Abstract
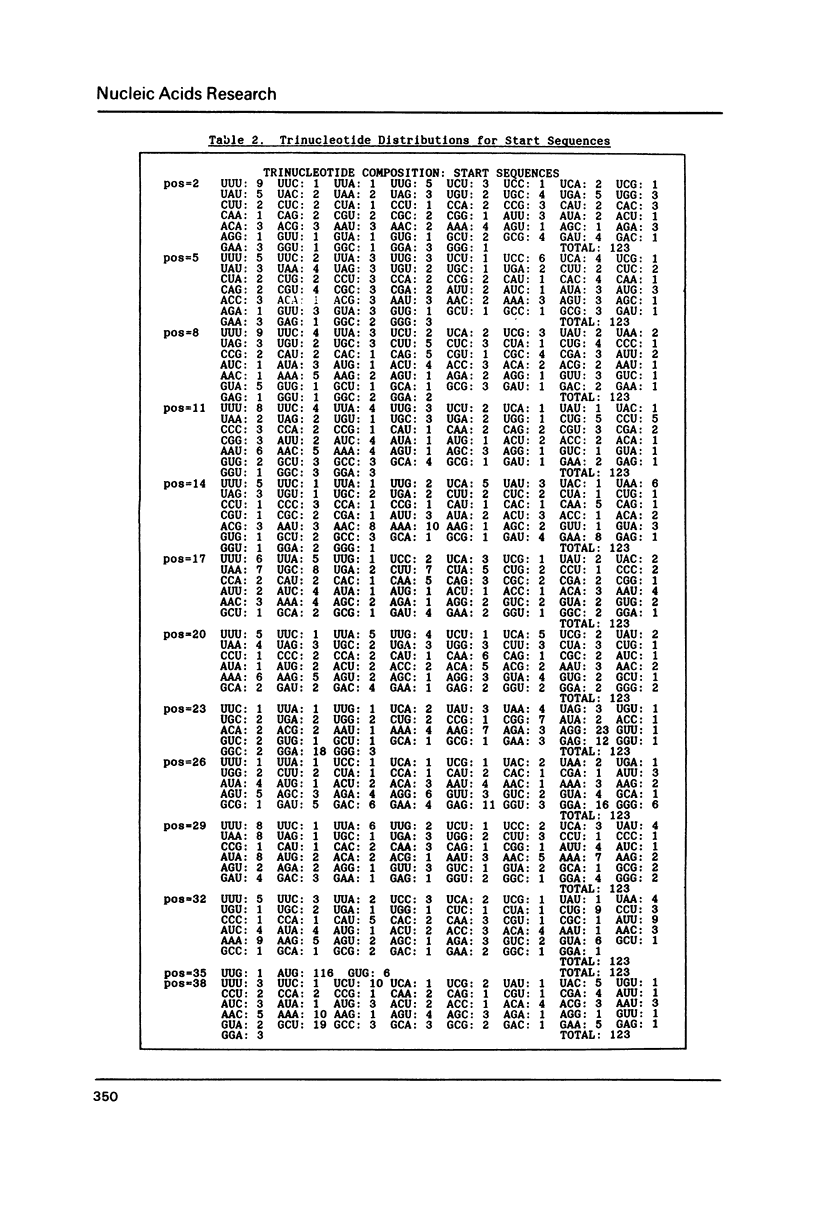
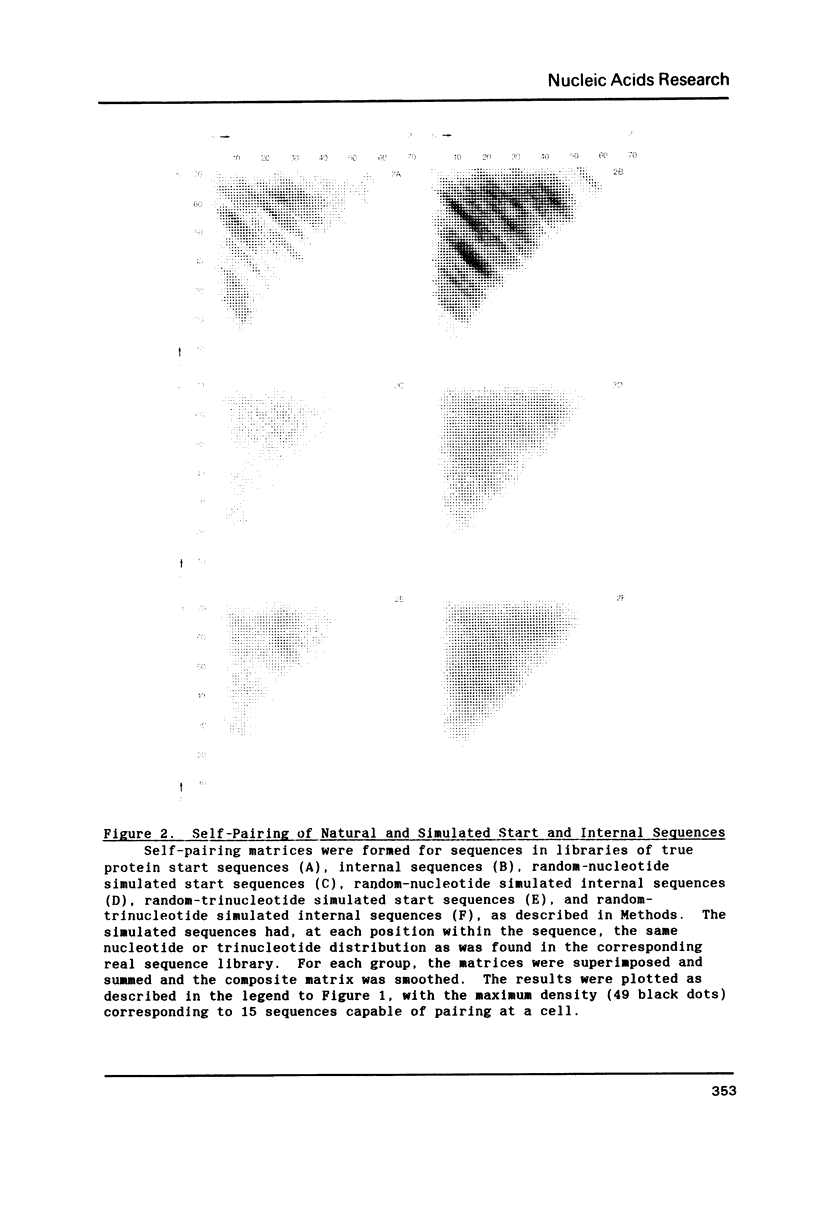
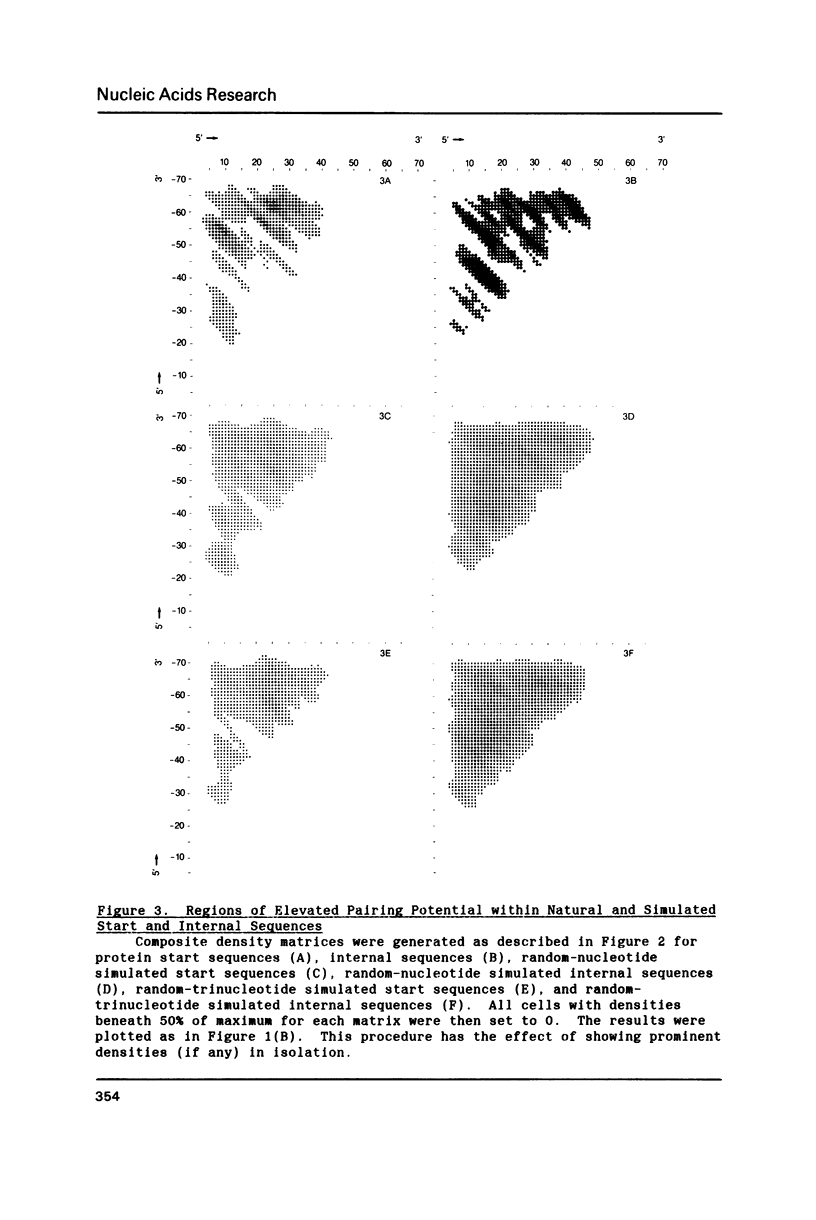
Since translational start codons also occur internally, more-complex features within mRNA must determine initiation. We compare the potential secondary structure of 123 prokaryotic mRNA start regions to that of regions coding for internal methionines. The latter display an unexpectedly-uniform, almost-periodic pattern of pairing potential. In contrast, sequences 5' to start codons have little self-pairing, and do not pair extensively with the proximal coding region. Pairing potential surrounding start codons was found to be less than half of that found near internal AUGs. In groups of random sequences where the distribution of nucleotides at each position, or of trinucleotides at each in-frame codon position, matched the observed natural distribution, there was no periodicity in the pairing potential of the internal sequences. Randomized internal sequences had less pairing: the ratio of pairing intensity between internals and starts was reduced from 2.0 to 1.6 by randomization. We propose that the transition from the relatively-unstructured start domains to the highly-structured internal sequences may be an important determinant of translational start-site recognition.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F. Is UAA or UGA part of the recognition signal for ribosomal initiation? Nucleic Acids Res. 1979 Oct 25;7(4):1035–1041. doi: 10.1093/nar/7.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahramian M. B. How bacterial ribosomes select translation initiation sites. J Theor Biol. 1980 May 7;84(1):103–118. doi: 10.1016/s0022-5193(80)81039-3. [DOI] [PubMed] [Google Scholar]
