Abstract
The Rtf1 subunit of the Paf1 complex is required for specific histone modifications, including histone H2B lysine 123 monoubiquitylation. In Saccharomyces cerevisiae, deletion of RTF1 is lethal in the absence of Rkr1, a ubiquitin-protein ligase involved in the destruction of nonstop proteins, which arise from mRNAs lacking stop codons or translational readthrough into the poly(A) tail. We performed a transposon-based mutagenesis screen to identify suppressors of rtf1Δ rkr1Δ lethality and found that a mutation in the gene encoding the protein chaperone Hsp104 rescued viability. Hsp104 plays a role in prion propagation, including the maintenance of [PSI+], which contributes to the synthesis of nonstop proteins. We demonstrate that rtf1Δ and rkr1Δ are synthetically lethal only in the presence of [PSI+]. The deletion, inactivation, and overexpression of HSP104 or the overexpression of prion-encoding genes URE2 and LSM4 clear [PSI+] and rescue rtf1Δ rkr1Δ lethality. In addition, the presence of [PSI+] decreases the fitness of rkr1Δ strains. We investigated whether the loss of RTF1 exacerbates an overload in nonstop proteins in rkr1Δ [PSI+] strains but, using reporter plasmids, found that rtf1Δ decreases nonstop protein levels, indicating that excess nonstop proteins may not be the cause of synthetic lethality. Instead, our data suggest that the loss of Rtf1-dependent histone modifications increases the burden on quality control pathways in cells lacking Rkr1 and containing [PSI+].
Keywords: [PSI+], prions, RKR1, RTF1, nonstop proteins
DURING transcription elongation, various proteins modify chromatin in coordination with RNA polymerase II (Pol II) to ensure accurate and efficient transcription of nucleosomal templates (Li et al. 2007). Changes in chromatin include nucleosome remodeling, the exchange of histone variants for canonical histones, and histone modifications such as the methylation, ubiquitylation, and acetylation of lysine (K) residues. The conserved Paf1 complex (Paf1C), which consists of Paf1, Ctr9, Leo1, Cdc73, and Rtf1, associates with Pol II on all actively transcribed genes (Mayer et al. 2010) and couples the modification of histones to transcription elongation (reviewed in Crisucci and Arndt 2011; Jaehning 2010). Paf1C is required for multiple histone modifications associated with active genes, including the monoubiquitylation of H2B K123, a modification for which the Rtf1 subunit of Paf1C plays a prominent role (Ng et al. 2003; Wood et al. 2003; Warner et al. 2007; Tomson et al. 2011). Rad6 is the ubiquitin-conjugating enzyme (E2) for H2B K123 ubiquitylation, while Bre1 is the ubiquitin-protein ligase (E3) (Hwang et al. 2003). This modification is a prerequisite for downstream histone H3 methylation (Dover et al. 2002; Sun and Allis 2002; Ng et al. 2003; Wood et al. 2003). In yeast, loss of H2B K123 ubiquitylation broadly impacts gene expression and chromatin structure (Mutiu et al. 2007; Batta et al. 2011). In humans, errors in Paf1C-dependent histone modifications can lead to aberrant gene expression and tumorigenesis (reviewed in Crisucci and Arndt 2011).
Paf1C has several functions in addition to promoting specific histone modifications, including directing the proper 3′-end formation of transcripts (Mueller et al. 2004; Penheiter et al. 2005; Sheldon et al. 2005; Nordick et al. 2008; Nagaike et al. 2011). Depletion of human Paf1C (hPaf1C) subunits impairs mRNA cleavage, polyadenylation, and export to the cytoplasm (Nagaike et al. 2011). Additionally, loss of hCdc73 results in aberrantly processed and polyadenylated histone mRNAs (Farber et al. 2010). In yeast, deletion of Paf1C subunits leads to decreased poly(A) tail length and alternative poly(A) site usage (Mueller et al. 2004; Strawn et al. 2009). These observations indicate that Paf1C is essential for both the proper expression and the processing of a subset of RNAs and that loss of Paf1C can result in aberrant transcripts, which are inefficiently exported or translated. In support of this idea, erroneous transcripts resulting from loss of Paf1C are substrates for mRNA quality control pathways, including nonsense mRNA decay (Penheiter et al. 2005; reviewed in Jaehning 2010 and Crisucci and Arndt 2011).
RKR1, which is required for viability in Saccharomyces cerevisiae strains lacking the Paf1C subunit Rtf1, encodes a conserved RING finger-containing ubiquitin-protein ligase (Braun et al. 2007). Deletion of RKR1 also causes severe growth defects in strains lacking PAF1 or CTR9 (Braun et al. 2007). Given that rkr1Δ causes severe synthetic growth defects in strains with an htb1-K123R mutation, Rkr1 most likely functions in a pathway parallel to the histone modification functions of Paf1C to promote an important cellular process (Braun et al. 2007). Interestingly, Rkr1 is required for the proper ubiquitylation and degradation of nonstop proteins in yeast and physically associates with ribosomes (Fleischer et al. 2006; Wilson et al. 2007; Bengtson and Joazeiro 2010). Nonstop proteins can result from mRNAs lacking stop codons (nonstop mRNAs), and both the mRNAs that encode these nonstop proteins and the resulting nonstop proteins themselves are targeted for degradation (van Hoof et al. 2002; Wilson et al. 2007; Bengtson and Joazeiro 2010), thus implicating Rkr1 in a protein quality control pathway (Bengtson and Joazeiro 2010). Importantly, mutations in RKR1 homologs in higher eukaryotes are associated with neurodegeneration and colon cancer (Ivanov et al. 2007; Chu et al. 2009).
To investigate the relationship between the transcription factor Rtf1 and the protein quality control factor Rkr1, we performed a transposon-based mutagenesis screen to identify suppressors of rtf1Δ rkr1Δ synthetic lethality in S. cerevisiae. We found that mutations in the gene encoding the Hsp104 chaperone rescue lethality of an rtf1Δ rkr1Δ strain. Enhanced or depleted levels of Hsp104 alter [PSI+] prion propagation (Chernoff et al. 1995), suggesting a role for this prion in rtf1Δ rkr1Δ synthetic lethality. Consistent with this idea, we found that conditions for curing [PSI+] restore viability of rtf1Δ rkr1Δ strains and that transfer of [PSI+] to rtf1Δ rkr1Δ [psi−] cells causes lethality. Additionally, we found that the presence of [PSI+], presumably through increased nonstop proteins, negatively influences the fitness of rkr1Δ strains even in the presence of Rtf1. Unexpectedly, the absence of RTF1 in a rkr1Δ background causes a decrease in the levels of nonstop reporter proteins. Our results suggest that Rtf1 and its H2B ubiquitylation function protect cells against the combined deleterious effects of [PSI+] and defects in Rkr1-mediated protein quality control.
Materials and Methods
Yeast strains and standard growth conditions
KY S. cerevisiae strains are isogenic with FY2, a GAL2+ derivative of S288C and are listed in the Supporting Information, Table S1 (Winston et al. 1995). Yeast deletion mutants, crosses, and transformants were created using standard protocols (Ausubel et al. 1988; Rose et al. 1991). Yeast were grown on rich (YPD), synthetic complete (SC), synthetic minimal (SD), 5-fluoroorotic acid (5-FOA), or sporulation media as specified and prepared as previously described (Rose et al. 1991). Strains were typically cured of prions by streaking for single colonies onto YPD supplemented with 5 mM guanidine hydrochloride. For creating [prion−] strains from diploids, tetrad dissections were performed on YPD containing 2.5 mM guanidine hydrochloride.
Plasmids
The his3 nonstop plasmid, pAV240 (LEU2-marked), and protein A nonstop plasmid, pAV184 (URA3-marked), were gifts from Ambro van Hoof (Wilson et al. 2007). The sup35NM-GFP (URA3- or LEU2-marked) and RNQ1-GFP plasmids were gifts from Susan Liebman (Zhou et al. 2001). The URA3-marked plasmid carrying RTF1, pKA69, was used to maintain rtf1Δ rkr1Δ viability (Stolinski et al. 1997). HSP104 was driven by a GPD promoter on a 2µ pRS424 (TRP1) plasmid (Mumberg et al. 1995; Rubel et al. 2008). This plasmid was used to derive plasmids for overexpression of URE2, LSM4, LSM2, and RNQ1. The open reading frames and 3′ UTR sequences of URE2, LSM4, LSM2, and RNQ1 were amplified by PCR from a plasmid source and inserted in place of HSP104 using the SacI and BamHI sites (Nagalakshmi et al. 2008). The ade1-14 and sup35-Y351C alleles (Bradley et al. 2003) were amplified from strains provided by Susan Liebman and cloned using XmaI and SacI sites into pRS306 for two-step gene replacement of the ADE1 or SUP35 gene, respectively (Scherer and Davis 1979).
