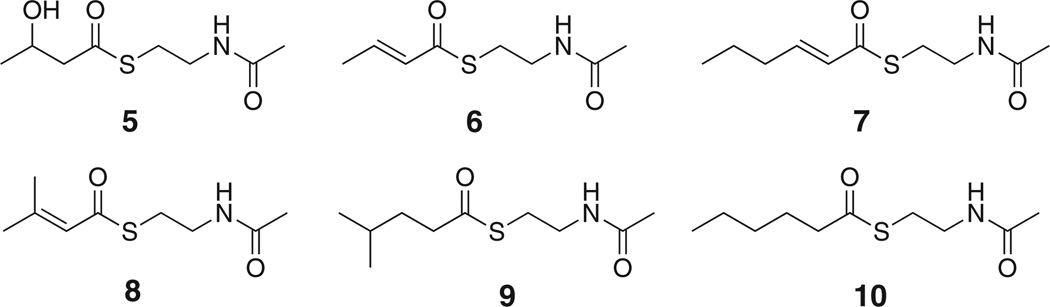
Suspecting that the release factor is clustered with other PKS genes, we first focused on the previously uncharacterized gene baeB of the bacillaene (1) pathway. Its translated product resembles thioester hydrolases, such as glyoxalase II, and therefore represented a good candidate for intermediate release. To investigate its function in vivo, we constructed the B. amyloliquefaciens mutant JM157 lacking baeB. Analysis of the extracts by liquid chromatography-high resolution mass spectrometry (LC-HRMS) showed that 1 was produced at significantly reduced (68%–87%) titers as compared to the wild-type strain FZB42 (Figure S1 available online), indicating that BaeB does not catalyze an essential biosynthetic step. Since no intermediates are observed in strains harboring an intact TE domain, the influence of BaeB on thioester hydrolysis could not be examined in this mutant. Therefore, we created the double mutant JM157-54 that lacked both the TE and BaeB to allow for direct metabolic comparison with the TE single mutant JM54-2. While JM54-2 produced the full series of intermediates, only traces of two compounds lacking three elongation units and one elongation unit, respectively, were detected in extracts of the double mutant. This result may suggest that BaeB is the off-loading factor. However, we noticed that, for unknown reasons, all baeB deletion mutants exhibited strongly diminished growth. Thus, although relative polyketide yields from both TE deletion strains had been normalized to an internal standard (the PKS-independent compound bacillibactin), we could not exclude the possibility that the decreased biosynthetic activity of the bae pathway was an indirect effect of the altered physiology. This issue was addressed by constructing a pHis8-based expression plasmid containing baeB for in vitro enzymatic studies. Expression resulted in soluble, N-terminally octahistidyl-tagged BaeB that was incubated with a range of N-acetylcysteamine (SNAC) conjugates of short-chain carboxylic acids (5–9; Figure 2). SNAC thioesters are known to be accepted as surrogates of ACP-bound intermediates by various PKS components (Kinoshita et al., 2001; Pohl et al., 1998; Vergnolle et al., 2011). To detect thioester cleavage, we monitored hydrolyzed free thiols in a continuous assay using Ellman’s reagent (DTNB, 5,5′-dithio-bis(2-nitrobenzoic acid)), which reacts with thiols to the yellow cleavage product 2-nitro-5-thiobenzoate. However, no conversion was detected for any tested compound or condition.
Figure 2. SNAC Derivatives Tested in the Hydrolysis Assays.
For the preparation of compounds 6–9 see Experimental Procedures. See also Figure S1.
Differential Roles of the Pederin AT-like Enzymes in Malonyl Transfer
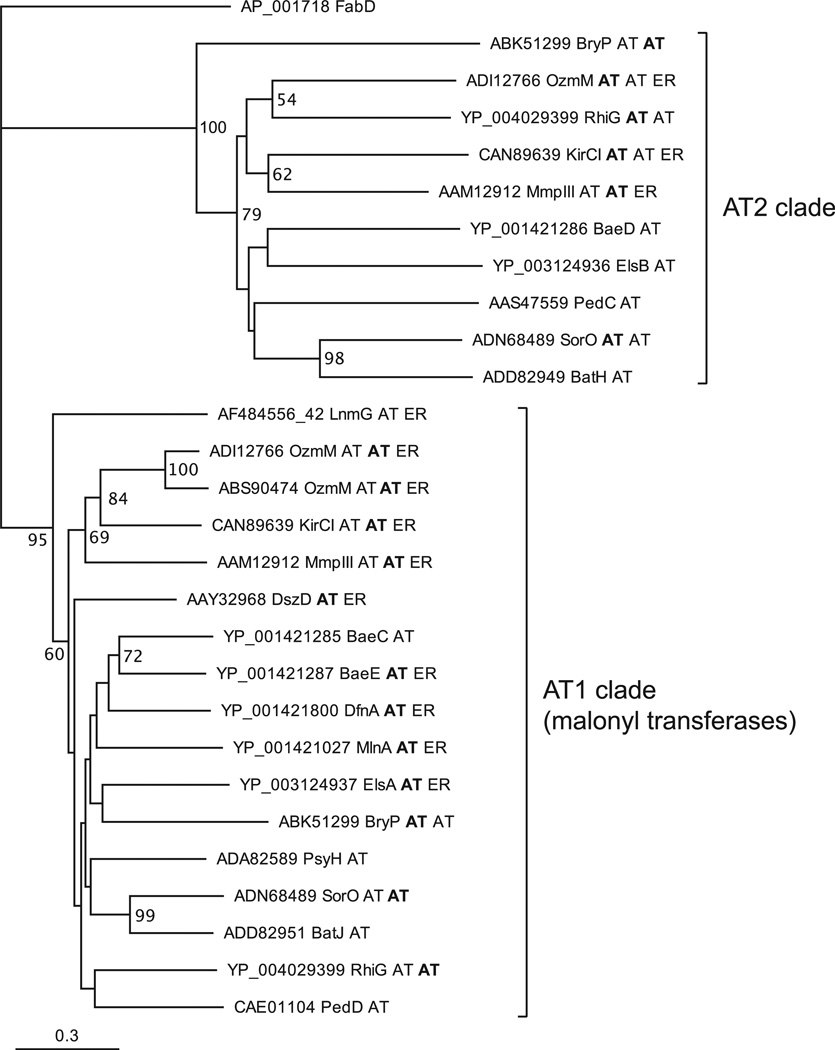
During these studies, it was found that TE deletion in the rhizoxin (3) producer Burkholderia rhizoxinica B1 resulted in a similar release of intermediates as that known for bacillaene (Kusebauch et al., 2009). The fact that neither the rhizoxin (rhi) PKS cluster nor the entire genome (Lackner et al., 2011a, 2011b) encoded a homolog of BaeB, combined with the negative results of the BaeB hydrolysis assay, suggested that a different enzyme is responsible for thioester cleavage. The AT2 present in many trans-AT systems was a reasonable candidate because homologs also occur in the bacillaene (1) and rhizoxin (4) pathways. Moreover, thioester hydrolysis conforms to the mechanism of ATs, which involves transfer of a thioester acyl moiety to a serine residue followed by nucleophilic displacement (Wong et al., 2011). Since the bacillaene AT2 from B. amyloliquefaciens could not be expressed as a soluble protein, and the presence of five AT-like genes within this strain complicates any in vivo knockout studies, we selected the AT homologs PedC and PedD of the pederin (1) pathway from a Pseudomonas sp. symbiont of Paederus spp. beetles (Piel, 2002) for further studies. These enzymes offered the distinct advantage of being among the very few known ATs that are not fused to additional domains, thus simplifying functional analysis.
