Abstract
Pseudomonas syringae is a member of an important group of Gram-negative bacterial pathogens of plants and animals that depend on a type III secretion system to inject virulence effector proteins into host cells. In P. syringae, hrp/hrc genes encode the Hrp (type III secretion) system, and avirulence (avr) and Hrp-dependent outer protein (hop) genes encode effector proteins. The hrp/hrc genes of P. syringae pv syringae 61, P. syringae pv syringae B728a, and P. syringae pv tomato DC3000 are flanked by an exchangeable effector locus and a conserved effector locus in a tripartite mosaic Hrp pathogenicity island (Pai) that is linked to a tRNALeu gene found also in Pseudomonas aeruginosa but without linkage to Hrp system genes. Cosmid pHIR11 carries a portion of the strain 61 Hrp pathogenicity island that is sufficient to direct Escherichia coli and Pseudomonas fluorescens to inject HopPsyA into tobacco cells, thereby eliciting a hypersensitive response normally triggered only by plant pathogens. Large deletions in strain DC3000 revealed that the conserved effector locus is essential for pathogenicity but the exchangeable effector locus has only a minor role in growth in tomato. P. syringae secretes HopPsyA and AvrPto in culture in a Hrp-dependent manner at pH and temperature conditions associated with pathogenesis. AvrPto is also secreted by Yersinia enterocolitica. The secretion of AvrPto depends on the first 15 codons, which are also sufficient to direct the secretion of an Npt reporter from Y. enterocolitica, indicating that a universal targeting signal is recognized by the type III secretion systems of both plant and animal pathogens.
Type III protein secretion systems underlie the pathogenicity of many Gram-negative bacteria, including important animal pathogens in the genera Yersinia, Salmonella, Shigella, and Escherichia, and plant pathogens in the genera Pseudomonas, Erwinia, Xanthomonas, and Ralstonia (1, 2). The plant pathogens cause diverse diseases in hosts that range from apple trees to the model weed Arabidopsis thaliana, and they all share an ability to colonize the intercellular spaces of plant tissues and to cause death (sometimes delayed) in plant cells. A fundamental difference between this group of plant pathogens and most of the animal pathogens is that the former do not enter living host cells, but rather interact with the host cytoplasm from outside of an approximately 200-nm-thick plant cell wall. The ability to deliver effector proteins across this barrier via the type III secretion system is likely to be unique to plant pathogens, and it is key to their pathogenicity.
Pseudomonas syringae is a widespread and representative plant pathogen. It is host specific and elicits leaf spots and other foliar necroses in host plants and the hypersensitive response (HR) in nonhosts (3). In host plants, disease symptoms typically develop after several days of bacterial growth in leaf intercellular spaces. In nonhosts, the defense-associated programmed cell death that characterizes the HR occurs within 24 h in plant cells that are in contact with the bacterium (4). Underlying both types of P. syringae interactions with plants are hrp (HR and pathogenicity) and hrc (HR and conserved) genes that encode the type III secretion system and avirulence (avr) and Hrp-dependent outer protein (hop) genes that encode effector proteins injected into plant cells by the system (three-letter suffixes often indicate the strain of origin for the effector) (5). Avr proteins are so named because in some potential hosts they betray the parasite to the R (resistance) gene surveillance system of plants, thereby triggering the HR (6).
P. syringae is divided into more than 40 pathovars based on pathogenic specificity for various plant species, and some pathovars are further divided into races on the basis of host range among differential cultivars of the host species. Although the basis for host range at the pathovar–plant species level has not been established, host range at the race-plant cultivar level is determined by combinations of Avr-R genes interacting in a gene-for-gene manner (6). That is, if the interactants contain corresponding Avr and R genes, then HR-associated defenses will be triggered. The R gene-encoded surveillance system, which appears to be arrayed primarily against the antihost proteins of parasites, is a key determinant of defense against highly adapted “stealth” pathogens like P. syringae. Similar gene-for-gene pathosystems involving multiple races and cultivars occur with many pathogenic fungi, nematodes, and viruses. The HR is similarly triggered in “incompatible” interactions with many of these parasites, and it is noteworthy that the HR is typically triggered in plants only by potential pathogens, not by encounters with the far more numerous nonpathogenic microbes in the environment.
Our research has focused on three strains of P. syringae: (i) P. syringae pv syringae (Psy) 61 is a weak pathogen of bean and is the source of the hrp/hrc gene cluster cloned on cosmid pHIR11 that contains all of the genes necessary for nonpathogenic bacteria like Pseudomonas fluorescens and Escherichia coli to elicit the HR in tobacco (7). (ii) Psy B728a is closely related to strain 61 but is a highly virulent model for studying epiphytic fitness and pathogenicity (brown spot of bean) in the field (8, 9). (iii) P. syringae pv. tomato (Pto) DC3000 is a well-studied pathogen of tomato and Arabidopsis (bacterial speck) that is taxonomically quite divergent from pathovar syringae (10), and it produces AvrPto, one of the best-studied Avr proteins (11). Thus, we can compare two closely related strains and one highly divergent strain in our investigation of the evolution and function of Hrp systems.
In the last decade, research on the evolution and function of type III secretion systems in Salmonella and Yersinia spp. has yielded two revolutionary insights. First, genes associated with pathogenicity, such as those encoding type III secretion systems, are often clustered in horizontally acquired pathogenicity islands (Pais) that may enable the evolution of virulence in “quantum leaps” (12, 13). Second, type III secretion systems have the remarkable ability to inject bacterial proteins into the cytoplasm of eukaryote host cells (14, 15). In this article, we will describe our progress in understanding how the P. syringae Hrp system expressed from pHIR11 enables a nonpathogen like E. coli to make a quantum leap in its ability to interact with plants by eliciting the HR, how hrp/hrc genes are arranged in Hrp Pais that also encode a variety of putative effectors, and how universal targeting signals and genetically dissectable secretion mechanisms underlie effector protein traffic through the pathway.
HopPsyA, pHIR11, and the Minimum Genetic Unit for Bacterial Elicitation of the Hypersensitive Response
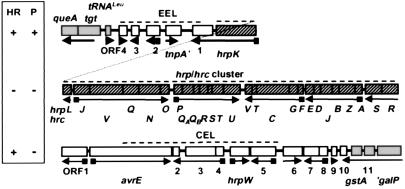
Cosmid pHIR11 was seminal in establishing the minimum genetic requirements and relative role of the Hrp system and effectors in HR elicitation. pHIR11 was cloned from Psy 61 on the basis of its ability to complement several hrp∷Tn5 mutations in that strain (7). It also enables P. fluorescens, P. putida, and E. coli (and probably many other Gram-negative bacteria) to elicit the HR in tobacco. However, pHIR11 does not enable nonpathogens to multiply or cause disease in any plants tested. For example, P. fluorescens (pHIR11) does not cause any symptoms in tobacco leaves unless inoculated at a very high level (≥5 × 106 cell/ml), such that enough individual plant cells undergo the HR to produce a confluent collapse. The DNA sequence of pHIR11 reveals a 25-kb cluster of hrp/hrc genes linked to an apparent operon encoding hopPsyA (hrmA) and ORF1 (16–22) (Fig. 1). The hrp/hrc clusters of Psy B728a and Pto DC3000 are arranged similarly (further discussed below), but HopPsyA is unique to Psy 61 (18, 23). Three proteins, the HrpZ and HrpW harpins and HrpA pilin, are secreted by the P. syringae Hrp pathway in culture more abundantly than other Hrp-dependent proteins (24–27). Harpins are glycine-rich cysteine-lacking proteins that possess heat-stable HR elicitor activity when infiltrated at relatively high concentration into the intercellular leaf spaces of many plants (5, 28). However, in P. syringae their HR-elicitation activity does not correlate with bacterial host range, and these proteins appear to have an ancillary role in plant interactions (21). HrpA forms a Hrp-specific pilus that is 6–8 nm in diameter and is essential for all Hrp phenotypes (26).
