Abstract
The outer membrane of microbial cells forms an effective barrier for hydrophobic compounds, potentially causing an uptake limitation for hydrophobic substrates. Low bioconversion activities (1.9 U gcdw−1) have been observed for the ω-oxyfunctionalization of dodecanoic acid methyl ester by recombinant Escherichia coli containing the alkane monooxygenase AlkBGT of Pseudomonas putida GPo1. Using fatty acid methyl ester oxygenation as the model reaction, this study investigated strategies to improve bacterial uptake of hydrophobic substrates. Admixture of surfactants and cosolvents to improve substrate solubilization did not result in increased oxygenation rates. Addition of EDTA increased the initial dodecanoic acid methyl ester oxygenation activity 2.8-fold. The use of recombinant Pseudomonas fluorescens CHA0 instead of E. coli resulted in a similar activity increase. However, substrate mass transfer into cells was still found to be limiting. Remarkably, the coexpression of the alkL gene of P. putida GPo1 encoding an outer membrane protein with so-far-unknown function increased the dodecanoic acid methyl ester oxygenation activity of recombinant E. coli 28-fold. In a two-liquid-phase bioreactor setup, a 62-fold increase to a maximal activity of 87 U gcdw−1 was achieved, enabling the accumulation of high titers of terminally oxyfunctionalized products. Coexpression of alkL also increased oxygenation activities toward the natural AlkBGT substrates octane and nonane, showing for the first time clear evidence for a prominent role of AlkL in alkane degradation. This study demonstrates that AlkL is an efficient tool to boost productivities of whole-cell biotransformations involving hydrophobic aliphatic substrates and thus has potential for broad applicability.
INTRODUCTION
The outer membrane of Gram-negative bacteria serves as an efficient barrier for hydrophobic molecules (12, 36, 44). Different approaches have been described to overcome uptake limitations in whole-cell biotransformations. The two-liquid-phase concept applied in stirred-tank reactors can increase mass transfer by ensuring maximal substrate availability and typically allows in situ product extraction (13, 20, 33, 37, 40, 47, 53, 72). Next to that, the addition of rhamnolipids, synthetic surfactants, and cosolvents has been described as enhancing biotransformation rates. Rhamnolipids are synthesized by several Pseudomonas species in order to facilitate the uptake of hydrophobic compounds (46). Rhamnolipids as well as synthetic surfactants (e.g., Triton X-100) solubilize hydrophobic substrates in the aqueous phase and interact with bacterial membranes (1, 39). Similarly, cosolvents, such as dimethyl sulfoxide (DMSO), or chelating agents, such as EDTA, can be used to enhance substrate solubility and/or membrane permeability (44). Further strategies to improve rates for hydrophobic substrate bioconversions include the use of host strains capable of growth on and thus efficient uptake of hydrophobic substrates, such as hydrocarbons, or the transfer of respective properties, e.g., uptake systems, into production strains. Here, favorable lipid and membrane protein (facilitators and transporters) compositions of Pseudomonas strains (61) may be exploited to enhance substrate uptake.
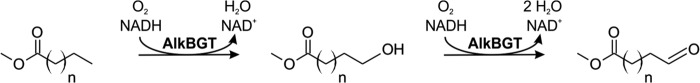
Uptake has recently been found to limit the selective ω-oxyfunctionalization of renewable fatty acid methyl esters (FAMEs) catalyzed by recombinant Escherichia coli W3110 (pBT10), when the fatty acid chain length exceeded nine carbon atoms (55). The alkane monooxygenase complex AlkBGT catalyzing this reaction (Fig. 1) originates from the OCT plasmid of Pseudomonas putida GPo1 (11) and is responsible for the first enzymatic step in alkane degradation. AlkBGT catalyzes the NADH-dependent ω-hydroxylation of a large variety of alkyl substrates and also was reported to catalyze alcohol oxidation (38, 55, 66). Whereas the oxygenation activity of AlkBGT-containing E. coli W3110 (pBT10) toward dodecanoic acid methyl ester (DAME) (1.9 U gcdw−1) and nonanoic acid methyl ester (NAME) (104 U gcdw−1) differed by a factor of 55, activities obtained with an enriched enzyme preparation differed only by a factor of 2.6, indicating poor uptake of the more hydrophobic substrate DAME into E. coli cells (55).
Fig 1.
Scheme of the NADH-dependent terminal oxyfunctionalization of fatty acid methyl esters catalyzed by the alkane monooxygenase system AlkBGT (n = 6 for NAME and n = 9 for DAME).
In this study, different approaches to enhance hydrophobic substrate uptake were investigated and compared, with the ω-oxyfunctionalization of DAME serving as a model reaction. Several additives were tested for their effect on mass transfer of DAME from an organic bulk phase into AlkBGT-containing E. coli W3110 (pBT10), applying a two-liquid-phase setup. Furthermore, the performance of Pseudomonas species as alternative host strains was investigated. Finally, coexpression of alkL, encoding an uncharacterized outer membrane protein, was investigated. The selection of AlkL was based on earlier observations showing a positive effect of an unidentified component of the alkane degradation pathway on the biotransformation of fatty acids (54), FAMEs (55), and alkanes (22). Via the characterization and comparison of factors influencing microbial uptake of hydrophobic molecules, this work identifies AlkL as a factor boosting the intracellular availability of long-chain alkylic substrates.
MATERIALS AND METHODS
Chemicals, bacterial strains, and media.
DAME (99.8% purity), 12-hydroxydodecanoic acid methyl ester (HDAME), and 12-oxododecanoic acid methyl ester (ODAME) (the last two with a purity of ≥95%) were obtained from Evonik Degussa GmbH (Marl, Germany). Nine-hydroxynonanoic acid methyl ester (HNAME) (purity, ≥95%) was obtained from TCI Europe N.V. (Zwijndrecht, Belgium). All other chemicals used in this study were purchased from Sigma-Aldrich (Steinheim, Germany) or Carl-Roth (Karlsruhe, Germany) with the highest purity available. The strains and plasmids used in this study are listed in Table 1. For cloning purposes, E. coli DH5α was used. For expression and biotransformation experiments, E. coli W3110 and the Pseudomonas strains were transformed with different plasmids following standard procedures (52). Strains were cultivated either in LB (52), in M9 medium (52), or in modified M9 medium (M9*) (48). For reactor cultivation of Pseudomonas fluorescens CHA0, aqueous batch medium (ABM) (30) was used. Mineral media were supplemented with 1 ml liter−1 USFe trace element solution (8). Antibiotics (50 mg liter−1 kanamycin or 12.5 mg liter−1 tetracycline) were added to all media if appropriate. Glucose and citrate were added as carbon sources for E. coli and Pseudomonas, respectively.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| E. coli W3110 | F− λ− rph-1, IN(rrnD-rrnE)1 | 2 |
| E. coli DH5α | supE44 ΔlacU169 (Φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 24 |
| P. putida KT2440 | P. putida mt-2 cured of the TOL plasmid | 3 |
| P. putida GPo1 | Contains the OCT plasmid | 11 |
| P. putida GPo12 | P. putida GPo1 cured of the OCT plasmid | 31 |
| P. fluorescens ATCC 15453 | 70 | |
| P. fluorescens CHA0 | Grows on C12–C32 alkanes | 60 |
| Pseudomonas sp. strain VLB120 | Wild-type Pseudomonas; styrene prototroph | 49 |
| Plasmids | ||
| pCom10 | Alkane-responsive broad-host-range vector, Kmr | 59 |
| pCom10_alkL | Contains alkL in pCom10, Kmr | This study |
| pBT10 | Contains alkBFG and alkST in pCom10, Kmr | 55 |
| pBTL10 | pBT10 with alkL in the operon alkBFGL, Kmr | This study |
| pGEc47 | Contains genes necessary for growth on alkanes (alkBFGHJKL and alkST) in broad-host-range vector pLAFR1, Tcr | 19 |
| pGEc47ΔB | pGEc47 without gene for the alkane monooxygenase AlkB, Tcr | 67 |
| pSMART-HCKan | Cloning vector, Kmr | Lucigen Corporation, Middleton, WI |
| pSMART-alkL | pSMART-HCKan containing alkL PCR fragment, Kmr | This study |
Tcr, tetracycline resistance; Kmr, kanamycin resistance.
