Abstract
Cytoplasmic dynein transports cargoes for a variety of crucial cellular functions. However, since dynein is essential in most eukaryotic organisms, the in-depth study of the cellular function of dynein via genetic analysis of dynein mutations has not been practical. Here, we identify and characterize 34 different dynein heavy chain mutations using a genetic screen of the ascomycete fungus Neurospora crassa, in which dynein is nonessential. Interestingly, our studies show that these mutations segregate into five different classes based on the in vivo localization of the mutated dynein motors. Furthermore, we have determined that the different classes of dynein mutations alter vesicle trafficking, microtubule organization, and nuclear distribution in distinct ways and require dynactin to different extents. In addition, biochemical analyses of dynein from one mutant strain show a strong correlation between its in vitro biochemical properties and the aberrant intracellular function of that altered dynein. When the mutations were mapped to the published dynein crystal structure, we found that the three-dimensional structural locations of the heavy chain mutations were linked to particular classes of altered dynein functions observed in cells. Together, our data indicate that the five classes of dynein mutations represent the entrapment of dynein at five separate points in the dynein mechanochemical and transport cycles. We have developed N. crassa as a model system where we can dissect the complexities of dynein structure, function, and interaction with other proteins with genetic, biochemical, and cell biological studies.
Keywords: cytoplasmic dynein, dynactin, intracellular transport, microtubule, Neurospora crassa
THE organization, survival, and function of eukaryotic cells depend on intracellular transport governed by the microtubule-based molecular motors cytoplasmic dynein and kinesin. Dynein carries out the inward transport of cargos whereas kinesins are responsible for the outward movement. These motors, in addition to the transport and distribution of a wide variety of cargos, are also responsible for vital cellular processes ranging from mitosis to organelle positioning to embryonic development (Schroer et al. 1989; Burkhardt et al. 1997; Rana et al. 2004; Kardon and Vale 2009). Although intracellular transport is necessary for the function of all cells, polarized cells in particular have specific transport needs due to their asymmetry and elongated shape. These extraordinary requirements necessitate efficient long-range microtubule-based transport mechanisms (Hirokawa and Takemura 2005; Zheng et al. 2008; Harada 2010). The anterograde transport needs in these cells are satisfied by a variety of kinesins but only a single cytoplasmic dynein fulfills the retrograde transport requirements.
Cytoplasmic dynein is a megadalton-sized, multiprotein complex composed of two heavy chains (DHCs), and varying numbers of dynein intermediate chains (DICs), light intermediate chains, and light chains. The DHCs perform the ATPase motor and microtubule-binding functions and the other subunits couple dynein to dynactin and to cargoes (Bielli et al. 2001; Traer et al. 2007; Cai et al. 2010). The DHC is an ∼4300- to 4600-amino-acid-long polypeptide and is a member of the hexameric AAA (ATPase Associated with various cellular Activities) family of ATPases (Iyer et al. 2004). The DHC can be further divided into two regions, a tail and a motor domain. The tail is involved in homodimerization of DHC and interaction with the other dynein subunits whereas the motor domain performs ATP hydrolysis-mediated microtubule translocation (Habura et al. 1999; King 2000; Tynan et al. 2000).
The motor domain is composed of six repeats of ATP modules designated as AAA1–AAA6. The first four AAA modules contain a conserved phosphate binding loop (p-loop) motif that is implicated in nucleotide binding and hydrolysis functions (Gibbons et al. 1991; Ogawa 1991; Koonce et al. 1992; Mocz and Gibbons 1996; Mocz et al. 1998). The AAA1 module is the principal site of ATP hydrolysis as revealed by vanadate-mediated photocleavage and mutational analyses (Gibbons et al. 1987; Silvanovich et al. 2003; Kon et al. 2004). Furthermore, data from the mutational analyses of AAA2–AAA4 p-loop motifs have revealed that these domains affect the microtubule (MT) interaction and ATP hydrolysis function of the motor to varying degrees, which implicates the nucleotide binding/hydrolysis functions of these domains in dynein regulation (Silvanovich et al. 2003; Kon et al. 2004; Cho et al. 2008). DHC binds to microtubules via a microtubule-binding stalk that extends from AAA4 (Carter et al. 2011; Kon et al. 2011, 2012). In addition, multiple studies have revealed the modulation of dynein activity by a variety of factors such as nucleotide status at different AAA modules, microtubule interaction, applied load, etc., indicating that different functions must be coupled through a less-understood, long-range intraprotein communication mechanism (Gee et al. 1997; Burgess et al. 2003, 2004; Kon et al. 2004, 2011; Mallik et al. 2004; Hook et al. 2005; Imamula et al. 2007; Roberts et al. 2009; Carter et al. 2011).
To execute many of its cellular functions, dynein interacts with other cellular factors such as dynactin (King et al. 2003; Schroer 2004). Dynactin interacts with dynein and acts as a cargo adapter (Holleran et al. 1996; Steffen et al. 1997) and as a molecular tether that enhances the processivity of the motor (King and Schroer 2000; Culver-Hanlon et al. 2006). In addition to dynactin, kinesin has also been known to cooperate with dynein to implement various cellular functions (Januschke et al. 2002; Schuster et al. 2011b). The role of dynein in intracellular transport, the coordination of dynein’s own multi-domain structure for efficient transport, and dynein’s regulation by dynactin and kinesin is not completely understood despite several genetic, biochemical, and cell biological studies.
Cytoplasmic dynein in Neurospora crassa plays an important role in a range of processes such as hyphal morphogenesis (Riquelme et al. 2000), microtubule organization (Riquelme et al. 2002), retrograde organelle transport (Seiler et al. 1999), and nuclear distribution (Plamann et al. 1994; Bruno et al. 1996). Despite dynein’s involvement in such key processes, it is nonessential for the viability of N. crassa, which makes large-scale mutational analysis practical. N. crassa grows by apical extension of the hyphae, which is achieved by the regulated delivery of membranous organelles to the hyphal tip and the recycling of membranes as well as the transport of endosomal cargoes to distal regions. These delivery mechanisms rely on microtubule-based motor transport (Seiler et al. 1999; Steinberg 2007; Riquelme et al. 2011). In the hyphal tips of filamentous fungi, the microtubules are arranged parallel along the longitudinal axis of the hyphae with the majority of plus ends closer to the tips than the minus ends (Mourino-Perez et al. 2006; Steinberg and Perez-Martin 2008). Dynein motors utilize this polarity to transport cargoes from the apical regions at the tips to the distal regions farther from the tip. Previous studies (Plamann et al. 1994; Bruno et al. 1996) have isolated mutant strains of N. crassa defective in dynein/dynactin function. These mutant strains exhibit very distinct curled hyphal growth morphology referred to as the ropy phenotype.
Large-scale mutational analyses of dynein have not been previously feasible in most eukaryotes due to its indispensable role in a variety of cellular processes. To overcome this limitation, we utilized the filamentous fungus N. crassa as our model system to characterize dynein in wild type and an array of ropy strains carrying DHC mutations. Our large-scale genetic study provides an important in-depth view of the complex links between specific dynein motor domains and particular functions of cytoplasmic dynein.
Materials and Methods
Isolation and initial genetic characterizations of DHC mutant strains
DHC mutant strains were isolated by employing a previously described genetic screen where mutations resulting in the loss of dynein/dynactin function were identified as partial suppressors of the cot-1 temperature-sensitive (cot-1ts) mutation (Plamann et al. 1994; Bruno et al. 1996; Tinsley et al. 1996; Minke et al. 1999). The N. crassa cot-1 gene encodes a serine/threonine protein kinase required for hyphal elongation (Yarden et al. 1992). The cot-1ts mutation results in normal radial colony growth similar to wild-type strains at permissive temperatures (25°) and in small (<1 mm) colonies at restrictive temperatures (37°) (Supporting Information, Figure S1). Mutations resulting in a loss of dynein/dynactin function partially suppress the cot-1ts growth defect (Plamann et al. 1994; Bruno et al. 1996; Tinsley et al. 1996; Minke et al. 1999). In brief, 105 conidia from the cot-1ts strain were suspended in 30 ml of cooled Vogel’s minimal agar media (50°), plated, and incubated for 3–4 days at 37°. Spontaneous revertants displaying increased radial growth (3- to 5-mm colonies) with slightly curled hyphae were identified, picked to slants, and incubated at 25°. Typically, one to five partially suppressed mutants were identified per plate. In this study, 690 independent ropy mutants were isolated and backcrossed to wild type. As described previously, complementation analysis was conducted by the formation of heterokaryons with known dynein/dynactin mutants to tentatively identify the dynein/dynactin gene likely to carry a mutation resulting in the ropy growth phenotype (Plamann et al. 1994; Bruno et al. 1996). The dynein heavy chain gene (ro-1) is tightly linked with the cot-1 gene, and our analysis of 690 ropy mutants backcrossed to wild type indicated that 290 of these ropy mutants were dynein heavy chain (ro-1) mutants.
The goal of this study was to examine the effects of DHC mutations where the DHC polypeptide is produced but has deficient function. Therefore, anti-DHC antibodies were used to screen by Western analysis the 290 ro-1 mutants for those that still appeared to produce full-length protein (Minke et al. 1999, 2000; Kumar et al. 2000; Lee et al. 2001). Seventy-six of the 290 ro-1 mutants produced what appeared to be full-length DHC protein. However, the DHC is a very large polypeptide (∼500 kDa), and it was likely that frameshift or nonsense mutations that generate slightly truncated proteins would be present within these 76 ro-1 mutants. To identify and eliminate these mutants from future consideration, we performed DNA sequence analysis of the last 1500 bases of the ro-1 structural gene. Of the 76 mutants, 44 were identified as having frameshift or nonsense mutation in this region, and we discarded all of these mutants but 2, which contained nonsense mutations that resulted in a slightly truncated DHC polypeptide (Figure 3). We sequenced the entire ∼14-kb ro-1 gene in all 34 strains used in this study, and we identified a single genetic lesion in each of the 34 ro-1 mutants (Figure 3, Table S2). Western blot analyses of a representative sample of the 34 mutant strains show that the mutant strains express varying amounts of DHC (Figure S2). We did not see a strong correlation between the amount of DHC expressed and any particular class of dynein mutant strains.
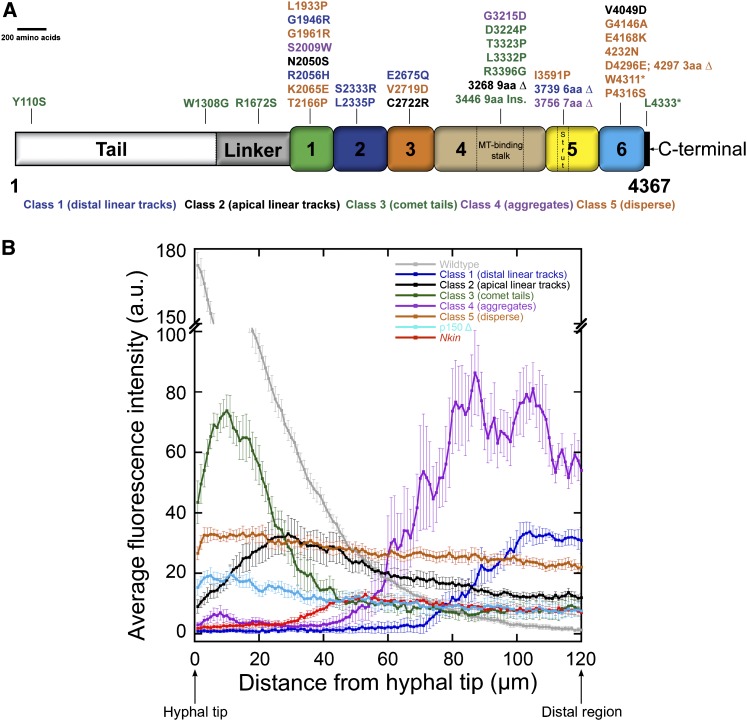
Figure 3 .
(A) Schematic representation of cytoplasmic dynein heavy chain from N. crassa showing the mutations analyzed. The relative positions of dynein heavy chain mutations are shown with corresponding amino acid changes. Amino acids (aa) are represented by single-letter codes. A “Δ” represents a deletion; “ins.” refers to insertion; and nonsense mutations are indicated by an asterisk. Numbers 1–6 indicate respective AAA domains. Color coding of the mutations represents grouping of different mutations to classes 1–5. (B) DIC-mCherry fluorescence intensity is plotted as a function of distance from the hyphal tip. Fluorescence signals in the wild-type strains (gray) exhibited an initial maximum closer to the hyphal tips and gradually tapered off at regions far from the tip. In class 1 (distal linear tracks) strains, fluorescence signals (medium blue) were low or absent at regions closer to tips but displayed increased signal intensities at distal regions from the tips. The class 2 (apical linear tracks) strains exhibited elevated fluorescence intensities (black) closer to the tips that gradually tapered off as a function of distance from the hyphal tip. Class 3 (comet tail) strains also exhibited increased fluorescence intensities (green) closer to the tips. Class 4 (aggregate) strains show irregular increased fluorescence (purple) at distal regions. The fluorescence intensity in class 5 (disperse) strains (brown) is slightly higher than the fluorescence signals of p150 Δ strains (aqua), but both profiles show a uniform distribution along the entire length of the hyphae. Nkin mutant strains exhibit faint fluorescence (red) at distal regions. Data are shown as mean ± SE (n = 15 for all classes).
Strains, media, and growth conditions
Table S3 shows a list of primary N. crassa strains used in these experiments and their genotypes. The DHC mutant strains examined in this study are presented in Figure 3 and Table S2. N. crassa strains were grown on Vogel’s minimal medium, supplemented with 1.5% sucrose (VSM). All reagents for media, supplements, and buffers used were purchased from Fisher Scientific unless otherwise indicated. Radial growth rates were determined from appropriate strains that were centrally inoculated on 150- × 15-mm petri dishes and grown for 16 hr at 25°. After this initial growth period, fungal colony diameters were measured at 8-hr intervals over the next 48 hr.
Generation of constructs and strains with dynein and dynactin fluorescent fusions
DIC-mCherry was created by fusing mCherry (CLONTECH) to the C terminus of the N. crassa dynein intermediate chain (ro-6) by a combination of PCR techniques and cloned into pBluescript to generate pDIC-mCherry. A 10-amino-acid (GGGGS)2 flexible linker (Wriggers et al. 2005) was added between the DIC and mCherry coding sequences. Enhanced green fluorescent protein (EGFP)-p150 was created by fusing EGFP (CLONTECH) to the N terminus of N. crassa dynactin p150 (ro-3). All fusion protein constructs were cloned in-frame and confirmed by DNA sequencing.
Transformation
We PCR-amplified a 2.7-kb fragment encoding 1.8-kb DIC sequence, mCherry fluorescent tag, and a 0.8-kb DIC downstream sequence from the pDIC-mCherry to generate a linear transformable DNA. This DNA fragment was integrated into the native DIC locus (ro-6) by homologous recombination (Figure S1). The DNA was transformed into a N. crassa DIC mutant ropy strain that carried a 7-bp deletion at nucleotide position 1695 (ro-6Δ1695–1702) in the DIC-coding sequence (Davis 2000). DIC-mCherry positive colonies were identified on the basis of (1) transformation-mediated rescue of the ropy phenotype of the DIC mutant strain leading to wild-type growth and (2) detection of mCherry fluorescence. The correct integration of DIC-mCherry onto the native locus was verified by genome sequencing. The colony morphology and radial growth rates of the DIC-mCherry strain were indistinguishable from the untransformed wild-type strain (Figure S1). A similar approach was used to generate an N-terminal EGFP fusion of the p150 subunit of dynactin. The DIC-mCherry strain was crossed with all the 34 DHC mutant strains using standard procedures (Davis 2000). Once the 34 DHC mutant strains with DIC-mCherry were isolated, genetic crosses were made with strains carrying EGFP-p150, β-tubulin-GFP [Fungal Genetics Stock Center (FGSC) #9520] and histone H1-GFP (FGSC #9518) to generate progeny that carried DHC mutations and were double-labeled (Table S2). A sequence encoding dual affinity tags comprising a hexahistidine (HHHHHH) and a Strep-tag II (WSHPQFEK) tag (Schmidt and Skerra 2007) was fused to the C terminus of DIC using a strategy similar to the one employed to generate the DIC-mCherry strain.