- Beck E., Sommer R., Auerswald E. A., Kurz C., Zink B., Osterburg G., Schaller H., Sugimoto K., Sugisaki H., Okamoto T. Nucleotide sequence of bacteriophage fd DNA. Nucleic Acids Res. 1978 Dec;5(12):4495–4503. doi: 10.1093/nar/5.12.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Borisova G. P., Volkova T. M., Berzin V., Rosenthal G., Gren E. J. The regulatory region of MS2 phage RNA replicase cistron. IV. Functional activity of specific MS2 RNA fragments in formation of the 70 S initiation complex of protein biosynthesis. Nucleic Acids Res. 1979;6(5):1761–1774. doi: 10.1093/nar/6.5.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone K. C., Steege D. A. Functional analysis of lac repressor restart sites in translational initiation and reinitiation. J Mol Biol. 1985 Dec 20;186(4):733–742. doi: 10.1016/0022-2836(85)90393-6. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Buzash-Pollert E., Studier F. W. Mutations of bacteriophage T7 that affect initiation of synthesis of the gene 0.3 protein. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2741–2745. doi: 10.1073/pnas.75.6.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt H., Lührmann R. Recognition by initiator transfer ribonucleic acid of a uridine 5' adjacent to the AUG codon: different conformational states of formylatable methionine-accepting transfer ribonucleic acid at the ribosomal peptidyl site. Biochemistry. 1981 Apr 14;20(8):2075–2080. doi: 10.1021/bi00511a002. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J. Sequence of the lacI gene. Nature. 1978 Aug 24;274(5673):765–769. doi: 10.1038/274765a0. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Duerinck F., Haegeman G., Iserentant D., Merregaert J., Min Jou W., Molemans F., Raeymaekers A., Van den Berghe A. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976 Apr 8;260(5551):500–507. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- Ganoza M. C., Fraser A. R., Neilson T. Nucleotides contiguous to AUG affect translational initiation. Biochemistry. 1978 Jul 11;17(14):2769–2775. doi: 10.1021/bi00607a011. [DOI] [PubMed] [Google Scholar]
