Abstract
The mechanisms that maintain the stability of chromosome ends have broad impact on genome integrity in all eukaryotes. Budding yeast is a premier organism for telomere studies. Many fundamental concepts of telomere and telomerase function were first established in yeast and then extended to other organisms. We present a comprehensive review of yeast telomere biology that covers capping, replication, recombination, and transcription. We think of it as yeast telomeres—soup to nuts.
Eukaryotic chromosomes are linear DNA molecules with physical ends, called telomeres. It is estimated that as many as 10,000 DNA damaging events occur each day in every cell in the human body (Loeb 2011). Perhaps the most hazardous of these events are double-stranded DNA breaks (DSBs), which create chromosome ends at internal sites on chromosomes. Thus, a central question is how cells distinguish natural ends or telomeres from DSBs. Telomeres on one hand are essential for the stable maintenance of chromosomes: they must be retained—they cannot be lost by degradation or fused with other ends. Exactly the opposite applies to DSBs: they must be repaired by either homologous or nonhomologous recombination, and this repair often involves regulated degradation of the DSB. In fact, unrepaired DSBs lead to cell cycle arrest to provide time for their repair. Capping is used to describe how telomeres prevent their degradation and recombinational fusion (Muller 1938; McClintock 1939). Perhaps as a consequence of capping, the regions near telomeres are gene poor. In many organisms, telomere proximal genes are subjected to a special type of transcriptional regulation called telomere position effect (TPE), where transcription of genes near telomeres is metastably repressed. Another key role for telomeres is to provide the substrate for a special mechanism of replication. Telomere replication is carried out by telomerase, a specialized ribonucleoprotein complex that is mechanistically related to reverse transcriptases (Greider and Blackburn 1987).
The biology of telomerase has broad ramifications for human health and aging. Therefore, the discovery of telomerase and studies on telomere capping by Elizabeth Blackburn, Carol Greider, and Jack Szostak, were honored with the 2009 Nobel Prize in Medicine. All three prize winners carried out research in single-cell organisms, including budding yeast. As described in this review, Saccharomyces cerevisiae continues to be a premier organism for telomere research.
Sequence and Structure of Telomeric Regions
Like most organisms whose telomeres are maintained by telomerase, the ends of S. cerevisiae chromosomes consist of nonprotein coding repeated DNA (Figure 1A). There are 300 ± 75 bp of simple repeats, typically abbreviated C1-3A/TG1-3. S. cerevisiae telomeric DNA is unusual, although not unique, in being heterogeneous. This sequence heterogeneity is due to a combination of effects: in a given extension cycle, only a portion of the RNA template is used and/or the RNA template and telomeric DNA align in different registers in different extension cycles (Forstemann and Lingner 2001). The heterogeneity of yeast telomeric DNA is experimentally useful as it makes it possible to distinguish newly synthesized from preexisting telomeric DNA (Wang and Zakian 1990; Teixeira et al. 2004). When many copies of the same telomere are sequenced from a given colony, the exact sequence of the internal half is the same from telomere to telomere while the terminal half turns over much more rapidly (Wang and Zakian 1990). Thus, under most conditions, only the terminal half of the telomere is subject to degradation and/or telomerase lengthening. These repeats in conjunction with the proteins that bind them are necessary and sufficient for telomere function.
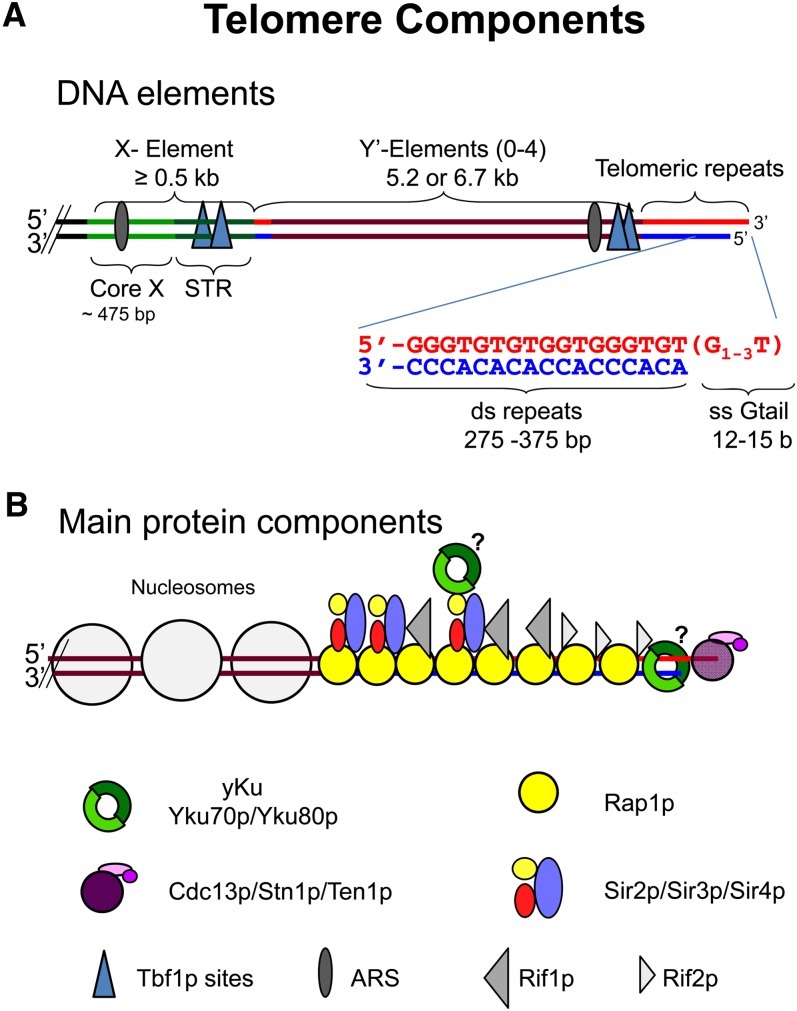
Figure 1 .
DNA structure and major protein components of telomeres. (A) DNA arrangement at telomeres indicating the subtelomeric X and Y′ elements as well as the terminal repeat sequences. Red strand, G-rich strand with 3′ overhanging end and blue strand, C-rich strand with 5′ end. Core X and STR (subtelomeric repeated elements; Louis et al. 1994) represent subareas in the X element. (B) Proteins are schematically positioned on the telomere drawing and the identity of the symbols explained on the bottom. Open circles represent nucleosomes (not to scale).
As in most eukaryotes, the very ends of S. cerevisiae chromosomes are not blunt ends. Rather the G-rich strand extends to form a 3′ single strand tail or G tail (Figure 1A). Throughout most of the cell cycle, G tails are short, only 12 to 15 nucleotides (nt) (Larrivee et al. 2004). However, G tails are much longer, ≥30–100 nt in size, during a short period in late S/G2 phase when they can be detected readily by nondenaturing Southern hybridization (Wellinger et al. 1993a,b). Long G tails are not due solely to telomerase-mediated lengthening as they are seen in late S/G2 phase even in telomerase-deficient cells (Wellinger et al. 1996; Dionne and Wellinger 1998). G tails are generated by cell-cycle–regulated C-strand degradation, which is dependent on the kinase activity of Cdk1p (Cdc28p; Frank et al. 2006; Vodenicharov and Wellinger 2006). This generation is obligatorily linked to semiconservative DNA replication, which occurs prior to C-strand degradation (Wellinger et al. 1993a; Dionne and Wellinger 1998).
Also similar to most organisms, yeast telomeric regions contain subtelomeric, middle, repetitive elements, often called TAS elements (telomere associated sequences; Figure 1A and http://www.nottingham.ac.uk/biology/people/louis/telomere-data-sets.aspx). S. cerevisiae has two classes of TAS elements, X and Y′. Y′ is found in zero to four tandem copies immediately internal to the telomeric repeats (Chan and Tye 1983a,b). About half of the telomeres in a given strain lack Y′, and the identity of Y′-less telomeres differs from strain to strain (Horowitz et al. 1984; Zakian et al. 1986). Y′ comes in two sizes, Y′ long (6.7 kb) and Y′ short (5.2 kb) (Chan and Tye 1983a,b), which differ from each other by multiple small insertions/deletions (Louis and Haber 1992). X is present at virtually all telomeres and is much more heterogeneous in sequence and size. Although X is found on all telomeres, it is composed of a series of repeats, many of which are present on only a subset of telomeres. When telomeres contain both X and Y′, X is centromere proximal to Y′. Short tracts of telomeric DNA are sometimes found at the Y′–X and Y′–Y′ junctions (Walmsley et al. 1984; Figure 1A).
Subtelomeric regions are dynamic, undergoing frequent recombination (Horowitz et al. 1984; Louis and Haber 1990). Moreover, subtelomeric repeats diverge rapidly even among related yeast strains (Chan and Tye 1983a,b). X and Y′ both contain potential replication origins or ARS elements (autonomously replicating sequences) whose presence probably contributes to the dynamic nature of subtelomeric regions. X and Y′ have binding sites for multiple transcription factors, whose identity differs from telomere to telomere (Mak et al. 2009). Because the sequence of subtelomeric regions and the proteins that bind them are variable, their presence can confer distinct behaviors on individual telomeres.
Whereas complete loss of the C1-3A/TG1-3 telomeric repeats from a chromosome end results in extremely high loss rates for the affected chromosome, chromosomes that lack X and Y′ at one (Sandell and Zakian 1993) or even both ends have normal mitotic stability and go through meiosis with ease (S. S. Wang and V. A. Zakian, unpublished results). However, Y′ amplification by recombination can provide a telomere maintenance function to cells lacking telomerase (Lundblad and Blackburn 1993). Y′ can also move by a transposition-like RNA-mediated process (Maxwell et al. 2004).
Ty5 is a transposable element found only in heterochromatin, including subtelomeric DNA. The number of Ty5 elements varies from strain to strain. The S288C strain has eight Ty5 insertions: six near telomeres and two near the HMR silent mating type locus (Zou et al. 1995). This chromosomal distribution is quite different from that of other classes of Ty elements, which are found close to tRNA genes. Movement of Ty5 to telomeres and HMR is regulated by the targeting domain of the Ty5-encoded integrase that interacts directly with Sir4p, one of the silencing proteins found in telomeric regions (Xie et al. 2001).
Telomeric Chromatin
Telomere binding proteins: direct binders and associated proteins
Table 1 presents a list of proteins that act at telomeres, divided into functional categories. Many of these proteins have multiple roles and could be listed in more than one category. The protein complexes associated with telomeres can be subdivided according to the three regions to which they bind: (A) subtelomeric areas containing Y′ and X, (B) double-stranded terminal repeat area, and (C) the 3′ G tail (Figure 1).
Table 1 . Major genes affecting Saccharomyces cerevisiae telomeres.
| Gene name | aa/MW (KD) | Essential (yes/no) | Function(s)a | |
|---|---|---|---|---|
| Structural proteins | ||||
| RAP1 | 827/92.4 | Yes | Sequence-specific double-strand DNA binding telomere capping and length regulation, TPE, major transcription factor. | |
| CDC13 | Cdc 13 complex | 924/104.9 | Yes | Three-protein complex comprised of Cdc13p, Stn1p, and Ten1p, which binds single-strand TG1-3 DNA in sequence-specific manner, capping, telomerase recruitment. |
| STN1 | 494/57.5 | No | ||
| TEN1 | 160/18.6 | No | ||
| RIF1 | 1916/217.9 | No | Interacts w. Rap1p; telomerase regulator. | |
| RIF2 | 395/45.6 | No | Interacts w. Rap1p; telomerase regulator, capping. | |
| YKU70 | YKu complex | 602/70.6 | No | Interacts w. TLC1; telomere length regulation; capping; TPE; telomere positioning; nonhomologous end joining. |
| YKU80 | 629/71.2 | No | ||
| SIR2 | 562/63.2 | No | Interact w. Sir4p; essential for TPE and HM silencing, histone deacetylase. | |
| SIR3 | 978/111.3 | No | Interacts with w. Rap1p, Sir4p, histone tails; essential for TPE and HM silencing. | |
| SIR4 | 1358/152.0 | No | Interacts with Yku80p, Sir2p, Rap1p, and histone tails; essential for TPE and HM silencing; telomere positioning. | |
| TBF1 | 562/62.8 | Yes | TPE boundary function, telomerase recruitment to short telomeres; transcription factor. | |
| NDJ1 | 352/40.8 | No | Meiosis specific, telomere binding, essential for bouquet formation. | |
| Telomere replication | ||||
| EST1 | 699/81.7 | No | Protein subunit of telomerase; recruitment, activation. | |
| EST2 | 884/102.6 | No | Protein subunit of telomerase; catalytic reverse transcriptase. | |
| EST3 | 181/20.5 | No | Protein subunit of telomerase. | |
| TLC1 | 1157 nt | No | Telomerase RNA; repeat templating. | |
| PIF1 | 859/87.6 | No | DNA helicase, removes telomerase from DNA, also required for maintenance of mitochondrial and nontelomeric nuclear DNA. | |
| TEL1 | 2787/321.5 | No | Interacts w. Xrs2p, telomere length regulation; telomerase recruitment; S phase checkpoint kinase. | |
| MRE11 | MRX Complex | 692/77.6 | No | Acts as complex in same pathway as TEL1; recruits telomerase; required for type II survivors and other homologous recombination events; Mre 11p is a nuclease, Rad50p has ATPase and DNA binding activity; Xrs2p interacts with Tel1p. |
| RAD50 | 1312/152.5 | |||
| XRS2 | 854/96.3 | |||
| MEC1 | 2368/273.3 | Yes | Major DNA damage checkpoint kinase; partially redundant function with Tel1p in telomerase recruitment; activated when very short or no telomere; mec1Δ sml1Δ cells are viable but deficient for both telomere and checkpoint functions of Mec1p | |
| RRM3 | 723/81.5 | No | DNA helicase, semiconservative telomere replication; promotes replication at many nontelomeric sites. | |
| Processing and recombination | ||||
| SGS1 | 1447/163.8 | No | DNA helicase, end processing DSBs and telomeres, required for type II survivors; rDNA recombination. | |
| RAD52 | 471/52.4 | No | Essential for all homologous recombination, including type I and type II survivors. | |
| RAD51 | 400/42.9 | No | Homologous recombination, required for type I survivors. | |
| SAE2 | 345/40.0 | No | 5′ strand resection at DSBs and telomeres. | |
| EXO1 | 702/80.1 | No | Nuclease, 5′ end resection at DSBs and telomeres. | |
| DNA2 | 1522/171.6 | Yes | Helicase-nuclease; 5′ end resection at telomeres and DSBs; Okazaki fragment maturation. | |
| POL32 | 350/40.3 | No | Subunit of DNA pol δ; required for break-induced replication and both type I and type II survivors. | |
See text for details and references; although many genes are involved in more than one process, each is listed here under only one heading. Essential/nonessential refers to viability, not telomere maintenance. aa, number of amino acids; MW sizes are from the SGD website http:/www.yeastgenome.org/.
(A) Subtelomeric regions are classified into XY′ and X-only ends. While most of the subtelomeric DNA is likely organized in nucleosomes (Wright et al. 1992), the cores of the X elements have a low histone content, and nucleosomes near the Xs have histone modifications characteristic of silenced regions such as unacetylated lysine 16 on histone H4 (H4K16) (Zhu and Gustafsson 2009). Consistent with these data, the NAD+-dependent histone deacetylase Sir2p, a H4K16 deacetylase, as well as Sir3p, are also enriched over X repeats (Imai et al. 2000; Zhu and Gustafsson 2009; Takahashi et al. 2011), and the area around many X elements is transcriptionally silent (Pryde and Louis 1999). The X elements on XY′ telomeres are organized similarly as on X-only telomeres (Takahashi et al. 2011). However, on the distal Y′ elements, the overall density of nucleosomes as well as the occurrence of H4K16ac is similar to euchromatic areas. In addition, Sir2p and Sir3p are not detected in this region (Zhu and Gustafsson 2009; Takahashi et al. 2011). Collectively, these data suggest that on X-only telomeres, the subtelomeric DNA elements are organized into silenced chromatin that demarcates the terminal area from more internal regions. On XY′ telomeres, the distal Y′ area is organized into chromatin that resembles that of expressed areas with the X element, again acting as a demarcation zone (Fourel et al. 1999; Pryde and Louis 1999; Takahashi et al. 2011). Thus, emerging evidence points toward differences in behavior depending on subtelomeric repeat content and perhaps even individual chromosomal context.
γ-H2A, which is generated by Mec1p/Tel1p-dependent phosphorylation, is also enriched in subtelomeric chromatin (Kim et al. 2007; Szilard et al. 2010). Since this modification normally marks damaged DNA, which activates checkpoints, it is unclear why it persists on telomeres and whether its occurrence has functional consequences. Finally, nucleosomes in certain areas within subtelomeric DNA contain the histone H2A variant H2A.Z. Nucleosomes containing H2A.Z often mark gene promoters for efficient activation and perhaps also function as heterochromatin–euchromatin boundary elements (Guillemette et al. 2005; Albert et al. 2007).
Remarkably, there are a few precise matches to the vertebrate telomeric repeat sequence, (TTAGGG)n, within X and Y′ DNA, and the essential transcription factor Tbf1p (Brigati et al. 1993) binds these repeats in vitro (Liu and Tye 1991) and in vivo (Koering et al. 2000; Preti et al. 2010; Figure 1A). This Tbf1p binding is functionally significant as it participates in telomerase recruitment (Arneric and Lingner 2007). Tbf1p can also provide Rap1p-independent capping on artificial telomeres consisting solely of vertebrate repeats (Alexander and Zakian 2003; Berthiau et al. 2006; Bah et al. 2011; Ribaud et al. 2011; Fukunaga et al. 2012). The boundary between subtelomeric DNA and telomeric repeats appears special as it is preferentially accessible to DNases, restriction enzymes, and DNA modifying enzymes (Conrad et al. 1990; Gottschling 1992; Wright et al. 1992; Wright and Zakian 1995). This behavior suggests a short stretch of DNA that is not strongly associated with proteins. Given this property, limited nuclease digestion can release the distalmost portion of chromosomes containing all telomeric repeat DNA in a soluble and protein bound form called the telosome (Conrad et al. 1990; Wright et al. 1992). This telosome appears to be histone free and should contain all telomeric repeat binding proteins (Wright and Zakian 1995).
(B) Double-stranded telomeric repeat DNA contains high-affinity Rap1p binding sites every ∼20 bp, which correlates well with the estimate that in vitro assembled Rap1 telomeric DNA contains 1 bound Rap1p molecule in 18 (±4) bp (Gilson et al. 1993; Ray and Runge 1999a,b; Figure 1B). Therefore, given an average telomere length of 300 bp, individual telomeres are probably covered by 15–20 Rap1p molecules (Wright and Zakian 1995). Rap1p is an abundant nuclear protein of 827 amino acids that was first discovered by its ability to repress or activate gene expression (repressor activator protein 1) (Shore and Nasmyth 1987). Indeed, given its abundance and the number of telomeric Rap1 binding sites, most (∼90%) Rap1p is not telomere associated. DNA consensus sites for Rap1p binding are quite heterogeneous, but those within telomeric DNA are among the highest affinity sites (Buchman et al. 1988; Longtine et al. 1989; Lieb et al. 2001). Genetic evidence, chromatin immunoprecipitation (ChIP) and in vivo localization leave little doubt that Rap1p covers telomeric DNA in living cells (Conrad et al. 1990; Lustig et al. 1990; Wright and Zakian 1995; Gotta et al. 1996; Bourns et al. 1998). Indeed, the amount of telomere bound Rap1p, along with its binding partners Rif1/2 somehow establishes the actual telomere length (Marcand et al. 1997; Levy and Blackburn 2004).
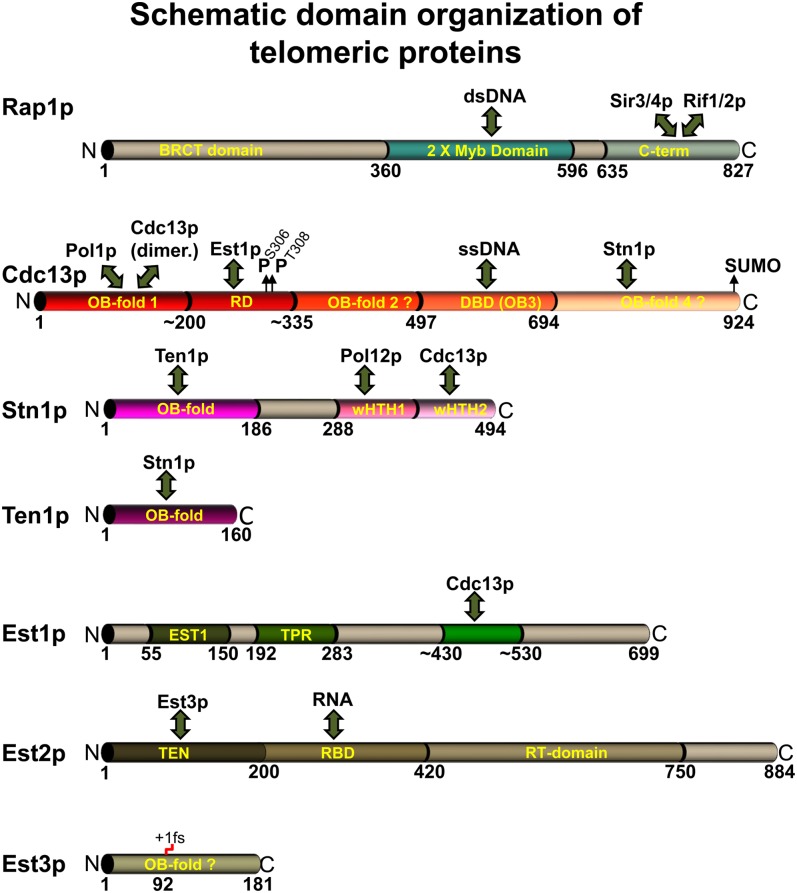
Although studied extensively, the functional domains for Rap1p are not completely defined (Figure 2). Loss of up to 340 amino acids from the N-terminal region, which has a BRCT domain, is well tolerated (Moretti et al. 1994; Graham et al. 1999). However, the double myb domain DNA binding module in the middle of Rap1p is essential for all functions of the protein, including those at telomeres (Graham et al. 1999). For example, temperature-sensitive alleles of RAP1 can cause telomere shortening and telomere-bound Rap1p is required to prevent telomere fusions (Conrad et al. 1990; Lustig et al. 1990; Marcand et al. 2008). The C terminus of Rap1p is key for its telomere functions as both the silencing proteins Sir3p/Sir4p and the length regulatory Rif1p/Rif2p bind this region (Hardy et al. 1992a,b; Moretti et al. 1994; Buck and Shore 1995; Wotton and Shore 1997; Figure 2).
Figure 2 .
Overall domain organizations and interaction areas for major telomeric proteins. Shown are Rap1p, members of the Cdc13 complex, and three protein subunits of the telomerase holoenzyme. Due to the paucity of information for Rif1p or Rif2p, they are omitted. For details on domain definitions, see text. Known interaction domains with other proteins, RNA, or DNA are indicated with a double arrow. Below the proteins, numbers define amino acid positions. Small up arrow indicates known amino acid modifications that affect functions and the red step on Est3p denotes a required +1 frameshift in protein translation.