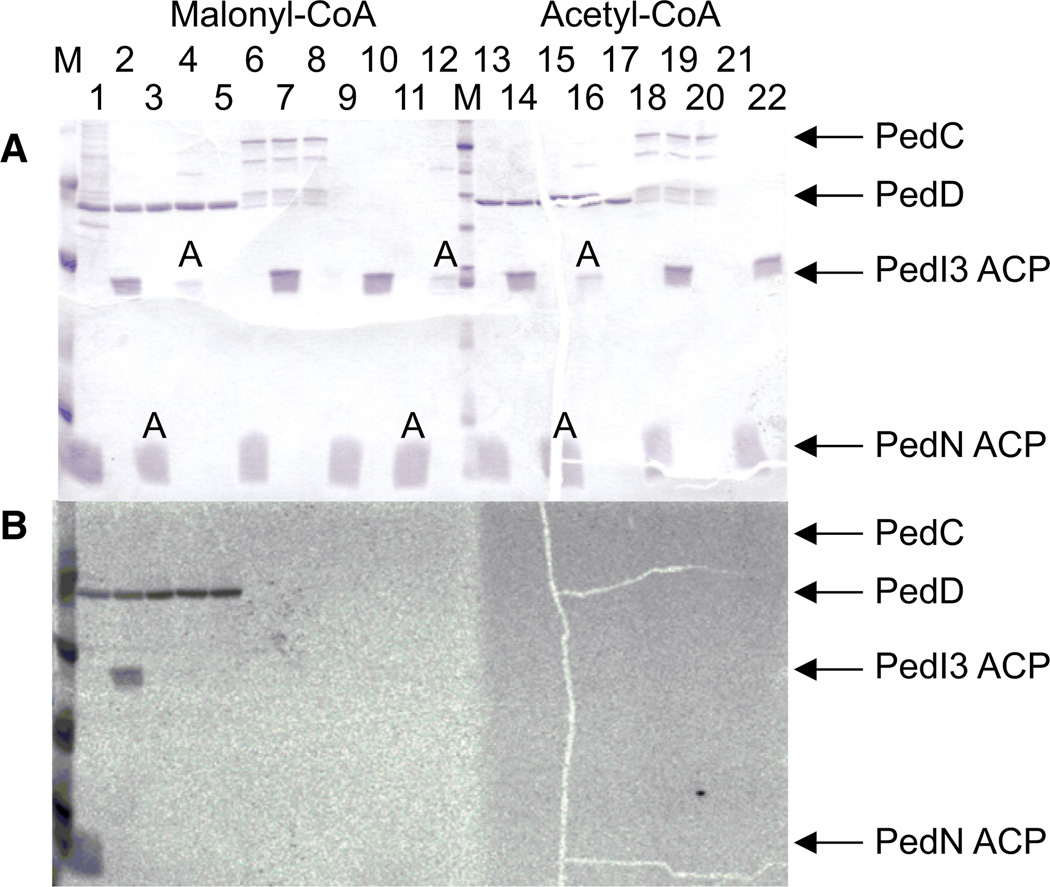
In a phylogenetic analysis (Figure 3), PedD grouped with members of the AT1 clade, including the previously characterized malonylating ATs LnmG of the leinamycin route (Cheng et al., 2003) and BryP (N-terminal domain) of the bryostatin pathway (Lopanik et al., 2008). PedC fell into the AT2 group. To functionally test the roles of ped ATs suggested by the phylogenetic data, we produced both enzymes for in vitro analyses. Initial difficulties in obtaining soluble PedC by a range of expression strategies were overcome by expressing the protein with an N-terminally fused maltose-binding protein or a C-terminal Strep-tag in the presence of the chaperones GroES and GroEL, which yielded low amounts of soluble enzyme. PedC and PedD were then subjected to a transfer assay using [2-14C]-malonyl- or [1-14C]-acetyl-CoA as test substrate with an ACP as acceptor. For our investigation of whether the two ATs might be specific for different types of ACPs, the following proteins were expressed: the free-standing variant PedN (Piel et al., 2004a), the integrated tandem domain PedI-ACP3 from module 3 of the pederin PKS (Piel et al., 2004b), and the integrated monodomain ACPs PsyA-ACP3 and PsyD-ACP1 from elongation modules 3 and 4 of the psymberin PKS (Figure 4; Figure S2) (Fisch et al., 2009). PedN is expected to be involved in the generation of the exomethylene carbon in pederin; PedI3 and PsyA3, to participate in the chain elongation process; and PsyD1, to belong to a non-elongating module with an inactive ketosynthase domain. ACPs were expressed in the apo form as well as in the holo form carrying the 4′-phosphopantetheinyl moiety that serves as anchor for the acyl units. After incubating the ATs, ACPs, and radiolabeled acyl-CoA species in different combinations, we monitored the labeling of proteins by electrophoretic separation and autoradiography. The analysis revealed that PedD self-loads the malonyl radiolabel and efficiently transfers it to all types of holo-ACPs but not to their apo forms. In contrast, ACPs remained unlabeled in the PedC assay. With [1-14C]-acetyl-CoA used as test substrate, neither AT or ACP was labeled. These results confirmed our in silico prediction that PedD is the AT responsible for substrate loading and identified PedC as a candidate for intermediate release.
Figure 3. Phylogram of AT-like Enzymes from Various Trans-AT PKS Pathways.
Tip labels consist of accession numbers, protein name, and domain architecture. For multidomain proteins, the relevant domain is shown in bold. Sequences from the following pathways were used: Bae, bacillaene (B. amyloliquefaciens); Bat, batumin/kalimantacin (Pseudomonas fluorescens); Bry, bryostatin (“Candidatus Endobugula sertula”); Dfn, difficidin (B. amyloliquefaciens); Dsz, disorazol (Sorangium cellulosum); Els, elansolid (Chitinophaga pinensis); Kir, kirromycin (Streptomyces collinus); Lnm, leinamycin (Streptomyces atroolivaceus); Mmp, mupirocin (Pseudomonas fluorescens); Ozm, oxazolomycin (ADI12766 is from Streptomyces bingchenggensis and ABS90474 from Streptomyces albus); Psy, psymberin (uncultivated symbiont of sponge Psammocinia aff. bulbosa); Rhi, rhizoxin (Burkholderia rhizoxina); Sor, soraphen (Sorangium cellulosum). Outgroup is the E. coli AT from fatty acid biosynthesis (FabD).
Figure 4. Acyltransferase Assay Using the Pederin ATs.
(A) SDS-PAGE analysis of incubations containing PedD (lanes 1–5, 13–17) or PedC (lanes 6–8, 18–20), radiolabeled malonyl-CoA (lanes 1–12) or acetyl-CoA (lanes 13–21), and the ACPs of PedI3 (tandem ACP) or PedN. ACPs were in the holo form or the apo form, the latter indicated by an A. Protein markers are indicated with an M.
(B) Autoradiogram of the same gel. See also Figure S2.
The AT2 PedC Hydrolyzes Acyl N-acetylcysteamine Thioesters
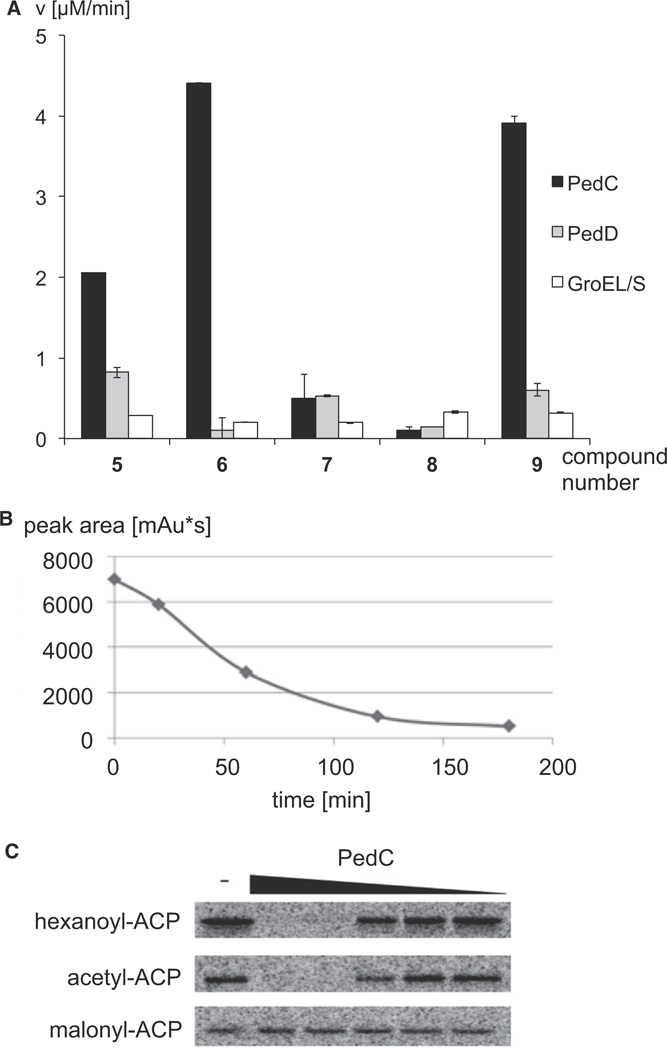
We next investigated hydrolysis of short-chain intermediate mimics by PedC and PedD using the SNAC derivatives 5–9 and Ellman’s reagent. In agreement with its function as a malonyl transferase, PedD did not have significant hydrolysis activity toward any of the thioesters tested (Figure 5A). In contrast, assays with the AT2 PedC showed conversion within minutes for reactions with the acyl donors 5, 6, and 9. On the other hand, hydrolysis was negligible for 7 and 8. We also tested hydrolysis of the hexanoyl thioester 10 by removing aliquots at different time points and quantifying the SNAC derivative after high-pressure liquid chromatography (HPLC) separation. This analysis revealed that 10 is hydrolyzed by PedC, albeit at a lower rate than 5, 6, and 9 (Figure 5B). Expression levels of PedC were very low, and even a 20 L culture provided just sufficient enzyme amounts to determine kinetic parameters for substrate 5 (KM of 88.48 mM and a kcat/KM of 14.41 M−1s−1) (Figure S4). These should be regarded as preliminary, since SNAC derivatives instead of PKS-bound acyl units were used and the true acyl substrates could be fully elongated intermediates. All incubations with PedC also contained the chaperones GroEL/ES, from which the enzyme could not be separated (Figure S3). Prior to kinetic characterization, background hydrolysis activity assays were therefore conducted using purified protein from parallel expressions containing the chaperones with and without PedC coexpression. The full range of substrate concentrations used for kinetic characterization were tested with both enzyme purifications, where minimal background hydrolysis was detected in the protein preparation lacking PedC. These control experiments link the SNAC hydrolysis activity solely to PedC. These results thus support a role for PedC as a hydrolase for acyl-SNAC thioesters carrying various functional groups that typically occur during polyketide biosynthesis; i.e., β-hydroxyl groups (2), α,β-double bonds (6), and α,β-reduced units (9,10). It should be noted that the β-keto variant could not be tested, since it was spontaneously hydrolyzed in the assay medium. Using the same assay, we also studied a PedC homolog from the rhizoxin (3) pathway. In this PKS system, the AT2 is fused to a C-terminal AT1 on the bidomain protein RhiG (Partida-Martinez and Hertweck, 2007). To uncouple both activities, three protein variants were expressed, in which none or either one of the AT domains carried an inactivating Ser-to-Ala mutation at the acylbinding site. However, neither protein displayed activity with any of the SNAC derivatives 5–9. One reason for this unexpectedly differential behavior of the ped and rhi AT2s might be that inactive forms of RhiG were obtained during protein expression or purification. Since SNAC mimics were used instead of acyl-ACPs, we could not exclude the possibility that the observed PedC activity was an artifact unrelated to PKS offloading.