- Ganoza M. C., Marliere P., Kofoid E. C., Louis B. G. Initiator tRNA may recognize more than the initiation codon in mRNA: a model for translational initiation. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4587–4591. doi: 10.1073/pnas.82.14.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganoza M. C., Sullivan P., Cunningham C., Hader P., Kofoid E. C., Neilson T. Effect of bases contiguous to AUG on translation initiation. J Biol Chem. 1982 Jul 25;257(14):8228–8232. [PubMed] [Google Scholar]
- Ganoza M. C. The Ayerst Award Lecture 1976. Novel factors in protein biosynthesis. Can J Biochem. 1977 Apr;55(4):267–281. doi: 10.1139/o77-038. [DOI] [PubMed] [Google Scholar]
- Gauss D. H., Sprinzl M. Compilation of tRNA sequences. Nucleic Acids Res. 1983 Jan 11;11(1):r1–53. [PMC free article] [PubMed] [Google Scholar]
- Gheysen D., Iserentant D., Derom C., Fiers W. Systematic alteration of the nucleotide sequence preceding the translation initiation codon and the effects on bacterial expression of the cloned SV40 small-t antigen gene. Gene. 1982 Jan;17(1):55–63. doi: 10.1016/0378-1119(82)90100-7. [DOI] [PubMed] [Google Scholar]
- Godson G. N., Barrell B. G., Staden R., Fiddes J. C. Nucleotide sequence of bacteriophage G4 DNA. Nature. 1978 Nov 16;276(5685):236–247. doi: 10.1038/276236a0. [DOI] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Hall M. N., Gabay J., Débarbouillé M., Schwartz M. A role for mRNA secondary structure in the control of translation initiation. Nature. 1982 Feb 18;295(5850):616–618. doi: 10.1038/295616a0. [DOI] [PubMed] [Google Scholar]
- Iserentant D., Fiers W. Secondary structure of mRNA and efficiency of translation initiation. Gene. 1980 Apr;9(1-2):1–12. doi: 10.1016/0378-1119(80)90163-8. [DOI] [PubMed] [Google Scholar]
- Kastelein R. A., Berkhout B., Overbeek G. P., van Duin J. Effect of the sequences upstream from the ribosome-binding site on the yield of protein from the cloned gene for phage MS2 coat protein. Gene. 1983 Sep;23(3):245–254. doi: 10.1016/0378-1119(83)90015-x. [DOI] [PubMed] [Google Scholar]
