Atg9-containing vesicles assemble to the preautophagosomal structure and eventually are incorporated into the autophagosomal outer membrane.
Abstract
During the process of autophagy, cytoplasmic materials are sequestered by double-membrane structures, the autophagosomes, and then transported to a lytic compartment to be degraded. One of the most fundamental questions about autophagy involves the origin of the autophagosomal membranes. In this study, we focus on the intracellular dynamics of Atg9, a multispanning membrane protein essential for autophagosome formation in yeast. We found that the vast majority of Atg9 existed on cytoplasmic mobile vesicles (designated Atg9 vesicles) that were derived from the Golgi apparatus in a process involving Atg23 and Atg27. We also found that only a few Atg9 vesicles were required for a single round of autophagosome formation. During starvation, several Atg9 vesicles assembled individually into the preautophagosomal structure, and eventually, they are incorporated into the autophagosomal outer membrane. Our findings provide conclusive linkage between the cytoplasmic Atg9 vesicles and autophagosomal membranes and offer new insight into the requirement for Atg9 vesicles at the early step of autophagosome formation.
Introduction
Autophagy is a major cellular process, conserved in eukaryotic cells, that mediates bulk degradation of cytoplasmic proteins and organelles in response to starvation (Ohsumi, 2001; Klionsky, 2007; Nakatogawa et al., 2009). Autophagy is also involved in various cellular functions, including development, intracellular quality control, tumor suppression, and stress responses (Levine and Klionsky, 2004; Mizushima, 2005; Rubinsztein, 2006; Mizushima and Levine, 2010). Upon induction of autophagy, a cup-shaped membrane structure called the isolation membrane emerges in the cytoplasm, expands to sequester the cytoplasmic materials, and finally, seals to become a double membrane–bound autophagosome, which fuses with the lytic compartment, either a vacuole or lysosome (Levine and Klionsky, 2004; Klionsky, 2005; Nakatogawa et al., 2009).
Previous studies using yeast have identified >30 autophagy-related (Atg) proteins required for several types of autophagy and provided various insights into the molecular basis of autophagosome formation (Ohsumi, 2001; Klionsky, 2005; Nakatogawa et al., 2009). However, the most intriguing issues in autophagy remain to be elucidated, namely, the origin of the autophagosomal membranes and the molecular mechanisms underlying autophagosome formation.
Among the Atg proteins, Atg9 is the sole multispanning membrane protein essential for autophagosome formation (Lang et al., 2000; Noda et al., 2000; Young et al., 2006). In yeast cells, Atg9-GFP can be observed in several punctate structures (Noda et al., 2000; Reggiori et al., 2004), which are postulated to play a key role in the delivery of lipids required for autophagosome formation. To date, however, these puncta have not been well characterized, and their involvement in autophagosome formation at the preautophagosomal structure (PAS) is still poorly understood. Furthermore, the intracellular behavior of these puncta remains controversial: some studies reported that the Atg9 puncta are adjacent to mitochondria (Reggiori et al., 2005a; Mari et al., 2010), whereas others reported that the Atg9 puncta are somewhat mobile in the cytoplasm but not ordinarily adjacent to mitochondria (Sekito et al., 2009; Ohashi and Munro, 2010).
In this study, we developed a high-sensitivity microscopy system that allowed us to precisely analyze the behavior of Atg9-2×GFP. Using this system, we observed that most Atg9 is present on single-membrane vesicles that move the cytoplasm. Our findings will help settle the controversy concerning the intracellular behavior of Atg9 and shed light on the involvement of Atg9-containing vesicles in the process of autophagosome formation.
Results
Atg9-containing structures are highly mobile in the cytoplasm
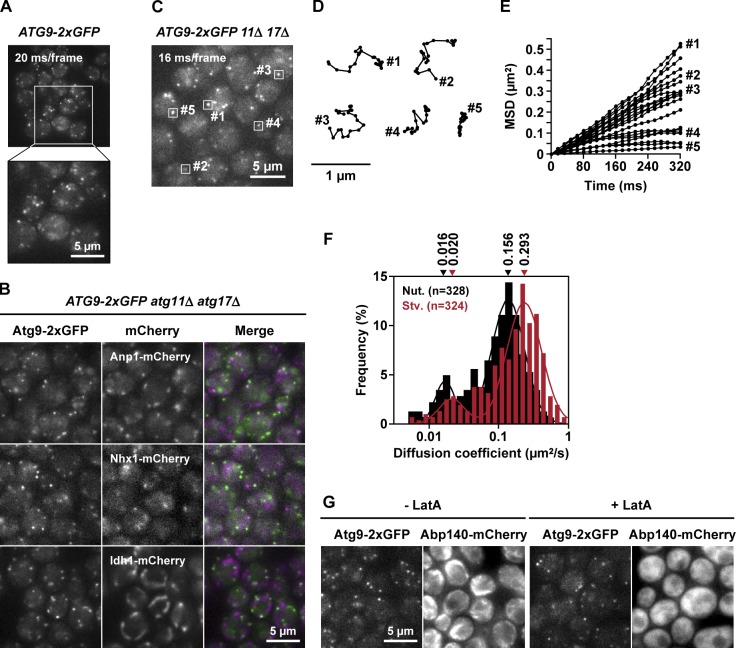
In yeast cells, Atg9-GFP has been observed on punctate structures in the cytoplasm; however, to date, the motion of these puncta has only been analyzed at relatively low temporal resolutions (>200 ms/frame; Reggiori et al., 2005a; Sekito et al., 2009; Mari et al., 2010; Ohashi and Munro, 2010) that may be insufficient to observe more rapid dynamic behavior. Therefore, we developed a high-sensitivity microscopy system (see Materials and methods) that enabled us to observe the Atg9 puncta with high temporal resolution (10–20 ms/frame). Data obtained using this system clearly revealed that Atg9-2×GFP, which is chromosomally expressed via its own promoter (Fig. S1, A and B), forms numerous punctate structures, most of which are highly mobile in the cytoplasm (Fig. 1 A and Video 1). Furthermore, the localization and motion of these puncta differ from those of the Golgi apparatus, endosomes, and mitochondria (Fig. 1 B); although the Golgi apparatus, endosomes, and mitochondria were immobile within the time span of our observations (unpublished data), the large majority of Atg9 puncta was highly mobile (Video 1).
Figure 1.
Atg9-containing structures are observed by high temporal resolution microscopy and analyzed by single-particle tracking. (A) ATG9-2×GFP cells were treated with rapamycin for 2 h and observed by fluorescence microscopy at 20 ms/frame (see also Video 1). Magnified view of the boxed area is shown. (B) ATG9-2×GFP atg11Δ atg17Δ cells were treated with rapamycin for 1 h and observed at 30 ms/frame. Anp1-mCherry (Golgi), Nhx1-mCherry (endosome), and Idh1-mCherry (mitochondria) were used as organelle markers. Green fluorescence and red fluorescence were acquired concurrently. (C) ATG9-2×GFP atg11Δ atg17Δ cells were observed at 16 ms/frame and subjected to single-particle tracking analysis. (D) Trajectories of the Atg9 puncta indicated in C. (E) 20 examples of the mean square displacement (MSD) curves calculated from traces of Atg9 puncta. (F) Histograms of the diffusion coefficients of the Atg9 puncta observed in cells grown to logarithmic phase (nutrient [Nut.]) or cells starved for 2 h (Stv.). The histograms were fitted to Gaussian distributions; medians of the fitting curves are indicated. The data shown are from a single representative experiment out of two repeats. (G) ATG9-2×GFP ABP140-mCherry cells were treated with 100 µg/ml latrunculin A (LatA) for 20 min and observed by fluorescence microscopy at 32 ms/frame (see also Video 2).
Next, we analyzed the mobility of Atg9 puncta in atg11Δ atg17Δ cells, in which the PAS (Suzuki et al., 2001) is not formed because of the absence of scaffold proteins (Cheong et al., 2008; Kawamata et al., 2008). We collected >300 trajectories by single-particle tracking and subjected them to statistical analysis. Diffusion coefficients were calculated from the mean square displacement (MSD) curves (Fig. 1, C–E). The mobility of Atg9 puncta could be roughly classified into two groups: a large population with high mobility and a very small population with low mobility (Fig. 1 F). These observations confirmed that most Atg9 resides on highly mobile homogeneous structures. In addition, the MSD curves indicated in Figs. 1 E and S1 C could fit with the liner function, which means free diffusion of particles (Anderson et al., 1992), suggesting that the large majority of mobile Atg9 puncta freely diffused within the cytoplasm. Although actin cytoskeleton has been reported to be involved in the dynamics of Atg9 movement (Reggiori et al., 2005b; Monastyrska et al., 2008), the motion of Atg9 puncta was not altered even after the treatment with the actin-depolymerizing reagent latrunculin A (Fig. 1 G and Video 2). Atg9 might interact only transiently with the actin cytoskeleton.
