Abstract
The ability of modular protein domains to independently fold and bind short peptide ligands both in vivo and in vitro has allowed a significant number of protein-protein interaction studies to take advantage of them as affinity and detection reagents. Here, we refer to modular domain based proteomics as “domainomics” to draw attention to the potential of using domains and their motifs as tools in proteomics. In this review we describe core concepts of domainomics, established and emerging technologies, and recent studies by functional category. Accumulation of domain-motif binding data should ultimately provide the foundation for domain-specific interactomes, which will likely reveal the underlying substructure of protein networks as well as the selectivity and plasticity of signal transduction.
Keywords: Modular protein domain, Proteomics, Domainomics, Motif scanning, Domain scanning, Multiplex scanning
1. Introduction
Eukaryotic proteins are modular in nature. Many proteins contain independently folding globular domains capable of binding short peptide motifs even when both domain and motif are removed from the context of their full-length protein [1, 2]. Modular protein interacting domains facilitate protein-protein interactions required for a diverse set of cellular processes including signal transduction and subcellular localization. Domains are categorized based on structural and sequence homology, with each domain family recognizing motifs with similar characteristics, such as phosphorylated tyrosine (pTyr) or proline rich sequences (Table 1). The combination of modular domains within a protein contributes to its biological function by defining its protein interaction network. The post-translational modification (PTM) of amino acid side chains within a peptide motif can modulate domain-motif binding, providing the basis for the elegant and complicated protein signaling networks required for life [3]. Over the past decade, exploitation of a number of high-throughput proteomic technologies including increasingly sensitive mass spectrometry (MS) and protein microarrays has led to the dissection of vast protein interaction networks and the role of PTMs in altering network topology [4]. Accurate quantification of protein-protein interactions and PTM is now possible [5].
Table 1.
Characteristics of modular binding domains. Representative modular protein domains grouped by functional categories are presented. Approximate amino acid sizes (a.a) and affinity ranges are based on our literature search. The number of human domains and domain-containing proteins were estimated using the SMART database.
| Recognition | Name | Size (a.a) | Domain | Protein | Affinity |
|---|---|---|---|---|---|
| Phosphotyrosine | PTB | 100–150 | 36 | 31 | nM ~ µM |
| SH2 | ~100 | 120 | 110 | nM ~ µM | |
| PTP*1 | 250–280 | 50 | 38 | nM ~ µM | |
| Phosphoserine & phosphothreonine | 14-3-3 | ~250 | 8 | 8 | nM ~ µM |
| BRCT | 90–100 | 46 | 24 | nM ~ µM | |
| FF | 50–60 | 25 | 6 | nM ~ µM | |
| FHA | 65–100 | 31 | 31 | nM ~ µM | |
| MH2/DWB | ~200 | 8 | 8 | nM ~ µM | |
| POLO-Box | ~200 | 5 | 5 | nM ~ µM | |
| Polyproline | EVH1/WH1 | ~115 | 8 | 8 | µM |
| GYF | ~60 | 3 | 3 | µM | |
| SH3 | ~60 | 291 | 217 | nM ~ µM | |
| WW*2 | 38–40 | 88 | 48 | µM | |
| Methyllysine | LRR | 22–28 | 1967 | 228 | µM |
| PHD | ~50 | 168 | 96 | µM | |
| Chromo | 30–70 | 43 | 31 | µM | |
| MBT | ~100 | 29 | 9 | µM | |
| Tudor | ~50 | 55 | 27 | µM | |
| PWWP | ~135 | 17 | 14 | µM | |
| Acetyllysine | BROMO | ~110 | 64 | 46 | µM |
| C-terminus | PDZ | ~90 | 264 | 151 | nM ~ µM |
| Miscellaneous: β-propeller family | WD40*3 | 40–60 | 1682 | 272 | nM ~ µM |
Catalytic domains of protein tyrosine phosphatases (PTP) are included because of their potential use in domainomics.
Pin 1 WW domain can bind phosphoserine and threonine motifs.
Typically seven WD40 repeats form a β-propeller module. These modules have been reported to recognize various ligand modifications including serine-phosphorylated, threonine-phosphorylated, lysine-methylated, and ubiquitinated residues.
Because of the role of modular domains in assembling protein complexes, a significant portion of proteomics studies take advantage of modular domains as a means to assess protein-protein interactions (e.g., as bait in pull-down or probes in microarray). Here we refer to this modular domain-based proteomics as “domainomics.” While this may be a somewhat artificial segmentation, it is meant to draw attention to the potential of domains and their motifs as tools in contemporary proteomics. The emergence of domainomics as a unique sub-genre of proteomics raises a number of important questions: 1) What are the characteristics of modular domains as a research tool? 2) How are assays tailored to address specific scientific questions? 3) What technologies are available for exploiting domains as tools? 4) What lessons can be learned from current domainomics studies? 5) Has in silico prediction become a reliable tool? 6) What insights can domainomics provide for the protein interactome? Providing comprehensive answers to all these questions would be a challenging task for any single review. However, we believe an overview of domainomics will provide some insight into these topics. We first describe the core concept of domainomics, then outline established and emerging technologies, and review recent studies by functional category. We finish with a perspective on the unique potential of domainomics and a discussion of how to enhance its role in proteomic studies.
2. Modular protein interacting domain as affinity reagents
Independent folding of domains, which preserves binding capabilities, allows for their use as affinity and detection reagents in a manner similar to antibodies. For example, in Western blotting, protein expression is visualized by probing a membrane-bound denatured lysate with a specific antibody. Similarly, a labeled domain probe is used to detect the presence of domain binding sites in far-Western blotting [6]. As an affinity reagent, an immobilized antibody can be incubated with a lysate to enrich for the target protein and its interacting partners (immunoprecipitation). Domains can also be used to pull-down binding partners within a lysate for identification by MS [7]. However, the functional characteristics of antibodies and domains differ in many ways. Antibodies are biochemically homogeneous, therefore procedures required in proteomics, such as purification, modification for labeling, and immobilization, can be shared. In contrast, each modular domain is a part of a different full-length protein, and as such, has distinct biochemical properties such as solubility and structural stability [8]. Thus, experimental procedures must be tailored to take advantage of each domain’s physiological binding activity. Further, the specificity spectrum of antibodies and domains is qualitatively different. Antibodies are meant to recognize an epitope on target molecules; so off target cross-reactivity can badly affect quantitative results. Therefore, multiple validation and normalization steps are necessary to eliminate false positive signals in antibody-based proteomics [9, 10]. On the other hand, modular domains naturally have wide-ranging specificity; promiscuity in ligand selection is considered a physiological propensity rather than experimental noise. Taken together, modular domains and antibodies both can serve as useful affinity reagents in biochemical research, though procedures and research applications are often quite different.
3. Application Design
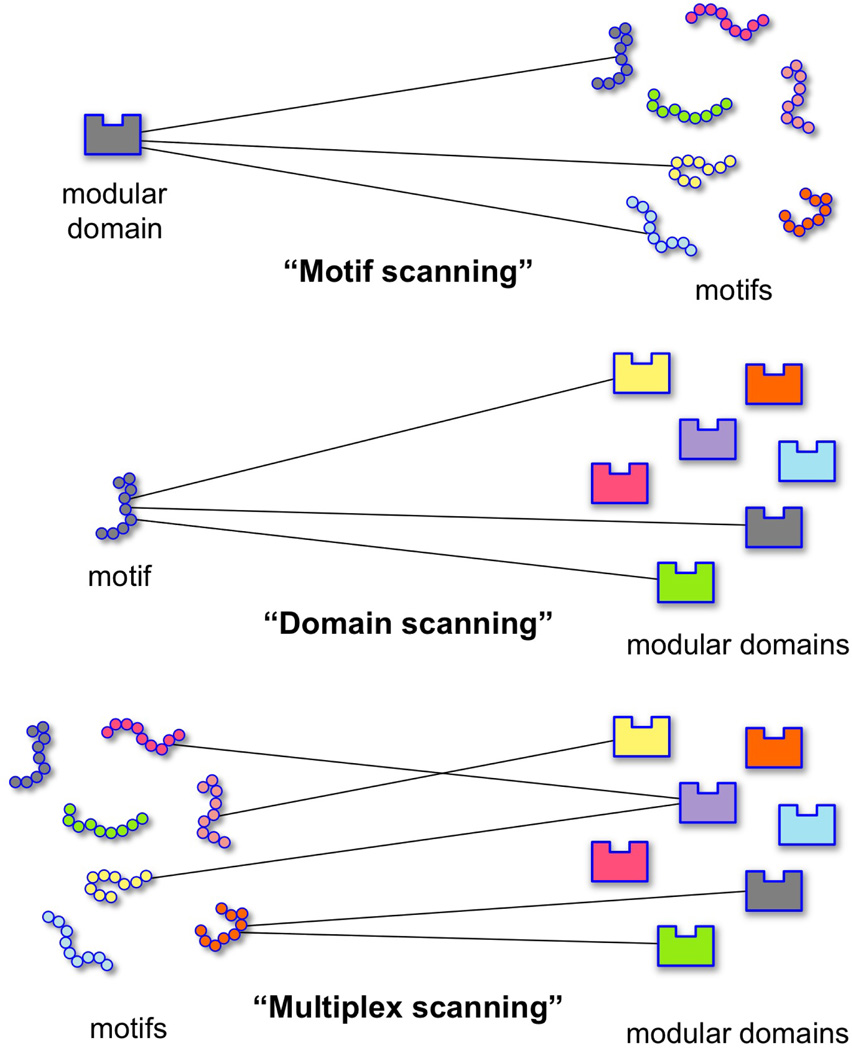
As affinity and detection reagents, modular domains and their short peptide ligands (motifs) can be used in three basic proteomic designs: motif scanning (motifs are scanned), domain scanning (domains are scanned), and multiplex scanning, each differing in their concept and execution (Fig. 1). “Motif scanning” surveys possible interaction partners containing a binding motif for a modular domain of interest. Typically, a modular domain is used as an affinity probe to either a library of synthesized peptides, e.g., SPOT arrays, or a whole proteome (far-Western and pull-down experiments). Motif scanning has been frequently used to define binding consensus motifs [11–13], to determine binding sites [14, 15], and to identify interacting proteins within a particular cellular environment, such as during growth factor treatment [7, 16]. In addition, a labeled domain can be used as a quantitative profiling tool for determining the presence or absence of modular domain binding sites in a group of cancer cell lysates or tissues [17].