Microscopy and imaging
For live-cell imaging, strains were grown on a slide coated with a thin film of VSM agar for 16–18 hr at 25°. Epifluorescence microscopy was performed using an Olympus BX50 microscope equipped with a mercury light source. Samples were visualized using U Plan FI dry ×20 [0.50 numerical aperture (na)] or oil immersion ×100 (1.3 na) objective lenses. The GFP and mCherry fluorescence was detected by appropriate emission and excitation filter sets. Images and movies were captured with a SPOT RT-SE 18 camera system (Diagnostic Instruments) using SPOT software (v3.5.9). The images were processed in Adobe Photoshop. Classifications of the dynein localization phenotypes were performed manually by visual examination of at least 50 intermediate magnifications as well as high-magnification fluorescence micrographs from each strain studied. Furthermore, the spatial variations in dynein fluorescence in relation to the hyphal structure were confirmed by fluorescence intensity profiles. The line-scan analysis tool in Metamorph software (Molecular Devices) was used for the measurement of fluorescence intensity (expressed as mean gray values) profiles. The area of analysis was defined by a line width that covered the width of the hyphae and a line length drawn from the hyphal tip up to 120 µm back from the tip. Fifteen hyphae from each class were randomly chosen from images taken at the same magnification and exposure settings for fluorescence intensity determination. Fluorescence intensity values from an untransfected control strain were used to determine the background fluorescence that was then subtracted from the values derived from other strains. The mean and the standard error were plotted using Kaleidagraph.
Distance measurements within hyphal tips were made using SPOT software. Data were expressed as the mean ± SD. Fluorescence intensity of the comet tails from wild-type and class 3 mutant strains were determined using National Institutes of Health (NIH) ImageJ software. The fluorescence intensity (expressed as mean gray values) of individual comet tails was determined by manually selecting a comet tail, measuring the mean gray value within the selection, and normalizing the data with the gray value measurements of regions close to corresponding comet tails. The final fluorescence intensity measurement from each strain was determined from the average of the gray values from measuring at least 40 individual comet tails from each strain.
Vesicle transport imaging experiments utilized N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide (trade name “FM 4-64,” Life Technologies), which has no appreciable fluorescence unless it is in a lipid membrane. Slide cultures of fungal strains were prepared in the same manner as described earlier. VSM liquid media containing FM 4-64 was added to a final concentration of 5 µM onto a growing colony, and a coverslip was placed on top. The hyphae were imaged in a time window between 5 and 15 min following dye treatment. Time-lapse image stacks (30-sec duration) were generated with 40-msec exposures captured every 200 msec. The individual image stacks were analyzed using Metamorph software to generate movement kymographs. The velocities, distances traveled, and the total number of the movements were determined from those kymographs using custom-designed software. Cytoplasmic streaming-based transport was identified by the slower movement of vesicles (<0.5 μm/sec) in unison. In this study, we did not further examine cytoplasmic streaming-based movement of vesicles. Individual vesicles were considered to have undergone a motility event if they exhibited a directed displacement at a velocity of at least 0.5 μm/sec. Student’s t-tests were used to examine significance between various experimental data sets. The motility index was calculated by taking the sum of the velocity multiplied by the distance of all motility events for a strain divided by the total length of the hyphae examined. Representative live-cell movies of FM 4-64-labeled vesicle trafficking in hyphal tips from wild type (File S1), DHC deletion (File S2), p150 deletion (File S3), Nkin (File S4), Class 1 (File S5), Class 2 (File S6), Class 3 (File S7), Class 4 (File S8), and Class 5 (File S9) strains are included.
Benomyl treatment experiments were performed by adding VSM liquid media containing 10 µM benomyl (DuPont, Wilmington, DE) to a colony on a slide culture at room temperature. Benomyl treatment led to the disruption of microtubules in ∼5 min after which the hyphae were imaged. For controls, media containing DMSO was added to the colony, and imaging was performed as for benomyl treatment.
Purification of dynein from N. crassa
N. crassa strains expressing DIC-His-Strep affinity tags were homogenized with buffer A (35 mM PIPES, pH 7.0, 5 mM MgSO4) supplemented with 1 mM EGTA, 0.5 mM EDTA, 1 mM DTT, 0.5 mM ATP, and protease inhibitor cocktail (0.25 mM phenylmethanesulfonyl fluoride, 0.5 μg/ml leupeptin, 1.0 µg/ml pepstatin A, 10 µg/ml tosylphenylalanine chloromethyl ketone, 10 µg/ml N-tosyl-l-lysine chloromethyl ketone, and 10 µg/ml p-toluenesulfonyl-l-arginine methyl ester). After ultracentrifugation supernatant was loaded onto a SP Sepharose column (GE Healthcare, Uppsala, Sweden), and bound protein was eluted with 500 mM KCl in buffer A. The eluted fraction was then loaded onto a HiTrap chelating HP column (GE Healthcare) charged with CoCl2, and the bound protein was eluted with 250 mM imidazole (Acros) in buffer A. The eluted protein was then loaded onto a StrepTrap column (GE Healthcare), and the bound protein was finally eluted with 2.5 mM d-desthiobiotin (Sigma) in buffer A supplemented with 1 mM EGTA, 0.5 mM EDTA, 1 mM DTT, and 10 µM ATP.
Microtubule cosedimentation assay
Purified N. crassa dynein was incubated at 32° for 20 min with 5 µM paclitaxel-stabilized microtubules in the presence of 1 mM ATP, ADP, AMPPNP, or nucleotide-free conditions. Samples were subject to centrifugation, and the supernatant and pellet fractions were resolved by electrophoresis through 7.5% acrylamide gel, stained with Coomassie Brilliant Blue, and digitized using a Hewlett Packard Scanjet 7400c scanner. Digital images were analyzed using NIH ImageJ software and dynein heavy chain bands in the supernatant, and pellet fractions were quantified densitometrically.
ATPase assay
The ATPase activity of N. crassa dynein was measured by a radio thin-layer chromatography (TLC) assay (Gilbert and Mackey 2000; Mesngon et al. 2006). The reactions were performed in BRB80 buffer (80 mM PIPES, 1 mM EGTA, 1 mM MgCl2, pH 6.9) supplemented with 1 mM ATP and 10 µCi α-32P-ATP (Perkin Elmer, Boston) in the absence or presence of 5 µM paclitaxel-stabilized microtubules. The amount of radiolabeled ADP generated after 60 min was determined to measure the rate of ATP hydrolysis. Reaction samples spotted onto PEI-Cellulose F TLC plates (EMD Chemicals, Gibbstown, NJ) were resolved by TLC that was subject to phosphorimaging (Molecular Dynamics, Sunnyvale, CA). ATP and ADP spots were quantified densitometrically using ImageQuant software (Molecular Dynamics). The specific activity was measured by quantifying the concentration of dynein isolated from wild-type and AAA3 E/Q strains relative to purified bovine dynein.
Vanadate-mediated UV photolysis of N. crassa dynein
N. crassa dynein was subject to vanadate-mediated photolysis using standard conditions (Gibbons et al. 1987). The major site of cleavage is termed as the V1 site, and the cleavage reaction components termed as the V1 condition demand specific requirements such as UV irradiation, Mg2+, and ATP. The V1 site cleavage leads to the formation of high- and low-molecular-weight UV fragments. Substitution of Mg2+ with Mn2+as a divalent metal ion suppresses V1 cleavage (Gibbons et al. 1987). The amount of cleavage products generated in the presence of magnesium (V1 Mg2+) or manganese ions (V1 Mn2+) was used to measure the extent of vanadate photolysis. Samples in cleavage buffer (80 mM PIPES, pH 6.9) supplemented with 1 mM ATP, 2 mM MgCl2 or MnCl2, 500 µM Na3VO4 were irradiated with UV (365 nm) light using UVL-56 BLAK-RAY lamp (UVP Inc., San Gabriel, CA) for 60 min on ice. Cleavage fragments were resolved by electrophoresis through 7.5% acrylamide gel, stained with Coomassie Brilliant Blue, and the gels were digitized. The UV fragments were quantified by densitometry using NIH ImageJ software.
Motility assay
In vitro bead-based motility assays were performed as reported earlier (King and Schroer 2000; Mallik et al. 2004; Culver-Hanlon et al. 2006). Purified tubulin was used to generate polymerized microtubules that were bound to a flow chamber coated with poly-lysine. Unbound microtubules were washed away by the addition of final dilution buffer (33 mM PIPES, pH 7.0, 0.7 mM EGTA, 0.3 mM MgSO4, 1 mM GTP, and 20 µM paclitaxel). Dynein purified from specified N. crassa strains was bound to polybead carboxylate microspheres (0.21-μm diameter; Polysciences, Warrington, PA) by nonspecific adsorption at varying molar ratios (20:1–40:1 dynein: bead) in the presence of 10 µm ATP. The beads were added to the flow chamber and viewed by video-enhanced differential interference contrast microscopy. Custom-built image-processing software (Labview 6.1, National Instruments) was used for tracking the beads that exhibit directional motion. The run lengths and velocities of motility events were determined from the tracking data.
Results
Localization of dynein molecules to the hyphal tip in N. crassa colonies
To better understand the roles that cytoplasmic dynein motors normally play in polarized growth, we utilized live-cell imaging approaches in N. crassa. We replaced the endogenous DIC with a version of the DIC that encoded a C-terminal mCherry tag (Figure S1). We then visualized DIC-mCherry fluorescence, which we will refer to as “dynein fluorescence” throughout this article. The replacement of the single copy of the DIC gene by homologous recombination in the haploid genome ensured that DIC-mCherry was expressed under the control of its native promoter and that all DIC present in the strain was tagged. The presence of the mCherry tag had no effect on the normal function of the DIC as determined by hyphal morphology and colony growth rates (Figure S1), confirming that DIC-mCherry-expressing cells have wild-type dynein function.
When we observed dynein localization in actively growing hyphae, we saw that the dynein fluorescence was predominantly found in the extreme tips of the hyphae (Figure 1A). The fluorescent signal tapered off dramatically within ∼50 μm (average 45 ± 11 µm) (Figure 3B), and the signal gradient maintained its location at the hyphal tip even though the hyphae itself was growing at several micrometers per minute. Very little substructure could be observed at intermediate magnification across the hyphal tube.
Figure 1 .
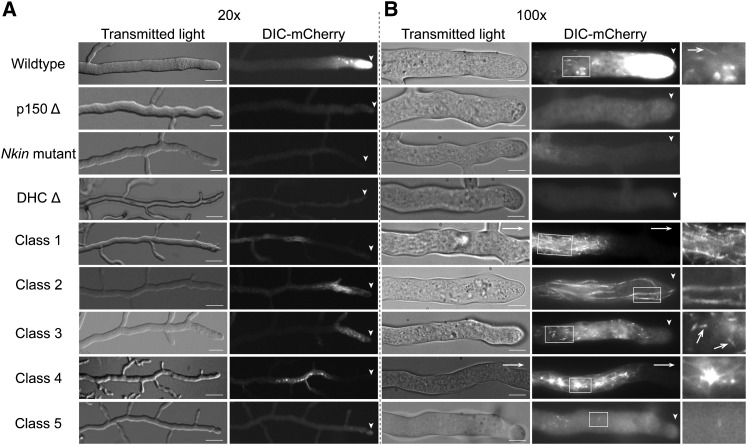
Localization of dynein in mutant strains of N. crassa. (A) Hyphae from the colony edge of different N. crassa strains were visualized at intermediate magnification to show the distance between dynein signal and the hyphal tip (arrowheads). (B) Hyphae visualized using higher magnification show details of dynein localization. The wild-type dynein localization includes a prominent hyphal-tip gradient as well as localization to short linear tracks (white arrows) and to other structures. In the mutant strains, dynein was mislocalized to particular hyphal structures and/or into a disperse signal. (A and B, left) Transmitted light. (A and B, right) mCherry fluorescence. Arrowheads indicate position of the hyphal tip. White arrows indicate that the hyphal tip was located outside the frame. Enlarged views of the boxed regions are shown on the right in B; white arrows indicate short linear tracks in wild-type and class 3 mutant strains. Bars in A, 50 µm; in B, 10 µm.
At higher magnification, additional details of the hyphal tip accumulation could be observed. Most images showed mCherry fluorescence in both short linear tracks and spherical structures in the region immediately following the hyphal tip (Figure 1B). The spherical structures were reminiscent of endomembrane organelles that have been described in previous studies (Bowman et al. 2009). The short linear tracks resembled the comet tail tip accumulation of dynein at microtubule plus ends that has been reported by studies in other systems ranging from fungi to mammalian neurons (Xiang et al. 1995; Vaughan et al. 1999; Xiang et al. 2000; Han et al. 2001; Ma and Chisholm 2002; Lee et al. 2003; Sheeman et al. 2003; Zhang et al. 2003, 2010, 2011; Lenz et al. 2006; Arimoto et al. 2011; Markus et al. 2011; Schuster et al. 2011a). Apart from the spherical structures and short linear tracks, no additional substructures were visible upon standard exposure settings or photobleaching the bright hyphal tip dynein fluorescence (Figure S3). As a control, we examined a DHC deletion strain and found that the specific localization of the DIC-mCherry signal was lost from hyphal tips, short linear tracks, and spherical structures (Figure 1).
To better determine if the short linear tracks were microtubule-based, we crossed the DIC-mCherry strain to a strain expressing β-tubulin-GFP (Freitag et al. 2004). Previous studies with these and similar strains have shown that abundant microtubules are predominantly arranged parallel to the longitudinal axis with plus ends extending into regions near the N. crassa hyphal tip. More complex arrangements with microtubules of mixed polarity are found around the nuclei in the syncytial cytosol further back from the hyphal tips (Riquelme et al. 2002; Freitag et al. 2004; Sampson and Heath 2005; Uchida et al. 2008). We found that dynein mCherry short linear track signals were co-linear with segments of the tubulin GFP signals near the hyphal tips (Figure 2A). To further examine dynein localization with respect to microtubules, we treated growing colonies with benomyl, a cell-permeable microtubule-depolymerizing drug. Treatment with 10 µM benomyl for 5 min depolymerized microtubules and resulted in a nonlinear, diffuse tubulin-GFP signal throughout the hyphae with occasional GFP puncta. Following microtubule depolymerization, the short linear tracks of the DIC-mCherry signal were abolished, and the high concentration of dynein at the hyphal tip began to expand distally along the hyphae (Figure 2B). Longer treatment times resulted in a loss of dynein signal at the hyphal tip, showing that the maintenance of the hyphal tip accumulation gradient requires a normal microtubule network. Our data confirm that the short linear tracks are associated with microtubule ends in a characteristic “comet-tail” pattern.
Figure 2 .