Figure 1.
The cluster of Psy 61 genes carried on pHIR11 that enables nonphytopathogenic bacteria to elicit the HR in tobacco. hopPsyA (checkered) encodes an effector protein that apparently is delivered into plant cells. Other genes encode regulatory factors (shaded), Hrc components associated with export across the inner membrane (diagonal hatching) or outer membrane (cross hatching), extracellular Hrp proteins (stippled), or proteins with unknown function (open boxes). Squares on arrows denote the presence of HrpL-activated promoters (55).
Through a series of observations, HopPsyA was identified as the HR-triggering effector that is injected into plant cells by the pHIR11 Hrp system, and it was simultaneously shown to have salient characteristics of known Avr proteins: (i) Mutations in hopPsyA abolish the ability of pHIR11 to direct HR elicitation without affecting HrpZ production or secretion, indicating that the essential role of HopPsyA is not as a component of the Hrp secretion system (29). (ii) HopPsyA travels the Hrp pathway, as demonstrated by its secretion in culture (discussed below) (30). (iii) HopPsyA has no apparent effect when delivered exogenously to tobacco cells but is lethal if expressed inside them via either biolistic- or Agrobacterium tumefaciens-mediated transient expression (23) (Fig. 2). (iv) Unlike tobacco (Nicotiana tabacum L. cv. Xanthi), Nicotiana benthamiana does not respond with the HR to either P. fluorescens (pHIR11) or hopPsyA transiently expressed inside plant cells after delivery by A. tumefaciens (Fig. 2). (v) When transformed into P. syringae pv tabaci, hopPsyA causes the tobacco wildfire pathogen to become avirulent in tobacco, as would be expected if tobacco possessed an R gene directing recognition of HopPsyA (23) (Fig. 2). (vi) A hopPsyA mutation in Psy 61 does not abolish virulence in bean or HR elicitation in tobacco, which is typical of known avr genes apparently because of their redundant contribution to parasitic fitness in hosts and HR elicitation in nonhost species (31). (vii) P. fluorescens and E. coli strains carrying pHIR11 fail to elicit the HR in soybean, Arabidopsis, and tomato, suggesting that those plants lack an R gene corresponding to HopPsyA; however, bacteria carrying pHIR11 do elicit an HR in those plants if transformed with avr genes that they recognize (32, 33) (Fig. 3). (viii) For AvrB and AvrPto, two of the Avr proteins demonstrated to work with pHIR11 in triggering an R gene-dependent HR, there are multiple lines of evidence that recognition by the R gene system occurs inside plant cells (33–36) (Fig. 3). Thus, pHIR11 directs heterologous HR elicitation in tobacco because it happens to encode a complete Hrp system plus an effector recognized by tobacco.
Figure 2.
Summary of evidence that HopPsyA functions like an avirulence protein that interacts inside plant cells with the product of an R gene present in N. tabacum but not N. benthamiana. The upper squares, labeled “pHIR11,” indicate the responses in leaves of N. tabacum (N.t.) and N. benthamiana (N.b.) to P. fluorescens 55 carrying pHIR11 (+) or a hopPsyA∷TnphoA derivative (−) after infiltration at a concentration of 5 × 107 cells/ml. “HR” indicates rapid confluent collapse of infiltrated tissue; “Null” indicates no visible response. The next two photographs, labeled “Agrobacterium,” show the effect in N.t. and N.b. of A. tumefaciens GV3101-mediated transient expression of hopPsyA via glucocorticoid-inducible expression vector pTA7002 (85). Plants receiving pTA7002 (−) or pTA7002∷hopPsyA (+) were sprayed with the glucocorticoid dexamethasone 48 h after infiltration and then photographed 24 h later. Note that the confluent tissue collapse indicative of the HR is observed only when hopPsyA is expressed in the N.t. leaf. The lower two photographs, labeled “P. s. tabaci,” show the effect in N.t. leaves of P. syringae pv tabaci 11528 carrying empty vector pDSK519 (−) or pCPP2349 (hopPsyA+) (+) at 1 and 5 days after inoculation (23). The level of inoculum was 5 × 108 in the lowest sector on each side of each leaf and 5 × 106 cell/ml in the next sectors up. Note that the HR developed by the end of day 1 in the sector infiltrated with 5 × 108 cells/ml of P. syringae pv tabaci (pCPP2349), whereas disease symptoms caused by P. syringae pv tabaci(pDSK519) developed later, with a lower level of inoculum, and were uniquely marked with the bright yellow chlorosis characteristic of wildfire. [Reproduced with permission from ref. 23 (Copyright 1997, Mol. Plant–Microbe Interact.).]
Figure 3.
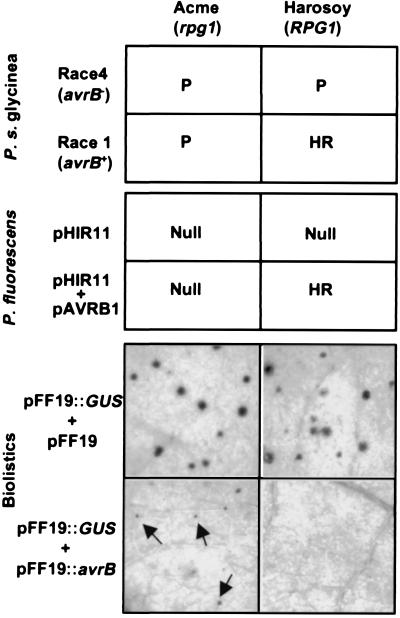
Summary of evidence that AvrB elicits an Rpg1-dependent HR whether delivered by P. syringae pv glycinea, P. fluorescens (pHIR11), or biolistic transformation. Top indicates the responses of soybean cultivars Acme and Harosoy to P. syringae pv glycinea and P. fluorescens (pHIR11) strains with or without avrB (33, 86). “P” indicates pathogenicity (bacterial blight symptoms); “HR” indicates rapid confluent collapse with an inoculum level of 5 × 107 cells/ml and no disease development at any inoculum level; “Null” indicates no visible response. Bottom shows the effects on β-glucuronidase (GUS) activity of transient coexpression of avrB in leaf cells of Acme and Harosoy. The leaves were biolistically cobombarded with tungsten particles coated with the indicated plasmids, incubated for 24 h, and then histochemically stained for GUS activity (23), which is an indicator of the viability of the transformed cells (87). Note that the histochemically stained spots are much smaller in the Acme leaves expressing avrB and completely absent in Harosoy leaves expressing avrB.