Fig. 1.
Domainomics assay designs. The three basic assay designs for studying interactions between modular protein domains and short peptide motifs are depicted. Top: In motif scanning, a domain of interest is used to probe a library of peptide motifs or proteins containing binding motifs, typically to define domain specificity or identify possible binding proteins. For example, an immobilized domain can be used as bait in pull-downs. Middle: In domain scanning, a motif of interest is used as a probe to screen a set of domains or domain-containing proteins. Bottom: Multiplex scanning simultaneously assesses interactions between many ligands and domains, providing the specificities of domains within a domain-motif interaction map. Multiplex scanning can be designed as an expanded version of domain or motif scanning, or as a “library to library pull-down” to screen for binding modules (see Section 5.3.).
“Domain scanning” uses a peptide binding motif as bait to screen a library of domains (e.g., domain microarray) or a proteome (e.g., pull-down experiments). This approach is often used to determine binding partners when a putative domain binding motif is known to play an important role. For example, downstream effector docking sites are often examined using domain scanning [18, 19].
In “multiplex scanning,” binary interactions between many domains and motifs are simultaneously analyzed in the same experimental system. By determining interactions between nearly all domains belonging to a functional subgroup and their putative peptide motifs in equivalent assay conditions, multiplex scanning allows for construction of domain interaction maps or classification of domain specificity [2, 20, 21].
Assays can be multiplexed by expanding the experimental scale of either motif scanning or domain scanning. For example, simultaneous analysis of 61 peptides and 159 domains using a microarrays-in-microplates system has been conducted [22]. Alternatively, use of one tube multiplex assays, in which multiple domains compete in binding, have also been reported [23–25].
In addition to these different designs, binding assays can be categorized as “in vivo or in vitro” and “direct or indirect.” Domain-motif interactions detected in vitro should be confirmed by in vivo methods, such as co-immunoprecipitation or in vivo pull-downs. Interactions observed in lysate-free systems, e.g., peptide arrays, and far-Westerns are considered direct, while immunoprecipitation or pull-down may detect direct, and indirect interactions due to bridging across precipitated proteins (e.g., Fig. 2E). Here, the presence of a known binding site for the bait within an identified protein suggests direct binding. In either case, direct binding should be confirmed using orthogonal assay systems. Importantly, indirect binding does not necessarily mean such an interaction is not specific; rather, it may represent an important functional subcellular complex [26].
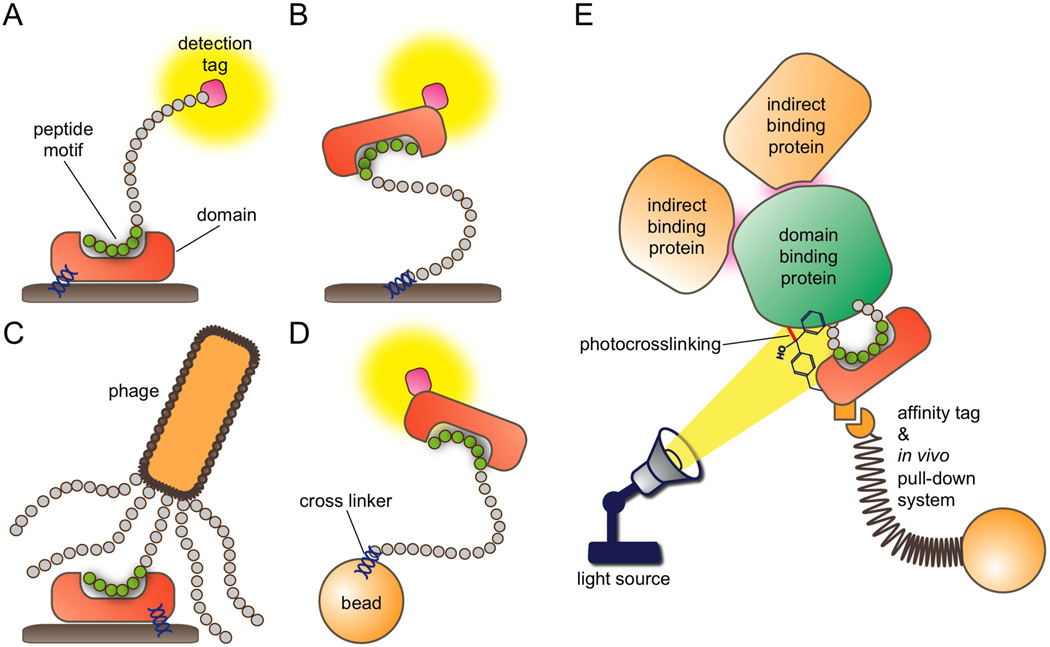
Fig. 2.
Domainomics technologies. (A) Forward phase array. Modular domains are immobilized on a solid support and probed with a peptide motif or protein (domain scanning). Duplicate arrays can be probed with different peptides (multiplex scanning). Binding is detected through a peptide-conjugated tag using fluorescence- or enzyme-based systems. (B) Reverse phase array. Peptides or proteins are immobilized on a solid support and probed with a tagged domain (motif scanning). Duplicate arrays can be probed with different domains (multiplex scanning). Binding is detected through a domain-conjugated tag using fluorescence- or enzyme-based systems. (C) Phage display. Peptide libraries are expressed as bacteriophage coat protein fusions and incubated with an immobilized domain (motif scanning). High affinity ligands are identified by sequencing of phage DNA. Multiple domains can be studied in separate wells with replicate libraries (multiplex scanning). Domains expressed on the phage surface can also be incubated with an immobilized peptide or protein (domain scanning). (D) Bead array. A bead-bound peptide library is probed with a tagged domain (motif scanning). Duplicate arrays can be probed with different domains (multiplex scanning). Binding is detected through a conjugated tag using fluorescence-, enzyme-, or mechanical-based systems. Peptide sequences are identified via mass spectrometry. Domains can also be immobilized on beads and probed with a motif (domain scanning). (E) In vivo photocrosslinking. This approach allows for incorporation of a photocrosslinking amino acid, e.g., p-benzoyl-L-phenylalanine (pBpa) into a modular domain expressed in vivo. Bound ligands are crosslinked to the domain under UV light, subjected to a pull-down assay using an affinity tag, and identified by MS. Covalent binding between the domain and ligands improves identification of weak transient binders, and allows for removal of indirect binding proteins by rigorous washing steps.
4. Domain-based Proteomics Assays
In this section we briefly describe core technologies for domainomics, namely, in silico screening, forward- and reverse-phase arrays, phage display, bead array, and photocrosslinking, as well as other methods (Fig. 2).
4.1. In silico screen
A number of searchable databases are available to obtain sequences, domain structures, PTM sites, and known interactions including NCBI gene, Human Protein Reference Database (HPRD), SMART, Pawson lab website, PROSITE, Domino, Phospho.ELM, PhosphoSitePlus, MmM, and PhosphoNET. Prior to either “domain scanning” or “motif scanning” studies (Fig. 1), it is advisable to use web-based prediction tools to test potential theoretical domain-motif interactions, possibly eliminating unnecessary experiments and helping efficient experimental design [27]. Prediction tools include Scansite, NetPhorest, SMALI, and DomPep [28–30]. Prediction models are typically based around position specific scoring matrices (PSSMs), which are trained using a set of experimentally determined interactions. Based on the training set, PSSMs define the likelihood of observing a particular amino acid at a specific position within a peptide that is known to bind a domain [2, 21]. Using this information, peptide binding scores can be calculated and used to predict the likelihood of novel peptide binding. However, the predictive ability of PSSM based models may be limited by interaction training sets that are insufficient to define every position within the matrix, the lack of sufficient negative interaction training data, and their inability to incorporate the effect of residue interdependence on peptide binding [31–33].
4.2. Forward phase array
Analogous to antibody microarrays, modular binding domains can be immobilized on a solid surface and probed with fluorophore/enzyme-labeled peptides or proteins (Fig. 2A)[2]. The labeling can be direct, or indirect via a biotin-streptavidin or epitope-tag system [34, 35]. Fluorescence-based detection systems are more amenable to quantitative analysis, while enzymatic detection may provide signal amplification [36]. Labels should be sufficiently spaced from motifs to minimize steric hindrance in ligand binding [37, 38]. However, a major concern for the forward phase format is whether immobilization of recombinant domain proteins might disrupt the conformation and orientation required for ligand binding [39–41]. For instance, covalent binding provides strong association to solid surfaces, but the modification of chemical groups which occurs could interfere with domain binding to specific ligands. Thus, the immobilization method for a given domain must be carefully optimized with an appropriate evaluation method that incorporates modular domain-specific positive and negative controls, e.g., modified and unmodified peptides, and wild type and mutated domains. Currently domain arrays are mainly used to survey synthetic peptides or purified proteins.
A prototypic domain array containing SH2, SH3, WW, PDZ, 14-3-3, FHA, PH, and EF domains on a nitrocellulose coated glass slide was introduced by Espejo et al. in 2002 [34]. In this assay, activity of immobilized SH3 domains was confirmed with peptides containing known SH3 motifs. In addition, they showed that endogenous proteins in cell lysates were bound by SH3 domains and could be detected using specific antibodies. Polyacrylamide hydro gel pads have also been used for protein domain immobilization [42]. Macbeath’s group introduced a multiplex forward-phase microarray by spotting the full complement of human SH2/PTB domains onto aldehyde-modified glass [22]. This system enabled affinity calculations for each domain-peptide pair based on the dependence of binding on ligand concentration, providing a more reliable measure than results obtained from a single ligand concentration.
4.3. Reverse phase array
In proteomics, reverse phase arrays are represented by lysate microarrays in which a large number of samples immobilized on a chip are probed with a specific antibody. Similarly, epitope-tagged or labeled domains can be used to probe a microarray or macroarray on which peptides, proteins, or lysates are immobilized (Fig. 2B). As opposed to forward-phase domain arrays, there is little concern associated with immobilization process as domains remain in solution. However, binding affinity may be affected by the type of labeling or epitope-tag used. Reverse phase arrays are particularly suitable to screen large-scale libraries of peptide motifs to determine consensus sequences for domains [43, 44]. Liu et al performed a SPOT based array analysis, probing a set of 192 phosphotyrosine peptides with 50 SH2 domains. They found that the selectivity of SH2 domains is driven not only by permissive (favorable) amino acid motifs but also non-permissive neighboring residues [45]. Our lab has generated a multiplex reverse phase array system in which a group of sample spots are probed with many SH2 domains in register within a 96-well plate apparatus, allowing for the assessment of ~2,000 domain-motif interactions in a single plate [8].