Construction of pBTL10 and pCom10_alkL.
In order to construct a plasmid for coexpression of alkL and alkBGT, alkL was amplified by PCR from pGEc47 (19) including the ribosome binding site upstream of alkL. Primers 5′-ACGCGTCGACCTGTAACGACAACAAAACGAGGGTAG-3′ and 5′-ACGCGTCGACCTGCGACAGTGACAGACCTG-3′ both contained a SalI restriction site (underlined) (Eurofins MWG, Ebersberg, Germany). Phusion high-fidelity DNA polymerase was obtained from Finnzymes Oy (Espoo, Finland). Restriction enzymes, T4 DNA ligase, and thermosensitive alkaline phosphatase were purchased from Fermentas GmbH (St. Leon-Rot, Germany). The PCR product was subcloned in pSMART-HCKan (Lucigen Corporation, Middleton, WI). After restriction with SalI, alkL was cloned downstream of alkG into pBT10 (55).
The alkL-containing vector pCom10_alkL was constructed by insertion of alkL amplified from pGEc47 using the primers 5′-CCGGAATTCCATATGAGTTTTTCTAATTATAA-3′ (EcoRI site underlined) and 5′-CGCGGATCCTTAGAAAACATATGACGCAC-3′ (BamHI site underlined) into the alkane-responsive expression vector pCom10 (59), using EcoRI and BamHI as restriction enzymes. Correct construction of pBTL10 and pCom10_alkL was confirmed by sequencing (Eurofins MWG, Ebersberg, Germany).
Toxicity studies.
A single colony from an LB agar plate was used to inoculate 3 ml LB medium, which was incubated for 16 h at 30°C and 200 rpm. One milliliter of the culture was used to inoculate 100 ml M9* medium containing 0.5% (wt/vol) carbon source in a 500-ml baffled shaking flask. The culture was incubated for 20 h at 30°C and 200 rpm and used to inoculate M9* medium (0.5% [wt/vol] carbon source) to an optical density at 450 nm (OD450) of 0.2. Growth experiments were carried out in multiwell plates in a Bioscreen C MBR (Oy Growth Curves Ab Ltd., Helsinki, Finland) with a culture volume of 195 μl at 30°C. After 2 h of growth, 5 μl of a stock solution containing NAME, DAME, HNAME, or HDAME in ethanol was added, and growth was followed for another 24 h. The growth rate μ (h−1) was determined from the average of 10 parallel cultures.
Whole-cell biotransformation of fatty acid methyl esters and alkanes.
Precultures were prepared as described above and used to inoculate 100 ml M9* medium containing 0.5% (wt/vol) carbon source in a 500-ml baffled shaking flask to an OD450 of 0.2. Induction, incubation, and resting-cell assays (1 ml) were performed as described before (55). Activities were calculated in U gcdw−1, where 1 U is defined as 1 μmol product formed per min. For this purpose, the formation of all oxygenation products was considered. For the alkanes, evaporation was considered based on a control experiment in an abiotic setup. Activities for nonane conversions were additionally calculated based on substrate consumption, since the mass balance could not be closed for this substrate.
Two-liquid-phase biotransformation of fatty acid methyl esters.
Precultures were prepared as described above. For E. coli, the M9* preculture was used to inoculate 250 ml M9 medium containing a 2-fold ammonium chloride concentration (2 g liter−1) and 1.5% (wt/vol) glucose in a 300-ml stirred-tank reactor (RALF; Bioengineering AG, Wald, Switzerland) at an OD450 of 0.3 (0.05 gcdw liter−1). The pH was controlled automatically at 7.4 using 15% (vol/vol) phosphoric acid and 12.5% (vol/vol) ammonia. Temperature, aeration rate, and agitation frequency were set to 30°C, 2 volume per volume per minute (vvm), and 1,500 rpm, respectively. After complete consumption of the carbon source, an additional 4 ml liter−1 USFe trace element solution was added and a feed (50% [wt/vol] glucose and 13.4 g liter−1 MgSO4 · 7H2O) was started and regulated for glucose-limited exponential growth at a rate of 0.15 h−1. Cultivation of P. fluorescens CHA0 was performed under the same conditions using ABM medium containing 1.5% (wt/vol) citrate. Fed-batch cultivation involved exponential feeding (for a growth rate of 0.15 h−1) of a solution containing 50% (wt/vol) citrate, 7.6 g liter−1 MgSO4 · 7H2O, and 2.3% (vol/vol) ammonia. For both E. coli W3110 and P. fluorescens CHA0, recombinant gene expression was induced by the addition of 0.025% (vol/vol) dicyclopropylketone (DCPK) at a cell density of 5 gcdw liter−1. Fed-batch cultivation was continued for 4 h. Then, the cells were harvested by centrifugation (4,600 × g, 4°C, 20 min) and immediately resuspended in 50 mM potassium phosphate buffer (pH 7.4) containing 1.0% (wt/vol) glucose to a cell density of ca. 10 gcdw liter−1 (cell densities are given in Results). Aliquots of 100 ml were transferred into a clean 300-ml reactor. To guarantee nitrogen limitation, 1.5 M NaOH was used as base for pH control. After 10 min of adaptation at 30°C, the biotransformation was started by the addition of 50 ml of the substrate, resulting in a two-liquid-phase system with a phase ratio of 1:3 (organic phase to total volume). The pH was adjusted to 7. In order to circumvent energy limitation, additional glucose was added to a concentration of 1.0% (wt/vol) before depletion occurred. During biotransformation, the glucose levels were checked by glucose test strips (Macherey-Nagel GmbH & CO. KG, Düren, Germany). Samples taken from the reactor were centrifuged for 5 min at 17,000 × g in 2-ml Eppendorf tubes. The upper organic phase was diluted 10- to 50-fold in diethyl ether containing 0.2 mM dodecane as an internal standard, dried over anhydrous sodium sulfate, and analyzed by gas chromatography (GC). The lower aqueous phase was sampled using needle and syringe and subjected to liquid chromatography analysis to quantify glucose and organic acid concentrations.
Analytical procedures.
FAMEs and the corresponding oxygenation products were quantified using a Trace GC UltraTM gas chromatograph as described before (55). Alkanes and corresponding oxygenation products were analyzed using the following temperature profile: 40°C for 3 min, 40 to 170°C at 15°C min−1, 170 to 300°C at 100°C min−1, 300°C for 3 min. Substrate and product quantifications were based on the analysis of standard solutions.
Glucose and organic acid concentrations in the aqueous phase were determined by high-performance liquid chromatography (HPLC) as described before (23).
Cell concentrations in the aqueous phase were determined by measuring the optical density at 450 nm (Libra S11 spectrophotometer; Biochrom Ltd., Cambridge, United Kingdom), whereby one OD450 unit corresponded to a cell concentration of 0.166 gcdw liter−1 (5).
SDS-PAGE was performed according to the method described by Laemmli (35). Total membrane fractions were obtained as described elsewhere (55).
RESULTS
The two-liquid-phase approach does not improve substrate mass transfer into microbial cells.