The minimum genetic requirements for being a bacterial parasite of plants are unknown. Parasitism apparently requires the delivery of multiple effector proteins that suppress general antibacterial defenses and/or promote nutrient release from plant cells. The number of effector proteins secreted by a given strain and the virulence targets of those proteins are unknown. More than 30 avr/hop genes from various P. syringae and Xanthomonas strains have been cloned and sequenced (37). Members of the AvrRxv/AvrBsT family are unique in being similar to animal pathogen effectors—the Yersinia YopJ/Salmonella AvrA family (38). The ability of YopJ to inhibit MAP kinase kinases suggests a potential role of the AvrRxv proteins in suppressing plant defenses (39). The Xanthomonas AvrBs2 is unique in having a sequence that predicts an enzymatic activity, and the similarity of AvrBs2 to A. tumefaciens agrocinopine synthase suggests a role in bacterial nutrition (40). Finally, the P. syringae AvrD protein family is unique in directing the synthesis of syringolide elicitors of an Rpg4-specific HR in soybean (41). Other Avr proteins offer little clue to their function as effectors, although it is noteworthy that many make a quantitative contribution to virulence (37), and they can be deleterious when overexpressed in plant cells lacking cognate R genes (33, 42) (Fig. 3).
Functions of Hrp System Components
P. syringae hrp genes were initially characterized on the basis of plant reaction phenotypes: typical mutants no longer elicited the HR in nonhosts or were pathogenic (or parasitic) in hosts (43, 44). Subsequently, levels of hrp expression and the secretion of HrpZ and Avr/Hop effectors provided phenotypes for dissecting the functions of Hrp system components. Table 1 summarizes the phenotypes of representative Hrp system mutants and indicates the following genetically distinguishable functions (Fig. 1): (i) positive and negative regulation of the Hrp regulon (hrp/hrc genes and known avr/hop genes); (ii) export of harpins and effectors across the inner membrane via a translocator apparently evolved from the flagellar biogenesis system; (iii) export of harpins and effectors across the outer membrane through a channel formed by secretin multimers; (iv) translocation of effectors across the plant cell wall and plasma membrane into the host cytoplasm by an unknown system.
Table 1.
Phenotypes resulting from mutating or overexpressing representative hrp/hrc genes or operons in P. syringae
| Gene/operon* | Mutant
phenotype
|
Constitutive expression phenotype | Comment and references | |||
|---|---|---|---|---|---|---|
| HR | Pathogenicity | HrpZ synthesis | HrpZ localization | |||
| hrpL | − | − | − | NA | Hrp regulon constitutively expressed | ECF family alternate sigma factor (31, 75, 79, 80) |
| hrpJ and hrpU operons | − | Virtually abolished† | + | Cytoplasmic only | Inner membrane translocator components (9, 17, 19, 20, 81) | |
| hrpV | + | + | + | Wild-type extracellular levels | Hrp regulon repressed in culture; but bacteria still Hrp+ | Negative regulator (22, 47) |
| hrcC | − | Virtually abolished† | + | Some periplasmic; none extracellular | Outer membrane “secretin” (9, 16, 22, 24, 81, 82) | |
| hrcJ | − | − | + | None extracellular | Associated with both inner and outer membranes (20, 82) | |
| hrpZ | + | + | − | NA | All HrpZ secretion blocked | Harpin (21, 24, 29) |
| hrpA | − | − | − | None extracellular | Hrp pilin (26, 46) | |
| hrpR-S | − | − | − | NA | Hrp regulon constitutively expressed | NtrC-family positive activators (75, 80, 83, 84) |
Genes and operons are presented in the order that they are arranged in the Hrp Pai (Fig. 1); mutations ablating operons resulted from insertion of interposons in hrpJ and hrpP.
Mutants deficient in the hrpJ operon and hrcC in Psy 61, Pto DC3000, and Psy B728a are nonpathogenic in standard assays on host plants; paradoxically Psy B728a mutants can still cause brown spot of bean in the field at a low frequency (≈5% of the wild type) when seeds are dipped in mutant inoculum before planting.
Recent observations highlight the complexity of Hrp system functions. Regulation involves not only the positive regulators HrpR, HrpS, and HrpL (45), but also HrpA and HrpV: multiple hrp genes are activated by the HrpA pilin and repressed by HrpV. Psy 61 and Pto DC3000 strains with nonpolar deletions of hrpA no longer express hrp genes in culture, and they have a Hrp− phenotype in plant bioassays (46). In contrast, constitutive expression of hrpV in Psy 61 represses the production of multiple Hrp components in culture, although it does not abolish HR elicitation, which suggests significant differences between Hrp regulation in culture and in planta (47). The repressive effects on hrp gene expression of deleting hrpA or overexpressing hrpV can be overcome by constitutive expression of hrpL or hrpRS, which suggests that HrpA and HrpV act upstream of the HrpRS-HrpL activation cascade (although effects on hrpRS expression have not yet been tested) (46, 47). The ability of constitutively produced HrpRS to restore the expression of the Hrp regulon in a Pto DC3000 hrpA mutant enabled testing of the role of HrpA in the secretion of the HrpW harpin and AvrPto (46). Surprisingly, HrpA is required for both of these proteins to be secreted in culture, which complicates genetic dissection of the role of HrpA in the translocation of effector proteins into plant cells. Furthermore, no mutations in P. syringae have identified factors specifically involved in the translocation step, as would be indicated by a block in effector protein translocation into plant cells (detectable by an R gene-dependent HR) without a block in secretion in culture. Mutants of this class have been extensively explored in Yersinia (yopB and yopD) (48).
Finally, it is noteworthy that Psy B728a hrpJ∷ΩSpR and hrcC∷nptII (nonpolar) mutants are strongly reduced in their ability to colonize bean leaves grown in the field from surface-inoculated seeds (9). The ability of Psy to achieve threshold population levels as an epiphyte on the surface of bean leaves has been shown to be important in pathogenesis in the field (8). Interestingly, B728a hrpJ∷ΩSpR and hrcC∷nptII mutants achieve high population levels on occasional leaves, and at a similarly low frequency they cause brown spot symptoms (9). This suggests that the Psy B728a Hrp system has a larger role in growth in planta than virulence per se, which is consistent with the finding of gacS (lemA) mutants of Psy B728a that do not produce disease lesions even though they grow to wild-type levels in bean and produce the HR in nonhost tobacco (49).