4.4. Phage Display
Phage display provides a renewable and scalable source of peptides or domains for proteomics screens using conventional tools available to most labs [46]. In this assay, foreign DNA fragments are expressed as fusions with coat proteins on the surface of the bacteria phage (Fig. 2C). Typically, 5 to 45 amino acid peptides are expressed as N-terminal fusions, however C-terminal fusions are also technically possible [47]. A process known as biopanning isolates high affinity clones through repetitive binding, washing, eluting and amplification [48]. High affinity clones are identified by sequencing of phage DNA and used to define the specificity of domain binding and natural domain ligands. Using this technology, large-scale multiplex scanning has been performed [49, 50]. For example, 1010 randomized peptides motifs were screened against 145 domains [21]. While phage display is typically used to express peptide motifs, Saksela’s group expressed a full complement of SH3 domains on the phage surface. Using this phage library, they revealed that some SH3 domains can bind ligands with nanomolar affinity, a much higher affinity than previously reported [51]. This library is now publicly available and widely used for domain scanning [52, 53].
4.5. Bead array
Bead-based assays have been used successfully for motif scanning of multiple domain families including PHD, SH2 and PTB domains [13, 54, 55]. These arrays provide an easy, cost-effective complement to microarrays. In bead based assays, a large number of pooled peptides can be screened simultaneously without the need for prior spatial separation of peptides [56]. Typically, combinatorial peptide libraries are synthesized on beads and probed with epitope-tagged modular domains (Fig. 2D). Interactions are detected using various methods such as “on bead western”, FACS, or magnetic nanoparticle pull-down; peptides are then identified by MS [13, 56, 57]. Generally, bead-based binding assays do not excel at quantification; as this requires a secondary assay in an orthogonal format using resynthesized peptides. To circumvent this, Astle et al. developed a bead-to-microarray screen. Peptides with positive interactions are cleaved from beads, divided into two fractions. One fraction is spotted on microarray for quantification by domain titration, and the other is used for sequence identification. By virtue of throughput and quantification capability, this system is thought to be suitable for screening of domain-peptide inhibitors [56].
4.6. In vivo photocrosslinking
In vitro substitution of a specific amino acid residue within a protein with an unnatural amino acid by manipulation of the ribosomal machinery was first described in late 80’s [58, 59]. Decades later, using bacterial amber suppressor tRNA and aminoacyl-tRNA synthetase specific for an unnatural amino acid, it is now possible to express mutated proteins containing unnatural residues harboring specific functional side chains in mammalian cells [60–62].
Incorporation of photo-reactive cross-linking amino acids such as p-benzoyl-L-phenylalanine (pBpa) into a modular domain of interest is particularly promising when studying transient domain-motif interactions (Fig. 2E) [63]. It allows for the capture of domain binding proteins whose interactions may not survive extensive washing during immunoprecipitation. Using this scheme, a photo-reactive domain and its ligands are cross-linked using UV-illumination, immunoprecipitated using an affinity tag, and subjected to LC-MS/MS. Recently, photocrosslinking methods were used to study in vitro SH2 or SH3 domain interactions [64, 65]. These studies led to the discovery of previously unidentified substrates. This approach is suitable for motif scanning by trapping proteome-wide interactors in vivo. However, a potential weakness of this technology is that mutation sites and cross-linker side-chain moieties may interfere with physiological ligand interactions [64]. The mutation site should be carefully determined so that the cross-linking site is close enough to the binding surface without causing steric interruption of ligand binding [65]. Because of the availability of a range of unnatural amino acids with various chemical functionalities and their potential capabilities in studying PTM-dependent protein-protein interactions in real time in vivo, studies using these technologies will clearly benefit domainomics [66]. For example, incorporation of a metabolically stable analog of phosphotyrosine could serve as a new tool in pTyr-interacting domain screening [67].
4.7. Other technologies
The yeast two-hybrid system (Y2H) has been a useful method for identifying novel domain interactions [68–70]. Modified Y2H assays provide a method for screening PTM-dependent interactions [71, 72]. However, some Y2H screens have high false positive and negative rates [73]. ELISA-based systems have also been used in domainomics [74, 75]. Fluorescent polarization has proven to be a useful method, capable of real-time in solution kinetic measurements, for drug screens or validation of microarray data [2, 76]. Alphascreen, a commercially available bead based proximity assay, also shows promise as a sensitive tool for assessment of domain-motif interactions and drug discovery [77]. In this assay, a pair of proteins are indirectly labeled with either donor or acceptor microbeads. If the proteins are interacting, irradiation of a donor bead triggers activation of a nearby acceptor bead, amplifying the chemiluminescence signal, which provides greater sensitivity and dynamic range [78, 79]. Proximity ligation assay is becoming a popular tool due to its high sensitivity and specificity in analyzing protein-protein interactions [80]. Commonly this approach is used in situ where cells or tissues are fixed and incubated with a pair of oligonucleotide-labeled antibodies for two proteins that are hypothesized to interact. Interactions are detected via rolling circle amplification [81]. An in-solution proximity ligation assay has also been reported as a multiplex protein detection tool requiring only 1 µl per sample [82].
5. Survey of domainomics
In this section we focus on representative domains in functional categories, how they are used in domainomics studies and what lessons can be learned from these applications.
5.1. Phosphotyrosine recognizing domains
At this time, there are 3,370 experimentally verified tyrosine phosphorylation sites in eukaryotic proteins according to Phospho.ELM [83]. The majority of pTyr-binding proteins contain SH2 domains, PTB domains, or both (Table 1) [84, 85]. SH2 domains are approximately 100 amino acids in length and bind in a pTyr specific manner via a conserved binding pocket [12, 86, 87]. PTB domains are 100–150 residues long, are found in scaffold proteins such as Shc and IRS-1, and vary in their dependence on phosphorylation [88]. Several other pTyr binding modules, such as the PKC delta C2 domain and pyruvate kinase M2, have also been reported [15, 89].
pTyr-dependent reversible interactions are crucial in propagating signals from activated receptor tyrosine kinases to downstream effectors. For example, upon stimulation, ErbB/EGF receptors autophosphorylate multiple Tyr sites on their cytoplasmic tails that serve as docking sites for SH2/PTB containing downstream effectors [90, 91]. To focus our methodological discussion, we outline independent domainomics studies on ErbB family receptors. Blagoev et al. used the Grb2-SH2 domain as bait to pull-down the EGFR interactome in HeLa cells [7]. They employed the SILAC method to quantify EGF-dependent interactions. Their study identified 228 proteins, of which 28 proteins, including 6 SH2 proteins and the novel interactor CD59, were selectively enriched upon stimulation. Schulze et al. conducted a SILAC pull-down with pTyr-peptide bait corresponding to all 89 ErbB cytosolic tyrosines, followed by MS [92]. Independent peptide pull-down MS identified 56 proteins, including 10 unique proteins (9 SH2/PTB proteins), most notably STAT5 as an EGFR interactor. EGFR and ErbB4 shared diverse interaction partners whereas ErbB2 and ErbB3 preferably bound Shc and PI3-Kinases, respectively. Jones et al. performed a SH2/PTB domain microarray with 33 tyrosine residues of ErbB receptors [22]. This approach identified numerous known and previously undocumented SH2/PTB-pTyr interactions, including 54 involving EGFR, 59 involving ErbB2, 37 involving ErbB3, and 8 involving ErbB4. Binding profiles of ErbB receptor sites were distinct: EGFR and ErbB2 binding became more promiscuous at high ligand concentration, while ErbB3 binding was independent of ligand concentration. A follow up study showed that the ErbB4 receptor displayed a more selective binding profile than the other receptors [93]. This observation appears to be inconsistent with the promiscuous profile of ErbB4 in Schulze’s pull-down data [92].
Reasons for this discrepancy are unknown, although the methodologies used in these three studies were quite distinct. First, the SH2 pull-down performed by Blagoev et al. is capable of detecting direct and indirect binding partners of EGFR in vivo. Second, while Schulze’s pTyr-peptide pull-down is a sensitive approach, some tyrosines used in the study may not be phosphorylated in vivo. Thus, observed binding may or may not be physiological. Third, while the sensitivity of the domain microarray approach should very high because domain-peptide interactions occur at fixed concentrations in a buffered solution without any competition or interference from other molecules, the activity of recombinant domain proteins can be lost in vitro due to misfolding, degradation, etc., resulting in poor reporting of physiological binding. These methodological differences should be taken into account in assay design and data interpretation. Limitations of these methodologies suggest that one approach is insufficient to correctly identify all physiological interactions. To illustrate this, we have summarized observed interactions from two studies mentioned above and Scansite predictions for four pYXN motifs of ErbB4 in Table 2. Shc binding to pY1188 is shared among three and Grb2 is prominently identified in pull-down. Further, microarray and Scansite indicate that more potential binding partners may exist in vivo. These differences may signify the complementarity of the three orthogonal methodologies.
Table 2.
ErbB4 pTyr site binding proteins. Four pYXN motifs of the ErbB4 cytosolic region were studied by pull-down MS and domain microarray. Identified binding proteins for each site are compared with Scansite predictions.
| ErbB4 site |
Sequence | Pull down | Microarray | Scansite |
|---|---|---|---|---|
| Y1162 | PMRDKPKQEpYLNPVEEN | Grb2 | Vav2; RASA1; Arg | PLCg; Grb2; Abl |
| Y1188 | DLQALDNPEpYHNASNGP | Grb2; Shc | Shc; Arg | Grb2; Shc |
| Y1202 | NGPPKAEDEpYVNEPLYL | Grb2 | Syk | Grb2; PLCg |
| Y1208 | EDEYVNEPLpYLNTFANT | Grb2 | - | - |
5.2. Proline rich sequence recognizing domains
Polyproline sequences are thought to provide a favorable domain-motif interface due to the stability of their helical confirmation, hydrophobicity, and presence on the surface of proteins [94, 95]. As a consequence, multiple domain families involved in subcellular localization and the assembly of multiprotein complexes recognize proline rich motifs [96]. SH3 domains were the first member of this group, discovered in 1988, followed by WW, EVH, and GYF domains (Table 1) [97–100]. It is worth noting that the first demonstration of polyproline recognition came from an early motif scanning experiment; a cDNA expression library was screened using a GST-tagged Abl SH3 domain as bait. Subsequent deletion analysis using the positive clones led to the finding [11].