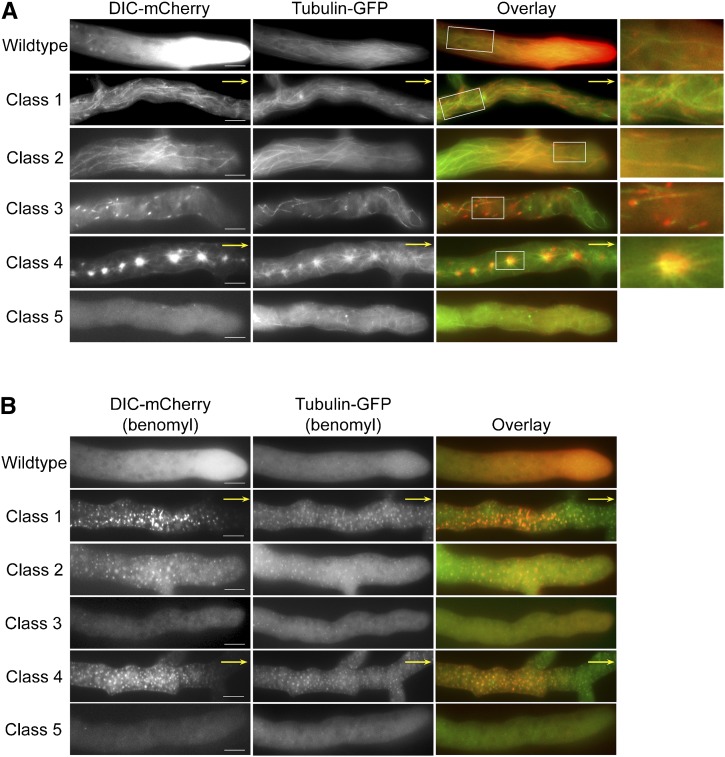
Localization of dynein and microtubules in wild-type and DHC mutant strains. (A) Epifluorescence images showing hyphal localization of DIC-mCherry (left panels) and β-tubulin-GFP (center panels) in wild-type and DHC mutant strains. Overlay images are shown in the right panels. Enlarged views of the boxed regions are shown on the far right. Yellow arrows indicate that the hyphal tips were located outside the frame. Dynein in wild-type, mutant class 1, 2, 3, and 4 strains was found along microtubule structures. Class 5 mutant strains did not show dynein colocalization with microtubules. Bars, 10 µm. (B) Dependence of dynein localization patterns on intact microtubule network. Treatment with benomyl depolymerizes microtubules and alters dynein localization in wild-type and DHC mutant strains expressing DIC-mCherry and β-tubulin-GFP. Dynein (red) is shown in left panels; microtubules (green) are shown in the center panels; overlay images are shown in the right panels. In wild-type and class 3 mutant strains, dynein localization to comet tails was disrupted immediately upon benomyl treatment. The cloud of dynein at the hyphal tips in wild-type strains disappeared after longer treatment times. Mutant class 1, class 2, and class 4 strains displayed punctate dynein signals that showed extensive overlap with microtubule remnants. Bars, 10 µm.
Dynein localization requires conventional kinesin and dynactin
Multiple studies have shown that kinesin and dynactin play essential roles in the localization of dynein within cells (Brady et al. 1990; Echeverri et al. 1996; Waterman-Storer et al. 1997; Martin et al. 1999; Vaughan et al. 1999; Duncan and Warrior 2002; Januschke et al. 2002; King et al. 2003; Zhang et al. 2003; Ligon et al. 2004; Theiss et al. 2005; Lenz et al. 2006; Arimoto et al. 2011). To determine if conventional kinesin (Nkin) or dynactin have similar functions in N. crassa, we crossed the DIC-mCherry strain to either a strain carrying repeat-induced point (RIP) mutations in conventional kinesin (Seiler et al. 1997) or a dynactin p150 (ro-3) null strain. In the Nkin mutant strain, hyphal tip accumulation of dynein was completely abolished; regions closer to the tips were devoid of any dynein signal but distal regions of the hyphae displayed a faint fluorescence (Figure 1, A and B; Figure 3B). Similar to the Nkin mutant strain, the p150 null strain also exhibited a loss of hyphal tip dynein accumulation. However, unlike the Nkin mutant strain, a diffuse dynein signal was found along the entire length of hyphae (Figure 1, A and B; Figure 3B). As an alternative to analyzing deletion strains, we also examined colocalization between dynein and dynactin in a strain expressing both DIC-mCherry and EGFP-p150. Dynactin exhibited almost complete colocalization with dynein to the hyphal tip, to the comet tails, and to spherical structures near the hyphal tip (Figure S4). These data together illustrate that the functions of both dynactin and kinesin are required for the correct cellular localization of dynein in N. crassa hyphal growth.
Mutations in the DHC lead to multiple dynein mislocalization phenotypes
To further characterize dynein function within the hyphal tip of N. crassa, we examined 34 DHC mutant strains isolated from a genetic screen (see Materials and Methods). In the current study, we crossed the mCherry-DIC allele into the 34 DHC mutant strains so that we could track dynein molecules in the mutant strains. All 34 strains lacked the bright hyphal tip accumulation observed in wild-type strains (Figure 1A). In addition, the remaining dynein molecules in the DHC mutant strains were mislocalized into one of five distinct classes of phenotypes that we termed classes 1–5 (Table 1).
Table 1 . Summary of dynein, dynactin, microtubule, and nuclear phenotypes in wild-type and mutant strains of N. crassa.
| Classification | Dynein and dynactin | Microtubules | Vesicle transport | Nuclei |
|---|---|---|---|---|
| Wild type | Bright hyphal-tip accumulation; comet tails | Long, parallel arrangement | Robust transport | Normal NEZs; random distribution |
| Class 1 | Distal: long linear tracks | Distal: bundled | Reduced transport | Reduced NEZs; random distribution |
| Class 2 | Apical: long linear tracks | Apical: bundled | Reduced transport | Reduced NEZs; random distribution |
| Class 3 | Comet tails | Disorganized; excessively curved | Reduced transport | Reduced NEZs; random distribution |
| Class 4 | Distal: aggregates | Distal: aster-like appearance | Reduced transport | Normal NEZs; clustered distribution |
| Class 5 | Diffuse | Disorganized | Reduced transport | Reduced NEZs; random distribution |
The dynein localization signal in six class 1 mutant strains was found in distal long linear tracks arranged mostly parallel to the hyphal axis (Figure 1B). This signal pattern was readily distinguishable from the comet tails described earlier for wild-type dynein localization in length of the track and location within the hyphae (Figure 2B). The dynein signal in these long linear tracks occurred in distal regions ∼100 ± 30 µm away from the tip and extended even farther into distal regions far from the tips (Figure 1, A and B; Figure 3B). The four class 2 DHC mutant strains, in contrast to class 1 mutant strains, showed long linear tracks that occurred at apical regions close to the tip of the hyphae that ran until ∼75 ± 15 µm back from the tip (Figure 1, A and B; Figure 3B). A mutant strain that exhibited one of these two mutant localization phenotypes (class 1 or class 2) consistently showed the specified localization pattern and did not exhibit the alternate class phenotype. The long linear tracks observed in both class 1 and class 2 likely reflect dynein associated with microtubules, given that microtubules are the only structures that exhibit long parallel arrangement inside the hyphae.
The class 3 mutations were composed of nine strains that each had residual DIC-mCherry fluorescence in hyphal tips that was concentrated in comet tail structures (Figure 1, A and B; Figure 3B). The comet tails in the class 3 mutant strains appeared more numerous than those in the wild-type strain. Furthermore, measurements of individual comet tail fluorescence intensities indicated a twofold increase in the mCherry fluorescence in class 3 mutant comet tails in comparison with wild-type comet tails. The localization pattern observed with class 3 mutant dynein was most similar to wild type and may indicate enhanced microtubule plus-end association at the expense of hyphal tip accumulation.
The three class 4 mutant strains displayed phenotypes where dynein accumulated distally as large “aggregates” starting ∼85 ± 20 µm back from the hyphal tips (Figure 1, A and B; Figure 3B). Interestingly, these aggregates were more or less evenly spaced, on average, 11 ± 5 µm from one another along the hyphae. At higher magnifications, these strains revealed the presence of “aster-like” dynein signals expanding from a central aggregate mass.
The remaining 12 DHC mutations in class 5 resulted in the “disperse” localization of dynein along the hyphae (Figure 1, A and B). In these strains, we saw diffuse dynein fluorescence of equal intensity throughout the hyphae (Figure 3B) that was reminiscent of what we observed with wild-type dynein in p150 deletion strains (Figure 1). We also observed a few punctate foci of dynein signal (Figure 1B).
To gain further insight into how particular DHC mutations could alter dynein function between different classes of DHC mutant strains, we examined the role of microtubules (Figure 2) and dynactin (Figure S4) in the altered dynein localization phenotypes. We tested the effect of dynein mutations with respect to microtubules by examining all the DHC mutant strains in a DIC-mCherry:tubulin-GFP background (Figure 2A; Table 1). In all classes of mutant strains, microtubules were less abundant in the apical regions in comparison to the wild-type strain, and the microtubules no longer showed the parallel arrangement seen in wild-type hyphae (Figure 2A). Prior studies that probed the effect of dynein disruption also observed altered microtubule dynamics and organization (Koonce et al. 1999; Riquelme et al. 2002; Uchida et al. 2008). In class 1 mutant strains, microtubules in the distal regions (where dynein was found) had a thicker appearance than microtubules in the wild-type strain. Similarly, in class 2 mutant strains, the apical microtubules along which mutant dynein were found appeared thicker than in the wild type and other classes of DHC mutant strains. The thickness of the microtubules in class 1 and 2 indicates that microtubule bundling may have occurred. Class 3 mutant strains had disorganized microtubule networks with the microtubules showing excessive curvature along the hyphal periphery. Class 4 mutant strains showed a unique aster-like microtubule pattern in regions away from the hyphal tip. Class 5 mutant strains had disorganized microtubule networks, and we observed many microtubules that were perpendicular to the hyphae in contrast to the normal parallel arrangement in wild-type hyphae.
We also found that the class 1, class 2, class 3, and class 4 mutant dynein localization patterns colocalized with subsets of visible microtubules (Figure 2A). The disperse distribution of dynein in class 5 DHC strains had no overlap with microtubules (Figure 2A), and the spots of dynein fluorescence present in the class 5 mutant strains were unaffected by benomyl treatment (Figure 2B). As expected for microtubule-dependent localization patterns, microtubule depolymerization with benomyl disrupted dynein localization to the long linear track, comet tail, and aggregate structures in mutant classes 1–4 strains (Figure 2B). After microtubule depolymerization, mutant class 1, class 2, and class 4 strains showed prominent punctate dynein signals that commonly overlapped with tubulin-GFP puncta. The presence of overlapping punctate fluorescence signals instead of diffuse signals as seen with wild type suggests that the mutant dynein altered microtubule-associated structures and remained bound to microtubule remnants in some way. In contrast, instead of comet tail localization, mutant class 3 strains showed diffuse dynein signals after benomyl treatment, suggesting complete dispersion of dynein in these strains. The variation in dynein dispersion upon microtubule depolymerization indicates that there is heterogeneity in how dynein is interacting with microtubules in the different mutant strains. Taken together, these data reiterate the importance of dynein function in microtubule organization and confirm our presumption that the dynein in mutant class 1, 2, 3, and 4 strains are indeed associated with microtubules.
We examined the DHC mutant strains in a DIC-mCherry:EGFP-p150 background to see if the mutations altered the localization of dynactin. In all classes of DHC mutant strains, the dynactin signal was disrupted and there was a loss of the bright cloud of accumulation at the hyphal tip (Figure S4A). In class 5 mutant strains, the dynein and dynactin signals were each diffuse; therefore it was impossible to determine the extent, if any, of colocalization between dynein and dynactin. In all other mutant strains, dynactin distribution and dynein distribution were indistinguishable (Figure S4A; Table 1).
To test the possibility that dynactin function is required for the five different localization patterns that we observed, we crossed representative strains from each of the five mutant heavy chain classes by the p150 deletion strain (Figure S4B). The distal linear track dynein localization pattern observed in class 1 strains was not altered by the loss of dynactin function, showing that this phenotype is dynactin independent. In contrast, the typical dynein localization phenotypes in class 2 and class 3 strains were abolished when those strains were examined in a p150 deletion background; instead, we observed the diffuse pattern normally found in class 5 strains. Interestingly, we found that the dynein localization pattern in class 4 strains was converted into a distal linear track phenotype similar to what was seen in class 1 strains. The diffuse dynein pattern observed in class 5 strains was not altered by the loss of dynactin function. These data indicate that dynactin has a complex role in the localization of dynein within cells.
Vesicle transport is altered in the DHC mutant strains
We examined the effect of the DHC mutations on vesicle transport in growing hyphae of N. crassa by utilizing the lipophilic styryl dye FM 4-64. In addition to representative strains from the five classes of DHC mutations, we also examined DHC deletion and dynactin p150 deletion strains as well as the RIP Nkin mutant strain utilized earlier. Colonies of wild-type and the various mutants strains were treated with FM 4-64, and hyphae at the colony edges were imaged with time-lapse microscopy to generate movies of endomembrane vesicle transport. The dye was quickly taken up by the hyphae into individual vesicles, and, in the majority of wild-type hyphae, many of the vesicles accumulated near the hyphal tip. Vesicle accumulation was more rarely observed in the hyphal body of wild-type strains. A similar pattern of vesicle accumulation at hyphal tips was seen in all the mutant strains except the p150 deletion strain, where instead we saw a reversal in the location of accumulated vesicles with a majority of hyphae showing vesicle accumulation in the hyphal body instead of the hyphal tip (Table S1).
The FM 4-64-positive vesicles underwent two different types of motility in all hyphae: cytoplasmic streaming that slowly propelled vesicles in unison toward the hyphal tip and individual motility events that were much faster and were directed in either an inward (toward the hyphal base) or an outward (toward the hyphal tip) trajectory. We generated motility kymographs of the time-lapse movies to determine the relative numbers, velocities, and displacements of the individual motility events for wild type and all the mutants strains (Figure 4). We did not see large differences in the percentage of inward or outward motility events in the DHC deletion, Nkin, or in any other mutant or wild-type strains (Table S1), as would be expected in a system where one direction of motor activity was lost when all the microtubules are aligned with the same polarity, as in an axon. This is likely due to the syncytial arrangement of scattered nuclei as foci for microtubule growth and organization in the hyphae, which means that microtubules that are not in the nuclear exclusion zone can be in either orientation (Freitag et al. 2004; Uchida et al. 2008). Similarly, we did not observe many differences between inward and outward transport in the wild-type or mutant strains (Table S1).
Figure 4 .
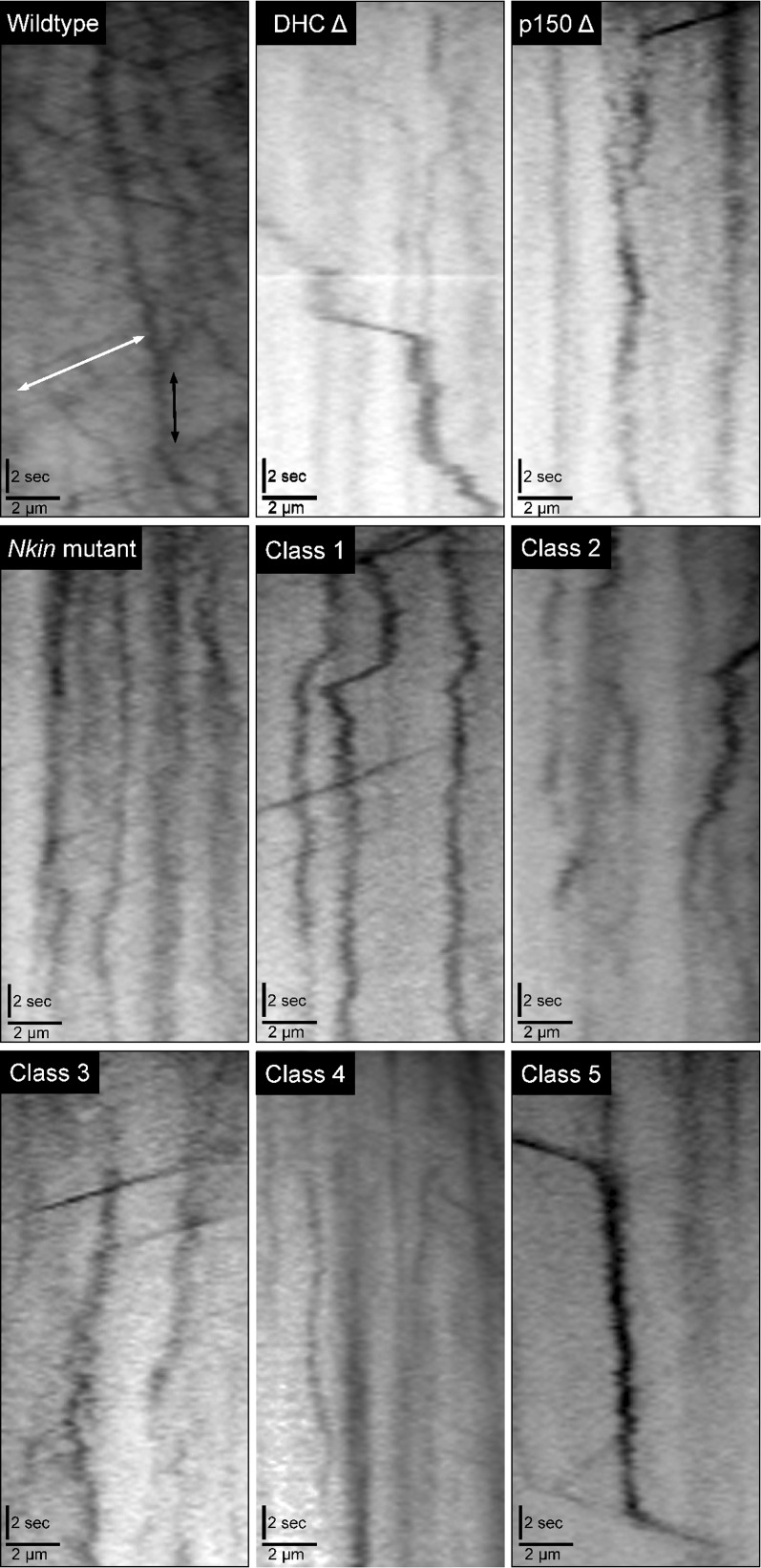
Kymograph analyses of vesicle transport in different strains. Representative kymographs of FM 4-64-positive vesicle transport are shown. All kymographs are oriented with the hyphal tip to the right, distance on the x-axis, and time on the y-axis. Motor-based movements are indicated by the higher velocity diagonal lines that are most abundant in the wild-type kymograph (white double-sided arrow). Cytoplasmic streaming causes the general back and forth drift of vesicles during the kymographs (solid double-sided arrow in wild type).
