Abstract
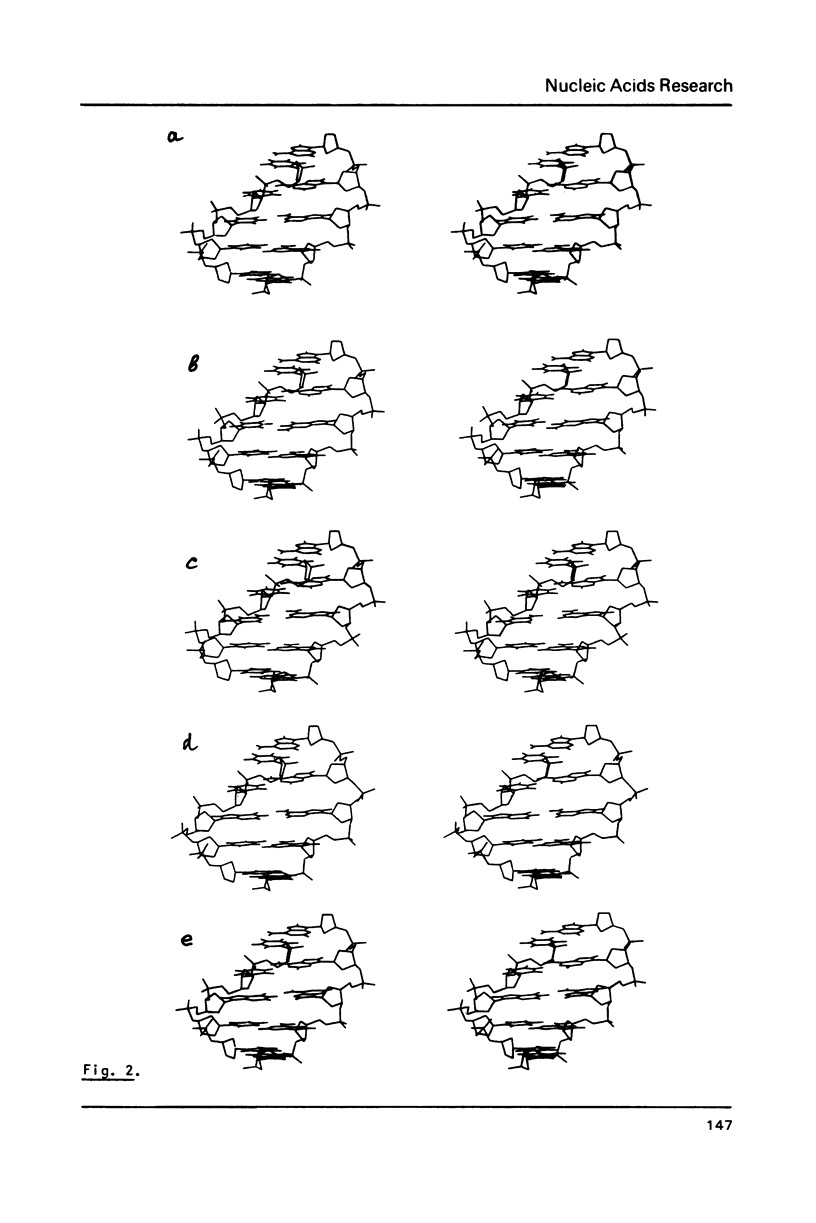
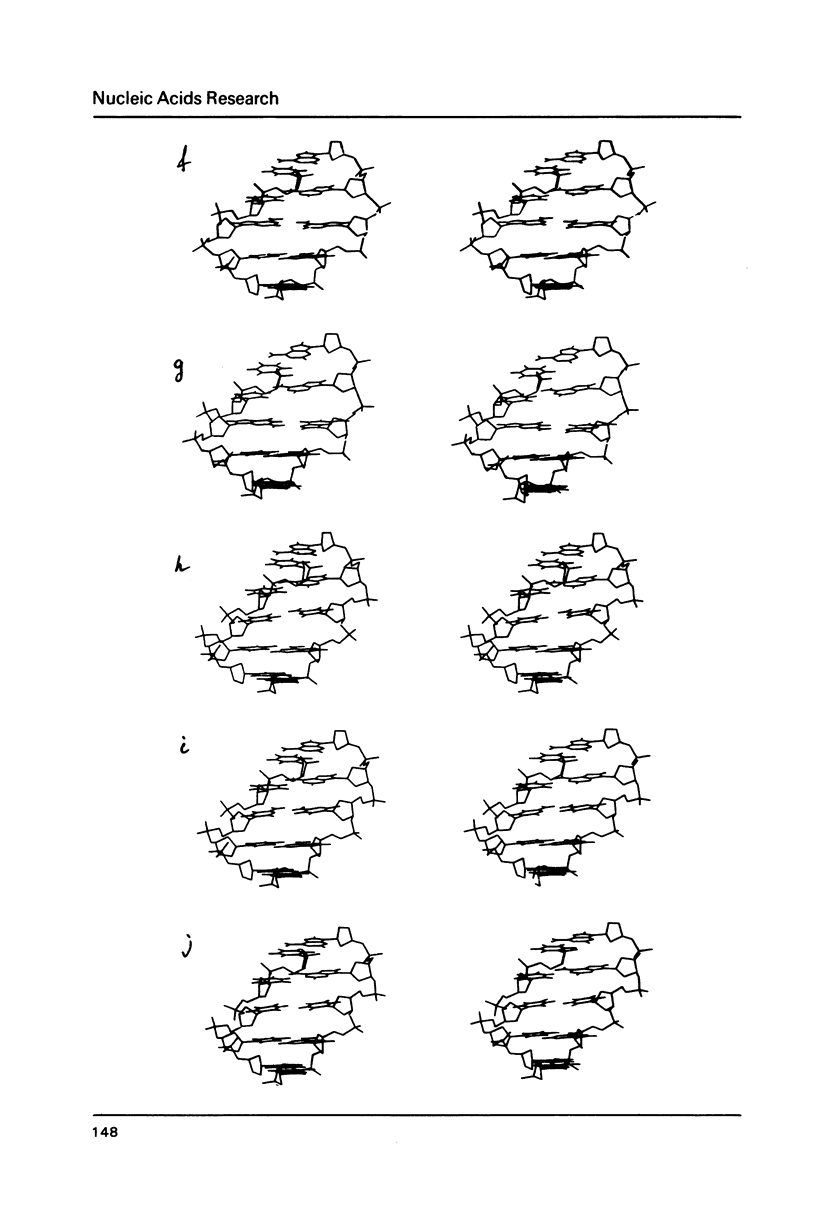
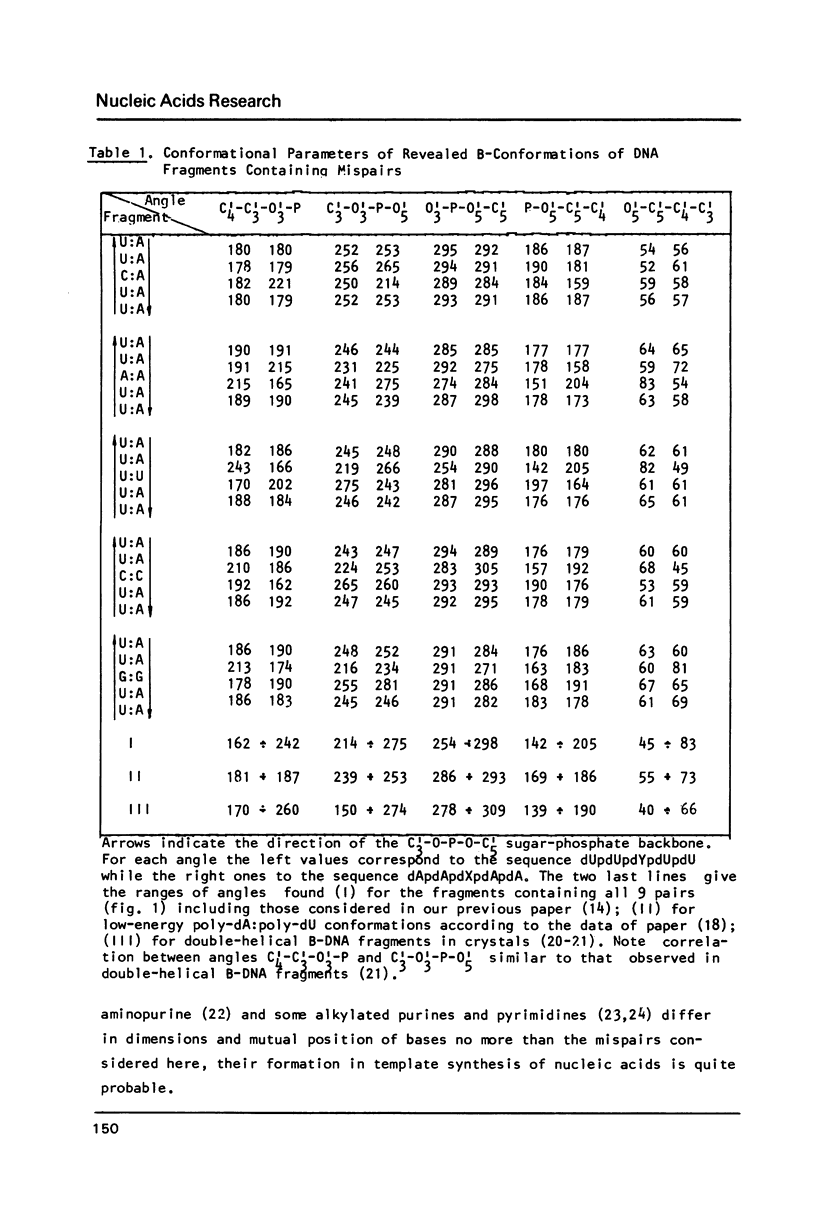
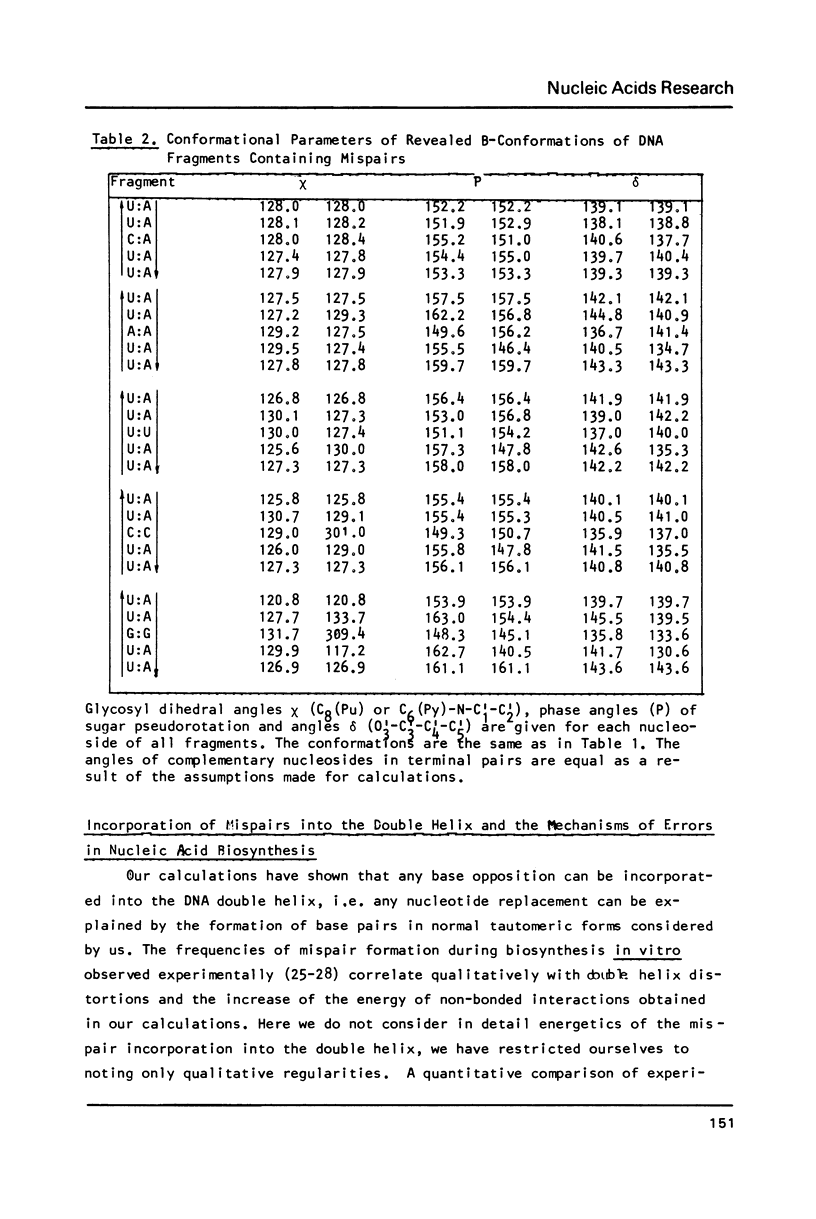
To explain biochemical and genetic data on spontaneous nucleotide replacements in nucleic acid biosynthesis all the 8 mispairs in normal tautomeric forms have been considered. Possible B-conformations of DNA fragments containing each of such mispairs incorporated between Watson-Crick pairs have been found using computations of the energy of non-bonded interactions via classical potential functions. These conformations have no reduced interatomic contacts. The values of each dihedral angle of the sugar-phosphate backbone fall within the limits of those of double-helical fragments of B-DNA in crystals. These values differ from those of the corresponding angles for the low-energy polynucleotide conformations consisting of canonical pairs by no more than 30 degrees (except for the fragment with the U:U pair for which the C4'-C3'-O-P angle differs by about 50 degrees). The difference in experimentally observed frequencies of various nucleotide replacements in DNA biosynthesis correlates with the difference in the energy of non-bonded interactions and with the extent of the sugar-phosphate backbone distortion for the fragments containing the mispairs which serve as intermediates for the replacements.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernardi F., Ninio J. The accuracy of DNA replication. Biochimie. 1978;60(10):1083–1095. doi: 10.1016/s0300-9084(79)80343-0. [DOI] [PubMed] [Google Scholar]