The Tripartite Mosaic Structure of the P. syringae Hrp Pai
To further characterize the Hrp system genes and any candidate effector genes linked to them, we have investigated the sequence of the hrp/hrc gene clusters and flanking regions of Psy B728a and Pto DC3000 (50). The hrp/hrc cluster resides at the center of a Hrp Pai with three distinct loci that make different contributions to pathogenicity. The hrp/hrc genes of the divergent strains 61 and DC3000 are similar in arrangement, although the hrpA genes are notably different (28% amino acid identity). In contrast, the hrpA genes of strains 61 and B728a are 100% identical. However, B728a is distinguished by a 3.6-kb insert containing homologs of bacteriophage λ genes Ea59 and Ea31 (50). The entire hrp/hrc cluster (hrpK-hrpR) was deleted from Pto DC3000 by a marker-exchange strategy using PCR-amplified DNA from the regions bordering hrpK and hrpR (Fig. 4). As expected, the mutant failed to grow significantly or to cause bacterial speck disease in tomato (Lycopersicon esculentum Mill. cv. Moneymaker) and Arabidopsis (Col-0), and it did not elicit the HR in tobacco (D.E.F. and A.C., unpublished data).
Figure 4.
The Hrp Pai of Pto DC3000 and the phenotype of large deletions affecting each of the three major regions. The shaded boxes denote genes considered to be outside of the Hrp Pai. Squares on arrows denote the presence of Hrp boxes. hrpK is presented on the same line as the EEL because ORF1 is in the same apparent operon. Open boxes denote genes in the EEL and CEL. The structure of the hrpJ and hrpU operons is based on partial sequence data and colinearity with this region in Psy 61. Dashed lines indicate the regions deleted, and the Inset (Left) shows the effect of each deletion on the ability of DC3000 to elicit the HR in tobacco or cause disease symptoms in tomato.
Three nucleotides downstream of the hrpK stop codon, the DNA sequence of Psy 61, Psy B728a, and Pto DC3000 is completely divergent (50). This divergent region, the exchangeable effector locus (EEL) further described below, has a significantly lower G + C content than the rest of the Hrp Pai and the P. syringae genome. The EELs have variable lengths of 2.5 kb (Psy 61), 7.3 kb (Psy B728a), and 5.9 kb (Pto DC3000), and they are bounded by hrpK and tRNALeu -queA-tgt sequences. The latter are also found in Pseudomonas aeruginosa but without linkage to any Hrp Pai genes. On the other side of the hrp/hrc cluster, beyond hrpR, resides a conserved effector locus (CEL) of ≈17-kb (further discussed below). Comparison of the CEL regions sequenced in the divergent strains B728a and DC3000 revealed that the first seven ORFs are arranged identically and have an average DNA sequence identity of 78% and a G + C content that is similar to that of the hrp/hrc region and the rest of the P. syringae genome. Overall, the Hrp Pai of P. syringae has key properties of Pais possessed by animal pathogens (13), including: (i) the presence of many virulence-associated genes (several with relatively low G + C content) in a large (≈50-kb) chromosomal region, (ii) linkage to the 3′ end of a tRNA gene, (iii) absence from the corresponding locus in a closely related species, and (iv) instability and possession of many sequences related to mobile genetic elements (specifically in the EEL, discussed below).
An EEL Makes a Small Contribution to Parasitic Fitness
The Psy B728a EEL possesses three ORFs predicting products similar to known Avr proteins: P. syringae pv phaseolicola AvrPphC and P. syringae pv glycinea AvrC (ORF1); P. syringae pv phaseolicola AvrPphE (ORF2); and Xanthomonas AvrBsT and AvrRxv (ORF5) (50). avrPphE illustrates the instability of the EEL region in being absent from the EELs of Psy 61 and Pto DC3000 and present in P. syringae pv phaseolicola 1302A but in a different location, immediately downstream of hrpK (hrpY) in that strain (51). Although Psy 61 and B728a are in the same pathovar, the strain 61 EEL is completely different and carries only hopPsyA and ORF1, which are present in only a few Psy strains (18, 23). The ORFs in the Pto DC3000 EEL predict no products with similarity to known Avr proteins; however, the ORF1 protein is secreted in a hrp-dependent manner by E. coli (pCPP2156), which expresses an Erwinia chrysanthemi Hrp system and secretes P. syringae Avr proteins (52) (J.R.A. and K.v.D., unpublished data). Several ORFs in these EELs are preceded by Hrp boxes indicative of HrpL-activated promoters (53–55).
The EELs of these three strains also contain sequences homologous to various mobile genetic elements (50). The Psy B728a EEL carries sequences similar to those in a P. syringae pv phaseolicola plasmid that harbors several avr genes (56) and to sequences homologous to insertion elements that are typically found on plasmids, which suggests plasmid integration via an insertion sequence element in this region (57). Psy B728a ORF3 and ORF4 show similarity to sequences implicated in the horizontal acquisition of the LEE Pai by pathogenic E. coli strains, and the Pto DC3000 EEL carries a TnpA′ fragment similar to Pseudomonas stutzeri TnpA1 (50). These ORFs are not preceded by Hrp boxes and are unlikely to encode effector proteins.
Cosmid pCPP2346, which carries the B728a hrp/hrc region and flanking sequences (4 kb on the left and 13 kb on the right), enabled P. fluorescens to secrete the B728a HrpZ harpin in culture and to elicit the HR in tobacco leaves. However, confluent necrosis developed more slowly than with P. fluorescens (pHIR11). These observations suggested that the product of at least one of the effector genes in the B728a EEL was recognized by an R gene in tobacco. In agreement with this hypothesis, a derivative of plasmid pCPP46 carrying the B728a EEL renders P. syringae pv tabaci avirulent in tobacco (W.-L.D. and A.C., unpublished data).
The contribution of the various EELs to the parasitic fitness of P. syringae strains was assayed with appropriate mutants (50). A hopPsyA∷TnphoA Psy 61 mutant had previously been shown to be only partially impaired in Hrp phenotypes (31). Deletions of the entire EEL regions of Pto DC3000 (50) and Psy B728a (W.-L.D, unpublished work) were constructed by marker exchange with appropriate border regions subcloned on either side of an ΩSpR cassette. The growth in host plants of mutant and wild-type strains was compared after inoculation by syringe infiltration. The Pto DC3000 ΔEEL mutant was slightly reduced in the final population it achieved in tomato (cv. Moneymaker) (50), but no significant reduction was observed with the Psy B728a ΔEEL mutant in bean (Phaseolus vulgaris L. cv. Eagle) (W.-L.D. and A.C., unpublished data). The mutants also retained the ability to elicit the HR on various nonhosts. Thus, additional effectors encoded elsewhere in the genome apparently contribute to parasitic fitness in hosts and betray the parasite to R gene surveillance in nonhosts.