When we examined all of the motility events that occurred within active hyphae, we saw that there was a 30–80% decrease in the number of motility events that occurred in the mutant strains compared to the wild-type strain. Furthermore, we determined that all of the mutant strains had a significant defect in the velocity of the motility events (Table S1). Class 1 and class 5 strains did not show a significant decrease in the distance traveled by FM 4-64 vesicles, but all other mutant strains exhibited significant defects in vesicle displacement. To provide a single-term overall assessment of vesicle transport that occurred in the hyphae from the different strains, we generated a motility index that is a function of the number of movements and the velocity and distance of those movements (see Materials and Methods and Table S1). All five classes of DHC mutant strains, as well as the DHC deletion, p150 deletion, and Nkin mutant strains exhibited two- to fivefold decreases in their motility index with respect to wild-type strains. Overall, the FM 4-64 experiments illustrate that each of the five classes of DHC mutant strains exhibits transport phenotypes that include decreases in the motility index and in the number of vesicles being actively transported by microtubule motors.
Nuclear distribution in strains expressing wild-type and mutant DHC
The role of dynein in the migration, distribution, and positioning of nuclei in cells has been well established (Plamann et al. 1994; Inoue et al. 1998; Alberti-Segui et al. 2001; Duncan and Warrior 2002). N. crassa hyphae are multinucleate, and these nuclei are normally found randomly distributed along the length of the hyphae except for a clear nuclear exclusion zone (NEZ) within 25 μm of the hyphal tip (average 25 ± 5 µm, n = 20) (Figure 5, A and B). The NEZ is a common phenomenon observed in filamentous fungi and has been reported previously in N. crassa (Freitag et al. 2004; Ramos-Garcia et al. 2009).
Figure 5 .
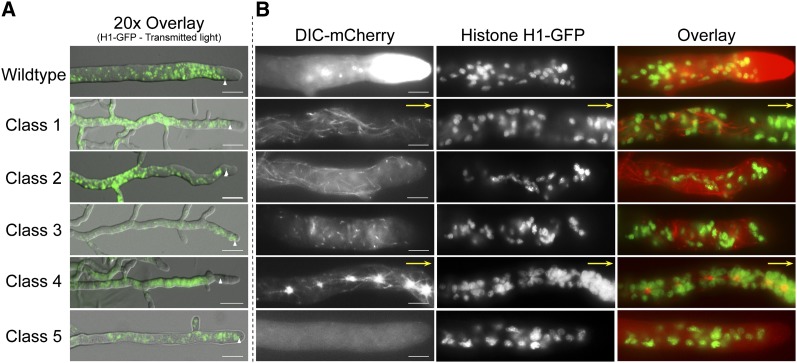
Nuclear distribution in wild-type and DHC mutant strains. (A) Intermediate magnification overlay images of hyphae from wild-type and DHC mutant N. crassa strains expressing histone H1-GFP. Upward pointing arrowheads indicate the position of the nucleus closest to the hyphal tip. Bars, 50 µm. (B) Epifluorescence images of hyphae from wild-type and DHC mutant strains expressing DIC-mCherry and histone H1-GFP. (Left panels) Dynein. (Center panels) Nuclei. (Right panels) Overlay. Nuclei are absent from the extreme tips of all hyphae and appear randomly dispersed in the remainder of the hyphae in wild-type, mutant class 1, 2, 3, and 5 strains. In class 4 mutant strains, there is abnormal clustering of nuclei around the aggregate dynein foci. Bars, 10 µm.
We examined the effect of DHC mutations on nuclear distribution in the mature hyphae of strains expressing DIC-mCherry and histone H1-GFP. Class 1, class 2, class 3, and class 5 mutant strains exhibited a reduction in the length of the NEZ (NEZ size for class 1 = 15 ± 5 µm; class 2 = 10 ± 4 µm; class 3 = 12 ± 5 µm; and class 5 = 11 ± 5 µm; n = 30 for all classes) (Figure 5A). It is highly likely that the altered microtubule organization observed with our DHC mutant strains contributed to NEZ size anomalies, as seen in another report (Ramos-Garcia et al. 2009). Although the NEZ of the mutant class 1, class 2, class 3, and class 5 strains was altered, nuclei in these strains were randomly distributed similar to wild-type strains (Figure 5). However, class 4 strains had a NEZ that was similar in size to that of wild-type strains yet the nuclei displayed dramatic clustering around the aggregates in distal regions with a few single nuclei extended into more apical regions up to the NEZ. This pattern suggests that the mutant dynein from class 4 strains may have a unique mechanism of interaction with both microtubules and nuclear cargoes. Our earlier experiments with class 4 mutations in combination with the loss of dynactin function also pointed out that class 4 strains behaved differently that the other classes of mutant heavy chain strains. Collectively, these observations indicate that the different DHC mutations affect nuclear positioning to varying degrees (Table 1).
Biochemical analyses of wild-type and mutant dynein motors
To determine if N. crassa dynein is functionally equivalent to dynein from other sources and to examine a possible mechanism of dynein mislocalization, we purified dynein from a wild type as well as a representative DHC mutant strain. We chose to study the E2675Q mutation (hereafter referred to as AAA3 E/Q) from a class 1 mutant strain that exhibits the distal long linear track microtubule-associated dynein localization phenotype. This mutant strain was chosen from among the other DHC mutations because (1) the mutation is located on the Walker B domain of AAA3 that is implicated in ATP hydrolysis on the basis of mutational analysis on various AAA family proteins (Whiteheart et al. 1994; Babst et al. 1998; Hartman and Vale 1999) and (2) previous studies have shown that the disruption of dynein AAA3 ATP-binding/hydrolysis functions can alter ATPase activity at AAA1, microtubule interaction, and motility properties of dynein (Silvanovich et al. 2003; Kon et al. 2004; Reck-Peterson and Vale 2004a,b; Cho et al. 2008). We sought to test if the cellular localization phenotype of this mutant strain is a reflection of such altered biochemical properties of dynein.
To study dynein motors from these strains, we developed a new purification protocol (Figure S5A) that did not rely upon nucleotide-dependent microtubule binding. This was because we expected that the AAA3 E/Q mutation would likely alter dynein mechanics in a way that would prevent efficient motor purification with such conventional approaches. Briefly, hexahistidine and Strep-tag II affinity tags were fused to the C terminus of the DIC by employing a strategy similar to DIC-mCherry generation. Dynein complexes were purified using this dual affinity tag (Figure S5B).
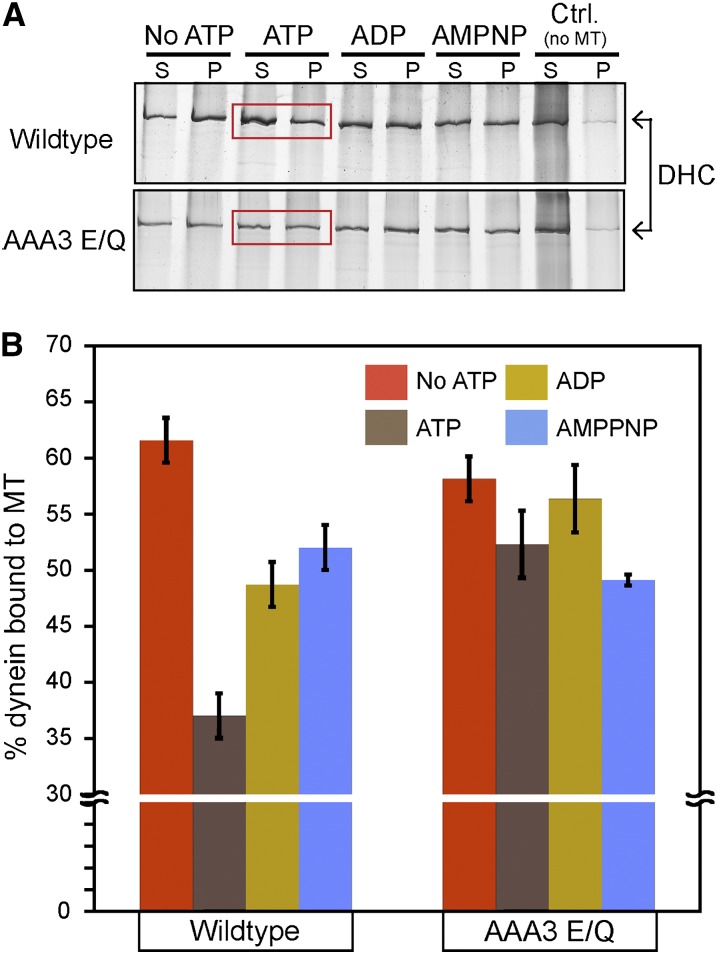
Dynein interaction with microtubules is normally modulated by different nucleotide conditions (Shpetner et al. 1988; Imamula et al. 2007; Mizuno et al. 2007). To determine if the AAA3 E/Q mutation affected this modulation, we examined the microtubule-binding behavior of wild-type and AAA3 E/Q dynein under varying nucleotide conditions using a microtubule-copelleting assay. As expected for wild-type dynein motors, the presence of 1 mM ATP resulted in a low percentage of dynein cosedimentation with microtubules (Figure 6, A and B). Under nucleotide-free, ADP, or AMPPNP conditions, we observed a significant increase in dynein cosedimentation with microtubules in comparison to the ATP condition (Figure 6, A and B). These observations are consistent with the nucleotide-sensitive microtubule-binding behavior exhibited by cytoplasmic dynein from other sources (Shpetner et al. 1988; Hays et al. 1994; Imamula et al. 2007). AAA3 E/Q dynein displayed a microtubule-dependent cosedimentation behavior similar to wild-type dynein under nucleotide-free, ADP, or AMPPNP conditions (Figure 6, A and B). However, in contrast to wild-type dynein, the presence of ATP did not significantly lower the cosedimentation of AAA3 E/Q dynein with microtubules (Figure 6, A and B). These results indicate that nucleotide-sensitive microtubule-binding behavior of N. crassa wild-type dynein is similar to microtubule-binding characteristics of cytoplasmic dynein from other sources and that the AAA3 E/Q mutation perturbs dynein so that its ability to bind microtubules is insensitive to the presence of ATP. The ATP-insensitive microtubule-binding ability of AAA3 E/Q is consistent with the rigor-like microtubule-binding reported for similar AAA3 mutants in previous studies (Silvanovich et al. 2003; Kon et al. 2004; Cho et al. 2008). The results from the AAA3 E/Q cosedimentation experiments suggest that the linear track phenotype observed in class 1 mutant strains may be a result of a rigor-like microtubule binding.
Figure 6 .
Effect of nucleotides on the cosedimentation of wild-type and AAA3 E/Q dynein with microtubules. (A) Representative silver-stained gel images showing microtubule cosedimentation of dynein isolated from wild-type and AAA3 E/Q strains under varying nucleotide conditions. Mixtures were centrifuged, and the supernatant (S) and pellet (P) fractions are shown. Regions boxed in red indicate dynein sedimentation in the presence of ATP. (B) Quantification of the percentage of dynein in pellet fraction in comparison to total dynein. Cosedimentation was quantified from three independent experiments. Data are shown as mean ± SEM. Wild-type dynein cosedimentation in the presence of 1 mM ATP was significantly less than wild-type dynein cosedimentation in the other conditions examined (t-test, P < 0.05). AAA3 E/Q dynein cosedimentation in the presence of ATP was not significantly different from that under the other conditions (t-test, P > 0.05)
Dynein has an intrinsic basal ATPase activity that is enhanced by the presence of microtubules (Shpetner et al. 1988; Silvanovich et al. 2003; Kon et al. 2004). We investigated the effect of the AAA3 E/Q mutation on both basal and microtubule-stimulated dynein ATPase activity. The basal ATPase activities of wild-type and AAA3 E/Q dynein motors were significantly different from one another (Table 2). The ATPase rate of wild-type N. crassa dynein is 115 nmol ATP/mg dynein/min and in range with reports that probed the ATPase activity of cytoplasmic dynein from mammalian sources (Ferro and Collins 1995; King and Schroer 2000; Mesngon et al. 2006). Surprisingly, the AAA3 E/Q dynein showed a basal ATPase activity that was approximately fourfold higher (Table 2). For both wild-type and AAA3 E/Q dynein, the addition of microtubules stimulated the ATPase activity approximately twofold in comparison to the corresponding basal activities (Table 2). This means that each motor sensed and responded to the presence of microtubules with a doubling in its own inherent ATPase rate. The enhanced ATPase rates (both basal and microtubule stimulated) observed with the AAA3 E/Q motor were unexpected on the basis of the nucleotide-insensitive microtubule-binding behavior that we observed (Figure 6, A and B).
Table 2 . ATPase activities of dynein isolated from wild-type and AAA3 E/Q strains.
| Dynein | Basal ATPase (nmol/min/mg dynein) | MT-stimulated ATPase (nmol/min/mg dynein) | Fold change |
|---|---|---|---|
| Wild type | 115 ± 7 | 201 ± 3 | ∼1.7 |
| AAA3 E/Q | 418 ± 35 | 729 ± 60 | ∼1.7 |
Basal and microtubule-stimulated ATPase activities are expressed as mean ± SD derived from three independent experiments. The basal and microtubule-stimulated ATPase rates are significantly different from each other in both wild-type and AAA3 E/Q dynein (t-test, P < 0.05).