The specificity of these domains to short proline-rich peptides is generally modest, with affinities in the low micromolar range (Table 1). However biochemical and structural studies suggest greater selectivity may be conferred by a larger binding surface [101, 102]. Given the large number of proline-rich sequence recognizing domains and the important role of selectivity in signaling (Table 1) [1], one might ask: What portion of these domains are selective or promiscuous? Additionally, do predicted peptide consensus classes, usually defined by in vitro screening, actually govern protein complexes in vivo? Several WW domainomics studies have provided partial answers to these intriguing questions.
Hu et al. generated a peptide-domain interaction map with 65 human WW domains and 1930 proline-rich peptides using an ELISA-type assay on a massive scale [74]. Interestingly, a number of WW domain binding sites showed selectivity. Of the 1056 proline-rich peptides which showed some binding, 33% bound to only 1–3 domains, 60% to 4–30 domains, and 7% to more than 30 domains. On the other hand, relative affinity comparisons based on signal intensity classified only 3.7% of interactions as strong while the remainder were medium or weak. In another study, Pawson and colleagues performed a pull-down experiment with a set of 10 WW domains, including all four in vitro defined consensus subclasses and identified 148 proteins by MS [26]. Hierarchical clustering segregated the 10 WW domains into three clusters based on the subset of proteins they bound. All 148 binding proteins were then categorized by the presence of previously defined WW subclass consensus motifs, e.g., PPXP, revealing good correlation between domain and motif subclasses. For example, PPXY motifs were preferentially enriched in a subgroup that contained WW domains with PPXY ligand specificity. Hence this correlation suggests consensus motifs identified in vitro indeed govern protein interactions in vivo. Further, functional analysis of WW domain pull-downs suggested that precipitated proteins represent physiological complexes in vivo. Taken together, these WW domain studies argue domainomics methods are sufficiently specific to provide a bottom up understanding of in vivo domain-based protein-protein interactions.
5.3. Phosphoserine/threonine recognizing domains
While it was surprising when dimeric 14-3-3 proteins were found to be genuine phosphoserine/threonine recognition modules [103], it quickly became apparent that other modular domains such as, FHA, WW and BRCT also share this behavior (Table 1) [104–106].
14-3-3 family proteins have attracted scientists’ attention because of their involvement in important cellular processes, such as signal transduction, and in disease [107]. 14-3-3 domainomics studies have been conducted for specificity mapping by peptide screening [108, 109], and protein identification by pull-down MS [110]. To date, several hundred interacting proteins have been reported including Raf and BCR [99], however direct binding has yet to be confirmed for many of them [100]. One perplexing problem, however, is that 14-3-3 interactomes produced by many studies display remarkably little overlap (20–40%). This may be attributed to differing experimental conditions such as enrichment methods, cell type, in vivo vs. in vitro, direct vs. indirect binding, or type of bait used [110–113].
Another plausible explanation is isoform dependence. 14-3-3 isoforms have been implicated to play unique roles in human disease, suggesting that each may bind a distinct set of ligands [114, 115]. Intriguingly, experimental data to date seem to be insufficient to explain apparent isoform selectivity in vivo. All isoforms share high affinity motifs RSXpSXP and RXF/YXpSXP with some variation [116], and a peptide scanning study failed to detect any isoform-specific phosphopeptide binding [117]. Structural observations also demonstrated that the conformation of 14-3-3 hetero- and homo-dimers were largely similar indicating their structural rigidity [116]. Thus, it has been hypothesized that 14-3-3 dimers act as rigid molecular scaffolds keeping an enzyme in its active conformation through a bidentate interaction [118]. Nevertheless, isoform specific ligands and minor conformational variability and flexibility have been reported [116, 119]. Therefore, it is likely 14-3-3 proteins largely share their ligands to exert broad regulatory functions, while playing selective roles by interacting with a few isoform-specific ligands. To address this issue, multiplex scanning domainomics, comparing all isoforms in equivalent conditions, would be informative.
BRCT, FHA, and 14-3-3 work cooperatively in the DNA damage response. DNA strand breaks activate the recruitment of serine/threonine kinases to the damage site, where they phosphorylate DNA repair proteins, transcription factors, protein kinases, and scaffolds [120, 121]. Phosphorylation signals are then “read” by proteins containing BRCT-1, FHA, and 14-3-3 domains. This process is organized as a hierarchy of domain-phosphosite binding interactions where specificity of kinases and domains are tightly regulated to prevent molecular signals from getting crossed [122]. For example, ATM kinase preferentially phosphorylates serine sites that are favored by MDC1 BRCT domains, while Chk kinase phosphorylates the motif recognized by 14-3-3 [123, 124]. In addition, other PTMs such as ubiquitination and methylation also play roles in the DNA damage response [125].
The link between BRCT domains and pS/pT-motifs was initially illuminated by a well-designed multiplex scanning study [106]. To identify novel phosphorylation-dependent domains involved in the DNA damage response, Manke et al. performed a pull-down assay against a cDNA expression library using an oriented phosphopeptide library mimicking ATM kinase substrates as bait. Proteins that bound in a phosphorylation-dependent manner were visualized by gel electrophoresis, which resulted in identification of the multi-BRCT domain-containing protein PTIP. Interestingly, they found that C-terminal tandem BRCT domains of PTIP function as a single module for phospho-specific binding. This “library to library pull-down” method is advantageous in that identification of PTM-recognizing domains is feasible without specifying a motif sequence and types of domains. Recently, this method has been combined with SILAC-MS technology leading to identification of a novel pTyr-binding module [89].
Highly dynamic and selective interactions at domain-PTM sites in the DNA damage response raise many questions: How many tandem BRCT domains are phosphorylation-dependent? How many domains are highly selective or promiscuous? Why are BRCT domains generally confined to DNA damage response signaling? While some studies have begun to address these questions [126], systematic domain binding analyses should provide more comprehensive answers.
5.4. Carboxyl terminus binding/PDZ domains
PDZ (PSD-95, DLG-4, ZO-1) domains are carboxyl terminal binding modules found in proteins generally considered to play a role in maintenance of cell polarity and adhesion [127]. Domains were initially divided into two major classes based on oriented peptide screening [128]. A third outlier class has also been proposed based on its lack of consensus with groups I and II [129]. Recently two large-scale studies have been undertaken to more explicitly define the specificity space of PDZ domains. Tonikian et al. screened a phage display library harboring 10 billion random C-terminal peptides against 82 PDZ domains from human and C. elegans, revealing that greater than 90% of PDZ domains fit into one of sixteen classes [21]. Stiffler et al. performed a domain microarray screen of 157 mouse PDZ domains probed with 217 C-terminal peptides. Surprisingly, they found that PDZ domains do not fall into discrete specificity classes; rather they lie on a continuum, evenly dispersed across the specificity space [2]. These data suggest that simplified classes based on consensus motifs are not suitable to precisely predict novel PDZ domain-ligand interactions.
Several groups have applied these binding datasets to develop prediction models. Using the microarray data, Chen et al. developed a modified PSSM that integrated domain/motif structural contact map information and was capable of predicting novel PDZ domain interactions across multiple species [31]. The sensitivity of this model was confirmed by another group using an independent human data set, although the false positive rate was shown to be higher than originally described [32]. Hui et al. incorporated Chen’s contact map information and trained a machine learning method using both phage and microarray data [130]. This approach significantly reduced the false positive rate. Instead of structural information, Li et al. used evolutionary relatedness of domain sequences and a PSSM to compare different domains, which resulted in increased predictive accuracy [30]. The model, referred to as DomPep, is publicly available online. Together, multiple efforts have been made to improve PDZ interaction prediction models. It is not possible to rank different prediction tools without a comparable value such as sensitivity or specificity, although we observed that ROC area under the curves for these models generally fell between 0.75–0.9 indicating good prediction accuracy. However, the obvious problem is that all models have been trained using two limited data sets based on PDZ domains from only three species. Because models are greatly affected by their training data set, broad validated experimental data is a requisite for further development. In addition, integration of protein expression and PTM data [131] could also provide more meaningful predictions of protein-protein interactions in vivo.
5.5. Methyllysine Binding Domains
PHD (Plant Homeodomain) fingers and Royal Family domains (Chromo, Tudor, MBT, and PWWP) recognize methyllysine and methylarginine on histone tails in the context of flanking residues [132]. These domains are thought to act as “readers” of the histone code which translate histone PTMs into biological outcomes in a process that in some ways parallels reading of tyrosine phosphorylation by SH2 domains [133, 134].
Intriguingly, over 392 proteins have been shown to contain at least one of these five domain families, yet only ~10 methylated residues have been identified on human histones [135, 136]. This raises the question: How do so many domains specifically interact with so few ligands? Several domainomics studies have provided some clues. A domain scanning approach employing a microarray of 109 PHD fingers and Royal Family domains, probed with histone tail peptides identified Tudor and MBT domains whose binding is dependent on the degree of lysine methylation (mono, di, tri) [20]. Further, peptide pull-down experiments employing a set of PHD finger, Tudor and WD40 domains showed that binding to methyllysine could be significantly affected by arginine methylation [137]. To test the effect of other PTMs, Garkse et al. performed a motif scanning assay in which a histone-3 N-terminal peptide bead library containing over 5,000 randomized PTM combinations was probed with five PHD fingers [13]. The screen identified a number of PTMs, including arginine methylation and threonine phosphorylation, which modulated PHD-methyllysine binding via switch (binary binding) and rheostat-like (variable binding) mechanisms. Subsequent SILAC-MS (domain scanning) and peptide array (motif scanning) experiments added support to the importance of phosphorylation in methyllysine binding [138, 139]. Taken together, these results suggest combinatorial PTMs provide specificity for domain binding to histone tails, adding credence to the theory that neighboring PTMs confer widespread regulation of histone tail effector binding [140]. A complete understanding of this mechanism will require further studies including a greater number of methyllysine binding domains.