- Kozak M., Nathans D. Translation of the genome of a ribonucleic acid bacteriophage. Bacteriol Rev. 1972 Mar;36(1):109–134. doi: 10.1128/br.36.1.109-134.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H. F. Secondary structure of bacteriophage f2 ribonucleic acid and the initiation of in vitro protein biosynthesis. J Mol Biol. 1970 Jun 28;50(3):689–702. doi: 10.1016/0022-2836(70)90093-8. [DOI] [PubMed] [Google Scholar]
- Munson L. M., Stormo G. D., Niece R. L., Reznikoff W. S. lacZ translation initiation mutations. J Mol Biol. 1984 Aug 25;177(4):663–683. doi: 10.1016/0022-2836(84)90043-3. [DOI] [PubMed] [Google Scholar]
- Neilson T., Kofoid E. C., Ganoza M. C. Association of a synthetic precistronic region with 70S ribosomes is enhanced by an intact initiation triplet and a sequence complementary to the 3'-terminus of 16S rRNA. Nucleic Acids Symp Ser. 1980;(7):313–323. [PubMed] [Google Scholar]
- Ninio J. Prediction of pairing schemes in RNA molecules-loop contributions and energy of wobble and non-wobble pairs. Biochimie. 1979;61(10):1133–1150. doi: 10.1016/s0300-9084(80)80227-6. [DOI] [PubMed] [Google Scholar]
- Papanicolaou C., Gouy M., Ninio J. An energy model that predicts the correct folding of both the tRNA and the 5S RNA molecules. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):31–44. doi: 10.1093/nar/12.1part1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Operators and promoters in the OR region of phage 434. Nucleic Acids Res. 1979 Apr;6(4):1495–1508. doi: 10.1093/nar/6.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M., Backman K., Humayun M. Z., Jeffrey A., Maurer R., Meyer B., Sauer R. T. Autoregulation and function of a repressor in bacteriophage lambda. Science. 1976 Oct 8;194(4261):156–161. doi: 10.1126/science.959843. [DOI] [PubMed] [Google Scholar]