- Bruskov V. I., Poltev V. I. On molecular mechanisms of nucleic acid synthesis. Fidelity aspects: 2. Contribution of protein-nucleotide recognition. J Theor Biol. 1979 May 7;78(1):29–41. doi: 10.1016/0022-5193(79)90323-0. [DOI] [PubMed] [Google Scholar]
- Chuprina V. P., Poltev V. I. Possible conformations of double-helical polynucleotides containing incorrect base pairs. Nucleic Acids Res. 1983 Aug 11;11(15):5205–5222. doi: 10.1093/nar/11.15.5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W., Baltz R. H. The biochemistry of mutagenesis. Annu Rev Biochem. 1976;45:11–37. doi: 10.1146/annurev.bi.45.070176.000303. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Wing R. M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R., Knill-Jones J. W. Fidelity of replication of bacteriophage phi X174 DNA in vitro and in vivo. J Mol Biol. 1983 Apr 25;165(4):633–654. doi: 10.1016/s0022-2836(83)80271-x. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Shi J. P., Tsui W. C. Kinetics of base misinsertion by DNA polymerase I of Escherichia coli. J Mol Biol. 1983 Apr 25;165(4):655–667. doi: 10.1016/s0022-2836(83)80272-1. [DOI] [PubMed] [Google Scholar]