Atg9 exists on 30–60-nm single-membrane vesicles
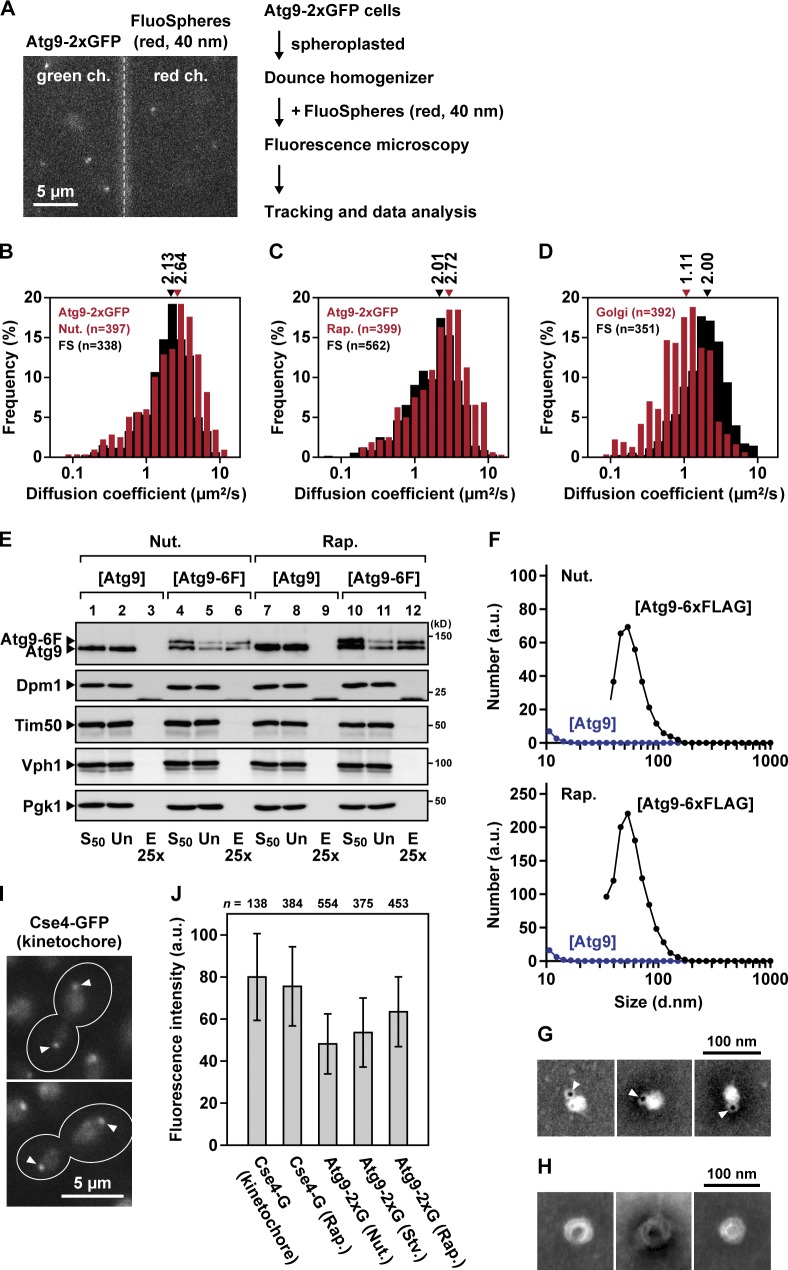
To estimate the size of the Atg9 puncta, their diffusion coefficients in cell homogenates were compared with those of monosized microbeads FluoSpheres (Fig. 2 A). The diffusion coefficients of FluoSpheres (44.1 nm in diameter; Fig. S1, D and E) were 2.00–2.13 µm2/s (Fig. 2, B–D), and those of the Atg9 puncta prepared from growing cells and rapamycin-treated cells were 2.64 and 2.72 µm2/s, respectively (Fig. 2, B and C). (The TOR [target of rapamycin] kinase inhibitor rapamycin was used to induce a starvation-like condition.) We further observed that the diffusion coefficients of Atg9 puncta were obviously different from those of the organelles, such as the Golgi apparatus (Figs. 2 D and S1 F). The diffusion coefficient is inversely proportional to the diameter of particles according to the Stokes–Einstein relation (Elsner et al., 2003). Therefore, we estimated that the mean diameters of Atg9 puncta prepared from growing cells and rapamycin-treated cells were 35.6 and 32.6 nm, respectively, by using the diameter of FluoSpheres as a standard.
Figure 2.
Atg9-containing structures are small single-membrane vesicles. (A) Mobility of Atg9 puncta in vitro. ATG9-2×GFP atg11Δ atg17Δ pep4Δ cells and ANP1-GFP atg11Δ atg17Δ pep4Δ cells were spheroplasted, ruptured with a Dounce homogenizer, and centrifuged at 15,000 g for 15 min. The supernatant fraction was mixed with 40-nm red FluoSpheres and then observed as in Fig. 1 C. Green fluorescence (green channel [ch.]) and red fluorescence (red channel [ch.]) were acquired concurrently. (B–D) Histograms of the diffusion coefficients of Atg9 puncta prepared from growing cells (B) or rapamycin-treated cells (C) and of the Golgi protein Anp1-GFP (D) are shown. FS, FluoSpheres. The mean size of the FluoSpheres used in this study was 44.1 nm (Fig. S1 D). The histograms were fitted to Gaussian distributions; medians of the fitting curves are indicated at the top. The data shown are from a single representative experiment out of three repeats. (E) Immunoisolation of Atg9-containing structures. atg11Δ atg17Δ pep4Δ cells expressing Atg9-6×FLAG were converted to spheroplasts (nutrient [Nut.]) and then treated with rapamycin for 2 h (Rap.). The spheroplasts were ruptured by passage through membrane filters with 5-µm pores. After a centrifugation at 50,000 g for 15 min, the supernatants (S50) were subjected to immunoisolation using the anti-FLAG antibody. The bound materials were eluted with 3×FLAG peptide and subjected to immunoblotting using antibodies against Atg9, Dpm1 (ER), Tim50 (mitochondria), Vph1 (vacuole), and Pgk1 (cytoplasm). Un, unbound fractions; E25×, eluted fractions concentrated 25-fold. (F) Size distribution profiles of the isolated Atg9-containing structures. The eluted fractions of E were subjected to DLS measurement. (G and H) Atg9-containing structures were immunoisolated from cells expressing both Atg9-6×FLAG and Atg9-3×BAP (3× biotinylated tag) and then subjected to negative staining EM. In G, the Atg9-containing structures were labeled with streptavidin-conjugated Qdots (Invitrogen). Arrowheads indicate Qdots. (I) CSE4-GFP cells were observed at 30 ms/frame. Arrowheads indicate kinetochore clusters consisting of ∼80 molecules of Cse4-GFP. Outlines indicate the edges of cells. (J) ATG9-2×GFP atg11Δ atg17Δ cells and CSE4-GFP cells were grown to logarithmic phase (nutrient) and then either starved for 2 h (Stv.) or treated with rapamycin for 2 h (Rap.). The fluorescence intensities of each GFP cluster (Atg9-2×GFP vesicle or Cse4-GFP kinetochore cluster) were quantified. Background signals of cytoplasmic regions were subtracted. The mean intensity of the Cse4-GFP kinetochore cluster is set to 80 U. Error bars indicate standard deviation. a.u., arbitrary unit.
In addition, we immunoisolated Atg9 puncta using anti-FLAG beads and analyzed them by dynamic light scattering (DLS). By the immunoisolation, Atg9-6×FLAG was recovered in eluted fractions together with nontagged Atg9 (Fig. 2 E, lanes 6 and 12), whereas nontagged Atg9 alone was not (Fig. 2 E, lanes 3 and 9). The DLS measurement revealed that the isolated Atg9 puncta were present in a single peak at 50–60 nm (Fig. 2 F). Next, the isolated Atg9 puncta were labeled with an electron-dense probe, Qdot, and analyzed by EM. In the EM images, we could observe spherical structures with a diameter of 30–50 nm attached with Qdots (Fig. 2 G); these Atg9-containing structures appeared to be single-membrane vesicles (Fig. 2 H). From the single-particle tracking, DLS, and EM analyses, we conclude that the cytoplasmic mobile Atg9 puncta are single-membrane vesicles with a diameter of 30–60 nm; hereafter, we designate them Atg9 vesicles.
We also quantitated the number of Atg9-2×GFP molecules on each Atg9 vesicle, using Cse4-GFP as a standard (Joglekar et al., 2006; Coffman et al., 2011; Lawrimore et al., 2011). A centromeric nucleosome contains approximately five molecules of Cse4-GFP; thus, a single kinetochore cluster represents ∼80 molecules of Cse4-GFP (Fig. 2 I). The fluorescence intensities of the Atg9-2×GFP vesicles in growing, starved, and rapamycin-treated cells corresponded to 48, 54, and 63 GFP units, respectively (Fig. 2 J), suggesting that each Atg9 vesicle contains 24–32 molecules of Atg9-2×GFP.
Atg9 vesicles are generated de novo during starvation
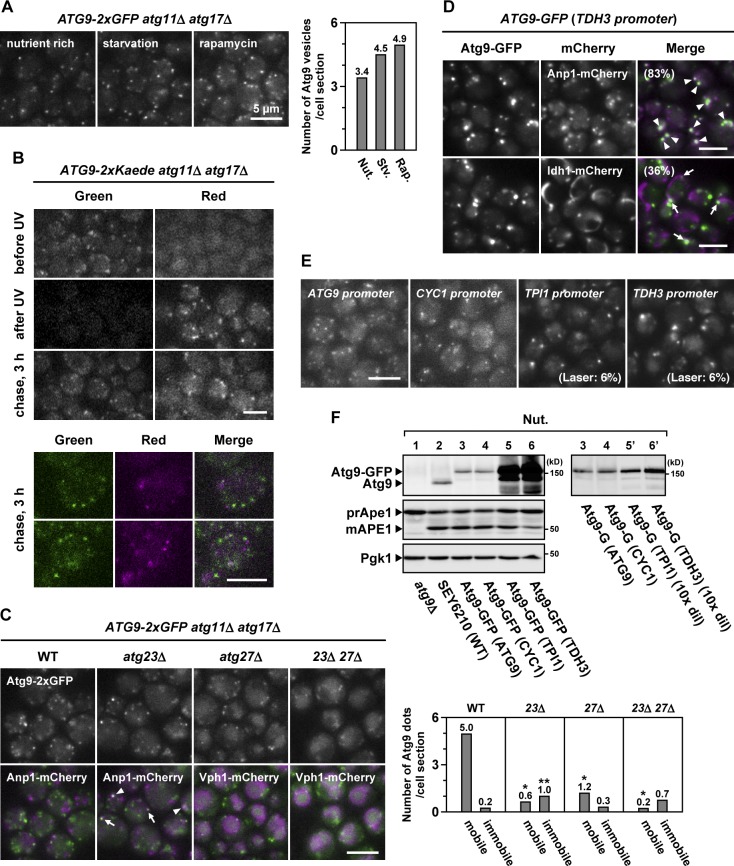
We also found that the Atg9 vesicles increased in number during starvation or rapamycin treatment (Fig. 3 A). To analyze the biogenesis of Atg9 vesicles, Atg9-2×Kaede cells were treated with rapamycin and UV irradiated to convert Kaede fluorescence from green to red and then subjected to chase incubation (Fig. 3 B, after UV). During the chase incubation, Atg9 vesicles were newly generated and visualized with green fluorescence. Their distribution was distinct from that of the preexisting (red) Atg9 vesicles (Fig. 3 B, chase). These results indicate that Atg9 vesicles are generated de novo during rapamycin treatment. Western blot analysis also revealed that Atg9 expression was up-regulated after treatment with rapamycin (Fig. S3 A). The increased synthesis of Atg9 vesicles during starvation or rapamycin treatment could contribute to up-regulation of autophagic activity.