Another key telomere binding protein is the yeast Ku complex (referred to as YKu), composed of Yku70p and Yku80p (Boulton and Jackson 1996; Porter et al. 1996; Gravel et al. 1998). Given that YKu is essential for DNA repair via non-homologous end joining (NHEJ) and telomeres are protected from NHEJ, the association of YKu with telomeres is counterintuitive. Nevertheless, this association is critical for telomere function (Gravel et al. 1998), not only in yeast but in many organisms (Fisher and Zakian 2005). It is still uncertain where and how Yku associates with chromosomal termini, but there is evidence for two pools, one bound directly to telomeric DNA in a mode similar to that used for the nonspecific DNA end binding in NHEJ and another being associated with telomeric chromatin via a Yku80p–Sir4p interaction (Martin et al. 1999; Roy et al. 2004). ChIP experiments suggest a Sir4p-independent association of YKu with some, but not all, core X sequences, and those bound areas also correlate with a high level of transcriptional and recombination repression (Marvin et al. 2009a,b). Furthermore, given the ability of YKu to associate with telomerase RNA, it has also been suggested that YKu functions to recruit telomerase to telomeres (Peterson et al. 2001; Stellwagen et al. 2003; Fisher et al. 2004; Chan et al. 2008) and/or telomerase trafficking from the cytoplasm to the nucleus (Gallardo et al. 2008, 2011). Consistent with that proposal, YKu association with telomeres is independent of its association with TLC1 RNA and occurs throughout the cell cycle (Fisher et al. 2004).
(C) The essential Cdc13p specifically and avidly binds single-stranded TG1-3 DNA of at least 11 nt in vitro (Lin and Zakian 1996; Nugent et al. 1996; Hughes et al. 2000) and is associated with telomeres in vivo (Bourns et al. 1998; Tsukamoto et al. 2001). The DNA binding domain (DBD) of Cdc13p is confined to amino acids 497–694 of this 924-amino-acid protein (Figure 2), and this domain reproduces the in vitro DNA binding characteristics of the full-length protein (Hughes et al. 2000). Furthermore, structure determinations of this DBD bound to a telomeric G strand provide a model for the very high affinity and specificity of this association (Mitton-Fry et al. 2002, 2004). The relatively large N-terminal region (amino acids 1–455) may contain two OB fold domains plus a region defining an interaction with Est1p that is involved in telomerase recruitment (recruitment domain, RD) (Nugent et al. 1996; Pennock et al. 2001; Figure 2). A direct Est1–RD interaction is shown by in vitro experiments (Wu and Zakian 2011). Finally, the N-terminal or first OB fold domain is important for an interaction with Pol1p and for Cdc13p dimerization (Grandin et al. 2000; Qi and Zakian 2000; Gelinas et al. 2009; Sun et al. 2011).
Two other essential proteins with genetic and biochemical interactions with Cdc13p, namely Stn1p and Ten1p, also have a potential for direct interactions with the single-stranded 3′ overhangs (Grandin et al. 1997, 2001; Gao et al. 2007). The three-member protein complex composed of Cdc13p/Stn1p/Ten1p has been referred to both as the CST complex or telomeric RPA. Herein, we refer to it as the Cdc13 complex. There are several structural similarities between the three members of the Cdc13 complex and the three proteins making up replication protein A (RPA) (Gao et al. 2007; Gelinas et al. 2009), and at least one essential OB fold domain can be swapped between Rpa2p and Stn1p (Gao et al. 2007).
Stn1p and Ten1p may also act independently of Cdc13p. For example, a Stn1p/Ten1p complex when overexpressed can act as a chromosome cap in the absence of Cdc13p (Petreaca et al. 2006, 2007; Sun et al. 2009). Stn1p can be divided roughly into two parts, an N-terminal and a C-terminal domain (Petreaca et al. 2006, 2007; Puglisi et al. 2008; Figure 2). The N-terminal domain, which is necessary for its interaction with Ten1p, is required for its essential functions (Petreaca et al. 2007; Puglisi et al. 2008). The C-terminal domain interacts with both Cdc13p and Pol12p, the latter protein a subunit of the DNA Polα complex that carries out lagging strand DNA replication (Grossi et al. 2004).
Telomere dedicated proteins vs. proteins doing double duty
Remarkably, the majority of telomeric proteins have both telomeric and nontelomeric functions (Table 1). For example, both Rap1p and Tbf1p are essential to regulate expression of a large number of genes, many of which are among the most highly transcribed genes in the genome (Pina et al. 2003; Preti et al. 2010). The Rap1p-associated proteins Sir2p, Sir3p, and Sir4p promote transcriptional silencing not only at telomeres but also at the silent mating type or HM loci (Rusche et al. 2003), and Rif1p has roles in establishing heterochromatin elsewhere than just at telomeres (Hardy et al. 1992b; Buck and Shore 1995; Buonomo 2010). The YKu complex is essential for NHEJ, in particular during G1 phase of the cell cycle (reviewed in Daley et al. 2005). The telomerase regulator Pif1p affects maintenance of mitochondrial DNA and replication of nontelomeric loci with the potential to form G-quadruplex structures (Foury and Kolodynski 1983; Schulz and Zakian 1994; Ivessa et al. 2000; Ribeyre et al. 2009; Paeschke et al. 2011). Taken together, at least for budding yeast, it looks as if the proteins important for telomere function by and large are doing double duty.
How many more genes affect telomere biology?
It is not surprising that a large number of additional genes affect telomere length as genes with general roles in DNA replication, recombination, intra S checkpoint, protein and RNA synthesis pathways would be expected to affect them (Dahlseid et al. 2003; Mozdy et al. 2008). Indeed, two systematic screens of the deletion collection of nonessential genes confirmed this idea (Askree et al. 2004; Gatbonton et al. 2006). Of some concern, the gene sets from the two screens show little overlap, and it is not yet clear how many of the genes act directly.
Screens for suppressors of telomere-capping defects also yielded numerous new interactions (Addinall et al. 2008, 2011). For example, members of the KEOPS complex (CGI121, KAE1, BUD32, and GON7) were linked to telomere biology because they were identified by their ability to suppress the growth defect of cells harboring the cdc13-1 allele incubated at slightly elevated temperatures (Downey et al. 2006). KEOPS genes were also identified via an unrelated screen looking for suppressors of a splicing defect (Kisseleva-Romanova et al. 2006), and one member of the KEOPS complex is linked to chromosome segregation (Ben-Aroya et al. 2008). It appears now that the primary function of the KEOPS complex is to add a specific base modification to certain tRNAs (t6A addition; Srinivasan et al. 2011). Similarly, SUA5, a gene first identified as a translational suppressor and then linked to telomere biology (Na et al. 1992; Meng et al. 2009) is required for the same tRNA modification as the KEOPS complex (Lin et al. 2010; Srinivasan et al. 2011). How this t6A tRNA modifying activity links with telomere biology is still a puzzle. In summary, with rare exceptions, we think it likely that all genes affecting yeast telomeres have been identified and would not be surprised if many of the genes identified by genome-wide approaches act indirectly.
The Capping Function
Classical chromosome capping
Arguably the most important function of a telomere is that of providing protection to the end of the chromosome. This capping function is the property that prompted chromosome researchers in the 1930s to name the ends of chromosomes telomeres (Muller 1938; McClintock 1939). Classically, the capping function prevents telomeres from being subject to DNA repair by homologous recombination or NHEJ. More recently, the capping function has expanded to include the concept of protecting telomeres from checkpoints as loss of a single telomere elicits a Rad9p-dependent cell cycle arrest (Sandell and Zakian 1993). Loss of these capping functions can be determined by monitoring the integrity of both strands of telomeric DNA, presence of fused chromosome ends, and/or cell cycle arrest. The conservation among eukaryotes of the underlying structure of telomeres, duplex telomeric DNA with G-rich 3′ overhangs and corresponding sequence-specific duplex and single-strand DNA binding proteins, suggests that the mechanisms of capping are based on conserved principles.
The earliest demonstration that Cdc13p functions in chromosome capping was the discovery that in cells with a temperature-sensitive cdc13-1 allele incubated at elevated temperatures, telomeres are degraded in a strand-specific manner such that their C strands are lost for many kilobases (Garvik et al. 1995). In addition, at nonpermissive temperatures, cdc13-1 cells arrest at the G2/M boundary of the cell cycle in a RAD9-dependent fashion (Weinert and Hartwell 1993). These phenotypes also occur in cdc13Δ cells (Vodenicharov and Wellinger 2006). Therefore, cells lacking Cdc13p display the two central hallmarks of telomere uncapping, unstable chromosome ends, and activation of a DNA damage checkpoint. Cdc13p undergoes cell cycle phase-specific post-translational modifications, including phosphorylation and SUMOylation that may affect capping (Tseng et al. 2006; Li et al. 2009; Hang et al. 2011). Genetic and biochemical data indicate that these capping activities of Cdc13p involve Stn1p and Ten1p, both of which are also essential for capping (Grandin et al. 1997, 2001; Gao et al. 2007; Petreaca et al. 2007; Xu et al. 2009).
An inducible degron allele of Cdc13p combined with cell cycle synchrony experiments demonstrated that the Cdc13 complex is only required for capping during late S and G2/M phases, but not in G1 or early S (Vodenicharov and Wellinger 2006, 2007, 2010). One might speculate that replication through the telomere would disrupt its capping function and therefore capping must be reassembled thereafter, creating a time-restricted situation of enhanced requirement for capping and hence Cdc13 complex function. This proposal is in line with the fact that during telomere replication, CDK-dependent end processing is at its peak (Ira et al. 2004; Frank et al. 2006; Vodenicharov and Wellinger 2006). However, given that members of the Cdc13 complex interact with components of the lagging strand machinery, it is also possible that the capping functions of the Cdc13 complex are directly associated with the passage of the replication fork (Nugent et al. 1996; Qi and Zakian 2000; Grossi et al. 2004; Vodenicharov and Wellinger 2010). In this context it is noteworthy that Cdc13p, although very sequence specific, does not require a physical 3′ end for its binding, as it can bind single-strand TG1-3 DNA even if the telomeric DNA is on a circular plasmid (Lin and Zakian 1996; Nugent et al. 1996). It thus remains unclear whether the C-strand–specific degradation of telomeres observed when Cdc13 complex-mediated capping is hampered is due to problems at the physical ends or problems associated with terminating replication of telomeric repeats (Figure 3; Anbalagan et al. 2011).
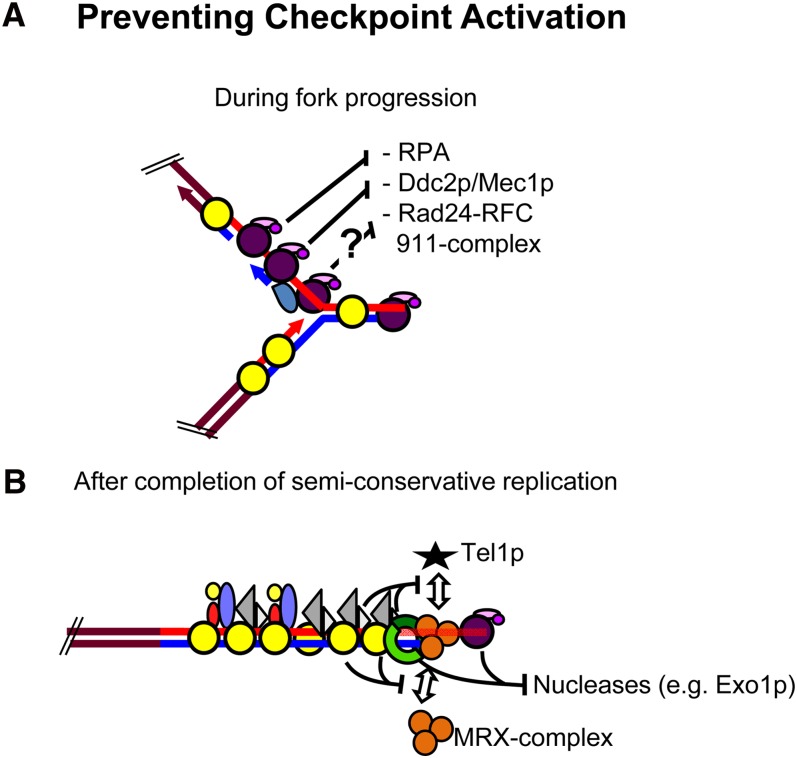
Figure 3 .
Preventing DNA damage checkpoint signaling at telomeres. Schematic of hypotheses for how DNA damage checkpoint signaling is prevented (A) during the passage of the replication fork through the double-stranded telomeric repeat area and (B) after having passed the end. Symbols are as in Figure 1.
Outside S phase, Rap1p is critical for capping. Rap1p with C terminus-associated Rif2p, and to a much lesser extent Rif1p, are important for preventing telomere fusions and limiting end resection (Marcand et al. 2008; Bonetti et al. 2010; Vodenicharov and Wellinger 2010). Furthermore, Rif2p (but not Rif1p) has a prominent role in preventing the association of Tel1p/MRX complex to telomeres (Hirano et al. 2009; Bonetti et al. 2010). MRX is a heterotrimeric complex composed of Mre11p, Rad50p, and Xrs2p that serves important roles in both DSB recognition, telomere capping, and checkpoint activation (Boulton and Jackson 1998; Nugent et al. 1998; Ritchie and Petes 2000; D’Amours and Jackson 2001; Grenon et al. 2001). Most likely there is a nucleolytic activity associated with the complex (Llorente and Symington 2004), and it appears the complex also has the capacity to hold broken chromosome ends in proximity for eventual repair (Kaye et al. 2004; Lobachev et al. 2004). On the other hand, Rif1p, and to a much lesser extent Rif2p, is important to maintain viability in cells where CDC13 capping is compromised (Addinall et al. 2011; Anbalagan et al. 2011). Thus, Rap1p and the associated Rif1p and Rif2p proteins have important capping functions outside of S phase with Rif1p and Rif2p making specific and separable contributions to this capping.
Finally, Yku affects capping in G1 phase (Vodenicharov and Wellinger 2007, 2010; Bonetti et al. 2010) as telomeres in ykuΔ cells are resected at this time, even when bound by the Cdc13 complex. However, the G1 resection in ykuΔ cells is much more modest than, for example, the resection that occurs during late S phase in cdc13-1 cells at elevated temperatures, and this limited resection does not activate a DNA damage checkpoint (Bonetti et al. 2010; Vodenicharov and Wellinger 2010).
It is unclear whether telomerase has a capping function that is independent from its telomere elongation activity. Physical assays do not reveal increased end degradation in tlc1Δ48 or yku80-135i cells (Vodenicharov and Wellinger 2010), mutations that result in reduced Est2p telomere binding (Fisher et al. 2004). However, cdc13-1 cells that also carry either the tlc1Δ48 or yku80-135i mutation are more temperature sensitive than cdc13-1 cells, suggesting that capping is compromised further by reduced Est2p telomere binding in these backgrounds (Vega et al. 2007). Moreover, cells lacking telomerase and the recombination protein Rad52p lose telomeric DNA more rapidly than if they lack telomerase alone (Lundblad and Blackburn 1993). One explanation for these data are that telomerase protects ends from recombinational lengthening (Lee et al. 2007).
Alternative ways of capping
While the Cdc13p-mediated capping of chromosome ends is essential, situations of telomere capping without Cdc13p have been described. In all such cases, chromosomes still end in canonical terminal TG1-3 sequences and in some cases, the repeat sequences are still maintained by telomerase (Larrivee and Wellinger 2006; Petreaca et al. 2006; Zubko and Lydall 2006; Dewar and Lydall 2010). In one particular case, capping requires the DNA polymerase α-associated Pol12p and overexpression of both an N-terminal part of Stn1p and Ten1p (Petreaca et al. 2006). In another case, cdc13Δ cells can be obtained by first deleting key genes involved in exonucleolytic degradation of DNA ends (EXO1, RAD24, and SGS1) and DNA damage checkpoints (RAD9 and PIF1) (Zubko and Lydall 2006; Dewar and Lydall 2010; Ngo and Lydall 2010). In these cases, telomeres are still maintained by telomerase, if homologous recombination is impossible due to a deletion of RAD52 (Zubko and Lydall 2006). Lastly, if telomere repeat maintenance is already accomplished by recombination, as in the survivors that arise in telomerase-deficient cells, then loss of Cdc13p can be tolerated in a small subset of cells. The fact that only a minor fraction of the culture survives suggests that additional events are required to maintain telomeres in such cells (Larrivee and Wellinger 2006).
DNA structures can also provide an alternative mode of capping. For example, cells that lack both major pathways for telomeric repeat maintenance, i.e., telomerase and homologous recombination, and that are also deficient in Exo1p, a 5′ to 3′ single-stranded exonuclease that processes DSBs, can divide and form colonies (Maringele and Lydall 2004b). Chromosomes in these survivor cells do not end in telomeric repeats but rather in DNA palindromes distal to the first essential gene on each chromosome arm.
Crosstalk between DNA damage checkpoint activation and DNA repair
Given that capping protects telomeres from repair and checkpoint activation, it seemed logical to think that proteins involved in DNA repair and checkpoints would not act at telomeres. Paradoxically, many checkpoint and DNA repair proteins associate with telomeres and contribute in important ways to telomeric functions, including capping. For example, the yeast YKu complex, which is critical for NHEJ, is telomere associated (Gravel et al. 1998), and in its absence, telomeres are very short and have long G tails throughout the cell cycle (Boulton and Jackson 1996; Porter et al. 1996; Gravel et al. 1998; Polotnianka et al. 1998). YKu contributes not only to capping but also protects telomeres from recombination, mediates nuclear import and/or retention of telomerase RNA, promotes TPE and telomere tethering (Polotnianka et al. 1998; Peterson et al. 2001; Stellwagen et al. 2003; Hediger et al. 2006; Ribes-Zamora et al. 2007; Gallardo et al. 2008; Marvin et al. 2009a) and is involved in telomere replication (Cosgrove et al. 2002; Gravel and Wellinger 2002).
Mec1p, the most important checkpoint kinase in yeast, has a minor role in telomere length regulation (Ritchie et al. 1999). Consistent with this, Mec1p binding is only detected at ultrashort telomeres that are probably already nonfunctional (Abdallah et al. 2009; McGee et al. 2010; Hector et al. 2012). In fact, Cdc13p inhibits Mec1p binding to a DSB (Hirano and Sugimoto 2007). Moreover, Mec1p prevents telomere formation at DSBs by phosphorylation of Cdc13p, which inhibits Cdc13p association with the DSB (Zhang and Durocher 2010; Ribaud et al. 2011). In addition, Mec1p phosphorylation of Pif1p inhibits telomere addition to DSBs (Makovets and Blackburn 2009). Normally, association of Mec1p to DSBs occurs after end processing and by binding to single-stranded DNA via the replication protein A heterotrimer (RPA) and Ddc2p (Zou and Elledge 2003). An important issue is whether or not RPA binds the single-stranded G tails generated at the end of S phase (Figure 3). RPA is detected transiently at telomeres at this time (Schramke et al. 2003; McGee et al. 2010), but this binding could be explained by the RPA that associates with telomeres during semiconservative replication (McGee et al. 2010). Mec1p binding is not detected at this time, suggesting that Cdc13p prevents RPA binding so that Mec1p-mediated DNA damage signaling is not elicited by the telomeric single-stranded G tails (Figure 3; Gao et al. 2007; Gelinas et al. 2009; McGee et al. 2010).
Although Tel1p associates with DSBs (Nakada et al. 2003; Shima et al. 2005), it has only minor functions in DNA repair. Rather, its major function is telomere length maintenance. Tel1p binds telomeres (Bianchi and Shore 2007b; Hector et al. 2007; Sabourin et al. 2007) via an interaction with the Xrs2p subunit of MRX. Indeed, Tel1p interacts preferentially with short telomeres and is thought to be involved in telomerase recruitment. However, in contrast to its binding at a DSB, its association with short telomeres does not elicit a checkpoint response, a difference that is not fully understood.
Other experiments involving the fate of DSBs made next to telomeric DNA emphasize the interconnections between telomeric DNA and checkpoints. For example, there is some evidence that a tract of telomeric DNA can affect cell cycle progression when it is adjacent to a DSB (Michelson et al. 2005; but note conflicting data in Hirano and Sugimoto 2007). In these experiments, an inducible DSB is created such that one of the ends exposes telomeric repeats and the other does not. The exposure of telomeric DNA does not affect the initial checkpoint response, but it allows for an accelerated recovery from the checkpoint arrest and resumption of cell cycle progression (Michelson et al. 2005). Intriguingly, this effect could be dependent on keeping the two ends created by the break in close proximity with Rif proteins at the DSB contributing to dampening of the checkpoint response (Ribeyre and Shore 2012).
Regulated resection
Given that G tails are an essential feature of chromosome ends, they must be regenerated after DNA replication. This processing is particularly a problem for the end replicated by the leading strand polymerase, which is predicted to produce a blunt end (Figure 4A). This problem is solved by postreplication C-strand degradation (Wellinger et al. 1996), which remarkably depends on the same genes that resect the ends of DSBs to generate the 3′ single-strand tails that initiate homologous recombination. This congruence is surprising as one of the key functions of telomeres is to prevent DNA repair at natural ends. Recent insights suggest a solution to this conundrum. C-strand resection at telomeres is strongly dependent on Sgs1p or Sae2p. Sgs1p is a 3′ to 5′ RecQ family DNA helicase, while Sae2p is an endonuclease whose phosphorylation by Cdk1 is critical for its activity (Huertas et al. 2008; Bonetti et al. 2009). Indeed, Cdk1 activity is required for cell-cycle–dependent telomere resection (Frank et al. 2006; Vodenicharov and Wellinger 2006). The MRX complex acts in the same pathway as Sae2p to generate G tails. Although G tails are shorter in mre11Δ cells, they still increase in length in late S/G2 phase in this background (Larrivee et al. 2004). However, the nuclease activity of Mre11 is not required to generate G tails (Tsukamoto et al. 2001). Thus, MRX is not as critical as Sae2p for G-tail generation. Likewise, in sae2Δ cells, C-strand degradation is not eliminated, as there is the second and partially overlapping degradation pathway that requires Sgs1p (Bonetti et al. 2009). The fact that multiple nucleases are involved in telomeric end processing is also true at DSBs (Zubko et al. 2004; Gravel et al. 2008; Mimitou and Symington 2008; Zhu et al. 2008). Indeed, on a DSB, a slow MRX-dependent and restrained resection soon gives way to fast and extensive resection carried out by Exo1p or Dna2p. At telomeres, the Cdc13 complex together with the YKu complex seems to inhibit this switch, as deep resection into telomere adjacent unique DNA rarely occurs. Consistent with this idea, there is rampant C-strand resection in cells expressing the temperature-sensitive cdc13-1 allele and growing at high temperatures. The YKu complex also contributes to limiting C-strand resection as cells lacking YKu have constitutively long G tails, and this phenotype is suppressed by deletion of EXO1 (Gravel et al. 1998; Polotnianka et al. 1998; Maringele and Lydall 2002). Furthermore, Rap1p and particularly the associated Rif2p act as inhibitors of MRX-dependent telomere resection (Bonetti et al. 2010). Taken together, these data suggest that telomere processing in late S phase, which occurs right after conventional DNA replication, is triggered similarly at telomeres and DSBs: a Cdk1p-stimulated Sae2p/MRX-mediated activity generates a short G tail. However, at telomeres, further resection is inhibited by a combination of YKu, the Cdc13 complex, and the Rif proteins such that resection is limited to ∼30–100 nt, occurring only in the distal half of the telomere. Since no deep resection occurs, no unique sequence single-stranded DNA is uncovered, and no DNA damage checkpoint activity or cell cycle arrest is elicited. In this scenario, YKu association to telomeres is the primary inhibitor of initiation of resection, while the other factors limit deep resection once resection has begun (Bonetti et al. 2010; Vodenicharov and Wellinger 2010). Telomeres on which resection generates G tails longer than the 10–15 nt must be processed prior to mitosis (Wellinger et al. 1993a,b). This processing probably involves C-strand resynthesis by conventional DNA replication, but there is also evidence for limited nucleolytic trimming of G tails (Diede et al. 2010).