- Grosse F., Krauss G., Knill-Jones J. W., Fersht A. R. Accuracy of DNA polymerase-alpha in copying natural DNA. EMBO J. 1983;2(9):1515–1519. doi: 10.1002/j.1460-2075.1983.tb01616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Orgel L. E. Oligomerization of (guanosine 5'-phosphor)-2-methylimidazolide on poly(C). An RNA polymerase model. J Mol Biol. 1982 Nov 25;162(1):201–217. doi: 10.1016/0022-2836(82)90169-3. [DOI] [PubMed] [Google Scholar]
- Kan L. S., Chandrasegaran S., Pulford S. M., Miller P. S. Detection of a guanine X adenine base pair in a decadeoxyribonucleotide by proton magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4263–4265. doi: 10.1073/pnas.80.14.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Meyer R. R., Loeb L. A. Single-strand binding protein enhances fidelity of DNA synthesis in vitro. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6331–6335. doi: 10.1073/pnas.76.12.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb L. A., Kunkel T. A. Fidelity of DNA synthesis. Annu Rev Biochem. 1982;51:429–457. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Dallas J., Itakura K., Breslauer K. J. Structure, dynamics, and energetics of deoxyguanosine . thymidine wobble base pair formation in the self-complementary d(CGTGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):437–444. doi: 10.1021/bi00532a003. [DOI] [PubMed] [Google Scholar]
- Poltev V. I., Bruskov V. I. O molekuliarnykh mekhanizmakh spontannykh transversiii i tranzitsii. Mol Biol (Mosk) 1977 May-Jun;11(3):661–670. [PubMed] [Google Scholar]
