Abstract
In vertebrates, mitotic and meiotic M phase is facilitated by the kinase Greatwall (Gwl), which phosphorylates a conserved sequence in the effector Endosulfine (Endos). Phosphorylated Endos inactivates the phosphatase PP2A/B55 to stabilize M-phase-specific phosphorylations added to many proteins by cyclin-dependent kinases (CDKs). We show here that this module functions essentially identically in Drosophila melanogaster and is necessary for proper mitotic and meiotic cell division in a wide variety of tissues. Despite the importance and evolutionary conservation of this pathway between insects and vertebrates, it can be bypassed in at least two situations. First, heterozygosity for loss-of-function mutations of twins, which encodes the Drosophila B55 protein, suppresses the effects of endos or gwl mutations. Several types of cell division occur normally in twins heterozygotes in the complete absence of Endos or the near absence of Gwl. Second, this module is nonessential in the nematode Caenorhaditis elegans. The worm genome does not contain an obvious ortholog of gwl, although it encodes a single Endos protein with a surprisingly well-conserved Gwl target site. Deletion of this site from worm Endos has no obvious effects on cell divisions involved in viability or reproduction under normal laboratory conditions. In contrast to these situations, removal of one copy of twins does not completely bypass the requirement for endos or gwl for Drosophila female fertility, although reducing twins dosage reverses the meiotic maturation defects of hypomorphic gwl mutants. These results have interesting implications for the function and evolution of the mechanisms modulating removal of CDK-directed phosphorylations.
Keywords: cell-cycle regulation, reversal of cyclin-dependent kinase (CDK)-driven phosphorylations, kinases and phosphatases, M-phase entry
ENTRY into M phase of the cell cycle is driven by the rapid activation of the kinase MPF (M phase promoting factor; Cdk1/cyclin B), which then phosphorylates hundreds of target proteins at proline-directed phosphosites (S/T P) (Dephoure et al. 2008; Lindqvist et al. 2009). These sites must remain phosphorylated during M phase, but then be dephosphorylated upon M-phase exit. It would make sense that the phosphatase(s) directed against mitotic phosphosites should be inhibited during M phase, because if not, these key phosphorylations would be prematurely removed, preventing cells from entering M phase.
Recent findings support this hypothesis. During M phase in Saccharomyces cerevisiae, the phosphatase Cdc14p, whose activity against MPF targets is required for mitotic exit, is sequestered in the nucleolus from its substrates (reviewed in Mocciaro and Schiebel 2010). In Xenopus egg extracts or in mammalian tissue culture cells, a different phosphatase, PP2A associated with a B55-type regulatory subunit, is primarily responsible for mitotic phosphosite dephosphorylation (Mochida et al. 2009). PP2A/B55 is inhibited during M phase by a pathway featuring Greatwall kinase (Gwl) and its substrates in the Endosulfine (Endos) family (Castilho et al. 2009; Mochida et al. 2009, 2010; Vigneron et al. 2009; Gharbi-Ayachi et al. 2010). Endos proteins phosphorylated by Gwl bind to PP2A/B55 and inhibit its phosphatase activity in Xenopus extracts (Gharbi-Ayachi et al. 2010; Mochida et al. 2010), and Drosophila melanogaster oocytes lacking Endos have reduced levels of phosphorylated epitopes despite normal Cdk1/cyclinB activity (Von Stetina et al. 2008). Xenopus Gwl is activated at least in part by MPF, ensuring PP2A/B55 inhibition specifically during M phase (Yu et al. 2006; Blake-Hodek et al. 2012). When Gwl or Endosulfine are depleted, frog egg extracts can neither enter nor maintain M phase (Yu et al. 2006; Zhao et al. 2008; Gharbi-Ayachi et al. 2010; Mochida et al. 2010), mammalian tissue culture cells exhibit delays at the G2/M transition (Burgess et al. 2010; Voets and Wolthuis 2010), and Drosophila oocytes fail to progress to metaphase I (Von Stetina et al. 2008).
In this report, we investigate the Gwl–Endos–PP2A/B55 pathway during several types of cell division in two invertebrate organisms, D. melanogaster and Caenorhabditis elegans. We anticipated that this pathway operates in the fruit fly, because gwl mutations cause defects in M phase entry in several tissue types (Yu et al. 2004; Archambault et al. 2007) and because hypomorphic mutation of the single endos gene in Drosophila prolongs prophase I and impairs oocyte meiotic maturation (Von Stetina et al. 2008). The situation in C. elegans was less predictable. The worm genome has no obvious gwl ortholog, but it does have a single endos-related gene whose protein product paradoxically retains the well-conserved phosphorylation site that Gwl targets in frog and fly Endosulfine.
Our results show that the Gwl–Endos–PP2A/B55 pathway is normally critical for many if not all types of cell division in Drosophila. Our observations further suggest that in the fly, Gwl is the predominant regulator of Endos, while Endos is the key target of Gwl. Surprisingly, however, many cell types in Drosophila can divide normally in the absence of Gwl or Endos if the dosage of twins, encoding the B55 regulatory subunit of PP2A, is halved. Thus, the only essential function of Gwl and Endos during mitosis and male meiotic divisions is PP2A regulation. In contrast, reducing twins function does not reverse the female sterility of endos or gwl mutations; however, twins heterozygosity can suppress some effects of weak, female sterile alleles of gwl on female meiosis. This specialized type of cell division is therefore particularly dependent on the Gwl–Endos–PP2A/B55 module; furthermore, Endos may have functions independent of Gwl or PP2A/B55 during female meiosis. We also found that the conserved site in Endos targeted by Gwl in other organisms is dispensable for cell division in C. elegans. Thus, even though the Gwl–Endos–PP2A/B55 mechanism influences the fate of hundreds of M-phase-specific phosphorylations, its importance to the development of the organism is surprisingly plastic.