A comparison of DAME conversion rates obtained with AlkBGT-containing E. coli W3110 (pBT10) and enriched enzyme preparations indicated that the biotransformation of DAME is limited mainly by substrate mass transfer into the cells (55). In order to evaluate if this limitation is related to interfacial areas and thus mass transfer of both oxygen and hydrophobic substrate, a two-liquid-phase reactor setup was applied with the substrate constituting a second liquid phase. Beside maximized mass transfer, this setup enables continuous substrate supply and efficient extraction of potentially toxic oxygenation products. First, the toxicity of the two liquid substrates NAME and DAME and of corresponding terminal alcohols was investigated (Table 2). FAMEs did not show any influence on the growth rate, also not at concentrations exceeding their solubility in water. For the corresponding alcohols, however, a concentration-dependent decrease in growth rate was observed. The strongest inhibition of growth was observed for 9-hydroxynonanoic acid methyl ester (HNAME), with the lowest logP (logarithm of the partition coefficient in an octanol/water system), i.e., 2.35, and the highest solubility in water (8.5 mM) of all tested compounds. Since both substrates, NAME and DAME, are not toxic to E. coli W3110, they were applied as bulk organic phase in the two-liquid-phase setup.
Table 2.
Growth rates of E. coli W3110 during batch growth on glucose in the presence of different concentrations of NAME, HNAME, DAME, and HDAMEa
| Concn of compound (mM) | Growth rate (μ) of E. coli W3110 (h−1) |
|||
|---|---|---|---|---|
| NAME (133 μM)b | HNAME (8,552 μM)b | DAME (5.3 μM)b | HDAME (234 μM)b | |
| 0 | 0.26 | 0.26 | 0.26 | 0.26 |
| 2.5% EtOH | 0.22 | 0.22 | 0.22 | 0.22 |
| 0.005 | NDc | ND | 0.21 | ND |
| 0.05 | 0.22 | 0.22 | ND | 0.22 |
| 0.13 | 0.22 | 0.22 | ND | ND |
| 0.23 | ND | ND | 0.21 | 0.21 |
| 2.5 | 0.23 | 0.14 | 0.21 | 0.13 |
| 5.0 | 0.23 | ND | 0.21 | 0.12 |
| 8.5 | ND | 0.03 | ND | ND |
| 50 | 0.23 | 0.06 | 0.20 | 0.17 |
All compounds were added via an ethanol (EtOH) stock solution resulting in a final ethanol concentration of 2.5% (vol/vol) for each culture.
Values in parentheses indicate solubility in water.
ND, not determined.
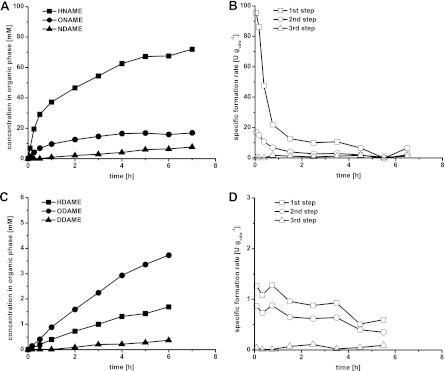
In a two-liquid-phase biotransformation of NAME at a phase ratio (organic to total volume) of 1:3 carried out with fully induced resting E. coli W3110 (pBT10) cells at a cell density of 8.9 gcdw liter−1, HNAME and 9-oxononanoic acid methyl ester (ONAME) initially accumulated with maximal specific activities for the first and second oxidation steps of 95 and 17 U gcdw−1, respectively (Fig. 2A and B). These activities decreased rapidly to levels below 10 and 3 U gcdw−1, respectively, after 2.5 h. Furthermore, slow nonanedioic acid monomethyl ester (NDAME) formation was observed. As for all biotransformations performed in this study, glucose and oxygen (dissolved oxygen tension [DOT] ≥ 20%) were present throughout the biotransformation, excluding their depletion as a possible reason for the activity decrease. Under identical conditions, DAME was converted with significantly lower but more constant specific activities, maximally amounting to 1.4 and 0.9 U gcdw−1 for the first and the second oxygenation steps, respectively (Fig. 2C and D). In this case, the aldehyde (12-oxododecanoic acid methyl ester [ODAME]) was the main product, which may be explained by the higher availability of 12-hydroxydodecanoic acid methyl ester (HDAME) due to its higher water solubility compared to DAME. Also here, slow acid (dodecanedioic acid monomethyl ester [DDAME]) formation was observed. Such acid formation can be due to overoxidation catalyzed by AlkBGT or host intrinsic enzymes, e.g., aldehyde dehydrogenases. A three-step oxygenation catalyzed by one single enzyme has been described for XylM originating from P. putida mt-2, an enzyme showing structural similarity to AlkB (9).
Fig 2.
Two-liquid-phase biotransformations of NAME and DAME using 8.9 gcdw liter−1 of resting E. coli W3110 (pBT10) cells fed with glucose. The substrates were applied as the organic phase at a phase ratio of 1:3 (organic phase to total volume). Product concentrations in the organic phase (A and C) and specific activities for the different oxygenation steps (B and D) are given. Specific activities are calculated based on product formation.
Although volumetric activities were improved in the two-liquid-phase setup due to the higher cell concentrations applied, the specific rates for NAME and DAME oxygenation were in the same range as activities found in small-scale resting-cell assays with low substrate concentrations (55) (see Table 5). Thus, the application of a two-liquid-phase setup in a stirred-tank reactor did not improve the substrate availability for the intracellular AlkBGT complex. Increasing the agitation rate and the phase ratio also did not result in higher specific DAME oxygenation rates (Table 3). An increase in cell density from 8.9 to 18 gcdw liter−1 resulted only in a minor specific activity decrease (Table 3), indicating that the substrate mass transfer over the organic-aqueous phase boundary and to the cells was not limiting at a biomass concentration of 8.9 gcdw liter−1. Since changes in the reaction setup with the aim to increase surface areas for organic-aqueous-phase and organic-phase cell mass transfer did not increase specific DAME oxygenation rates, substrate uptake into the cells was most likely causing a kinetic limitation of DAME conversion. Thus, further experiments in this study focused on the investigation of factors influencing mass transfer over the membrane.
Table 5.
Activities of different recombinant E. coli W3110 strains expressing alkBGT and optionally alkL for NAME, DAME, octane, and nonane oxygenation
| Plasmid | Expression level of: |
Maximal sp acta (U gcdw−1) |
||||
|---|---|---|---|---|---|---|
| AlkBGT | AlkL | NAMEb | DAMEb | Octanec | Nonane | |
| pBT10 | High | 104 | 1.9 | 24 | 2.4d | |
| pGEc47 | Low | Low | 71 | 31 | 54 | 72e (37) |
| pBT10;GEc47ΔB | High | Low | 125 | 23 | 77 | 88e (46) |
| pBTL10 | High | High | 128 | 53 | 100 | 97e (40) |
Calculated for the first oxygenation step based on product formation by resting cells; for nonane, substrate consumption also was considered (see Materials and Methods).
Cell concentration, 1.0 gcdw liter−1; initial substrate concentration, 2.5 mM; reaction time, 5 min.
Cell concentration, 0.5 gcdw liter−1; initial substrate concentration, 1 mM; reaction time, 6 min.
Cell concentration, 1.0 gcdw liter−1; initial substrate concentration, 1 mM; reaction time, 6 min.
Cell concentration, 0.5 gcdw liter−1; initial substrate concentration, 1 mM; reaction time, 6 min; activities based on substrate depletion as well as product formation (in parentheses) are given.
Table 3.
Maximal DAME oxygenation activities of resting E. coli W3110 (pBT10) cells in the presence of different surfactants and solvents in a two-liquid-phase reactor setup
| Parameter or additive | Setting or concn | Relative maximal hydroxylation activitiesa |
|---|---|---|
| Parameters | ||
| Cell density | 20 gcdw liter−1 | 0.9 |
| Stirring speed | 3,000 rpm | 1.0 |
| Phase ratio | 1:2 (organic/total vol) | 1.0 |
| Additives | ||
| Rhamnolipids | 0.0001% (wt/vol) | 0.8 |
| 0.001% (wt/vol) | 1.0 | |
| 0.01% (wt/vol) | 1.0 | |
| DMSO | 1% (vol/vol) | 1.0 |
| 2% (vol/vol) | 0.3 | |
| Triton X-100 | 0.05 (vol/vol) | 1.0 |
| EDTA | 1 mM | 1.5 |
| 2.5 mM | 2.8 |
For each biotransformation, maximal activities are normalized to the maximal activity achieved with untreated cells in a parallel experiment (see the text for details).