The CEL Is Important for Pathogenicity
The region to the right of hrpR in DC3000 had been known for several years to contain the avrE locus, which is comprised of two transcriptional units and, when heterologously expressed, causes P. syringae pv glycinea to become avirulent on all soybean cultivars tested (58). Also known to be in this region is the hrpW gene encoding a second harpin, which is distinguished by its C-terminal domain with homology to class III pectate lyases and its ability to bind to calcium pectate (27). A previous sequence analysis of the 5′ sequences for the first four transcriptional units beyond hrpR (58) was extended to include the first 14 ORFs to the right of hrpR in Pto DC3000 and a partial sequence of the corresponding region in Psy B728a (50). Unlike the EEL, this region contains no sequences similar to known mobile genetic elements, and it appears conserved between Psy and Pto because the first seven ORFs are arranged identically in these divergent strains and have an average DNA sequence identity of 78%. In Fig. 4, the outer border of the CEL is given tentatively as around ORF10. ORF8 is preceded by a Hrp box and is therefore a candidate effector. In contrast, the gene beyond ORF10 shows homology to a family of bacterial GstA proteins (50). Because glutathione S-transferase activity is common in nonpathogenic fluorescent pseudomonads (59), this gene is not likely to be an effector or part of the CEL. The ORF5 protein is secreted in a hrp-dependent manner by E. coli (pCPP2156), but mutation with an ΩSpr cassette has little effect on either HR elicitation in tobacco or pathogenicity in tomato (A.O.C., J.L.B, and A.C., unpublished data). Notably, six operons in this region are preceded by a Hrp box, which is characteristic of known avr genes in P. syringae (53–55, 58) (Fig. 4).
To assess the collective contribution of the CEL ORFs that were both partially characterized and likely to encode effectors, we constructed a mutation in Pto DC3000 that replaced avrE through ORF5 with an ΩSpr cassette (50). The ΔCEL mutant still elicited the HR in tobacco, but tissue collapse was delayed ≈5 h. The mutant no longer elicited disease symptoms in tomato when infiltrated at a concentration of 104 cfu/ml, and growth in planta was strongly reduced (50). Pathogenicity was restored to the ΔCEL mutant by a plasmid carrying ORF2 through ORF10, and the mutant was able to secrete AvrPto in culture. All of these observations suggest that the ΔCEL mutation does not interfere with Hrp secretion functions and that the loss of pathogenicity can be attributed to the loss of multiple effectors. Finally, although avrE and several other candidate effector genes are located in the Hrp Pai of Pto DC3000, additional effector genes, such as avrPto, are located somewhere else (60), and the complete inventory of effector genes in this strain remains unknown. Because most of the known P. syringae avr genes are associated with mobile genetic elements (61), the avr composition of various P. syringae strains may vary considerably, presumably as a result of opposing selection pressures to promote parasitism while evading host R gene surveillance.
HrpK and CEL ORF1
The hrpL and hrpR genes bracket a cluster of operons that contain both hrp and hrc genes and appear sufficient to encode a complete Hrp type III secretion system. hrpK and the CEL ORF1 reside in the two borders between this core hrp/hrc cluster and known effector genes, and the functions of these two genes are unknown. The HrpK proteins of Psy and Pto are 79% identical, which makes them more conserved than several of the proteins encoded by the core hrp/hrc cluster (50). hrpK mutants have a variable Hrp phenotype (51, 62), and a Psy B728a hrpK mutant still elicits the HR in tobacco and secretes HrpZ in culture (Fig. 5). These observations suggest that HrpK is a conserved effector rather than a component of the Hrp system. It is noteworthy that candidate effector genes appear to reside downstream of hrpK in the same operon in Psy B728a and Pto DC3000 (50). In contrast to HrpK, the CEL ORF1 is more likely to be an ancillary component of the Hrp system than an effector, because it is most similar to E. coli murein lytic transglycosylase MltD and shares a lysozyme-like domain with the product of ipgF (63), which is a Shigella flexneri gene linked to type III secretion system genes (64). Although mutations in these genes in Pto DC3000 and S. flexneri have no obvious phenotype (58, 64), other peptidoglycan hydrolases may mask the phenotype (65). The region to the right of hrpR in pHIR11 has not been sequenced and may harbor ORF1. However, TnPhoA mutations in this region have no apparent phenotype (31).
Figure 5.
Secretion of the HrpZ harpin by Psy B728a hrpK mutant CUCPB5092. Bacteria were grown under Hrp-inducing conditions and fractionated into cell-bound (C) and supernatant (S) fractions as previously described (30). Proteins were resolved by SDS/PAGE and analyzed by immunoblotting with anti-HrpZ antibodies.
Effector Protein Secretion and a Universal Type III Targeting Signal
The ability of pHIR11 to deliver the products of avr genes from other P. syringae pathovars suggested that the Hrp system recognizes a universal targeting signal in these proteins. Indeed, the cluster of hrp/hrc genes from the soft-rot pathogen E. chrysanthemi, cloned in cosmid pCPP2156, enables E. coli to secrete AvrB and AvrPto (52), and Erwinia amylovora and P. syringae can interchangeably deliver their respective DspE and AvrE proteins to plants, as indicated by appropriate plant reactions (66). Moreover, Xanthomonas campestris pv vesicatoria can secrete AvrB (67), Yersinia enterocolitica can secrete both AvrB and AvrPto (68), and X. campestris pv vesicatoria and E. coli (pCPP2156) can secrete YopE, a Yersinia effector (67, 68). These observations extend the original discovery of heterologous delivery of effectors by the type III secretion systems of Yersinia, Salmonella, and Shigella, and they strongly suggest the existence of universal targeting signals in proteins traveling all type III secretion pathways (69).
Yersinia secretes multiple Yops (Yersinia outer proteins) via the type III secretion pathway, and Yops carry an mRNA targeting signal in their first 15 codons (70, 71). Fusion of the first 15 codons of YopE to an Npt reporter is sufficient for type III secretion of the hybrid to the bacterial milieu, and mutations that shift the reading frame of these codons do not abolish secretion, which indicates that the targeting information resides in the mRNA rather than the peptide (71). Several Yops, including YopE, have a second targeting domain, which depends on a customized chaperone and is required for translocation into host cells (72). However, YopQ has only an mRNA targeting signal (73), and it is also secreted by E. coli (pCPP2156). Several observations support the hypothesis that the first 15 codons of avrPto similarly carry an mRNA targeting signal: (i) deletion of the first 10 codons abolishes the secretion of AvrPto by E. coli (pCPP2156) and Y. enterocolitica; (ii) fusion of the first 15 codons of AvrPto to an Npt reporter is sufficient for type III secretion of the hybrid to the Y. enterocolitica milieu; and (iii) mutations that shift the reading frame of the AvrPto codons (+1, +2, and −1) do not abolish secretion of the Npt reporter (68). Thus, the mRNA signal recognized by type III secretion systems appears to be shared by the effectors of both plant and animal pathogens.
The efficiency with which different P. syringae Avr proteins are secreted in culture by different type III systems varies considerably (as indicated by the proportion of total effector protein released to the medium), and secretion by native P. syringae Hrp systems has been reported only recently (30, 74). AvrB and AvrPto secretion illustrates this variability. Both proteins are secreted much more strongly by Y. enterocolitica than by E. coli (pCPP2156) (52, 68), and AvrPto is the only one of these two that is secreted by P. syringae (30). Heterologously expressed AvrRpt2 is similarly secreted by E. coli (pCPP2156) and Pto DC3000 (74), but much less efficiently than AvrPto is secreted by P. syringae (30). These differences in secretion behavior in culture bear no apparent relationship to the biological activity of the effectors. For example, although AvrB secretion from P. syringae has yet to be observed in culture, AvrB is almost certainly delivered into plant cells (33, 34). Thus, it remains unclear whether the differing secretion behaviors of AvrB and AvrPto reflect some form of effector sorting by the pathway or whether it is peculiar to secretion in culture.