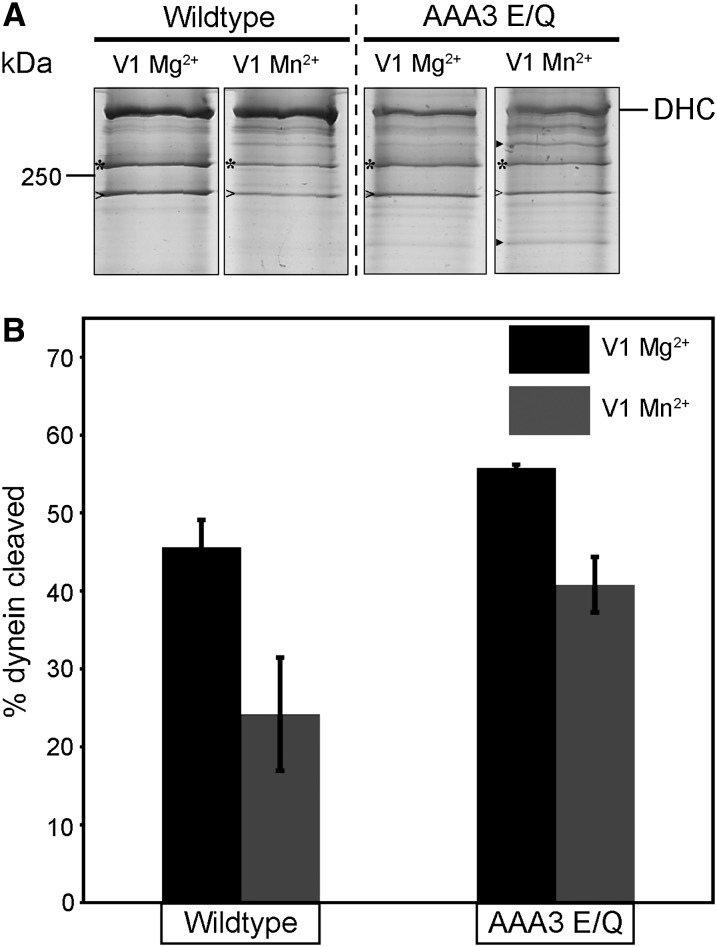
Dynein undergoes a signature, site-specific UV photolysis induced by vanadium ions (VO4)—a structural analog of inorganic phosphate (Gibbons et al. 1987). Vanadate-mediated photolysis has been considered a defining characteristic of dynein and can be applied as a tool to probe the conservation of active site (ATPase) conformation and any alterations to this conformation (Grammer et al. 1988; Gibbons and Mocz 1991). We performed vanadate-mediated photolysis experiments on N. crassa dynein and found that the wild-type dynein was susceptible to vanadate-mediated photocleavage using a standard Mg2+ condition (V1 Mg2+) (Figure 7A). Quantification of the cleavage products showed that almost 50% of the total dynein was cleaved (Figure 7B). When we substituted Mn2+ instead of Mg2+ under V1 conditions (V1 Mn2+), wild-type dynein vanadate cleavage was reduced approximately twofold in comparison to the V1 Mg2+ condition (Figure 7, A and B). The reduction in cleavage upon the substitution of the coordination metal (Mg2+) is in line with observations from previous studies (Gibbons et al. 1987). Collectively, these observations demonstrate that the structural features responsible for V1 site cleavage are conserved between N.crassa dynein and dynein studied from other sources (Gibbons et al. 1987; Schnapp and Reese 1989; Gatti et al. 1994; Hays et al. 1994; Allan 1995; Kon et al. 2004).
Figure 7 .
Effect of divalent metal ion on the vanadate-mediated V1 photocleavage pattern of wild-type and mutant dynein. (A) Representative silver-stained gel images showing vanadate photocleavage of DHC from wild-type (left panels) and AAA3 E/Q (right panels) under V1 Mg2+ and V1 Mn2+ conditions. High- and low-molecular-weight UV cleavage products are indicated by “*” and “>”, respectively. Additional prominent cleavage products found under AAA3 E/Q V1 Mn2+ conditions are indicated by “►”. (B) Quantification of the percentage of dynein cleaved in comparison to the total dynein in the sample. Percentage cleavage was quantified from three independent experiments. Data are shown as mean ± SEM.
AAA3 E/Q dynein showed a V1 Mg2+ cleavage pattern similar to wild-type dynein (Figure 7, A and B). In contrast, under V1 Mn2+ conditions, AAA3 E/Q dynein did not show as great a reduction in cleavage (Figure, 7 A and B). In addition, there was cleavage of AAA3 E/Q dynein at a second site under V1 Mn2+ conditions. The alteration of AAA3 E/Q dynein V1 Mn2+ cleavage patterns indicates that the AAA3 E/Q mutation alters the active site conformation of dynein. The alteration in vandate cleavage seen for AAA3 E/Q is related to the elevated ATPase activity that we observed (Table 2).
Next, we probed the motility properties of N. crassa wild-type and AAA3 E/Q dynein using an in vitro microtubule-based bead motility assay. Beads coated with wild-type dynein exhibited motility as seen by the directional motion of the beads on microtubules. Wild-type dynein moved with an average velocity of 0.7 µm/sec (Table 3), which is consistent with the dynein velocities observed in previous studies (King and Schroer 2000; Toba et al. 2006; Ori-McKenney et al. 2010). The mean run length of wild-type dynein (Table 3) was well within the range of in vivo dynein-based motility events (Ma and Chisholm 2002; Pilling et al. 2006; Ori-McKenney et al. 2010). When we examined beads with AAA3 E/Q dynein, we first noted enhanced binding of beads to microtubules in our motility chambers. Further analyses showed that none of these bound motors moved along microtubules. This inability to move is consistent with the in vivo microtubule decoration phenotype (Figure 1B; Figure 2A) and the nucleotide-insensitive microtubule-binding phenotypes (Figure 6, A and B).
Table 3 . In vitro motility characteristics of beads coated with dynein isolated from wild-type and AAA3 E/Q strains.
| Dynein | MT binding | Motility | Velocity (µm/sec−1) | Distance (µm) | n |
|---|---|---|---|---|---|
| Wild type | + | + | 0.7 ± 0.3 | 2.6 ± 1.7 | 104 |
| AAA3 E/Q | +++ | — | 0.0 | 0.0 | 155 |
Velocities and distances are expressed as mean ± SD; n = number of events tracked.
Our studies were the first to examine the AAA3 Walker B E/Q mutation in a whole dynein molecule, and our results are in general agreement with previous studies that showed increased MT-binding affinity and altered dynein localization in fruit fly and budding yeast (Silvanovich et al. 2003; Cho et al. 2008; Markus et al. 2009). For example, one study showed that UV-vanadate cleavage occurred at levels equal to wild-type strains (Silvanovich et al. 2003). Unfortunately, that study did not have the resolution to determine if the cleavage products included altered species such as we identified (Figure 7A). In contrast to our work, the ATPase activity of tail-less artificially dimerized yeast AAA3 E/Q dynein was reduced relative to wild type (Cho et al. 2008) whereas we saw an increase in the ATPase activity of purified native molecules (Table 2). This same report indicated that the artificially dimerized yeast AAA3 E/Q dynein exhibited extremely slow motility (∼0.0046 μm/sec) along axonemes while we failed to see any appreciable motility for AAA3 E/Q dynein along microtubules. The differences between our data and some of the yeast dynein data may reflect either the use of tail-less motors (Cho et al. 2008; Markus et al. 2009) or that yeast dynein is known to act quite differently from fungal and higher-order organisms. In yeast, dynein does not transport endomembrane vesicles inside cells and instead is predominantly attached to the cell cortex for nuclear-positioning roles (Carminati and Stearns 1997; Cottingham and Hoyt 1997; Sheeman et al. 2003; Markus et al. 2009).
The properties exhibited by N. crassa AAA3 E/Q dynein, such as the nucleotide-insensitive microtubule binding, enhanced ATPase activity, and inability to translocate along microtubules are analogous to characteristics observed with kinesin and myosin motors carrying uncoupling mutations (Ruppel and Spudich 1996; Song and Endow 1998; Brendza et al. 2000; Heuston et al. 2010). Previous studies have shown that uncoupling mutations are able to disengage a motor’s intradomain coordination mechanisms in a way that results in constitutive cytoskeletal filament binding as well as enzymatic and motility behaviors that are independent of each other (Ruppel and Spudich 1996; Song and Endow 1998; Brendza et al. 2000; Heuston et al. 2010). Our biochemical analyses of AAA3 E/Q dynein are consistent with those analogous studies and strongly suggest that the AAA3 E/Q mutation is an uncoupling mutation. Taken together, our results indicate that the N. crassa wild-type dynein exhibits motility properties comparable with dynein from other sources and that the AAA3 E/Q mutation uncouples the critical dynein motor functions of microtubule interaction and nucleotide hydrolysis.
Mapping of the DHC mutations onto the crystal structure
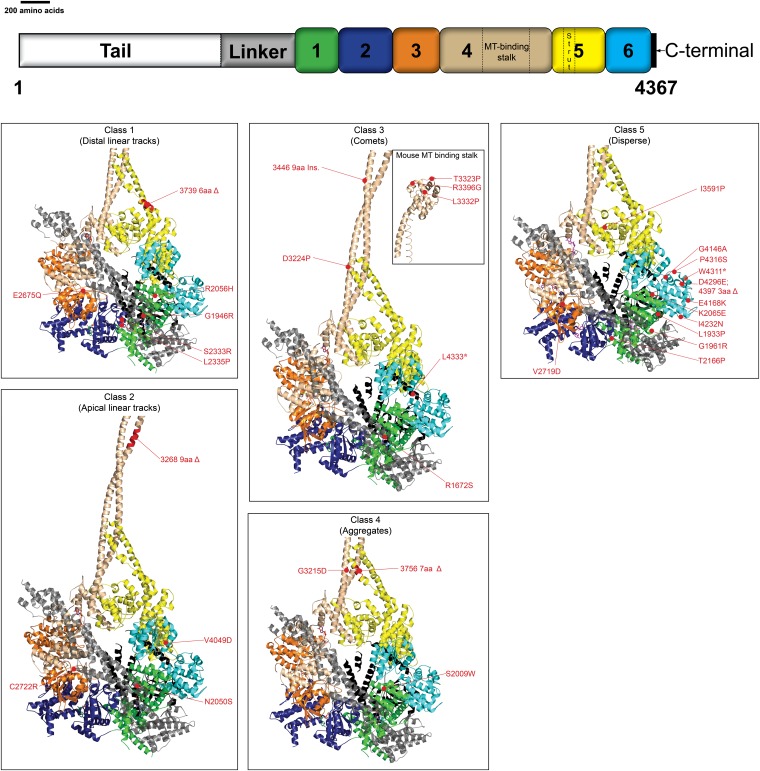
Recent crystal structures of the dynein heavy chain have shown many interesting features of the dynein molecules, including how the six AAA domains of the dynein ring are arranged with respect to each other, how the linker domain arches over the top of the motor ring, how the microtubule-binding domain (MTBD) stalk and its supporting strut interact at a specific interface, and how multiple regions of the AAA domains may be able to communicate nucleotide status and hydrolysis information to neighbor rings (Carter et al. 2011; Kon et al. 2011; Kon et al. 2012; Schmidt et al. 2012). These same structures provided us with the opportunity to map our mutations onto the DHC crystal structure (Kon et al. 2012) and to a portion of the microtubule-binding stalk closer to the MTBD (Carter et al. 2008) to gain a better understanding of the structure/function relationships within the DHC. Our intention was to obtain a glimpse into how different motor domains may be utilized for particular dynein functions. The structural mutation map (Figure 8) shows some intriguing correlations between the positions of the five classes of mutations and the dynein mislocalization phenotypes of those classes.
Figure 8 .
Mapping of DHC mutations onto Dictyostelium DHC crystal structure. Mapping of DHC mutations onto the crystal structure of Dictyostelium discoideum DHC [Protein Data Bank (PDB) ID 3VKG] and microtubule-binding domain from mouse cytoplasmic dynein (PDB ID 3ERR). Different structural domains of the motor are color-coded as in the linear representation shown at the top. The motor is shown as a ribbon molecular representation and visualized from the linker face. Each panel shows the position of a single class of DHC mutations. Mutation locations (red spheres) are labeled. Mutations were located on the basis of sequence alignment between the DHC sequences of N. crassa (XP_962616.1) and the corresponding D. discoideum DHC sequence retrieved from PDB. Ribbon representations were generated using PyMOL (http://www.pymol.org).
Class 1 mutations:
The six class 1 (distal linear tracks) mutations were mapped to the p-loop of AAA1 (G1946R), helix 4 (H4) of AAA1 (R2056H), the pre-sensor 1 insert of AAA2 (S2333R, L2335P), Walker B of AAA3 (E2675Q), and AAA5 strut coil 1 (3739 6-aa Δ) (Figure 8; Table S1). The AAA1 p-loop (G1946R) and AAA3 Walker B (E2675Q) mutations have been reported to inhibit ATP binding at AAA1 and ATP hydrolysis at AAA3, respectively (Silvanovich et al. 2003; Kon et al. 2004; Cho et al. 2008). The R2056H mutation is present in the second region of homology (SRH) domain, which has been implicated in sensing nucleotide status as well as in the stabilization of the transition state of ATP hydrolysis (Ogura et al. 2004). The S2233R and L3335P mutations are located on the pre-sensor I insert of the AAA2. Disruption of the pre-sensor I–linker interactions alters ATP-sensitive microtubule interaction as well as linker motions (Kon et al. 2012), and pre-sensor I insert mutations have been reported to affect ATPase kinetics (Burrows et al. 2009). The strut region has been proposed to relay rigid body motions between AAA4 and AAA5 domains into shear motions between the helices of the stalk coiled-coil. Therefore, it seems likely that the six-amino-acid deletion in the strut mutation (3739 6-aa Δ) could interfere with the normal communication of nucleotide status between AAA1 and the microtubule-binding domain. The strut mutation 3739 6-aa Δ is intriguing in that a similar alteration of the strut structure (Kon et al. 2012) results in a microtubule-independent, elevated ATPase activity similar to that observed in the AAA3 Walker B (E2675Q) mutation in our study. A common theme that can be seen in all six of these class 1 mutations is that they are each located in an area that is intimately involved in the sensing of nucleotide status, hydrolysis of nucleotides, or communication of information about nucleotide status.
Class 2 mutations:
The four class 2 (apical linear track) mutations are located proximal to the sensor 1 of AAA1 (N2050S) and AAA3 (C2722R), in coiled coil 1 of the AAA4 microtubule-binding stalk (3268 9-aa Δ) and H1 of the AAA6 domain (V4049D) domain (Figure 8; Table S1). Not much can be inferred from the N2050S and C2722R mutations. However, sensor 1, which is a part of the SRH domain, has been reported to contact the Walker B domain and the γ-phosphate of the ATP and has been proposed to act as a switch in mediating conformational changes (Davies et al. 2008). Mutations in the sensor 1 region in AAA family members have previously been shown to reduce the rate of ATP hydrolysis (Song et al. 2000; Hattendorf and Lindquist 2002). This suggests that the apical microtubule decoration phenotype observed in these strains may be caused by a decrease in the ATPase rates of the motor without the dramatic increase in the microtubule-binding behavior of dynein seen with class 1 mutations. A reduction in the ATPase activity could lead to significantly increased dwell times of dynein on microtubules (Figure 2A) and, by moving more slowly, could give rise to the observed apical microtubule mislocalization phenotype.
Class 3 mutations:
Of the nine class 3 mutations (comet tails), two were located in the tail domain where structural data are not available (Y110S, W1308G), one was found in H11 of the linker (R1672S), one was at the extreme C terminus of the molecule (L4333*), and the remaining five (D3224P, T3323P, L3332P, R3396G, and 3446 9-aa insertion) were present in the coiled coils or putative microtubule-binding interface of the microtubule-binding domain (Figure 8; Table S1). It is easy to envision that alterations in the putative microtubule-binding interface or in the coiled-coils that connect and control the register of the interface to the rest of the dynein molecule could alter the binding of dynein to microtubule ends, enhancing the comet tails of dynein. For example, the MTBD mutations could (1) alter dynein–microtubule interaction mechanisms at the interaction interface and/or (2) alter the helix-sliding or stalk-tilting mechanisms of allosteric communication (Carter et al. 2008; Kon et al. 2009, 2012). Although the tail domain is known to be involved in the homodimerization of DHCs as well as interaction with DIC, DLIC, and Lis1 (Pollock et al. 1998; Habura et al. 1999; Tynan et al. 2000; Tai et al. 2002; Ori-McKenney et al. 2010; Markus and Lee 2011), there is no structural information that can help us decipher how the Y110S or W1308G mutations alter dynein activity and cause class 3 mutant dynein mislocalization. In contrast, the linker domain (where the R1672S mutation is located) is known to make extensive contacts with AAA2 (Kon et al. 2012) and AAA5 (Schmidt et al. 2012) of the motor domain, and the L4333* mutation in the C terminus is nestled near AAA1. It is possible that, even though long distances occur between the tail, linker domain, or C-terminal and the MTBD, the class 3 mutations outside the MTBD may alter propagation of signals to or from the MTBD stalk, leading to an altered microtubule interaction behavior similar to that seen in the remaining MTBD class 3 mutations.