Chemical treatment of the biocatalyst to improve mass transfer over the membrane.
In order to increase substrate availability, either by an increased solubility of the substrate in the aqueous phase or permeabilization of the cells, different additives were applied during two-liquid-phase biotransformations, i.e., rhamnolipids and Triton X-100 serving as biosurfactants and synthetic surfactant, respectively; the cosolvent DMSO; and the chelating compound EDTA. The presence of rhamnolipids, Triton X-100, or DMSO did not increase DAME oxygenation activities (Table 3). The addition of 2% (vol/vol) DMSO even decreased the activity to one-third of the activity reached without DMSO, probably caused by cell permeabilization. The addition of EDTA, however, did show a positive effect on DAME oxygenation activities, which increased up to 2.8-fold (Table 3). EDTA is known to destabilize the lipopolysaccharide (LPS) layer by chelating divalent cations (45) and may thus facilitate hydrophobic substrate uptake. Still, DAME oxygenation rates remained 20-fold lower than NAME oxygenation rates. Considering that only a 2.6-fold-lower DAME oxygenation (compared to NAME oxygenation) for the intrinsic AlkB specificity was observed, as determined by means of enriched enzyme preparations (55), substrate mass transfer limitation into the cells was not completely relieved by EDTA addition. Except for EDTA addition, chemical treatment of the cells did not improve mass transfer of the hydrophobic substrate.
Pseudomonas spp. as alternative hosts.
Several Pseudomonas species are known for their ability to utilize hydrophobic compounds as sources for carbon and energy (4, 21). Efficient substrate uptake is a prerequisite for such growth. Following this approach, NAME and DAME oxygenation activities of different recombinant Pseudomonas strains were determined in small-scale resting-cell assays. The AlkB expression levels estimated from SDS-PAGE gels were lower than those of E. coli W3110 (pBT10), varied among the strains investigated (data not shown), and correlated with the NAME oxygenation activities observed (Table 4). In comparison to E. coli W3110 (pBT10), none of the Pseudomonas strains showed improved activities toward NAME. P. fluorescens CHA0 (pBT10), however, showed an increased activity of 5.2 U gcdw−1 for DAME conversion, being only a factor of 4 lower than the NAME oxygenation activity. Product formation caused by host intrinsic alkane hydroxylases could be excluded since no oxygenated products were detected during resting-cell assays with the wild-type strain.
Table 4.
NAME and DAME oxygenation activities of different alkBGT-expressing Pseudomonas strains
| Strain | Oxygenation activity (U gcdw−1)a |
|
|---|---|---|
| NAME | DAME | |
| P. putida KT2440 (pBT10) | 2.2 | 0 |
| P. putida GPo1 | 18 | 1.9b |
| P. putida GPo12 (pBT10) | 0 | 0 |
| P. fluorescens ATCC15453 (pBT10) | 3.3 | 1.8b |
| P. fluorescens CHA0 (pBT10) | 21 | 5.2 |
| Pseudomonas sp. strain VLB120 (pBT10) | 57 | 0.9 |
Calculated for the first oxygenation step based on product formation by resting cells: cell concentration, 1.0 gcdw liter−1; initial substrate concentration, 2.5 mM; reaction time, 5 min.
Reaction time, 15 min.
Resting cells of P. fluorescens CHA0 (pBT10) were used in the two-liquid-phase setup described above using DAME as the substrate. DAME was converted with an initial activity of 5.7 U gcdw−1 with the respective alcohol being the most prominent product (5.3 mM HDAME, 4.6 mM ODAME, and 1.9 mM DDAME after 4 h; see the supplemental material). As was also found for E. coli W3110 (pBT10), the application of resting cells of P. fluorescens CHA0 (pBT10) in a two-liquid-phase setup resulted in an improved volumetric activity without improving specific activities compared to small-scale resting-cell assays. However, despite lower AlkBGT expression levels, P. fluorescens CHA0 (pBT10) showed a DAME oxygenation activity 4-fold higher than that of E. coli W3110 (pBT10) in the two-liquid-phase setup.
The outer membrane protein AlkL enhances DAME oxygenation.
Interestingly, E. coli W3110 (pGEc47) expressing all genes of the alkane degradation pathway of P. putida GPo1 was found to catalyze DAME oxygenation at a rate of 31 U gcdw−1, which was only 2.3-fold lower than its NAME oxygenation activity (55) (Table 5). In comparison to E. coli W3110 (pBT10), E. coli W3110 (pGEc47) showed lower AlkBGT expression levels but higher oxygenation rates for long-chain substrates. This suggests that pGEc47 encodes an uptake system for hydrophobic substrates. Similarly, with the alk operon expressed in microbial whole cells, efficient conversion of n-dodecane and pentadecanoic acid has been reported with AlkBGT and cytochrome P450 BM3 as the respective oxygenating enzymes (22, 54). In this study, the outer membrane protein AlkL, with so-far-unknown function, was investigated regarding its role in the uptake and conversion of hydrophobic substrates. For this purpose, the alkL gene was coexpressed with alkBGT by means of the newly constructed expression plasmid pBTL10. Interestingly, E. coli W3110 (pBTL10) showed a DAME oxygenation activity as high as 53 U gcdw−1 in small-scale resting-cell assays, constituting a 28-fold activity increase compared to E. coli W3110 (pBT10) (Table 5). Hence, AlkL is able to enhance DAME availability for the monooxygenase AlkBGT in E. coli cells. NAME oxygenation activities were in the same range for E. coli W3110 (pBTL10) and E. coli W3110 (pBT10) (Table 5). Obviously, substrate mass transfer limitation into cells was not or less relevant for NAME bioconversion, given the similar apparent Ks (substrate uptake constant) values for both strains (data not shown).
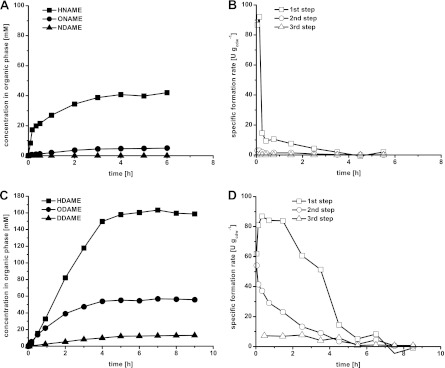
E. coli W3110 (pBTL10) also was investigated in the two-liquid-phase setup (Fig. 3). Using a cell concentration of 6.3 gcdw liter−1 and DAME as the organic phase, significantly increased product concentrations and specific activities were observed compared to the two-liquid-phase biotransformation with E. coli W3110 (pBT10). After 9 h of biotransformation, HDAME, ODAME, and DDAME accumulated at concentrations of up to 159, 56, and 13 mM in the organic phase (Fig. 3C). The initial activity for the first oxygenation step was 61 U gcdw−1 and increased to a maximum of 87 U gcdw−1 after 20 min of biotransformation (Fig. 3D), constituting a 62-fold increase compared to the strain without AlkL. After approximately 1.5 h of biotransformation, the activity started to decrease to reach levels below 10 U gcdw−1 after 5.5 h. The initial activity for the second oxygenation step was 54 U gcdw−1 and decreased from the beginning on. Thus, in the two-liquid-phase setup, the relieving of substrate uptake limitation by means of AlkL enabled not only increased volumetric activities but also increased specific activities for DAME oxygenation. However, for NAME conversion, the presence of AlkL in the outer membrane lowered the achieved product concentrations (46 mM HNAME, 5.0 mM ONAME, and traces of NDAME; Fig. 3A and B). This was due to a very fast activity decrease, whereas the initial activity of 92 U gcdw−1 for the first step was similar to the activity achieved without AlkL. In conclusion, AlkL tremendously improved mass transfer of the large and hydrophobic substrate DAME over the outer membrane, allowing activities in the same range as obtained for the smaller and more soluble substrate NAME. These activities range among the highest activities reported for monooxygenase-based whole-cell biocatalysis (17, 50).