Mutagenesis and confirmation of genetic suppressors
Transposon (Tn) mutagenesis to identify suppressors of rtf1Δ rkr1Δ synthetic lethality was performed by transforming a LEU2-marked set of integrating plasmids (described in Kumar et al. 2000) into an rtf1Δ rkr1Δ strain (KY1663) carrying an RTF1/URA3/CEN/ARS plasmid (pKA69) and selecting on SC-leucine (L) medium. Transformants were replica-plated onto SC-L containing 5-FOA to select for colonies that had lost the RTF1 plasmid. Fifteen thousand colonies were screened and 55 candidates were purified and analyzed further. Stable integration of the transposon was verified by streaking strains onto YPD and replica-plating onto SC-L and 5-FOA. Thirty-nine candidates passing this test were selected from 5-FOA plates. These strains, rtf1Δ rkr1Δ TnSup::LEU2, were then used in backcrosses with an rtf1Δ strain containing pKA69 because a functional copy of RTF1 is required for sporulation (data not shown). Tetrad analysis of these crosses confirmed that only one Tn was present per candidate by 2:2 sorting of LEU2. This cross also confirmed that every rtf1Δ rkr1Δ Leu− spore was 5-FOAS while every rtf1Δ rkr1Δ Leu+ spore was 5-FOAR, verifying linkage of the Tn to suppression of lethality. Fifteen rkr1Δ TnSup::LEU2 strains from these crosses were taken through a second backcross with an rtf1Δ strain to verify that the transposon rescued rtf1Δ rkr1Δ lethality independently of pKA69 and 5-FOA. In this case, we expected suppression of lethality only in rtf1Δ rkr1Δ TnSup::LEU2 strains. Three Tn mutants passed these genetic criteria, one of which is described in this article. The Tn insertion was recovered as previously described by rescuing the insertion in yeast with linearized pRSQ2-URA3, and the plasmid insert was sequenced with an M13 oligonucleotide primer (Burns et al. 1994). In the described candidate, the Tn mapped within HSP104, 68 bp from the 3′-end of the open reading frame. A mutation in HSP104 was verified to suppress rtf1Δ rkr1Δ lethality by creating a precise KanMX replacement of HSP104 and performing tetrad analysis of a triple heterozygous diploid strain (rtf1Δ/RTF1rkr1Δ/RKR1hsp104Δ/HSP104).
High-copy-number suppressor screen
To obtain high-copy-number suppressors of rtf1Δ rkr1Δ synthetic lethality, a 2μ LEU2-marked plasmid library of genomic fragments (Yoshihisa and Anraku 1989; Rubel et al. 2008) was transformed into an rtf1Δ rkr1Δ strain (KY2205) carrying pKA69. Approximately 13,500 Leu+ colonies were screened for the ability to grow on 5-FOA medium, indicating loss of pKA69. One hundred and thirty-one candidates were verified by testing on 5-FOA medium, and plasmids were isolated by standard extraction methods (Hoffman and Winston 1987). Plasmids were then retransformed into KY2205, and 48 exhibited the suppression phenotype. Of these, 21 contained either RTF1 or RKR1. Of the remaining candidates, six unique plasmids remained. One plasmid, which was isolated seven times, contained the gene LSM4. Another plasmid, which was isolated three times, contained multiple open reading frames, including URE2. A third plasmid, obtained twice, also contained multiple open reading frames, including HSP104.
Yeast dilution growth assays
Unless stated otherwise, yeast strains were grown to saturation at 30° in rich or selective media, washed with sterile water, and diluted into 1 × 108 cells/ml stocks from which 10-fold dilutions were made. Two microliters of cell suspension were plated on appropriate control and selective media, and plates were incubated at 30° for the specified number of days. Media for testing rtf1Δ and rkr1Δ phenotypes contained 0.8 µg/ml cycloheximide, 50 µM cadmium chloride, 10% ethanol, or 15 mM caffeine in YPD or SC as indicated.
Cytoduction
An rtf1Δ rkr1Δ ade1-14 [psi−] strain (KY2286), created by dissection onto YPD containing guanidine hydrochloride (GuHCl), was transformed with RTF1/URA3 (pKA69) and depleted of mitochondrial DNA (ρ0 conditions) by growth in liquid culture with ethidium bromide. Using previously described methods (Wickner et al. 2006), this recipient strain was used for cytoduction with two kar1 donor strains, L2261 ([PIN+] [psi−]) and L2265 ([PSI+] [pin−]) (Mathur et al. 2009). Transfer of cytoplasm to the recipient strain was confirmed by growth on YP medium containing 3% glycerol (YPG), and transfer of the donor prion was confirmed by live-cell confocal microscopy of plasmid-encoded GFP-tagged prion domains. The kar1 donor strains and GFP plasmids were gifts from Susan Liebman.
Live-cell confocal microscopy
Strains were transformed with sup35NM-GFP plasmid (Zhou et al. 2001) to test for the presence of [PSI+] or a RNQ1-GFP plasmid to test for the presence of [PIN+] and patched onto selective media containing 100 µM CuSO4. Plates were protected from light and incubated at 30° for several days. Live-cell imaging was performed on wet mounts using a Leica TCS SP5 confocal microscope (Leica Microsystems, Buffalo Grove, IL).
Immunofluorescence
A rkr1Δ strain (KY2289) was transformed with either an HA-RKR1 (pMB11) or an untagged RKR1 plasmid (pPC65) (Braun et al. 2007). These strains were grown to midlog phase and prepared as previously described (Amberg et al. 2006). Briefly, cells were fixed with formaldehyde, treated with zymolyase 20T, and adhered to a polylysine slide before overnight incubation with 1:500 anti-HA (Roche) and a 1-hr incubation with 1:250 Alexa 647 (Molecular Probes). Slides were mounted with ProLong GOLD Antifade DAPI reagent (Invitrogen) and imaged using a Leica TCS SP5 confocal microscope (Leica Microsystems, Buffalo Grove, IL).
Western analysis of nonstop protein levels
Strains transformed with pAV184 (Wilson et al. 2007) were grown at 30° in SC-uracil (U) liquid culture containing 2% galactose to an OD600 of 0.7–0.9. Cells were normalized to 10.8 OD600 units, and extracts were made using glass-bead lysis in 20% trichloroacetic acid as previously described (Cox et al. 1997; Zheng et al. 2010). An equal amount of each extract (5 µl) was run on a 15% SDS-polyacrylamide gel and transferred to nitrocellulose membrane for Western analysis using standard methods (Harlow and Lane 1988). The membrane was probed with peroxidase–anti-peroxidase (1:2000 dilution; Sigma) to assay levels of protein A and anti-G6PDH antibody (1:50,000 dilution; Sigma) as a loading control. Immunoreactivity was measured using chemiluminescence (Perkin-Elmer) and a 440 CF digital imaging station (Kodak).
Results
Genetic suppressors of rtf1Δ rkr1Δ synthetic lethality
To investigate the basis for the lethality of strains lacking Rtf1 and the ubiquitin-protein ligase Rkr1, we performed a Tn-based mutagenesis screen for suppressors of the rtf1Δ rkr1Δ synthetic lethal interaction. A library of plasmids containing yeast genomic DNA and LEU2-marked transposon insertions (Kumar et al. 2000) was transformed into an rtf1Δ rkr1Δ strain, which carried a URA3-marked RTF1 plasmid for viability. Approximately 15,000 transformants were screened for loss of the URA3-marked RTF1 plasmid on medium containing 5-FOA. Following phenotypic confirmation, rtf1Δ rkr1Δ TnSup::LEU2 candidates were crossed to an rtf1Δ strain containing a URA3-marked RTF1 plasmid to verify the 2:2 sorting of the LEU2-marked transposon, indicating only one insertion site, as well as linkage of the 5-FOA resistance to the LEU2 marker in rtf1Δ rkr1Δ colonies. Fifteen strains met these requirements, and rkr1Δ TnSup::LEU2 strains from those crosses were backcrossed to an rtf1Δ strain to verify that the Tn insertion rescued lethality of the rtf1Δ rkr1Δ double mutants in the absence of the RTF1-containing plasmid. One candidate that met these criteria is described here.