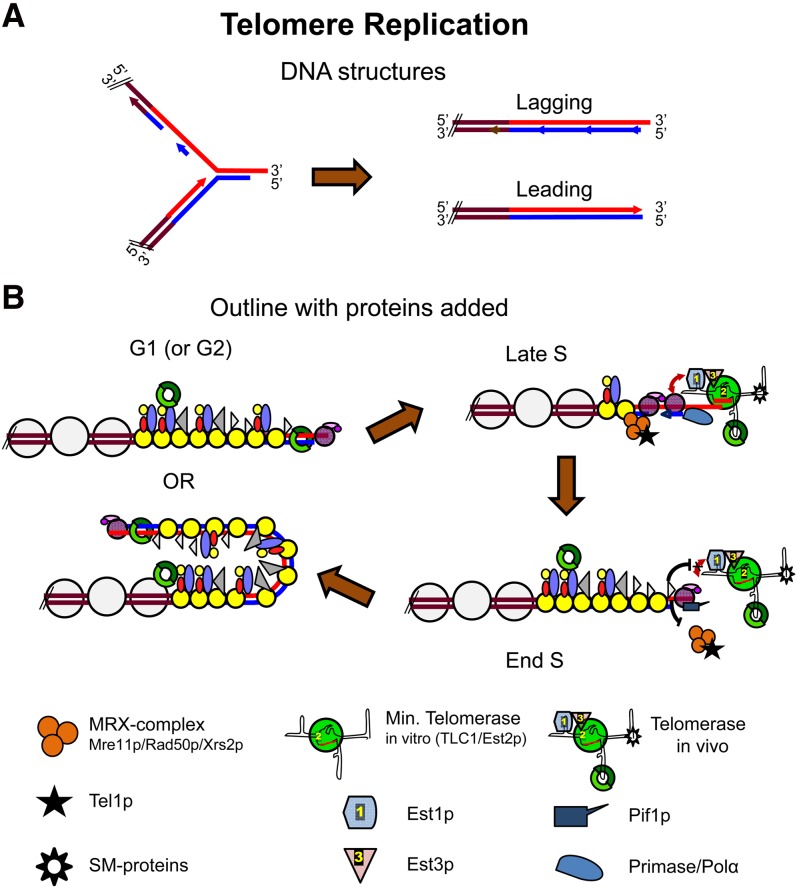
Figure 4 .
Molecular models for telomere replication. (A) DNA structures thought to be generated during telomere replication when the replication fork is still in the double-stranded telomeric repeats (left) and after having reached the physical end (right). Strand colors as in Figure 1. Brown, subtelomeric sequences. (B) Proposed telomeric chromatin changes during a cell cycle. Note that telomerase elongation drawn for late S does not occur on all telomeres in every cell cycle. This step occurs preferentially on short telomeres. Bottom shows involved proteins and complexes as well as a sketch of the proposed secondary structure of the TLC1 RNA with associated proteins (telomerase). Short red line in RNA indicates templating area. Symbols for other proteins are the same as in Figure 1.
Telomere Replication
Semiconservative replication of telomeric and subtelomeric DNA
Discussions of telomere replication usually focus on telomerase, a telomere-specific reverse transcriptase that replicates the very end of the chromosome. However, most of the telomeric repeats are replicated by standard semiconservative DNA replication. Conventional replication of telomeric DNA is one of the last events in S phase. Density transfer experiments reveal that Y′ repeats and the unique regions adjacent to telomeres replicate very late in S phase (McCarroll and Fangman 1988; Raghuraman et al. 2001). This late replication is due primarily to late activation of origins near telomeres, such as the late firing ARS501 (Ferguson and Fangman 1992). This late firing is independent of origin sequence as an origin that is normally activated in early S phase, such as ARS1 or the origin from the 2-μm plasmid, is activated late in S phase when placed near a telomere (Ferguson and Fangman 1992; Wellinger et al. 1993a). Likewise, ARS501 fires in early S phase when moved to a circular plasmid, while linearization of the ARS501 plasmid by telomere addition results again in its late activation (Ferguson and Fangman 1992). One possibility is that late origin firing results from the topological freedom enjoyed by unrestrained ends. This model is ruled out by the finding that when a DSB is induced next to an early firing origin, that origin still activates in early S phase (Raghuraman et al. 1994). Thus, telomeres exert a position effect on the timing of origin activation. Late activation of telomere adjacent origins is programmed in G1 phase. Thus, if a telomere proximal ARS is excised from the chromosome in late G1 phase, a circular plasmid containing it still replicates in late S phase (Raghuraman et al. 1997). Late firing of telomere adjacent origins is affected by telomere length as origins next to short telomeres fire earlier in S phase than origins near wild-type (WT)–length telomeres (Bianchi and Shore 2007a).
It is tempting to speculate that late activation of telomeric origins is due to the same heterochromatic chromatin structure that causes TPE. However, depleting cells of Sir3p, which eliminates TPE, has little effect on replication timing of telomere adjacent DNA (Stevenson and Gottschling 1999). In contrast, the YKu complex, whose absence causes telomere shortening, long G tails, and reduced TPE, is essential for late activation of telomeric origins yet it does not affect activation of more internal origins (Cosgrove et al. 2002). Deletion of Rif1p, which causes telomere lengthening, also results in early replication of telomeric regions (Lian et al. 2011).
Perhaps because of late replication, telomere length is particularly sensitive to mutations in conventional replication proteins. For example, telomeres lengthen in cells with temperature-sensitive alleles of several replication proteins, such as DNA polymerase α, DNA replication factor C, and Rad27p (Carson and Hartwell 1985; Adams and Holm 1996; Parenteau and Wellinger 1999, 2002; Adams Martin et al. 2000; Grossi et al. 2004). Since the telomere lengthening in these mutants is telomerase dependent (Adams Martin et al. 2000), it likely reflects a competition between semiconservative DNA replication and telomerase extension, both of which occur in late S phase. The key player in this competition is probably the Cdc13 complex, as two of its subunits interact with subunits of the DNA polymerase α complex, Cdc13p with the catalytic subunit of DNA polymerase α (Qi and Zakian 2000; Sun et al. 2011) and Stn1p with Pol12p (Grossi et al. 2004). Cdc13p also interacts with Est1p, a telomerase subunit (Qi and Zakian 2000; Pennock et al. 2001; Wu and Zakian 2011). Thus, when replication proteins are limiting, it may facilitate Cdc13p interaction with telomerase and promote telomere lengthening.
Semiconservative replication of telomeres is a prerequisite for the C-strand degradation that occurs in late S/G2 phase (Wellinger et al. 1993a; Dionne and Wellinger 1998). The two telomeres on each chromosome are synthesized differently, and these differences affect their need for C-strand degradation. At one end, the new strand is the product of leading strand synthesis while at the other end, it is the product of lagging strand synthesis (Figure 4A). Theoretically, the telomere replicated by leading strand synthesis can be replicated fully to generate a blunt end, while the other end will be left with a small gap at the 5′ end of the newly replicated strand after removal of the terminal RNA primer (Figure 4A). Although both ends of at least some DNA molecules are subject to C-strand degradation in a given cell cycle (Wellinger et al. 1996), the leading strand and lagging strand telomeres are treated differently (Parenteau and Wellinger 2002). While both bind Cdc13p, only the telomere replicated by the leading strand polymerase binds the MRX complex (Faure et al. 2010).
When most people think about difficulties replicating chromosome ends, they think about telomerase and its role in solving the “end replication” problem. However, even semiconservative replication of telomeric DNA poses problems, as replication forks in yeast and other organisms move more slowly through telomeric DNA than through most other regions of the genome (Ivessa et al. 2002; Miller et al. 2006; Sfeir et al. 2009). This difficulty is thought to arise from the GC-rich nature of telomeric DNA, which gives it a high thermal stability and also allows it to form stable secondary structures, such as G-quadruplex DNA, which can pose problems for DNA replication (Lopes et al. 2011; Paeschke et al. 2011).
The first evidence that telomeric DNA, even at nontelomeric sites, slows replication forks came from two-dimensional gel analyses (Ivessa et al. 2002). Additionally, there are multiple other sites in subtelomeric regions, such as inactive replication origins, that slow fork progression. Slow replication of telomeric regions is also seen in genome-wide studies that monitor DNA polymerase II occupancy (Azvolinsky et al. 2009). The yeast replication fork also moves slowly through human telomeric DNA (Bah et al. 2011).
Although fork slowing is detected in telomeric and subtelomeric DNA in wild-type cells, this slowing is 10-fold higher in the absence of Rrm3p, a 5′ to 3′ DNA helicase (Ivessa et al. 2002; Azvolinsky et al. 2009). The effects of Rrm3p on fork progression are not limited to telomeres (Ivessa et al. 2000; Ivessa et al. 2003) as it promotes fork progression at many nontelomeric loci, such as RNA polymerase III transcribed genes. All of the Rrm3p-sensitive sites are bound by stable protein–DNA complexes whose removal obviates the need for Rrm3p during DNA replication (Ivessa et al. 2003; Torres et al. 2004). Eliminating any of the silencing proteins Sir2p, Sir3p, or Sir4p reduces replication pausing within telomeres in RRM3 cells. However, when both Sir proteins and Rrm3p are absent, telomeric pausing is still high (Ivessa et al. 2003).Taken together, these data suggest that the sequence, as well as the chromatin structure, of telomeres contribute to their negative effects on fork progression.
Telomere maintenance via telomerase
End replication problems and the discovery of telomerase:
All DNA polymerases synthesize DNA only in the 5′ to 3′ direction and are unable to start replication de novo. Thus, DNA polymerases require a primer, which for eukaryotic chromosomes is a short 8–12 nt stretch of RNA. A DNA polymerase can theoretically extend this primer on the so-called leading strand, until it reaches the end of the chromosome to produce a blunt end. In contrast, the lagging strand is made discontinuously, and each Okazaki fragment starts with an RNA primer. Removal of the most distal RNA primer leaves a gap of 8–12 nt at the 5′ ends of newly replicated strands that cannot be filled in by a conventional DNA polymerase. In the absence of a special end replication mechanism, the product is shorter than the starting template. This dilemma is the so-called end-replication problem, as classically defined (Watson 1972).
Since eukaryotic chromosomes end with 3′ single-stranded G tails that are essential for chromosome stability, there is a second end-replication problem that affects leading strand replication (Lingner et al. 1995). The leading strand DNA polymerase should generate a blunt ended DNA terminus, rather than a G tail (Figure 4A). Postreplication C-strand degradation at both ends of chromosomes can solve this problem (Wellinger et al. 1996). In this scenario, the 5′ ends of the template for leading strand synthesis is degraded to generate long G tails. RNA primed C-strand resynthesis can fill in the C strand, but when the RNA that primes this synthesis is removed, a short G tail will be generated.
In the vast majority of eukaryotes, the continuous loss of DNA due to incomplete replication is solved by telomerase. This activity was first identified by a biochemical approach using extracts from the ciliate Tetrahymena (Greider and Blackburn 1985). Telomerase consists of both protein and RNA subunits (Greider and Blackburn 1987). During DNA extension, telomerase uses a short segment within its integral RNA subunit as the template to extend the 3′ end of the G-rich strand of the telomere (Greider and Blackburn 1989). Thus, telomerase-generated telomeric repeats are templated not by the chromosome but by telomerase RNA. Once telomerase extends the 3′ strand, RNA primed DNA replication by a conventional DNA polymerase can fill in the complementary C strand.
C-strand degradation makes a de facto lagging strand-like terminus at the telomere that was lengthened by the leading strand polymerase. This degradation has the benefit of generating a G tail, but it will magnify the first end-replication problem as now, in the absence of telomerase, both the leading and the lagging strand telomeres lose ∼10 nt per S phase (assuming that the average RNA primer is 10 nt). However, the measured loss rate is only half this rate (Lundblad and Szostak 1989; Singer and Gottschling 1994). A possible explanation for this discrepancy is that telomerase provides protection from a telomerase-independent lengthening activity, such as recombination. In this model, telomeres in telomerase-deficient cells would be lengthened by recombination that would partially compensate for sequence loss by incomplete replication. This proposal provides an explanation for why telomeric repeats are lost at a faster rate, ∼10 nt/generation, in strains that are both telomerase and recombination deficient compared to a strain deficient for telomerase alone (Lundblad and Szostak 1989; Singer and Gottschling 1994; Lee et al. 2007).
Telomerase does not act on blunt-ended DNA molecules. Thus, C-strand degradation of the blunt end produced by leading strand replication generates not only a G tail for binding of the Cdc13 complex, it also creates a potential substrate for telomerase. With G tails at both ends of a chromosome, telomerase could theoretically act on telomeres replicated by either the leading or lagging strand polymerase. However, MRX, which recruits Tel1p and hence telomerase to telomeres, binds preferentially to telomeres replicated by the leading strand polymerase (Faure et al. 2010), perhaps because MRX is needed to process blunt ends. MRX also binds preferentially to short telomeres (McGee et al. 2010) and to DSBs next to short (81 bp) but not long (162 bps) tracts of telomeric DNA (Negrini et al. 2007; Hirano et al. 2009). Since MRX is needed for efficient recruitment of telomerase, these data predict that telomerase acts preferentially at short telomeres replicated by the leading strand DNA polymerase.
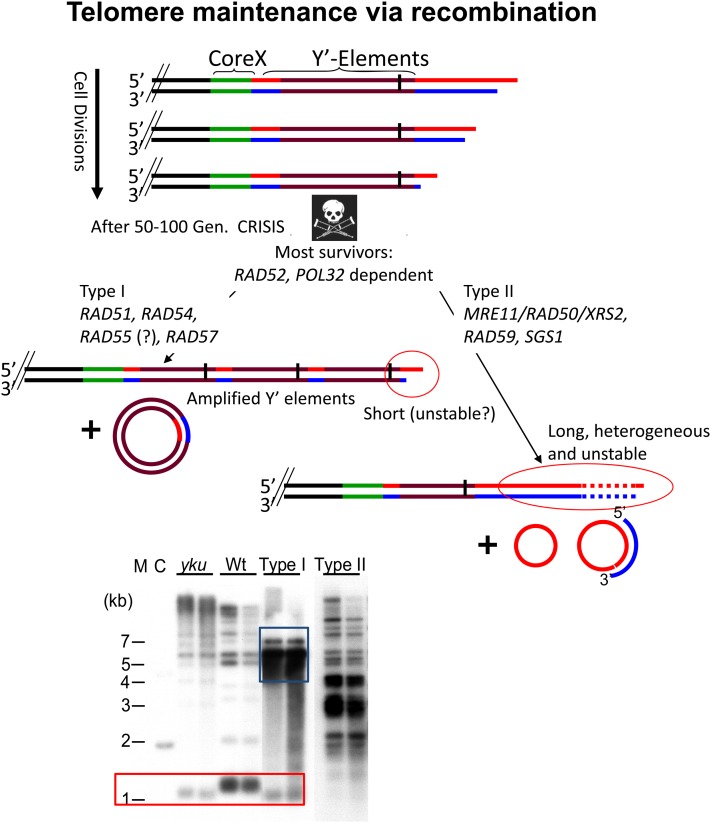
Biochemical characterization of S. cerevisiae telomerase was slow in coming, perhaps because the enzyme is not abundant. In contrast, genetic analysis of telomerase was pioneered in S. cerevisiae. The first known telomerase subunit, EST1 (ever shorter telomeres 1), was identified in a screen for genes with defective telomere function (Lundblad and Szostak 1989). Although est1Δ cells are viable, they slowly but progressively lose C1-3A/TG1-3 telomeric DNA. Once telomeres become very short, chromosome loss and cell cycle length go up dramatically. After 50–100 generations, most est1Δ cells die. The combination of progressive telomere loss and eventual chromosome instability and cell death is known collectively as the est phenotype (Lundblad and Szostak 1989).
A similar screen identified an additional three genes whose deletion (EST2 and EST3) or mutation (EST4) also yields an est phenotype (Lendvay et al. 1996). When the wild-type copy of est4 was cloned, it was found to be a separation-of-function allele of the previously identified essential CDC13 gene and renamed cdc13-2 (Nugent et al. 1996). Cells with the cdc13-2 allele are telomerase deficient but viable because the end protection function of Cdc13p is intact. A separate screen to identify genes whose overexpression interfered with TPE, unexpectedly identified another est gene, called TLC1 (telomerase component 1) (Singer and Gottschling 1994). TLC1 encodes a large RNA whose sequence has a 17-nt stretch complementary to the G strand of yeast telomeric DNA. Altering the putative template region in TLC1 produced mutant telomeric repeats in vivo, proving that TLC1 is indeed the templating RNA. Est2p was identified as the catalytic reverse transcriptase subunit of yeast telomerase when its sequence was found to be similar to that of the biochemically purified catalytic subunit of Euplotes aediculatus (a ciliated protozoan) telomerase (Lingner et al. 1997).
Now that the entire yeast genome has been evaluated for telomeric roles, it is clear that TLC1, EST1, EST2, EST3, and CDC13 are the only genes whose mutation yields a telomerase null phenotype. However, certain double mutations also have an est phenotype. TEL1 encodes an ATM-like checkpoint kinase, but its major function is in telomere length maintenance. A tel1Δ strain has very short but stable telomeres and does not senesce (Lustig and Petes 1986; Greenwell et al. 1995; Morrow et al. 1995). The kinase activity of Tel1p is required for its role in telomere length maintenance as a kinase dead allele has the same phenotype as tel1Δ (Mallory and Petes 2000). Cells deficient for Mec1p, the yeast ATR equivalent and the major checkpoint kinase in yeast, have a very modest decrease in telomere length (Ritchie et al. 1999). Although MEC1 is essential, both its checkpoint and telomere maintenance functions are dispensable for cell viability. Its essential function can be bypassed by deleting SML1, an inhibitor of ribonucleotide reductase (Zhao et al. 1998). Although neither tel1Δ nor mec1Δ sml1Δ cells senesce, cells deficient in both kinases have an est phenotype (Ritchie et al. 1999). Cells lacking any one (or all three) of the MRX subunits act in the same pathway as Tel1p to affect telomere length (Nugent et al. 1998). Thus, like tel1Δ cells, mrx mutants have short but stable telomeres and an est phenotype in combination with loss of Mec1p (Ritchie and Petes 2000). Likewise, mrx ykuΔ cells have an est phenotype (DuBois et al. 2002; Maringele and Lydall 2004a).
Tel1p and the MRX complex are not part of the telomerase holoenzyme but have important roles in recruiting telomerase to telomeres. Consistent with this interpretation, fusion of Cdc13p to Est2p allows telomere maintenance in tel1mec1 cells (Tsukamoto et al. 2001). Moreover, tel1mec1 cells have normal telomerase activity by in vitro assays and can maintain telomeres in a rif1Δ rif2Δ background (Chan et al. 2001).
Characteristics of components of the telomerase holoenzyme:
Est1p:
The EST1 ORF predicts a 699-amino-acid protein with no strong structural motifs (Figure 2; Lundblad and Szostak 1989). Est1 binds RNA and single-stranded TG1-3 DNA in vitro (Virta-Pearlman et al. 1996; DeZwaan and Freeman 2009). Unlike Cdc13p, Est1p binding to TG1-3 DNA requires a 3′ OH end. Although Est1p is conserved through mammals, its sequence is divergent, even in fungi (Beernink et al. 2003; Reichenbach et al. 2003; Snow et al. 2003). Unlike the other telomerase subunits, Est1p abundance is cell cycle regulated, low in G1 phase (∼20 molecules/cell) when telomerase is not active and higher in late S/G2 phase (∼110 molecules/cell) when it is (Taggart et al. 2002; Wu and Zakian 2011). This cell cycle pattern is due primarily to proteasome-dependent cell-cycle–regulated proteolysis (Osterhage et al. 2006), although Est1 mRNA degradation by Rnt1p also contributes to its cell-cycle–regulated abundance (Spellman et al. 1998; Larose et al. 2006).
Although est1Δ cells have a classic telomerase-deficient phenotype in vivo, standard primer extension assays for telomerase activity in vitro are not Est1p dependent (Cohn and Blackburn 1995). Nonetheless, Est1p immunoprecipitates with both TLC1 RNA and telomerase activity, suggesting that it is an integral part of the telomerase holoenzyme (Lin and Zakian 1995; Steiner et al. 1996). Est1p binds directly to a stem-bulge region in TLC1, and disruption of this interaction confers an est phenotype in vivo (Seto et al. 2002). The Est1p–TLC1 interaction is essential to bring both Est1p and Est2p to telomeres in late S/G2 phase (Chan et al. 2008).
Genetic evidence using fusion proteins provided the first evidence that a Cdc13p–Est1p interaction recruits the telomerase holoenzyme to telomeres. Est1p is dispensable for telomere maintenance in cells expressing a fusion of the DNA binding domain of Cdc13p (DBDCdc13) and Est2p (DBDCdc13–Est2) (Evans and Lundblad 1999). These results suggest that the critical function of Est1p is to mediate the interaction between telomerase and the telomere. Two-hybrid and coimmunoprecipitation studies support this hypothesis by providing physical evidence of an interaction between the two proteins (Qi and Zakian 2000). Moreover, this interaction is direct, as purified Cdc13p and Est1p interact in vitro to form a 1:1 complex (Wu and Zakian 2011). The interaction is also specific, as Cdc13p does not interact with Est3p and is sufficient for recruiting Est1p to Cdc13p-coated TG1-3 single-strand DNA in vitro.
The telomerase null phenotypes of certain mutations in CDC13 and EST1, such as cdc13-2 and est1-60, are proposed to be due to a disruption of the Cdc13p–Est1p interaction (Pennock et al. 2001). These particular mutations are charge swap alleles: while each mutation alone confers an est phenotype in vivo, cdc13-2 est1-60 cells have short, stable telomeres and do not senesce. Because the charge interaction between the two proteins is restored in the double mutant, the telomerase proficiency of the double mutant can be explained by restoration of a physical interaction between Cdc13p and Est1p. Consistent with this interpretation, cdc13-2 cells have low Est1p and Est2p binding to telomeres (Chan et al. 2008) and DSBs (Bianchi et al. 2004). However, the strengths of various combinations of interactions (i.e., Cdc13p–Est1p, Cdc13–2p–Est1p, and Cdc13p–Est1–60p) are indistinguishable in vitro (Wu and Zakian 2011). The best model to fit all of the data is that these charge swap mutants support wild-type levels of Cdc13p–Est1p interaction, but the resulting complex is somehow defective in vivo such that it is unable to support wild-type levels of telomerase–telomere interaction or telomerase extension. Indeed, visualization of telomerase RNA in living cells suggests that it associates with telomeres in cdc13-2 cells, but this association is transient (Gallardo et al. 2011).