Fig 3.
Two-liquid-phase biotransformations of NAME and DAME using 10.0 gcdw liter−1 and 6.3 gcdw liter−1, respectively, of resting E. coli W3110 (pBTL10) cells fed with glucose. The substrates were applied as the organic phase at a phase ratio of 1:3 (organic phase to total volume). Product concentrations in the organic phase (A and C) and specific activities for the different oxygenation steps (B and D) are given. Specific activities are calculated based on product formation.
Functionality of AlkL regarding uptake and conversion of FAMEs and alkanes.
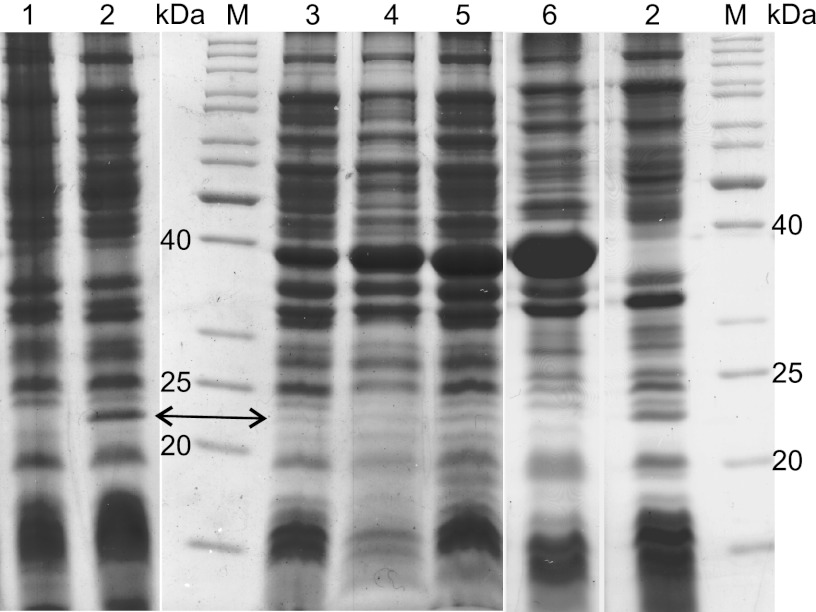
In order to get a better insight into the functionality of AlkL, strains expressing alkBGT and alkL at diverging levels, i.e., E. coli W3110 (pGEc47) and E. coli W3110 (pBT10;pGEc47ΔB), were tested for their oxygenation activities and compared with E. coli W3110 harboring pBT10 or pBTL10. Furthermore, the conversion of alkanes, the natural substrates of AlkB, was included in these studies. For induced E. coli W3110 carrying pGEc47 or pGEc47ΔB instead of pBTL10, SDS-PAGE analysis of membrane fractions showed lower AlkL expression levels (Fig. 4) as a result of the lower copy number and the alkL position further downstream of the alkB promoter. Similarly, cells carrying pGEc47 showed lower AlkB levels in the membrane fraction than cells carrying pBT10 or pBTL10. Consistent with the differences in AlkB levels, the last two plasmids enabled a 1.5- or 1.8-fold-higher NAME oxygenation activity, respectively, than that of pGEc47 (Table 5). In contrast, DAME oxygenation activities correlated with AlkL expression levels, being, in comparison to E. coli W3110 (pBT10), 16 and 12 times higher for E. coli W3110 (pGEc47) and E. coli W3110 (pBT10;pGEc47ΔB), respectively. The higher AlkL expression levels in E. coli W3110 (pBTL10) enabled even higher activities (Table 5), being, in comparison to E. coli W3110 (pBT10), 28- and 62-fold higher in activity assays and two-liquid-phase biotransformations, respectively. Thus, DAME oxygenation rates increased in correlation with alkL expression and were not influenced by the amount of monooxygenase present.
Fig 4.
SDS-PAGE (12%) of membrane fractions of E. coli W3110 carrying different plasmids. pCom10 (lane 1) and pCom10_alkL (lanes 2) served as negative and positive controls, respectively, whereas strains containing pGEc47 (lane 3), pGEc47ΔB/pBT10 (lane 4), and pBTL10 (lane 5) showed low but significant alkL expression, which was absent with pBT10 (lane 6). Lanes M, PageRuler SM26614 (Fermentas GmbH, St. Leon-Rot, Germany). The arrow indicates AlkL (23 kDa), whereas AlkB prominently runs at ∼40 kDa.
Besides the effect of AlkL via FAME uptake on AlkBGT activity, its effect on the uptake of alkanes was investigated. All strains converted octane, with 1-octanol and octanal as the only products accumulating. The strains expressing alkL showed significantly increased specific activities compared to the strain lacking alkL, with the activity again correlating with the alkL and alkBGT expression levels (Table 5). The supply of nonane as the substrate led to the formation of nonanoic acid in addition to 1-nonanol and nonanal. Furthermore, the mass balance could not be closed, pointing to nonanoic acid degradation via β-oxidation as reported for E. coli before (51). Considering product formation and substrate consumption, activities found for nonane oxygenation showed a dependency on AlkL and AlkBGT expression levels similar to that of DAME oxygenation activities, with a tremendous increase in activity, by a factor of up to 40, in the presence of AlkL (Table 5). Thus, the presence of AlkL in the outer membrane of E. coli strains also proved to be essential for the efficient uptake of alkanes.
DISCUSSION
Host selection and permeabilization of E. coli by chemical treatment.
The ability of several Pseudomonas species to grow on hydrocarbons, necessitating an efficient uptake system for such hydrophobic substrates, qualifies Pseudomonas strains as interesting host candidates for the bioconversion of hydrophobic substrates. Furthermore, the microbial origin of AlkBGT, P. putida GPo1 (11), makes the selection of Pseudomonas strains as alternative hosts an obvious first choice. Finally, some Pseudomonas strains are known to produce biosurfactants, which are supposed to increase the availability of hydrophobic substrates for microbial metabolization (46). Among the tested strains, P. fluorescens CHA0 (pBT10) gave the best results, with a 4-fold-higher DAME oxygenation activity than that of untreated E. coli W3110 (pBT10). Furthermore, this strain, with only 4-fold-higher activity toward NAME than toward DAME, comes close to the specificity of AlkB determined in vitro (55). These results are in accordance with the ability of P. fluorescens CHA0 to take up and grow on hydrophobic substrates such as C12 to C32 alkanes (58).
Except for EDTA, the addition of cell-permeabilizing and substrate-solubilizing chemicals did not result in accelerated DAME oxyfunctionalization using E. coli W3110 (pBT10). Treatment of E. coli with both organic solvents and detergents significantly improved biocatalytic l-carnitine formation (10). Chemical and physical treatments of cells also have been reported to be effective for hydrophobic substrates (16). However, such treatment apparently does not improve DAME oxygenation by AlkB in the cytoplasmic membrane of E. coli W3110. Especially for oxygenase catalysis, such treatment may generally be critical, as intended effects such as membrane permeabilization and destabilization can lead to cofactor leakage, enzyme destabilization, inhibition of metabolism, impeded cofactor regeneration, and thus reduced biocatalyst performance.
Improved substrate availability in the presence of the outer membrane protein AlkL.