In Yersinia, the type III pathway can be activated by growth at 37°C in low-calcium medium, and these conditions obviate the normal requirement for host cell contact (1). In P. syringae, the hrp/hrc genes are induced in minimal media that do not support rapid growth (75), and two environmental factors relevant to pathogenesis have been found to be critical for the secretion of HopPsyA and AvrPto by Psy 61 and Pto DC3000, respectively (30). That is, both proteins are secreted at pH 6.0 and 20°C but not at pH 7.0 or 30°C. These conditions correspond to the low pH of plant intercellular fluids and the cool temperatures that favor disease development with these bacteria (76). However, the secretion capacity of the type III systems of plant pathogens in culture, even under optimal conditions, seems much less than that of Yersinia, regardless of the effector protein.
One explanation is that the Hrp systems are not fully activated until contact with plant cells, and the appropriate mutants or signals needed to unlock that capacity have not been found. Alternatively, the secretion capacity of the Hrp system may be reduced by adaptations for delivery through the plant cell wall matrix. In support of the first hypothesis, the Hrp regulon in Ralstonia solanacearum is induced maximally in culture by cocultivation with plant cells (77, 78). We have similarly observed that a PhrpA-uidA fusion is induced ≈20-fold when suspension-cultured tobacco cells are added to Hrp-inducing minimal medium (W.-L.D. and A.C., unpublished data), which suggests that contact-dependent induction may be widespread with the Hrp systems of plant pathogenic bacteria.
The differing abilities of type III systems to translocate effector-reporter hybrid proteins provides support for the second hypothesis. Early evidence for the translocation of Yop proteins into host cells was obtained with a YopE-CyA hybrid that produced adenylate cyclase activity in a calmodulin-dependent manner (15). This reporter system is a powerful tool for investigating translocated proteins and their targeting signals. Unfortunately, translocation into host cells of effector-reporter hybrids has not been described for any plant pathogens, and fusion of the C terminus of AvrRpt2 with Myc6, Gfp, or CyA blocks avrRpt2-Rps2-dependent HR elicitation and diminishes the virulence of Pto DC3000 in Arabidopsis plants lacking the cognate Rps2 gene (74). It seems that the fusion of large polypeptides to the C terminus of Avr proteins disrupts Hrp functions. Thus, it appears that effector proteins can be targeted to the type III pathway by a universal mRNA targeting signal and secreted across inner and outer membranes by machinery that is common to all type III systems. However, translocation into host cells is likely to be unique because of adaptations to the fundamentally different surfaces of plant and animal cells.
Conclusions
Our investigation of the basis for P. syringae phytopathogenicity has focused on the mechanisms underlying elicitation of the HR, a signature of plant encounters with incompatible phytopathogens, and it has revealed the modular nature of the process and its underlying genetics. Thus, the requirements for HR elicitation can be reduced to two components: a functional Hrp type III secretion system and an injected effector protein that is recognized by the R-gene surveillance system of the test plant. Hrp protein secretion in culture can be further dissected genetically, revealing two operons directing export across the inner membrane and another directing export across the outer membrane. The effector proteins also appear modular in their possession of a universal type III targeting signal in the 5′ ends of their cognate mRNAs. This modularity has several experimental consequences: a cloned P. syringae Hrp system is sufficient to direct heterologous secretion and delivery of effector proteins by nonphytopathogenic bacteria; effector proteins from P. syringae can be heterologously delivered into plants by Erwinia Hrp systems or secreted in culture by the Yersinia type III system; and the need for any Hrp system for HR elicitation can be circumvented entirely by delivery of effector protein genes into plant cells by biolistics or Agrobacterium-mediated transformation. The modular nature of the Hrp/effector system is also seen in the tripartite mosaic architecture of the P. syringae Hrp Pai, which features both exchangeable and conserved effector loci. The EEL represents a region in flux because of its high frequency of recombination, and this probably allows fine tuning of pathogenicity. On the basis of its similar G + C content to the hrp/hrc cluster and the rest of the P. syringae chromosome, the CEL was probably acquired at the same time as the core hrp/hrc cluster, and it encodes effectors that contribute more significantly to pathogenicity than the EEL. The modular nature of the Hrp/effector system suggests that it functions universally in a broad range of potential plant hosts and with a frequently changing pool of effectors. Effector gene instability may be driven by the evolution of R gene surveillance systems and changes in effector targets in plants. The next challenge is to identify all of the effector proteins produced by model strains of P. syringae, to understand how these proteins promote parasitism, and to understand how the type III system of phytopathogens has been adapted to deliver these proteins across plant cell walls.
Acknowledgments
We thank Nam-Hai Chua (The Rockefeller University, New York, NY) for providing pTA7002. This work was supported by National Science Foundation grant MCB 97–35303-4488 (A.C.), National Research Initiative Competitive Grants Program United States Department of Agriculture grants 98–35303-6464 (J.R.A.) and 98–35303-6662 (D.E.F.), and United States Public Health Service Grant AI 42797 from the National Institutes of Health—National Institute of Allergy and Infectious Diseases Branch (O.S.).
Abbreviations
- Psy
Pseudomonas syringae pv syringae
- Pto
P.s. tomato
- HR
hypersensitive response
- hrp
HR and pathogenicity
- hrc
HR and conserved
- Pai
pathogenicity island
- EEL
exchangeable effector locus
- CEL
conserved effector locus
- Hop
Hrp-dependent outer protein
- Avr
avirulence
Footnotes
This paper was presented at the National Academy of Sciences colloquium “Virulence and Defense in Host–Pathogen Interactions: Common Features Between Plants and Animals,” held December 9–11, 1999, at the Arnold and Mabel Beckman Center in Irvine, CA.