In addition to its role in telomerase recruitment, Est1p is thought to activate telomerase. The best evidence for this model also comes from studies with fusion proteins. Cells expressing a DBDCdc13–Est2 fusion protein have hyperelongated telomeres, presumably because telomerase is always telomere associated (Evans and Lundblad 1999). However, telomeres are not hyperelongated in est1Δ cells expressing the fusion. In line with an activating role for Est1p, biochemical studies show that Est1p interacts directly with Est3p, an interaction that is required for Est3p telomere binding (Tuzon et al. 2011). The role of Est1p in recruiting Est3p might explain its activation function.
Est2p:
The EST2 ORF predicts an 884-amino-acid protein with motifs found in other reverse transcriptases including three invariant aspartate residues that are essential for catalysis (Lingner et al. 1997). Mutation of any one of the conserved aspartates leads to an est phenotype equivalent to that seen in est2Δ cells and also eliminates telomerase activity in vitro. Thus, Est2p is the catalytic reverse transcriptase subunit of S. cerevisiae telomerase.
Like other telomerase reverse transcriptases (TERTs), but unlike most other reverse transcriptases, Est2p contains a long basic N-terminal (TEN) domain that is essential for telomerase activity in vivo and in vitro (Friedman and Cech 1999; Figure 2). The TEN domain supports multiple interactions within the holoenzyme, including interactions with TLC1 (Friedman and Cech 1999) and Est3p (Friedman et al. 2003; Talley et al. 2011). Est2p is a low abundance protein (<40 molecules/cell; Tuzon et al. 2011), and its levels are TLC1 dependent (reduced by ∼50% in tlc1Δ cells; Taggart et al. 2002).
Est3p:
The EST3 ORF, which predicts an 181-amino-acid protein, has the unusual property of being generated by a programmed translation frameshift (Figure 2) (Morris and Lundblad 1997). Like Est1p, Est3p is essential for telomere maintenance in vivo but not for catalysis in vitro (Lendvay et al. 1996; Lingner et al. 1997). Nonetheless, coimmunoprecipitation shows that Est3p is part of the telomerase holoenzyme (Hughes et al. 2000). The association of Est3p with telomerase is Est1p dependent (Osterhage et al. 2006), consistent with the direct interaction of purified Est1p and Est3p seen in vitro (Tuzon et al. 2011). By genetic and biochemical criteria, Est3p also interacts with the TEN domain of Est2p (Friedman et al. 2003; Talley et al. 2011), and Est3p association with telomeres is also Est2p dependent, especially in G1 phase (Tuzon et al. 2011).
Although Est1p and Est3p have certain similarities, they do not have redundant functions. For example, a DBDCdc13–Est3 fusion protein can maintain telomeres in est3Δ but not est1Δ cells (Hughes et al. 2000). Likewise, an Est1–DBDCdc13 fusion protein does not rescue the telomerase defect of est3Δ cells, and a DBDCdc13–Est2 fusion bypasses the need for Est1p, but not Est3p (Evans and Lundblad 1999).
So far Est3p is found only in budding yeasts. However, a possible key to its function comes from a predicted structural similarity between it and a mammalian telomere structural protein TPP1 (Lee et al. 2008; Yu et al. 2008). Unlike Est3p, TPP1 is not a telomerase subunit but rather part of the multiprotein shelterin complex that protects telomeric DNA. However, TPP1 affects telomerase by cooperating with Pot1, a mammalian G-strand binding protein, to increase telomerase processivity (Wang et al. 2007; Xin et al. 2007).
TLC1:
Like the Est proteins, the TLC1 RNA is not abundant, present in ∼30 molecules/cell (Mozdy and Cech 2006). Transcription of TLC1 RNA by RNA polymerase II generates two populations, a slightly longer polyadenylated form (5–10% of total) and a polyA minus form (> 90%), the version in active telomerase (Chapon et al. 1997; Bosoy et al. 2003). Akin to snRNAs and snoRNAs, the 5′ end of the TLC1 RNA has a trimethylguanosine cap (Seto et al. 1999; Franke et al. 2008), while generation of the mature nonpolyadenylated 3′ end occurs via the Nrd1p-dependent noncoding RNA termination pathway (Jamonnak et al. 2011; Noel et al. 2012). Similar to several fungal telomerase RNAs, TLC1 is >1000 nt in size, much larger than its ciliate (∼160 nt) or mammalian (∼450 nt) counterparts (Singer and Gottschling 1994). However, a TLC1 RNA derivative that reduces the native RNA from 1157 to 384 nt is sufficient to maintain short, but stable yeast telomeres in vivo and to support catalysis in vitro (Zappulla et al. 2005). Thus, much of TLC1 RNA is dispensable for enzyme activity.
Although the sequence and size of telomerase RNAs evolve rapidly, conserved secondary structures have been deduced. The structure predicted for the S. cerevisiae TLC1 RNA centers about a conserved pseudoknot domain that contains the templating region of the RNA and interacts with Est2p (Livengood et al. 2002; Dandjinou et al. 2004; Lin et al. 2004; Zappulla and Cech 2004; Qiao and Cech 2008). The remainder of the RNA forms three largely duplex arms that are proposed to act as a flexible scaffold to organize TLC1 RNA interacting proteins (Figure 4). One arm binds Est1p, and this binding is essential for telomerase activity in vivo (Seto et al. 2002). One arm binds Yku80p, an interaction that is not essential for telomere maintenance but brings TLC1 to the nucleus and recruits Est2p to telomeres in G1 phase (Stellwagen et al. 2003; Fisher et al. 2004; Vega et al. 2007; Gallardo et al. 2008). The third arm binds the seven-member Sm protein ring, an association that is dispensable for activity but important for TLC1 accumulation (Seto et al. 1999).
Regulation of telomerase by the cell cycle:
Two experiments using quite different approaches show that telomerase-mediated lengthening is cell-cycle regulated. The first experiment followed telomerase action at a DSB induced next to a short stretch of telomeric repeats (Diede and Gottschling 1999). When this break is made in G2/M arrested cells, it is lengthened by telomerase. However, the break is not lengthened in G1-arrested cells, suggesting that telomerase does not act at this time. However, in vitro assays show similar levels of telomerase activity in extracts prepared from cells arrested at these two points in the cell cycle.
The second assay studied the fate of a short telomere in cells with otherwise wild-type length telomeres by using site-specific recombination to generate a single short telomere (Marcand et al. 2000). The resulting short telomere is preferentially lengthened by telomerase (Marcand et al. 1999), but this lengthening does not occur in G1 or early S phase but rather only in late S/G2 phase (Marcand et al. 2000).
One way to reconcile the finding that telomerase is active in vitro in extracts from G1-phase cells with its inability to lengthen telomeres in vivo in G1 phase is if the telomere is inaccessible to telomerase in G1 phase. An obvious way to test this model is to use chromatin immunoprecipitation (ChIP) to detect the presence of telomerase at telomeres as a function of position in the cell cycle. This type of experiment yields support both for and against regulated accessibility (Taggart et al. 2002). Cdc13p is telomere associated throughout the cell cycle, but its binding increases dramatically in late S phase, as expected by the occurrence of long G tails at this time (Wellinger et al. 1993b). The telomere binding of Est1p (Taggart et al. 2002) and Est3p (Tuzon et al. 2011) is largely limited to late S/G2 phase, consistent with regulated accessibility. However, Est2p is telomere associated throughout most of the cell cycle, including in G1 and early S phase when telomerase does not act (Taggart et al. 2002). Nonetheless, Est2p binding is not constitutive as there is a second peak of Est2p binding in late S/G2 phase.
The two peak pattern of Est2p telomere binding reflects two independent pathways of telomerase recruitment. Both pathways are TLC1 dependent as there is no telomere-associated Est2p in tlc1Δ cells (Taggart et al. 2002). However, Est2p telomere association in G1 phase requires a specific interaction between Yku80p and a 48-bp stem-loop structure in TLC1 RNA (Fisher et al. 2004) while the late S/G2 phase binding requires Est1p binding to a stem-bulge region in TLC1 as well as its interaction with Cdc13p (Chan et al. 2008). The Est2p that is telomere associated in G1 phase is likely not engaged with the very end of the chromosome as expected for active telomerase as much of it is bound >100 bp from the chromosome end (Sabourin et al. 2007). Consistent with this view, the G1-phase association is not necessary for telomerase action as mutations that eliminate it (tlc1Δ48; yku80-135i) (Fisher et al. 2004) result in only modest telomere shortening (Peterson et al. 2001). Even this small reduction in telomere length may not be due to lack of G1-phase telomerase binding as nuclear levels of TLC1 are reduced in the absence of the TLC1–Ku interaction (Gallardo et al. 2008; Pfingsten et al. 2012). Thus, the short telomeres in tlc1Δ48 and yku80-135i cells could be a consequence of reduced amounts of holoenzyme being imported and/or retained in the nucleus. Recent data indicate that Yku binding to DNA and RNA are mutually exclusive (Pfingsten et al. 2012). Since the binding of Est2p to telomeres in G1 phase requires a Yku80p–TLC1 interaction, it is likely that the Yku that is involved in this interaction associates with the telomere via protein–protein interactions, not by direct DNA binding.
Cell-cycle–limited telomerase activity at telomeres is also inferred from results in which TLC1 RNA is visualized in individual cells in real time (Gallardo et al. 2011). Telomerase RNA marked with GFP is much more mobile than telomeres in G1 and G2 phases, whereas in late S phase, telomerase RNA movement slows. Thus, TLC1 association with telomeres is more transient in G1 and G2 phases than in late S phase. Genetic experiments argue that the more stably associated TLC1 reflects active telomerase, as these associations are less frequent in genetic backgrounds where telomerase recruitment is impaired. Thus, results with live cell imaging support previous findings that the association of telomerase with telomeres can occur throughout the cell cycle (Taggart et al. 2002), but only the late S phase telomere-associated Est2p is important for telomere length regulation (Fisher et al. 2004). This study also suggests that more than one telomerase complex is present on elongating telomeres as the TLC1 complexes, dubbed T-Recs (telomerase recruitment clusters), are brighter and larger in late S phase (Gallardo et al. 2011).
Est1p is cell cycle regulated with peak abundance in late S/G2 phase (Taggart et al. 2002; Osterhage et al. 2006). Moreover, Est3p telomere binding is Est1p dependent, so its telomere binding also occurs mainly in late S/G2 phase (Tuzon et al. 2011). Thus, telomerase is cell cycle limited at least in part because the telomerase holoenzyme is assembled only during a narrow window in the cell cycle (Osterhage et al. 2006). However, even when Est1p is expressed in G1 phase, which results in both Est1p and Est3p being Est2p–TLC1 associated, telomerase is still not active on telomeres in G1 phase (Osterhage et al. 2006). Thus, Est1p abundance is not the whole answer to cell-cycle–regulated activity. Rif proteins also contribute to limiting telomerase action to late S phase as in the absence of either protein, short telomeres can be lengthened in G1 phase (Gallardo et al. 2011). Cell-cycle–regulated changes in telomere structure, such as C-strand degradation, which is Cdk1 dependent, may also contribute to cell cycle limited telomerase action (Frank et al. 2006; Vodenicharov and Wellinger 2006).
Regulation of telomerase by telomere length:
Two types of experiments indicate that short telomeres are preferentially lengthened by yeast telomerase. The first evidence comes from experiments where lengthening of a single short telomere is followed over time (Marcand et al. 1999). It takes ∼50 generations to return a short telomere to a wild-type length. However, its rate of lengthening changes as it lengthens. When the telomere is at its shortest, it lengthens by ∼15 nt/generation. This rate progressively decreases until it is only ∼1 nt/generation when the once short telomere approaches wild-type length.
The preferential lengthening of short telomeres is best illustrated using the single telomere extension assay (STEX) that monitors lengthening of individual telomeres in a single S phase at nucleotide resolution (Teixeira et al. 2004). STEX is particularly informative because it monitors events at individual telomeres rather than being a population average. In this assay, telomerase-deficient cells (recipient cells) are mated to telomerase proficient cells (donor cells). Telomere extension is monitored in the first generation after mating. The recipient cells contain marked telomere(s) that can be examined specifically by PCR because of differences in subtelomeric DNA from the same telomere in donor cells. Because the yeast telomeric sequence is heterogeneous, the starting telomeric DNA can be distinguished from newly added telomeric repeats simply by lining up telomeres and comparing their sequences.
In a given cell cycle, STEX finds that <10% of wild-type–length (∼300 bp) telomeres are lengthened by telomerase, while a 100-bp telomere is lengthened ∼50% of the time (Teixeira et al. 2004). Thus, length-dependent extension is not an all or none event: many short telomeres are not lengthened while some long telomeres are. Although the frequency of telomerase action is dependent on length, the amount of telomeric DNA added is not until telomeres are very short (≤100 bp). On these very short telomeres, telomerase appears to be more processive. STEX is also useful to determine how different proteins affect telomerase. By STEX, Rif1p and Rif2p inhibit the frequency of telomere lengthening but not the amount of telomeric DNA added per S phase (Teixeira et al. 2004). The preference for telomerase action at short telomeric tracts is also reflected during de novo telomere addition. A DSB induced next to an 81-bp stretch of telomeric DNA is more efficiently elongated than a break next to 162 bp of telomeric DNA (Negrini et al. 2007; Hirano et al. 2009).
ChIP is useful to determine the protein content of short vs. wild-type–length telomeres. Using inducible short telomere assays (Marcand et al. 1999), Est2p and Est1p have approximately fourfold higher binding at short telomeres specifically in late S/G2 phase, when telomerase is active (Bianchi and Shore 2007b; Sabourin et al. 2007). The similar level of increase for Est1p and Est2p argues against the idea that an elongation-incompetent Est2p binds all telomeres in G1 phase and then is activated in late S/G2 phase by Est1p binding.
Short telomere assays have also helped us understand how telomerase is targeted to short telomeres. Since Est2p recruitment to telomeres requires a Yku80p–TLC1 interaction in G1 phase and a Cdc13p–Est1p interaction in late S/G2 phase, if Yku80p and/or Cdc13p bound better to short telomeres, it could explain why short telomeres bind more telomerase. However, Yku80p and Cdc13p bind to similar extents at short and WT length telomeres (Bianchi and Shore 2007b; Sabourin et al. 2007). Another possibility is that Yku80p or Cdc13p is preferentially modified at short telomeres. For example, Cdc13p is phosphorylated by Cdk1p late in the cell cycle, and in the absence of this phosphorylation, Est1p telomere binding and telomere length are modestly reduced (Li et al. 2009). Cdc13p is also sumoylated in early to mid S phase, a modification that limits telomerase action probably by increasing the Cdc13p–Stn1p interaction (Hang et al. 2011).
In tel1Δ cells, telomeres are very short (Lustig and Petes 1986; Greenwell et al. 1995; Morrow et al. 1995) yet unlike other short telomeres, tel1Δ telomeres bind very little Est1p or Est2p (Goudsouzian et al. 2006). These data suggest that Tel1p might affect preferential lengthening of short telomeres. Indeed, while Tel1p binding to wild-type–length telomeres is low, transient, and limited to late S/G2 phase, Tel1p binding is about 10 times higher at short telomeres (Sabourin et al. 2007). Tel1p binding to short telomeres is detectable even in early S phase, increases in magnitude as cells progress through the cell cycle, and persists for at least two cell cycles. In contrast, Mec1p telomere binding is extremely low, even in tel1Δ cells where it is required for telomere elongation. Preferential binding of Tel1p to short telomeres is also seen when short telomeres are generated by deleting YKu or by deleting a telomerase subunit (Hector et al. 2007).
Tel1p binding to telomeres is dependent on an interaction between Tel1p and the carboxyl terminus of Xrs2p (Hector et al. 2007; Sabourin et al. 2007), just as it is at DSBs. Moreover, each of the three MRX subunits binds preferentially to short telomeres, and like high Tel1p binding, this high binding persists for at least two cell cycles (McGee et al. 2010). MRX binding occurs mainly at telomeres that have been replicated by the leading strand DNA polymerase (Faure et al. 2010). A unifying model for these data are that MRX binds preferentially to short telomeres replicated by the leading strand DNA polymerase; this binding recruits Tel1p, Tel1p phosphorylates one or more telomere proteins, and these changes in telomeric chromatin result in higher telomerase recruitment. Indeed, deleting TEL1 eliminates preferential binding of telomerase to short telomeres that lack subtelomeric repeats (Arneric and Lingner 2007; Sabourin et al. 2007). However, when telomeres contain subtelomeric binding sites for Tbf1p, short telomeres are still preferentially lengthened in tel1Δ cells, and tethering Tbf1p to a short telomere with no natural Tbf1p binding sites allows its preferential elongation in tel1Δ cells (Arneric and Lingner 2007). Thus, Tbf1p and Tel1p act in a partially redundant manner to distinguish short from wild-type–length telomeres. Like Rif2p, Tbf1p inhibits MRX, and hence telomerase, binding to short telomeres (Fukunaga et al. 2012).
Although the kinase activity of Tel1p is required for its role in telomere length maintenance (Mallory and Petes 2000), its critical targets at telomeres are not yet identified. Like other ATM-like kinases, Tel1p phosphorylates SQ/TQ motifs (Mallory and Petes 2000; Tseng et al. 2006). Xrs2p and Mre11p are phosphorylated in a Tel1p-dependent process in response to DNA damage, but mutation of the phosphorylated residues in these proteins to nonphosphorylatable amino acids does not result in short telomeres (Mallory et al. 2003). Cdc13p is another candidate for a Tel1p target as it contains 11 SQ/TQ motifs. Clusters of these motifs are in functionally important regions of Cdc13p, one in the DBD and one in the RD (Figure 2; Tseng et al. 2006). In vitro, Tel1p phosphorylates Cdc13p on three RD serine residues (S225, 249, and and 255). Moreover, simultaneous mutation of two of these residues (S249 and S255) results in an est phenotype. In addition, if Est1p is targeted to the telomere using a DBDCdc13–Est1 fusion protein, phosphorylation of these residues is no longer required for telomerase action.
The phenotype of cdc13-S249A, S255A cells makes a strong argument that their phosphorylation is critical for Cdc13p–Est1p interaction (Tseng et al. 2006). However, cells expressing a cdc13 allele in which each of the S/TQ motifs is mutated to AQ supports near wild-type–length telomeres (Gao et al. 2010). In addition, exhaustive mass spectrophotometric analysis of purified Cdc13p detects 21 sites of phosphorylation but no phosphorylation at S249 or S255, even in G2/M phase (W. Yun, P. A. DiMaggio, Jr., D. H. Perlman, V. A. Zakian, and B. A. Garcia, unpublished results). One possible way to reconcile these data are if cdc13-S249A, S255A cells are telomerase defective because these mutations destabilize the RD domain (rather than preventing it from being phosphorylated). In any case, the key sites of Tel1p phosphorylation relevant to telomerase recruitment have probably not been identified.
Telomere length is proportional to the number of Rap1p binding sites at a given telomere (Lustig et al. 1990). Rap1p recruits Rif1p, Rif2p, Sir3p, and Sir4p, each of which binds to the C terminus of Rap1p (Figure 2). Sir proteins function mainly in TPE, not telomere length control, but both Rif proteins negatively regulate telomerase (Teng et al. 2000). Deletion of either RIF1 or RIF2 results in telomere elongation (Hardy et al. 1992b; Wotton and Shore 1997). Since deletion of both proteins results in synergistic lengthening, Rif1p and Rif2p do not act redundantly (Wotton and Shore 1997). Rif1p and Rif2p also act synergistically to inhibit telomere addition at DSBs induced near a 162-bp tract of telomeric DNA (Negrini et al. 2007; Hirano et al. 2009).
By definition, telomeres progressively lose Rap1p binding sites concomitant with loss of telomeric DNA. Thus, an appealing model is that short telomeres are marked for MRX binding by their low content of Rif1p and Rif2p. Surprisingly, short telomeres have about the same amount of Rif1p binding as wild-type–length telomeres (Sabourin et al. 2007; McGee et al. 2010). Thus, short telomeres are not distinguished from long telomeres by their Rif1p content, although Rif1p may be selectively modified at short telomeres. In contrast, Rif2p content is lower at short telomeres so its absence could mark short telomeres for elongation. Consistent with this possibility, Tel1p no longer binds preferentially to short telomeres in rif2Δ cells (McGee et al. 2010). The observation that shortening telomeres lose Rif2p before losing Rif1p suggests that the two proteins are distributed nonrandomly along the telomere with Rif2p being closer to the chromosome terminus than Rif1p (Figure 1).
A mechanistic explanation for the effects of Rif2p on Tel1p binding comes from studies on DSBs made adjacent to telomeric DNA (Hirano et al. 2009). Tel1p binding to these DSBs is increased in rif2Δ cells, suggesting that Rif2p inhibits Tel1p association with the break. Moreover, tethering Rif2p to a nontelomeric DSB decreases Tel1p but not MRX binding to the break. Rif1 has similar but much smaller effects in these assays. Finally, coimmunoprecipitation shows that the N terminus of Rif2p, but not Rif1p, interacts with the C terminus of Xrs2p. Since Tel1p also binds this portion of Xrs2p, Rif2p, and Tel1p probably compete with each other for binding to MRX. If these results are applicable to telomeres, Rif2p could sequester Xrs2p in a manner that prevents its interaction with Tel1p.
Like Rif proteins, Pif1p, a 5′ to 3′ DNA helicase, is a negative regulator of telomerase. However, Rif proteins and Pif1p inhibit telomere elongation by different mechanisms as their absence has additive effects on telomere length (Schulz and Zakian 1994). Genetic data suggest that Pif1p interacts with the finger domain of Est2p (Eugster et al. 2006). Pif1p also reduces the number of gross chromosomal rearrangements, complex genetic changes of the type seen in cancers, by channelling DSBs toward recombination, rather than telomere addition (Myung et al. 2001). Lengthening of existing telomeres by telomerase is also inhibited by Pif1p, and its effects at both DSBs and telomeres require its ATPase activity (Schulz and Zakian 1994; Zhou et al. 2000). Although Pif1p inhibits telomerase at both telomeres and DSBs (Schulz and Zakian 1994), it may help distinguish the two as Pif1p phosphorylation by Mec1p is required for its inhibition of telomerase at DSBs but not at telomeres (Makovets and Blackburn 2009). Pif1p also appears to contribute to the preferential lengthening of short telomeres since in its absence, Est2p binds equally well to short and wild-type–length telomeres. This behavior can be explained by the finding that Pif1p itself binds preferentially to long telomeres. Like Est1p, Pif1p is cell cycle regulated by proteasome-dependent proteolysis such that nuclear Pif1p peaks in abundance in S/G2 phase (Vega et al. 2007).