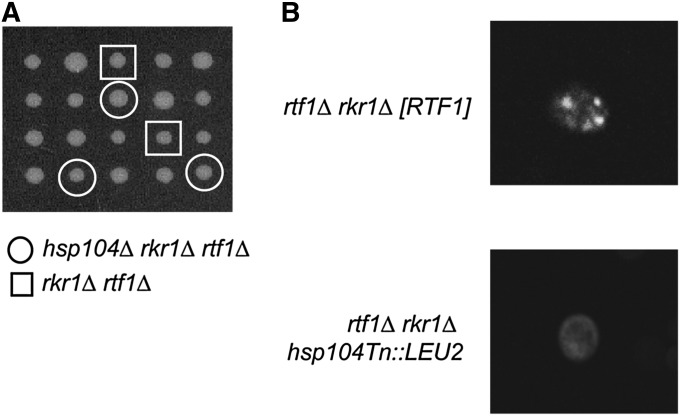
Following the plasmid rescue of the transposon insertion (Burns et al. 1994), DNA sequencing revealed a transposon insertion in the 3′-coding region of HSP104. Hsp104 encodes a heat-shock protein that can disrupt aggregated proteins (Parsell et al. 1994) and is involved in the maintenance and propagation of yeast prions (reviewed in Grimminger-Marquardt and Lashuel 2010). To confirm that the Tn mutation rescued rtf1Δ rkr1Δ synthetic lethality by disrupting Hsp104 function, we generated an hsp104Δ strain and crossed it to a rkr1Δ strain. Double mutants from this cross were then mated with an rtf1Δ strain. The diploids, which are heterozygous for three genes (rtf1Δ, rkr1Δ, and hsp104Δ), were subjected to tetrad analysis. Surprisingly, the rtf1Δ rkr1Δ double mutant segregants were alive and healthy, indicating dominant suppression by hsp104Δ (Figure 1A). In addition, the hsp104Δ rtf1Δ rkr1Δ triple mutants were viable, independently confirming the identification of an hsp104 mutation in our suppressor screen (Figure 1A).
Figure 1 .
Mutation of HSP104 suppresses rtf1Δ rkr1Δ synthetic lethality and cures [PSI+]. (A) Tetrad dissections of crosses between an rtf1Δ strain (KY958) and a rkr1Δ hsp104Δ strain. Dissections were done on YPD and incubated at 30° for 3 days. Double mutant rtf1Δ rkr1Δ segregants are highlighted by boxes and triple mutants are highlighted by circles. Note that suppression of rtf1Δ rkr1Δ synthetic lethality occurs with or without hsp104Δ cosegregation, indicating that hsp104Δ acts as a dominant suppressor in the diploid. (B) The strain originally used for transposon-mediated mutagenesis (KY1663) and the hsp104Tn::LEU2 mutant recovered from the transposon-based suppressor screen were transformed with a LEU2-marked or a URA3-marked pCUP1-SUP35NM-GFP plasmid. Strains were patched onto SC-L or SC-U plates containing 100 µM CuSO4 and incubated in the dark at 30° before live-cell imaging was performed by confocal microscopy. Observations were made of at least three transformants per strain and 100 cells per transformant. Representative images are shown. No variability was seen among cells with respect to the GFP pattern.
Transposon insertion in HSP104 cures cells of [PSI+]
Deletions in the C-terminal domain of Hsp104 have been shown to weaken its ATPase activity and ability to propagate prions, particularly [PSI+], an aggregate of the translation termination factor Sup35 (Chernoff et al. 1995; Mackay et al. 2008). Because the transposon insertion disrupted the C-terminal domain of Hsp104, we investigated if the rtf1Δ rkr1Δ strain used in the suppressor screen was [PSI+] and whether the hsp104Tn::LEU2 suppressor mutation cleared [PSI+] from this strain. A plasmid expressing the GFP-tagged prion domain of Sup35 was transformed into the original rtf1Δ rkr1Δ [RTF1, URA3, CEN/ARS] strain used in our transposon mutagenesis screen, and transformants were visualized by live-cell imaging using a confocal microscope (Zhou et al. 2001). Previous studies have shown that this GFP-tagged Sup35 protein appears as small fluorescent puncta in [PSI+] cells and as diffuse fluorescence in [psi−] cells (Zhou et al. 2001). Using this method, we found that the original strain used in our screen was indeed [PSI+] and that a transposon insertion within HSP104 resulted in [psi−] conditions in all cells examined (Figure 1B). Given the importance of Hsp104 in prion propagation, we next investigated the role of prions in the genetic interaction between RTF1 and RKR1.
RKR1 genetic interactions are rescued by curing strains of [PSI+]
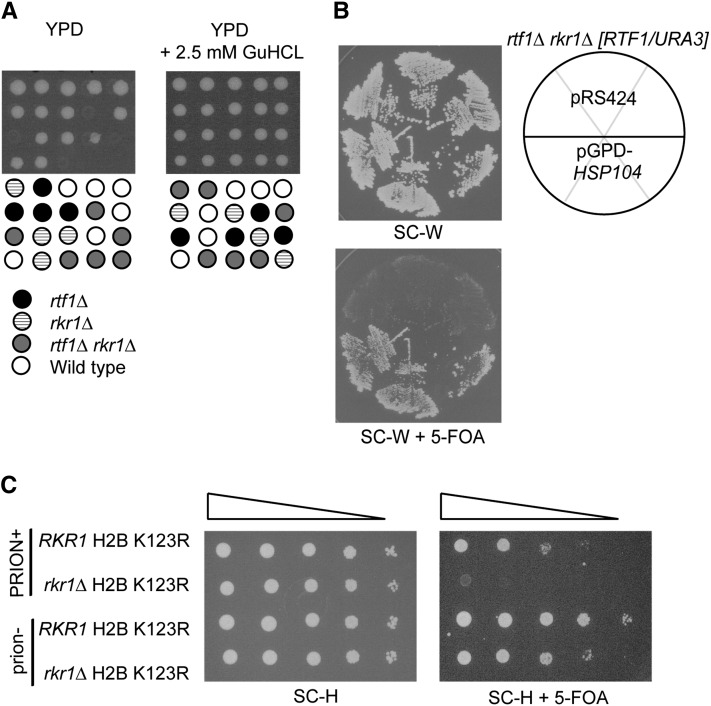
As noted above, HSP104 is required for the propagation of yeast prions (Chernoff et al. 1995; Shorter and Lindquist 2006). Therefore, the loss of prions in an hsp104Δ/HSP104 heterozygous diploid strain likely rescued lethality between rtf1Δ and rkr1Δ, independently of whether the hsp104Δ mutation actually segregated with the rtf1Δ and rkr1Δ mutations (Figure 1A). Growth on media containing GuHCl has been shown to cure yeast of prions by inactivating the ATPase domain of Hsp104 (Ferreira et al. 2001; Jung and Masison 2001). To determine if clearing prions through this method could also rescue rtf1Δ rkr1Δ lethality, an rtf1Δ/RTF1rkr1Δ/RKR1 heterozygous diploid was sporulated and tetrads were dissected onto YPD or YPD containing 2.5 mM GuHCl. Strikingly, tetrad analysis revealed that rtf1Δ rkr1Δ double mutants grew as well as wild-type strains on YPD containing GuHCl but were inviable on YPD alone (Figure 2A). Therefore, inactivation of HSP104 by mutation or treatment with GuHCl rescues rtf1Δ rkr1Δ synthetic lethality.
Figure 2 .
Inactivation or overexpression of HSP104 rescues rkr1Δ synthetic genetic interactions. (A) Heterozygous rtf1Δ/RTF1 rkr1Δ/RKR1 diploids (KY2202 mated by KY453) were dissected onto YPD or YPD containing 2.5 mM GuHCl and incubated at 30° for 3 days. (B) An rtf1Δ rkr1Δ [RTF1/URA3] strain (KY1663) was transformed with either a 2µ TRP1-marked pGPD-HSP104 plasmid or a TRP1-marked empty vector (pRS424), and transformants were purified on SC-W and replica-plated to SC-W + 5-FOA (W = tryptophan). Plates were incubated at 30° for 3 days. (C) Wild-type (KY2203) or rkr1Δ (KY2204) strains, lacking both endogenous histone H2A and H2B gene copies and containing a URA3-marked wild-type copy of HTA1-HTB1, were transformed with a HIS3-marked HTA1-htb1 K123R plasmid. Strains were cured by streaking onto YPD + 5 mM GuHCl. Dilution growth assays were performed on SC-H or SC-H + 5-FOA, and cells were incubated at 30° for 2 days (H = histidine). “PRION+” indicates uncured cells; “prion−” indicates cells passaged on medium containing GuHCl.