Figure 3.
Atg9 vesicles are generated de novo during starvation. (A) ATG9-2×GFP atg11Δ atg17Δ cells were either starved for 2 h or treated with rapamycin for 2 h. The number of Atg9 vesicles per cell section was counted. This experiment was completed once (n > 135). Nut., nutrient; Stv., starved; Rap., rapamycin. (B) ATG9-2×Kaede atg11Δ atg17Δ cells were treated with rapamycin for 1 h (before UV) and then irradiated with 365-nm UV (after UV). The cells were subjected to chase incubation for 3 h (chase, 3 h). (C) ATG9-2×GFP atg11Δ atg17Δ cells lacking Atg23 and/or Atg27 were treated with rapamycin for 3 h (see also Video 5). Anp1-mCherry (Golgi) and Vph1-mCherry (vacuole) were used as organelle markers. Arrowheads and arrows indicate Atg9-GFP clusters accumulated at and adjacent to the Golgi apparatus, respectively. The number of mobile Atg9 dots and immobile Atg9 dots per cell section was counted. This experiment was completed once (n > 250). Single asterisks indicate, in these mutant cells, very small mobile Atg9 dots that were observed but not counted. The double asterisk indicates, in atg23Δ cells, immobile Atg9 dots that were occasionally located at or adjacent to the Golgi apparatus (42%). (D) Cells expressing Atg9-GFP via the TDH3 promoter were observed at 32 ms/frame. Anp1-mCherry (Golgi) and Idh1-mCherry (mitochondria) were used as organelle markers. Arrowheads and arrows indicate immobile Atg9-GFP clusters adjacent to the Golgi apparatus and mitochondria, respectively. (E) Cells expressing Atg9-GFP via the ATG9 promoter, CYC1 promoter, TPI1 promoter, or TDH3 promoter were observed at 32 ms/frame (see also Video 6). When Atg9-GFP was expressed via the TPI1 promoter or TDH3 promoter, the excitation laser intensity was lowered to 6% with a neutral density filter. (F) Total cell lysates were prepared from the cells used in E and subjected to immunoblotting. Some samples were diluted 10-fold (10× dil). WT, wild type.
Atg9 vesicles are derived from the Golgi apparatus in a process involving Atg23 and Atg27
Previous studies have reported that Atg9-containing structures are generated via the Golgi-related secretory pathway (Mari et al., 2010; Ohashi and Munro, 2010). We confirmed that Atg9 was partially accumulated at the trans-Golgi network in sec7 mutant cells (Fig. S2 and Videos 3 and 4). In addition, we observed that the Atg9 vesicles are generated normally in vps17Δ cells and snx4/atg24Δ cells (unpublished data), which are deficient for retrograde trafficking from the endosomes (Seaman, 2008). These data suggest that endosome to Golgi trafficking is not required for the generation of Atg9 vesicles and support the model that the Atg9 vesicles are derived from the Golgi apparatus.
Next, we investigated the biogenesis of Atg9 vesicles in various atg mutant cells. Atg9 vesicles were generated normally in the absence of Atg1, Atg2, Atg7, Atg8, or Atg14 (unpublished data), suggesting that these Atg proteins essential for autophagosome formation are not required for the biogenesis of Atg9 vesicles. In contrast, cytoplasmic mobile Atg9 vesicles were severely reduced in atg23Δ or atg27Δ cells (Figs. 3 C and S3, A–D; and Video 5). In atg23Δ cells, immobile Atg9-containing structures were frequently observed at or near the Golgi apparatus (42%; Fig. 3 C). In contrast, in atg27Δ cells and atg23Δ atg27Δ cells, Atg9-2×GFP was mislocalized into the vacuolar lumen (Figs. 3 C and S3, E and F), probably via the multivesicular body pathway. Consistent with this, in atg23Δ atg27Δ vps4Δ cells defective of the endosomal sorting complex required for transport pathway (Babst et al., 1997), Atg9-2×GFP accumulated at the endosomes but not in the vacuolar lumen (Fig. S3 E), and the processed GFP moiety was not formed (Fig. S3 F, lanes 4–6). These mislocalizations could be caused by missorting of Atg9 in the process of vesicle formation via the Golgi apparatus. Although Atg23 and Atg27 have been proposed to be involved in the assembly of Atg9 to the PAS (Legakis et al., 2007; Yen et al., 2007), our findings suggest instead that Atg23 and Atg27 are involved in the biogenesis of Atg9 vesicles.
Overexpressed Atg9 aberrantly accumulates at the Golgi apparatus
Recently, novel Atg9-containing tubulovesicular structures (Atg9 reservoirs) have been proposed to exist (Mari et al., 2010). In cells expressing Atg9-GFP via the TPI1 promoter, the Atg9 reservoirs have been observed adjacent to mitochondria (in these cells, Atg9-GFP was expressed eightfold higher than endogenous Atg9; Mari et al., 2010). However, the reported features of the Atg9 reservoirs were significantly inconsistent with our observations. Furthermore, we have noticed that when Atg9-GFP is overexpressed via the TDH3 promoter, immobile Atg9-containing structures were observed frequently at the Golgi apparatus (83%; Figs. 3 D and S4 A) and occasionally adjacent to mitochondria (36%; Fig. 3 D) and endosomes (Fig. S4 A). Therefore, the TPI1 promoter was used to express Atg9-GFP, and we found that immobile Atg9-containing structures, which are similar to those observed when Atg9 was overexpressed under control of the TDH3 promoter, were formed in these cells (Fig. 3 E and Video 6). These aberrant accumulations were likely caused by excess expression of Atg9 (Fig. 3 F) but not by expression via the exogenous promoter because the Atg9 vesicles were substantially generated with low expression via the CYC1 promoter, and such aberrant accumulations were not observed (Video 6). We also found that, even in cells overexpressing Atg9-GFP, a significant number of mobile Atg9 vesicles were still observed in addition to the immobile Atg9 clusters accumulated at the Golgi apparatus (Fig. S4 B), which may explain why these cells exhibit normal autophagic activities (Fig. S4 C; Mari et al., 2010). These results suggest that overproduction of Atg9 leads to the aberrant accumulation of Atg9-containing structures at the Golgi apparatus; therefore, we should use the endogenous promoter to express Atg9-2×GFP for characterization of Atg9 vesicles (and Atg9-containing structures).
Cytoplasmic Atg9 vesicles individually assemble to the PAS
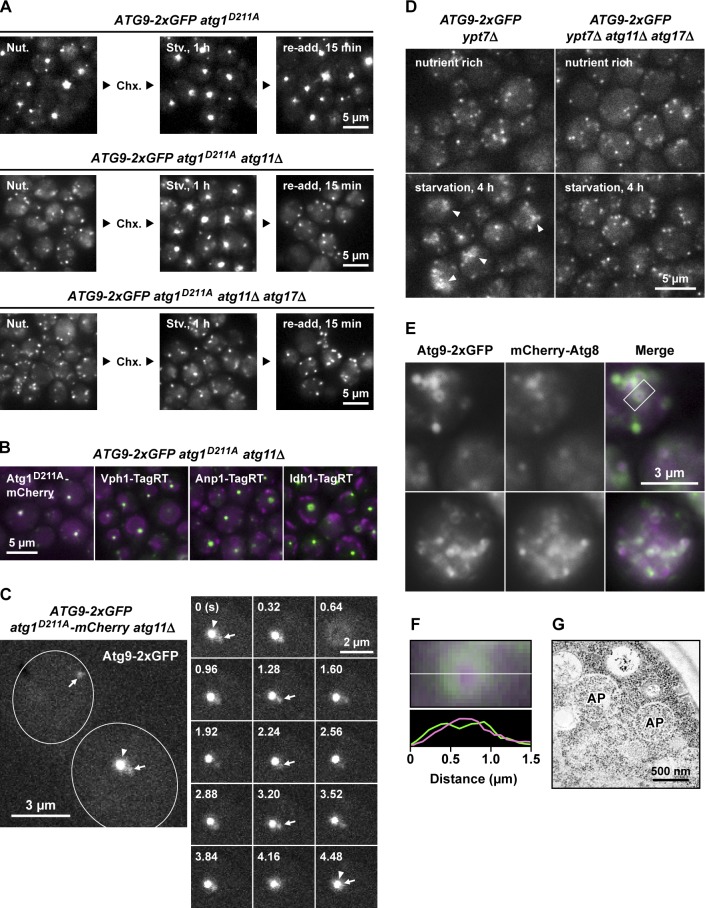
To determine whether the Atg9 vesicle itself possesses the ability to localize to the PAS, we analyzed the behavior of Atg9 vesicles in atg1D211A atg11Δ cells, in which the PAS is not formed under nutrient-rich conditions (Kawamata et al., 2008). In response to starvation of these cells, the large majority of Atg9 vesicles accumulated at the PAS (Figs. 4 A, middle; and S5 A) because of the defect in Atg1 kinase activity (Shintani and Klionsky, 2004; Sekito et al., 2009). According to the assembly at the PAS, the number of cytoplasmic mobile Atg9 vesicles significantly decreased (Figs. 4 A, middle; and S5 B; and Video 7). Furthermore, in response to readdition of nutrients, the accumulated Atg9 vesicles dispersed immediately, and mobile Atg9 vesicles were again observed (Fig. 4 A, middle). In contrast, in atg11Δ atg17Δ cells deficient for PAS formation, Atg9 vesicles were still mobile even under starvation conditions, and no clusters of them were formed (Figs. 4 A, bottom; and S5 B; and Video 7). These observations explain the transition of Atg9 vesicles from the cytoplasmic pool to the PAS (Reggiori et al., 2004) and suggest that the cytoplasmic Atg9 vesicles assemble individually at the PAS. Furthermore, we occasionally observed in time-lapse videos that a single Atg9 punctum, with fluorescence intensity comparable to a single Atg9 vesicle but smaller than that of a cluster of numerous Atg9 vesicles, such as the Atg9 reservoirs, localized to the PAS (Fig. 4 C and Video 8). This observation also supports the model that the Atg9 vesicles are able to assemble individually at the PAS.
Figure 4.