5.6. β-propeller family
WD40 repeats are one of the most abundant domain families found in eukaryotic proteomes (Table 1). They rank as one of the top interacting domains in human interactome data sets [141], and are found in proteins which play roles in a diverse set of biological functions, including signal transduction and vesicular trafficking [142, 143]. It is hypothesized that WD40 domains act as a scaffolds for multiprotein complexes. These domains differ from “classical” modular protein domains in multiple ways: Typically WD40 repeats fold into a non-catalytic, seven blade β-propeller that functions as an independent module. These propellers share little sequence conservation, and studies have shown they have PTM-dependent (e.g., phosphorylation, ubiquitination, and methylation) and independent substrates [144, 145]. In this sense, WD40 propellers behave in a similar manner as antibodies, providing a framework on which specificity for different ligands can be built.
The unique features of WD repeats could impede application of conventional domainomics. For example, motif scanning, to define binding specificity for poorly understood WD40 proteins, would require an enormous peptide library containing unrelated motifs and different types of PTMs. Further, recombinant WD propeller proteins tend to be insoluble proving a hurdle for in vitro studies [146]. As a result, a systematic inspection of WD40 ligand selectivity has yet to be undertaken to date. Nevertheless, it has been shown that SPOT array scanning was able to define the consensus motif of the Cdc4 WD40 propeller, suggesting a multi-domain study is feasible. In addition, their deep involvement in the human interactome and multiple PMT-dependency suggests a possible role as signaling gatekeepers. For instance, there is a switch-like interaction between WD40 repeats of Cdc4 and multiple phosphorylation sites of Sic1; only when all six sites are phosphorylated, can the WD40 repeats bind Sic1 leading to cell-cycle progression [147]. Thus, it would be interesting to exploit WD40 repeats as molecular probes to distinguish fluctuations in cellular signaling pathways, e.g., as biosensors for a multiply phosphorylated site.
6. Perspectives
6.1. Domainomics-reducing the complexity
The major premise of domainomics is that in vivo protein networks are generally governed by interactions between modular protein domains and short peptide motifs, and that these interactions are generally reproducible in an in vitro binding assay. Unfortunately, some information is lost at the expense of the convenience provided by this overly simplified view. First, non-domain-dependent binding that relies on the reciprocal large surface interactions between proteins will be missed [148]. Second, cooperative interactions involving multiple domains might also be missed [149]. Third, transient enzyme-substrate interactions may not be captured. Thus domainomics does not provide a complete picture of regulatory protein networks. Nevertheless, domains and motifs are valuable tools for surveying specific domain-motif interactions, and thus widely used in motif and domain scanning, as outlined above.
We believe, however, the power of domainomics can be fully exerted in multiplex scanning formats, especially when many domains are employed. By incorporating all functionally similar domains into a high-throughput system, it is possible to dissect individual layers of a complex full-length protein interaction network. For example, the SH3/WW domain-ligand map is a subset of the interactome involving only proline-rich sequences [43, 150]. Such a sub-network map is not only useful to overview the distribution of binding specificity and promiscuity, but also to predict effects caused by interruption or overexpression at a particular interaction node and in rewiring networks [151, 152]. Likewise, functional layers based on phosphotyrosine, phosphoserine, and methyllysine dependent domains should provide distinct interaction maps. Usefulness of this approach is visible even when using a subset of domains or motifs. For example a SH2 and PTB domain macroarray provided a pTyr-dependent effector map for ErbB/EGFR family members, revealing a qualitative difference among them [22]. Another study compared interaction profiles of selected WW domains and suggested a connection may exist between binding motifs and specific subcellular systems [26]. Taken together, multiplex scanning domainomics offers a unique tool to dissect interactomes in a way not obtainable through non-domain proteomics.
6.2. Genome-wide domainomics
Given the potential of domainomics and the availability of advanced technologies and bioinformatics, is it now possible to construct many different interactomes based on modular protein domains and their binding proteins? Unfortunately, because the domainomics toolbox is insufficient, the answer is no. Publicly available complete domain libraries are limited to only a few domains, e.g., SH2 and SH3, precluding the construction of comprehensive domain-interaction maps. There are, however, independent efforts to create such libraries and interaction maps for a number of domains [2, 43, 74]. In the long term though, redundant collection of clones by individual research groups is highly inefficient. Ideally, the modular domain research community should work together to bank domain constructs.
Along with modular interaction-dedicated domains, enzyme catalytic domains may be a potential addition to the toolbox. PTM-dependent protein interactions are governed by the specificities of both binding domains (readers, e.g., SH2s) and enzymes (writers and erasers, e.g., tyrosine kinases and phosphatases) [153]. Although enzymes bind substrates transiently, making them inappropriate for use as affinity reagents, stabilization of binding by mutagenesis or cross-linking is feasible at least in part [154, 155]. These enzymes, as well as modified interaction domains [155, 156] and domain-specific antibodies [157, 158], may strengthen domainomics by revealing further functional layers of protein-protein networks.
7. Concluding remarks
In this review, we highlighted a portion of proteomics (domainomics) focusing on how modular protein interacting domains are popularly used as a means to address specific scientific questions. Methods involving domains and their motifs are increasingly playing a role in the proteomics era, powered by the availability of genome data, domain prediction tools, and high-throughput technologies. Accumulation of domain-motif binding data will ultimately lead to a domain-based interactome, providing insights into the underlying structure of protein networks and the selectivity and plasticity of signal transduction.
Acknowledgements
The authors gratefully acknowledge Jonathon Ditlev, Peter Nollau, and Bruce Mayer for critical reading of the manuscript, and Joshua Kritzer for helpful discussions. This work was supported by Neag Comprehensive Cancer Center, American Cancer Society Institutional Research Grant and NIH grants RC1 CA146843 and U01 CA154966.
Abbreviations
- pTyr
phosphotyrosine
- PTM
post-translational modification
- MS
mass spectrometry
- PSSM
position specific scoring matrix
- SH2
Src homology 2
- SH3
Src homology 3
- WW
tryptophan tryptophan
- PDZ
Post synaptic density protein 95-Disks large protein 4-Zonula occludens 1
- FHA
forkhead-associated
- PH
pleckstrin homology
- PHD
plant homeo domain
- PTB
phosphotyrosine binding
- FACS
fluorescence-activated cell sorting
- pBpa
p-benzoyl-L-phenylalanine
- LC-MS/MS
liquid chromatography followed by tandem mass spectrometry
- Y2H
yeast-two-hybrid
- ELISA
enzyme linked immunosorbent assay
- IRS-1
insulin receptor substrate 1
- PKC
protein kinase C
- SILAC
stable isotope labeling by amino acids in cell culture
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- EVH
Ena/Vasp homology
- GYF
glycine-tyrosine-phenylalanine
- GST
glutathione S-transferase
- BRCT
BRCA1 C-terminus
- BCR
breakpoint cluster
- ATM
ataxia telangiectasia mutated
- MDC1
mediator of DNA damage checkpoint protein 1
- pS
phosphoserine
- pT
phosphothreonine
- PTIP
Pax interacting protein 1
- ROC
receiver operating characteristic
- MBT
malignant brain tumor
- PWWP
proline-tryptophan-tryptophan-proline
- WD40
tryptophan-aspartic acid 40
- SPOT
peptide synthesis on membrane
- STAT
Signal transducers and activators of transcription
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 2.Stiffler MA, Chen JR, Grantcharova VP, Lei Y, Fuchs D, Allen JE, Zaslavskaia LA, MacBeath G. PDZ domain binding selectivity is optimized across the mouse proteome. Science. 2007;317:364–369. doi: 10.1126/science.1144592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nature reviews Molecular cell biology. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 4.Wolf-Yadlin A, Sevecka M, MacBeath G. Dissecting protein function and signaling using protein microarrays. Current opinion in chemical biology. 2009;13:398–405. doi: 10.1016/j.cbpa.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vermeulen M, Hubner NC, Mann M. High confidence determination of specific protein-protein interactions using quantitative mass spectrometry. Current opinion in biotechnology. 2008;19:331–337. doi: 10.1016/j.copbio.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Machida K, Mayer BJ. Detection of protein-protein interactions by far-western blotting. Methods Mol Biol. 2009;536:313–329. doi: 10.1007/978-1-59745-542-8_34. [DOI] [PubMed] [Google Scholar]