HSP104 is required for the propagation of several yeast prions, including [PSI+], [URE3], and [PIN+] (reviewed in Haslberger et al. 2010). Similar to a loss of HSP104 function, overexpression of HSP104 can also alleviate yeast of [PSI+], but it has not been shown to affect the propagation of other yeast prions (Chernoff et al. 1995; Shorter and Lindquist 2006). Therefore, to investigate if rtf1Δ rkr1Δ lethality might be rescued by loss of [PSI+] or of yeast prions in general, we overexpressed HSP104 to test for rescue of synthetic lethality. An rtf1Δ rkr1Δ strain, which carried an RTF1URA3-marked plasmid to allow growth, was transformed with a TRP1-marked HSP104 overexpression plasmid. Transformants were grown under selective conditions and replica-plated to medium containing 5-FOA to select for loss of the RTF1 plasmid. Interestingly, the HSP104 high-copy plasmid allowed for growth of the rtf1Δ rkr1Δ double mutants (Figure 2B). Taken together, our results indicate that deletion, inactivation, or overexpression of HSP104 suppresses rtf1Δ rkr1Δ synthetic lethality by clearing [PSI+].
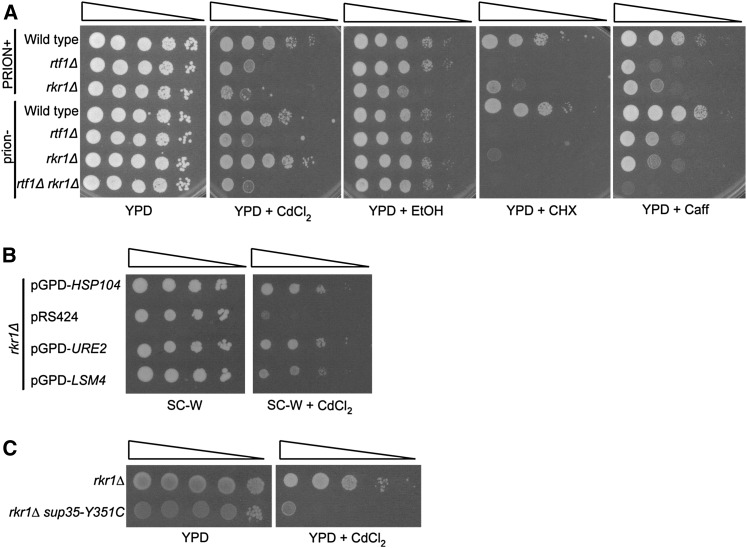
A defect in H2B K123 ubiquitylation can phenocopy an rtf1Δ mutation with respect to rkr1Δ synthetic growth defects (Braun et al. 2007). We therefore investigated if the inactivation of Hsp104 could also rescue the genetic interaction between rkr1Δ and htb1-K123R, a derivative of H2B that lacks the ubiquitylation site for Rad6-Bre1. To answer this question, we performed a plasmid shuffle experiment with RKR1 and rkr1Δ strains, which carried a URA3-marked HTA1-HTB1 plasmid and a HIS3-marked HTA1-htb1K123R plasmid and were deleted for the chromosomal H2A and H2B genes. Serial dilution analysis was conducted on 5-FOA medium to select for cells that had lost the URA3-marked wild-type HTA1-HTB1 plasmid and retained the HIS3-marked HTA1-htb1K123R plasmid. As previously shown, the rkr1Δ mutation causes a strong synthetic growth defect in combination with the H2B K123R substitution by this assay (Figure 2C) (Braun et al. 2007). However, if these strains were first cured of prions by passaging on medium containing 5 mM GuHCl prior to plating on 5-FOA medium, rkr1Δ htb1K123R strains were viable (Figure 2C).
Overexpression of the prion-coding genes URE2 and LSM4 rescues rtf1Δ rkr1Δ lethality
Over 20 potential or verified prions have been identified in budding yeast, with [PSI+] being one of the best characterized (Alberti et al. 2009). Interactions between different prions can be both positive and negative. The de novo formation and propagation of some prions require the presence of other prions (Derkatch et al. 2001), while the maintenance of some prions may be negatively impacted by the presence of other prions, possibly by affecting chaperone activity (reviewed in Crow and Li 2011).
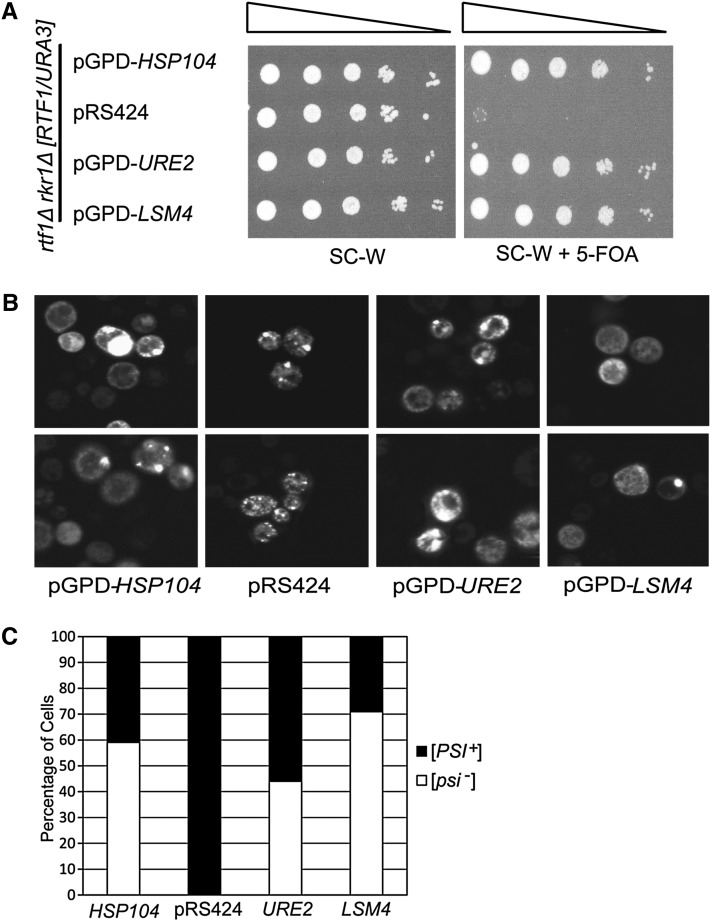
In a second, unbiased suppression screen (see Materials and Methods), we identified high-copy-number plasmids that rescued rtf1Δ rkr1Δ synthetic lethality. Confirming earlier results, one plasmid suppressor contained HSP104. Interestingly, several other suppressor plasmids contained URE2, which encodes the prion [URE3] (reviewed in Masison et al. 2000), or LSM4, which codes for a protein with a prion-forming domain (Alberti et al. 2009). The overexpression of [URE3] has been shown to antagonize the propagation of [PSI+] (Schwimmer and Masison 2002). Therefore, to further test the hypothesis that the clearance of [PSI+] suppresses rtf1Δ rkr1Δ lethality, we transformed an rtf1Δ rkr1Δ double mutant strain carrying an RTF1URA3-marked plasmid with overexpression plasmids for the prions [URE3] or [LSM4] and assessed the growth of these strains on 5-FOA medium (Derkatch et al. 2001; Alberti et al. 2009). HSP104 overexpression served as a positive control. As expected, the rtf1Δ rkr1Δ strain was unable to grow without a plasmid source of RTF1. However, the overexpression of the prion-encoding genes URE2 and LSM4 suppressed rtf1Δ rkr1Δ synthetic lethality to the same degree as overexpression of HSP104 (Figure 3A). We also investigated the effects of overexpressing the gene RNQ1, which encodes the prion [PIN+] (Derkatch et al. 2001), on rtf1Δ rkr1Δ synthetic lethality. Although some transformants revealed suppression of rtf1Δ rkr1Δ synthetic lethality by RNQ1 overexpression, others did not, possibly because [PSI+] and [PIN+] variants can differentially affect each other’s propagation (Figure S1) (Bradley and Liebman 2003).
Figure 3 .