In vitro telomerase assays:
Although yeast telomerase is constitutively expressed, it is present at low levels, which probably explains why its detection by in vitro assays lagged behind other organisms. Even after in vitro assays were established, they were (and still are) inefficient, generating only short extension products (Cohn and Blackburn 1995). Furthermore, although these assays require TLC1 and Est2p, they are not Cdc13p, Est1p, or Est3p dependent (Cohn and Blackburn 1995; Lingner et al. 1997). However, as it is not clear whether the fractionated telomerase prepared from extracts from wild-type cells even contained Est1p, Est3p, or Cdc13p, their absence might explain why the in vitro reactions are not robust. Molecular chaperones such as Hsp82p affect telomerase activity in vitro as well as having modest effects on telomere length in vivo (Toogun et al. 2008).
Definitive answers to the mechanistic contributions of Est proteins to telomerase activity will almost surely require purified components. Recombinant Cdc13p, Est1p, and Est3p (but not Est2p) have recently been purified by multiple labs. All three proteins are reported to influence primer extension assays, although these analyses are in early stages and are sometimes contradictory. Cdc13p has been reported to inhibit (Zappulla et al. 2009) and to stimulate in vitro telomerase activity (DeZwaan and Freeman 2009). The reasons for this difference are not clear, but there are multiple experimental differences in the two studies. Another in vitro study found that, in the context of Stn1p and Ten1p, Hsp82p can modulate Cdc13p DNA binding and thereby its effects on telomerase activity (DeZwaan et al. 2009). Purified Est1p is reported to increase the amount of product in a primer extension assay by up to 14-fold. Surprisingly, this stimulation does not require that Est1p be able to interact with the stem-bulge region in TLC1 RNA or to bind TG1-3 single-strand DNA. An earlier report using a PCR rather than primer extension assay found that long extension products were Est1p dependent, perhaps providing additional evidence for an activating role for this subunit (Lin and Zakian 1995). Finally, two groups report that purified Est3p from S. cerevisiae (Talley et al. 2011) or the related S. castelli (Lee et al. 2010) stimulates telomerase two- to threefold. This stimulation requires interaction of Est3p with the TEN domain of Est2p (Talley et al. 2011). It is not clear whether the Est3p stimulation is Est1p dependent. Although more experiments are needed, the availability of in vitro assays should provide more detailed mechanistic information on the telomerase reaction.
S. cerevisiae telomerase is not very processive in vitro. The enzyme pauses after each nucleotide addition and rarely translocates on the DNA template as required for multiple rounds of synthesis. This lack of processivity is not due to enzyme falling off the DNA primer. Rather, after elongation, yeast telomerase remains tightly bound to its DNA substrate (Prescott and Blackburn 1997). However, if the Pif1p DNA helicase is added to the in vitro reaction, Est2p is released into the supernatant (Boule et al. 2005). As a result of this release, Pif1p reduces telomerase processivity in vitro. Likewise, in vivo, telomerase can dissociate and then reassociate with a given telomere in a single cell cycle (Chang et al. 2007). The effects of Pif1p on telomerase require its enzymatic activity as Walker A box Pif1 mutant proteins bind single-stranded DNA as well as wild-type Pif1p but do not displace telomerase from DNA or reduce telomerase processivity (Boule et al. 2005).
Telomere maintenance via recombination
Telomerase is not the only activity that can maintain telomeric DNA. Although discovered in S. cerevisiae (Lundblad and Blackburn 1993), telomere maintenance by recombination is widespread occurring from yeasts to mammals. Recombinational maintenance of telomeres was detected by the finding that a small fraction of est1Δ cells survive senescence and form viable colonies. The importance of recombination was inferred from the virtual absence of survivors in est1Δ rad52Δ strains. All est strains, except mec1tel1, produce survivors via recombination (Figure 5).
Figure 5 .
Outline of the proposed sequence of events leading to telomere maintenance via recombination after telomerase loss. DNA strand coloring is as above. Tick marks on the brown sequence indicate a conserved XhoI restriction enzyme site. Most cells die after ∼50–100 generations of growth, but rare cells with the indicated two types of DNA arrangements can continue to divide. Virtually all events are dependent on RAD52 and POL32. Bottom: Typical southern blot analysis using XhoI-digested DNA derived from indicated strains. The probe consisted of a 32P labeled DNA fragment specific for telomeric repeat sequences. M, molecular size standards; yku, DNA derived from a strain lacking YKU80 and harboring short terminal repeat tracts. WT, DNA from a wild-type strain; type I, DNA from type I survivors; type II, DNA derived from type II survivors. Red square, location of terminal XhoI fragments. Blue square, signal for the amplified Y′ elements in type I survivors. Note that the fragment pattern for type II survivors is highly variable and unstable; thus the patterns shown in the last two lanes should be taken as an example for illustration purposes only.
Even in very early telomerase negative cultures, some cells stop growing when the average telomere length is expected to be near wild type (Lundblad and Blackburn 1993; Enomoto et al. 2002; Khadaroo et al. 2009). Thus, the progressive shortening of the majority of telomeres is probably not the major determinant for growth arrest (Abdallah et al. 2009; Khadaroo et al. 2009; Noel and Wellinger 2011). Most likely, this arrest is due to an occasional short telomere that arises during DNA replication and that cannot be relengthened by telomerase (Hackett et al. 2001; Hackett and Greider 2003). Indeed, only one chromosome end that lacks a telomere is sufficient to trigger growth arrest, and this arrest occurs even in telomerase proficient cells (Sandell and Zakian 1993; Abdallah et al. 2009; Khadaroo et al. 2009). The survivors that emerge from the arrested cultures continue to divide. However, individual survivor clones often grow considerably slower than wild-type cells, and some may even go through additional growth arrests.
Survivors have one of two different arrangements of telomeric DNA (Lundblad and Blackburn 1993), now dubbed type I and type II survivors (Teng and Zakian 1999; Figure 5). In addition to RAD52, generation of both classes requires the replication protein Pol32p (Lydeard et al. 2007), suggesting that the recombination that maintains telomeric DNA involves replication. Type I survivors are more common than type II survivors. For example, in one strain background, 90% of survivors have type I telomeres (Teng et al. 2000). However, type I survivors are not stable and easily convert to type II cells, which owing to their faster growth rate, take over liquid cultures. This effect shows that the two major survivor pathways are not mutually exclusive.
Type I survivors:
These cells grow slowly with intermittent periods of growth arrest. The vast majority of telomeres in these cells contain multiple tandem Y′ repeats, but the very ends still have short (50–150 bp) tracts of duplex telomeric DNA and normal G tails (Lundblad and Blackburn 1993; Larrivee and Wellinger 2006; Figure 5). The terminal arrays of Y′ repeats can be so substantial that individual cells can have up to 70-fold increase in Y′ elements (Lundblad and Blackburn 1993). Type I survivors also contain extrachromosomal circular Y′ elements that are proposed to serve as substrates for Y′ recombination (Larrivee and Wellinger 2006). Chromosomes of type I survivors do not enter agarose gels that are used to separate very large DNA molecules, probably because they contain a high fraction of highly structured recombination intermediates (Liti and Louis 2003; E. Louis, personal communication).
About half of the Y′ repeats contain an ORF encoding a potential helicase called Y′-Help1 (Louis and Haber 1992; Yamada et al. 1998). Expression of this ORF is greatly increased during growth arrest in telomerase lacking cells (Yamada et al. 1998). Although amplification of Y′ usually occurs by recombination (Lundblad and Blackburn 1993), in cells lacking telomerase, Y′ can also move by a transposition-like RNA-mediated process that relies on the Ty1 retrotransposon (Maxwell et al. 2004). In addition to RAD52 and POL32, the RAD51, RAD54, RAD57, and presumably RAD55 genes are also required to generate type I survivors (Le et al. 1999; Chen et al. 2001).
Type II survivors:
Telomeres in type II survivors show only minor amplifications of subtelomeric repeats but rather large increases in C1-3A/TG1-3 telomeric repeats (Figure 5). Telomeres in type II survivors are highly heterogeneous with some exceeding 12 kb in size and others being very short (Teng and Zakian 1999; Teng et al. 2000; Figure 5). The long telomeres are not stable but progressively shrink during outgrowth and then are subject to stochastic and dramatic lengthening events, consistent with rolling circle replication as an initiating event (Teng et al. 2000). In agreement with this hypothesis, circles of telomeric DNA are detected in type II survivors but not in wild-type cells (Lin et al. 2005; Larrivee and Wellinger 2006). Unlike type I survivors, the generation of type II survivors requires the MRX complex, RAD59 and SGS1, the yeast RecQ helicase, which is an ortholog of the Blm helicase (Le et al. 1999; Teng et al. 2000; Chen et al. 2001; Huang et al. 2001; Johnson et al. 2001).
Telomeric length control by telomeric rapid deletions:
In wild type, telomerase positive cells, over-elongated telomeres can be shortened to approximately normal length via a single intrachromosomal recombination event between telomeric repeats, a mechanism dubbed telomeric rapid deletion (TRD) (Li and Lustig 1996; Bucholc et al. 2001). This process contributes to keeping the average telomere length within a normal range (Bucholc et al. 2001). As a side product of this reaction, extrachromosomal circular DNA molecules with telomeric repeats are generated. TRD could produce the circular telomeric DNA molecules necessary for rolling circle replication during generation of type II survivors (Lustig 2003) as demonstrated in Kluyveromyces lactis (Natarajan and McEachern 2002; McEachern and Haber 2006) and proposed for human cells (Pickett et al. 2009; Cesare and Reddel 2010).
Transcription at Telomeres
Telomere-associated RNA
Despite having hallmarks of heterochromatin, subtelomeric sequences are actually transcribed to yield a new class of noncoding RNAs called telomeric repeat-containing RNA (TERRA) (Azzalin et al. 2007; Luke et al. 2008; Iglesias et al. 2011). TERRA occurs widely in eukaryotes as it has been detected in yeasts, plants, and vertebrates, including mammals, suggesting conserved functions (Luke and Lingner 2009; Feuerhahn et al. 2010). For budding yeast, earlier investigations had already shown that an artificially constructed telomere where the C strand of the telomere is transcribed at high levels shortens by ∼25% of its overall initial length (Sandell et al. 1994). However, transcription on a telomere per se did not induce signs of telomere dysfunction, such as high levels of chromosome loss, except that silencing of the adjacent gene was lost (Sandell et al. 1994).
Naturally occurring TERRA is composed of composite RNAs containing both subtelomeric sequences, such as Y′ and X, and telomeric G-strand transcripts. The size of TERRAs range from 100 to 1200 nt, they are generated by RNA polymerase II, and are polyadenylated. In wild-type cells, TERRA is rapidly degraded by the essential RNA exonuclease Rat1p, which also functions in processing standard mRNAs (Rosonina et al. 2006; Luke et al. 2008; Rondon et al. 2010). TERRA probably regulates telomere length and replication. For example, rat1-1 cells grown at semipermissive temperatures and having increased levels of TERRA have shorter telomeres, and this telomere shortening is due to impairment of the telomerase pathway (Luke et al. 2008). However, the telomere shortening due to reduced Rat1p levels can be reversed by overexpression of RNaseH. Since RNaseH removes RNA that is basepaired to DNA, this finding suggests that TERRA is associated with telomeric DNA when it inhibits telomerase. TERRAs transcribed from X telomeres vs. XY′ telomeres are subject to different regulation (Iglesias et al. 2011). Both are repressed via a Rap1p-mediated pathway, but only the X TERRA is repressed by Sir proteins. X and Y′ TERRAs are repressed by Rif1p and to a lesser extent, Rif2p.
Telomere silencing or TPE
Telomeric silencing (or TPE) was discovered serendipitously in S. cerevisiae during attempts to generate a uniquely marked telomere that could be used for chromatin studies (Gottschling et al. 1990). To mark the telomere, URA3 was inserted immediately adjacent to the left telomere of chromosome VII, in the process deleting the TAS sequences that are normally present at this telomere. Cells carrying URA-TEL, the URA3 marked telomere, are Ura+ as expected but unexpectedly, many of them are also FOA resistant (FOAR; FOA is a drug that kills cells expressing Ura3p). The FOAR cells have not lost or mutated URA3 as the FOAR phenotype is reversible. These effects correlate with URA3 mRNA levels: cells growing on medium lacking uracil have ∼10 times more URA3 mRNA than FOA-grown cells (Gottschling et al. 1990). Thus, TPE is due to repression of transcription, but this repression is reversible.
TPE is gene and telomere nonspecific. Expression of multiple RNA Pol II transcribed genes are repressed when they are near a telomere, and TPE is detected at other truncated telomeres (Gottschling et al. 1990). The metastable nature of TPE is easily visualized when ADE2 is the telomeric marker, as Ade2+ cells produce white colonies while Ade2− cells generate red colonies. A large fraction of ADE2-TEL cells produce largely red colonies (ADE2 expression repressed), while about an equal number produce largely white colonies (ADE2 expressed). However, red colonies have many white sectors, and white colonies have many red sectors. These sectors reflect phenotypic switches in transcription state within individual cells during the ∼25 divisions it takes to generate a colony. This altered state is then inherited by their progeny to produce a sector of opposite color.
Over 50 genes affect TPE, although the effects of many are relatively minor, suggesting that some may act indirectly. Moreover, FOA medium can affect ribonucleotide reductase expression in such a way that assays using URA3 as a TPE reporter can misidentify genes, such as POL30 and DOT1, as having roles in TPE when their effects are more likely due to metabolic changes (Rossmann et al. 2011; Takahashi et al. 2011). In contrast, Sir2p, Sir3p, Sir4p (Aparicio et al. 1991), and the YKu complex (Boulton and Jackson 1998) are all essential for TPE, although ykuΔ cells are TPE proficient if they also lack RIF1 and RIF2 (Mishra and Shore 1999). Since Rif1p, Rif2p, Sir3p, and Sir4p all interact with the C terminus of Rap1p, the absence of the two Rif proteins probably reduces their competition with Sir3p and Sir4p for the Rap1p interaction, which brings them to telomeres.
Sir2, 3, 4, and YKu bind telomeres and thus act directly to promote TPE. The three Sir (silence information regulator) proteins are part of the Sir silencing complex, which is also needed for transcriptional repression at the two silent mating type loci, HML and HMR. The carboxyl terminus of Rap1p, the major sequence-specific telomeric binding protein, interacts with both Sir3p and Sir4p, while Sir4p interacts with Sir2p (Moretti et al. 1994; Moretti and Shore 2001). Thus, Sir3p/Sir4p–Rap1p and Sir2p/Sir4p interactions recruit these silencing proteins to telomeres. Sir4p also interacts with YKu (Tsukamoto et al. 1997), which provides a Rap1p independent pathway to recruit silencing proteins to telomeres (Martin et al. 1999; Luo et al. 2002). Both recruitment pathways are essential for TPE.
After TPE is initiated at telomeres, it spreads several kilobases from the Rap1p-bound telomeric repeats into subtelomeric nucleosomes. This spread is mediated by protein–protein interactions between Sir3p and Sir4p with the N-terminal tails of histones H3 and H4 (Hecht et al. 1995; Strahl-Bolsinger et al. 1997). Thus, deleting the amino terminal tails of histones H3 and H4 abolishes TPE (Aparicio et al. 1991; Mann and Grunstein 1992; Thompson et al. 1994). Spreading also requires the histone deacetylase activity of Sir2p, as acetylation, especially of histone H4 K16, decreases Sir3/4p–histone interactions (Hoppe et al. 2002). Many other genes that modify histones or that regulate these modifications also affect TPE.
Early studies suggested that TPE requires proximity to a telomere, not just telomeric sequence, as an 81-bp internal tract of telomeric DNA does not silence an adjacent gene (Gottschling et al. 1990). However, long (≥300 bp) internal tracts of telomeric DNA can repress transcription, even if the affected gene and adjacent tract are on a circular chromosome (Stavenhagen and Zakian 1994). This phenomenon is called C1-3A-based silencing, CBS. The fraction of cells exhibiting CBS increases with the length of telomeric sequence, but CBS is never as effective as TPE. However, CBS acts synergistically with TPE as the closer an internal tract is to a telomere, the more effectively it silences. This synergism suggests a higher order chromatin structure, such as looping, that brings internal telomeric tracts close to chromosome ends. Consistent with their ability to silence, internal tracts of telomeric DNA efficiently bind Rap1p and Sir proteins (Bourns et al. 1998).
TPE was discovered using truncated telomeres that lack X and Y′, so it was not clear from early studies if this regulation affects genes that reside near native telomeres. This possibility was first tested by inserting a marker gene near a telomere without deleting its subtelomeric repeats. By this assay, only 6 of 17 telomeres (only half of the telomeres were tested) are subject to TPE (Pryde and Louis 1999). X-only telomeres are more likely than XY′ telomeres to silence. However, there is enormous variation in the TPE phenotypes of different telomeres. These differences are largely due to telomere-to-telomere variation in the identity and precise sequence of X and Y′. Subtelomeric sequences, especially X, contain recognition sites for different transcription factors such as Reb1p, Tbf1p, and Abf1p. Indeed, computational analysis of genome-wide ChIP data for 203 transcription factors finds that >10% of these show preferential association with the 25-kb regions next to telomeres (Mak et al. 2009). These enrichments are particularly high in stressed cells. Of these transcription factors, some activate and others repress TPE while others contribute to boundary activity, which limits the spread of silencing. The effects of these transcription factors on TPE may differ from their effects on transcription at nontelomeric loci. For example, Reb1p promotes transcription of ribosomal RNA (Morrow et al. 1989), but in subtelomeric DNA, Reb1p has boundary activity (Fourel et al. 1999). The binding of transcription factors to subtelomeric repeats explains why silencing is propagated differently at truncated vs. natural telomeres. At telomeres like URA-TEL, TPE extends inward continuously but dissipates quickly as the marker gene is moved further from the telomere (Renauld et al. 1993). However, at natural telomeres, domains of silencing are discontinuous.
TPE in the context of native telomeres is also assessed by examining mRNA levels and the effects of Sir3p on these levels for genes that are naturally located near telomeres. Thus, transcription of a Ty5 transposon near the III-L telomere is low in wild-type cells but higher in sir3 cells (Vega-Palas et al. 1997). Genome-wide studies also provide insights into the biological importance of TPE. For example, the 267 yeast genes that are within 20 kb of a telomere produce about five times fewer mRNA molecules (average of 0.5/cell) than nontelomeric genes, providing support for the repressive effects of telomere proximity (Wyrick et al. 1999). However, transcription of only 20 of these genes is Sir3p inhibited, and almost all such genes are very close (≤8 kb) to a telomere. Thus, from the classical view of TPE as a Sir-dependent phenomenon, very few genes are regulated by TPE. However, if criteria other than Sir3p dependence are used, many more genes are affected by telomere proximity. For example, in hda1Δ cells, which lack a histone deacetylase, genes that are 10–25 kb from telomeres are specifically derepressed (Robyr et al. 2002). Thus, Hda1p-sensitive regions are near telomeres but are distinct from Sir3p sensitive regions, which are even closer to telomeres. Unfortunately, none of these studies include the classic test to establish a position effect, which requires moving the gene away from the telomere and showing that its expression pattern is telomere dependent.
Many of the genes located near telomeres are members of multigene families. The functions of these telomere-regulated genes include rapamycin resistance (Ai et al. 2002), stress responsiveness, and ability to grow in nonstandard carbon sources (Robyr et al. 2002). For example, four of five members of the FLO gene family are near telomeres and are usually repressed except under nutrient conditions that promote pseudohyphal growth (Guo et al. 2000; Halme et al. 2004). Many of the transcription factors that bind X repeats (e.g., Rox1p, Gzf3p, and Oaf1p) regulate TPE in response to either stress or metabolic change (Smith et al. 2011). Thus, almost all genes that are naturally regulated by TPE, whether or not this regulation is Sir3p or Hda1p dependent, are genes that are not expressed under standard growth conditions. This pattern suggests a genomic logic where rarely or situationally expressed genes are located near telomeres where transcription is usually low.
Telomeres and Nuclear Organization
Higher order chromatin structure and telomere folding
In some organisms, including mammals (Griffith et al. 1999) and plants (Cesare et al. 2003), telomeres end in t-loops. T-loops are formed by G tails looping back and invading the duplex region of the telomere. This invasion displaces the G-rich strand to form a single-stranded displacement (D)-loop. T-loops are thought to contribute to telomere capping by sequestering the 3′ end of the chromosome within the telomeric tract (Griffith et al. 1999). Alternatively (or in addition), t-loops may be recombination intermediates (Cesare and Griffith 2004).
Throughout most of the cell cycle, G tails on S. cerevisiae telomeres are probably too short to form t-loops. However, yeast telomeres do form a higher order fold-back structure or telomere loop (Strahl-Bolsinger et al. 1997). Unlike t-loops, which are held together by DNA base pairing, the yeast telomere loop is maintained by protein–protein interactions. Telomere looping was proposed as an explanation for why Rap1p can be cross-linked in vivo not only to the telomeric repeats but also to subtelomeric DNA. As subtelomeric DNA is histone (not Rap1p) associated, and Rap1p does not interact with histones, its detection in these regions is explained by a fold-back structure that puts the Rap1p-bound telomeric repeats in proximity to subtelomeric chromatin. Yku80p binds both C1-3A/TG1-3 repeats and X elements, and it too is proposed to contribute to telomere folding (Marvin et al. 2009a). High rates of transcription through a telomere eliminate TPE in cis (Sandell et al. 1994) as well as telomere looping, suggesting that TPE might require this fold-back structure (de Bruin et al. 2000). Probably the best evidence for the importance of telomere looping comes from gene expression studies (de Bruin et al. 2001). A yeast upstream activating sequence (UAS) is similar to enhancers in other organisms, except that it affects transcription only when it is upstream of a gene. However, a downstream UAS can activate transcription if the affected gene is next to a telomere. Telomere looping, which is expected to bring the downstream UAS close to the gene’s promoter, is a possible explanation for why the UAS works in the downstream context. In addition to a possible role in TPE, telomere folding is proposed to protect the telomere from ectopic recombination (Marvin et al. 2009a,b).
Telomere organization in mitotic cells
Telomeres are clustered and at the nuclear periphery in many organisms. However, in most cases, it is not clear whether this pattern is functionally important or is just a passive consequence of the way chromosomes segregate at mitosis with centromeres leading the way and telomeres lagging behind. Yeast chromosomes are small and thus hard to visualize. As a result, the first suggestion for their nonrandom localization came from the subnuclear distribution of telomere binding proteins. Rap1p concentrates in 7–8 spots (called Rap1p foci) on the nuclear periphery (Palladino et al. 1993). These Rap1p foci also contain Sir2p, Sir3p, Sir4p, and the YKu complex and ∼70% of the Y′ repeats (Palladino et al. 1993; Gotta et al. 1996; Laroche et al. 1998). As these studies were done in diploid cells where there are 68 telomeres, the much larger number of telomeres compared to the number of Rap1p foci suggests that individual Rap1p foci contain many telomeres.
Although yeast chromosomes are too small to localize by fluorescent in situ hybridization (FISH), they can be visualized if they are marked with multiple binding sites for a GFP–DNA binding protein expressed in the same cells (Robinett et al. 1996). This system confirmed that the VII-L telomere is located at the nuclear periphery in ∼50% of cells (Tham et al. 2001). This fraction changes throughout the cell cycle, being particularly low after DNA replication. Peripheral localization of the VII-L telomere does not require Sir3p or Yku70p and thus is independent of both Rap1p foci and TPE. Other telomeres are also at the periphery but the fraction localized and their requirements for localization vary from telomere to telomere (Hediger et al. 2002). Thus, as with TPE, individual telomeres have different behaviors in terms of subnuclear localization. This variation is explained in part by differences in the TAS content of different telomeres (Mondoux and Zakian 2007). The GFP–DNA binding protein visualization method also confirmed telomere clustering, but found that the clusters are quite transient and do not involve specific subsets of telomeres (Therizols et al. 2010).