Atg9 localizes around the autophagosome in ypt7Δ cells. (A) ATG9-2×GFP atg1D211A cells (top), ATG9-2×GFP atg1D211A atg11Δ VPH1-TagRFPT cells (middle), and ATG9-2×GFP atg1D211A atg11Δ atg17Δ cells (bottom) were starved for 1 h (Stv., 1 h) in the presence of 1 mg/ml cycloheximide (Chx.). After starvation, nutrient-rich medium was added to the culture and incubated for 15 min (re-add, 15 min). Nut., nutrient. (B) ATG9-2×GFP atg1D211A atg11Δ cells were treated with rapamycin for 1 h. Atg1D211A-mCherry (PAS), Vph1-TagRFPT (vacuole), Anp1-TagRFPT (Golgi), and Idh1-TagRFPT (mitochondria) were used as organelle markers. The Atg9-2×GFP clusters observed in these cells represent the PAS (labeled with Atg1D211A-mCherry) but not clusters accumulated at the Golgi apparatus (labeled with Anp1-TagRFPT). (C) Cytoplasmic Atg9 vesicles individually assemble to the PAS. ATG9-2×GFP atg1D211A-mCherry atg11Δ cells were treated with rapamycin for 1 h and observed at 32 ms/frame (see also Video 8). Arrows and arrowheads indicate cytoplasmic mobile Atg9 vesicles and Atg9 clusters assembled at the PAS, respectively. Outlines indicate the edges of cells. (D) Intracellular behavior of Atg9-2×GFP is altered in a manner dependent on autophagosome formation. ATG9-2×GFP ypt7Δ cells and ATG9-2×GFP ypt7Δ atg11Δ atg17Δ cells were starved for 4 h and observed at 32 ms/frame (see also Video 9). Arrowheads indicate immobile Atg9-positive structures. (E) Atg9 localizes around the autophagosome. ATG9-2×GFP mCherry-Atg8 ypt7Δ atg11Δ cells were starved for 4 h and observed at 3,000 ms/frame. The rectangle indicates the autophagosome analyzed in F. (F) The autophagosome highlighted in E was analyzed using the line scan analysis function of MetaMorph software. (G) The cells used in E were starved for 4 h and subjected to EM. AP, autophagosomes.
Atg9 localizes around the autophagosome in ypt7Δ cells
To address how the Atg9 vesicles contribute to autophagosome formation, we investigated the relationship between the cytoplasmic Atg9 vesicles and autophagosomes. To this end, we analyzed the distribution of Atg9 in ypt7Δ cells, in which autophagosomes accumulate in the cytoplasm (Kim et al., 1999; Ishihara et al., 2001). Under nutrient-rich conditions, Atg9 vesicles were generated normally in the absence of Ypt7; however, after prolonged starvation, immobile Atg9-positive structures were formed concomitant with a decrease in the number of cytoplasmic mobile Atg9 vesicles (Fig. 4 D and Video 9). These immobile structures were not observed in ypt7Δ atg11Δ atg17Δ cells (Fig. 4 D and Video 9). In long-exposure observations, these structures could be visualized as ring-shaped objects surrounding the autophagosomal content marker mCherry-Atg8 (Fig. 4 E). The ring-shaped structure highlighted in Fig. 4 E was ∼500 nm in diameter, consistent with the size of the autophagosomes observed by EM (Fig. 4, F and G). In addition, the ring-shaped Atg9 signals were not dispersed even after nutrient readdition (Fig. S5 C). These observations raised the possibility that Atg9 is localized to the autophagosomal membranes.
Atg9 is embedded in the autophagosomal outer membrane
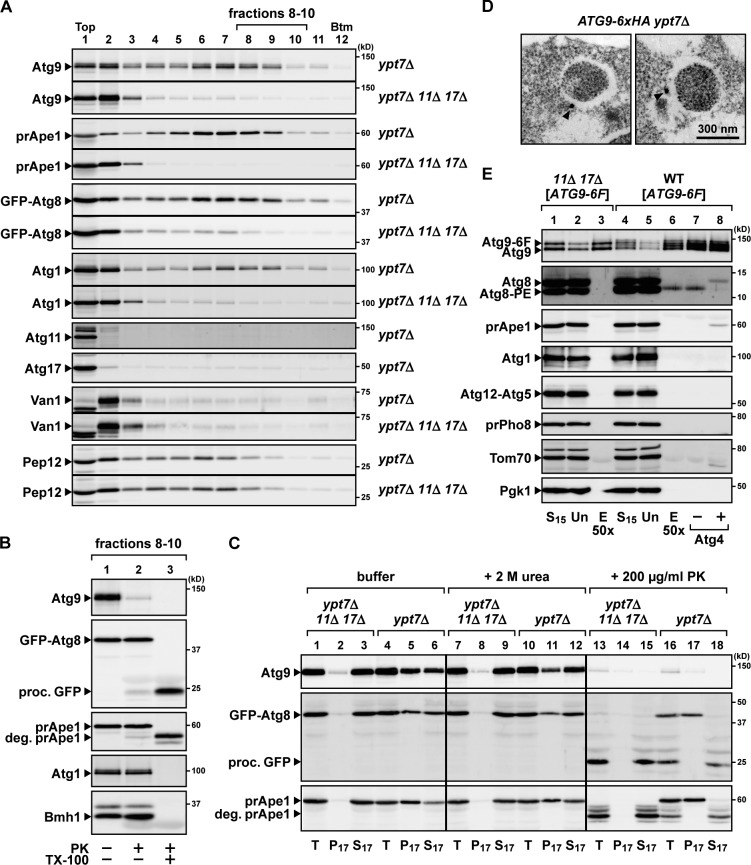
To further investigate whether Atg9 is present on the autophagosomal membranes, total cell lysates prepared from starved ypt7Δ cells were subjected to density gradient centrifugation (Fig. 5 A). Upon autophagosome formation, GFP-Atg8, Atg1, and a proform of Ape1 (prApe1), all of which are enclosed within autophagosomes (Suzuki et al., 2001), were distributed to fractions 4–10 (Fig. 5 A, ypt7Δ). Under these conditions, Atg9 co-migrated with the autophagosomal contents (GFP-Atg8, Atg1, and prApe1) in a manner dependent on autophagosome formation (as when comparing ypt7Δ with ypt7Δ atg11Δ atg17Δ). These results suggest that Atg9 was localized in or on the autophagosomes. Next, we harvested autophagosome-enriched fractions and subjected them to proteinase K treatment. As shown in Fig. 5 B, a large population of Atg9 was proteolytically degraded even in the absence of a detergent, whereas the autophagosomal contents (GFP-Atg8, prApe1, Atg1, and Bmh1, a nonselective cargo) were resistant to proteinase K treatment. Thus, Atg9 is exposed outside the autophagosomal membranes.
Figure 5.
Atg9 is embedded in the autophagosomal outer membrane. (A) Atg9 co-migrates with autophagosomes. ypt7Δ cells and ypt7Δ atg11Δ atg17Δ cells expressing GFP-Atg8 were converted to spheroplasts, treated with rapamycin for 5 h, and then ruptured by passage through a membrane filter with 5-µm pores. After removal of cell debris, the total lysates were loaded onto 8–30% (vol/vol) OptiPrep linear gradients and centrifuged at 200,000 g for 1 h. Van1 (Golgi) and Pep12 (endosome) were used as organelle markers. prApe1, a proform of Ape1; Btm, bottom. (B) Atg9 is exposed outside the autophagosomal membrane. After density gradient centrifugation, the autophagosome-enriched fractions (fractions 8–10 in A) were mixed, divided into three aliquots, and then treated with 100 µg/ml proteinase K (PK) for 20 min on ice with or without 0.3% Triton X-100 (TX-100). To exclude free Atg9 vesicles, fractions 8–10 were used as autophagosome-enriched fractions. proc. GFP, the processed form of GFP moiety; deg. prApe1, the degraded form of prApe1. (C) Total lysates were prepared as in A from ypt7Δ cells and ypt7Δ atg11Δ atg17Δ cells treated with rapamycin for 5 h. The total lysates (T) were either treated with 2 M urea or 200 µg/ml proteinase K for 20 min on ice or mock treated (buffer) and then separated into pellet (P17) and supernatant (S17) fractions by centrifugation at 17,400 g for 15 min. (D) ATG9-6×HA ypt7Δ cells were treated with rapamycin for 3 h and then subjected to pre-embedding immuno-EM using anti-HA antibody. Arrowheads indicate gold-enhanced NanoGold particles. (E) Wild-type (WT) cells and atg11Δ atg17Δ cells expressing Atg9-6×FLAG were converted to spheroplasts and treated with rapamycin for 2 h. Total lysates were prepared as in A. Total lysates were centrifuged at 15,000 g for 10 min; supernatant fractions (S15) were subjected to immunoisolation using the anti-FLAG antibody and eluted with the 3×FLAG peptide. The eluted fraction (lane 6) was treated with 1 µM recombinant Atg4 for 10 min at 30°C (lane 8) or mock treated (lane 7). Atg9-6F, Atg9-6×FLAG; prPho8, a proform of Pho8; Un, unbound fractions; E50×, eluted fractions concentrated 50-fold.
Next, we investigated whether Atg9 is embedded in the autophagosomal outer membrane or merely associated with its surface. When total cell lysate was prepared from the cells deficient in autophagosome formation (ypt7Δ atg11Δ atg17Δ), the large majority of Atg9 was recovered in the low-speed supernatant (Fig. 5 C, lane 3), indicating that the Atg9 vesicles did not pellet under these conditions. In contrast, upon autophagosome formation, about one half of Atg9 was recovered to the pellet fraction together with GFP-Atg8 and prApe1 (Fig. 5 C, lane 5), suggesting that Atg9 was precipitated with the autophagosomes, again consistent with the results of the density gradient centrifugation (Fig. 5 A). Furthermore, a significant portion of Atg9 was recovered in the pellet fraction even after treatment with a denaturing agent, 2 M urea (Fig. 5 C, lane 11) or 5 M urea (not depicted). These results suggest that Atg9 is not merely associated with the autophagosome surface via protein–protein interaction but is rather embedded in the autophagosomal outer membrane. Immuno-EM analysis also supported these results; gold particles representing Atg9-6×HA were detected in close proximity to the autophagosomal outer membrane (Fig. 5 D). Therefore, we conclude that in ypt7Δ cells, Atg9 is embedded in the autophagosomal outer membrane.