Overexpression of HSP104, URE2, or LSM4 rescues rtf1Δ rkr1Δ lethality and clears [PSI+]. (A) An rtf1Δ rkr1Δ strain (KY2205) carrying a URA3-marked RTF1 plasmid was transformed with 2µ TRP1-marked pGPD-HSP104, pGPD-URE2, pGPD-LSM4, or empty vector. Dilution assays were performed on SC-W or SC-W + 5-FOA, and plates were incubated at 30° for 2 days. (B) Strains from A were transformed with LEU2-marked pCUP1-SUP35NM-GFP. Transformants were patched onto SC-L-W containing 100 µM CuSO4 and incubated in the dark at 30° before live-cell imaging was performed using confocal microscopy. Two representative images are shown for each strain. (C) The percentage of [PSI+] and [psi−] cells from among 50–100 cells from three separate transformants.
Collectively, our results indicate that [PSI+] is negatively impacted by overexpression of [URE3] and [LSM4] prions and that [PSI+] causes rtf1Δ rkr1Δ mutants to be inviable. To investigate if the aggregation propensity of [PSI+] is negatively affected under each of these conditions, we used live-cell confocal microscopy to image the presence of [PSI+] in strains overexpressing HSP104, URE2, or LSM4. As expected, rtf1Δ rkr1Δ [RTF1, URA3, CEN/ARS] cells expressing the Sup35NM-GFP 2μ plasmid contained fluorescent puncta, confirming the presence of [PSI+] (Figure 3B). In strains that overexpressed Sup35NM-GFP and also HSP104, URE2, or LSM4, we observed a large number of cells that exhibited diffuse fluorescence and were apparently cured of [PSI+] (Figure 3, B and C).
The recovery of URE2 and LSM4 from an independent genetic screen as suppressors of rtf1Δ rkr1Δ lethality further indicated the importance of [PSI+] to this genetic interaction. However, due to the known roles of Lsm4 in regulating RNA processing and degradation as part of the Lsm complex (reviewed in Beggs 2005), we asked whether the suppression of rtf1Δ rkr1Δ lethality by LSM4 overexpression could be explained by a disruption of Lsm complex function. To this end, we constructed a plasmid to overexpress another member of the Lsm complex, Lsm2, which is not known to have a prion-forming domain. Unlike the case for LSM4, the LSM2 overexpression plasmid did not rescue rtf1Δ rkr1Δ synthetic lethality (Figure S1). Collectively, our data indicate a positive correlation between overexpression of certain prions, clearance of [PSI+], and suppression of rtf1Δ rkr1Δ inviability.
[PSI+] causes rtf1Δ rkr1Δ synthetic lethality
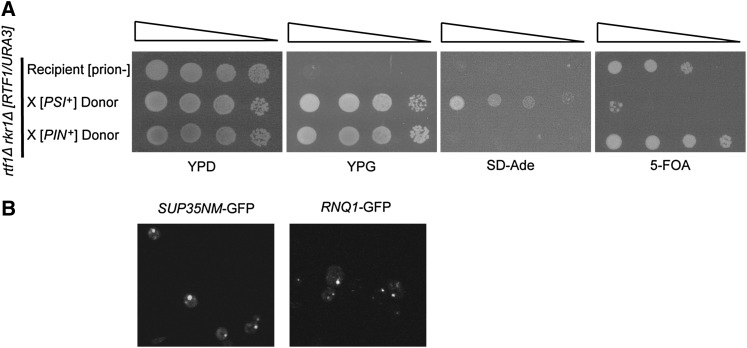
To confirm that the combined loss of RTF1 and RKR1 causes synthetic lethality only in [PSI+] conditions, we performed cytoduction experiments using a viable, cured rtf1Δ rkr1Δ strain transformed with a URA3-marked RTF1 plasmid as a recipient strain and specific prion-containing donor strains. The recipient also carried the ade1-14 nonsense allele to monitor the transfer of [PSI+], which causes translational read-through of ade1-14 and production of functional Ade1 protein (Chernoff et al. 1995). Cytoductions were performed with two donor strains (Mathur et al. 2009), one carrying [PSI+] and the other carrying [PIN+]. Only cytoduction of [PSI+] resulted in the inability to lose the RTF1 plasmid, as indicated by growth on SD-Ade and death on 5-FOA media (Figure 4A). We confirmed transfer of [PSI+] or [PIN+] by live-cell confocal microscopy with GFP tagged-prion domains (Figure 4B). These results confirm that the prion [PSI+] causes rtf1Δ rkr1Δ lethality.
Figure 4 .
Cytoduction of [PSI+] into cured rtf1Δ rkr1Δ strains causes synthetic lethality. (A) An rtf1Δ rkr1Δ ade1-14 strain (KY2286) cured of prions by dissection on medium containing GuHCl was transformed with a URA3-marked RTF1 plasmid and made ρ0 by growth in ethidium bromide-containing medium. This strain served as a recipient for cytoduction with donor kar1 strains containing only [PSI+] (L2265) or [PIN+] (L2261). A dilution growth assay is shown with the recipient and cytoductants on YPD and YPG to show successful cytoduction of recipients, on SD-Ade to show transfer of [PSI+], and on 5-FOA to score for loss of the RTF1/URA3 plasmid. Plates were incubated at 30° for 2–6 days. (B) Cytoductants from A were transformed with a HIS3-marked pCUP1-SUP35NM-GFP or pCUP1-RNQ1-GFP plasmid, patched on selective plates containing 100 µM CuSO4, and incubated in the dark at 30° before live-cell imaging was performed by confocal microscopy. Observations were made of at least three transformants per strain and 100 cells per transformant. Representative images are shown. No variability was seen among cells with respect to the GFP pattern.
Synthetic lethality is due in part to [PSI+]-mediated nonsense suppression
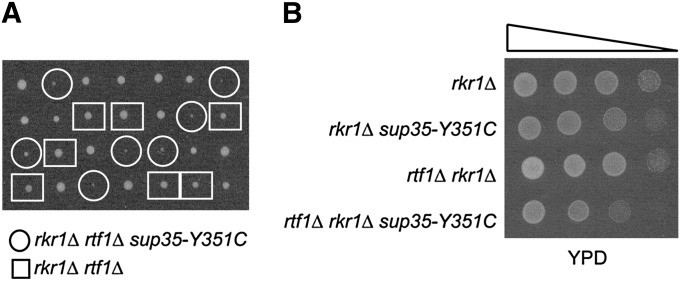
To investigate if the lethal effect of [PSI+] on rtf1Δ rkr1Δ double mutants is due to the presence of a prion or to a reduction in Sup35 function, we introduced the sup35-Y351C mutation into our genetic background and asked whether this mutation could inhibit the growth of rtf1Δ rkr1Δ [psi−] double mutants. The sup35-Y351C mutation was previously shown to increase readthrough of stop codons (Bradley et al. 2003) and impair the growth of a rkr1Δ strain (Bengtson and Joazeiro 2010). Interestingly, rtf1Δ rkr1Δ sup35-Y351C [psi−] strains grew more slowly than rtf1Δ rkr1Δ SUP35 [psi−] cells (Figure 5). The increased nonsense suppression due to sup35-Y351C did not fully recapitulate the effects of [PSI+] in rtf1Δ rkr1Δ strains; however, this may be due to the effect of the sup35-Y351C mutation in our strain background, as [PSI+] and sup35 alleles can cause different phenotypes in different strain backgrounds (True et al. 2004). Additionally, our results may indicate that Sup35 aggregates or that additional consequences of [PSI+], not duplicated by the sup35-Y351C allele, contribute to the rtf1Δ rkr1Δ genetic interaction. Regardless, the slow growth of rtf1Δ rkr1Δ sup35-Y351C cells suggests that an increase in nonsense suppression plays at least a partial role in rtf1Δ rkr1Δ [PSI+] lethality.
Figure 5 .
Nonsense suppression impairs growth of rtf1Δ rkr1Δ cells. (A) Tetrad dissections of crosses between a [psi−] rkr1Δ sup35-Y351C strain (KY2292) and a [psi−] rtf1Δ rkr1Δ SUP35 strain (KY2286). Dissections were done on YPD and incubated at 30° for 2 days. Double-mutant rtf1Δ rkr1Δ SUP35 segregants are highlighted by boxes while rtf1Δ rkr1Δ sup35-Y351C triple mutants are highlighted by circles. (B) A tetrad from A was further analyzed by fivefold serial dilution analysis on YPD and incubated at 30° for 1 day. Growth differences are less apparent on YPD on later days.