References
- 1.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galán J E, Collmer A. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 3.Hirano S S, Upper C D. Annu Rev Phytopathol. 1990;28:155–177. [Google Scholar]
- 4.Richael C, Gilchrist D. Physiol Mol Plant Pathol. 1999;55:5–12. [Google Scholar]
- 5.Alfano J R, Collmer A. J Bacteriol. 1997;179:5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keen N T. Annu Rev Genet. 1990;24:447–463. doi: 10.1146/annurev.ge.24.120190.002311. [DOI] [PubMed] [Google Scholar]
- 7.Huang H-C, Schuurink R, Denny T P, Atkinson M M, Baker C J, Yucel I, Hutcheson S W, Collmer A. J Bacteriol. 1988;170:4748–4756. doi: 10.1128/jb.170.10.4748-4756.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirano S S, Rouse D I, Clayton M K, Upper C D. Plant Dis. 1995;79:1085–1093. [Google Scholar]
- 9.Hirano S, Charkowski A O, Collmer A, Willis D K, Upper C D. Proc Natl Acad Sci USA. 1999;96:9851–9856. doi: 10.1073/pnas.96.17.9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manceau C, Horvais A. Appl Environ Microbiol. 1997;63:498–505. doi: 10.1128/aem.63.2.498-505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin G B. Curr Opin Plant Biol. 1999;2:273–279. doi: 10.1016/S1369-5266(99)80049-1. [DOI] [PubMed] [Google Scholar]
- 12.Groisman E A, Ochman H. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 13.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 14.Rosqvist R, Magnusson K E, Wolf-Watz H. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sory M-P, Cornelis G R. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 16.Huang H-C, He S Y, Bauer D W, Collmer A. J Bacteriol. 1992;174:6878–6885. doi: 10.1128/jb.174.21.6878-6885.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang H-C, Xiao Y, Lin R-H, Lu Y, Hutcheson S W, Collmer A. Mol Plant-Microbe Interact. 1993;6:515–520. doi: 10.1094/mpmi-6-515. [DOI] [PubMed] [Google Scholar]
- 18.Heu S, Hutcheson S W. Mol Plant–Microbe Interact. 1993;6:553–564. doi: 10.1094/mpmi-6-553. [DOI] [PubMed] [Google Scholar]
- 19.Lidell M C, Hutcheson S W. Mol Plant–Microbe Interact. 1994;7:488–497. doi: 10.1094/mpmi-7-0488. [DOI] [PubMed] [Google Scholar]
- 20.Huang H-C, Lin R-W, Chang C-J, Collmer A, Deng W-L. Mol Plant-Microbe Interact. 1995;8:733–746. doi: 10.1094/mpmi-8-0733. [DOI] [PubMed] [Google Scholar]
- 21.Preston G, Huang H-C, He S Y, Collmer A. Mol Plant-Microbe Interact. 1995;8:717–732. doi: 10.1094/mpmi-8-0717. [DOI] [PubMed] [Google Scholar]
- 22.Deng W-L, Preston G, Collmer A, Chang C-J, Huang H-C. J Bacteriol. 1998;180:4523–4531. doi: 10.1128/jb.180.17.4523-4531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alfano J R, Kim H-S, Delaney T P, Collmer A. Mol Plant–Microbe Interact. 1997;10:580–588. doi: 10.1094/MPMI.1997.10.5.580. [DOI] [PubMed] [Google Scholar]
- 24.He S Y, Huang H-C, Collmer A. Cell. 1993;73:1255–1266. doi: 10.1016/0092-8674(93)90354-s. [DOI] [PubMed] [Google Scholar]
- 25.Yuan J, He S Y. J Bacteriol. 1996;178:6399–6402. doi: 10.1128/jb.178.21.6399-6402.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roine E, Wei W, Yuan J, Nurmiaho-Lassila E-L, Kalkkinen N, Romantschuk M, He S Y. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charkowski A O, Alfano J R, Preston G, Yuan J, He S Y, Collmer A. J Bacteriol. 1998;180:5211–5217. doi: 10.1128/jb.180.19.5211-5217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei Z-M, Laby R J, Zumoff C H, Bauer D W, He S Y, Collmer A, Beer S V. Science. 1992;257:85–88. doi: 10.1126/science.1621099. [DOI] [PubMed] [Google Scholar]
- 29.Alfano J R, Bauer D W, Milos T M, Collmer A. Mol Microbiol. 1996;19:715–728. doi: 10.1046/j.1365-2958.1996.415946.x. [DOI] [PubMed] [Google Scholar]
- 30.van Dijk K, Fouts D E, Rehm A H, Hill A R, Collmer A, Alfano J R. J Bacteriol. 1999;181:4790–4797. doi: 10.1128/jb.181.16.4790-4797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang H-C, Hutcheson S W, Collmer A. Mol Plant–Microbe Interact. 1991;4:469–476. doi: 10.1094/mpmi-6-515. [DOI] [PubMed] [Google Scholar]
- 32.Pirhonen M U, Lidell M C, Rowley D L, Lee S W, Jin S, Liang Y, Silverstone S, Keen N T, Hutcheson S W. Mol Plant-Microbe Interact. 1996;9:252–260. doi: 10.1094/mpmi-9-0252. [DOI] [PubMed] [Google Scholar]
- 33.Gopalan S, Bauer D W, Alfano J R, Loniello A O, He S Y, Collmer A. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leister R T, Ausubel F M, Katagiri F. Proc Natl Acad Sci USA. 1996;93:15497–15502. doi: 10.1073/pnas.93.26.15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang X, Frederick R D, Zhou J, Halterman D A, Jia Y, Martin G B. Science. 1996;274:2060–2062. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 36.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 37.Leach J E, White F F. Annu Rev Phytopathol. 1996;34:153–179. doi: 10.1146/annurev.phyto.34.1.153. [DOI] [PubMed] [Google Scholar]
- 38.Ciesiolka L D, Hwin T, Gearlds J D, Minsavage G V, Saenz R, Bravo M, Handley V, Conover S M, Zhang H, Caporgno J, et al. Mol Plant–Microbe Interact. 1999;12:35–44. doi: 10.1094/MPMI.1999.12.1.35. [DOI] [PubMed] [Google Scholar]
- 39.Orth K, Palmer L E, Bao Z Q, Stewart S, Rudolph A E, Bliska J B, Dixon J E. Science. 1999;285:1920–1923. doi: 10.1126/science.285.5435.1920. [DOI] [PubMed] [Google Scholar]
- 40.Swords K M M, Dahlbeck D, Kearney B, Roy M, Staskawicz B J. J Bacteriol. 1996;178:4661–4669. doi: 10.1128/jb.178.15.4661-4669.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yucel I, Midland S L, Sims J J, Keen N T. Mol Plant–Microbe Interact. 1994;7:148–150. doi: 10.1094/mpmi-7-0148. [DOI] [PubMed] [Google Scholar]
- 42.McNellis T W, Mudgett M B, Li K, Aoyama T, Horvath D, Chua N-H, Staskawicz B J. Plant J. 1998;14:247–257. doi: 10.1046/j.1365-313x.1998.00106.x. [DOI] [PubMed] [Google Scholar]
- 43.Lindgren P B, Peet R C, Panopoulos N J. J Bacteriol. 1986;168:512–522. doi: 10.1128/jb.168.2.512-522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindgren P B, Panopoulos N J, Staskawicz B J, Dahlbeck D. Mol Gen Genet. 1988;211:499–506. [Google Scholar]
- 45.Hutcheson S W. In: Plant–Microbe Interactions, Vol. 3. Stacey G, Keen N T, editors. New York: Chapman & Hall; 1997. pp. 145–179. [Google Scholar]