Next, we performed immunoisolation using wild-type cells, in which autophagosomes do not accumulate in the cytoplasm. By the immunoisolation of Atg9-6×FLAG, a lipidated form of Atg8 (Atg8-phosphatidylethanolamine [PE]; Ichimura et al., 2000), but not a free from of Atg8, was coisolated with Atg9-6×FLAG in a manner dependent on autophagy (Fig. 5 E, lane 6). The coisolated Atg8-PE was completely converted to a free from of Atg8 by an externally added recombinant Atg4, a deconjugation enzyme of Atg8-PE (Fig. 5 E, lanes 6–8), indicating that the Atg8-PE was not enclosed by a membrane. These results suggest that Atg9 exists on unsealed membranes enriched in Atg8-PE, i.e., the isolation membranes. Based on these results, we conclude that Atg9 is embedded in the isolation membrane and is ultimately localized to the autophagosomal outer membrane. These observations suggest that the Atg9 vesicles themselves become part of the isolation membranes and the autophagosomal membranes.
Only a few Atg9 vesicles assemble to the PAS and contribute to a single round of autophagosome formation
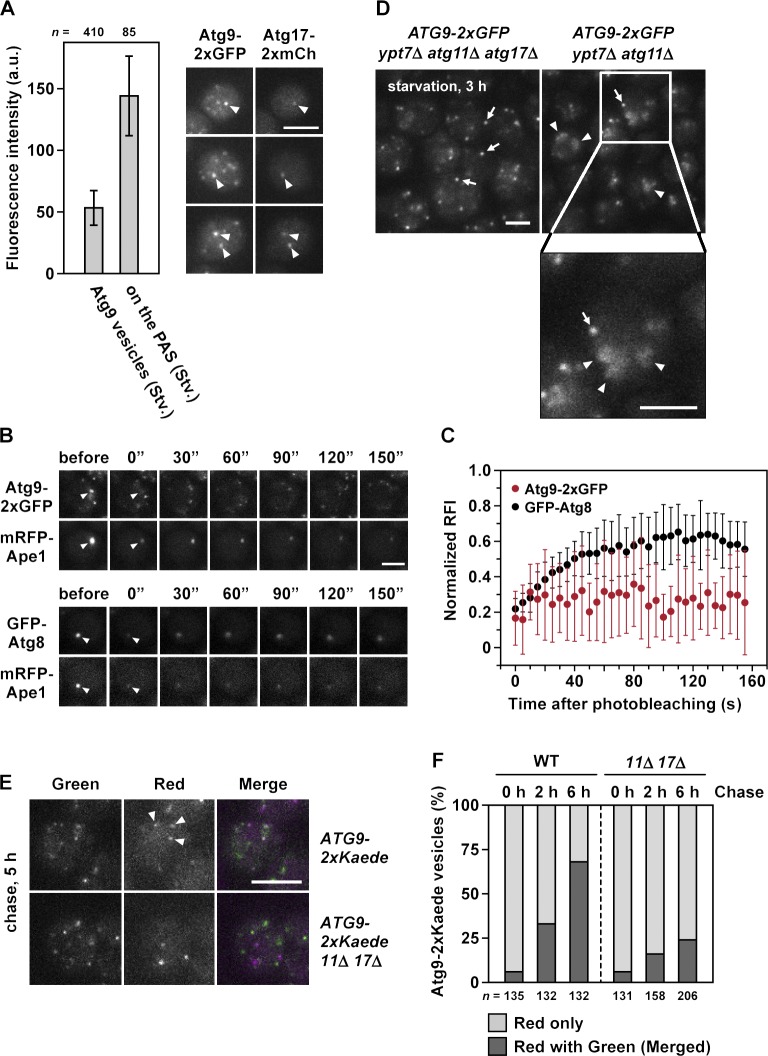
To estimate how many Atg9 vesicles are involved in a single round of autophagosome formation, we investigated the number of Atg9 vesicles assembling at the PAS by quantitative fluorescence microscopy. Atg9-2×GFP signals merged with the PAS marker Atg17-2×mCherry had an intensity of 144 GFP units (corresponding to 72 molecules of Atg9-2×GFP) in the starvation condition (Fig. 6 A). This quantitation revealed that during starvation, approximately three Atg9 vesicles (each Atg9 vesicle contains ∼27 molecules of Atg9-2×GFP as shown in Figs. 2 F and 6 A) assemble at the PAS.
Figure 6.
Only a small number of Atg9 vesicles are involved in a single round of autophagosome formation. (A) Wild-type cells expressing both Atg9-2×GFP and Atg17-2×mCherry were starved for 2 h and observed at 30 ms/frame. GFP fluorescence intensities of cytoplasmic mobile Atg9 vesicles and PAS-assembled Atg9 vesicles were quantitated as in Fig. 2 J. Error bars indicate standard deviation. Arrowheads indicate Atg9-2×GFP vesicles and Atg17-2×mCherry assembling at the PAS. a.u., arbitrary unit; Stv., starved. (B) FRAP analysis of Atg proteins at the PAS. ATG9-2×GFP mRFP-APE1 cells and GFP-ATG8 mRFP-APE1 cells were treated with rapamycin for 30 min and then subjected to FRAP analysis. Arrowheads indicate positions of photobleaching. (C) Quantitation of FRAP observations in B; normalized relative fluorescence intensities (RFI) after photobleaching of Atg9-2×GFP (n = 9) and GFP-Atg8 (n = 10). Error bars indicate standard deviation. (D) ypt7Δ atg11Δ cells and ypt7Δ atg11Δ atg17Δ cells expressing Atg9-2×GFP were starved for 3 h and observed at 30 ms/frame (see also Video 10). Arrows and arrowheads indicate cytoplasmic Atg9-2×GFP vesicles and Atg9-2×GFP clusters located on the autophagosomal membranes, respectively. (E) Multiple Atg9 vesicles are mixed during autophagy. Wild-type cells and atg11Δ atg17Δ cells expressing Atg9-2×Kaede were treated with rapamycin for 1 h. After UV irradiation (0 h), the cells were subjected to prolonged chase incubation for 2, 5, or 6 h. Arrowheads indicate Atg9-2×Kaede vesicles observed with both red and green fluorescence. (F) The number of mobile Atg9-2×Kaede vesicles observed with either red fluorescence alone or with both red and green fluorescence in E were counted. To exclude Atg9 signals colocalized at the PAS, immobile Atg9 puncta were not counted. The data shown are from a single representative experiment out of three repeats. WT, wild type. Bars: (A, D, and E) 5 µm; (B) 3 µm.
Next, we investigated whether only a small number of Atg9 vesicles are required for autophagosome formation or whether, instead, a larger population of Atg9 vesicles engages in repeated cycles of fusion with and retrieval from the isolation membrane. To this end, we analyzed the flux of Atg9 vesicles at the PAS using FRAP analysis. After photobleaching, GFP-Atg8 fluorescence recovered to a plateau with an ∼23-s half-recovery time, but Atg9-2×GFP fluorescence did not recover at all within 150 s (Fig. 6, B and C), indicating that no additional assembly of Atg9 vesicles to the PAS occurred at the later step of autophagosome formation. These results suggest that the initially assembled Atg9 vesicles are sufficient to accomplish a single round of autophagosome formation.
Next, we compared Atg9-2×GFP fluorescence intensity between Atg9 vesicles and autophagosomal membranes. As shown in Fig. 4 E, the Atg9-2×GFP signals on the autophagosomal membranes could be observed as ring-shaped objects in long exposures (3,000 ms/frame); however, short-exposure observations at 30 ms/frame revealed that Atg9-2×GFP formed several clusters moving on the autophagosomal membranes (Fig. 6 D and Video 10). The intensities of these clusters were comparable to those of the cytoplasmic Atg9 vesicles (Fig. 6 D), suggesting that even after the completion of autophagosome formation, Atg9 molecules derived from the initially assembled several Atg9 vesicles are retained on the autophagosomal membrane.
Atg9 vesicles are recycled after autophagosome formation
As represented in Fig. 7, after the fusion of the autophagosomal membrane with the vacuolar membrane, the Atg9 molecules retained on the autophagosomal outer membrane would be located onto the vacuolar membrane; however, Atg9 was not detected on the vacuolar membrane even after prolonged starvation (not depicted). This result raised the possibility that the Atg9 vesicles used for autophagosome formation are recycled back to the cytoplasm. To examine whether the Atg9 vesicles are recycled, we used Atg9-2×Kaede cells. We hypothesized that if more than two Atg9 vesicles (for example, the green fluorescent Atg9 vesicle and the red fluorescent Atg9 vesicle) are used for a single round of autophagosome formation and eventually recycled as new Atg9 vesicles, Atg9 vesicles exhibiting merged fluorescence will be detected after autophagosome formation. As shown in Fig. 6 (E and F), after prolonged incubation for 6 h, the number of merged Atg9 vesicles increased significantly in an autophagy-dependent manner (as when comparing wild-type cells with atg11Δ atg17Δ cells; Fig. 6, E and F). These results suggest that Atg9 molecules derived from multiple vesicles comingle during the process of autophagosome formation and are eventually recycled as new Atg9 vesicles.
Figure 7.
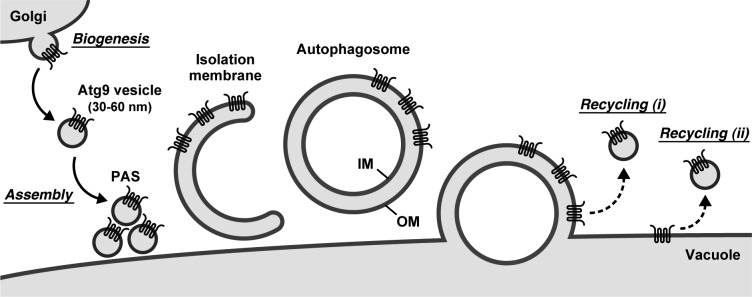
Involvement of Atg9-containing structures in the membrane dynamics of autophagy. Atg9 vesicles are derived from the Golgi apparatus as single-membrane structures with a diameter of 30–60 nm. During starvation, a small number of Atg9 vesicles assemble to the PAS to become part of the isolation membrane and ultimately part of the autophagosomal outer membrane. After autophagosome formation, Atg9 clusters remaining on the outer membrane are recycled back to the cytoplasm: (i) Atg9 vesicle recycling is coupled with fusion of the autophagosomal outer membrane with the vacuolar membrane; or (ii) the Atg9 vesicle is recycled from the vacuolar membrane (alternatively, Atg9 is translocated to the endosome or the Golgi apparatus by retrograde transport and generated via the Golgi-related secretory system as well as the biogenesis of Atg9 vesicles). The asymmetrical distribution of Atg9 on the outer membrane would allow Atg9 to avoid degradation and to be recycled back to the cytoplasm. IM and OM, autophagosomal inner and outer membranes, respectively.