- Ray P. N., Pearson M. L. Functional inactivation of bacteriophage lambda morphogenetic gene in RNA. Nature. 1975 Feb 20;253(5493):647–650. doi: 10.1038/253647a0. [DOI] [PubMed] [Google Scholar]
- Roberts T. M., Kacich R., Ptashne M. A general method for maximizing the expression of a cloned gene. Proc Natl Acad Sci U S A. 1979 Feb;76(2):760–764. doi: 10.1073/pnas.76.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M., Manderschied U., Kyriatsoulis A., Brinckmann U., Gassen H. G. Tetranucleotides as effectors for the binding of initiator tRNA to Escherichia coli ribosomes. Eur J Biochem. 1980 Aug;109(1):291–299. doi: 10.1111/j.1432-1033.1980.tb04794.x. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Singleton C. K., Roeder W. D., Bogosian G., Somerville R. L., Weith H. L. DNA sequence of the E. coli trpR gene and prediction of the amino acid sequence of Trp repressor. Nucleic Acids Res. 1980 Apr 11;8(7):1551–1560. doi: 10.1093/nar/8.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J. A., Jakes K. How ribosomes select initiator regions in mRNA: base pair formation between the 3' terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4734–4738. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N., Bolshoi G. Open and closed 5 S ribosomal RNA, the only two universal structures encoded in the nucleotide sequences. J Mol Biol. 1983 Sep 5;169(1):1–13. doi: 10.1016/s0022-2836(83)80172-7. [DOI] [PubMed] [Google Scholar]
- Wahba A. J., Iwasaki K., Miller M. J., Sabol S., Sillero M. A., Vasquez C. Initiation of protein synthesis in Escherichia coli. II. Role of the initiation factors in polypeptide synthesis. Cold Spring Harb Symp Quant Biol. 1969;34:291–299. doi: 10.1101/sqb.1969.034.01.035. [DOI] [PubMed] [Google Scholar]
- Wood C. R., Boss M. A., Patel T. P., Emtage J. S. The influence of messenger RNA secondary structure on expression of an immunoglobulin heavy chain in Escherichia coli. Nucleic Acids Res. 1984 May 11;12(9):3937–3950. doi: 10.1093/nar/12.9.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]