- 7.Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nature biotechnology. 2003;21:315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 8.Machida K, Thompson CM, Dierck K, Jablonowski K, Karkkainen S, Liu B, Zhang H, Nash PD, Newman DK, Nollau P, Pawson T, Renkema GH, Saksela K, Schiller MR, Shin DG, Mayer BJ. High-throughput phosphotyrosine profiling using SH2 domains. Molecular cell. 2007;26:899–915. doi: 10.1016/j.molcel.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 9.Hause RJ, Kim HD, Leung KK, Jones RB. Targeted protein-omic methods are bridging the gap between proteomic and hypothesis-driven protein analysis approaches. Expert review of proteomics. 2011;8:565–575. doi: 10.1586/epr.11.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sevecka M, Wolf-Yadlin A, MacBeath G. Lysate microarrays enable high-throughput, quantitative investigations of cellular signaling. Molecular & cellular proteomics : MCP. 2011;10 doi: 10.1074/mcp.M110.005363. M110 005363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren R, Mayer BJ, Cicchetti P, Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 12.Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 13.Garske AL, Oliver SS, Wagner EK, Musselman CA, LeRoy G, Garcia BA, Kutateladze TG, Denu JM. Combinatorial profiling of chromatin binding modules reveals multisite discrimination. Nature chemical biology. 2010;6:283–290. doi: 10.1038/nchembio.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mongiovi AM, Romano PR, Panni S, Mendoza M, Wong WT, Musacchio A, Cesareni G, Di Fiore PP. A novel peptide-SH3 interaction. The EMBO journal. 1999;18:5300–5309. doi: 10.1093/emboj/18.19.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benes CH, Wu N, Elia AE, Dharia T, Cantley LC, Soltoff SP. The C2 domain of PKCdelta is a phosphotyrosine binding domain. Cell. 2005;121:271–280. doi: 10.1016/j.cell.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 16.Nakamoto T, Yamagata T, Sakai R, Ogawa S, Honda H, Ueno H, Hirano N, Yazaki Y, Hirai H. CIZ, a zinc finger protein that interacts with p130(cas) and activates the expression of matrix metalloproteinases. Molecular and cellular biology. 2000;20:1649–1658. doi: 10.1128/mcb.20.5.1649-1658.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machida K, Eschrich S, Li J, Bai Y, Koomen J, Mayer BJ, Haura EB. Characterizing tyrosine phosphorylation signaling in lung cancer using SH2 profiling. PloS one. 2010;5:e13470. doi: 10.1371/journal.pone.0013470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Xu X, Wang J, Ma J, Fan Y, Rakestraw KM, Webster RG, Hoffmann E, Krauss S, Zheng J, Zhang Z, Naeve CW. Large-scale sequence analysis of avian influenza isolates. Science. 2006;311:1576–1580. doi: 10.1126/science.1121586. [DOI] [PubMed] [Google Scholar]
- 19.Hanke S, Mann M. The phosphotyrosine interactome of the insulin receptor family and its substrates IRS-1 and IRS-2. Molecular & cellular proteomics : MCP. 2009;8:519–534. doi: 10.1074/mcp.M800407-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO reports. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tonikian R, Zhang Y, Sazinsky SL, Currell B, Yeh JH, Reva B, Held HA, Appleton BA, Evangelista M, Wu Y, Xin X, Chan AC, Seshagiri S, Lasky LA, Sander C, Boone C, Bader GD, Sidhu SS. A specificity map for the PDZ domain family. PLoS biology. 2008;6:e239. doi: 10.1371/journal.pbio.0060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 23.Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 24.Dierck K, Machida K, Voigt A, Thimm J, Horstmann M, Fiedler W, Mayer BJ, Nollau P. Quantitative multiplexed profiling of cellular signaling networks using phosphotyrosine-specific DNA-tagged SH2 domains. Nature methods. 2006;3:737–744. doi: 10.1038/nmeth917. [DOI] [PubMed] [Google Scholar]
- 25.Christofk HR, Wu N, Cantley LC, Asara JM. Proteomic screening method for phosphopeptide motif binding proteins using peptide libraries. Journal of proteome research. 2011;10:4158–4164. doi: 10.1021/pr200578n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingham RJ, Colwill K, Howard C, Dettwiler S, Lim CS, Yu J, Hersi K, Raaijmakers J, Gish G, Mbamalu G, Taylor L, Yeung B, Vassilovski G, Amin M, Chen F, Matskova L, Winberg G, Ernberg I, Linding R, O'Donnell P, Starostine A, Keller W, Metalnikov P, Stark C, Pawson T. WW domains provide a platform for the assembly of multiprotein networks. Molecular and cellular biology. 2005;25:7092–7106. doi: 10.1128/MCB.25.16.7092-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Z, Hou T, Li N, Xu Y, Wang W. Proteome-wide detection of Abl1 SH3-binding peptides by integrating computational prediction and peptide microarray. Molecular & cellular proteomics : MCP. 2012;11 doi: 10.1074/mcp.O111.010389. O111 010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic acids research. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller ML, Jensen LJ, Diella F, Jorgensen C, Tinti M, Li L, Hsiung M, Parker SA, Bordeaux J, Sicheritz-Ponten T, Olhovsky M, Pasculescu A, Alexander J, Knapp S, Blom N, Bork P, Li S, Cesareni G, Pawson T, Turk BE, Yaffe MB, Brunak S, Linding R. Linear motif atlas for phosphorylation-dependent signaling. Science signaling. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Zhao B, Du J, Zhang K, Ling CX, Li SS. DomPep--a general method for predicting modular domain-mediated protein-protein interactions. PloS one. 2011;6:e25528. doi: 10.1371/journal.pone.0025528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen JR, Chang BH, Allen JE, Stiffler MA, MacBeath G. Predicting PDZ domain-peptide interactions from primary sequences. Nature biotechnology. 2008;26:1041–1045. doi: 10.1038/nbt.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luck K, Fournane S, Kieffer B, Masson M, Nomine Y, Trave G. Putting into practice domain-linear motif interaction predictions for exploration of protein networks. PloS one. 2011;6:e25376. doi: 10.1371/journal.pone.0025376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomovic A, Oakeley EJ. Position dependencies in transcription factor binding sites. Bioinformatics. 2007;23:933–941. doi: 10.1093/bioinformatics/btm055. [DOI] [PubMed] [Google Scholar]
- 34.Espejo A, Cote J, Bednarek A, Richard S, Bedford MT. A protein-domain microarray identifies novel protein-protein interactions. The Biochemical journal. 2002;367:697–702. doi: 10.1042/BJ20020860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He J, Bellini M, Inuzuka H, Xu J, Xiong Y, Yang X, Castleberry AM, Hall RA. Proteomic analysis of beta1-adrenergic receptor interactions with PDZ scaffold proteins. The Journal of biological chemistry. 2006;281:2820–2827. doi: 10.1074/jbc.M509503200. [DOI] [PubMed] [Google Scholar]
- 36.Espina V, Woodhouse EC, Wulfkuhle J, Asmussen HD, Petricoin EF, Liotta LA. Protein microarray detection strategies: focus on direct detection technologies. J Immunol Methods. 2004;290:121–133. doi: 10.1016/j.jim.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 37.Koman A, Terenius L. Bifunctional enkephalin analogues for affinity separation purposes. FEBS letters. 1980;118:293–295. doi: 10.1016/0014-5793(80)80241-9. [DOI] [PubMed] [Google Scholar]
- 38.Sun YS, Landry JP, Fei YY, Zhu XD, Luo JT, Wang XB, Lam KS. Effect of fluorescently labeling protein probes on kinetics of protein-ligand reactions. Langmuir : the ACS journal of surfaces and colloids. 2008;24:13399–13405. doi: 10.1021/la802097z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asbach B, Kolb M, Liss M, Wagner R, Schaferling M. Protein microarray assay for the screening of SH3 domain interactions. Analytical and bioanalytical chemistry. 2010;398:1937–1946. doi: 10.1007/s00216-010-4202-x. [DOI] [PubMed] [Google Scholar]
- 40.Yeo DS, Panicker RC, Tan LP, Yao SQ. Strategies for immobilization of biomolecules in a microarray. Combinatorial chemistry & high throughput screening. 2004;7:213–221. doi: 10.2174/1386207043328823. [DOI] [PubMed] [Google Scholar]
- 41.Hu S, Xie Z, Qian J, Blackshaw S, Zhu H. Functional protein microarray technology. Wiley interdisciplinary reviews Systems biology and medicine. 2011;3:255–268. doi: 10.1002/wsbm.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brueggemeier SB, Wu D, Kron SJ, Palecek SP. Protein-acrylamide copolymer hydrogels for array-based detection of tyrosine kinase activity from cell lysates. Biomacromolecules. 2005;6:2765–2775. doi: 10.1021/bm050257v. [DOI] [PubMed] [Google Scholar]
- 43.Landgraf C, Panni S, Montecchi-Palazzi L, Castagnoli L, Schneider-Mergener J, Volkmer-Engert R, Cesareni G. Protein interaction networks by proteome peptide scanning. PLoS biology. 2004;2:E14. doi: 10.1371/journal.pbio.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li L, Wu C, Huang H, Zhang K, Gan J, Li SS. Prediction of phosphotyrosine signaling networks using a scoring matrix-assisted ligand identification approach. Nucleic acids research. 2008;36:3263–3273. doi: 10.1093/nar/gkn161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu BA, Jablonowski K, Shah EE, Engelmann BW, Jones RB, Nash PD. SH2 domains recognize contextual peptide sequence information to determine selectivity. Molecular & cellular proteomics : MCP. 2010;9:2391–2404. doi: 10.1074/mcp.M110.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jespers LS, Messens JH, De Keyser A, Eeckhout D, Van den Brande I, Gansemans YG, Lauwereys MJ, Vlasuk GP, Stanssens PE. Surface expression and ligand-based selection of cDNAs fused to filamentous phage gene VI. Biotechnology (N Y) 1995;13:378–382. doi: 10.1038/nbt0495-378. [DOI] [PubMed] [Google Scholar]
- 47.Fuh G, Pisabarro MT, Li Y, Quan C, Lasky LA, Sidhu SS. Analysis of PDZ domain-ligand interactions using carboxyl-terminal phage display. The Journal of biological chemistry. 2000;275:21486–21491. doi: 10.1074/jbc.275.28.21486. [DOI] [PubMed] [Google Scholar]
- 48.Smith GP, Petrenko VA. Phage Display. Chemical reviews. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 49.Pande J, Szewczyk MM, Grover AK. Phage display: concept, innovations, applications and future. Biotechnology advances. 2010;28:849–858. doi: 10.1016/j.biotechadv.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 50.Zucconi A, Dente L, Santonico E, Castagnoli L, Cesareni G. Selection of ligands by panning of domain libraries displayed on phage lambda reveals new potential partners of synaptojanin 1. Journal of molecular biology. 2001;307:1329–1339. doi: 10.1006/jmbi.2001.4572. [DOI] [PubMed] [Google Scholar]
- 51.Karkkainen S, Hiipakka M, Wang JH, Kleino I, Vaha-Jaakkola M, Renkema GH, Liss M, Wagner R, Saksela K. Identification of preferred protein interactions by phage-display of the human Src homology-3 proteome. EMBO reports. 2006;7:186–191. doi: 10.1038/sj.embor.7400596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kesti T, Ruppelt A, Wang JH, Liss M, Wagner R, Tasken K, Saksela K. Reciprocal regulation of SH3 and SH2 domain binding via tyrosine phosphorylation of a common site in CD3epsilon. J Immunol. 2007;179:878–885. doi: 10.4049/jimmunol.179.2.878. [DOI] [PubMed] [Google Scholar]
- 53.Voss M, Lettau M, Janssen O. Identification of SH3 domain interaction partners of human FasL (CD178) by phage display screening. BMC immunology. 2009;10:53. doi: 10.1186/1471-2172-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sweeney MC, Wavreille AS, Park J, Butchar JP, Tridandapani S, Pei D. Decoding protein-protein interactions through combinatorial chemistry: sequence specificity of SHP-1, SHP-2, and SHIP SH2 domains. Biochemistry. 2005;44:14932–14947. doi: 10.1021/bi051408h. [DOI] [PubMed] [Google Scholar]
- 55.Yaoi T, Chamnongpol S, Jiang X, Li X. Src homology 2 domain-based high throughput assays for profiling downstream molecules in receptor tyrosine kinase pathways. Molecular & cellular proteomics : MCP. 2006;5:959–968. doi: 10.1074/mcp.T600002-MCP200. [DOI] [PubMed] [Google Scholar]