Class 4 mutations:
The three class 4 (aggregate) mutations were found in H3 of the AAA1 domain (S2009W), coiled-coil 1 of the stalk (in an invariant glycine, G3215D), and the strut coil 1 (3756 7-aa Δ) (Figure 8; Table S1). Two of these mutations are located in positions at an interface between the stalk and the strut (Carter et al. 2011; Kon et al. 2011, 2012; Schmidt et al. 2012). The strut/stalk interface is an ideal location for a mutation to lock the conformation of the dynein. The location of the S2009W AAA1 domain mutation between AAA1 Walker B and the pre-sensor 1 insert suggests that this region of AAA1 communicates with or relays information through the strut/stalk interface to the MTBD. Additionally, only 12 amino acids within strut coil 1 separate the 3756 7-aa Δ class 4 mutation from the 3739 6-aa Δ class 1 mutation. The identification of two different dynein localization phenotypes by strut mutations located so close to each other suggests that the strut can be altered to disrupt different dynein motor functions.
Class 5 mutations:
The 12 class 5 (diffuse) mutations were located throughout the primary structure of the dynein motor (AAA1, AAA3, AAA5, and AAA6) (Figure 8; Table S1). However, the structural location of these mutations in three-dimensional space is remarkable as 10 of the 12 are near each other, with most of those occurring at the interfaces of AAA1 and AAA6 (Figure 8; Table S1). The locations of these mutations within the AAA domains do not provide further information on the potential relationship to alterations in ATP binding, hydrolysis, the relay of information, or other dynein functions.
Relationships between different classes of mutations
Examining the positions of the DHC mutations in relation to each other, it is possible to see that there is a range of possible relationships that can exist between mutations in the same or similar structures of the DHC. There are examples of mutations in the same structure having similar phenotypes (the majority of the MTBD mutations have class 3 phenotypes; S2333R and L2335P in the pre-sensor 1 insert of AAA2 have class 1 phenotypes). There are also examples of similar regions from different AAA domains having identical phenotypes. Mutations in helix 0 of AAA1 (L1933P) and AAA5 (I3591P) each have class 5 phenotypes, and mutations near sensor 1 of AAA1 (N2050S) and AAA3 (C2722R) each have class 2 phenotypes. In contrast, we also found examples of mutations in the same structure of the same domain having different phenotypes. We identified two mutations where the defect lies just proximal to sensor 1, yet the mutations can cause either class 2 (C2722R) or class 5 (V2719D) phenotypes. Similarly, we identified two mutations that were each in-frame deletions of the strut region yet resulted in either class 1 (3756 7-aa deletion) or class 4 (3739 6-aa deletion) phenotypes. We also note that the phenotypes that we observed in the N. crassa AAA3 E/Q strain are not similar to the phenotypes observed in the corresponding Aspergillus nidulans AAA1 E/Q strain (Zhang et al. 2010). Surprising results like these will be important reminders of the complexity of dynein motors as comparisons are made between studies in different organisms and/or different AAA domains.
Discussion
In this study of dynein function in N. crassa, we utilized a fluorescent imaging approach to observe wild-type dynein in growing polarized hyphae, we performed initial characterizations of >30 DHC mutant strains, and we used biochemical analyses to analyze in detail one of those mutant strains. The combination of these approaches has provided us with multiple new insights into dynein function.
Wild-type dynein accumulated most prominently at the extreme tip of the hyphae; the dynein signal dropped as a gradient at positions farther away from the tip. This accumulation most likely represents a reservoir of dynein being retained at the hyphal tip for future cargo transport. In addition, wild-type dynein was also found on the ends of microtubules near the hyphal apex in the classic comet-tail structures described in a variety of past studies (Xiang et al. 1995, 2000; Vaughan et al. 1999; Han et al. 2001; Ma and Chisholm 2002; Zhang et al. 2003, 2010, 2011; Lenz et al. 2006; Arimoto et al. 2011; Markus et al. 2011; Schuster et al. 2011a).
By examining wild-type dynein in the absence of kinesin or dynactin function, we were able to determine how those factors interact with dynein in N. crassa. We found that kinesin and dynactin have different roles that affect dynein’s ability to accumulate at hyphal tips and comet tails. The loss of kinesin function resulted in a diffuse dynein presence in distal regions of the hyphae with a concomitant absence of dynein in apical regions. Our data with defective kinesin strains show that dynein is actively recruited to the hyphal tips by the action of kinesin motors and that the absence of kinesin prevents dynein from effectively getting to the hyphal tip region. This interpretation of our data is in agreement with work from others that illustrates an essential role for kinesin in the plus-end accumulation of dynein (Brady et al. 1990; Echeverri et al. 1996; Waterman-Storer et al. 1997; Martin et al. 1999; Vaughan et al. 1999; Duncan and Warrior 2002; Januschke et al. 2002; King et al. 2003; Zhang et al. 2003; Ligon et al. 2004; Theiss et al. 2005; Lenz et al. 2006; Arimoto et al. 2011).
The loss of dynactin function in a wild-type DHC strain resulted in a subtly different phenotype where diffuse dynein signal was present throughout both apical and distal regions of the hyphae. Our data support a model where dynein motors can be transported by kinesin to the hyphal tip in the absence of dynactin function (as seen by the presence of diffuse dynein signal along both apical and distal regions of the hyphae), but the dynein motors cannot then accumulate or interact with cargoes or microtubule ends in the absence of functional dynactin (Quintyne et al. 1999; Quintyne and Schroer 2002). Double-mutant analyses with the dynactin p150 deletion allele in combination with each of the five classes of DHC mutations were able provide more information about the role of dynactin in dynein function and to show differences between the five classes of DHC mutations. We found that DHC strains with class 1 mutations (distal microtubule tracks) had no change in dynein localization patterns in the absence of dynactin. On the basis of the data that we have from in vitro characterizations of one class 1 mutation (E2675Q), it seems likely that the localization of class 1 DHC mutant dynein motors is a result of direct but aberrant binding of the mutant motors to microtubules in distal regions of the hyphae and is independent of dynactin function. In contrast, dynein localization in DHC strains with class 2 (apical microtubule tracks) and class 3 (comet tails) mutations altered to a diffuse class 5-like phenotype in combination with a mutation deleting dynactin p150. The conversion into the class 5 phenotype could be a result of several different mechanisms, including (1) epistasis of the dynactin deletion to the class 2 and class 3 mutations and/or (2) a necessary interaction between mutant dynein and dynactin to exhibit the apical microtubule track and comet-tail phenotypes. Furthermore, the alteration of class 2 and class 3 phenotypes into class 5 diffuse staining presents the possibility that an underlying defect in class 5 mutant heavy chain strains is an inability of dynein molecules to effectively interact with dynactin. Finally, the class 4 DHC mutation mislocalization of dynein to large aggregates was altered and became identical to the class 1 linear track phenotype in combination with the dynactin deletion allele. This again raises the possibility that the interaction between dynein and dynactin is altered in class 4 mutations.
Our screen of dynein heavy chain mutations was quite successful at identifying specific amino acid changes associated with altered dynein activity. The mutant alleles examined in this study all resulted in nonlethal colony growth phenotypes (classic ropy phenotypes) as seen in a variety of recessive dynein mutations (Plamann et al. 1994; Bruno et al. 1996; Tinsley et al. 1996; Minke et al. 1999). Altogether, we were able to examine the cellular phenotypes of more independent mutant dynein alleles than any other previous screen of any other organism. Factors that aided our ability to identify such a large number of mutant dynein alleles in N. crassa include the haploid genome, the nonlethal nature of the mutations, and the particular temperature-sensitive screen utilized (Plamann et al. 1994; Bruno et al. 1996; Tinsley et al. 1996).
All the ropy phenotype DHC mutant strains examined in this study demonstrated two common features: a loss of the cloud of dynein that normally accumulated at the hyphal tip and defects in endomembrane transport. We propose that the cloud of dynein acts as a reservoir and that the loss of such a pool in ropy strains is a useful cellular indicator of altered dynein function. In addition to the loss of dynein tip accumulation, all of the DHC mutant strains exhibited defects in the transport of FM 4-64-labeled endomembrane vesicles. Defects in vesicle transport are likely the direct cause of the ropy-altered hyphal growth phenotype seen in our mutant strains because the wild-type pattern of straight hyphal growth is dependent upon the correct localization of the particular Spitzenkörper “organelle” and correct vesicle transport to and from that organelle during hyphal growth (Bartnicki-Garcia et al. 1995; Reynaga-Pena et al. 1997; Riquelme et al. 2000; Harris 2009). Our DHC mutant strains also exhibited varying degrees of altered microtubule networks and displayed one of five distinct classes of dynein mislocalization phenotypes. The observation that there are multiple mislocalization phenotypes in the collection of DHC mutations suggests that there must be at least five distinct ways that dynein function can be perturbed yet still impart ropy colony morphology. The range of mutant mislocalization phenotypes observed in conjunction with the number and placement of DHC mutations (Figure 1; Figure 8; Table 1) was initially surprising, as this phenotypic diversity had not previously been described for dynein. However, the complexity of the results from our study appears quite similar to the initial genetic and cell biological studies that investigated key cellular processes such as the cell cycle (Hartwell et al. 1974) or secretory pathway (Novick et al. 1981). In these two classic studies, large numbers of mutant alleles were required to point out and define discrete steps in those respective pathways. Our study is different in that all the mutant alleles occur within the same polypeptide. However, further analysis of the location of specific classes of the mutations within the dynein three-dimensional structure (Figure 8) provides evidence that defects in particular dynein substructures may manifest in the entrapment of dynein to specific substeps of the dynein mechanochemical cycle, with a concomitant entrapment of the mutant motors to one step of the intracellular dynein transport cycle. The complex interactions between specific dynein heavy chain mutations, structural domains of the dynein motor, and the cellular function of dynein begin to be revealed by analyses of the five classes of DHC mutant strains.
Class 1 mutant strains
We understand more about the class 1 DHC mutant strains than the other classes because we have in vitro data from purified motors in addition to the in vivo studies that were performed on all the mutant classes. In all class 1 mutant strains, we found that the mutant dynein molecules were mislocalized to microtubules in regions distal to the hyphal tip. In addition, we purified and performed several in vitro characterizations of a representative class 1 mutation, AAA3 E2675Q. The range of phenotypes that we saw with the AAA3 E2675Q mutant strain (mutant dynein bound to distal microtubule in vivo, enhanced binding of dynein to microtubules in vitro, uncoupling of ATPase activity from microtubule binding, and lack of motility) are all consistent with each other and with the possibility that the AAA3 E/Q mutant dynein may actually be cycling through the ATP hydrolysis cycle while bound in a rigor-like association with microtubules (Figure 2A; Figure 6, A and B).
We speculate that this type of rigor complex with microtubules is what causes the mislocalization of dynein molecules to distal microtubules in class 1 mutant strains. Normally, we would expect that kinesin is preferentially active compared to dynein in these distal regions, as has been described for many cargoes (Valetti et al. 1999; Gross et al. 2002; Fridolfsson and Starr 2010; Encalada et al. 2011). We believe this transport by kinesin is what generates the large reservoir of dynein at the hyphal tip. In the absence of kinesin function, we saw that wild-type dynein tip accumulation was abolished (Figure 1). Furthermore, there was actually a clear zone in the apical regions of the hyphae in kinesin-deficient strains that ended at approximately the same distance back from the tip as we began to see class 1 mislocalized dynein (Figure 1). It seems reasonable that, instead of being transported by kinesin, altered AAA3 E/Q motors could bind microtubules in a rigor state with sufficiently high affinity to prevent kinesin-based transport. The binding of the mutant class 1 motors to microtubules is not accomplished through dynactin, as the loss of dynactin function had no effect on that phenotype (Figure S4B). This uncoupled binding to microtubules could effectively trap dynein, leading to the long linear track patterns that we observed and blocking progression of the dynein motors to the hyphal tip. If this model is true, then dynein from all class 1 mutant strains should have increased microtubule binding that could override the normal cycling of dynein from distal regions to the hyphal tip.
Class 2 mutant strains
In class 2 DHC strains, we found that the mutant dynein motors were persistently mislocalized to microtubules present in apical regions of the growing hyphae. Even in newly elongated hyphae, these microtubule tracks are found spanning the regions (∼75 μm) closest to the tip, suggesting that dynein molecules in these strains retain a mechanism for getting to the apical regions. We propose that the dynein molecules in these class 2 mutant strains were delivered to the hyphal tips via kinesin, were trapped after binding to microtubules in apical regions, and became a dynein sink in these strains as the hyphae continued to grow. This dynein behavior is in contrast to the long linear tracks in distal regions observed in the class 1 mutant strains such as AAA3 E/Q where it seems most plausible that the dynein molecules were never delivered to the apical regions by kinesin. The similarity in microtubule localization phenotypes between class 1 and class 2 mutant strains (both being preferentially bound to microtubules in specific regions of the hyphae) makes it initially appealing to suggest that the uncoupling phenotype of AAA3 E/Q may manifest in some similar way in the class 2 mutant strains. However, in contrast to the class 1 mutant strains, the binding to microtubules by class 2 mutant dynein was dependent upon the presence of dynactin, as seen by the complete loss of microtubule binding by the class 2 mutant dynein in a p150 deletion double-mutant strain (Figure S4B). Furthermore, the mapping of two of the class 2 mutations to sites that are expected to significantly lower ATPase activity when altered suggests that a significant decrease in ATPase activity, and maybe a significantly slower motor, may be the root cause of the apical microtubule-binding phenotype in growing hyphae.
Class 3 mutant strains
The class 3 mutant DHC strains were the only DHC mutant class where dynein was mislocalized to structures or regions where it was normally found in wild-type strains; in this case, we found an overabundance of dynein in comet-tail structures. There is a strong correlation between class 3 phenotypes and mutations in the MTBD as five of the seven mutations that occur within the dynein MTBD result in class 3 dynein phenotypes and five of the nine class 3 strains have defects in the MTBD. On the basis of that correlation, it seems reasonable to expect that further characterization of the class 3 strains will show that dynein motors from the mutant strains have a particular alteration in how the motors bind to microtubule ends, somehow enhancing that interaction at the expense of other dynein functions. Furthermore, we expect that the enhanced binding to microtubule plus ends will not utilize a rigor-like binding similar to what we characterized in class 1 E2675Q mutant dynein. Several studies have illustrated that the normal dynein localization to growing plus ends of microtubule involves a complicated interplay of a variety of cellular proteins, of which dynactin is one member (Gouveia and Akhmanova 2010). The absolute requirement for dynactin function in the class 3 mislocalization phenotypes (Figure S4B) indicates that any putative alterations of the mutant class 3 dynein motors that could enhance microtubule-end binding likely act in some way through, or in concert with, dynactin. Another possibility that we favor less is that an essential role of dynactin that precedes dynein’s binding to microtubule ends is disrupted in the dynactin p150 double-mutant strains. In either case, the binding of dynein to microtubule ends is likely an initial step in the transport process as data from recent studies in Ustilago maydis have shown that endosomal cargoes recruit dynein motors located in the comet tails rather than dynein from the soluble pool (Schuster et al. 2011a,c).
Class 4 mutant strains
The discovery that two of the three class 4 mutations occur at the strut/stalk interface is very exciting, as the presence of the strut is one of the most intriguing features of the recently solved dynein structures (Carter et al. 2011; Kon et al. 2011, 2012; Schmidt et al. 2012). With the long-distance communication that has to occur within dynein, it is possible to see how even minimal alterations of the strut/stalk interface could have profound effects on conformation changes within the motor. The three class 4 mutations all caused the formation of massive complex assemblies of dynein, dynactin, nuclei, and aster-like microtubule structures into characteristic aggregate foci. The stellate patterns of nuclei around the aster-like microtubule network were quite striking and suggested that the nuclei were bound to the microtubules that were themselves cross-linked into aster-like arrays. Both microtubule motors (dynein and kinesin) can cause microtubule aster formation in vivo and in vitro under specific conditions (Verde et al. 1991; Vaisberg et al. 1993; Gaglio et al. 1996; Heald et al. 1996; Inoue et al. 1998; Nilsson and Wallin 1998). All of these previous studies point to a few key necessities for motor-dependent aster formation, such as (1) accumulation of motors to one of the microtubule ends and (2) motor-dependent cross-linking and sliding of the microtubules. The double-mutant analysis with class 4 and dynactin mutations was of special interest as this experiment showed that dynactin was required to make the aggregates yet the motors retain microtubule binding in the absence of dynactin. In addition, on the basis of the analogy to the rigor-like binding of class 1 E2675Q mutant dynein, we expect that the class 4 mutations are able to bind tightly to microtubules in a comparable manner to class 1 mutant dynein, albeit with the added ability to form the aggregate structures in combination with dynactin.