[PSI+] impacts rkr1Δ phenotypes
Our results demonstrate that the presence of [PSI+] greatly affects rkr1Δ genetic interactions. To further assess the physiological impact of [PSI+] on these strains, we assayed the growth of rtf1Δ and rkr1Δ strains in the presence or absence of prions under conditions of cell stress. Both rtf1Δ and rkr1Δ strains are sensitive to caffeine and cadmium chloride (Figure 6A). Interestingly, the CdCl2 sensitivity of rkr1Δ strains was strongly alleviated by curing prions through GuHCl treatment (Figure 6A). In addition, a slight ethanol sensitivity of rkr1Δ strains was detected only in uncured conditions. Together, these results suggest that prions influence the fitness of strains lacking RKR1, particularly under conditions of cell stress. We verified that [PSI+] influenced rkr1Δ phenotypes by transforming a rkr1Δ strain with the HSP104, URE2, and LSM4 overexpression plasmids and by measuring cadmium chloride sensitivity. Overexpressing these genes suppressed the sensitivity of rkr1Δ cells to cadmium chloride, similarly to the effects seen in a cured strain obtained by passage on GuHCl (Figure 6, A and B). Additionally, we tested the effect of the sup35-Y351C mutation on the phenotype of a rkr1Δ [psi−] strain and found that it causes sensitivity to cadmium chloride, as seen for rkr1Δ [PSI+] cells (Figure 6, A and C). This result demonstrates that [PSI+]-mediated nonsense suppression causes this rkr1Δ phenotype and affects the fitness of strains lacking RKR1.
Figure 6 .
The presence of [PSI+] affects rkr1Δ phenotypes. (A) Wild-type (KY761), rtf1Δ (KY2211), and rkr1Δ (KY2236) strains, or these strains first passaged onto YPD + 5 mM GuHCl, as well as an rtf1Δ rkr1Δ [psi−] strain (KY2209), were used for dilution growth assays on YPD or YPD containing 50 µM CdCl2, 10% EtOH, 15 mM caffeine, or 0.8 µg/ml cycloheximide and incubated at 30° for 2–6 days. “PRION+” indicates uncured cells; “prion−” indicates cells passaged on medium containing GuHCl. (B) A rkr1Δ strain (KY2236) was transformed with 2µ TRP1-marked pGPD-HSP104, pGPD-URE2, pGPD-LSM4, or empty vector (pRS424). Fivefold serial dilution analysis of these transformants was performed on SC-W or SC-W containing 50 µM CdCl2 and incubated at 30° for 3 days. (C) rkr1Δ (KY2309) and rkr1Δ sup35-Y351C (KY2306) strains were used for 10-fold dilution analysis on YPD and YPD containing 50 µM CdCl2 and incubated at 30° for 3 days.
While the curing of prions did not suppress the caffeine sensitivity of rkr1Δ strains, it did partially suppress the caffeine sensitivity of an rtf1Δ strain (Figure 6A). Interestingly, double-mutant rtf1Δ rkr1Δ strains obtained by dissection onto medium containing GuHCl were more sensitive to caffeine than either cured single mutant strain (Figure 6A). These data demonstrate that both RTF1 and RKR1 are required for cell viability under certain growth conditions even in the absence of prions. These results also correlate with our observation that a residual growth defect is apparent for GuHCl-treated rkr1Δ htb1-K123R double mutants compared to GuHCl-treated RKR1htb1-K123R strains (Figure 2C).
rtf1Δ suppresses the elevated levels of nonstop proteins in rkr1Δ cells
In our phenotypic analyses, we found that rkr1Δ mutants are sensitive to cycloheximide (CHX) (Figure 6A). Furthermore, the RKR1 genetic interactors rtf1Δ and htb1-K123R also confer sensitivity to CHX (Figure 6A and Figure S2A), and this phenotype is not rescued by clearing cells of prions (Figure 6C). CHX inhibits ribosome translocation during protein synthesis (Schneider-Poetsch et al. 2010) as well as the decay of nonsense and nonstop mRNA (Frischmeyer et al. 2002; Wagner and Lykke-Andersen 2002). Therefore, the CHX sensitivity of cells lacking Rkr1 or Rtf1 may indicate a requirement for these proteins under conditions of impaired translation and/or mRNA quality control.
In previous studies, cells lacking Rkr1 exhibited an increase in nonstop protein levels without affecting the levels of nonstop mRNA (Wilson et al. 2007; Bengtson and Joazeiro 2010). As the prion form of Sup35, [PSI+] results in suppression of nonsense codons in nonsense mRNAs as well as readthrough of normal stop codons (Paushkin et al. 1996; Wilson et al. 2005). Rkr1 has been reported to interact with ribosomes and be localized in the cytoplasm, where it is then necessary for nonstop protein degradation (Fleischer et al. 2006; Bengtson and Joazeiro 2010). Because our previous studies indicated nuclear localization of Rkr1 (Braun et al. 2007), we decided to re-examine the localization of Rkr1 using different strains, an N-terminally tagged HA-Rkr1 construct, and better visualization using confocal microscopy. Here, we found that Rkr1 is predominantly, although not exclusively, cytoplasmic, thus supporting its role in nonstop protein degradation in our strains (Figure S3).
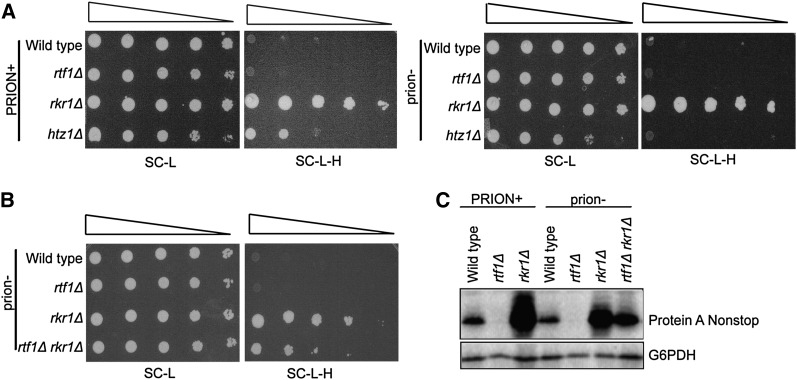
Given the role of Rkr1 in degrading nonstop proteins and the role of [PSI+] in generating nonstop proteins, we hypothesized that the lethality of rtf1Δ rkr1Δ [PSI+] cells might be due to an overabundance of these proteins. To test this idea and examine if RTF1 plays a role in regulating nonstop protein levels, we used a nonstop reporter plasmid containing the HIS3 gene without a stop codon (Wilson et al. 2007). Because wild-type cells carrying the reporter efficiently degrade the his3 nonstop transcript and protein, they fail to grow on media lacking histidine (Wilson et al. 2007). However, cells lacking RKR1 are unable to degrade the His3 nonstop protein and therefore grow on media lacking histidine (Figure 7A) (Wilson et al. 2007). Cells lacking RTF1 alone did not exhibit a his3 nonstop phenotype in the presence or absence of prions (Figure 7A). Interestingly, however, absence of the histone H2A variant Htz1, which was previously shown to increase nonstop transcript levels (Wilson et al. 2007), exhibited only a his3 nonstop phenotype in [PSI+] conditions (Figure 7A). This result indicates that Sup35 aggregation exacerbates the nonstop phenotype in some strains, although not detectably in rkr1Δ strains by this assay. As for rtf1Δ cells, we did not observe a his3 nonstop phenotype for H2B K123R strains, suggesting that loss of H2B ubiquitylation does not cause an increase in nonstop RNA or protein levels as measured by this reporter (Figure S2B).
Figure 7 .
rtf1Δ rkr1Δ strains exhibit a decrease in nonstop reporter proteins. (A and B) Wld-type (KY1030), rtf1Δ (KY564), rkr1Δ (KY2202), rtf1Δ rkr1Δ (KY2210), and htz1Δ (KY1404) strains were transformed with a LEU2-marked plasmid containing a his3 nonstop reporter. Tenfold serial dilutions were plated on SC-L or SC-L-H medium and incubated at 30° for 2 or 3 days, respectively. Prion− strains were generated prior to transformation by curing on medium containing 2.5 or 5 mM GuHCl. (C) Wild-type (KY307), rtf1Δ (KY2211), rkr1Δ (KY2236), and rtf1Δ rkr1Δ (KY2209) were transformed with a URA3-marked plasmid containing a protein A nonstop reporter and grown to early log phase in SC-U containing 2% galactose. prion− strains were generated as in A. Trichloroacetic acid extracts were analyzed by Western blotting using antibodies against protein A or G6PDH, which served as a loading control. “PRION+” indicates uncured cells; “prion−” indicates passage on medium containing GuHCl.