A priori, association with the nuclear periphery requires at least two proteins, one that is telomere associated and one located at the nuclear periphery. There are at least two nuclear envelope proteins that affect telomere tethering, Esc1p (establishes silent chromatin; Andrulis et al. 2002) and Mps3p (monopolar spindle; Bupp et al. 2007). Esc1p resides at the inner face of the nuclear envelope and interacts with the C-terminal portion of Sir4p (called the PAD4 domain; Andrulis et al. 2002). The Esc1p–Sir4p interaction can tether plasmid and chromosomal telomeres to the nuclear periphery (Andrulis et al. 2002; Taddei et al. 2004). However, the PAD4 domain also interacts with Yku80p, and this interaction also affects telomere tethering (Taddei et al. 2004). Although Mps3p was discovered as an essential subunit of the spindle pole body (yeast centrosome), a fraction of Mps3p is in the nuclear envelope (Jaspersen et al. 2002). Mps3p spans the inner nuclear envelope with its nonessential N terminus extending into the nucleoplasm where it can interact with telomere bound Sir4p or Yku. These interactions are important for telomere positioning as cells expressing the N-terminally truncated mps3Δ75-150 allele are viable but unable to tether telomeres (Bupp et al. 2007).
Sumoylation is also important for telomere tethering. The two known telomere parts of the tether, Sir4p and Yku80p, are both sumoylated in vivo by the SUMO E3 ligase SIZ2, and this modification affects their tethering functions (Zhao and Blobel 2005; Ferreira et al. 2011; Hang et al. 2011). In siz2Δ cells, tethering is lost, but TPE and Rap1p foci are unaffected. For Yku80p, loss of tethering is probably a direct result of loss of sumoylation, as an Yku80p–SUMO fusion increases tethering, and this tethering is now Siz2p independent (Ferreira et al. 2011).
Telomerase has also been implicated in telomere tethering. By two-hybrid and coimmunoprecipitation, Mps3p interacts with Est1p (Antoniacci et al. 2007). In early S phase, tethering requires a specific interaction between Yku80p and TLC1 (Schober et al. 2009), the same interaction needed for Est2p telomere binding in G1 phase (Fisher et al. 2004). The Mps3p–Est1p interaction raises the possibility that telomere tethering might regulate telomerase. However, while mps3Δ75-150 cells are tethering deficient (Bupp et al. 2007), they have wild-type–length telomeres (M. Paul and V. A. Zakian, unpublished results). Nonetheless, telomeres in siz2Δ cells are modestly longer than wild-type telomeres, and this lengthening is telomerase dependent (Ferreira et al. 2011; Hang et al. 2011). Moreover, epistasis analysis suggests that Siz2p and Pif1p act in the same pathway to affect telomere length as telomeres are no longer in pif1siz2 cells than in the absence of Pif1 alone (Ferreira et al. 2011). Because pif1 cells have higher levels of telomere-bound telomerase (Boule et al. 2005), this result led to the hypothesis that siz2Δ telomeres are longer because they bind more telomerase. Furthermore, pif1Δ restores telomerase-dependent tethering in siz2Δ cells, presumably by increasing the amount of telomere-bound Est2p/Est1p (Ferreira et al. 2011). These data lead to the somewhat contradictory view that telomerase tethers telomeres to the periphery in a manner that is not permissive for telomere lengthening, while release of telomeres from the periphery promotes telomerase lengthening of the released telomere. Perhaps, when telomeres are bound at the periphery by an Mps3p–Est1p interaction, Est1p cannot interact with Cdc13p in a productive way. Consistent with this view, artificially tethering a telomere to the periphery results in telomere shortening without affecting the lengths of other telomeres in the cell (Mondoux et al. 2007). However, given that many telomere proteins are modified by sumoylation (Hang et al. 2011), it is probably wise to be cautious in attributing the modest telomere lengthening seen in siz2Δ cells to telomeres being lengthened preferentially when released from the nuclear envelope.
Tethering is lost in siz2Δ cells, yet TPE and Rap1p foci are normal. These results seem to rule out a critical role for tethering in TPE (Ferreira et al. 2011). This conclusion is consistent with experiments indicating that TPE and tethering are separable phenotypes (Tham et al. 2001; Mondoux and Zakian 2007). However, this conclusion is still surprising, given numerous examples in diverse organisms for a connection between the nuclear periphery, heterochromatin formation, and gene silencing. Perhaps the importance of concentrating silencing proteins at the periphery is not to support silencing but to sequester silencing proteins from the rest of the genome so that actively transcribed genes are not inadvertently repressed (Taddei et al. 2009).
Finally, telomere tethering has been suggested to affect recombination and repair of telomeric regions. Deleting YKU80, which reduces the association of some telomeres with the periphery, increases recombination between telomeres and nontelomeric sites in a pathway that acts through Yku80p-associated X elements (Marvin et al. 2009a,b). These data suggest that telomere tethering suppresses ectopic recombination within telomeric regions. In contrast, efficient repair of subtelomeric DSBs may require telomere localization at the periphery as such breaks within the XI-L telomere are less often repaired in genetic backgrounds where telomere tethering is lost (Therizols et al. 2006).
Telomeres in meiosis
It has been known for many years that meiotic chromosomes in most organisms assume a characteristic conformation called the bouquet in early prophase of the first meiotic division with telomeres clustered at the nuclear periphery at a position near the spindle pole body. Progress has been made in S. cerevisiae in learning how the bouquet is set up, although its functional significance is still being established. The S. cerevisiae NDJ1 was discovered in a screen for genes whose overexpression causes mis-segregation of meiotic chromosomes (Conrad et al. 1997). Ndj1p expression is limited to meiosis, and it localizes to meiotic telomeres in vivo. Cytological studies show that telomere clustering is Ndj1p dependent, making NDJ1 the first gene linked to bouquet formation in any organism. Meiotic chromosome segregation is also defective in ndj1 cells, suggesting that the bouquet configuration is important for normal meiotic chromosome behavior. Ndj1p interacts with nuclear envelope-localized Mps3p (Conrad et al. 2007, 2008). Mps3p is the yeast member of the conserved SUN family of inner nuclear membrane proteins. SUN proteins interact with chromosomal binding proteins in the nuclear interior and with outer nuclear membrane proteins in the space between the inner and outer nuclear membranes. Because the outer nuclear membrane protein can directly or indirectly bind to the cytoskeleton, the formation of a linker complex involving Mps3p and Ndj1p is able to move chromosome ends within the nucleus into a bouquet using energy derived from the cytoplasmic cytoskeleton. In contrast to Schizosaccharomyces pombe and multicellular eukaryotes, meiotic bouquet formation in S. cerevisiae is actin-, not tubulin dependent (Scherthan et al. 2007; Koszul et al. 2008).
Time-lapse imaging reveals that meiotic chromosomes engage in rapid and sustained movements throughout prophase in virtually all eukaryotes. These rapid meiotic chromosome movements were first documented in fission yeast where they are particularly dramatic (Chikashige et al. 1994). However, fission yeast meiosis is unusual in that it occurs in the absence of synaptonemal complexes. Therefore, the discovery of similar movements during S. cerevisiae meiosis was important because it made clear that these movements are not restricted to organisms with an atypical meiosis (Trelles-Sticken et al. 2005; Scherthan et al. 2007; Conrad et al. 2008; Koszul et al. 2008). In S. cerevisiae, meiotic chromosome movements are often rapid, in excess of 1 μm/sec and are dependent on the nuclear envelope protein, Csm4p, whose expression is also limited to meiosis (Conrad et al. 2008). The outcome of meiosis in csm4Δ cells suggests that meiotic chromosome movements are important for meiotic progression, in part by preventing spindle checkpoint activation (Zanders et al. 2011). However, since Csm4p is also needed for bouquet formation, it is not clear whether its effects on meiotic progression are due to its role in telomere clustering or chromosome movement (Wanat et al. 2008). Current models suggest that meiotic chromosome movements test homology between chromosomes to facilitate pairing and synapsis of homologous chromosomes.
Classical genetic studies showed that meiotic recombination occurs at lower levels near telomeres than in the rest of the genome (Barton et al. 2003). Genome-wide mapping of meiotic DSB positions confirms that DSBs are infrequent near telomeres with the best estimate being that they occur 3.5-fold less often in the 20 kb closest to telomeres than in most other genomic regions (Pan et al. 2011). Although the mechanistic basis for the relative paucity of meiotic recombination in telomeric regions is not known, a plausible explanation for its significance is to prevent exchanges between nonhomologous chromosomes.
Outlook
Given the multiple genome-wide approaches available in S. cerevisiae, it is likely that most genes affecting telomeres are identified. However, the functions of many of these genes have not been explored. Moreover, we lack mechanistic information even for well-studied telomere proteins. For example, although Rif1p has been known for years to act in cis to inhibit telomerase-mediated telomere lengthening, the mechanism(s) by which it does so is not understood. Likewise, the Tel1p kinase is critical for telomere length regulation yet there is no consensus on its phosphorylation targets nor information on how these targets differ at telomeres vs. DSBs. It is not known how the highly abundant RPA complex, which binds in a sequence nonspecific manner to single-stranded DNA, is excluded from TG1-3 tails, a binding that is expected to trigger a checkpoint-mediated arrest. Even though Est1p was the first identified telomerase subunit, its exact role and that of Est3p are only beginning to be understood. The recently discovered TERRA opens up a whole new area of possibilities for telomerase regulation. There is a lot of information on telomere tethering, yet its functional role is not resolved. Of particular interest is to establish how telomere tethering can be telomerase dependent while at the same time telomere release from the periphery promotes telomerase action. Research over the past years demonstrates that telomeres have individual personalities: they differ by their subtelomeric repeats, TPE behavior, and nuclear localization, suggesting that we will only fully understand the impact of genes and conditions on telomere behavior if we study individual chromosome ends. The list of important unanswered questions goes on and on, making it clear that yeast telomere biologists still have a lot to do.
What do we think will be particularly important for future advances? Biochemical approaches for yeast telomerase have long been thwarted by difficulties isolating telomerase proteins and reconstituting telomerase. Given recent success with in vitro assays, this is an area that will likely yield new insights in the near future. The impact of in vitro studies will be increased enormously by the large number of mutants that have been generated and characterized in vivo, reagents that are not available to anywhere near the same extent in other organisms. So far, there is no successful mass spectrometry on yeast telomerase, yet this approach has been extremely fruitful for both mammalian and ciliate research. The small size of yeast chromosomes has limited cell biological approaches in telomere research. However, the ability to visualize specific telomere regions with GFP technology has largely solved this problem. We anticipate that soon these chromosome visualization techniques will be combined with new methods to visualize telomerase itself to yield important information on telomerase dynamics vis a vis nuclear organization at the single-cell level. TERRA is so newly discovered that it seems inescapable that its continued analysis will provide new and perhaps unanticipated findings.
Research in ciliates and to a lesser extent yeasts, pioneered the field of telomere biology. In the past decade or so, there has been a lamentable decline in the number of labs doing telomere research in ciliates. As a consequence, the importance of yeast as a model organism for telomere research has, if anything, become more apparent. Many important discoveries on mammalian telomerase were inspired by work in yeast. We expect this trend to continue. Given the clear links between telomere biology and human aging and cancer, there is little doubt that basic research in yeast telomere biology has an important place in biomedical research.
Acknowledgments
We are grateful to B. Brewer, I. Dionne, K. Friedman, S. Jasperson, J. Lingner, D. Lydall, C. Nugent, and Y. Wu for their reading of the manuscript and for courageously providing valuable input. The V.A.Z. lab is funded by the National Institutes of Health (RO1’s GM43265 and GM026938); the R.J.W. lab has funding from the Canadian Institutes for Health Research (grants MOP97874 and MOP110982). We thank the considerable physical distance separating our two universities, which prevented us from killing each other over minor details during the writing.
Footnotes
Communicating editor: R. Rothstein
Literature Cited
- Abdallah P., Luciano P., Runge K. W., Lisby M., Geli V., et al. , 2009. A two-step model for senescence triggered by a single critically short telomere. Nat. Cell Biol. 11: 988–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A. K., Holm C., 1996. Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 4614–4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams Martin A., Dionne I., Wellinger R. J., Holm C., 2000. The function of DNA polymerase alpha at telomeric G tails is important for telomere homeostasis. Mol. Cell. Biol. 20: 786–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addinall S. G., Downey M., Yu M., Zubko M. K., Dewar J., et al. , 2008. A genomewide suppressor and enhancer analysis of cdc13–1 reveals varied cellular processes influencing telomere capping in Saccharomyces cerevisiae. Genetics 180: 2251–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addinall S. G., Holstein E. M., Lawless C., Yu M., Chapman K., et al. , 2011. Quantitative fitness analysis shows that NMD proteins and many other protein complexes suppress or enhance distinct telomere cap defects. PLoS Genet. 7: e1001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai W., Bertram P. G., Tsang C. K., Chan T. F., Zheng X. F., 2002. Regulation of subtelomeric silencing during stress response. Mol. Cell 10: 1295–1305 [DOI] [PubMed] [Google Scholar]
- Albert I., Mavrich T. N., Tomsho L. P., Qi J., Zanton S. J., et al. , 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 [DOI] [PubMed] [Google Scholar]
- Alexander M. K., Zakian V. A., 2003. Rap1p telomere association is not required for mitotic stability of a C(3)TA(2) telomere in yeast. EMBO J. 22: 1688–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbalagan S., Bonetti D., Lucchini G., Longhese M. P., 2011. Rif1 supports the function of the CST complex in yeast telomere capping. PLoS Genet. 7: e1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis E. D., Zappulla D. C., Ansari A., Perrod S., Laiosa C. V., et al. , 2002. Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. Mol. Cell. Biol. 22: 8292–8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniacci L. M., Kenna M. A., Skibbens R. V., 2007. The nuclear envelope and spindle pole body-associated Mps3 protein bind telomere regulators and function in telomere clustering. Cell Cycle 6: 75–79 [DOI] [PubMed] [Google Scholar]
- Aparicio O. M., Billington B. L., Gottschling D. E., 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66: 1279–1287 [DOI] [PubMed] [Google Scholar]
- Arneric M., Lingner J., 2007. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep. 8: 1080–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askree S. H., Yehuda T., Smolikov S., Gurevich R., Hawk J., et al. , 2004. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. USA 101: 8658–8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A., Giresi P., Lieb J., Zakian V., 2009. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol. Cell 34: 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin C. M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J., 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 [DOI] [PubMed] [Google Scholar]
- Bah A., Gilson E., Welllinger R. J., 2011. Telomerase is required to protect chromosomes with vertebrate-type T2AG3 3′-ends in S. cerevisiae. J. Biol. Chem. 286: 27132–27138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton A. B., Su Y., Lamb J., Barber D., Kaback D. B., 2003. A function for subtelomeric DNA in Saccharomyces cerevisiae. Genetics 165: 929–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beernink H. T., Miller K., Deshpande A., Bucher P., Cooper J. P., 2003. Telomere maintenance in fission yeast requires an est1 ortholog. Curr. Biol. 13: 575–580 [DOI] [PubMed] [Google Scholar]
- Ben-Aroya S., Coombes C., Kwok T., O’Donnell K. A., Boeke J. D., et al. , 2008. Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol. Cell 30: 248–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthiau A. S., Yankulov K., Bah A., Revardel E., Luciano P., et al. , 2006. Subtelomeric proteins negatively regulate telomere elongation in budding yeast. EMBO J. 25: 846–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A., Shore D., 2007a. Early replication of short telomeres in budding yeast. Cell 128: 1051–1062 [DOI] [PubMed] [Google Scholar]
- Bianchi A., Shore D., 2007b. Increased association of telomerase with short telomeres in yeast. Genes Dev. 21: 1726–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A., Negrini S., Shore D., 2004. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol. Cell 16: 139–146 [DOI] [PubMed] [Google Scholar]
- Bonetti D., Martina M., Clerici M., Lucchini G., Longhese M. P., 2009. Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol. Cell 35: 70–81 [DOI] [PubMed] [Google Scholar]
- Bonetti D., Clerici M., Anbalagan S., Martina M., Lucchini G., et al. , 2010. Shelterin-like proteins and Yku inhibit nucleolytic processing of Saccharomyces cerevisiae telomeres. PLoS Genet. 6: e1000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosoy D., Peng Y., Mian I. S., Lue N. F., 2003. Conserved N-terminal motifs of telomerase reverse transcriptase required for ribonucleoprotein assembly in vivo. J. Biol. Chem. 278: 3882–3890 [DOI] [PubMed] [Google Scholar]
- Boule J., Vega L., Zakian V., 2005. The yeast Pif1p helicase removes telomerase from DNA. Nature 438: 57–61 [DOI] [PubMed] [Google Scholar]
- Boulton S. J., Jackson S. P., 1996. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 24: 4639–4648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton S. J., Jackson S. P., 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17: 1819–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourns B. D., Alexander M. K., Smith A. M., Zakian V. A., 1998. Sir proteins, Rif proteins, and Cdc13p bind Saccharomyces telomeres in vivo. Mol. Cell. Biol. 18: 5600–5608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigati C., Kurtz S., Balderes D., Vidali G., Shore D., 1993. An essential yeast gene encoding a TTAGGG repeat-binding protein. Mol. Cell. Biol. 13: 1306–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Lue N. F., Kornberg R. D., 1988. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol. Cell. Biol. 8: 5086–5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholc M., Park Y., Lustig A. J., 2001. Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 6559–6573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck S. W., Shore D., 1995. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 9: 370–384 [DOI] [PubMed] [Google Scholar]
- Buonomo S. B., 2010. Heterochromatin DNA replication and Rif1. Exp. Cell Res. 316: 1907–1913 [DOI] [PubMed] [Google Scholar]
- Bupp J. M., Martin A. E., Stensrud E. S., Jaspersen S. L., 2007. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 179: 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson M. J., Hartwell L., 1985. CDC17: an essential gene that prevents telomere elongation in yeast. Cell 42: 249–257 [DOI] [PubMed] [Google Scholar]
- Cesare A. J., Griffith J. D., 2004. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell. Biol. 24: 9948–9957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare A. J., Reddel R. R., 2010. Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 11: 319–330 [DOI] [PubMed] [Google Scholar]
- Cesare A. J., Quinney N., Willcox S., Subramanian D., Griffith J. D., 2003. Telomere looping in P. sativum (common garden pea). Plant J. 36: 271–279 [DOI] [PubMed] [Google Scholar]
- Chan A., Boule J. B., Zakian V. A., 2008. Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet. 4: e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. S., Tye B. K., 1983a. A family of Saccharomyces cerevisiae repetitive autonomously replicating sequences that have very similar genomic environments. J. Mol. Biol. 168: 505–523 [DOI] [PubMed] [Google Scholar]
- Chan C. S., Tye B. K., 1983b. Organization of DNA sequences and replication origins at yeast telomeres. Cell 33: 563–573 [DOI] [PubMed] [Google Scholar]
- Chan S. W., Chang J., Prescott J., Blackburn E. H., 2001. Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr. Biol. 11: 1240–1250 [DOI] [PubMed] [Google Scholar]
- Chang M., Arneric M., Lingner J., 2007. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 21: 2485–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapon C., Cech T. R., Zaug A. J., 1997. Polyadenylation of telomerase RNA in budding yeast. RNA 3: 1337–1351 [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Ijpma A., Greider C. W., 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21: 1819–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y., Ding D. Q., Funabiki H., Haraguchi T., Mashiko S., et al. , 1994. Telomere-led premeiotic chromosome movement in fission yeast. Science 264: 270–273 [DOI] [PubMed] [Google Scholar]
- Cohn M., Blackburn E. H., 1995. Telomerase in yeast. Science 269: 396–400 [DOI] [PubMed] [Google Scholar]
- Conrad M. N., Wright J. H., Wolf A. J., Zakian V. A., 1990. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell 63: 739–750 [DOI] [PubMed] [Google Scholar]
- Conrad M. N., Dominguez A. M., Dresser M. E., 1997. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science 276: 1252–1255 [DOI] [PubMed] [Google Scholar]
- Conrad M. N., Lee C. Y., Wilkerson J. L., Dresser M. E., 2007. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 104: 8863–8868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M. N., Lee C. Y., Chao G., Shinohara M., Kosaka H., et al. , 2008. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell 133: 1175–1187 [DOI] [PubMed] [Google Scholar]
- Cosgrove A. J., Nieduszynski C. A., Donaldson A. D., 2002. Ku complex controls the replication time of DNA in telomere regions. Genes Dev. 16: 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amours D., Jackson S. P., 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15: 2238–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlseid J. N., Lew-Smith J., Lelivelt M. J., Enomoto S., Ford A., et al. , 2003. mRNAs encoding telomerase components and regulators are controlled by UPF genes in Saccharomyces cerevisiae. Eukaryot. Cell 2: 134–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley J. M., Palmbos P. L., Wu D., Wilson T. E., 2005. Nonhomologous end joining in yeast. Annu. Rev. Genet. 39: 431–451 [DOI] [PubMed] [Google Scholar]
- Dandjinou A. T., Levesque N., Larose S., Lucier J. F., Abou Elela S., et al. , 2004. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr. Biol. 14: 1148–1158 [DOI] [PubMed] [Google Scholar]
- de Bruin D., Kantrow S. M., Liberatore R. A., Zakian V. A., 2000. Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol. Cell. Biol. 20: 7991–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin D., Zaman Z., Liberatore R. A., Ptashne M., 2001. Telomere looping permits gene activation by a downstream UAS in yeast. Nature 409: 109–113 [DOI] [PubMed] [Google Scholar]
- Dewar J. M., Lydall D., 2010. Pif1- and Exo1-dependent nucleases coordinate checkpoint activation following telomere uncapping. EMBO J. 29: 4020–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan D. C., Freeman B. C., 2009. The conserved Est1 protein stimulates telomerase DNA extension activity. Proc. Natl. Acad. Sci. USA 106: 17337–17342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan D. C., Toogun O. A., Echtenkamp F. J., Freeman B. C., 2009. The Hsp82 molecular chaperone promotes a switch between unextendable and extendable telomere states. Nat. Struct. Mol. Biol. 16: 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede S. J., Gottschling D. E., 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99: 723–733 [DOI] [PubMed] [Google Scholar]
- Diede S. J., Gottschling D. E., 2001. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr. Biol. 11: 1336–1340 [DOI] [PubMed] [Google Scholar]
- Dionne I., Wellinger R. J., 1998. Processing of telomeric DNA ends requires the passage of a replication fork. Nucleic Acids Res. 26: 5365–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey M., Houlsworth R., Maringele L., Rollie A., Brehme M., et al. , 2006. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell 124: 1155–1168 [DOI] [PubMed] [Google Scholar]
- DuBois M. L., Haimberger Z. W., McIntosh M. W., Gottschling D. E., 2002. A quantitative assay for telomere protection in Saccharomyces cerevisiae. Genetics 161: 995–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto S., Glowczewski L., Berman J., 2002. MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 2626–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugster A., Lanzuolo C., Bonneton M., Luciano P., Pollice A., et al. , 2006. The finger subdomain of yeast telomerase cooperates with Pif1p to limit telomere elongation. Nat. Struct. Mol. Biol. 13: 734–739 [DOI] [PubMed] [Google Scholar]