Figure 5. Hydrolysis Assays Using Acyl-SNAC and -ACP Test Substrates.
(A) Reaction rates observed for various SNAC derivatives. Error bars specify standard deviations.
(B) Consumption of hexanoyl-SNAC by PedC. After treatment of the thioester with PedC, aliquots were removed and separated by HPLC. The diagram shows the amount of hexanoyl-SNAC determined by manual integration at 232 nm at different reaction time points.
(C) Phosphor image of PksA acyl-ACP hydrolysis assays with PedC. The signal is emitted from radiolabeled acyl-ACP after electrophoretic separation of proteins. Acyl-ACP (10 µM) was treated for 10 min with 0 (minus sign indicates negative control), 5, 0.83, 0.14, 0.023, or 0.005 µM PedC. See also Figures S3 and S4.
The AT2 PedC Releases ACP-Bound Acyl Units
This issue was addressed in a further series of experiments using ACP-bound 14C-radiolabeled acyl variants. Based on the ability of PedC to hydrolyze hexanoyl-SNAC (10), [1-14C]-hexanoyl-ACP was selected as an initial test substrate. Hexanoyl is the native starter unit used in aflatoxin biosynthesis by Aspergillus spp. fungi (Minto and Townsend, 1997), thus allowing us to test an extended acyl unit with its cognate ACP. Moreover, since this ACP is derived from a fungal iterative PKS, we hoped to obtain insights into whether PedC would act on enzymes other than trans-AT PKSs. To generate the hexanoyl-loaded protein, the N-terminally His6-tagged ACP monodomain from the iterative megasynthase PksA of Aspergillus parasiticus was expressed in its apo form and loaded using [1-14C]-hexanoyl-CoA by the phosphopantetheinyl transferase Svp (Crawford et al., 2006). In an analogous way, we also prepared [1-14C]-acetyl- and [2-14C]-malonyl-ACP as further test substrates. These acyl-ACP variants were individually incubated with a serial dilution of PedC in a fixed time point assay. Subsequently, aliquots from incubations were separated by polyacrylamide electrophoresis, and radiolabeled proteins were visualized by phosphorimaging (Figure 5C). The data clearly show that hexanoyl- and acetyl-ACP are completely hydrolyzed within 10 min at higher PedC concentrations while malonyl-ACP remained intact.
DISCUSSION
Our studies on SNAC and ACP derivatives demonstrate that PedC hydrolyzes structurally diverse thioesters. Acyl units accepted as substrates include completely reduced, α,β-unsaturated, β-hydroxylated, and branched moieties of chain lengths between two and at least six carbon atoms. None of the tested compounds were cleaved by PedD. Furthermore, PedD, but not PedC, exhibited malonyl transfer activity. These data are consistent with differential roles for the two AT homologs in pederin biosynthesis, during which PedD loads all ACPs with malonyl units, and PedC acts as a proofreading enzyme that removes stalled acyl units from blocked modules. In agreement, PedC did not cleave malonyl-ACP, which is an essential precursor for polyketide biosynthesis. As PedC does not display detectable AT activity, we propose to rename this enzyme acyl hydrolase (AH). PedC homologs occur in many trans-AT PKS systems (Piel, 2010), suggesting that AH-mediated proofreading is widespread in these pathways.
To date, AT-like editing enzymes are only known from few type II PKSs, where they participate in the selection of non-acetyl starter units (Tang et al., 2004). A different type of editing enzyme is known for cis-AT PKSs (Heathcote et al., 2001) and nonribosomal peptide synthetases (Schwarzer et al., 2002; Yeh et al., 2004). Here, free-standing (type II) TEs act as release factors for the removal of stalled aberrant units. These units are likely short-chain (amino-)acyl moieties that can arise from premature decarboxylation of PKS extenders (Heathcote et al., 2001) or from misloading of carrier proteins by either phosphopantetheinyl transferases (Schwarzer et al., 2002) or building block-selecting domains (Yeh et al., 2004). In agreement, almost all examined type II TEs preferred short-chain reduced acyl units in in vitro assays, while more extended moieties and those carrying hydroxyl or double bond functionality were poor or not substrates (Heathcote et al., 2001; Koglin et al., 2008; Schwarzer et al., 2002; Yeh et al., 2004). This selectivity appears in contrast to PedC, which cleaves acyl donors ranging from two (acyl-ACP) to six (9 and hexanoyl-ACP) carbon atoms and exhibits tolerance for various functional groups occurring in polyketide biosynthesis. The data therefore suggest that PedC and other AHs might not only restore PKS modules blocked with decarboxylated monomers but also play a role in the release of stalled oligoketides. In agreement, all cases of intermediate release involving trans-AT PKSs, i.e., for bacillaene (Moldenhauer et al., 2007), rhizoxin (Kusebauch et al., 2009), and mupirocin (Wu et al., 2008), have been reported from assembly lines containing AH-type enzymes. In trans-AT systems, this function could be of particular significance since phylogenetic (Irschik et al., 2010; Nguyen et al., 2008; Teta et al., 2010) and preliminary unpublished functional data indicate that KSs are highly selective for the α,β-functionalization of incoming substrates. Thus, if β-processing is faulty, further elongation of incorrectly processed intermediates would not occur, resulting in biosynthetic blocks. This issue should be of less importance for cis-AT PKSs, in which KS domains are rather promiscuous (Chen et al., 2006a; Menzella et al., 2007). Nevertheless, hydrolysis of long-chain intermediates was also reported for a small number of cis-AT systems, an important example being rifamycin biosynthesis (Yu et al., 1999). In this case, however, functional data argued against an involvement of the type II TE, leaving the underlying biochemistry unclear (Floss and Yu, 1999).
In conclusion, our data show that PedC, a member of a clade of previously uncharacterized AT-like enzymes ubiquitously encoded in trans-AT PKS gene clusters, is a thioester hydrolase that could play a key role in biosynthetic proofreading. Such a function provides interesting perspectives for engineering and studying PKS pathways. Addition of AHs to trans-AT PKS pathways lacking these enzymes could result in enhanced polyketide yields. With PKSs carrying engineered loss-of-function mutations in their TE domains, AHs could be effective tools to study entire PKS pathways by observing released intermediates in a similar way as previously performed for bacillaene and rhizoxin biosynthesis. Such potential applications are currently under investigation in our laboratory.