To test if deletion of RTF1 and RKR1 additively elevate nonstop protein levels, we measured expression of the his3 nonstop reporter in viable, cured rtf1Δ rkr1Δ strains. Surprisingly, deletion of RTF1 suppressed the his3 nonstop phenotype of a rkr1Δ mutant, as indicated by reduced growth on –His medium (Figure 7B). We confirmed this result using a second nonstop reporter in which the protein A gene lacks a stop codon (Wilson et al. 2007). Total protein A levels were measured by Western analysis (Figure 7C). Using this assay, we observed that the curing of prions caused a decrease in nonstop protein A levels in the rkr1Δ strain (Figure 7C). Also, in agreement with results obtained with the his3 nonstop reporter, rtf1Δ rkr1Δ double mutants had reduced nonstop protein A levels compared to rkr1Δ cells (Figure 7C). These results argue against a simple model in which a combinatorial increase in nonstop protein levels causes rtf1Δ rkr1Δ synthetic growth defects.
Discussion
We investigated the genetic relationship between the Paf1C subunit Rtf1 and the ubiquitin-protein ligase Rkr1 to better understand the interaction between their transcription and protein quality-control functions. A transposon mutagenesis screen for genetic suppressors of rtf1Δ rkr1Δ synthetic lethality identified a mutation in the gene encoding Hsp104. Further investigation into the suppression mechanism of an hsp104 mutation showed that rtf1Δ rkr1Δ strains are inviable only in the presence of [PSI+]. The overexpression, deletion, or GuHCl-mediated inactivation of HSP104, as well as the overexpression of the prion-coding genes URE2 and LSM4, all rescue rtf1Δ rkr1Δ synthetic lethality and clear cells of [PSI+]. [PSI+], the prion aggregate of Sup35, a translation termination factor necessary for proper stop codon recognition, results in readthrough of normal stop codons (Paushkin et al. 1996; Wilson et al. 2005). In turn, Rkr1 is required for the efficient ubiquitylation and degradation of nonstop proteins by recognition of a polylysine tract resulting from translation through the poly(A) tail (Bengtson and Joazeiro 2010). We have shown that [PSI+] and the resulting nonsense suppression conditions exacerbate rkr1Δ phenotypes, suggesting that the presence of [PSI+] and excess nonstop proteins is detrimental in the absence of RKR1. However, it is only in the absence of Rtf1 that this increased burden on protein quality-control machinery causes inviability.
Improper recognition of stop codons leads to readthrough of both normal and premature stop codons, resulting in nonstop proteins and nonsense suppression, respectively, and explaining the multiple phenotypic effects of [PSI+] (True et al. 2004; Wilson et al. 2005). In addition, because degradation of many aberrant mRNAs depends on proper translation termination, the presence of [PSI+] also affects the degradation of these transcripts (Wilson et al. 2005). Therefore, in combination, the presence of [PSI+] and the absence of RKR1 likely results in increased levels of nonstop proteins that cannot be efficiently recognized and degraded, as well as in an increased burden on mRNA quality control. Although we have shown that [PSI+] influences the phenotypic consequences of deleting RKR1 in yeast, added stress to protein or mRNA quality-control systems could also impact the severity of rkr1 mutations in higher eukaryotes and contribute to the development of diseases, such as neurodegeneration (Chu et al. 2009).
In addition to mechanisms that recognize and degrade aberrant proteins, such as that involving Rkr1, several mRNA surveillance pathways also prevent the translation of erroneous transcripts. Two such quality-control pathways are nonsense-mediated decay, which recognizes transcripts with premature termination codons, and nonstop decay, which recognizes transcripts without stop codons (reviewed in Fasken and Corbett 2005). Cells deficient in Paf1C exhibit transcript-specific increases or decreases in mRNAs targeted for quality control, presumably due to errors in RNA processing (Penheiter et al. 2005; Strawn et al. 2009). For example, mutations in yeast PAF1 result in shortened poly(A) tails and in altered poly(A) site utilization, which produce substrates for mRNA surveillance pathways (Mueller et al. 2004; Penheiter et al. 2005; Nordick et al. 2008), and defects in hPaf1C give rise to aberrant transcripts, which are inefficiently processed or exported (Farber et al. 2010; Nagaike et al. 2011). In agreement with these earlier observations, we have shown that deletion of RTF1 results in decreased protein product from two nonstop reporters (Figure 7), presumably because the absence of Rtf1-dependent histone modifications leads to transcriptional alterations and/or effects on RNA export, stability, or translation.
The mechanism by which Rtf1 and H2B ubiquitylation protects cells against the combined lethal effects of [PSI+] and rkr1Δ is not clear. Our initial hypothesis was that deletion of RTF1 elevates nonstop protein synthesis or stability to levels that are intolerable in [PSI+] rkr1Δ strains; however, our nonstop reporter assays detected decreased, not increased, nonstop protein levels in rtf1Δ strains. Therefore, the synthetic lethality between rtf1Δ and rkr1Δ is not easily explained by an elevation in nonstop protein levels. An alternative explanation, based on the importance of Paf1C in transcription and RNA processing (reviewed in Jaehning 2010 and Crisucci and Arndt 2011), is that rtf1Δ leads to a spectrum of aberrant transcripts that may impose stress on the cell or impair the expression of specific genes whose products play a role in RNA surveillance or protein quality control. These products would be especially important in cells lacking Rkr1 and containing [PSI+]. Our observation that rtf1Δ and rkr1Δ mutants have similar stress-related phenotypes (Figure 6) further supports a role for both proteins in preventing the accumulation of quality-control substrates. Finally, our data do not preclude the possibility that Rkr1 possesses alternate activities, which remain to be identified, and it is the absence of these functions that elevates the need for Paf1C and its associated histone modifications. Regardless of the precise mechanism, our results reveal a previously unrecognized requirement for a functional Paf1C and its associated histone modifications in protecting cells from the adverse effects of [PSI+] in the context of impaired protein quality control.
Supplementary Material
Acknowledgments
We thank Brett Tomson, Greg Prelich, Rich Gardner, and Jeff Brodsky for careful reading of the manuscript and helpful comments. We also thank Mark Sullivan for help in constructing the URE2 and LSM4 overexpression plasmids. Additionally, we thank Susan Liebman and Ambro Van Hoof for helpful strains and plasmids. This work was supported by National Institutes of Health grant GM52593 to K.M.A. and an Andrew Mellon Predoctoral Fellowship to K.M.K.