- 56.Astle JM, Simpson LS, Huang Y, Reddy MM, Wilson R, Connell S, Wilson J, Kodadek T. Seamless bead to microarray screening: rapid identification of the highest affinity protein ligands from large combinatorial libraries. Chemistry & biology. 2010;17:38–45. doi: 10.1016/j.chembiol.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muller K, Gombert FO, Manning U, Grossmuller F, Graff P, Zaegel H, Zuber JF, Freuler F, Tschopp C, Baumann G. Rapid identification of phosphopeptide ligands for SH2 domains. Screening of peptide libraries by fluorescence-activated bead sorting. The Journal of biological chemistry. 1996;271:16500–16505. [PubMed] [Google Scholar]
- 58.Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG. A general method for site-specific incorporation of unnatural amino acids into proteins. Science. 1989;244:182–188. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- 59.Benner SA. Expanding the genetic lexicon: incorporating non-standard amino acids into proteins by ribosome-based synthesis. Trends in biotechnology. 1994;12:158–163. doi: 10.1016/0167-7799(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 60.Young TS, Schultz PG. Beyond the canonical 20 amino acids: expanding the genetic lexicon. The Journal of biological chemistry. 2010;285:11039–11044. doi: 10.1074/jbc.R109.091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chin JW. Reprogramming the genetic code. The EMBO journal. 2011;30:2312–2324. doi: 10.1038/emboj.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu W, Brock A, Chen S, Chen S, Schultz PG. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nature methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 63.Hino N, Okazaki Y, Kobayashi T, Hayashi A, Sakamoto K, Yokoyama S. Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid. Nature methods. 2005;2:201–206. doi: 10.1038/nmeth739. [DOI] [PubMed] [Google Scholar]
- 64.Hino N, Oyama M, Sato A, Mukai T, Iraha F, Hayashi A, Kozuka-Hata H, Yamamoto T, Yokoyama S, Sakamoto K. Genetic incorporation of a photo-crosslinkable amino acid reveals novel protein complexes with GRB2 in mammalian cells. Journal of molecular biology. 2011;406:343–353. doi: 10.1016/j.jmb.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 65.Okada H, Uezu A, Mason FM, Soderblom EJ, Moseley MA, 3rd, Soderling SH. SH3 domain-based phototrapping in living cells reveals Rho family GAP signaling complexes. Science signaling. 2011;4:rs13. doi: 10.1126/scisignal.2002189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davis L, Chin JW. Designer proteins: applications of genetic code expansion in cell biology. Nature reviews Molecular cell biology. 2012;13:168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 67.Xie J, Supekova L, Schultz PG. A genetically encoded metabolically stable analogue of phosphotyrosine in Escherichia coli. ACS chemical biology. 2007;2:474–478. doi: 10.1021/cb700083w. [DOI] [PubMed] [Google Scholar]
- 68.Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. The EMBO journal. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J, Rotin D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. The EMBO journal. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 70.Tong AH, Drees B, Nardelli G, Bader GD, Brannetti B, Castagnoli L, Evangelista M, Ferracuti S, Nelson B, Paoluzi S, Quondam M, Zucconi A, Hogue CW, Fields S, Boone C, Cesareni G. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science. 2002;295:321–324. doi: 10.1126/science.1064987. [DOI] [PubMed] [Google Scholar]
- 71.Wang B, Lemay S, Tsai S, Veillette A. SH2 domain-mediated interaction of inhibitory protein tyrosine kinase Csk with protein tyrosine phosphatase-HSCF. Molecular and cellular biology. 2001;21:1077–1088. doi: 10.1128/MCB.21.4.1077-1088.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liao J, Fu Y, Shuai K. Distinct roles of the NH2- and COOH-terminal domains of the protein inhibitor of activated signal transducer and activator of transcription (STAT) 1 (PIAS1) in cytokine-induced PIAS1-Stat1 interaction. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5267–5272. doi: 10.1073/pnas.97.10.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ito T, Ota K, Kubota H, Yamaguchi Y, Chiba T, Sakuraba K, Yoshida M. Roles for the two-hybrid system in exploration of the yeast protein interactome. Molecular & cellular proteomics : MCP. 2002;1:561–566. doi: 10.1074/mcp.r200005-mcp200. [DOI] [PubMed] [Google Scholar]
- 74.Hu H, Columbus J, Zhang Y, Wu D, Lian L, Yang S, Goodwin J, Luczak C, Carter M, Chen L, James M, Davis R, Sudol M, Rodwell J, Herrero JJ. A map of WW domain family interactions. Proteomics. 2004;4:643–655. doi: 10.1002/pmic.200300632. [DOI] [PubMed] [Google Scholar]
- 75.Cui H, Hayashi A, Sun HS, Belmares MP, Cobey C, Phan T, Schweizer J, Salter MW, Wang YT, Tasker RA, Garman D, Rabinowitz J, Lu PS, Tymianski M. PDZ protein interactions underlying NMDA receptor-mediated excitotoxicity and neuroprotection by PSD-95 inhibitors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:9901–9915. doi: 10.1523/JNEUROSCI.1464-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reindl W, Strebhardt K, Berg T. A high-throughput assay based on fluorescence polarization for inhibitors of the polo-box domain of polo-like kinase 1. Analytical biochemistry. 2008;383:205–209. doi: 10.1016/j.ab.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 77.Wigle TJ, Herold JM, Senisterra GA, Vedadi M, Kireev DB, Arrowsmith CH, Frye SV, Janzen WP. Screening for inhibitors of low-affinity epigenetic peptide-protein interactions: an AlphaScreen-based assay for antagonists of methyl-lysine binding proteins. Journal of biomolecular screening. 2010;15:62–71. doi: 10.1177/1087057109352902. [DOI] [PubMed] [Google Scholar]
- 78.Taouji S, Dahan S, Bosse R, Chevet E. Current Screens Based on the AlphaScreen Technology for Deciphering Cell Signalling Pathways. Current genomics. 2009;10:93–101. doi: 10.2174/138920209787847041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eglen RM, Reisine T, Roby P, Rouleau N, Illy C, Bosse R, Bielefeld M. The use of AlphaScreen technology in HTS: current status. Current chemical genomics. 2008;1:2–10. doi: 10.2174/1875397300801010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Blokzijl A, Friedman M, Ponten F, Landegren U. Profiling protein expression and interactions: proximity ligation as a tool for personalized medicine. Journal of internal medicine. 2010;268:232–245. doi: 10.1111/j.1365-2796.2010.02256.x. [DOI] [PubMed] [Google Scholar]
- 81.Leuchowius KJ, Jarvius M, Wickstrom M, Rickardson L, Landegren U, Larsson R, Soderberg O, Fryknas M, Jarvius J. High content screening for inhibitors of protein interactions and post-translational modifications in primary cells by proximity ligation. Molecular & cellular proteomics : MCP. 2010;9:178–183. doi: 10.1074/mcp.M900331-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fredriksson S, Dixon W, Ji H, Koong AC, Mindrinos M, Davis RW. Multiplexed protein detection by proximity ligation for cancer biomarker validation. Nature methods. 2007;4:327–329. doi: 10.1038/nmeth1020. [DOI] [PubMed] [Google Scholar]
- 83.Dinkel H, Chica C, Via A, Gould CM, Jensen LJ, Gibson TJ, Diella F. Phospho.ELM: a database of phosphorylation sites--update 2011. Nucleic acids research. 2011;39:D261–D267. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yaffe MB. Phosphotyrosine-binding domains in signal transduction. Nature reviews Molecular cell biology. 2002;3:177–186. doi: 10.1038/nrm759. [DOI] [PubMed] [Google Scholar]
- 85.Pawson T, Gish GD, Nash P. SH2 domains, interaction modules and cellular wiring. Trends in cell biology. 2001;11:504–511. doi: 10.1016/s0962-8924(01)02154-7. [DOI] [PubMed] [Google Scholar]
- 86.Waksman G, Shoelson SE, Pant N, Cowburn D, Kuriyan J. Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: crystal structures of the complexed and peptide-free forms. Cell. 1993;72:779–790. doi: 10.1016/0092-8674(93)90405-f. [DOI] [PubMed] [Google Scholar]
- 87.Machida K, Mayer BJ. The SH2 domain: versatile signaling module and pharmaceutical target. Biochimica et biophysica acta. 2005;1747:1–25. doi: 10.1016/j.bbapap.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 88.Smith MJ, Hardy WR, Murphy JM, Jones N, Pawson T. Screening for PTB domain binding partners and ligand specificity using proteome-derived NPXY peptide arrays. Molecular and cellular biology. 2006;26:8461–8474. doi: 10.1128/MCB.01491-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 90.Dengjel J, Kratchmarova I, Blagoev B. Receptor tyrosine kinase signaling: a view from quantitative proteomics. Molecular bioSystems. 2009;5:1112–1121. doi: 10.1039/b909534a. [DOI] [PubMed] [Google Scholar]
- 91.Wilson KJ, Gilmore JL, Foley J, Lemmon MA, Riese DJ., 2nd Functional selectivity of EGF family peptide growth factors: implications for cancer. Pharmacology & therapeutics. 2009;122:1–8. doi: 10.1016/j.pharmthera.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Molecular systems biology. 2005;1 doi: 10.1038/msb4100012. 2005 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kaushansky A, Gordus A, Budnik BA, Lane WS, Rush J, MacBeath G. System-wide investigation of ErbB4 reveals 19 sites of Tyr phosphorylation that are unusually selective in their recruitment properties. Chemistry & biology. 2008;15:808–817. doi: 10.1016/j.chembiol.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2000;14:231–241. [PubMed] [Google Scholar]
- 95.Li SS. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. The Biochemical journal. 2005;390:641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zarrinpar A, Bhattacharyya RP, Lim WA. The structure and function of proline recognition domains. Science's STKE : signal transduction knowledge environment. 2003;2003:RE8. doi: 10.1126/stke.2003.179.re8. [DOI] [PubMed] [Google Scholar]
- 97.Mayer BJ. SH3 domains: complexity in moderation. Journal of cell science. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 98.Bork P, Sudol M. The WW domain: a signalling site in dystrophin? Trends in biochemical sciences. 1994;19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 99.Ball LJ, Jarchau T, Oschkinat H, Walter U. EVH1 domains: structure, function and interactions. FEBS letters. 2002;513:45–52. doi: 10.1016/s0014-5793(01)03291-4. [DOI] [PubMed] [Google Scholar]
- 100.Kofler MM, Freund C. The GYF domain. The FEBS journal. 2006;273:245–256. doi: 10.1111/j.1742-4658.2005.05078.x. [DOI] [PubMed] [Google Scholar]
- 101.Ghose R, Shekhtman A, Goger MJ, Ji H, Cowburn D. A novel, specific interaction involving the Csk SH3 domain and its natural ligand. Nature structural biology. 2001;8:998–1004. doi: 10.1038/nsb1101-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang X, Poy F, Zhang R, Joachimiak A, Sudol M, Eck MJ. Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan. Nature structural biology. 2000;7:634–638. doi: 10.1038/77923. [DOI] [PubMed] [Google Scholar]