Class 5 mutant strains
The presence of class 5 mutant dynein molecules throughout the entire hyphal region suggested that dynein was not able to interact with some essential partner molecule. We saw similar patterns with wild-type dynein if we removed dynactin (via mutation) or microtubules (via the depolymerization drug benomyl). On the basis of the similarities found in those three conditions, the diffuse pattern of dynein in class 5 strains could reasonably be due either to a loss in the ability to bind dynactin or to a loss in the ability to bind microtubules. If the diffuse dynein localization pattern in class 5 mutant strains occurs because dynein molecules in those strains are unable to bind microtubules, then the inability to bind microtubules could potentially prevent kinesin-mediated apical transport of dynein, plus-end comet-tail binding of dynein at the apical tip, and transport of cargoes away from the hyphal tip by dynein. Thus, dynein molecules that were unable to bind to any microtubule-based cytoskeletal structures would be left to diffuse freely in the cytosol, as we see for class 5 mutant strains. However, dynein can bind to dynactin, and dynactin can itself bind microtubules, which would seem to indicate that even in a loss of direct dynein/microtubule interactions, dynein would still be able to interact with microtubules via dynactin. An alternative possibility is that the class 5 mutations alter the ability of the dynein motor to effectively interact with dynactin. First, the loss of dynactin caused wild-type dynein motors to lose their normal accumulation patterns and to become diffuse through the hyphae. Second, the loss of dynactin did not alter the diffuse dynein pattern in class 5 dynactin p150 double-mutant strains. Third, the class 2 and class 3 mutant strain dynein phenotypes were altered into the diffuse pattern in the absence of dynactin in the double-mutant experiments. In either case, the mechanism for class 5 alterations to dynein function appears to be centered around the AAA1 and AAA6 interfaces, as seen by the preponderance of mutation in those regions.
Future work will be needed to determine the exact mechanisms by which these specific mutations in the dynein heavy chain disrupt dynein function and entrap motors within the dynein transport cycle. We believe that the N. crassa dynein system that we have developed is a powerful system to utilize because of the ability to perform genetic, cell biological, biochemical, and in vitro motility studies on dynein. As we continue to characterize these mutations and to add to our mutant strain collection, we will be in a much stronger position to understand dynein function in other complex situations such as along the long axonal processes of neurons.
Supplementary Material
Acknowledgments
Strains and plasmids were obtained from the Fungal Genetics Stock Center (Kansas City). We thank Linda King for critical reading of the manuscript. This work was supported by grants to S.J.K. [National Institutes of Health (NIH) NS048501] and to M.D.P. (National Science Foundation MCB0235871 and NIH P01 GM069087).
Footnotes
Communicating editor: D. Lew
Literature Cited
- Alberti-Segui C., Dietrich F., Altmann-Johl R., Hoepfner D., Philippsen P., 2001. Cytoplasmic dynein is required to oppose the force that moves nuclei towards the hyphal tip in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 114: 975–986 [DOI] [PubMed] [Google Scholar]
- Allan V., 1995. Protein phosphatase 1 regulates the cytoplasmic dynein-driven formation of endoplasmic reticulum networks in vitro. J. Cell Biol. 128: 879–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto M., Koushika S. P., Choudhary B. C., Li C., Matsumoto K., et al. , 2011. The Caenorhabditis elegans JIP3 protein UNC-16 functions as an adaptor to link kinesin-1 with cytoplasmic dynein. J. Neurosci. 31: 2216–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E. J., Emr S. D., 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17: 2982–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartnicki-Garcia S., Bartnicki D. D., Gierz G., Lopez-Franco R., Bracker C. E., 1995. Evidence that Spitzenkorper behavior determines the shape of a fungal hypha: a test of the hyphoid model. Exp. Mycol. 19: 153–159 [DOI] [PubMed] [Google Scholar]
- Bielli A., Thornqvist P. O., Hendrick A. G., Finn R., Fitzgerald K., et al. , 2001. The small GTPase Rab4A interacts with the central region of cytoplasmic dynein light intermediate chain-1. Biochem. Biophys. Res. Commun. 281: 1141–1153 [DOI] [PubMed] [Google Scholar]
- Bowman B. J., Draskovic M., Freitag M., Bowman E. J., 2009. Structure and distribution of organelles and cellular location of calcium transporters in Neurospora crassa. Eukaryot. Cell 8: 1845–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady S. T., Pfister K. K., Bloom G. S., 1990. A monoclonal antibody against kinesin inhibits both anterograde and retrograde fast axonal transport in squid axoplasm. Proc. Natl. Acad. Sci. USA 87: 1061–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza K. M., Sontag C. A., Saxton W. M., Gilbert S. P., 2000. A kinesin mutation that uncouples motor domains and desensitizes the gamma-phosphate sensor. J. Biol. Chem. 275: 22187–22195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno K. S., Tinsley J. H., Minke P. F., Plamann M., 1996. Genetic interactions among cytoplasmic dynein, dynactin, and nuclear distribution mutants of Neurospora crassa. Proc. Natl. Acad. Sci. USA 93: 4775–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S. A., Walker M. L., Sakakibara H., Knight P. J., Oiwa K., 2003. Dynein structure and power stroke. Nature 421: 715–718 [DOI] [PubMed] [Google Scholar]
- Burgess S. A., Walker M. L., Sakakibara H., Oiwa K., Knight P. J., 2004. The structure of dynein-c by negative stain electron microscopy. J. Struct. Biol. 146: 205–216 [DOI] [PubMed] [Google Scholar]
- Burkhardt J. K., Echeverri C. J., Nilsson T., Vallee R. B., 1997. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J. Cell Biol. 139: 469–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows P. C., Schumacher J., Amartey S., Ghosh T., Burgis T. A., et al. , 2009. Functional roles of the pre-sensor I insertion sequence in an AAA+ bacterial enhancer binding protein. Mol. Microbiol. 73: 519–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Lu L., Tian J. H., Zhu Y. B., Qiao H., et al. , 2010. Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron 68: 73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati J. L., Stearns T., 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138: 629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A. P., Garbarino J. E., Wilson-Kubalek E. M., Shipley W. E., Cho C., et al. , 2008. Structure and functional role of dynein's microtubule-binding domain. Science 322: 1691–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A. P., Cho C., Jin L., Vale R. D., 2011. Crystal structure of the dynein motor domain. Science 331: 1159–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C., Reck-Peterson S. L., Vale R. D., 2008. Regulatory ATPase sites of cytoplasmic dynein affect processivity and force generation. J. Biol. Chem. 283: 25839–25845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham F. R., Hoyt M. A., 1997. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J. Cell Biol. 138: 1041–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver-Hanlon T. L., Lex S. A., Stephens A. D., Quintyne N. J., King S. J., 2006. A microtubule-binding domain in dynactin increases dynein processivity by skating along microtubules. Nat. Cell Biol. 8: 264–270 [DOI] [PubMed] [Google Scholar]
- Davies J. M., Brunger A. T., Weis W. I., 2008. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure 16: 715–726 [DOI] [PubMed] [Google Scholar]
- Davis R. H., 2000. Neurospora: Contributions of a Model Organism. Oxford University Press, New York
- Duncan J. E., Warrior R., 2002. The cytoplasmic dynein and kinesin motors have interdependent roles in patterning the Drosophila oocyte. Curr. Biol. 12: 1982–1991 [DOI] [PubMed] [Google Scholar]
- Echeverri C. J., Paschal B. M., Vaughan K. T., Vallee R. B., 1996. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 132: 617–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encalada S. E., Szpankowski L., Xia C. H., Goldstein L. S., 2011. Stable kinesin and dynein assemblies drive the axonal transport of mammalian prion protein vesicles. Cell 144: 551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro K. L., Collins C. A., 1995. Microtubule-independent phospholipid stimulation of cytoplasmic dynein ATPase activity. J. Biol. Chem. 270: 4492–4496 [DOI] [PubMed] [Google Scholar]
- Freitag M., Hickey P. C., Raju N. B., Selker E. U., Read N. D., 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41: 897–910 [DOI] [PubMed] [Google Scholar]
- Fridolfsson H. N., Starr D. A., 2010. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J. Cell Biol. 191: 115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T., Saredi A., Bingham J. B., Hasbani M. J., Gill S. R., et al. , 1996. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J. Cell Biol. 135: 399–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti J. L., Nicolle J. C., Dacheux J. L., 1994. Characterisation of boar sperm dynein heavy chains by UV-vanadate dependent photocleavage. Biol. Cell 82: 203–210 [DOI] [PubMed] [Google Scholar]
- Gee M. A., Heuser J. E., Vallee R. B., 1997. An extended microtubule-binding structure within the dynein motor domain. Nature 390: 636–639 [DOI] [PubMed] [Google Scholar]
- Gibbons I. R., Mocz G., 1991. Photocatalytic cleavage of proteins with vanadate and other transition metal complexes. Methods Enzymol. 196: 428–442 [DOI] [PubMed] [Google Scholar]
- Gibbons I. R., Lee-Eiford A., Mocz G., Phillipson C. A., Tang W. J., et al. , 1987. Photosensitized cleavage of dynein heavy chains. Cleavage at the “V1 site” by irradiation at 365 nm in the presence of ATP and vanadate. J. Biol. Chem. 262: 2780–2786 [PubMed] [Google Scholar]
- Gibbons I. R., Gibbons B. H., Mocz G., Asai D. J., 1991. Multiple nucleotide-binding sites in the sequence of dynein beta heavy chain. Nature 352: 640–643 [DOI] [PubMed] [Google Scholar]
- Gilbert S. P., Mackey A. T., 2000. Kinetics: a tool to study molecular motors. Methods 22: 337–354 [DOI] [PubMed] [Google Scholar]
- Gouveia S. M., Akhmanova A., 2010. Cell and molecular biology of microtubule plus end tracking proteins: end binding proteins and their partners. Int. Rev. Cell Mol. Biol. 285: 1–74 [DOI] [PubMed] [Google Scholar]
- Grammer J. C., Cremo C. R., Yount R. G., 1988. UV-induced vanadate-dependent modification and cleavage of skeletal myosin subfragment 1 heavy chain. 1. Evidence for active site modification. Biochemistry 27: 8408–8415 [DOI] [PubMed] [Google Scholar]
- Gross S. P., Welte M. A., Block S. M., Wieschaus E. F., 2002. Coordination of opposite-polarity microtubule motors. J. Cell Biol. 156: 715–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habura A., Tikhonenko I., Chisholm R. L., Koonce M. P., 1999. Interaction mapping of a dynein heavy chain. Identification of dimerization and intermediate-chain binding domains. J. Biol. Chem. 274: 15447–15453 [DOI] [PubMed] [Google Scholar]
- Han G., Liu B., Zhang J., Zuo W., Morris N. R., et al. , 2001. The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr. Biol. 11: 719–724 [DOI] [PubMed] [Google Scholar]
- Harada A., 2010. Molecular mechanism of polarized transport. J. Biochem. 147: 619–624 [DOI] [PubMed] [Google Scholar]
- Harris S. D., 2009. The Spitzenkorper: A signalling hub for the control of fungal development? Mol. Microbiol. 73: 733–736 [DOI] [PubMed] [Google Scholar]
- Hartman J. J., Vale R. D., 1999. Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science 286: 782–785 [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Culotti J., Pringle J. R., Reid B. J., 1974. Genetic control of the cell division cycle in yeast. Science 183: 46–51 [DOI] [PubMed] [Google Scholar]
- Hattendorf D. A., Lindquist S. L., 2002. Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. EMBO J. 21: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays T. S., Porter M. E., McGrail M., Grissom P., Gosch P., et al. , 1994. A cytoplasmic dynein motor in Drosophila: identification and localization during embryogenesis. J. Cell Sci. 107: 1557–1569 [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., et al. , 1996. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382: 420–425 [DOI] [PubMed] [Google Scholar]
- Heuston E., Bronner C. E., Kull F. J., Endow S. A., 2010. A kinesin motor in a force-producing conformation. BMC Struct. Biol. 10: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N., Takemura R., 2005. Molecular motors and mechanisms of directional transport in neurons. Nat. Rev. Neurosci. 6: 201–214 [DOI] [PubMed] [Google Scholar]
- Holleran E. A., Tokito M. K., Karki S., Holzbaur E. L., 1996. Centractin (ARP1) associates with spectrin revealing a potential mechanism to link dynactin to intracellular organelles. J. Cell Biol. 135: 1815–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook P., Mikami A., Shafer B., Chait B. T., Rosenfeld S. S., et al. , 2005. Long range allosteric control of cytoplasmic dynein ATPase activity by the stalk and C-terminal domains. J. Biol. Chem. 280: 33045–33054 [DOI] [PubMed] [Google Scholar]
- Imamula K., Kon T., Ohkura R., Sutoh K., 2007. The coordination of cyclic microtubule association/dissociation and tail swing of cytoplasmic dynein. Proc. Natl. Acad. Sci. USA 104: 16134–16139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S., Turgeon B. G., Yoder O. C., Aist J. R., 1998. Role of fungal dynein in hyphal growth, microtubule organization, spindle pole body motility and nuclear migration. J. Cell Sci. 111(Pt. 11): 1555–1566 [DOI] [PubMed] [Google Scholar]
- Iyer L. M., Leipe D. D., Koonin E. V., Aravind L., 2004. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146: 11–31 [DOI] [PubMed] [Google Scholar]
- Januschke J., Gervais L., Dass S., Kaltschmidt J. A., Lopez-Schier H., et al. , 2002. Polar transport in the Drosophila oocyte requires Dynein and Kinesin I cooperation. Curr. Biol. 12: 1971–1981 [DOI] [PubMed] [Google Scholar]
- Kardon J. R., Vale R. D., 2009. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 10: 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S. J., Schroer T. A., 2000. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat. Cell Biol. 2: 20–24 [DOI] [PubMed] [Google Scholar]
- King S. J., Brown C. L., Maier K. C., Quintyne N. J., Schroer T. A., 2003. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol. Biol. Cell 14: 5089–5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S. M., 2000. AAA domains and organization of the dynein motor unit. J. Cell Sci. 113: 2521–2526 [DOI] [PubMed] [Google Scholar]
- Kon T., Nishiura M., Ohkura R., Toyoshima Y. Y., Sutoh K., 2004. Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry 43: 11266–11274 [DOI] [PubMed] [Google Scholar]
- Kon T., Imamula K., Roberts A. J., Ohkura R., Knight P. J., et al. , 2009. Helix sliding in the stalk coiled coil of dynein couples ATPase and microtubule binding. Nat. Struct. Mol. Biol. 16: 325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T., Sutoh K., Kurisu G., 2011. X-ray structure of a functional full-length dynein motor domain. Nat. Struct. Mol. Biol. 18: 638–642 [DOI] [PubMed] [Google Scholar]
- Kon T., Oyama T., Shimo-Kon R., Imamula K., Shima T., et al. , 2012. The 2.8 A crystal structure of the dynein motor domain. Nature 484: 345–350 [DOI] [PubMed] [Google Scholar]
- Koonce M. P., Grissom P. M., McIntosh J. R., 1992. Dynein from Dictyostelium: primary structure comparisons between a cytoplasmic motor enzyme and flagellar dynein. J. Cell Biol. 119: 1597–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonce M. P., Kohler J., Neujahr R., Schwartz J. M., Tikhonenko I., et al. , 1999. Dynein motor regulation stabilizes interphase microtubule arrays and determines centrosome position. EMBO J. 18: 6786–6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Lee I. H., Plamann M., 2000. Cytoplasmic dynein ATPase activity is regulated by dynactin-dependent phosphorylation. J. Biol. Chem. 275: 31798–31804 [DOI] [PubMed] [Google Scholar]