Discussion
Atg9-containing structures have been predicted to be a source of the autophagosomal membranes, but until now, the property of these structures has remained poorly understood. In this study, we identified specific Atg9-containing structures as single-membrane vesicles of 30–60-nm diameter (Figs. 1 and 2) and designated them Atg9 vesicles. Furthermore, by use of the improved microscopy, we succeeded in the observation of their intracellular dynamics with high temporal resolution (10–20 ms/frame) and demonstrated that the Atg9 vesicles are highly mobile within the cytoplasm, whereas in the previous analyses, their spatiotemporal information could be lost with a long-exposure observation or acquisition of a set of z-plane images (deconvolution). The improved microscopy allowed us to investigate in detail the biogenesis of Atg9 vesicles and their subsequent assembly to the PAS as well as the novel roles of Atg23 and Atg27 (Fig. 3 C). We also revealed that after assembly to the PAS, the Atg9 vesicles are incorporated into the isolation membrane and subsequently the autophagosomal outer membrane (Figs. 5 and 6). These findings provide the first experimental evidence that the cytoplasmic mobile Atg9 vesicles directly contribute to autophagosome formation as at least one of the sources for the autophagosomal membranes.
An important question addressed by our results is the number of Atg9 vesicles involved in autophagosome formation. During starvation, approximately three Atg9 vesicles initially assemble to the PAS (Fig. 6 A), but no additional Atg9 vesicles are required for later steps of autophagosome formation (Fig. 6, B and C). From these findings, we propose a model wherein only a small number of Atg9 vesicles are required for a single round of autophagosome formation; these initially assembled Atg9 vesicles themselves become part of the isolation membrane (Fig. 7). Our findings also indicate the existence of a vesicle-mediated process at the early step of autophagosome formation.
Furthermore, we suggest that although Atg9 vesicles certainly supply lipid bilayers to the autophagosomal membranes, only a small quantity of lipids would be supplied via these vesicles. Thus, other lipid sources should still be considered (Tooze and Yoshimori, 2010). In mammalian cells, the ER membrane (Hayashi-Nishino et al., 2009; Ylä-Anttila et al., 2009), the mitochondrial membrane (Hailey et al., 2010), and the plasma membrane (Ravikumar et al., 2010) have been proposed as the sources for the autophagosomal membranes.
The notion that the Atg9 vesicles are unlikely to be major lipid suppliers for the autophagosomal membranes raises the possibility that these vesicles possess another vital function in autophagosome formation. In response to starvation, Atg9 vesicles assemble to the PAS just after the PAS scaffold proteins but before other Atg proteins (Suzuki et al., 2007), probably via the direct interaction with Atg11 and/or Atg17 (He et al., 2006; Sekito et al., 2009). In addition, after the assembly to the PAS, the Atg9 vesicles become part of the isolation membrane (Fig. 5 E). These observations strengthen the idea that the Atg9 vesicles play an essential role in an early event of autophagosome formation, such as nucleation of the isolation membrane. The Atg9 vesicles could supply unidentified proteins that are required for subsequent membrane expansion. Alternatively, Atg9 itself could possess essential functions, such as recruitment of other Atg proteins to the PAS or onto the isolation membrane. Furthermore, we found that Atg9 is localized to the autophagosomal outer membrane but not the inner membrane (Fig. 5 B). This asymmetrical distribution could be related to the function of Atg9, for example, sensing or regulation of membrane curvature. Proteomic and lipidomic analyses will provide important insights into the function of these vesicles. Comparison of the protein and lipid compositions of these vesicles with those of the isolation membrane (or other organellar membranes) will lead to a better understanding of membrane flow during autophagosome formation.
Another open question is the molecular mechanism of the transition undergone by Atg9 vesicles as they become part of the isolation membrane, a process that likely involves fusion of Atg9 vesicles. As shown in Fig. 4 A, even though Atg9 vesicles accumulate highly at the PAS, they do not fuse with each other in atg1D211A atg11Δ cells, in which other Atg proteins are poorly recruited to the PAS (Suzuki et al., 2007). Presumably, one or more downstream effectors are required for the fusion of Atg9 vesicles. Atg8-PE, which mediates tethering and hemifusion of liposomes in vitro (Nakatogawa et al., 2007), is a candidate for the downstream effector that promotes membrane fusion. Other candidates include SNARE complexes or TRAPP-III tethering machinery, which are also involved in autophagy (Lynch-Day et al., 2010; Nair et al., 2011).
Atg9 is highly conserved from yeast to mammals (mAtg9/Atg9A/Atg9L1 in mammals; Young et al., 2006). In mammalian cells, mAtg9 cycles between the trans-Golgi network and endosomes in response to nutritional changes, and a subpopulation of mAtg9 colocalizes with both LC3, a mammalian homologue of Atg8, and Rab7, a late endosomal protein (Young et al., 2006). These observations imply that mAtg9-containing structures are directly implicated in the formation of autophagosomes; however, a recent study reported that mAtg9 interacts only transiently with autophagosomes, and mAtg9 is not incorporated into the autophagosomal membranes (Orsi et al., 2012). One possibility is that in mammalian cells, mAtg9-containing structures used for autophagosome formation would be recycled immediately after the early step that requires mAtg9 function. The differences between the yeast Atg9 vesicles and mAtg9-containing structures should be elucidated in the future. Furthermore, the mechanisms whereby mAtg9 is translocated from the trans-Golgi network or endosomes to the autophagosomes are poorly understood. Unidentified mAtg9-containing structures, such as the Atg9 vesicles in yeast, could also exist in mammalian cells. Our findings will facilitate advances in elucidating the molecular basis of the initial step of autophagosome formation in both yeast and mammals.
Materials and methods
Yeast strains and media
Saccharomyces cerevisiae strains used in this study are listed in Table S1. Standard protocols were used for yeast manipulations (Kaiser et al., 1994). Cells were cultured at 30°C in SD/CA medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, 0.5% casamino acids, and 2% glucose) supplemented with appropriate nutrients. Autophagy was induced by transferring the cells to SD(–N) medium (0.17% yeast nitrogen base, without amino acids and ammonium sulfate, and 2% glucose). Otherwise, to induce autophagy, cells and spheroplasts were treated with 0.2 and 0.5 µg/ml rapamycin (Sigma-Aldrich), respectively.
Cells expressing GFP-, mCherry-, TagRFPT (TagRFP with S158T mutation; Evrogen)-, Kaede-, 6×HA-, or 3× biotin acceptor peptide (BAP; 3× biotinylated tag; van Werven and Timmers, 2006)–tagged proteins with a 17-residue linker (GGAAGGSSASGASGASG) were generated using the pYM series (Janke et al., 2004) with minor modifications by a PCR-based gene modification method (Janke et al., 2004). Gene deletions were performed using the pFA6a-kanMX6 series (Janke et al., 2004) as described previously (Janke et al., 2004). Cells expressing Atg9 via the TDH3 promoter and the CYC1 promoter were generated by a PCR-based gene modification method using pYM-N15 and pYM-N11, respectively (Janke et al., 2004). Cells expressing Atg9 under control of the TPI1 promoter were generated as follows: a DNA fragment including the TPI1 promoter (900-bp upstream region of the TPI1 gene) was amplified from yeast genomic DNA (primers 5′-GGCCGGATCCTCTTCAGATTCCCTCATGGAGAAAG-3′ and 5′-GGCCAAGCTTTTTTAGTTTATGTATGTGTTTTTTG-3′), digested with BamHI and HindIII, and inserted into the BamHI–HindIII site of pFA6a-hphNT1 (Janke et al., 2004). A DNA fragment including the TPI1 promoter and hphNT1 drug resistance cassette was amplified from the resultant plasmid (primers 5′-GAGTATCTCACTTTGATAAGAAGACATTTAAGAACAGCCTGAAATATCAAAATCACGGAATATCGATGAATTCGAGCTCG-3′ and 5′-CGCGATAAGAAAGTATTCTTCCCATGAGAGTTGGGTAACTGGTATTCATCTCTCTCCATTTTTAGTTTATGTATGTGTTT-3′) and integrated into the chromosome upstream of the ATG9 gene.
Plasmids and other materials
The low-copy plasmid for expression of Atg9 under control of the ATG9’s own promoter was constructed as follows: a DNA fragment including the ATG9 promoter, the ATG9 gene, and the ATG9 terminator (from a 500-bp upstream region of the initiation codon to a 270-bp downstream region of the termination codon of the ATG9 gene) was amplified from yeast genomic DNA (primers 5′-GGGCCCGGTACCCTCGAGGTCTATACGGGTTCACAAATCTGG-3′ and 5′-GGCCGAGCTCAAAGAGAAAGAAAACTACGTGGCGG-3′), digested with KpnI and SacI, and inserted into the KpnI–SacI site of pRS316 (Sikorski and Hieter, 1989), yielding pRS316-ATG9. The low-copy plasmid for expression of Atg9-6×FLAG under control of the ATG9’s own promoter was constructed as follows: to introduce the 6×FLAG DNA fragment, a BamHI–HindIII site was inserted into pRS316-ATG9 just upstream of the stop codon of the ATG9 gene by site-directed mutagenesis (QuikChange; Agilent Technologies; primers 5′-CAAGAAGTCTGACGTCGGAAGAGGATCCTAATGATAGCCCAAGCTTGGCAGACTGTGTCGTGTACATC-3′ and 5′-GATGTACACGACACAGTCTGCCAAGCTTGGGCTATCATTAGGATCCTCTTCCGACGTCAGACTTCTTG-3′), resulting in pRS316-ATG9 (BamHI). A DNA fragment encoding 3×FLAG was amplified separately (primers 5′-GGGAAAGGGCCGGATCCGACTACAAAGACCATGACGGTGACTACAAAGATCATGATATCGATTATAAGG-3′ and either 5′-CCCTTTGGGCCAAGCTTTTACTTATCATCATCATCCTTATAATCGATATCATGATCTTTGTAGTCACCG-3′ or 5′-GGGGATCCCCTCCTCCTCCTCCACCACCACCAGGAGGATGGAGTCATCCACAATTTGAAAAAGGGGGGTGGTCCCACCCC-3′). The amplified fragment was digested with BamHI and HindIII and inserted into the BamHI–HindIII site of pRS316-ATG9(BamHI), yielding pRS316-ATG9-3×FLAG. Furthermore, a DNA fragment encoding 3×FLAG (BamHI-3×FLAG-BamHI) was amplified (primers 5′-GGGAAAGGGCCGGATCCGACTACAAAGACCATGACGGTGACTACAAAGATCATGATATCGATTATAAGG-3′ and 5′-CCCTTTGGGCCGGATCCTTACTTATCATCATCATCCTTATAATCGATATCATGATCTTTGTAGTCACCG-3′), digested with BamHI, and inserted into the BamHI site of pRS316-ATG9-3×FLAG, yielding pRS316-ATG9-6×FLAG.