Footnotes
Communicating editor: M. Hampsey
Literature Cited
- Alberti S., Halfmann R., King O., Kapila A., Lindquist S., 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137: 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg D. C., Burke D. J., Strathern J. N., 2006. Yeast immunofluorescence. CSH Protoc, pii
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., et al. , 1988. Current Protocols in Molecular Biology. Green Publishing Associates and Wiley-Interscience, New York [Google Scholar]
- Batta K., Zhang Z., Yen K., Goffman D. B., Pugh B. F., 2011. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 25: 2254–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs J. D., 2005. Lsm proteins and RNA processing. Biochem. Soc. Trans. 33: 433–438 [DOI] [PubMed] [Google Scholar]
- Bengtson M. H., Joazeiro C. A., 2010. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467: 470–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley M. E., Liebman S. W., 2003. Destabilizing interactions among [PSI(+)] and [PIN(+)] yeast prion variants. Genetics 165: 1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley M. E., Bagriantsev S., Vishveshwara N., Liebman S. W., 2003. Guanidine reduces stop codon read-through caused by missense mutations in SUP35 or SUP45. Yeast 20: 625–632 [DOI] [PubMed] [Google Scholar]
- Braun M. A., Costa P. J., Crisucci E. M., Arndt K. M., 2007. Identification of Rkr1, a nuclear RING domain protein with functional connections to chromatin modification in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 2800–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns N., Grimwade B., Ross-Macdonald P. B., Choi E. Y., Finberg K., et al. , 1994. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 8: 1087–1105 [DOI] [PubMed] [Google Scholar]
- Chernoff Y. O., Lindquist S. L., Ono B., Inge-Vechtomov S. G., Liebman S. W., 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268: 880–884 [DOI] [PubMed] [Google Scholar]
- Chu J., Hong N. A., Masuda C. A., Jenkins B. V., Nelms K. A., et al. , 2009. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Natl. Acad. Sci. USA 106: 2097–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. S., Chapman R. E., Walter P., 1997. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell 8: 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisucci E. M., Arndt K. M., 2011. The roles of the Paf1 complex and associated histone modifications in regulating gene expression. Genet. Res. Int.,15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow E. T., Li L., 2011. Newly identified prions in budding yeast, and their possible functions. Semin. Cell Dev. Biol. 22: 452–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I. L., Bradley M. E., Hong J. Y., Liebman S. W., 2001. Prions affect the appearance of other prions: the story of [PIN(+)]. Cell 106: 171–182 [DOI] [PubMed] [Google Scholar]
- Dover J., Schneider J., Tawiah-Boateng M. A., Wood A., Dean K., et al. , 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277: 28368–28371 [DOI] [PubMed] [Google Scholar]
- Farber L. J., Kort E. J., Wang P., Chen J., Teh B. T., 2010. The tumor suppressor parafibromin is required for posttranscriptional processing of histone mRNA. Mol. Carcinog. 49: 215–223 [DOI] [PubMed] [Google Scholar]
- Fasken M. B., Corbett A. H., 2005. Process or perish: quality control in mRNA biogenesis. Nat. Struct. Mol. Biol. 12: 482–488 [DOI] [PubMed] [Google Scholar]
- Ferreira P. C., Ness F., Edwards S. R., Cox B. S., Tuite M. F., 2001. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol. 40: 1357–1369 [DOI] [PubMed] [Google Scholar]
- Fleischer T. C., Weaver C. M., McAfee K. J., Jennings J. L., Link A. J., 2006. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 20: 1294–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer P. A., van Hoof A., O’Donnell K., Guerrerio A. L., Parker R., et al. , 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295: 2258–2261 [DOI] [PubMed] [Google Scholar]
- Grimminger-Marquardt V., Lashuel H. A., 2010. Structure and function of the molecular chaperone Hsp104 from yeast. Biopolymers 93: 252–276 [DOI] [PubMed] [Google Scholar]
- Harlow E., Lane D., 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Haslberger T., Bukau B., Mogk A., 2010. Towards a unifying mechanism for ClpB/Hsp104-mediated protein disaggregation and prion propagation. Biochem. Cell Biol. 88: 63–75 [DOI] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F., 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57: 267–272 [DOI] [PubMed] [Google Scholar]
- Hwang W. W., Venkatasubrahmanyam S., Ianculescu A. G., Tong A., Boone C., et al. , 2003. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11: 261–266 [DOI] [PubMed] [Google Scholar]
- Ivanov I., Lo K. C., Hawthorn L., Cowell J. K., Ionov Y., 2007. Identifying candidate colon cancer tumor suppressor genes using inhibition of nonsense-mediated mRNA decay in colon cancer cells. Oncogene 26: 2873–2884 [DOI] [PubMed] [Google Scholar]
- Jaehning J. A., 2010. The Paf1 complex: Platform or player in RNA polymerase II transcription? Biochim. Biophys. Acta 1799: 379–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G., Masison D. C., 2001. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr. Microbiol. 43: 7–10 [DOI] [PubMed] [Google Scholar]
- Kumar A., des Etages S. A., Coelho P. S., Roeder G. S., Snyder M., 2000. High-throughput methods for the large-scale analysis of gene function by transposon tagging. Methods Enzymol. 328: 550–574 [DOI] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J. L., 2007. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Mackay R. G., Helsen C. W., Tkach J. M., Glover J. R., 2008. The C-terminal extension of Saccharomyces cerevisiae Hsp104 plays a role in oligomer assembly. Biochemistry 47: 1918–1927 [DOI] [PubMed] [Google Scholar]
- Masison D. C., Edskes H. K., Maddelein M. L., Taylor K. L., Wickner R. B., 2000. [URE3] and [PSI] are prions of yeast and evidence for new fungal prions. Curr. Issues Mol. Biol. 2: 51–59 [PubMed] [Google Scholar]
- Mathur V., Hong J. Y., Liebman S. W., 2009. Ssa1 overexpression and [PIN(+)] variants cure [PSI(+)] by dilution of aggregates. J. Mol. Biol. 390: 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., Lidschreiber M., Siebert M., Leike K., Soding J., et al. , 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17: 1272–1278 [DOI] [PubMed] [Google Scholar]
- Mueller C. L., Porter S. E., Hoffman M. G., Jaehning J. A., 2004. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell 14: 447–456 [DOI] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M., 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Mutiu A. I., Hoke S. M., Genereaux J., Liang G., Brandl C. J., 2007. The role of histone ubiquitylation and deubiquitylation in gene expression as determined by the analysis of an HTB1(K123R) Saccharomyces cerevisiae strain. Mol. Genet. Genomics 277: 491–506 [DOI] [PubMed] [Google Scholar]
- Nagaike T., Logan C., Hotta I., Rozenblatt-Rosen O., Meyerson M., et al. , 2011. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol. Cell 41: 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., et al. , 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H. H., Dole S., Struhl K., 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278: 33625–33628 [DOI] [PubMed] [Google Scholar]
- Nordick K., Hoffman M. G., Betz J. L., Jaehning J. A., 2008. Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot. Cell 7: 1158–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell D. A., Kowal A. S., Singer M. A., Lindquist S., 1994. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372: 475–478 [DOI] [PubMed] [Google Scholar]
- Paushkin S. V., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D., 1996. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15: 3127–3134 [PMC free article] [PubMed] [Google Scholar]
- Penheiter K., Washburn T., Porter S., Hoffman M., Jaehning J., 2005. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol. Cell 20: 213–223 [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P., 1991. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Rubel A. A., Saifitdinova A. F., Lada A. G., Nizhnikov A. A., Inge-Vechtomov S. G., et al. , 2008. Yeast chaperone Hspl04 regulates gene expression on the posttranscriptional level. Mol. Biol. (Mosk.) 42: 123–130 (in Russian) [DOI] [PubMed] [Google Scholar]
- Scherer S., Davis R. W., 1979. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc. Natl. Acad. Sci. USA 76: 4951–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Poetsch T., Ju J., Eyler D. E., Dang Y., Bhat S., et al. , 2010. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6: 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwimmer C., Masison D. C., 2002. Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 22: 3590–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon K. E., Mauger D. M., Arndt K. M., 2005. A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol. Cell 20: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J., Lindquist S., 2006. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol. Cell 23: 425–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolinski L. A., Eisenmann D. M., Arndt K. M., 1997. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 4490–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn L. A., Lin C. A., Tank E. M., Osman M. M., Simpson S. A., et al. , 2009. Mutants of the Paf1 complex alter phenotypic expression of the yeast prion [PSI+]. Mol. Biol. Cell 20: 2229–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z. W., Allis C. D., 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108 [DOI] [PubMed] [Google Scholar]
- Tomson B. N., Davis C. P., Warner M. H., Arndt K. M., 2011. Identification of a role for histone H2B ubiquitylation in noncoding RNA 3′-end formation through mutational analysis of Rtf1 in Saccharomyces cerevisiae. Genetics 188: 273–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- True H. L., Berlin I., Lindquist S. L., 2004. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 431: 184–187 [DOI] [PubMed] [Google Scholar]
- van Hoof A., Frischmeyer P. A., Dietz H. C., Parker R., 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262–2264 [DOI] [PubMed] [Google Scholar]
- Wagner E., Lykke-Andersen J., 2002. mRNA surveillance: the perfect persist. J. Cell Sci. 115: 3033–3038 [DOI] [PubMed] [Google Scholar]
- Warner M. H., Roinick K. L., Arndt K. M., 2007. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol. Cell. Biol. 27: 6103–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., Edskes H. K., Shewmaker F., 2006. How to find a prion: [URE3], [PSI+] and [beta]. Methods 39: 3–8 [DOI] [PubMed] [Google Scholar]
- Wilson M. A., Meaux S., Parker R., van Hoof A., 2005. Genetic interactions between [PSI+] and nonstop mRNA decay affect phenotypic variation. Proc. Natl. Acad. Sci. USA 102: 10244–10249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. A., Meaux S., van Hoof A., 2007. A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics 177: 773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Dollard C., Ricupero-Hovasse S. L., 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53–55 [DOI] [PubMed] [Google Scholar]
- Wood A., Schneider J., Dover J., Johnston M., Shilatifard A., 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278: 34739–34742 [DOI] [PubMed] [Google Scholar]
- Yoshihisa T., Anraku Y., 1989. Nucleotide sequence of AMS1, the structure gene of vacuolar alpha-mannosidase of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 163: 908–915 [DOI] [PubMed] [Google Scholar]
- Zheng S., Wyrick J. J., Reese J. C., 2010. Novel trans-tail regulation of H2B ubiquitylation and H3K4 methylation by the N terminus of histone H2A. Mol. Cell. Biol. 30: 3635–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Derkatch I. L., Liebman S. W., 2001. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI(+)] and [PIN(+)]. Mol. Microbiol. 39: 37–46 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.