Reported host cell engineering approaches to improve substrate availability include knockout strategies involving a smaller lipopolysaccharide layer or the absence of Braun's lipoprotein in the outer membrane. Such knockout strategies enabled increased whole-cell bioconversion rates with a tetrapeptide, nitrofecin, or aromatic hydrocarbons as substrates (42, 43). However, these strategies may influence cell metabolism and/or cell viability (12, 26, 64). The present study shows that the sole introduction of an outer membrane protein, i.e., AlkL, functioning as an uptake facilitator, vastly increases resting-cell-based bioconversion rates of hydrophobic substrates such as DAME without affecting growth characteristics in LB and minimal medium. Therefore, alkL coexpression can overcome disadvantages of nonspecific permeabilization methods. The alkL gene is located on the alkane degradation operon on the catabolic OCT plasmid of P. putida GPo1 (65), suggesting a role of AlkL in the conversion of alkanes as it was confirmed for octane and nonane (Table 5). The AlkL-related tremendous increase in specific hydrophobic substrate oxygenation rates and their observed correlation with alkL expression levels provide clear evidence for the role of AlkL in hydrophobic substrate transfer over the outer membrane. The ratio of 2.4 between NAME and DAME oxygenation activities of E. coli W3110 (pBTL10) in resting-cell assays (Table 5) correlates well with the respective ratio of 2.6 found with enriched enzyme preparations (55), which is another indication that the presence of AlkL in E. coli relieved substrate uptake limitation. Interestingly, AlkL-containing cells reached even higher DAME oxygenation activities when applied in a two-liquid-phase bioreactor setup (87 compared to 53 U gcdw−1), which may be explained by better mass transfer to and saturation of AlkL due to better mixing and a larger interface area between the aqueous and organic phases. Such an effect was not observed for NAME oxygenation but, in this case, may have been masked by the fast activity decrease discussed below.
AlkL shows homology to the outer membrane proteins OmpW (29%) from E. coli and OprG (23%) from Pseudomonas aeruginosa (see the supplemental material), which might be involved in the uptake of hydrophobic molecules (27, 63). OmpW and OprG structures involve an eight-stranded β-barrel forming a hydrophobic channel. As a mechanism for hydrophobic substrate uptake, this channel was proposed to enable the transfer of hydrophobic molecules through the LPS layer, followed by their release into the outer membrane through a lateral opening (27, 63, 69). A similar mechanism has been described for the uptake of long-chain fatty acids by FadL (25, 68). Outer membrane proteins with a possible role in substrate uptake have also been reported for catabolic pathways for the degradation of naphthalene (15, 18), xylenes (29), and toluene (28, 71).
The positive influence of AlkL on hydrophobic substrate bioconversion also explains respective beneficial effects of the plasmid pGEc47 observed in earlier studies. For example, the presence of pGEc47 effected a 3-fold increase in the specific rate for subterminal pentadecanoic acid hydroxylation catalyzed by cytochrome P450 BM3 in recombinant E. coli W3110 (54). Furthermore, E. coli W3110 (pGEc47), containing the complete alkane degradation pathway, was applied to produce octanoic acid from n-octane (20). A recombinant Pseudomonas strain containing AlkBGT but lacking AlkL was used to produce 1-octanol from n-octane (6) but did not show n-dodecane hydroxylation activity, whereas such activity of AlkB was reported for enriched enzyme preparations (66). This discrepancy can be explained by insufficient uptake of the hydrophobic substrate n-dodecane into Pseudomonas cells lacking AlkL. Successful in vivo n-dodecane hydroxylation was achieved using E. coli GEC137 (pGEc47ΔJ) containing AlkBGT and AlkL (22). The results of the present study are in agreement with these reported observations. E. coli W3110 (pBT10) was able to convert octane with considerable activities despite the lack of AlkL. In contrast, nonane was converted at very low rates (Table 5). As also seen for FAMEs, the activity increase caused by AlkL becomes more prominent with increasing hydrophobicity and size of the substrate. This study for the first time shows that AlkL efficiently facilitates hydrophobic substrate uptake into living cells, making AlkL an invaluable tool to overcome hydrophobic substrate uptake limitation via microbial biocatalyst engineering.
Influence of facilitated solvent uptake on solvent toxicity and whole-cell biocatalyst stability.
Notwithstanding the tremendous activity increase obtained for DAME conversion, AlkL appeared to affect the stability of the resting cell biocatalyst. During two-liquid-phase biotransformations, AlkL-containing cells lost their activity faster than did AlkL-negative strains (Fig. 2 and 3). These results indicate a negative correlation between facilitated substrate uptake and biocatalyst stability. The latter may be reduced due to increased intracellular concentrations of hydrophobic substrates and/or products in the presence of AlkL, which in turn may lead to toxic effects on the host cell and/or AlkBGT inhibition or deactivation. SDS-PAGE analysis showed that AlkB degradation did not occur during the biotransformations and thus can be excluded as a reason for the observed activity decrease (data not shown).
In two-liquid-phase biotransformations, recombinant E. coli strains expressing the alk gene cluster recently have been reported to be less stable with n-octane than with n-dodecane as the substrate (22). This behavior was assigned to a higher product and substrate toxicity with n-octane as the substrate. Similarly, biocatalyst stabilities were lower during the bioconversion of the less-hydrophobic and more-water-soluble substrate NAME than during that of DAME (Fig. 2 and 4), correlating with the lower toxicity of HDAME than of HNAME (Table 2). In general, organic solvents with an octanol-water partition coefficient logP of <4 are considered to be toxic to microbial cells, preventing their application as carrier solvents in whole-cell biotransformations (7, 14, 34, 57). Solvents with a logP of >4 are considered to be suitable for two-liquid-phase biocatalysis based on microbial cells. Low-logP solvents are relatively water soluble and readily diffuse into cells and accumulate in microbial membranes, which increases membrane fluidity and permeability (41, 56), thus leading to leakage of ions and macromolecules, interference with energy metabolism, and finally membrane deterioration and cell death. Reported logP values for NAME and DAME are 4.32 (62) and 5.41 (32), respectively. Although the logP value for both substrates is above 4 and both compounds did not affect growth of E. coli W3110, inhibiting effects were observed in the presence of AlkL, as can be expected for a transport facilitator alleviating the barrier function of the outer membrane for hydrophobic molecules. For HNAME and HDAME, the logP values were calculated to be 2.35 and 3.82, respectively (data from ChemSpider), and both compounds were found to affect growth of E. coli W3110. Products of further oxygenation steps have similarly low logP values and, although accumulating to lower concentrations, may contribute to reduced viability and biocatalytic activity. Thus, facilitated uptake and toxicity of solvents seem to be interconnected, necessitating a fine-tuned regulation of systems involved in the uptake of and the defense against solvents. Given this background, it will be interesting to investigate the effect of alkL expression on microbial solvent tolerance and whole-cell biocatalyst stabilities as well as the molecular mechanism of AlkL operation.
In conclusion, this study for the first time shows direct evidence for the role of AlkL in hydrophobic substrate mass transfer over the outer membrane, enabling efficient AlkBGT catalysis. The 62-fold increase in biotransformation rates achieved for DAME and the 40-fold increase for the natural substrate nonane qualify alkL coexpression as an efficient strategy to overcome hydrophobic substrate uptake limitation in whole-cell biocatalysis. More specifically, the findings now enable the development of a productive process for DAME ω-oxyfunctionalization.
Supplementary Material
ACKNOWLEDGMENTS
We express our gratitude to Christina Münnig and Christian David for supporting growth experiments and two-liquid-phase biotransformations.
This study was financially supported by the German Federal Ministry of Education and Research (BMBF, grant number 0315205).