- Poltev V. I., Bruskov V. I. On molecular mechanisms of nucleic acid synthesis fidelity aspects. 1. Contribution of base interactions. J Theor Biol. 1978 Jan 7;70(1):69–83. doi: 10.1016/0022-5193(78)90303-x. [DOI] [PubMed] [Google Scholar]
- Poltev V. I., Shuliupina N. V., Bruskov V. I. Molekuliarnye mekhanizmy oshibok biosinteza nukleinovykh kislot, indutsirovannykh alkilirovannem azotistykh osnovanii. Mol Biol (Mosk) 1981 Nov-Dec;15(6):1286–1294. [PubMed] [Google Scholar]
- Quigley G. J., Seeman N. C., Wang A. H., Suddath F. L., Rich A. Yeast phenylalanine transfer RNA: atomic coordinates and torsion angles. Nucleic Acids Res. 1975 Dec;2(12):2329–2341. doi: 10.1093/nar/2.12.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankoff D., Cedergren R. J., Lapalme G. Frequency of insertion-deletion, transversion, and transition in the evolution of 5S ribosomal RNA. J Mol Evol. 1976 Mar 29;7(2):133–149. doi: 10.1007/BF01732471. [DOI] [PubMed] [Google Scholar]
- Sinha N. K., Haimes M. D. Molecular mechanisms of substitution mutagenesis. An experimental test of the Watson-Crick and topal-fresco models of base mispairings. J Biol Chem. 1981 Oct 25;256(20):10671–10683. [PubMed] [Google Scholar]
- Topal M. D., Fresco J. R. Complementary base pairing and the origin of substitution mutations. Nature. 1976 Sep 23;263(5575):285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953 May 30;171(4361):964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]