SIGNIFICANCE
Trans-acyltransferase polyketide synthases (trans-AT PKSs) are an important but still poorly understood group of biosynthetic enzymes involved in the production of bioactive bacterial polyketides. About half of all known trans-AT PKS gene clusters encode, in addition to the trans-acting AT, another AT-like protein with a previously unknown function. We have identified for the first time an enzymatic function for a member of this group. Our data show that the pederin homolog from an uncultivated beetle symbiont hydrolyzes SNAC- and ACP-bound acyl units with various functional groups and chain lengths. In contrast, neither the malonyl-transferring AT nor the bacillaene-specific enzyme BaeB with an as yet unknown function displayed measurable hydrolase activity in vitro. These results suggest the existence of a group of editing hydrolases, here termed AHs, that remove aberrant or stalled acyl units from blocked trans-AT PKS modules and might be responsible for intermediate release observed in several polyketide routes. The pederin AH or its homologs could be useful tools for studying PKS functionality or, as inactivation of AHs has been shown to reduce polyketide titers (Lopanik et al., 2008), for increasing compound yields by adding AH genes to routes lacking them. In addition to attributing function to a core component of trans-AT PKSs, this study represents one of the few examples (Lopanik et al., 2008; Zimmermann et al., 2009) that provide biochemical insights into polyketide pathways of uncultivated bacterial symbionts associated with invertebrates. As trans-AT PKSs are a major group of enzymes responsible for natural product biosynthesis in these organisms (Piel, 2009), knowledge about their enzymology is an important prerequisite for developing heterologous expression systems for rare animal-derived polyketides.
EXPERIMENTAL PROCEDURES
General
[1-14C]-hexanoyl-CoA, [1-14C]-acetyl-CoA, and [2-14C]-malonyl-CoA were purchased from American Radiolabeled Chemicals, Perkin-Elmer or NEN Radiochemicals, and Morovek, respectively. Unlabeled acyl-CoAs were purchased from Sigma.
Bacterial Strains and Culture Conditions
Escherichia coli BL21(DE3) (Invitrogen) or E. coli Rosetta-gami 2 (DE3) pLysS (Novagen) served as the host strain for protein expression experiments. E. coli strain XL1 blue (Stratagen) served as host for routine subcloning. E. coli strains were grown in Luria-Bertani (LB) medium (Sambrook and Russell, 2000). Detailed information regarding antibiotics, additions, and inducer of the single strains can be found in Table S1. Because it was not possible to obtain soluble proteins without chaperones in some cases, pKJE7, pGro7, and pG-Tf2 (Takara) were used that produced DnaK-DnaJ-GrpE, GroEL-GroES, and GroES-GroEL-Tig, respectively.
Plasmids and General DNA Procedures
DNA isolation, plasmid preparation, restriction digests, gel electrophoresis, and ligation reactions were conducted according to standard methods (Sambrook and Russell, 2000). pBluescript II SK(+) was used for subcloning. All primer sequences are listed in Table S2. Final constructs were verified by sequencing. If not mentioned otherwise, PCR products were cloned into pBluescript II SK(+) by using the method of Marchuk et al. (1991).
Generation of Bacillus amyloliquefaciens Mutants and Polyketide Analysis
Generation of B. amyloliquefaciens strain JM54-2 (the TE single mutant) and the conditions for polyketide analysis were published previously (Moldenhauer et al., 2010). Sequences of PCR primers can be seen in Table S2.
To generate JM157-54 (lacking the TE and BaeB), we used a more complex cloning strategy. A PCR product carrying the chloramphenicol resistance gene was amplified from pDG268 (Antoniewski et al., 1990) using primers FP-cat-BamHI and RP-cat-XbaI. This PCR product was cloned into pBluescript II SK(+) to obtain pJM128. This plasmid was digested with XbaI/BamHI, and the insert was ligated into the corresponding restriction sites of pJM135 to obtain pJM140. To construct pJM135, a PCR product was amplified from B. amyloliquefaciens genomic DNA using primers RP-H-vor-Promotor-XbaI and FP-H-vor-Promotor-NotI and ligated into pBluescript II SK(+) to yield pJM133. This plasmid was digested with NotI/XbaI, and the resulting insert was ligated into the corresponding restriction sites of pBluescript II SK(+) to obtain pJM135. For the generation of pJM137, a PCR product was amplified from B. amyloliquefaciens genomic DNA using primers RP-nach-BaeB-2-KpnI and FP-nach-BaeB-2-ApaI. This PCR product was ligated into pBluescript II SK(+) to obtain pJM137. pJM137 was then digested with ApaI/KpnI, and the insert was ligated into pJM140 to obtain pJM155. A PCR product was amplified from B. amyloliquefaciens genomic DNA using primers RP-Promotor-ApaI and FP-Promotor-BamHI and ligated into pBluescript II SK(+) to give pJM139. The insert of pJM139 was digested with BamHI/ApaI and inserted into the corresponding site of pJM155 to obtain pJM157. After linearization of pJM157 with KpnI, the DNA was integrated into the genome of B. amyloliquefaciens JM161 (construction described later) by double crossover to yield the baeB deletion mutant JM157. In the last step, pJM54 (Moldenhauer et al., 2010) was linearized with SacI and transformed into B. amyloliquefaciens JM157 to obtain the baeB-TE double mutant JM157-54.
The following strategy was used to generate JM161. A PCR product was amplified from pUCemr (gift from R. Borriss, Humboldt University, Berlin, Germany) using primers FP-erm-XbaI and RP-erm-ApaI and ligated into pBluescript II SK(+) to yield pJM136. The plasmids pJM135 and pJM136 were both digested with XbaI and ApaI, and the insert of pJM136 was ligated into pJM135 to obtain pJM143. pJM137 was digested with ApaI and KpnI, and this insert was ligated into the corresponding restriction sites of pJM143 to obtain pJM145. The erythromycin antibiotic cassette was replaced with the pIC333 (Steinmetz and Richter, 1994) spectinomycin cassette through a blunt-end/XbaI ligation into pJM145 to obtain pJM161. The resulting construct was linearized and transformed into the B. amyloliquefaciens wild-type strain FZB42 to obtain strain JM161.
Construction of Expression Plasmids
All primer sequences to generate inserts of expression plasmids are provided in Table S2.
To construct the BaeB expression plasmid pKJ7, a 0.7 kb PCR product was amplified from B. amyloliquefaciens genomic DNA using primers BaeB_low-GC_pHis8-3_for and BaeB_low-GC_pHis8-3_rev. This PCR product was ligated into pBluescript II SK(+) to obtain pKJ1. pKJ1 was then digested with BamHI and HindIII, and the 0.7 kb fragment was inserted into the corresponding sites of pHis8 (Jez et al., 2000).
To generate the pedC C-terminal strep-tagged fusion construct, a 1 kb PCR product was amplified from pPD23E7 (Piel, 2002) using primers pedC_FP and pedC_RP. This PCR product was ligated into pBluescript II SK(+) to obtain pHN37. pHN37 was then digested with BsaI, and the 1 kb fragment was inserted into the corresponding site of pASK-IBA3 (IBA Biotagnology) to yield pHN38.
The pedC C-terminal hexahistidine-tagged fusion construct pET-pedC was generated by PCR amplification of the gene using pHN38 as template DNA and pedC-5 and pedC-3 as primer. The PCR product was ligated into pET-24a(+) (Novagen) at the NdeI and XhoI restriction sites after digestion of the insert with the same enzymes.
To construct the PedD expression plasmid pKZ178, a 1.1 kb PCR product was amplified from pPD23E7 (Piel et al., 2004b) using primers pedD_FP and pedD_RP. This PCR product was ligated into pBluescript II SK(+) to obtain pKZ177. pKZ177 was then digested with EcoRI and HindIII, and the 1.1 kb fragment was inserted into the corresponding sites of pHis8 to yield pKZ178.
To construct the holo-PedI3 expression plasmid pKZ123, a 550 bp PCR product was amplified from pS9D2 (Piel et al., 2004b) using primers ped I3_pRSETf and pedI3_pRSETr. This PCR product was ligated into pBluescript II SK(+) to obtain pKZ57. pKZ57 was then digested with BglII und HindIII, and the 550 bp fragment was inserted into the corresponding site of pHis8 svp to yield pKZ123. To construct the apo-PedI3 expression plasmid pKZ176, pKZ123 was cut with NotI to remove the phosphopantetheinyl transferase gene svp and then religated to obtain pKZ176.
To construct the holo-PedN expression plasmid pKZ124, a 240 bp PCR product was amplified from Paederus fuscipes metagenomic DNA using primers pedN_pRSETf and pedN_pRSETr. This PCR product was ligated into pBluescript II SK(+) to obtain pKZ32. pKZ32 was then digested with BglII and HindIII, and the 240 bp fragment was inserted into the corresponding site of pHis8 svp to yield pKZ124. To construct the apo-PedN expression plasmid pKZ161, pKZ124 was cut with NotI to remove the svp gene and then religated to obtain pKZ161.
To construct the holo-PsyA expression plasmid pHN77, a 300 bp PCR product was amplified from pPSKF1 (Fisch et al., 2009) using primers PsyA-ACP3 for and PsyA-ACP3 rev. This PCR product was ligated into pBluescript II SK(+) to obtain pHN75. pHN75 was then digested with BamHI and HindIII, and the 300 bp fragment was inserted into the corresponding site of pHis8 svp to yield pHN77. To construct the apo-PsyA expression plasmid pHN60, a 300 bp PCR product was amplified from pPSKF1 (Fisch et al., 2009) using primers PsyA-ACP3 for and PsyA-ACP3 rev. This PCR product was ligated into pBluescript II SK(+) to yield pHN59. pHN59 was then digested with BamHI/EagI, and the 300 bp fragment was inserted into the corresponding site of pHis8 to yield pHN60.