- Evans S. K., Lundblad V., 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120 [DOI] [PubMed] [Google Scholar]
- Faure V., Coulon S., Hardy J., Geli V., 2010. Cdc13 and telomerase bind through different mechanisms at the lagging- and leading-strand telomeres. Mol. Cell 38: 842–852 [DOI] [PubMed] [Google Scholar]
- Ferguson B. M., Fangman W. L., 1992. A position effect on the time of replication origin activation in yeast. Cell 68: 333–339 [DOI] [PubMed] [Google Scholar]
- Ferreira H. C., Luke B., Schober H., Kalck V., Lingner J., et al. , 2011. The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat. Cell Biol. 13: 867–874 [DOI] [PubMed] [Google Scholar]
- Feuerhahn S., Iglesias N., Panza A., Porro A., Lingner J., 2010. TERRA biogenesis, turnover and implications for function. FEBS Lett. 584: 3812–3818 [DOI] [PubMed] [Google Scholar]
- Fisher T. S., Zakian V. A., 2005. Ku: a multifunctional protein involved in telomere maintenance. DNA Repair (Amst.) 4: 1215–1226 [DOI] [PubMed] [Google Scholar]
- Fisher T. S., Taggart A. K. P., Zakian V. A., 2004. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 11: 1198–1205 [DOI] [PubMed] [Google Scholar]
- Forstemann K., Lingner J., 2001. Molecular basis for telomere repeat divergence in budding yeast. Mol. Cell. Biol. 21: 7277–7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G., Revardel E., Koering C. E., Gilson E., 1999. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 18: 2522–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F., Kolodynski J., 1983. pif mutation blocks recombination between mitochondrial rho+ and rho- genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 80: 5345–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank C. J., Hyde M., Greider C. W., 2006. Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol. Cell 24: 423–432 [DOI] [PubMed] [Google Scholar]
- Franke J., Gehlen J., Ehrenhofer-Murray A. E., 2008. Hypermethylation of yeast telomerase RNA by the snRNA and snoRNA methyltransferase Tgs1. J. Cell Sci. 121: 3553–3560 [DOI] [PubMed] [Google Scholar]
- Friedman K. L., Cech T. R., 1999. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 13: 2863–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman K. L., Heit J. J., Long D. M., Cech T. R., 2003. N-terminal domain of yeast telomerase reverse transcriptase: recruitment of Est3p to the telomerase complex. Mol. Biol. Cell 14: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga K., Hirano Y., Sugimoto K., 2012. Subtelomere-binding protein Tbf1 and telomere-binding protein Rap1 collaborate to inhibit localization of the Mre11 complex to DNA ends in budding yeast. Mol. Biol. Cell 23: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo F., Olivier C., Dandjinou A. T., Wellinger R. J., Chartrand P., 2008. TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J. 27: 748–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo F., Laterreur N., Cusanelli E., Querido E., Wellinger R. J., et al. , 2011. Live cell imaging of telomerase RNA dynamics reveals cell cycle-dependent clustering of telomerase at elongating telomeres. Mol. Cell 44: 819–827 [DOI] [PubMed] [Google Scholar]
- Gao H., Cervantes R. B., Mandell E. K., Otero J. H., Lundblad V., 2007. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 14: 208–214 [DOI] [PubMed] [Google Scholar]
- Gao H., Toro T. B., Paschini M., Braunstein-Ballew B., Cervantes R. B., et al. , 2010. Telomerase recruitment in Saccharomyces cerevisiae is not dependent on Tel1-mediated phosphorylation of Cdc13. Genetics 186: 1147–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B., Carson M., Hartwell L., 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15: 6128–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatbonton T., Imbesi M., Nelson M., Akey J. M., Ruderfer D. M., et al. , 2006. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelinas A. D., Paschini M., Reyes F. E., Heroux A., Batey R. T., et al. , 2009. Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc. Natl. Acad. Sci. USA 106: 19298–19303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Roberge M., Giraldo R., Rhodes D., Gasser S. M., 1993. Distortion of the DNA double helix by RAP1 at silencers and multiple telomeric binding sites. J. Mol. Biol. 231: 293–310 [DOI] [PubMed] [Google Scholar]
- Gotta M., Laroche T., Formenton A., Maillet L., Scherthan H., et al. , 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134: 1349–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D. E., 1992. Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc. Natl. Acad. Sci. USA 89: 4062–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D. E., Aparicio O. M., Billington B. L., Zakian V. A., 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762 [DOI] [PubMed] [Google Scholar]
- Goudsouzian L., Tuzon C., Zakian V. A., 2006. S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol. Cell 24: 603–610 [DOI] [PubMed] [Google Scholar]
- Graham I. R., Haw R. A., Spink K. G., Halden K. A., Chambers A., 1999. In vivo analysis of functional regions within yeast Rap1p. Mol. Cell. Biol. 19: 7481–7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N., Reed S. I., Charbonneau M., 1997. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 11: 512–527 [DOI] [PubMed] [Google Scholar]
- Grandin N., Damon C., Charbonneau M., 2000. Cdc13 cooperates with the yeast Ku proteins and stn1 To regulate telomerase recruitment. Mol. Cell. Biol. 20: 8397–8408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N., Damon C., Charbonneau M., 2001. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 20: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., Wellinger R. J., 2002. Maintenance of double-stranded telomeric repeats as the critical determinant for cell viability in yeast cells lacking Ku. Mol. Cell. Biol. 22: 2182–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., Larrivee M., Labrecque P., Wellinger R. J., 1998. Yeast Ku as a regulator of chromosomal DNA end structure. Science 280: 741–744 [DOI] [PubMed] [Google Scholar]
- Gravel S., Chapman J. R., Magill C., Jackson S. P., 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22: 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwell P. W., Kronmal S. L., Porter S. E., Gassenhuber J., Obermaier B., et al. , 1995. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82: 823–829 [DOI] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H., 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43: 405–413 [DOI] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H., 1987. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 51: 887–898 [DOI] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H., 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337: 331–337 [DOI] [PubMed] [Google Scholar]
- Grenon M., Gilbert C., Lowndes N. F., 2001. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat. Cell Biol. 3: 844–847 [DOI] [PubMed] [Google Scholar]
- Griffith J. D., Comeau L., Rosenfield S., Stansel R. M., Bianchi A., et al. , 1999. Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Grossi S., Puglisi A., Dmitriev P. V., Lopes M., Shore D., 2004. Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev. 18: 992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B., Bataille A. R., Gevry N., Adam M., Blanchette M., et al. , 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3: e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Styles C. A., Feng Q., Fink G. R., 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 97: 12158–12163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett J. A., Greider C. W., 2003. End resection initiates genomic instability in the absence of telomerase. Mol. Cell. Biol. 23: 8450–8461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett J. A., Feldser D. M., Greider C. W., 2001. Telomere dysfunction increases mutation rate and genomic instability. Cell 106: 275–286 [DOI] [PubMed] [Google Scholar]
- Halme A., Bumgarner S., Styles C., Fink G. R., 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116: 405–415 [DOI] [PubMed] [Google Scholar]
- Hang L. E., Liu X., Cheung I., Yang Y., Zhao X., 2011. SUMOylation regulates telomere length homeostasis by targeting Cdc13. Nat. Struct. Mol. Biol. 18: 920–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy C. F., Balderes D., Shore D., 1992a. Dissection of a carboxy-terminal region of the yeast regulatory protein RAP1 with effects on both transcriptional activation and silencing. Mol. Cell. Biol. 12: 1209–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy C. F., Sussel L., Shore D., 1992b. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 6: 801–814 [DOI] [PubMed] [Google Scholar]
- Hecht A., Laroche T., Strahl-Bolsinger S., Gasser S. M., Grunstein M., 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80: 583–592 [DOI] [PubMed] [Google Scholar]
- Hector R. E., Shtofman R. L., Ray A., Chen B.-R., Nyun T., et al. , 2007. Tel1p Preferentially Associates with Short Telomeres to Stimulate Their Elongation. Mol. Cell 27: 851–858 [DOI] [PubMed] [Google Scholar]
- Hector R. E., Ray A., Chen B. R., Shtofman R., Berkner K. L., et al. , 2012. Mec1p associates with functionally compromised telomeres. Chromosoma (in press, PMID: 22289863). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F., Neumann F. R., Van Houwe G., Dubrana K., Gasser S. M., 2002. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 12: 2076–2089 [DOI] [PubMed] [Google Scholar]
- Hediger F., Berthiau A. S., van Houwe G., Gilson E., Gasser S. M., 2006. Subtelomeric factors antagonize telomere anchoring and Tel1-independent telomere length regulation. EMBO J. 25: 857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y., Sugimoto K., 2007. Cdc13 telomere capping decreases Mec1 association but does not affect Tel1 association with DNA ends. Mol. Biol. Cell 18: 2026–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y., Fukunaga K., Sugimoto K., 2009. Rif1 and Rif2 inhibit localization of Tel1 to DNA ends. Mol. Cell 33: 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe G. J., Tanny J. C., Rudner A. D., Gerber S. A., Danaie S., et al. , 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22: 4167–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz H., Thorburn P., Haber J. E., 1984. Rearrangements of highly polymorphic regions near telomeres of Saccharomyces cerevisiae. Mol. Cell. Biol. 4: 2509–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Pryde F. E., Lester D., Maddison R. L., Borts R. H., et al. , 2001. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 11: 125–129 [DOI] [PubMed] [Google Scholar]
- Huertas P., Cortes-Ledesma F., Sartori A. A., Aguilera A., Jackson S. P., 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455: 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T. R., Weilbaecher R. G., Walterscheid M., Lundblad V., 2000. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc. Natl. Acad. Sci. USA 97: 6457–6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N., Redon S., Pfeiffer V., Dees M., Lingner J., et al. , 2011. Subtelomeric repetitive elements determine TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO Rep. 12: 587–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Armstrong C. M., Kaeberlein M., Guarente L., 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800 [DOI] [PubMed] [Google Scholar]
- Ira G., Pellicioli A., Balijja A., Wang X., Fiorani S., et al. , 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa A. S., Zhou J. Q., Zakian V. A., 2000. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100: 479–489 [DOI] [PubMed] [Google Scholar]
- Ivessa A. S., Zhou J. Q., Schulz V. P., Monson E. K., Zakian V. A., 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 16: 1383–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa A. S., Lenzmeier B. A., Bessler J. B., Goudsouzian L. K., Schnakenberg S. L., et al. , 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell 12: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Jamonnak N., Creamer T. J., Darby M. M., Schaughency P., Wheelan S. J., et al. , 2011. Yeast Nrd1, Nab3, and Sen1 transcriptome-wide binding maps suggest multiple roles in post-transcriptional RNA processing. RNA 17: 2011–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S. L., Giddings T. H., Jr, Winey M., 2002. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J. Cell Biol. 159: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F. B., Marciniak R. A., McVey M., Stewart S. A., Hahn W. C., et al. , 2001. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 20: 905–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J. A., Melo J. A., Cheung S. K., Vaze M. B., Haber J. E., et al. , 2004. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr. Biol. 14: 2096–2106 [DOI] [PubMed] [Google Scholar]
- Khadaroo B., Teixeira M. T., Luciano P., Eckert-Boulet N., Germann S. M., et al. , 2009. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat. Cell Biol. 11: 980–987 [DOI] [PubMed] [Google Scholar]
- Kim J. A., Kruhlak M., Dotiwala F., Nussenzweig A., Haber J. E., 2007. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J. Cell Biol. 178: 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseleva-Romanova E., Lopreiato R., Baudin-Baillieu A., Rousselle J. C., Ilan L., et al. , 2006. Yeast homolog of a cancer-testis antigen defines a new transcription complex. EMBO J. 25: 3576–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koering C. E., Fourel G., Binet-Brasselet E., Laroche T., Klein F., et al. , 2000. Identification of high affinity Tbf1p-binding sites within the budding yeast genome. Nucleic Acids Res. 28: 2519–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R., Kim K. P., Prentiss M., Kleckner N., Kameoka S., 2008. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell 133: 1188–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T., Martin S. G., Gotta M., Gorham H. C., Pryde F. E., et al. , 1998. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 8: 653–656 [DOI] [PubMed] [Google Scholar]
- Larose S., Laterreur N., Ghazal G., Gagnon J., Wellinger R. J., et al. , 2006. RNase III-dependent regulation of yeast telomerase. J. Biol. Chem. 282: 4373–4381 [DOI] [PubMed] [Google Scholar]
- Larrivee M., LeBel C., Wellinger R. J., 2004. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 18: 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrivee M., Wellinger R. J., 2006. Telomerase- and capping-independent yeast survivors with alternate telomere states. Nat. Cell Biol. 8: 741–747 [DOI] [PubMed] [Google Scholar]
- Le S., Moore J. K., Haber J. E., Greider C. W., 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152: 143–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Mandell E. K., Tucey T. M., Morris D. K., Lundblad V., 2008. The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat. Struct. Mol. Biol. 15: 990–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Mandell E. K., Rao T., Wuttke D. S., Lundblad V., 2010. Investigating the role of the Est3 protein in yeast telomere replication. Nucleic Acids Res. 38: 2279–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Kozak M., Martin J. D., Pennock E., Johnson F. B., 2007. Evidence that a RecQ helicase slows senescence by resolving recombining telomeres. PLoS Biol. 5: e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay T. S., Morris D. K., Sah J., Balasubramanian B., Lundblad V., 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144: 1399–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D. L., Blackburn E. H., 2004. Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol. Cell. Biol. 24: 10857–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Lustig A. J., 1996. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 10: 1310–1326 [DOI] [PubMed] [Google Scholar]
- Li S., Makovets S., Matsuguchi T., Blethrow J. D., Shokat K. M., et al. , 2009. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle Progression. Cell 136: 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H. Y., Robertson E. D., Hiraga S., Alvino G. M., Collingwood D., et al. , 2011. The effect of Ku on telomere replication time is mediated by telomere length but is independent of histone tail acetylation. Mol. Biol. Cell 22: 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb J. D., Liu X., Botstein D., Brown P. O., 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28: 327–334 [DOI] [PubMed] [Google Scholar]
- Lin C. A., Ellis S. R., True H. L., 2010. The Sua5 protein is essential for normal translational regulation in yeast. Mol. Cell. Biol. 30: 354–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. Y., Chang H. H., Wu K. J., Tseng S. F., Lin C. C., et al. , 2005. Extrachromosomal telomeric circles contribute to Rad52-, Rad50-, and polymerase delta-mediated telomere-telomere recombination in Saccharomyces cerevisiae. Eukaryot. Cell 4: 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Ly H., Hussain A., Abraham M., Pearl S., et al. , 2004. A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. Proc. Natl. Acad. Sci. USA 101: 14713–14718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.-J., Zakian V. A., 1995. An in vitro assay for Saccharomyces telomerase requires EST1. Cell 81: 1127–1135 [DOI] [PubMed] [Google Scholar]
- Lin J.-J., Zakian V. A., 1996. The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl. Acad. Sci. USA 93: 13760–13765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Cooper J. P., Cech T. R., 1995. Telomerase and DNA end replication: no longer a lagging strand problem? Science 269: 1533–1534 [DOI] [PubMed] [Google Scholar]
- Lingner J., Cech T. R., Hughes T. R., Lundblad V., 1997. Three ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. USA 94: 11190–11195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Louis E. J., 2003. NEJ1 prevents NHEJ-dependent telomere fusions in yeast without telomerase. Mol. Cell 11: 1373–1378 [DOI] [PubMed] [Google Scholar]
- Liu Z. P., Tye B. K., 1991. A yeast protein that binds to vertebrate telomeres and conserved yeast telomeric junctions. Genes Dev. 5: 49–59 [DOI] [PubMed] [Google Scholar]
- Livengood A. J., Zaug A. J., Cech T. R., 2002. Essential regions of Saccharomyces cerevisiae telomerase RNA: separate elements for Est1p and Est2p interaction. Mol. Cell. Biol. 22: 2366–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B., Symington L. S., 2004. The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol. Cell. Biol. 24: 9682–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev K., Vitriol E., Stemple J., Resnick M. A., Bloom K., 2004. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr. Biol. 14: 2107–2112 [DOI] [PubMed] [Google Scholar]
- Loeb L. A., 2011. Human cancers express mutator phenotypes: origin, consequences and targeting. Nat. Rev. Cancer 11: 450–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., Wilson N. M., Petracek M. E., Berman J., 1989. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAP1. Curr. Genet. 16: 225–239 [DOI] [PubMed] [Google Scholar]
- Lopes J., Piazza A., Bermejo R., Kriegsman B., Colosio A., et al. , 2011. G-quadruplex-induced instability during leading-strand replication. EMBO J. 30: 4033–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis E. J., Haber J. E., 1990. Mitotic recombination among subtelomeric Y’ repeats in Saccharomyces cerevisiae. Genetics 124: 547–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis E. J., Haber J. E., 1992. The structure and evolution of subtelomeric Y’ repeats in Saccharomyces cerevisiae. Genetics 131: 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis E. J., Naumova E. S., Lee A., Naumov G., Haber J. E., 1994. The chromosome end in yeast: its mosaic nature and influence on recombinational dynamics. Genetics 136: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B., Lingner J., 2009. TERRA: telomeric repeat-containing RNA. EMBO J. 28: 2503–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B., Panza A., Redon S., Iglesias N., Li Z., et al. , 2008. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell 32: 465–477 [DOI] [PubMed] [Google Scholar]
- Lundblad V., Blackburn E. H., 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73: 347–360 [DOI] [PubMed] [Google Scholar]
- Lundblad V., Szostak J. W., 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643 [DOI] [PubMed] [Google Scholar]
- Luo K., Vega-Palas M. A., Grunstein M., 2002. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 16: 1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig A. J., 2003. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat. Rev. Genet. 4: 916–923 [DOI] [PubMed] [Google Scholar]
- Lustig A. J., Petes T. D., 1986. Identification of yeast mutants with altered telomere structure. Proc. Natl. Acad. Sci. USA 83: 1398–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig A. J., Kurtz S., Shore D., 1990. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science 250: 549–553 [DOI] [PubMed] [Google Scholar]
- Lydeard J. R., Jain S., Yamaguchi M., Haber J. E., 2007. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448: 820–823 [DOI] [PubMed] [Google Scholar]
- Mak H. C., Pillus L., Ideker T., 2009. Dynamic reprogramming of transcription factors to and from the subtelomere. Genome Res. 19: 1014–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovets S., Blackburn E. H., 2009. DNA damage signalling prevents deleterious telomere addition at DNA breaks. Nat. Cell Biol. 11: 1383–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory J. C., Petes T. D., 2000. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc. Natl. Acad. Sci. USA 97: 13749–13754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory J. C., Bashkirov V. I., Trujillo K. M., Solinger J. A., Dominska M., et al. , 2003. Amino acid changes in Xrs2p, Dun1p, and Rfa2p that remove the preferred targets of the ATM family of protein kinases do not affect DNA repair or telomere length in Saccharomyces cerevisiae. DNA Repair (Amst.) 2: 1041–1064 [DOI] [PubMed] [Google Scholar]
- Mann R. K., Grunstein M., 1992. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J. 11: 3297–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S., Gilson E., Shore D., 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275: 986–990 [DOI] [PubMed] [Google Scholar]
- Marcand S., Brevet V., Gilson E., 1999. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 18: 3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S., Brevet V., Mann C., Gilson E., 2000. Cell cycle restriction of telomere elongation. Curr. Biol. 10: 487–490 [DOI] [PubMed] [Google Scholar]
- Marcand S., Pardo B., Gratias A., Cahun S., Callebaut I., 2008. Multiple pathways inhibit NHEJ at telomeres. Genes Dev. 22: 1153–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L., Lydall D., 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 16: 1919–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L., Lydall D., 2004a. EXO1 plays a role in generating type I and type II survivors in budding yeast. Genetics 166: 1641–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L., Lydall D., 2004b. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 18: 2663–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. G., Laroche T., Suka N., Grunstein M., Gasser S. M., 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97: 621–633 [DOI] [PubMed] [Google Scholar]
- Marvin M. E., Becker M. M., Noel P., Hardy S., Bertuch A. A., et al. , 2009a. The association of yKu with subtelomeric core X sequences prevents recombination involving telomeric sequences. Genetics 183: 453–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin M. E., Griffin C. D., Eyre D. E., Barton D. B., Louis E. J., 2009b In Saccharomyces cerevisiae, yKu and subtelomeric core X sequences repress homologous recombination near telomeres as part of the same pathway. Genetics 183: 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell P. H., Coombes C., Kenny A. E., Lawler J. F., Boeke J. D., et al. , 2004. Ty1 mobilizes subtelomeric Y’ elements in telomerase-negative Saccharomyces cerevisiae survivors. Mol. Cell. Biol. 24: 9887–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll R. M., Fangman W. L., 1988. Time of replication of yeast centromeres and telomeres. Cell 54: 505–513 [DOI] [PubMed] [Google Scholar]
- McClintock B., 1939. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc. Natl. Acad. Sci. USA 25: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern M. J., Haber J. E., 2006. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 75: 111–135 [DOI] [PubMed] [Google Scholar]
- McGee J. S., Phillips J. A., Chan A., Sabourin M., Paeschke K., et al. , 2010. Reduced Rif2 and lack of Mec1 target short telomeres for elongation rather than double-strand break repair. Nat. Struct. Mol. Biol. 17: 1438–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F. L., Hu Y., Shen N., Tong X. J., Wang J., et al. , 2009. Sua5p a single-stranded telomeric DNA-binding protein facilitates telomere replication. EMBO J. 28: 1466–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelson R. J., Rosenstein S., Weinert T., 2005. A telomeric repeat sequence adjacent to a DNA double-stranded break produces an anticheckpoint. Genes Dev. 19: 2546–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. M., Rog O., Cooper J. P., 2006. Semi-conservative DNA replication through telomeres requires Taz1. Nature 440: 824–828 [DOI] [PubMed] [Google Scholar]
- Mimitou E. P., Symington L. S., 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra K., Shore D., 1999. Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by rif proteins. Curr. Biol. 9: 1123–1126 [DOI] [PubMed] [Google Scholar]
- Mitton-Fry R. M., Anderson E. M., Hughes T. R., Lundblad V., Wuttke D. S., 2002. Conserved structure for single-stranded telomeric DNA recognition. Science 296: 145–147 [DOI] [PubMed] [Google Scholar]
- Mitton-Fry R. M., Anderson E. M., Theobald D. L., Glustrom L. W., Wuttke D. S., 2004. Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J. Mol. Biol. 338: 241–255 [DOI] [PubMed] [Google Scholar]