The low-copy plasmid for expression of GFP-Atg8 under control of the ATG8’s own promoter (pRS316-GFP-ATG8/AUT7) was described previously (Suzuki et al., 2001). The monomeric RFP (mRFP)-Ape1 expression construct was a gift from D.J. Klionsky (University of Michigan, Ann Arbor, MI). The auxin-inducible degron (AID) construct was a gift from M. Kanemaki (National Institute of Genetics, Shizuoka, Japan). Anti-Atg9 polyclonal antibodies were raised against synthetic peptides corresponding to residues 1–15, 74–89, and 138–156 of Atg9. Anti-Atg23 polyclonal antibodies were raised against synthetic peptides corresponding to residues 253–266 of Atg23. Anti-Atg27 polyclonal antibodies were raised against synthetic peptides corresponding to residues 40–54 of Atg27. Anti-Van1 polyclonal antibodies were a gift from K. Yoda (The University of Tokyo, Tokyo, Japan).
Fluorescence microscopy
Fluorescence microscopy was performed at room temperature using an inverted fluorescence microscope (IX71; Olympus) equipped with an electron-multiplying charge-coupled device camera (ImagEM C9100-13; Hamamatsu Photonics) and 150× total internal reflection fluorescence objective (UAPON 150×OTIRF, NA 1.45; Olympus). A 488-nm blue laser (20 mW; Spectra-Physics) and a 561-nm yellow laser (25 mW; Cobalt) were used for excitation of GFP and mCherry/TagRFPT, respectively. To increase image intensity and decrease background intensity, specimens were illuminated with a highly inclined laser beam (Tokunaga et al., 2008). For simultaneous observation of GFP and mCherry/TagRFPT, both lasers were combined and guided without an excitation filter. Fluorescence was filtered with a dichroic mirror (Di01-R488/561-25; Semrock) and a band pass filter (Em01-R488/568-25; Semrock) and separated into two channels using a splitter (U-SIP; Olympus) equipped with a dichroic mirror (DM565HQ; Olympus). The fluorescence was further filtered with a band pass filter (FF02-525/50-25; Semrock) for the GFP channel and a band pass filter (FF01-624/40-25; Semrock) for the mCherry/TagRFPT channel. Images were acquired by AQUACOSMOS software (Hamamatsu Photonics) and processed by AQUACOSMOS software or MetaMorph software (Molecular Devices).
Single-particle tracking data analysis
Precise positions of individual puncta were determined with an ∼2-nm accuracy in 2D by fitting the 13 × 13–pixel region fluorescent image using the 2D Gaussian function (Yildiz et al., 2003) f(x, y, z) = I0 exp[−{(x − x0)2+(y − y0)2}/σ1] + (αx + βy + C), in which I0 is the peak intensity, (x0, y0) is the position of the punctum, and (αx + βy + C) is the background intensity. When I0 was larger than the arbitrary set value we determined for each experiment, the position (x0, y0) was used for the analysis of the next frame, otherwise the tracking of the position was terminated. When more than six subsequent frames were successfully tracked, the MSD was determined by the equation
in which (x(jδt + nδt), y(jδt + nδt)) describes the particle position following a time interval of nδt after the start position (x(jδt), y(jδt)), N is the total number of data points, n and j are positive integers, and δt is the frame rate of the acquisition camera. The diffusion coefficient of individual puncta was calculated by fitting the MSD curves with a linear function as previously reported (Kusumi et al., 1993). In brief, the MSD calculated by this function was plotted against δt and fitted the plots with f (δt) = 4D × δt, in which D is the diffusion coefficient. Because fitting precision is greatly influenced by the number of points, four data points were used to fit the MSD curves.
Immunoisolation
For immunoisolation of the Atg9 vesicles, atg11Δ atg17Δ cells expressing Atg9-6×FLAG were treated with 10 mM DTT for 10 min at 30°C and then with 100 µg/ml Zymolyase 100T for 30 min at 30°C in spheroplasting buffer (50 mM Hepes-KOH, pH 7.2, 1 M sorbitol, 1 mM DTT, 0.5% yeast extract, 1% bacto-peptone, and 1% glucose). For rapamycin treatments, the spheroplasts were cultured for 20 min at 30°C in spheroplasting buffer containing protease inhibitor cocktail (P8340; Sigma-Aldrich) and then treated with 0.5 µg/ml rapamycin for 2 h. The spheroplasts were washed twice with HSE buffer (25 mM Hepes-KOH, pH 7.2, 750 mM sorbitol, and 5 mM EDTA) and then ruptured by passage through a membrane filter with 5-µm pores (Millipore) in HSE buffer. After removal of cell debris by centrifugation at 700 g for 5 min, the total lysate was centrifuged at 50,000 g for 15 min. The resulting supernatant was incubated with the anti-FLAG antibody (M2; Sigma-Aldrich) and protein G–Dynabeads (Invitrogen) for 3 h at 4°C in HSE buffer. The bound materials were washed twice with HSE buffer and then eluted for 1 h at 16°C with 2 mg/ml 3×FLAG peptide (Sigma-Aldrich) in HSE buffer containing 0.1% BSA (Sigma-Aldrich).
For immunoisolation of the isolation membrane, total lysate was prepared from wild-type cells expressing Atg9-6×FLAG as described in the previous paragraph. The total lysate was brought to a final concentration of 125 mM KCl and then centrifuged at 15,000 g for 10 min. The resulting supernatant fraction was incubated for 5 h at 4°C with the anti-FLAG antibody and protein G–Dynabeads in HSE buffer. The bound materials were washed four times with HSE buffer and then eluted for 1 h at 16°C with 2 mg/ml 3×FLAG peptide in HSE buffer.
DLS measurement
The size distribution of the Atg9 vesicles and FluoSpheres (Invitrogen) was analyzed using a size measurement system (Zetasizer Nano S; Malvern Instruments). Viscosity of the buffer used in Fig. 2 B was determined to be 1.365 cP using a viscometer system (AMVn; Anton Paar).
FRAP analysis
ATG9-2×GFP mRFP-APE1 cells or GFP-ATG8 mRFP-APE1 cells were loaded onto Concanavalin A–coated glass-bottom dishes (Asahi Glass), and the dishes were set on a heated stage (30°C). Cells were observed with a microscope (LSM 510 META; Carl Zeiss) combined with imaging device (ConfoCor 3) using a C-Apochromat 40×, 1.2 NA UV-visible infrared differential interference contrast water immersion objective (Carl Zeiss). A 488-nm Argon+ laser and a 594-nm He-Ne laser were used for excitation of GFP and mRFP, respectively. The fluorescence signals were selected using BP505-540 IR (GFP channel; Carl Zeiss) and BP615-680 IR (mRFP channel; Carl Zeiss) filters after separation through a dichroic mirror (NFT 600; Carl Zeiss). Two independent avalanche photodiodes in the ConfoCor3 system were used for detection of each fluorescence signal. After a PAS region was chosen for photobleaching using the mRFP channel, GFP molecules in the region of interest were photobleached by one iteration of irradiation with a 488-nm Argon+ laser. Fluorescent images were captured at 5-s intervals, and fluorescence intensities were determined using ImageJ version 1.43 software (National Institutes of Health).
EM
EM was conducted by Tokai-EMA, Inc. Cells were sandwiched between copper grids and rapidly frozen in liquid propane (−175°C) using a cryopreparation chamber (EM CPC; Leica) followed by substitution fixation in 2% osmium tetroxide dissolved in acetone containing 3% distilled water. Samples were embedded in Quetol-651. EM was performed using a transmission electron microscope (JEM-1200EX; JEOL). Negative staining EM was performed as follows: isolated Atg9 vesicles or FluoSpheres were applied to a carbon-coated 400-mesh copper grid and stained with 1% phosphotungstic acid for 5 min at room temperature.
Pre-embedding immuno-EM was performed as follows: cells were spheroplasted as described in the Immunoisolation section, treated with 0.2 µg/ml rapamycin for 3 h, and then fixed for 30 min at room temperature with 3% paraformaldehyde and 0.05% glutaraldehyde in 25 mM Hepes-KOH, pH 7.2, 750 mM sorbitol, and 100 mM NaCl. The spheroplasts were permeabilized for 5 min at 4°C with 0.05% saponin followed by treatment for 30 min at 4°C with 0.5% BSA and 10 mM glycine and then incubated for 2 h at 4°C with the anti-HA antibody (HA-7; Sigma-Aldrich). The samples were incubated for 1 h at 4°C with the Nanogold-labeled Fab′ fragment of goat anti–mouse IgG (Nanoprobes) and then fixed for 10 min at room temperature with 1.5% glutaraldehyde followed by treatment for 10 min with 0.5% BSA and 10 mM glycine. After signal enhancement using an EM kit (GoldEnhance; Nanoprobes), the samples were fixed with 0.5% osmium tetroxide, dehydrated, and embedded in Quetol-651. Negative staining EM and pre-embedding immuno-EM were performed using a transmission electron microscope (H-7500; Hitachi).