To construct the holo-PsyD expression plasmid pHN78, a 320 bp PCR product was amplified from pPSKF1 using primers PsyD-ACP1 for and PsyD-ACP1 rev. This PCR product was ligated into pBluescript II SK(+) to obtain pHN76. pHN76 was then digested with BamHI and HindIII, and the 320 bp fragment was inserted into the corresponding site of pHis8 svp to yield pHN78. To construct the apo-PsyD expression plasmid pHN66, a 320 bp PCR product was amplified from pPSKF1 using primers PsyDACP1 for and PsyD-ACP1 rev. This PCR product was ligated into pBluescript II SK(+) to obtain pHN65. pHN65 was then digested with BamHI and EagI, and the 320 bp fragment was inserted into the corresponding site of pHis8 to yield pHN66.
Construction of the expression plasmid for the PksA ACP was reported earlier (Crawford et al., 2006).
The gene coding for RhiG was amplified by PCR from cosmid 20G04 (Partida-Martinez and Hertweck, 2007) with the primers Expr_tAT_F and Expr_tAT_R. Ligation into the EcoRV site of the pGEM-T Easy cloning vector (Promega) yielded pNB118. The NdeI/HindIII fragment of pNB118 was ligated into the NdeI/HindIII sites of pET28a(+) expression vector, which carries an N-terminal His6-Tag, resulting in pNB121.
To introduce point mutations into active site of RhiG, we used the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene) on pNB121 (AT1°AT2–S106A and AT1AT2°–S468A). The plasmid pNB146 contains a rhiG variant coding for AT1°AT2, and the plasmid pNB147 contains a rhiG variant coding for AT1AT2°.
Expression and Purification of Proteins
To prepare sufficient amounts of PedC for kinetic studies, large-scale fermentation was needed. A colony of E. coli cells freshly transformed with pHN38 and pGro7 was cultivated up to an optical density (OD600) of about 4 at 37°C in 2 ml LB medium. The grown biomass was used to inoculate the second preculture in 1 l baffled shake flasks with 150 ml culture volume at 120 rpm min−1 and 37°C. The second preculture medium contained 5 g/l−1 yeast extract, 10 g/l−1 tryptone, 2.5 g/l−1 NaCl, 0.25 g/l−1 MgSO4 · 7 H2O, 1.26 g/l−1 NaH2PO4, and 10 g/l−1 glucose, adjusted to pH = 7.2 with 4 N NaOH. Main cultivation was performed in a 30 l scale bioreactor (Chemap) with 20 l initial culture volume at 37°C. Dissolved oxygen concentration was controlled at ≥40% of air saturation, and the pH was kept at 7.0 with 25% ammonia solution. Medium was similar as for the second preculture, except that 20 g/l−1 glucose and 5 g/l−1 arabinose were present. A total of 900 ml of the second preculture was used to inoculate the main culture, resulting in an initial OD600 = 0.25. At OD600 = 4.5 (after about 3 hr), 4 mg of anhydrotetracycline (IBA Biotagnology) was added (resulting in 200 ng/ml), and the temperature was decreased to 25°C in less than 10 min. After 7 hr cultivation time, a glucose feed (600 g/l−1 glucose monohydrate) was started to keep the glucose concentration in a range of 13–17 g/l throughout the cultivation process. Glucose measurement was performed using an AkkuCheck glucose analyzer (Roche Diagnostics). After 24 hr of cultivation time, the bioreactor was cooled to 10°C, and the cell suspension (OD600 = 35) was harvested. Biomass separation was achieved by centrifugation at 8,000 rpm (rotor type JLA8.1000, Avanti J-20 XP, Beckman), and the biomass was stored at −80°C prior to further protein purification. The wet cell weight was 500 g. For kinetic measurements of PedC, elution fractions were dialyzed three times for at least 3 hr against 200 mM potassium phosphate buffer (KPi), pH 7.4 containing 10% (v/v) glycerol. If necessary, fractions were concentrated with Vivaspin columns (VWR). Protein concentration was determined either by Bradford assay or, if expressed with chaperones, with SDS-PAGE-absorption measurements. Specifically, SDS-PAGE was performed with all protein samples along with BSA samples of known concentration. The band intensities were integrated using the program GeneTools (Syngene). A calibration curve was established, and the concentration of the protein samples was determined. Proteins were flash frozen in liquid nitrogen after adding 10% (v/v) glycerol and stored at −80°C.
For the overexpression of all other proteins, except for pET-PedC, 1 l cultures were grown to an OD600 of 0.7 (37°C, 200 rpm), induced with either isopropyl β-d-1-thiogalactopyranoside (IPTG) or anhydrotetracycline, and grown for approximately 20 hr at 16°C. Cells of expression strains were harvested by centrifugation (11,000 × g, 7 min) at 4°C. For pET-PedC, 1 l expression cultures were supplemented with 50 ng/ml−1 tetracycline prior to inoculation with a 10 ml saturated starter culture. Cells were grown at 37°C 250 rpm, and growth was followed by monitoring OD600. The cultures, at OD600 ~0.4, were cooled on ice for 15 min, induced with 1 mM IPTG, and expressed at 18°C for 16 hr.
Cells expressing proteins with His6- or His8-Tag were resuspended in lysis buffer (25 mM Tris-HCl, pH 7.6, 0.5 M NaCl, 10 mM imidazole, and 0.1% (v/v) Triton X-100). The pH was adjusted to 7.6 in the case of PedD, RhiG, and their derivatives. A pH of 8 was chosen for all other proteins. The cell suspension was kept on ice and sonicated in 10 s intervals. The cell debris was removed by centrifugation at 23,500 × g for 30 min at 4°C. The supernatant was incubated with 1.0 ml Ni-NTA agarose (QIAGEN) for at least 1 hr at 4°C and washed with 10 ml lysis buffer. His-tagged proteins were eluted with lysis buffer containing increasing concentrations of imidazole up to 250 mM. For the expression using pET-pedC, a modified protocol was used. Cells were harvested by centrifugation (4,100 × g, 20 min) at 4°C, resuspended (1 g per 4 ml) in Ni-buffer (50 mM KPi pH 7.6, 300 mM NaCl, 10% glycerol, 20 mM imidazole), and lysed by treatment with 1 mg/ml lysozyme (Sigma) for 30 min, followed by sonication in 10 s intervals. The lysate was clarified by centrifugation (27,000 × g, 30 min), decanted, and bound to 1 ml Ni-NTA resin (QIAGEN) for 1 hr rotating at 4°C. The resin was washed three times with 10 ml Ni-buffer and eluted three times with 0.5 ml Ni-buffer containing 250 mM imidazole. Fractions were analyzed by SDS-PAGE. The flowthrough contained a large amount of unbound PedC, which was subsequently collected by a second round of Ni2+ affinity chromatography. Fractions containing PedC were pooled, concentrated to ~7 mg/ml−1 using a 10,000 MWCO Amicon Ultra Centrifugal Filter Unit (Millipore), and dialyzed against storage buffer containing 100 mM KPi pH 7.0, 10% glycerol, and 2 mM dithiothreitol (DTT). The concentration of PedC was determined by Bradford assay in duplicate and adjusted for GroEL content.
Cells containing expression constructs with Strep tags were resuspended in Buffer W (100 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA). After sonication and centrifugation as described earlier, the supernatant was applied to a column with 1 ml Strep-Tactin-Sepharose (IBA Biotagnology). The column was washed twice with 5 ml Buffer W and eluted with six fractions of 0.5 ml Buffer E (100 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 2.5 mM desthiobiotin). Columns were used up to five times and regenerated according to manufacturer’s specifications.
Acyltransferase Assays
To study acyl transfer from ATs to ACPs, we performed assays with [2-14C]-malonyl-CoA and [1-14C]-acetyl-CoA (Perkin-Elmer). Assay mixtures consisted of 4 µl DTT (10 mM), 5 µl AT, 9 µl ACP, and either 2 µl [malonyl-2-14C-CoA] (0.01 mCi/ml−1) or 1 µl acetyl-CoA [acetyl-1-14C-CoA] (0.02 mCi/ml−1). Lysis buffer, instead of buffer containing the enzyme, was used in the negative controls. Everything was mixed on ice, and the reaction was started with the addition of the substrate. Samples were incubated on ice for various times, followed by the addition of equal volumes of 23 SDS-sample buffer (0.09 M Tris-HCl, pH 6.8, 20% (v/v) glycerol, 2%(v/v) SDS, 0.02% (v/v) bromophenol blue, 0.1 M DTT) and incubation at 99°C for 3 min. After separation by SDS-PAGE, the gels were dried and exposed to X-ray film. Band quantification was performed with an Image Reader and the software Bas Reader (Raytest). The autoradiogram was edited with the software TINA 2.09d (Raytest). To differentiate between self-loading of ACPs and AT-mediated loading, we also performed measurements without AT.