- Lee I. H., Kumar S., Plamann M., 2001. Null mutants of the Neurospora actin-related protein 1 pointed-end complex show distinct phenotypes. Mol. Biol. Cell 12: 2195–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. L., Oberle J. R., Cooper J. A., 2003. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J. Cell Biol. 160: 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz J. H., Schuchardt I., Straube A., Steinberg G., 2006. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 25: 2275–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon L. A., Tokito M., Finklestein J. M., Grossman F. E., Holzbaur E. L., 2004. A direct interaction between cytoplasmic dynein and kinesin I may coordinate motor activity. J. Biol. Chem. 279: 19201–19208 [DOI] [PubMed] [Google Scholar]
- Ma S., Chisholm R. L., 2002. Cytoplasmic dynein-associated structures move bidirectionally in vivo. J. Cell Sci. 115: 1453–1460 [DOI] [PubMed] [Google Scholar]
- Mallik R., Carter B. C., Lex S. A., King S. J., Gross S. P., 2004. Cytoplasmic dynein functions as a gear in response to load. Nature 427: 649–652 [DOI] [PubMed] [Google Scholar]
- Markus S. M., Lee W. L., 2011. Regulated offloading of cytoplasmic dynein from microtubule plus ends to the cortex. Dev. Cell. 20: 639–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus S. M., Punch J. J., Lee W. L., 2009. Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr. Biol. 19: 196–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus S. M., Plevock K. M., St. Germain B. J., Punch J. J., Meaden C. W., et al. , 2011. Quantitative analysis of Pac1/LIS1-mediated dynein targeting: implications for regulation of dynein activity in budding yeast. Cytoskeleton 68: 157–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Iyadurai S. J., Gassman A., Gindhart J. G., Jr, Hays T. S., et al. , 1999. Cytoplasmic dynein, the dynactin complex, and kinesin are interdependent and essential for fast axonal transport. Mol. Biol. Cell 10: 3717–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesngon M. T., Tarricone C., Hebbar S., Guillotte A. M., Schmitt E. W., et al. , 2006. Regulation of cytoplasmic dynein ATPase by Lis1. J. Neurosci. 26: 2132–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minke P. F., Lee I. H., Tinsley J. H., Bruno K. S., Plamann M., 1999. Neurospora crassa ro-10 and ro-11 genes encode novel proteins required for nuclear distribution. Mol. Microbiol. 32: 1065–1076 [DOI] [PubMed] [Google Scholar]
- Minke P. F., Lee I. H., Tinsley J. H., Plamann M., 2000. A Neurospora crassa Arp1 mutation affecting cytoplasmic dynein and dynactin localization. Mol. Gen. Genet. 264: 433–440 [DOI] [PubMed] [Google Scholar]
- Mizuno N., Narita A., Kon T., Sutoh K., Kikkawa M., 2007. Three-dimensional structure of cytoplasmic dynein bound to microtubules. Proc. Natl. Acad. Sci. USA 104: 20832–20837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocz G., Gibbons I. R., 1996. Phase partition analysis of nucleotide binding to axonemal dynein. Biochemistry 35: 9204–9211 [DOI] [PubMed] [Google Scholar]
- Mocz G., Helms M. K., Jameson D. M., Gibbons I. R., 1998. Probing the nucleotide binding sites of axonemal dynein with the fluorescent nucleotide analogue 2’(3’)-O-(-N-methylanthraniloyl)-adenosine 5’-triphosphate. Biochemistry 37: 9862–9869 [DOI] [PubMed] [Google Scholar]
- Mourino-Perez R. R., Roberson R. W., Bartnicki-Garcia S., 2006. Microtubule dynamics and organization during hyphal growth and branching in Neurospora crassa. Fungal Genet. Biol. 43: 389–400 [DOI] [PubMed] [Google Scholar]
- Nilsson H., Wallin M., 1998. Microtubule aster formation by dynein-dependent organelle transport. Cell Motil. Cytoskeleton 41: 254–263 [DOI] [PubMed] [Google Scholar]
- Novick P., Ferro S., Schekman R., 1981. Order of events in the yeast secretory pathway. Cell 25: 461–469 [DOI] [PubMed] [Google Scholar]
- Ogawa K., 1991. Four ATP-binding sites in the midregion of the beta heavy chain of dynein. Nature 352: 643–645 [DOI] [PubMed] [Google Scholar]
- Ogura T., Whiteheart S. W., Wilkinson A. J., 2004. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J. Struct. Biol. 146: 106–112 [DOI] [PubMed] [Google Scholar]
- Ori-McKenney K. M., Xu J., Gross S. P., Vallee R. B., 2010. A cytoplasmic dynein tail mutation impairs motor processivity. Nat. Cell Biol. 12: 1228–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling A. D., Horiuchi D., Lively C. M., Saxton W. M., 2006. Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol. Biol. Cell 17: 2057–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann M., Minke P. F., Tinsley J. H., Bruno K. S., 1994. Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J. Cell Biol. 127: 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock N., Koonce M. P., de Hostos E. L., Vale R. D., 1998. In vitro microtubule-based organelle transport in wild-type Dictyostelium and cells overexpressing a truncated dynein heavy chain. Cell Motil. Cytoskeleton 40: 304–314 [DOI] [PubMed] [Google Scholar]
- Quintyne N. J., Schroer T. A., 2002. Distinct cell cycle-dependent roles for dynactin and dynein at centrosomes. J. Cell Biol. 159: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne N. J., Gill S. R., Eckley D. M., Crego C. L., Compton D. A., et al. , 1999. Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 147: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Garcia S. L., Roberson R. W., Freitag M., Bartnicki-Garcia S., Mourino-Perez R. R., 2009. Cytoplasmic bulk flow propels nuclei in mature hyphae of Neurospora crassa. Eukaryot. Cell 8: 1880–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana A. A., Barbera J. P., Rodriguez T. A., Lynch D., Hirst E., et al. , 2004. Targeted deletion of the novel cytoplasmic dynein mD2LIC disrupts the embryonic organiser, formation of the body axes and specification of ventral cell fates. Development 131: 4999–5007 [DOI] [PubMed] [Google Scholar]
- Reck-Peterson S. L., Vale R. D., 2004a. Molecular dissection of the roles of nucleotide binding and hydrolysis in dynein’s AAA domains in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101: 1491–1495 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Reck-Peterson S. L., Vale R. D., 2004b. Molecular dissection of the roles of nucleotide binding and hydrolysis in dynein’s AAA domains in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101: 14305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaga-Pena C. G., Gierz G., Bartnicki-Garcia S., 1997. Analysis of the role of the Spitzenkorper in fungal morphogenesis by computer simulation of apical branching in Aspergillus niger. Proc. Natl. Acad. Sci. USA 94: 9096–9101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme M., Gierz G., Bartnicki-Garcia S., 2000. Dynein and dynactin deficiencies affect the formation and function of the Spitzenkorper and distort hyphal morphogenesis of Neurospora crassa. Microbiology 146: 1743–1752 [DOI] [PubMed] [Google Scholar]
- Riquelme M., Roberson R. W., McDaniel D. P., Bartnicki-Garcia S., 2002. The effects of ropy-1 mutation on cytoplasmic organization and intracellular motility in mature hyphae of Neurospora crassa. Fungal Genet. Biol. 37: 171–179 [DOI] [PubMed] [Google Scholar]
- Riquelme M., Yarden O., Bartnicki-Garcia S., Bowman B., Castro-Longoria E., et al. , 2011. Architecture and development of the Neurospora crassa hypha: a model cell for polarized growth. Fungal Biol. 115: 446–474 [DOI] [PubMed] [Google Scholar]
- Roberts A. J., Numata N., Walker M. L., Kato Y. S., Malkova B., et al. , 2009. AAA+ ring and linker swing mechanism in the dynein motor. Cell 136: 485–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppel K. M., Spudich J. A., 1996. Structure-function studies of the myosin motor domain: importance of the 50-kDa cleft. Mol. Biol. Cell 7: 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson K., Heath I. B., 2005. The dynamic behaviour of microtubules and their contributions to hyphal tip growth in Aspergillus nidulans. Microbiology 151: 1543–1555 [DOI] [PubMed] [Google Scholar]
- Schmidt H., Gleave E. S., Carter A. P., 2012. Insights into dynein motor domain function from a 3.3-Å crystal structure. Nat. Struct. Mol. Biol. 19: 492–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T. G., Skerra A., 2007. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2: 1528–1535 [DOI] [PubMed] [Google Scholar]
- Schnapp B. J., Reese T. S., 1989. Dynein is the motor for retrograde axonal transport of organelles. Proc. Natl. Acad. Sci. USA 86: 1548–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer T. A., 2004. Dynactin. Annu. Rev. Cell Dev. Biol. 20: 759–779 [DOI] [PubMed] [Google Scholar]
- Schroer T. A., Steuer E. R., Sheetz M. P., 1989. Cytoplasmic dynein is a minus end-directed motor for membranous organelles. Cell 56: 937–946 [DOI] [PubMed] [Google Scholar]
- Schuster M., Kilaru S., Ashwin P., Lin C., Severs N. J., et al. , 2011a. Controlled and stochastic retention concentrates dynein at microtubule ends to keep endosomes on track. EMBO J. 30: 652–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Kilaru S., Fink G., Collemare J., Roger Y., et al. , 2011b. Kinesin-3 and dynein cooperate in long-range retrograde endosome motility along a nonuniform microtubule array. Mol. Biol. Cell 22: 3645–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Lipowsky R., Assmann M. A., Lenz P., Steinberg G., 2011c. Transient binding of dynein controls bidirectional long-range motility of early endosomes. Proc. Natl. Acad. Sci. USA 108: 3618–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler S., Nargang F. E., Steinberg G., Schliwa M., 1997. Kinesin is essential for cell morphogenesis and polarized secretion in Neurospora crassa. EMBO J. 16: 3025–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler S., Plamann M., Schliwa M., 1999. Kinesin and dynein mutants provide novel insights into the roles of vesicle traffic during cell morphogenesis in Neurospora. Curr. Biol. 9: 779–785 [DOI] [PubMed] [Google Scholar]
- Sheeman B., Carvalho P., Sagot I., Geiser J., Kho D., et al. , 2003. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 13: 364–372 [DOI] [PubMed] [Google Scholar]
- Shpetner H. S., Paschal B. M., Vallee R. B., 1988. Characterization of the microtubule-activated ATPase of brain cytoplasmic dynein (MAP 1C). J. Cell Biol. 107: 1001–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvanovich A., Li M. G., Serr M., Mische S., Hays T. S., 2003. The third P-loop domain in cytoplasmic dynein heavy chain is essential for dynein motor function and ATP-sensitive microtubule binding. Mol. Biol. Cell 14: 1355–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Endow S. A., 1998. Decoupling of nucleotide- and microtubule-binding sites in a kinesin mutant. Nature 396: 587–590 [DOI] [PubMed] [Google Scholar]
- Song H. K., Hartmann C., Ramachandran R., Bochtler M., Behrendt R., et al. , 2000. Mutational studies on HslU and its docking mode with HslV. Proc. Natl. Acad. Sci. USA. 97: 14103–14108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen W., Karki S., Vaughan K. T., Vallee R. B., Holzbaur E. L., et al. , 1997. The involvement of the intermediate chain of cytoplasmic dynein in binding the motor complex to membranous organelles of Xenopus oocytes. Mol. Biol. Cell 8: 2077–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G., 2007. Hyphal growth: a tale of motors, lipids, and the Spitzenkorper. Eukaryot. Cell 6: 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G., Perez-Martin J., 2008. Ustilago maydis, a new fungal model system for cell biology. Trends Cell Biol. 18: 61–67 [DOI] [PubMed] [Google Scholar]
- Tai C. Y., Dujardin D. L., Faulkner N. E., Vallee R. B., 2002. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J. Cell Biol. 156: 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theiss C., Napirei M., Meller K., 2005. Impairment of anterograde and retrograde neurofilament transport after anti-kinesin and anti-dynein antibody microinjection in chicken dorsal root ganglia. Eur. J. Cell Biol. 84: 29–43 [DOI] [PubMed] [Google Scholar]
- Tinsley J. H., Minke P. F., Bruno K. S., Plamann M., 1996. p150Glued, the largest subunit of the dynactin complex, is nonessential in Neurospora but required for nuclear distribution. Mol. Biol. Cell 7: 731–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba S., Watanabe T. M., Yamaguchi-Okimoto L., Toyoshima Y. Y., Higuchi H., 2006. Overlapping hand-over-hand mechanism of single molecular motility of cytoplasmic dynein. Proc. Natl. Acad. Sci. USA 103: 5741–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traer C. J., Rutherford A. C., Palmer K. J., Wassmer T., Oakley J., et al. , 2007. SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat. Cell Biol. 9: 1370–1380 [DOI] [PubMed] [Google Scholar]
- Tynan S. H., Gee M. A., Vallee R. B., 2000. Distinct but overlapping sites within the cytoplasmic dynein heavy chain for dimerization and for intermediate chain and light intermediate chain binding. J. Biol. Chem. 275: 32769–32774 [DOI] [PubMed] [Google Scholar]
- Uchida M., Mourino-Perez R. R., Freitag M., Bartnicki-Garcia S., Roberson R. W., 2008. Microtubule dynamics and the role of molecular motors in Neurospora crassa. Fungal Genet. Biol. 45: 683–692 [DOI] [PubMed] [Google Scholar]
- Vaisberg E. A., Koonce M. P., McIntosh J. R., 1993. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J. Cell Biol. 123: 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valetti C., Wetzel D. M., Schrader M., Hasbani M. J., Gill S. R., et al. , 1999. Role of dynactin in endocytic traffic: effects of dynamitin overexpression and colocalization with CLIP-170. Mol. Biol. Cell 10: 4107–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan K. T., Tynan S. H., Faulkner N. E., Echeverri C. J., Vallee R. B., 1999. Colocalization of cytoplasmic dynein with dynactin and CLIP-170 at microtubule distal ends. J. Cell Sci. 112: 1437–1447 [DOI] [PubMed] [Google Scholar]
- Verde F., Berrez J. M., Antony C., Karsenti E., 1991. Taxol-induced microtubule asters in mitotic extracts of Xenopus eggs: requirement for phosphorylated factors and cytoplasmic dynein. J. Cell Biol. 112: 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer C. M., Karki S. B., Kuznetsov S. A., Tabb J. S., Weiss D. G., et al. , 1997. The interaction between cytoplasmic dynein and dynactin is required for fast axonal transport. Proc. Natl. Acad. Sci. USA 94: 12180–12185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteheart S. W., Rossnagel K., Buhrow S. A., Brunner M., Jaenicke R., et al. , 1994. N-ethylmaleimide-sensitive fusion protein: a trimeric ATPase whose hydrolysis of ATP is required for membrane fusion. J. Cell Biol. 126: 945–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wriggers W., Chakravarty S., Jennings P. A., 2005. Control of protein functional dynamics by peptide linkers. Biopolymers 80: 736–746 [DOI] [PubMed] [Google Scholar]
- Xiang X., Roghi C., Morris N. R., 1995. Characterization and localization of the cytoplasmic dynein heavy chain in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 92: 9890–9894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X., Han G., Winkelmann D. A., Zuo W., Morris N. R., 2000. Dynamics of cytoplasmic dynein in living cells and the effect of a mutation in the dynactin complex actin-related protein Arp1. Curr. Biol. 10: 603–606 [DOI] [PubMed] [Google Scholar]
- Yarden O., Plamann M., Ebbole D. J., Yanofsky C., 1992. cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J. 11: 2159–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Li S., Fischer R., Xiang X., 2003. Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol. Biol. Cell 14: 1479–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhuang L., Lee Y., Abenza J. F., Penalva M. A., et al. , 2010. The microtubule plus-end localization of Aspergillus dynein is important for dynein-early-endosome interaction but not for dynein ATPase activation. J. Cell Sci. 123: 3596–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Yao X., Fischer L., Abenza J. F., Penalva M. A., et al. , 2011. The p25 subunit of the dynactin complex is required for dynein-early endosome interaction. J. Cell Biol. 193: 1245–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Wildonger J., Ye B., Zhang Y., Kita A., et al. , 2008. Dynein is required for polarized dendritic transport and uniform microtubule orientation in axons. Nat. Cell Biol. 10: 1172–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.