- Mondoux M. A., Zakian V. A., 2007. Subtelomeric elements influence but do not determine silencing levels at Saccharomyces cerevisiae telomeres. Genetics 177: 2541–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondoux M. A., Scaife J. G., Zakian V. A., 2007. Differential nuclear localization does not determine the silencing status of Saccharomyces cerevisiae telomeres. Genetics 177: 2019–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P., Shore D., 2001. Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol. Cell. Biol. 21: 8082–8094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P., Freeman K., Coodly L., Shore D., 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 8: 2257–2269 [DOI] [PubMed] [Google Scholar]
- Morris D. K., Lundblad V., 1997. Programmed translational frameshifting in a gene required for yeast telomere replication. Curr. Biol. 7: 969–976 [DOI] [PubMed] [Google Scholar]
- Morrow B. E., Johnson S. P., Warner J. R., 1989. Proteins that bind to the yeast rDNA enhancer. J. Biol. Chem. 264: 9061–9068 [PubMed] [Google Scholar]
- Morrow D. M., Tagle D. A., Shiloh Y., Collins F. S., Hieter P., 1995. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell 82: 831–840 [DOI] [PubMed] [Google Scholar]
- Mozdy A. D., Cech T. R., 2006. Low abundance of telomerase in yeast: implications for telomerase haploinsufficiency. RNA 12: 1721–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy A. D., Podell E. R., Cech T. R., 2008. Multiple yeast genes, including Paf1 complex genes, affect telomere length via telomerase RNA abundance. Mol. Cell. Biol. 28: 4152–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1938. The remaking of chromosomes. The Collecting Net 13: 181–195, 198. [Google Scholar]
- Myung K., Chen C., Kolodner R. D., 2001. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073–1076 [DOI] [PubMed] [Google Scholar]
- Na J. G., Pinto I., Hampsey M., 1992. Isolation and characterization of SUA5, a novel gene required for normal growth in Saccharomyces cerevisiae. Genetics 131: 791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D., Matsumoto K., Sugimoto K., 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 17: 1957–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan S., Mc Eachern M. J., 2002. Recombinational telomere elongation promoted by DNA circles. Mol. Cell. Biol. 22: 4512–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S., Ribaud V., Bianchi A., Shore D., 2007. DNA breaks are masked by multiple Rap1 binding in yeast: implications for telomere capping and telomerase regulation. Genes Dev. 21: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo H.-P., Lydall D., 2010. Survival and growth of yeast without telomere capping by Cdc13 in the absence of Sgs1, Exo1, and Rad9. PLoS Genet. 6: e1001072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel J. F., Wellinger R. J., 2011. Abrupt telomere losses and reduced end-resection can explain accelerated senescence of Smc5/6 mutants lacking telomerase. DNA Repair (Amst.) 10: 271–282 [DOI] [PubMed] [Google Scholar]
- Noel J. F., Larose S., Abou Elela S., Wellinger R. J., 2012. Budding yeast telomerase RNA transcription termination is dictated by the Nrd1 non-coding RNA termination pathway. Nucleic Acids Res. (in press, PMID: 22379137). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent C. I., Hughes T. R., Lue N. F., Lundblad V., 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249–252 [DOI] [PubMed] [Google Scholar]
- Nugent C. I., Bosco G., Ross L. O., Evans S. K., Salinger A. P., et al. , 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8: 657–660 [DOI] [PubMed] [Google Scholar]
- Osterhage J. L., Talley J. M., Friedman K. L., 2006. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 13: 720–728 [DOI] [PubMed] [Google Scholar]
- Paeschke K., Capra J. A., Zakian V. A., 2011. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145: 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino F., Laroche T., Gilson E., Axelrod A., Pillus L., et al. , 1993. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75: 543–555 [DOI] [PubMed] [Google Scholar]
- Pan J., Sasaki M., Kniewel R., Murakami H., Blitzblau H. G., et al. , 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144: 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenteau J., Wellinger R. J., 1999. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol. 19: 4143–4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenteau J., Wellinger R. J., 2002. Differential processing of leading- and lagging-strand ends at Saccharomyces cerevisiae telomeres revealed by the absence of Rad27p nuclease. Genetics 162: 1583–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennock E., Buckley K., Lundblad V., 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104: 387–396 [DOI] [PubMed] [Google Scholar]
- Peterson S. E., Stellwagen A. E., Diede S. J., Singer M. S., Haimberger Z. W., et al. , 2001. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 27: 64–67 [DOI] [PubMed] [Google Scholar]
- Petreaca R. C., Chiu H.-C., Eckelhoefer H. A., Chuang C., Xu L., et al. , 2006. Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat. Cell Biol. 8: 748–755 [DOI] [PubMed] [Google Scholar]
- Petreaca R. C., Chiu H. C., Nugent C. I., 2007. The role of Stn1p in Saccharomyces cerevisiae telomere capping can be separated from its interaction with Cdc13p. Genetics 177: 1459–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten J. S., Goodrich K. J., Taabazuing C., Ouenzar F., P. Chartrand et al, 2012. Mutually exclusive binding of telomerase RNA and DNA by yeast Ku: a new model for telomerase recruitment. Cell 148: 922–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett H. A., Cesare A. J., Johnston R. L., Neumann A. A., Reddel R. R., 2009. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 28: 799–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina B., Fernandez-Larrea J., Garcia-Reyero N., Idrissi F. Z., 2003. The different (sur)faces of Rap1p. Mol. Genet. Genomics 268: 791–798 [DOI] [PubMed] [Google Scholar]
- Polotnianka R. M., Li J., Lustig A. J., 1998. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 8: 831–834 [DOI] [PubMed] [Google Scholar]
- Porter S. E., Greenwell P. W., Ritchie K. B., Petes T. D., 1996. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 24: 582–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J., Blackburn E., 1997. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 11: 2790–2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preti M., Ribeyre C., Pascali C., Bosio M. C., Cortelazzi B., et al. , 2010. The telomere-binding protein Tbf1 demarcates snoRNA gene promoters in Saccharomyces cerevisiae. Mol. Cell 38: 614–620 [DOI] [PubMed] [Google Scholar]
- Pryde F. E., Louis E. J., 1999. Limitations of silencing at native yeast telomeres. EMBO J. 18: 2538–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi A., Bianchi A., Lemmens L., Damay P., Shore D., 2008. Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J. 27: 2328–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Zakian V. A., 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 14: 1777–1788 [PMC free article] [PubMed] [Google Scholar]
- Qiao F., Cech T. R., 2008. Triple-helix structure in telomerase RNA contributes to catalysis. Nat. Struct. Mol. Biol. 15: 634–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman M. K., Brewer B. J., Fangman W. L., 1994. Activation of a yeast replication origin near a double-stranded DNA break. Genes Dev. 8: 554–562 [DOI] [PubMed] [Google Scholar]
- Raghuraman M. K., Brewer B. J., Fangman W. L., 1997. Cell cycle-dependent establishment of a late replication program. Science 276: 806–809 [DOI] [PubMed] [Google Scholar]
- Raghuraman M. K., Winzeler E. A., Collingwood D., Hunt S., Wodicka L., et al. , 2001. Replication dynamics of the yeast genome. Science 294: 115–121 [DOI] [PubMed] [Google Scholar]
- Ray A., Runge K. W., 1999a. Varying the number of telomere-bound proteins does not alter telomere length in tel1Delta cells. Proc. Natl. Acad. Sci. USA 96: 15044–15049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., Runge K. W., 1999b. The yeast telomere length counting machinery is sensitive to sequences at the telomere-nontelomere junction. Mol. Cell. Biol. 19: 31–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach P., Hoss M., Azzalin C. M., Nabholz M., Bucher P., et al. , 2003. A human homolog of yeast est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr. Biol. 13: 568–574 [DOI] [PubMed] [Google Scholar]
- Renauld H., Aparicio O. M., Zierath P. D., Billington B. L., Chhablani S. K., et al. , 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7: 1133–1145 [DOI] [PubMed] [Google Scholar]
- Ribaud V., Ribeyre C., Damay P., Shore D., 2011. DNA-end capping by the budding yeast transcription factor and subtelomeric binding protein Tbf1. EMBO J. 31: 138–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes-Zamora A., Mihalek I., Lichtarge O., Bertuch A. A., 2007. Distinct faces of the Ku heterodimer mediate DNA repair and telomeric functions. Nat. Struct. Mol. Biol. 14: 301–307 [DOI] [PubMed] [Google Scholar]
- Ribeyre C., Shore D., 2012. Anticheckpoint pathways at telomeres in yeast. Nat. Struct. Mol. Biol. 19: 307–313 [DOI] [PubMed] [Google Scholar]
- Ribeyre C., Lopes J., Boule J. B., Piazza A., Guedin A., et al. , 2009. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 5: e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K. B., Petes T. D., 2000. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics 155: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K. B., Mallory J. C., Petes T. D., 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 6065–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett C. C., Straight A., Li G., Willhelm C., Sudlow G., et al. , 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135: 1685–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D., Suka Y., Xenarios I., Kurdistani S. K., Wang A., et al. , 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109: 437–446 [DOI] [PubMed] [Google Scholar]
- Rondon A. G., Jimeno S., Aguilera A., 2010. The interface between transcription and mRNP export: from THO to THSC/TREX-2. Biochim. Biophys. Acta 1799: 533–538 [DOI] [PubMed] [Google Scholar]
- Rosonina E., Kaneko S., Manley J. L., 2006. Terminating the transcript: breaking up is hard to do. Genes Dev. 20: 1050–1056 [DOI] [PubMed] [Google Scholar]
- Rossmann M. P., Luo W., Tsaponina O., Chabes A., Stillman B., 2011. A common telomeric gene silencing assay is affected by nucleotide metabolism. Mol. Cell 42: 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R., Meier B., McAinsh A. D., Feldmann H. M., Jackson S. P., 2004. Separation-of-function mutants of yeast Ku80 reveal a Yku80p-Sir4p interaction involved in telomeric silencing. J. Biol. Chem. 279: 86–94 [DOI] [PubMed] [Google Scholar]
- Rusche L. N., Kirchmaier A. L., Rine J., 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72: 481–516 [DOI] [PubMed] [Google Scholar]
- Sabourin M., Tuzon C., Zakian V., 2007. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol. Cell 27: 550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell L. L., Zakian V. A., 1993. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75: 729–739 [DOI] [PubMed] [Google Scholar]
- Sandell L. L., Gottschling D. E., Zakian V. A., 1994. Transcription of a yeast telomere alleviates telomere position effect without affecting chromosome stability. Proc. Natl. Acad. Sci. USA 91: 12061–12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H., Wang H., Adelfalk C., White E. J., Cowan C., et al. , 2007. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 104: 16934–16939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H., Ferreira H., Kalck V., Gehlen L. R., Gasser S. M., 2009. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 23: 928–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramke V., Luciano P., Brevet V., Guillot S., Corda Y., et al. , 2003. RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat. Genet. 36: 46–54 [DOI] [PubMed] [Google Scholar]
- Schulz V. P., Zakian V. A., 1994. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76: 145–155 [DOI] [PubMed] [Google Scholar]
- Seto A. G., Zaug A. J., Sobel S. G., Wolin S. L., Cech T. R., 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401: 177–180 [DOI] [PubMed] [Google Scholar]
- Seto A. G., Livengood A. J., Tzfati Y., Blackburn E. H., Cech T. R., 2002. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 16: 2800–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A., Kosiyatrakul S. T., Hockemeyer D., MacRae S. L., Karlseder J., et al. , 2009. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima H., Suzuki M., Shinohara M., 2005. Isolation and characterization of novel xrs2 mutations in Saccharomyces cerevisiae. Genetics 170: 71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D., Nasmyth K., 1987. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell 51: 721–732 [DOI] [PubMed] [Google Scholar]
- Singer M. S., Gottschling D. E., 1994. TLC1, the template RNA component of the Saccharomyces cerevisiae telomerase. Science 266: 404–409 [DOI] [PubMed] [Google Scholar]
- Smith J. J., Miller L. R., Kreisberg R., Vazquez L., Wan Y., et al. , 2011. Environment-responsive transcription factors bind subtelomeric elements and regulate gene silencing. Mol. Syst. Biol. 7: 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow B. E., Erdmann N., Cruickshank J., Goldman H., Gill R. M., et al. , 2003. Functional conservation of the telomerase protein est1p in humans. Curr. Biol. 13: 698–704 [DOI] [PubMed] [Google Scholar]
- Spellman P. T., Sherlock G., Zhang M. Q., Iyer V. R., Anders K., et al. , 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9: 3273–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan M., Mehta P., Yu Y., Prugar E., Koonin E. V., et al. , 2011. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J. 30: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavenhagen J. B., Zakian V. A., 1994. Internal tracts of telomeric DNA act as silencers in Saccharomyces cerevisiae. Genes Dev. 8: 1411–1422 [DOI] [PubMed] [Google Scholar]
- Steiner B. R., Hidaka K., Futcher B., 1996. Association of the Est1 protein with telomerase activity in yeast. Proc. Natl. Acad. Sci. USA 93: 2817–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A. E., Haimberger Z. W., Veatch J. R., Gottschling D. E., 2003. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 17: 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson J., Gottschling D., 1999. Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev. 15: 146–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Hecht A., Luo K., Grunstein M., 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11: 83–93 [DOI] [PubMed] [Google Scholar]
- Sun J., Yu E. Y., Yang Y., Confer L. A., Sun S. H., et al. , 2009. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 23: 2900–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Yang Y., Wan K., Mao N., Yu T. Y., et al. , 2011. Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase alpha. Cell Res. 21: 258–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilard R. K., Jacques P. E., Laramee L., Cheng B., Galicia S., et al. , 2010. Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nat. Struct. Mol. Biol. 17: 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Hediger F., Neumann F. R., Bauer C., Gasser S. M., 2004. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 23: 1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Van Houwe G., Nagai S., Erb I., van Nimwegen E., et al. , 2009. The functional importance of telomere clustering: global changes in gene expression result from SIR factor dispersion. Genome Res. 19: 611–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart A. K., Teng S. C., Zakian V. A., 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297: 1023–1026 [DOI] [PubMed] [Google Scholar]
- Takahashi Y. H., Schulze J. M., Jackson J., Hentrich T., Seidel C., et al. , 2011. Dot1 and histone H3K79 methylation in natural telomeric and HM silencing. Mol. Cell 42: 118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talley J. M., DeZwaan D. C., Maness L. D., Freeman B. C., Friedman K. L., 2011. Stimulation of yeast telomerase activity by the ever shorter telomere 3 (est3) subunit is dependent on direct interaction with the catalytic protein est2. J. Biol. Chem. 286: 26431–26439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M. T., Arneric M., Sperisen P., Lingner J., 2004. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- Teng S. C., Zakian V. A., 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 8083–8093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S. C., Chang J., McCowan B., Zakian V. A., 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6: 947–952 [DOI] [PubMed] [Google Scholar]
- Tham W. H., Wyithe J. S., Ferrigno P. K., Silver P. A., Zakian V. A., 2001. Localization of yeast telomeres to the nuclear periphery is separable from transcriptional repression and telomere stability functions. Mol. Cell 8: 189–199 [DOI] [PubMed] [Google Scholar]
- Therizols P., Fairhead C., Cabal G. G., Genovesio A., Olivo-Marin J. C., et al. , 2006. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J. Cell Biol. 172: 189–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P., Duong T., Dujon B., Zimmer C., Fabre E., 2010. Chromosome arm length and nuclear constraints determine the dynamic relationship of yeast subtelomeres. Proc. Natl. Acad. Sci. USA 107: 2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. S., Ling X., Grunstein M., 1994. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature 369: 245–247 [DOI] [PubMed] [Google Scholar]
- Toogun O. A., Dezwaan D. C., Freeman B. C., 2008. The hsp90 molecular chaperone modulates multiple telomerase activities. Mol. Cell. Biol. 28: 457–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J. Z., Bessler J. B., Zakian V. A., 2004. Local chromatin structure at the ribosomal DNA causes replication fork pausing and genome instability in the absence of the S. cerevisiae DNA helicase Rrm3p. Genes Dev. 18: 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelles-Sticken E., Adelfalk C., Loidl J., Scherthan H., 2005. Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. J. Cell Biol. 170: 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng S. F., Lin J. J., Teng S. C., 2006. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 34: 6327–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y., Kato J., Ikeda H., 1997. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature 388: 900–903 [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y., Taggart A. K., Zakian V. A., 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase- mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11: 1328–1335 [DOI] [PubMed] [Google Scholar]
- Tuzon C. T., Wu Y., Chan A., Zakian V. A., 2011. The Saccharomyces cerevisiae telomerase subunit Est3 binds telomeres in a cell cycle- and Est1-dependent manner and interacts directly with Est1 in vitro. PLoS Genet. 7: e1002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Palas M. A., Venditti S., Di Mauro E., 1997. Telomeric transcriptional silencing in a natural context. Nat. Genet. 15: 232–233 [DOI] [PubMed] [Google Scholar]
- Vega L. R., Phillips J. A., Thornton B. R., Benanti J. A., Onigbanjo M. T., et al. , 2007. Sensitivity of yeast strains with long G-tails to levels of telomere-bound telomerase. PLoS Genet. 3: e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virta-Pearlman V., Morris D. K., Lundblad V., 1996. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 10: 3094–3104 [DOI] [PubMed] [Google Scholar]
- Vodenicharov M. D., Wellinger R. J., 2006. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol. Cell 24: 127–137 [DOI] [PubMed] [Google Scholar]
- Vodenicharov M. D., Wellinger R. J., 2007. The cell division cycle puts up with unprotected telomeres: cell cycle regulated telomere uncapping as a means to achieve telomere homeostasis. Cell Cycle 6: 1161–1167 [DOI] [PubMed] [Google Scholar]
- Vodenicharov M. D., Wellinger R. J., 2010. Telomere capping in non-dividing yeast cells requires Yku and Rap1. EMBO J. 29: 3007–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walmsley R. W., Chan C. S., Tye B. K., Petes T. D., 1984. Unusual DNA sequences associated with the ends of yeast chromosomes. Nature 310: 157–160 [DOI] [PubMed] [Google Scholar]
- Wanat J. J., Kim K. P., Koszul R., Zanders S., Weiner B., et al. , 2008. Csm4, in collaboration with Ndj1, mediates telomere-led chromosome dynamics and recombination during yeast meiosis. PLoS Genet. 4: e1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Podell E. R., Zaug A. J., Yang Y., Baciu P., et al. , 2007. The POT1–TPP1 telomere complex is a telomerase processivity factor. Nature 445: 506–510 [DOI] [PubMed] [Google Scholar]
- Wang S. S., Zakian V. A., 1990. Sequencing of Saccharomyces telomeres cloned using T4 DNA polymerase reveals two domains. Mol. Cell. Biol. 10: 4415–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J. D., 1972. Origin of concatemeric T7 DNA. Nat. New Biol. 239: 197–201 [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H., 1993. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics 134: 63–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger R. J., Wolf A. J., Zakian V. A., 1993a. Origin activation and formation of single-strand TG1–3 tails occur sequentially in late S phase on a yeast linear plasmid. Mol. Cell. Biol. 13: 4057–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger R. J., Wolf A. J., Zakian V. A., 1993b. Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell 72: 51–60 [DOI] [PubMed] [Google Scholar]
- Wellinger R. J., Ethier K., Labrecque P., Zakian V. A., 1996. Evidence for a new step in telomere maintenance. Cell 85: 423–433 [DOI] [PubMed] [Google Scholar]
- Wotton D., Shore D., 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11: 748–760 [DOI] [PubMed] [Google Scholar]
- Wright J. H., Gottschling D. E., Zakian V. A., 1992. Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev. 6: 197–210 [DOI] [PubMed] [Google Scholar]
- Wright J. H., Zakian V. A., 1995. Protein-DNA interactions in soluble telosomes from Saccharomyces cerevisiae. Nucleic Acids Res. 23: 1454–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zakian V., 2011. Cdc13 interacts directly with telomerase recruiter-activator Est1 to bring it to telomeric DNA ends in vitro. Proc. Natl. Acad. Sci. USA 108: 20362–20369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick J. J., Holstege F. C., Jennings E. G., Causton H. C., Shore D., et al. , 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402: 418–421 [DOI] [PubMed] [Google Scholar]
- Xie W., Gai X., Zhu Y., Zappulla D. C., Sternglanz R., et al. , 2001. Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol. Cell. Biol. 21: 6606–6614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H., Liu D., Wan M., Safari A., Kim H., et al. , 2007. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature 445: 559–562 [DOI] [PubMed] [Google Scholar]
- Xu L., Petreaca R. C., Gasparyan H. J., Vu S., Nugent C. I., 2009. TEN1 is essential for CDC13-mediated telomere capping. Genetics 183: 793–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Hayatsu N., Matsuura A., Ishikawa F., 1998. Y’-Help1, a DNA helicase encoded by the yeast subtelomeric Y’ element, is induced in survivors defective for telomerase. J. Biol. Chem. 273: 33360–33366 [DOI] [PubMed] [Google Scholar]
- Yu E. Y., Wang F., Lei M., Lue N. F., 2008. A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat. Struct. Mol. Biol. 15: 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakian V. A., Blanton H. M., Wetzel L., 1986. Distribution of telomere-associated sequences in yeast. Basic Life Sci. 40: 493–498 [DOI] [PubMed] [Google Scholar]
- Zanders S., Brown M. S., Chen C., Alani E., 2011. Pch2 modulates chromatid partner choice during meiotic double-strand break repair in Saccharomyces cerevisiae. Genetics 188: 511–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla D. C., Cech T. R., 2004. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc. Natl. Acad. Sci. USA 101: 10024–10029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla D. C., Goodrich K., Cech T. R., 2005. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat. Struct. Mol. Biol. 12: 1072–1077 [DOI] [PubMed] [Google Scholar]
- Zappulla D. C., Roberts J. N., Goodrich K. J., Cech T. R., Wuttke D. S., 2009. Inhibition of yeast telomerase action by the telomeric ssDNA-binding protein, Cdc13p. Nucleic Acids Res. 37: 354–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Durocher D., 2010. De novo telomere formation is suppressed by the Mec1-dependent inhibition of Cdc13 accumulation at DNA breaks. Genes Dev. 24: 502–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Blobel G., 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 102: 4777–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Muller E. G., Rothstein R., 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2: 329–340 [DOI] [PubMed] [Google Scholar]
- Zhou J.-Q., Monson E. M., Teng S.-C., Schulz V. P., Zakian V. A., 2000. The Pif1p helicase, a catalytic inhibitor of telomerase lengthening of yeast telomeres. Science 289: 771–774 [DOI] [PubMed] [Google Scholar]
- Zhu X., Gustafsson C. M., 2009. Distinct differences in chromatin structure at subtelomeric X and Y’ elements in budding yeast. PLoS ONE 4: e6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G., 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C., Diffley J. F., 2008. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 27: 1875–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Elledge S. J., 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
- Zou S., Wright D. A., Voytas D. F., 1995. The Saccharomyces Ty5 retrotransposon family is associated with origins of DNA replication at the telomeres and the silent mating locus HMR. Proc. Natl. Acad. Sci. USA 92: 920–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko M. K., Lydall D., 2006. Linear chromosome maintenance in the absence of essential telomere-capping proteins. Nat. Cell Biol. 8: 734–740 [DOI] [PubMed] [Google Scholar]
- Zubko M. K., Guillard S., Lydall D., 2004. Exo1 and Rad24 differentially regulate generation of ssDNA at telomeres of Saccharomyces cerevisiae cdc13–1 mutants. Genetics 168: 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]