Online supplemental material
Fig. S1 shows the statistical analyses of the Atg9 vesicles. Fig. S2 shows the accumulation of Atg9 at the Golgi apparatus in sec7 mutant cells. Fig. S3 shows the mislocalization of Atg9 in atg23Δ or atg27Δ cells. Fig. S4 shows the aberrant accumulation of Atg9 in cells overexpressing Atg9. Fig. S5 shows that Atg9 vesicles assembled to the PAS and were eventually incorporated into the autophagosomal membranes. Video 1 shows that Atg9 vesicles were highly mobile in the cytoplasm. Video 2 shows that the motion of Atg9 vesicles was not altered after the treatment with latrunculin A. Video 3 shows the accumulation of Atg9 at the Golgi apparatus in cells depleted of Sec7. Video 4 shows the accumulation of Atg9 at the Golgi apparatus in sec7ts cells. Video 5 shows the mislocalization of Atg9 in atg23Δ or atg27Δ cells. Video 6 shows the aberrant accumulation of Atg9 in cells overexpressing Atg9. Video 7 shows that in response to starvation, cytoplasmic Atg9 vesicles assembled to the PAS. Video 8 shows that Atg9 vesicles assembled individually to the PAS. Video 9 shows that Atg9 localized onto the autophagosomal membranes. Video 10 shows that the intensity of Atg9-2×GFP clusters on the autophagosomal membrane was apparently comparable to that of the cytoplasmic mobile Atg9-2×GFP vesicles. Table S1 lists S. cerevisiae strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201202061/DC1.
Supplementary Material
Acknowledgments
We are grateful to Dr. Daniel J. Klionsky for the mRFP-Ape1 construct, Dr. Masato Kanemaki for the AID system, and Dr. Koji Yoda for anti-Van1 antibodies. We thank the members of Ohsumi’s laboratory for materials and helpful discussions. We also thank the Bio-Technical Center in the Technical Department, Tokyo Institute of Technology, and the Center for Analytical Instruments of the National Institute for Basic Biology for technical assistance.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Abbreviations used in this paper:
- AID
- auxin-inducible degron
- ALP
- alkaline phosphatase
- Atg
- autophagy related
- BAP
- biotin acceptor peptide
- DLS
- dynamic light scattering
- IAA
- indole-3-acetic acid
- mRFP
- monomeric RFP
- MSD
- mean square displacement
- PAS
- preautophagosomal structure
- PE
- phosphatidylethanolamine
- prApe1
- proform of Ape1
References
- Anderson C.M., Georgiou G.N., Morrison I.E., Stevenson G.V., Cherry R.J. 1992. Tracking of cell surface receptors by fluorescence digital imaging microscopy using a charge-coupled device camera. Low-density lipoprotein and influenza virus receptor mobility at 4 degrees C. J. Cell Sci. 101:415–425 [DOI] [PubMed] [Google Scholar]
- Babst M., Sato T.K., Banta L.M., Emr S.D. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16:1820–1831 10.1093/emboj/16.8.1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Nair U., Geng J., Klionsky D.J. 2008. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 19:668–681 10.1091/mbc.E07-08-0826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman V.C., Wu P., Parthun M.R., Wu J.Q. 2011. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J. Cell Biol. 195:563–572 10.1083/jcb.201106078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsner M., Hashimoto H., Simpson J.C., Cassel D., Nilsson T., Weiss M. 2003. Spatiotemporal dynamics of the COPI vesicle machinery. EMBO Rep. 4:1000–1004 10.1038/sj.embor.embor942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey D.W., Rambold A.S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P.K., Lippincott-Schwartz J. 2010. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 141:656–667 10.1016/j.cell.2010.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11:1433–1437 10.1038/ncb1991 [DOI] [PubMed] [Google Scholar]
- He C., Song H., Yorimitsu T., Monastyrska I., Yen W.L., Legakis J.E., Klionsky D.J. 2006. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J. Cell Biol. 175:925–935 10.1083/jcb.200606084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., et al. 2000. A ubiquitin-like system mediates protein lipidation. Nature. 408:488–492 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A., Yoshimori T., Noda T., Ohsumi Y. 2001. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 12:3690–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Joglekar A.P., Bouck D.C., Molk J.N., Bloom K.S., Salmon E.D. 2006. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 8:581–585 10.1038/ncb1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., Mitchell A. 1994. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York: 234 pp [Google Scholar]
- Kawamata T., Kamada Y., Kabeya Y., Sekito T., Ohsumi Y. 2008. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell. 19:2039–2050 10.1091/mbc.E07-10-1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Dalton V.M., Eggerton K.P., Scott S.V., Klionsky D.J. 1999. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol. Biol. Cell. 10:1337–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J. 2005. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118:7–18 10.1242/jcs.01620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J. 2007. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8:931–937 10.1038/nrm2245 [DOI] [PubMed] [Google Scholar]
- Kusumi A., Sako Y., Yamamoto M. 1993. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys. J. 65:2021–2040 10.1016/S0006-3495(93)81253-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T., Reiche S., Straub M., Bredschneider M., Thumm M. 2000. Autophagy and the cvt pathway both depend on AUT9. J. Bacteriol. 182:2125–2133 10.1128/JB.182.8.2125-2133.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore J., Bloom K.S., Salmon E.D. 2011. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J. Cell Biol. 195:573–582 10.1083/jcb.201106036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legakis J.E., Yen W.L., Klionsky D.J. 2007. A cycling protein complex required for selective autophagy. Autophagy. 3:422–432 [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D.J. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 6:463–477 10.1016/S1534-5807(04)00099-1 [DOI] [PubMed] [Google Scholar]
- Lynch-Day M.A., Bhandari D., Menon S., Huang J., Cai H., Bartholomew C.R., Brumell J.H., Ferro-Novick S., Klionsky D.J. 2010. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. USA. 107:7811–7816 10.1073/pnas.1000063107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari M., Griffith J., Rieter E., Krishnappa L., Klionsky D.J., Reggiori F. 2010. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 190:1005–1022 10.1083/jcb.200912089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. 2005. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 12(Suppl. 2):1535–1541 10.1038/sj.cdd.4401728 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B. 2010. Autophagy in mammalian development and differentiation. Nat. Cell Biol. 12:823–830 10.1038/ncb0910-823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastyrska I., He C., Geng J., Hoppe A.D., Li Z., Klionsky D.J. 2008. Arp2 links autophagic machinery with the actin cytoskeleton. Mol. Biol. Cell. 19:1962–1975 10.1091/mbc.E07-09-0892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U., Jotwani A., Geng J., Gammoh N., Richerson D., Yen W.L., Griffith J., Nag S., Wang K., Moss T., et al. 2011. SNARE proteins are required for macroautophagy. Cell. 146:290–302 10.1016/j.cell.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Ichimura Y., Ohsumi Y. 2007. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 130:165–178 10.1016/j.cell.2007.05.021 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10:458–467 10.1038/nrm2708 [DOI] [PubMed] [Google Scholar]
- Noda T., Kim J., Huang W.P., Baba M., Tokunaga C., Ohsumi Y., Klionsky D.J. 2000. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 148:465–480 10.1083/jcb.148.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y., Munro S. 2010. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol. Biol. Cell. 21:3998–4008 10.1091/mbc.E10-05-0457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y. 2001. Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2:211–216 10.1038/35056522 [DOI] [PubMed] [Google Scholar]
- Orsi A., Razi M., Dooley H.C., Robinson D., Weston A.E., Collinson L.M., Tooze S.A. 2012. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell. 23:1860–1873 10.1091/mbc.E11-09-0746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B., Moreau K., Jahreiss L., Puri C., Rubinsztein D.C. 2010. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 12:747–757 10.1038/ncb2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Tucker K.A., Stromhaug P.E., Klionsky D.J. 2004. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell. 6:79–90 10.1016/S1534-5807(03)00402-7 [DOI] [PubMed] [Google Scholar]
- Reggiori F., Shintani T., Nair U., Klionsky D.J. 2005a. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 1:101–109 10.4161/auto.1.2.1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Shintani T., Klionsky D.J. 2005b. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 16:5843–5856 10.1091/mbc.E05-07-0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein D.C. 2006. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 443:780–786 10.1038/nature05291 [DOI] [PubMed] [Google Scholar]
- Seaman M.N.J. 2008. Endosome protein sorting: motifs and machinery. Cell. Mol. Life Sci. 65:2842–2858 10.1007/s00018-008-8354-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekito T., Kawamata T., Ichikawa R., Suzuki K., Ohsumi Y. 2009. Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells. 14:525–538 10.1111/j.1365-2443.2009.01299.x [DOI] [PubMed] [Google Scholar]
- Shintani T., Klionsky D.J. 2004. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279:29889–29894 10.1074/jbc.M404399200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. 2001. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20:5971–5981 10.1093/emboj/20.21.5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. 2007. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 12:209–218 10.1111/j.1365-2443.2007.01050.x [DOI] [PubMed] [Google Scholar]
- Tokunaga M., Imamoto N., Sakata-Sogawa K. 2008. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods. 5:159–161 10.1038/nmeth1171 [DOI] [PubMed] [Google Scholar]
- Tooze S.A., Yoshimori T. 2010. The origin of the autophagosomal membrane. Nat. Cell Biol. 12:831–835 10.1038/ncb0910-831 [DOI] [PubMed] [Google Scholar]
- van Werven F.J., Timmers H.T. 2006. The use of biotin tagging in Saccharomyces cerevisiae improves the sensitivity of chromatin immunoprecipitation. Nucleic Acids Res. 34:e33 10.1093/nar/gkl003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen W.L., Legakis J.E., Nair U., Klionsky D.J. 2007. Atg27 is required for autophagy-dependent cycling of Atg9. Mol. Biol. Cell. 18:581–593 10.1091/mbc.E06-07-0612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A., Forkey J.N., McKinney S.A., Ha T., Goldman Y.E., Selvin P.R. 2003. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 300:2061–2065 10.1126/science.1084398 [DOI] [PubMed] [Google Scholar]
- Ylä-Anttila P., Vihinen H., Jokitalo E., Eskelinen E.L. 2009. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 5:1180–1185 10.4161/auto.5.8.10274 [DOI] [PubMed] [Google Scholar]
- Young A.R., Chan E.Y., Hu X.W., Köchl R., Crawshaw S.G., High S., Hailey D.W., Lippincott-Schwartz J., Tooze S.A. 2006. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119:3888–3900 10.1242/jcs.03172 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.