Acyl-SNAC Hydrolysis Assays
After desalting in 200 mM KPi, pH 7.4, and 10% (v/v) glycerol, proteins were incubated with different test substrates at a range of concentrations. In these assays, PedC was used only as purified, Strep-tagged protein (Figure S3). A typical assay, adapted from Heathcote et al. (2001), consisted of 250 nM AT, 200 mM KPi, pH 7.4, 0.2 mM DTNB (Alfa Aesar), and 3% (v/v) dimethylsufoxide (DMSO) at 30°C in a total volume of 1 mL. All test substrates were dissolved as 2 M stocks in DMSO except compound 5, which was dissolved in 200 mM KPi, pH 7.4. A 20 mM DTNB stock solution was resuspended in 200 mM KPi, pH 7.4. Samples containing 7 and 9 were incubated for 2 min at room temperature (RT) prior to measurement, allowing better solubilization. Enzymatic activities were monitored by measuring the absorbance increase at 412 nm resulting from hydrolysis of Ellman’s regent (ε = 14,150 M−1 cm−1) (Frank et al., 2007) for 18 min using a BioMate 3 photometer (Thermo). All measurements were carried out in at least duplicate and corrected for the background rate of hydrolysis without enzyme. For the determination of kinetic parameters, initial rates were plotted versus the substrate concentration and fitted to the Michaelis-Menten equation with GraphPad Prism 4.0 (GraphPad Software). For those enzymes, a Michaelis-Menten fit was not possible; only the initial velocities were plotted.
Hydrolysis of hexanoyl-SNAC (mimicking the hexanoyl starter unit of PksA from Aspergillus parasiticus involved in norsolorinic acid biosynthesis) was determined using an HPLC-based assay. Hexanoyl-SNAC (1 mM) was treated with 5 µM PedC in 100 mM KPi, pH 7.0, 10% glycerol, and 1 mM DTT. At each time point, 150 µl reaction aliquots were passed through a 10,000 MWCO Microcon centrifugal filter device (Millipore) at 14,000 × g for 10 min to remove the protein. Fifty microliter injections of microcon flowthrough were separated over a Prodigy 5µ ODS3 100 Å, 250 × 4.6 mm column (Phenomenex) using an Agilent Technologies 1200 series HPLC and a 40:60:0.1 acetonitrile:water:formic acid isocratic method at 1 ml/min−1. The peak area of hexanoyl-SNAC (RT for 12.0 min) was determined by manual integration at 232 nm wavelength. Controls with no enzyme or BSA showed no loss in hexanoyl-SNAC signal, indicating that hydrolysis was catalyzed by PedC.
Acyl-ACP Hydrolysis Assays
To determine whether PedC can hydrolyze acyl-ACP intermediates, we used a radiochemical hydrolysis assay. The PksA ACP monodomain was activated with various 14C-labeled CoAs and treated with a serial dilution of PedC in a fixed time point assay.
The N-terminally hexahistidine-tagged PksA apo-ACP monodomain (excised from the six domain iterative megasynthase) was purified as previously described and activated by an amended protocol described later (Crawford et al., 2006). 14C-labeled acyl-ACP was generated by treating 100 µM apo-ACP with 5 µMSvp (phosphopantetheinyl transferase from Streptomyces verticillus), and 400 µMtotal acyl-CoA (mixture of unlabeled and 14C-labeled to achieve 5.5 µCi/ml−1) in 100 mM KPi, 10 mM MgCl2, 1 mM TCEP, and 10% glycerol for 45 min at RT. Acyl-ACP can be frozen for later use or diluted to be used immediately.
PedC was thawed and serially diluted (1:5) to five different concentrations ranging from 20 to 0.02 µM 43 stock solutions in 100 mM KPi, pH 7.0, with 10% glycerol. Twelve microliters of each PedC stock was pre-aliquoted, and the assay was initiated by addition of 36 µl of 13.3 µM 14C-acyl-ACP. After 10 min at room temperature, 20 µl aliquots of each reaction were quenched into 10 µl of 43 SDS loading dye. Final reaction conditions were 10 µM acyl-ACP treated with 0 (negative control), 5, 0.83, 0.14, 0.023, or 0.005 µM PedC. Samples were separated by 16% SDS-PAGE, dried on filter paper with a slab gel dryer (Hoeffer Scientific), exposed overnight to a storage phosphor screen (Amersham), and imaged with a Typhoon 9410 Imager (Johns Hopkins Integrated Imaging Center).
Phylogenetic Analysis
Sequences of AT-like proteins were aligned using MUSCLE. Phylogenetic analysis was performed by maximum likelihood inference using PhyML with a Jones-Taylor-Thornten amino acid replacement model.
Synthesis of Acyl-SNAC Derivatives 5–10
Synthesis of the known compounds 5 and 10 was performed as published previously (Crawford et al., 2006; Grüschow et al., 2007) The general procedure for compounds 6–9 was as follows. A solution of the corresponding acid (5 mmol) in dichloromethane (15 ml) was cooled down to 0°C for 15 min. 4-Dimethylaminopyridine (DMAP, 1 mmol), 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide (EDC, 6 mmol), and N-acetylcysteamine (6 mmol) were added, and the mixture was stirred overnight at 25°C. The reaction mixture was quenched with saturated aqueous ammonium chloride and extracted with dichloromethane. The combined organic phases were dried with MgSO4, filtered, concentrated, and subjected to silica gel column chromatography as eluant to provide the respective thioester.
S-Crotonoyl-N-acetylcysteamine
Yield was as follows: 54%, column chromatography with ethylacetate. 1HNMR: (400 MHz, CDCl3, RT) δ [ppm] = 6.95 (dq, 1H, 3JH-H = 15.4 Hz, 3JH-H = 6.9 Hz), 6.16 (dq, 1H, 3JH-H = 15.4 Hz, 4JH-H = 1.6 Hz), 5.95 (brs, 1H), 3.46 (q, 2H, 3JH-H = 6.6 Hz), 3.08 (t, 2H, 3JH-H = 6.6 Hz), 1.93 (s, 3H), 1.88 (dd, 3H, 3JH-H = 6.9 Hz, 4JH-H = 1.6 Hz); 13C-NMR: (100 MHz, CDCl3, RT) δ [ppm] = 190.5, 170.6, 142.1, 130.1, 40.1, 28.4, 23.4, 18.2; MS: (ESI, 10 eV, m/z (%)) 224.1 (100) [M + Na]+; HR-MS (ESI, 10 eV, m/z): C8H13NO2SH 188.0740; measured: 188.0743 [M + H]+.
(E)-S-(Hex-2-enoyl)-N-acetylcysteamine
Yield was as follows: 43%, column chromatography with cylcohexane/ethylacetate 2:1. 1H-NMR: (400 MHz, CDCl3, RT) δ [ppm] = 6.92 (dt, 1H, 3JH-H = 15.5 Hz, 3JH-H = 7.0 Hz), 6.13 (td, 1H, 3JH-H = 15.5 Hz, 4JH-H = 1.5 Hz), 5.88 (bs; 1H), 3.49-3.44 (pq, 2H, 3JH-H = 6.5 Hz), 3.09 (t, 2H, 3JH-H = 6.5 Hz), 2.19 (ddt, 2H, 4JH-H = 1.5 Hz, 3JH-H = 7.0 Hz, 3JH-H = 7.3 Hz), 1.96 (s; 3H; H-1), 1.50 (tq, 3H, 3JH-H = 7.4 Hz), 0.94 (t, 6H, 3JH-H = 7.4 Hz); 13C-NMR: (100 MHz, CDCl3, RT) δ [ppm] = 190.4, 170.2, 146.5, 128.4, 39.8, 34.2, 28.2, 23.2, 21.2, 13.7; MS: (ESI, 10 eV, m/z) 120.0 (8) [NAC + H]+, 216.1 [M + H]+, 238.1 [M + Na]+.
S-(3-Methylcrotonyl)-N-acetylcysteamine
Yield was as follows: 40%, column chromatography with ethylacetate. 1HNMR: (400 MHz, CDCl3, RT) δ [ppm] = 5.98 (s, 1H), 5.98 (brs, 1H), 3.45 (q, 2H, 3JH-H = 6.0 Hz), 3.04 (t, 2H, 3JH-H = 6.0 Hz), 2.13 (s, 3H), 1.93 (s, 3H), 1.86 (s, 3H); 13C-NMR: (100 MHz, CDCl3, RT) δ [ppm] = 189.8, 170.5, 155.1, 123.2, 40.2, 28.6, 27.5, 23.4, 21.5; MS: (ESI, 10 eV, m/z (%)) 210.1 (100) [M + Na]+; HR-MS (ESI, 10 eV, m/z): C9H15NO2SH 202.0896; measured: 202.0893 [M + H]+.