- 103.Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 104.Durocher D, Henckel J, Fersht AR, Jackson SP. The FHA domain is a modular phosphopeptide recognition motif. Molecular cell. 1999;4:387–394. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- 105.Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 106.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 107.Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annual review of pharmacology and toxicology. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 108.Lu CH, Sun H, Abu Bakar FB, Uttamchandani M, Zhou W, Liou YC, Yao SQ. Rapid affinity-based fingerprinting of 14-3-3 isoforms using a combinatorial peptide microarray. Angew Chem Int Ed Engl. 2008;47:7438–7441. doi: 10.1002/anie.200801395. [DOI] [PubMed] [Google Scholar]
- 109.Chan PM, Ng YW, Manser E. A robust protocol to map binding sites of the 14-3-3 interactome: Cdc25C requires phosphorylation of both S216 and S263 to bind 14-3-3. Molecular & cellular proteomics : MCP. 2011;10 doi: 10.1074/mcp.M110.005157. M110 005157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, Metalnikov P, O'Donnell P, Taylor P, Taylor L, Zougman A, Woodgett JR, Langeberg LK, Scott JD, Pawson T. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Current biology : CB. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 111.Pozuelo Rubio M, Geraghty KM, Wong BH, Wood NT, Campbell DG, Morrice N, Mackintosh C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. The Biochemical journal. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meek SE, Lane WS, Piwnica-Worms H. Comprehensive proteomic analysis of interphase and mitotic 14-3-3-binding proteins. The Journal of biological chemistry. 2004;279:32046–32054. doi: 10.1074/jbc.M403044200. [DOI] [PubMed] [Google Scholar]
- 113.Johnson C, Tinti M, Wood NT, Campbell DG, Toth R, Dubois F, Geraghty KM, Wong BH, Brown LJ, Tyler J, Gernez A, Chen S, Synowsky S, MacKintosh C. Visualization and biochemical analyses of the emerging mammalian 14-3-3-phosphoproteome. Molecular & cellular proteomics : MCP. 2011;10 doi: 10.1074/mcp.M110.005751. M110 005751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wilker EW, Grant RA, Artim SC, Yaffe MB. A structural basis for 14-3-3sigma functional specificity. The Journal of biological chemistry. 2005;280:18891–18898. doi: 10.1074/jbc.M500982200. [DOI] [PubMed] [Google Scholar]
- 115.Yacoubian TA, Slone SR, Harrington AJ, Hamamichi S, Schieltz JM, Caldwell KA, Caldwell GA, Standaert DG. Differential neuroprotective effects of 14-3-3 proteins in models of Parkinson's disease. Cell death & disease. 2010;1:e2. doi: 10.1038/cddis.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gardino AK, Smerdon SJ, Yaffe MB. Structural determinants of 14-3-3 binding specificities and regulation of subcellular localization of 14-3-3-ligand complexes: a comparison of the Xray crystal structures of all human 14-3-3 isoforms. Seminars in cancer biology. 2006;16:173–182. doi: 10.1016/j.semcancer.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 117.Panni S, Montecchi-Palazzi L, Kiemer L, Cabibbo A, Paoluzi S, Santonico E, Landgraf C, Volkmer-Engert R, Bachi A, Castagnoli L, Cesareni G. Combining peptide recognition specificity and context information for the prediction of the 14-3-3-mediated interactome in S. cerevisiae and H. sapiens. Proteomics. 2011;11:128–143. doi: 10.1002/pmic.201000030. [DOI] [PubMed] [Google Scholar]
- 118.Yaffe MB. How do 14-3-3 proteins work?-- Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS letters. 2002;513:53–57. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
- 119.Yang X, Lee WH, Sobott F, Papagrigoriou E, Robinson CV, Grossmann JG, Sundstrom M, Doyle DA, Elkins JM. Structural basis for protein-protein interactions in the 14-3-3 protein family. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17237–17242. doi: 10.1073/pnas.0605779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes & development. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 121.Huen MS, Chen J. The DNA damage response pathways: at the crossroad of protein modifications. Cell research. 2008;18:8–16. doi: 10.1038/cr.2007.109. [DOI] [PubMed] [Google Scholar]
- 122.Mohammad DH, Yaffe MB. 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA repair. 2009;8:1009–1017. doi: 10.1016/j.dnarep.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rodriguez M, Yu X, Chen J, Songyang Z. Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains. The Journal of biological chemistry. 2003;278:52914–52918. doi: 10.1074/jbc.C300407200. [DOI] [PubMed] [Google Scholar]
- 124.Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 125.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes & development. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Leung CC, Glover JN. BRCT domains: easy as one, two, three. Cell Cycle. 2011;10:2461–2470. doi: 10.4161/cc.10.15.16312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lee HJ, Zheng JJ. PDZ domains and their binding partners: structure, specificity, and modification. Cell communication and signaling : CCS. 2010;8:8. doi: 10.1186/1478-811X-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 129.Nourry C, Grant SG, Borg JP. PDZ domain proteins: plug and play! Science's STKE : signal transduction knowledge environment. 2003;2003:RE7. doi: 10.1126/stke.2003.179.re7. [DOI] [PubMed] [Google Scholar]
- 130.Hui S, Bader GD. Proteome scanning to predict PDZ domain interactions using support vector machines. BMC bioinformatics. 2010;11:507. doi: 10.1186/1471-2105-11-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell research. 2011;21:564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lim WA, Pawson T. Phosphotyrosine signaling: evolving a new cellular communication system. Cell. 2010;142:661–667. doi: 10.1016/j.cell.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 135.Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. In vivo residue-specific histone methylation dynamics. The Journal of biological chemistry. 2010;285:3341–3350. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic acids research. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Iberg AN, Espejo A, Cheng D, Kim D, Michaud-Levesque J, Richard S, Bedford MT. Arginine methylation of the histone H3 tail impedes effector binding. The Journal of biological chemistry. 2008;283:3006–3010. doi: 10.1074/jbc.C700192200. [DOI] [PubMed] [Google Scholar]
- 138.Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 139.Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of combinatorial histone modifications on antibody and effector protein recognition. Current biology : CB. 2011;21:53–58. doi: 10.1016/j.cub.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- 141.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 142.Xu C, Min J. Structure and function of WD40 domain proteins. Protein & cell. 2011;2:202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Stirnimann CU, Petsalaki E, Russell RB, Muller CW. WD40 proteins propel cellular networks. Trends in biochemical sciences. 2010;35:565–574. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 144.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Molecular cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 145.Pashkova N, Gakhar L, Winistorfer SC, Yu L, Ramaswamy S, Piper RC. WD40 repeat propellers define a ubiquitin-binding domain that regulates turnover of F box proteins. Molecular cell. 2010;40:433–443. doi: 10.1016/j.molcel.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rao PN, Gullipalli D, Bhuyan AK. Bacterially expressed recombinant WD40 domain of human Apaf-1. Protein expression and purification. 2009;67:53–60. doi: 10.1016/j.pep.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 147.Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 148.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. Journal of molecular biology. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 149.Feng W, Zhang M. Organization and dynamics of PDZ-domain-related supramodules in the postsynaptic density. Nature reviews Neuroscience. 2009;10:87–99. doi: 10.1038/nrn2540. [DOI] [PubMed] [Google Scholar]
- 150.Carducci M, Perfetto L, Briganti L, Paoluzi S, Costa S, Zerweck J, Schutkowski M, Castagnoli L, Cesareni G. The protein interaction network mediated by human SH3 domains. Biotechnology advances. 2012;30:4–15. doi: 10.1016/j.biotechadv.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 151.Bashor CJ, Horwitz AA, Peisajovich SG, Lim WA. Rewiring cells: synthetic biology as a tool to interrogate the organizational principles of living systems. Annual review of biophysics. 2010;39:515–537. doi: 10.1146/annurev.biophys.050708.133652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shou C, Bhardwaj N, Lam HY, Yan KK, Kim PM, Snyder M, Gerstein MB. Measuring the evolutionary rewiring of biological networks. PLoS computational biology. 2011;7 doi: 10.1371/journal.pcbi.1001050. e1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pincus D, Letunic I, Bork P, Lim WA. Evolution of the phospho-tyrosine signaling machinery in premetazoan lineages. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9680–9684. doi: 10.1073/pnas.0803161105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Flint AJ, Tiganis T, Barford D, Tonks NK. Development of "substrate-trapping" mutants to identify physiological substrates of protein tyrosine phosphatases. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Statsuk AV, Maly DJ, Seeliger MA, Fabian MA, Biggs WH, 3rd, Lockhart DJ, Zarrinkar PP, Kuriyan J, Shokat KM. Tuning a three-component reaction for trapping kinase substrate complexes. Journal of the American Chemical Society. 2008;130:17568–17574. doi: 10.1021/ja807066f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Huang J, Koide A, Makabe K, Koide S. Design of protein function leaps by directed domain interface evolution. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6578–6583. doi: 10.1073/pnas.0801097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pershad K, Pavlovic JD, Graslund S, Nilsson P, Colwill K, Karatt-Vellatt A, Schofield DJ, Dyson MR, Pawson T, Kay BK, McCafferty J. Generating a panel of highly specific antibodies to 20 human SH2 domains by phage display. Protein engineering, design & selection : PEDS. 2010;23:279–288. doi: 10.1093/protein/gzq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Colwill K, Graslund S. A roadmap to generate renewable protein binders to the human proteome. Nature methods. 2011;8:551–558. doi: 10.1038/nmeth.1607. [DOI] [PubMed] [Google Scholar]