- 46.Wei W, Plovanich-Jones A, Deng W-L, Collmer A, Huang H-C, He S Y. Proc Natl Acad Sci USA. 2000;97:2247–2252. doi: 10.1073/pnas.040570097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Preston G, Deng W-L, Huang H-C, Collmer A. J Bacteriol. 1998;180:4532–4537. doi: 10.1128/jb.180.17.4532-4537.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cornelis, G. R. & Van Gijsegem, F. (2000) Annu. Rev. Microbiol, in press. [DOI] [PubMed]
- 49.Willis D K, Hrabak E M, Rich J J, Barta T M, Lindow S E, Panopoulos N J. Mol Plant–Microbe Interact. 1990;3:149–156. [Google Scholar]
- 50.Alfano J R, Charkowski A O, Deng W-L, Badel J L, Petnicki-Ocwieja T, van Dijk K, Collmer A. Proc Natl Acad Sci USA. 2000;97:4856–4861. doi: 10.1073/pnas.97.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mansfield J, Jenner C, Hockenhull R, Bennett M A, Stewart R. Mol Plant–Microbe Interact. 1994;7:726–739. doi: 10.1094/mpmi-7-0726. [DOI] [PubMed] [Google Scholar]
- 52.Ham J H, Bauer D W, Fouts D E, Collmer A. Proc Natl Acad Sci USA. 1998;95:10206–10211. doi: 10.1073/pnas.95.17.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen H, Keen N T. J Bacteriol. 1993;175:5916–5924. doi: 10.1128/jb.175.18.5916-5924.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Innes R W, Bent A F, Kunkel B N, Bisgrove S R, Staskawicz B J. J Bacteriol. 1993;175:4859–4869. doi: 10.1128/jb.175.15.4859-4869.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao Y, Hutcheson S. J Bacteriol. 1994;176:3089–3091. doi: 10.1128/jb.176.10.3089-3091.1994. , and correction (1994) 176, 6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jackson R W, Athanassopoulos E, Tsiamis G, Mansfield J W, Sesma A, Arnold D L, Gibbon M J, Murillo J, Taylor J D, Vivian A. Proc Natl Acad Sci USA. 1999;96:10875–10880. doi: 10.1073/pnas.96.19.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szabo L J, Mills D. J Bacteriol. 1984;157:821–827. doi: 10.1128/jb.157.3.821-827.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lorang J M, Keen N T. Mol Plant–Microbe Interact. 1995;8:49–57. doi: 10.1094/mpmi-8-0049. [DOI] [PubMed] [Google Scholar]
- 59.Zablotowicz R M, Hoagland R E, Locke M A, Hickey W J. Appl Environ Microbiol. 1995;61:1054–1060. doi: 10.1128/aem.61.3.1054-1060.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salmeron J M, Staskawicz B J. Mol Gen Genet. 1993;239:6–10. doi: 10.1007/BF00281595. [DOI] [PubMed] [Google Scholar]
- 61.Kim J F, Charkowski A O, Alfano J R, Collmer A, Beer S V. Mol Plant–Microbe Interact. 1998;11:1247–1252. [Google Scholar]
- 62.Bozso Z, Ott P G, Kecskes M L, Klement Z. Physiol Mol Plant Pathol. 1999;55:215–223. [Google Scholar]
- 63.Mushegian A R, Fullner K J, Koonin E V, Nester E W. Proc Natl Acad Sci USA. 1996;93:7321–7326. doi: 10.1073/pnas.93.14.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allaoui A, Menard R, Sansonetti P J, Parsot P. Infect Immun. 1993;61:1707–1714. doi: 10.1128/iai.61.5.1707-1714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dijkstra A J, Keck W. J Bacteriol. 1996;178:5555–5562. doi: 10.1128/jb.178.19.5555-5562.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bogdanove A J, Kim J F, Wei Z, Kolchinsky P, Charkowski A O, Conlin A K, Collmer A, Beer S V. Proc Natl Acad Sci USA. 1998;95:1325–1330. doi: 10.1073/pnas.95.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rossier O, Wengelnik K, Hahn K, Bonas U. Proc Natl Acad Sci USA. 1999;96:9368–9373. doi: 10.1073/pnas.96.16.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson D M, Fouts D E, Collmer A, Schneewind O. Proc Natl Acad Sci USA. 1999;96:12839–12843. doi: 10.1073/pnas.96.22.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rosqvist R, Hakansson S, Forsberg A, Wolf-Watz H. EMBO J. 1995;14:4187–4195. doi: 10.1002/j.1460-2075.1995.tb00092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anderson D M, Schneewind O. Curr Opin Microbiol. 1999;2:18–24. doi: 10.1016/s1369-5274(99)80003-4. [DOI] [PubMed] [Google Scholar]
- 71.Anderson D M, Schneewind O. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 72.Wattiau P, Woestyn S, Cornelis G R. Mol Microbiol. 1996;20:255–262. doi: 10.1111/j.1365-2958.1996.tb02614.x. [DOI] [PubMed] [Google Scholar]
- 73.Anderson D M, Schneewind O. Mol Microbiol. 1999;31:1139–1148. doi: 10.1046/j.1365-2958.1999.01254.x. [DOI] [PubMed] [Google Scholar]
- 74.Mudgett M B, Staskawicz B J. Mol Microbiol. 1999;32:927–941. doi: 10.1046/j.1365-2958.1999.01403.x. [DOI] [PubMed] [Google Scholar]
- 75.Huynh T V, Dahlbeck D, Staskawicz B J. Science. 1989;245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 76.Rudolph K, Burr T J, Mansfield J W, Stead D, Vivian A, von Kietzell J. Pseudomonas syringae Pathovars and Related Pathogens. Dordrecht, The Netherlands: Kluwer; 1997. p. 663. [Google Scholar]
- 77.Marenda M, Brito B, Callard D, Genin S, Barberis P, Boucher C, Arlat M. Mol Microbiol. 1998;27:437–453. doi: 10.1046/j.1365-2958.1998.00692.x. [DOI] [PubMed] [Google Scholar]
- 78.Brito B, Marenda M, Barberis P, Boucher C, Genin S. Mol Microbiol. 1999;31:237–251. doi: 10.1046/j.1365-2958.1999.01165.x. [DOI] [PubMed] [Google Scholar]
- 79.Xiao Y, Lu Y, Heu S, Hutcheson S W. J Bacteriol. 1992;174:1734–1741. doi: 10.1128/jb.174.6.1734-1741.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiao Y, Heu S, Yi J, Lu Y, Hutcheson S W. J Bacteriol. 1994;176:1025–1036. doi: 10.1128/jb.176.4.1025-1036.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Charkowski A O, Huang H-C, Collmer A. J Bacteriol. 1997;179:3866–3874. doi: 10.1128/jb.179.12.3866-3874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deng W-L, Huang H-C. J Bacteriol. 1998;181:2298–2301. doi: 10.1128/jb.181.7.2298-2301.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grimm C, Panopoulos N J. J Bacteriol. 1989;171:5031–5038. doi: 10.1128/jb.171.9.5031-5038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rahme L G, Mindrinos M N, Panopoulos N J. J Bacteriol. 1991;173:575–586. doi: 10.1128/jb.173.2.575-586.1991. , erratum (1992) 174, 3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aoyama T, Chua N-H. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- 86.Staskawicz B, Dahlbeck D, Keen N, Napoli C. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mindrinos M, Katagiri F, Yu G-L, Ausubel F M. Cell. 1994;78:1089–1099. doi: 10.1016/0092-8674(94)90282-8. [DOI] [PubMed] [Google Scholar]