S-(4-Methylvaleryl)-N-acetylcysteamine
Yield was as follows: 90%, column chromatography with cylcohexane/ethylacetate; gradient: 1:1 to 1:3. 1H-NMR: (400 MHz, CDCl3, RT) δ [ppm] = 5.82 (bs, 1H), 3.43 (pq, 2H, 3JH-H = 6.4 Hz), 3.02 (t, 2H, 3JH-H = 6.4 Hz), 2.60-2.56 (m, 2H), 1.96 (s, 3H), 1.62-1.53 (m, 3H), 0.90 (d, 6H); 13C-NMR: (100 MHz, CDCl3, RT) δ [ppm] = 200.3, 170.2, 42.2, 39.7, 34.3, 28.4, 27.6, 23.2, 22.2; MS: (ESI, 10 eV, m/z) 218.1 [M + H]+, 240.1 [M + Na]+. For details, see Supplemental Information.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. Chang, M. Engeser, C. Sondag, and H.-J. Brandt for technical assistance; R. Borriss, B.S. Moore, and M. Marahiel for plasmids; and M.F. Freeman for experimental suggestions. This work was financially supported by the DFG (German Research Foundation) (PI 430/1-3, PI 430/6-1, PI 430/9-1, SFB 624, and GRK 804 to J.P.).
K.J., K.Z., H.N., A.L.V., N.B., and M.O. expressed proteins; J.M. constructed the B. amyloliquefaciens mutants and analyzed metabolites of the bacillaene pathway; K.Z. and H.N. conducted the acyl transfer assays; K.J. conducted acyl-SNAC hydrolysis assays; A.L.V. conducted acyl-ACP hydrolysis assays; S.F., P.P., and C.K. synthesized SNAC derivatives; and J.P. performed the phylogenetic analysis. All authors designed the experiments, analyzed the data, and wrote the manuscript.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and two tables and can be found with this article online at doi:10.1016/j.chembiol.2012.01.005.
REFERENCES
- Antoniewski C, Savelli B, Stragier P. The spoIIJ gene, which regulates early development steps in Bacillus subtilis, belongs to a class of environmentally responsive genes. J. Bacteriol. 1990;17284:86–93. doi: 10.1128/jb.172.1.86-93.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumpus SB, Magarvey NA, Kelleher NL, Walsh CT, Calderone CT. Polyunsaturated fatty-acid-like trans-enoyl reductases utilized in polyketide biosynthesis. J. Am. Chem. Soc. 2008;130:11614–11616. doi: 10.1021/ja8040042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher RA, Schroeder FC, Fischbach MA, Straight PD, Kolter R, Walsh CT, Clardy J. The identification of bacillaene, the product of the PksX megacomplex in Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 2007;104:1506–1509. doi: 10.1073/pnas.0610503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone CT. Isoprenoid-like alkylations in polyketide biosynthesis. Nat. Prod. Rep. 2008;25:845–853. doi: 10.1039/b807243d. [DOI] [PubMed] [Google Scholar]
- Calderone CT, Kowtoniuk WE, Kelleher NL, Walsh CT, Dorrestein PC. Convergence of isoprene and polyketide biosynthetic machinery: isoprenyl-S-carrier proteins in the pksX pathway of Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 2006;103:8977–8982. doi: 10.1073/pnas.0603148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone CT, Bumpus SB, Kelleher NL, Walsh CT, Magarvey NA. A ketoreductase domain in the PksJ protein of the bacillaene assembly line carries out both alpha- and beta-ketone reduction during chain growth. Proc. Natl. Acad. Sci. USA. 2008;105:12809–12814. doi: 10.1073/pnas.0806305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AY, Schnarr NA, Kim CY, Cane DE, Khosla C. Extender unit and acyl carrier protein specificity of ketosynthase domains of the 6-deoxyerythronolide B synthase. J. Am. Chem. Soc. 2006a;128:3067–3074. doi: 10.1021/ja058093d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XH, Vater J, Piel J, Franke P, Scholz R, Schneider K, Koumoutsi A, Hitzeroth G, Grammel N, Strittmatter AW, et al. Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J. Bacteriol. 2006b;188:4024–4036. doi: 10.1128/JB.00052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XH, Koumoutsi A, Scholz R, Schneider K, Vater J, Süssmuth R, Piel J, Borriss R. Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J. Biotechnol. 2009;140:27–37. doi: 10.1016/j.jbiotec.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Cheng YQ, Tang GL, Shen B. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc. Natl. Acad. Sci. USA. 2003;100:3149–3154. doi: 10.1073/pnas.0537286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichewicz RH, Valeriote FA, Crews P. Psymberin, a potent sponge-derived cytotoxin from Psammocinia distantly related to the pederin family. Org. Lett. 2004;6:1951–1954. doi: 10.1021/ol049503q. [DOI] [PubMed] [Google Scholar]
- Crawford JM, Dancy BCR, Hill EA, Udwary DW, Townsend CA. Identification of a starter unit acyl-carrier protein transacylase domain in an iterative type I polyketide synthase. Proc. Natl. Acad. Sci. USA. 2006;103:16728–16733. doi: 10.1073/pnas.0604112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrestein PC, Bumpus SB, Calderone CT, Garneau-Tsodikova S, Aron ZD, Straight PD, Kolter R, Walsh CT, Kelleher NL. Facile detection of acyl and peptidyl intermediates on thiotemplate carrier domains via phosphopantetheinyl elimination reactions during tandem mass spectrometry. Biochemistry. 2006;45:12756–12766. doi: 10.1021/bi061169d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du LC, Lou LL. PKS and NRPS release mechanisms. Nat. Prod. Rep. 2010;27:255–278. doi: 10.1039/b912037h. [DOI] [PubMed] [Google Scholar]
- Fisch KM, Gurgui C, Heycke N, van der Sar SA, Anderson SA, Webb VL, Taudien S, Platzer M, Rubio BK, Robinson SJ, et al. Polyketide assembly lines of uncultivated sponge symbionts from structure-based gene targeting. Nat. Chem. Biol. 2009;5:494–501. doi: 10.1038/nchembio.176. [DOI] [PubMed] [Google Scholar]
- Floss HG, Yu TW. Lessons from the rifamycin biosynthetic gene cluster. Curr. Opin. Chem. Biol. 1999;3:592–597. doi: 10.1016/s1367-5931(99)00014-9. [DOI] [PubMed] [Google Scholar]
- Frank B, Knauber J, Steinmetz H, Scharfe M, Blöcker H, Beyer S, Müller R. Spiroketal polyketide formation in Sorangium: identification and analysis of the biosynthetic gene cluster for the highly cytotoxic spirangienes. Chem. Biol. 2007;14:221–233. doi: 10.1016/j.chembiol.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Grüschow S, Buchholz TJ, Seufert W, Dordick JS, Sherman DH. Substrate profile analysis and ACP-mediated acyl transfer in Streptomyces coelicolor Type III polyketide synthases. ChemBioChem. 2007;8:863–868. doi: 10.1002/cbic.200700026. [DOI] [PubMed] [Google Scholar]
- Heathcote ML, Staunton J, Leadlay PF. Role of type II thioesterases: evidence for removal of short acyl chains produced by aberrant decarboxylation of chain extender units. Chem. Biol. 2001;8:207–220. doi: 10.1016/s1074-5521(01)00002-3. [DOI] [PubMed] [Google Scholar]
- Hertweck C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. Engl. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- Irschik H, Kopp M, Weissman KJ, Buntin K, Piel J, Müller R. Analysis of the sorangicin gene cluster reinforces the utility of a combined phylogenetic/retrobiosynthetic analysis for deciphering natural product assembly by trans-AT PKS. ChemBioChem. 2010;11:1840–1849. doi: 10.1002/cbic.201000313. [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Namikoshi M, Kobayashi H, Furukawa J, Okuda S, Itai A, Kasuya A, Iitaka Y, Sato Z. Studies on macrocyclic lactone antibiotics. VIII. Absolute structures of rhizoxin and a related compound. J. Antibiot. (Tokyo) 1986;39:424–429. doi: 10.7164/antibiotics.39.424. [DOI] [PubMed] [Google Scholar]
- Jez JM, Ferrer JL, Bowman ME, Dixon RA, Noel JP. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry. 2000;39:890–902. doi: 10.1021/bi991489f. [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Williard PG, Khosla C, Cane DE. Precursor-directed biosynthesis of 16-membered macrolides by the erythromycin polyketide synthase. J. Am. Chem. Soc. 2001;123:2495–2502. doi: 10.1021/ja004139l. [DOI] [PubMed] [Google Scholar]