Footnotes
Published ahead of print 8 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Al-Tahhan RA, Sandrin TR, Bodour AA, Maier RM. 2000. Rhamnolipid-induced removal of lipopolysaccharide from Pseudomonas aeruginosa: effect on cell surface properties and interaction with hydrophobic substrates. Appl. Environ. Microbiol. 66:3262–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bachmann BJ. 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 36:525–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bagdasarian M, et al. 1981. Specific-purpose plasmid cloning vectors II: broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237–247 [DOI] [PubMed] [Google Scholar]
- 4. Baptist JN, Gholson RK, Coon MJ. 1963. Hydrocarbon oxidation by a bacterial enzyme system. I. Products of octane oxidation. Biochim. Biophys. Acta 69:40–47 [DOI] [PubMed] [Google Scholar]
- 5. Blank LM, Ebert BE, Bühler B, Schmid A. 2008. Metabolic capacity estimation of Escherichia coli as a platform for redox biocatalysis: constraint-based modeling and experimental verification. Biotechnol. Bioeng. 100:1050–1065 [DOI] [PubMed] [Google Scholar]
- 6. Bosetti A, van Beilen JB, Preusting H, Lageveen RG, Witholt B. 1992. Production of primary aliphatic alcohols with a recombinant Pseudomonas strain, encoding the alkane hydroxylase enzyme system. Enzyme Microb. Technol. 14:702–708 [Google Scholar]
- 7. Bruce LJ, Daugulis AJ. 1991. Solvent selection strategies for extractive biocatalysis. Biotechnol. Prog. 7:116–124 [DOI] [PubMed] [Google Scholar]
- 8. Bühler B, Bollhalder I, Hauer B, Witholt B, Schmid A. 2003. Use of the two-liquid phase concept to exploit kinetically controlled multistep biocatalysis. Biotechnol. Bioeng. 81:683–694 [DOI] [PubMed] [Google Scholar]
- 9. Bühler B, Schmid A, Hauer B, Witholt B. 2000. Xylene monooxygenase catalyzes the multistep oxygenation of toluene and pseudocumene to corresponding alcohols, aldehydes, and acids in Escherichia coli JM101. J. Biol. Chem. 275:10085–10092 [DOI] [PubMed] [Google Scholar]
- 10. Canovas M, Torroglosa T, Iborra JL. 2005. Permeabilization of Escherichia coli cells in the biotransformation of trimethylammonium compounds into L-carnitine. Enzyme Microb. Technol. 37:300–308 [Google Scholar]
- 11. Chakrabarty AM, Chou G, Gunsalus IC. 1973. Genetic regulation of octane dissimilation plasmid in Pseudomonas. Proc. Natl. Acad. Sci. U. S. A. 70:1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen RRZ. 2007. Permeability issues in whole-cell bioprocesses and cellular membrane engineering. Appl. Microbiol. Biotechnol. 74:730–738 [DOI] [PubMed] [Google Scholar]
- 13. Cornelissen S, Liu S, Deshmukh AT, Schmid A, Bühler B. 2011. Cell physiology rather than enzyme kinetics can determine the efficiency of cytochrome P450-catalyzed C-H-oxyfunctionalization. J. Ind. Microbiol. Biotechnol. 38:1359–1370 [DOI] [PubMed] [Google Scholar]
- 14. de Bont JAM. 1998. Solvent-tolerant bacteria in biocatalysis. Trends Biotechnol. 16:493–499 [Google Scholar]
- 15. Denome SA, Stanley DC, Olson ES, Young KD. 1993. Metabolism of dibenzothiophene and naphthalene in Pseudomonas strains: complete DNA sequence of an upper naphthalene catabolic pathway. J. Bacteriol. 175:6890–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doig SD, Simpson H, Alphand V, Furstoss R, Woodley JM. 2003. Characterization of a recombinant Escherichia coli TOP10 [pQR239] whole-cell biocatalyst for stereoselective Baeyer-Villiger oxidations. Enzyme Microb. Technol. 32:347–355 [Google Scholar]
- 17. Duetz WA, van Beilen JB, Witholt B. 2001. Using proteins in their natural environment: potential and limitations of microbial whole-cell hydroxylations in applied biocatalysis. Curr. Opin. Biotechnol. 12:419–425 [DOI] [PubMed] [Google Scholar]
- 18. Eaton RW. 1994. Organization and evolution of naphthalene catabolic pathways: sequence of the DNA encoding 2-hydroxychromene-2-carboxylate isomerase and trans-o-hydroxybenzylidenepyruvate hydratase-aldolase from the NAH7 plasmid. J. Bacteriol. 176:7757–7762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eggink G, Lageveen RG, Altenburg B, Witholt B. 1987. Controlled and functional expression of the Pseudomonas oleovorans alkane utilizing system in Pseudomonas putida and Escherichia coli. J. Biol. Chem. 262:17712–17718 [PubMed] [Google Scholar]
- 20. Favre-Bulle O, Schouten T, Kingma J, Witholt B. 1991. Bioconversion of n-octane to octanoic acid by a recombinant Escherichia coli cultured in a two-liquid phase bioreactor. Biotechnology (NY) 9:367–371 [DOI] [PubMed] [Google Scholar]
- 21. Gibson J, Harwood SC. 2002. Metabolic diversity in aromatic compound utilization by anaerobic microbes. Annu. Rev. Microbiol. 56:345–369 [DOI] [PubMed] [Google Scholar]
- 22. Grant M, Woodley JM, Baganz F. 2011. Whole-cell bio-oxidation of n-dodecane using the alkane hydroxylase system of P. putida GPo1 expressed in E. coli. Enzyme Microb. Technol. 48:480–486 [DOI] [PubMed] [Google Scholar]
- 23. Gross R, Lang K, Buhler K, Schmid A. 2010. Characterization of a biofilm membrane reactor and its prospects for fine chemical synthesis. Biotechnol. Bioeng. 105:705–717 [DOI] [PubMed] [Google Scholar]
- 24. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 25. Hearn EM, Patel DR, Lepore BW, Indic M, van den Berg B. 2009. Transmembrane passage of hydrophobic compounds through a protein channel wall. Nature 458:367–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heipieper HJ, Neumann G, Cornelissen S, Meinhardt F. 2007. Solvent-tolerant bacteria for biotransformations in two-phase fermentation systems. Appl. Microbiol. Biotechnol. 74:961–973 [DOI] [PubMed] [Google Scholar]
- 27. Hong H, Patel DR, Tamm LK, van den Berg B. 2006. The outer membrane protein OmpW forms an eight-stranded beta-barrel with a hydrophobic channel. J. Biol. Chem. 281:7568–7577 [DOI] [PubMed] [Google Scholar]
- 28. Kahng HY, Byrne AM, Olsen RH, Kukor JJ. 2000. Characterization and role of tbuX in utilization of toluene by Ralstonia pickettii PKO1. J. Bacteriol. 182:1232–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kasai Y, Inoue J, Harayama S. 2001. The TOL plasmid pWW0 xylN gene product from Pseudomonas putida is involved in m-xylene uptake. J. Bacteriol. 183:6662–6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kellerhals MB, Hazenberg W, Witholt B. 1999. High cell density fermentations of Pseudomonas oleovorans for the production of mcl-PHAs in two-liquid phase media. Enzyme Microb. Technol. 24:111–116 [Google Scholar]
- 31. Kok M, et al. 1989. The Pseudomonas oleovorans alkBAC operon encodes two structurally related rubredoxins and an aldehyde dehydrogenase. J. Biol. Chem. 264:5442–5451 [PubMed] [Google Scholar]
- 32. Krop HB, van Velzen MJM, Parsons JR, Govers HAJ. 1997. Determination of environmentally relevant physical-chemical properties of some fatty acid esters. J. Am. Oil Chem. Soc. 74:309–315 [Google Scholar]
- 33. Kuhn D, Kholiq MA, Heinzle E, Bühler B, Schmid A. 2010. Intensification and economic and ecological assessment of a biocatalytic oxyfunctionalization process. Green Chem. 12:815–827 [Google Scholar]
- 34. Laane C, Boeren S, Vos K, Veeger C. 1987. Rules for optimization of biocatalysis in organic solvents. Biotechnol. Bioeng. 30:81–87 [DOI] [PubMed] [Google Scholar]
- 35. Laemmli UK. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 36. Leive L. 1974. The barrier function of the gram-negative envelope. Ann. N. Y. Acad. Sci. 235:109–129 [DOI] [PubMed] [Google Scholar]
- 37. Mathys RG, Schmid A, Witholt B. 1999. Integrated two-liquid phase bioconversion and product-recovery processes for the oxidation of alkanes: process design and economic evaluation. Biotechnol. Bioeng. 64:459–477 [DOI] [PubMed] [Google Scholar]
- 38. May SW, Katopodis AG. 1986. Oxygenation of alcohol and sulfide substrates by a prototypical non-heam iron monooxygenase: catalysis and biotechnological potential. Enzyme Microb. Technol. 8:17–21 [Google Scholar]
- 39. Miozzari GF, Niederberger P, Hutter R. 1978. Permeabilization of microorganisms by Triton X-100. Anal. Biochem. 90:220–233 [DOI] [PubMed] [Google Scholar]
- 40. Morrish JL, Brennan ET, Dry HC, Daugulis AJ. 2008. Enhanced bioproduction of carvone in a two-liquid-phase partitioning bioreactor with a highly hydrophobic biocatalyst. Biotechnol. Bioeng. 101:768–775 [DOI] [PubMed] [Google Scholar]
- 41. Neumann G, et al. 2005. Prediction of the adaptability of Pseudomonas putida DOT-T1E to a second phase of a solvent for economically sound two-phase biotransformations. Appl. Environ. Microbiol. 71:6606–6612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ni Y, Chen RR. 2004. Accelerating whole-cell biocatalysis by reducing outer membrane permeability barrier. Biotechnol. Bioeng. 87:804–811 [DOI] [PubMed] [Google Scholar]
- 43. Ni Y, Chen RR. 2005. Lipoprotein mutation accelerates substrate permeability-limited toluene dioxygenase-catalyzed reaction. Biotechnol. Prog. 21:799–805 [DOI] [PubMed] [Google Scholar]
- 44. Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nikaido H, Vaara M. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Noordman WH, Janssen DB. 2002. Rhamnolipid stimulates uptake of hydrophobic compounds by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 68:4502–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Panke S, Held M, Wubbolts MG, Witholt B, Schmid A. 2002. Pilot-scale production of (S)-styrene oxide from styrene by recombinant Escherichia coli synthesizing styrene monooxygenase. Biotechnol. Bioeng. 80:33–41 [DOI] [PubMed] [Google Scholar]
- 48. Panke S, Meyer A, Huber CM, Witholt B, Wubbolts MG. 1999. An alkane-responsive expression system for the production of fine chemicals. Appl. Environ. Microbiol. 65:2324–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Panke S, Witholt B, Schmid A, Wubbolts MG. 1998. Towards a biocatalyst for (S)-styrene oxide production: characterization of the styrene degradation pathway of Pseudomonas sp. strain VLB120. Appl. Environ. Microbiol. 64:2032–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Park JB. 2007. Oxygenase-based whole-cell biocatalysis in organic synthesis. J. Microbiol. Biotechnol. 17:379–392 [PubMed] [Google Scholar]
- 51. Salanitro JP, Wegener WS. 1971. Growth of Escherichia coli on short-chain fatty acids: growth characteristics of mutants. J. Bacteriol. 108:885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sambrook J, Russell DW. 2001. Molecular cloning—a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 53. Schewe H, Holtmann D, Schrader J. 2009. P450(BM-3)-catalyzed whole-cell biotransformation of alpha-pinene with recombinant Escherichia coli in an aqueous-organic two-phase system. Appl. Microbiol. Biotechnol. 83:849–857 [DOI] [PubMed] [Google Scholar]
- 54. Schneider S, Wubbolts MG, Sanglard D, Witholt B. 1998. Biocatalyst engineering by assembly of fatty acid transport and oxidation activities for in vivo application of cytochrome P-450BM-3 monooxygenase. Appl. Environ. Microbiol. 64:3784–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schrewe M, Magnusson A, Willrodt C, Bühler B, Schmid A. 2011. Kinetic analysis of terminal and unactivated C-H bond oxyfunctionalization in fatty acid methyl esters by monooxygenase-based whole-cell biocatalysis. Adv. Synth. Catal. 353:3485–3495 [Google Scholar]
- 56. Sikkema J, de Bont JA, Poolman B. 1994. Interactions of cyclic hydrocarbons with biological membranes. J. Biol. Chem. 269:8022–8028 [PubMed] [Google Scholar]
- 57. Sikkema J, de Bont JA, Poolman B. 1995. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 59:201–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Smits TH, Balada SB, Witholt B, van Beilen JB. 2002. Functional analysis of alkane hydroxylases from gram-negative and gram-positive bacteria. J. Bacteriol. 184:1733–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Smits TH, Seeger MA, Witholt B, van Beilen JB. 2001. New alkane-responsive expression vectors for Escherichia coli and Pseudomonas. Plasmid 46:16–24 [DOI] [PubMed] [Google Scholar]
- 60. Stutz EW, Defago G, Kern H. 1986. Naturally-occurring fluorescent pseudomonads involved in suppression of black root-rot of tobacco. Phytopathology 76:181–185 [Google Scholar]
- 61. Tamber S, Hancock REW. 2004. The outer membranes of pseudomonads, p 575–601 In Ramos JL. (ed), Pseudomonas, 1st ed, vol 1 Kluwer Academic/Plenum Publishers, New York, NY [Google Scholar]
- 62. Tewari YB, Miller MM, Wasik SP, Martire DE. 1982. Aqueous solubility and octanol/water partition coefficient of organic compounds at 25.0°C. J. Chem. Eng. Data 27:451–454 [Google Scholar]
- 63. Touw DS, Patel DR, van den Berg B. 2010. The crystal structure of OprG from Pseudomonas aeruginosa, a potential channel for transport of hydrophobic molecules across the outer membrane. PLoS One 5:e15016 doi:10.1371/journal.pone.0015016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Upadhya R, Nagajyothi Bhat SG. 2000. Stabilization of D-amino acid oxidase and catalase in permeabilized Rhodotorula gracilis cells and its application for the preparation of α-ketoacids. Biotechnol. Bioeng. 68:430–436 [DOI] [PubMed] [Google Scholar]
- 65. van Beilen JB, Eggink G, Enequist H, Bos R, Witholt B. 1992. DNA sequence determination and functional characterization of the OCT-plasmid-encoded alkJKL genes of Pseudomonas oleovorans. Mol. Microbiol. 6:3121–3136 [DOI] [PubMed] [Google Scholar]
- 66. van Beilen JB, Kingma J, Witholt B. 1994. Substrate specificity of the alkane hydroxylase system of Pseudomonas oleovorans GPo1. Enzyme Microb. Technol. 16:904–911 [Google Scholar]
- 67. van Beilen JB, Penninga D, Witholt B. 1992. Topology of the membrane-bound alkane hydroxylase of Pseudomonas oleovorans. J. Biol. Chem. 267:9194–9201 [PubMed] [Google Scholar]
- 68. van den Berg B. 2005. The FadL family: unusual transporters for unusual substrates. Curr. Opin. Struct. Biol. 15:401–407 [DOI] [PubMed] [Google Scholar]
- 69. van den Berg B. 2010. Going forward laterally: transmembrane passage of hydrophobic molecules through protein channel walls. Chembiochem 11:1339–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wagner F, et al. 1983. Production and chemical characterization of surfactants from Rhodococcus erythropolis and Pseudomonas sp. MUB grown on hydrocarbons, p 55–60 In Zajic IE, Cooper DG, Jack TR, Kosaric N. (ed), Microbial enhanced oil recovery. Penwell Books, Tulsa, OK [Google Scholar]
- 71. Wang Y, et al. 1995. Identification of a membrane protein and a truncated LysR-type regulator associated with the toluene degradation pathway in Pseudomonas putida F1. Mol. Gen. Genet. 246:570–579 [DOI] [PubMed] [Google Scholar]
- 72. Wubbolts MG, Favre-Bulle O, Witholt B. 1996. Biosynthesis of synthons in two-liquid-phase media. Biotechnol. Bioeng. 52:301–308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.