- Koglin A, Löhr F, Bernhard F, Rogov VV, Frueh DP, Strieter ER, Mofid MR, Güntert P, Wagner G, Walsh CT, et al. Structural basis for the selectivity of the external thioesterase of the surfactin synthetase. Nature. 2008;454:907–911. doi: 10.1038/nature07161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusebauch B, Busch B, Scherlach K, Roth M, Hertweck C. Polyketide-chain branching by an enzymatic Michael addition. Angew. Chem. Int. Ed. Engl. 2009;48:5001–5004. doi: 10.1002/anie.200900277. [DOI] [PubMed] [Google Scholar]
- Lackner G, Moebius N, Partida-Martinez L, Hertweck C. Complete genome sequence of Burkholderia rhizoxinica, an Endosymbiont of Rhizopus microsporus. J. Bacteriol. 2011a;193:783–784. doi: 10.1128/JB.01318-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner G, Moebius N, Partida-Martinez LP, Boland S, Hertweck C. Evolution of an endofungal lifestyle: Deductions from the Burkholderia rhizoxinica genome. BMC Genomics. 2011b;12:210. doi: 10.1186/1471-2164-12-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopanik NB, Shields JA, Buchholz TJ, Rath CM, Hothersall J, Haygood MG, Håkansson K, Thomas CM, Sherman DH. In vivo and in vitro trans-acylation by BryP, the putative bryostatin pathway acyltransferase derived from an uncultured marine symbiont. Chem. Biol. 2008;15:1175–1186. doi: 10.1016/j.chembiol.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchuk D, Drumm M, Saulino A, Collins FS. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 1991;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzella HG, Carney JR, Santi DV. Rational design and assembly of synthetic trimodular polyketide synthases. Chem. Biol. 2007;14:143–151. doi: 10.1016/j.chembiol.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Minto RE, Townsend CA. Enzymology and molecular biology of aflatoxin biosynthesis. Chem. Rev. 1997;97:2537–2556. doi: 10.1021/cr960032y. [DOI] [PubMed] [Google Scholar]
- Moldenhauer J, Chen XH, Borriss R, Piel J. Biosynthesis of the antibiotic bacillaene, the product of a giant polyketide synthase complex of the trans-AT family. Angew. Chem. Int. Ed. Engl. 2007;46:8195–8197. doi: 10.1002/anie.200703386. [DOI] [PubMed] [Google Scholar]
- Moldenhauer J, Götz DCG, Albert CR, Bischof SK, Schneider K, Süssmuth RD, Engeser M, Gross H, Bringmann G, Piel J. The final steps of bacillaene biosynthesis in Bacillus amyloliquefaciens FZB42: direct evidence for beta,gamma dehydration by a trans-acyltransferase polyketide synthase. Angew. Chem. Int. Ed. Engl. 2010;49:1465–1467. doi: 10.1002/anie.200905468. [DOI] [PubMed] [Google Scholar]
- Musiol EM, Härtner T, Kulik A, Moldenhauer J, Piel J, Wohlleben W, Weber T. Supramolecular templating in kirromycin biosynthesis: the acyltransferase KirCII loads ethylmalonyl-CoA extender onto a specific ACP of the trans-AT PKS. Chem. Biol. 2011;18:438–444. doi: 10.1016/j.chembiol.2011.02.007. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Ishida K, Jenke-Kodama H, Dittmann E, Gurgui C, Hochmuth T, Taudien S, Platzer M, Hertweck C, Piel J. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat. Biotechnol. 2008;26:225–233. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]
- Partida-Martinez LP, Hertweck C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature. 2005;437:884–888. doi: 10.1038/nature03997. [DOI] [PubMed] [Google Scholar]
- Partida-Martinez LP, Hertweck C. A gene cluster encoding rhizoxin biosynthesis in “Burkholderia rhizoxina”, the bacterial endosymbiont of the fungus Rhizopus microsporus. ChemBioChem. 2007;8:41–45. doi: 10.1002/cbic.200600393. [DOI] [PubMed] [Google Scholar]
- Pettit GR, Xu JP, Chapuis JC, Pettit RK, Tackett LP, Doubek DL, Hooper JNA, Schmidt JM. Antineoplastic agents. 520. Isolation and structure of irciniastatins A and B from the Indo-Pacific marine sponge Ircinia ramosa. J. Med. Chem. 2004;47:1149–1152. doi: 10.1021/jm030207d. [DOI] [PubMed] [Google Scholar]
- Piel J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl. Acad. Sci. USA. 2002;99:14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel J. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 2009;26:338–362. doi: 10.1039/b703499g. [DOI] [PubMed] [Google Scholar]
- Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 2010;27:996–1047. doi: 10.1039/b816430b. [DOI] [PubMed] [Google Scholar]
- Piel J, Hui DQ, Wen GP, Butzke D, Platzer M, Fusetani N, Matsunaga S. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl. Acad. Sci. USA. 2004a;101:16222–16227. doi: 10.1073/pnas.0405976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel J, Wen GP, Platzer M, Hui DQ. Unprecedented diversity of catalytic domains in the first four modules of the putative pederin polyketide synthase. ChemBioChem. 2004b;5:93–98. doi: 10.1002/cbic.200300782. [DOI] [PubMed] [Google Scholar]
- Pohl NL, Gokhale RS, Cane DE, Khosla C. Synthesis and incorporation of an N-acetylcysteamine analogue of methylmalonyl-CoA by a modular polyketide synthase. J. Am. Chem. Soc. 1998;120:11206–11207. [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Schwarzer D, Mootz HD, Linne U, Marahiel MA. Regeneration of misprimed nonribosomal peptide synthetases by type II thioesterases. Proc. Natl. Acad. Sci. USA. 2002;99:14083–14088. doi: 10.1073/pnas.212382199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- Steinmetz M, Richter R. Easy cloning of mini-Tn10 insertions from the Bacillus subtilis chromosome. J. Bacteriol. 1994;176:1761–1763. doi: 10.1128/jb.176.6.1761-1763.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Koppisch AT, Khosla C. The acyltransferase homologue from the initiation module of the R1128 polyketide synthase is an acyl-ACP thioesterase that edits acetyl primer units. Biochemistry. 2004;43:9546–9555. doi: 10.1021/bi049157k. [DOI] [PubMed] [Google Scholar]
- Teta R, Gurgui M, Helfrich EJN, Künne S, Schneider A, Van Echten-Deckert G, Mangoni A, Piel J. Genome mining reveals trans-AT polyketide synthase directed antibiotic biosynthesis in the bacterial phylum bacteroidetes. ChemBioChem. 2010;11:2506–2512. doi: 10.1002/cbic.201000542. [DOI] [PubMed] [Google Scholar]
- Vergnolle O, Hahn F, Baerga-Ortiz A, Leadlay PF, Andexer JN. Stereoselectivity of isolated dehydratase domains of the borrelidin polyketide synthase: implications for cis double bond formation. ChemBioChem. 2011;12:1011–1014. doi: 10.1002/cbic.201100011. [DOI] [PubMed] [Google Scholar]
- Wong FT, Jin X, Mathews II, Cane DE, Khosla C. Structure and mechanism of the trans-acting acyltransferase from the disorazole synthase. Biochemistry. 2011;50:6539–6548. doi: 10.1021/bi200632j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Hothersall J, Mazzetti C, O’Connell Y, Shields JA, Rahman AS, Cox RJ, Crosby J, Simpson TJ, Thomas CM, Willis CL. In vivo mutational analysis of the mupirocin gene cluster reveals labile points in the biosynthetic pathway: the “leaky hosepipe” mechanism. ChemBioChem. 2008;9:1500–1508. doi: 10.1002/cbic.200800085. [DOI] [PubMed] [Google Scholar]
- Yeh E, Kohli RM, Bruner SD, Walsh CT. Type II thioesterase restores activity of a NRPS module stalled with an aminoacyl-S-enzyme that cannot be elongated. ChemBioChem. 2004;5:1290–1293. doi: 10.1002/cbic.200400077. [DOI] [PubMed] [Google Scholar]
- Yu TW, Shen YM, Doi-Katayama Y, Tang L, Park C, Moore BS, Richard Hutchinson C, Floss HG. Direct evidence that the rifamycin polyketide synthase assembles polyketide chains processively. Proc. Natl. Acad. Sci. USA. 1999;96:9051–9056. doi: 10.1073/pnas.96.16.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann K, Engeser M, Blunt JW, Munro MHG, Piel J. Pederin-type pathways of uncultivated bacterial symbionts: analysis of o-methyltransferases and generation of a biosynthetic hybrid. J. Am. Chem. Soc. 2009;131:2780–2781. doi: 10.1021/ja808889k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.