Materials and Methods
Drosophila strains
Fly stocks were maintained at 22–25° on standard medium. Canton-S or y w served as wild-type controls in different experiments. The endosEY04709 allele used for production of the endos null allele was generated by the BDGP gene disruption project (http://flypush.imgen.bcm.tmc.edu/pscreen/). It contains a P{EPgy2}-element (Bellen et al. 2004) inserted 512 genomic bp upstream of the translational start of endos, in the 5′-untranslated region. Imprecise excisions of the EY04709 P-element insertion were generated by temporarily introducing Δ2–3 as a source of transposase using standard genetic procedures. The endos215-4 excision eliminates the P element and deletes ∼580 bp of genomic DNA, from the original EY04709 insertion site to 71 bp downstream of the translational start site.
The endos00003 female sterile allele (Drummond-Barbosa and Spradling 2004; Von Stetina et al. 2008); the strongly hypomorphic gwl alleles gwlSR18, gwl180, gwl716, gwl1080, and gwl2790 (Yu et al. 2004); the strong loss-of-function mutation twinsP (Uemura et al. 1993); and Df(3L)fzGF3b, a deletion of the region containing endos, have been previously described and are listed in Flybase (http://www.flybase.org). Two other strong loss-of-function twins mutations, twins196 and twins430, were isolated in the laboratory of R. Booker (Cornell University, Ithaca, NY). In several of these gwl and twins stocks, the mutation-bearing third chromosomes accumulated over time second-site lethal mutations, necessitating the use of trans-heterozygous combinations of alleles to obtain mutant third instar larvae for cytological analysis. Mutation-bearing third chromosomes were balanced over TM6C, Tb Sb to allow the identification of mutant larvae and/or adults.
For in vivo time-lapse imaging of larval brains, strains of the appropriate genotypes were created using an insertion of P{H2AvD-RFP} on the second chromosome that expressed red fluorescent protein-labeled histone H2AvD (http://flybase.org/reports/FBtp0056035.html).
Drosophila transgenic line generation
The Drosophila endos coding region (including 21 bp immediately upstream of the initiation codon) amplified from LD19034 (Drosophila Genomics Resource Center) was subcloned into pGem-T Easy (Promega, Madison, WI). Mutagenesis to substitute amino acids at Serine 68 (predicted Gwl phosphorylation site) or serine 107 (predicted PKA phosphorylation site) was performed using the QuikChange site-directed mutagenesis kit (Stratagene, Santa Clara, CA) and confirmed by sequencing. EcoRI-digested inserts were subcloned in pUASpI (Von Stetina et al. 2008). Transgenic lines were generated as described (Spradling and Rubin 1982), either in house or by BestGene, Inc. (Chino Hills, CA).
Cytology of Drosophila larval neuroblasts
Procedures for the cytological analysis of fixed larval brains have previously been described (Yu et al. 2004). For detailed observations of chromosomes in larval neuroblasts, the brains were treated for 1 hr at room temperature with colchicine and subsequently with hypotonic solution as described (Gatti et al. 1994).
In vivo time-lapse imaging of larval brains was carried out as described (Rahmani et al. 2009). Cells were examined with a Zeiss Axiovert 20 microscope equipped with an HBO 50-W mercury lamp and a filter wheel combination (ChromaTechnology, Bellows Falls, VT). The objective used was 63 × 3 (NA = 1.3). Images were acquired with a CoolSnap HQ camera (Photometrics, Tucson, AZ) with a 2 3 2 bin. Image acquisition was controlled through the Metamorph software package (Universal Imaging, Downing Town, PA). Images were collected at 2-min intervals, and eight fluorescence optical sections were captured at 1-μm z steps. Movies were created with Metamorph software; each fluorescent image shown is the maximum-intensity projection of all the sections.
RNAi and cytology of Drosophila tissue culture cells
Protocols for RNAi, chromosome squashes, and immunostaining of S2 tissue culture cells are detailed in Somma et al. (2008). Antibodies used in immunostaining include rabbit polyclonal anti-phosphoHistone H3 (pH3) (Millipore, Billerica, MA) and a mouse monoclonal anti-lamin antibody from clone ADL67.10 (developed by P.A. Fisher under the auspices of the National Institute of Child Health and Human Development (NICHD) and obtained from the Developmental Studies Hybridoma Bank maintained by The University of Iowa, Department of Biology, Iowa City, IA).
Drosophila oocyte DNA analyses and egg counts
Egg chamber staging and analysis of oocyte meiotic maturation were performed as described (Von Stetina et al. 2008). Results were subjected to χ2 tests.
To measure egg production, five pairs of 0- to 2-day-old flies were maintained in triplicate in plastic bottles containing molasses plates with a layer of wet yeast paste at 25°. The number of eggs laid per group was counted every 24 hr for 5 days. Results were subjected to Student’s t-tests.
RT–PCR
endos00003/TM3 P{ActGFP}JMR2, Ser1 (control) and endos00003 homozygous late third-instar larvae were collected under a Leica M165 FC fluorescent stereo microscope. Larval RNA extracted using RNAqueous-4PCR was reverse transcribed (RT) using oligo(dT)16 for priming and SuperScript II reverse transcriptase (all reagents from Life Technologies, Grand Island, NY). PCR was performed using undiluted or diluted (1:10) RT reactions and the following primer pairs: DDB103 (5′-ATGTTCCGAAATAACGCCTTCTGC-3′) and DDB83 (5′-GCTCTGCAAGCAGCAGTACC-3′) for endos, DDB421 (5′-AACCAATCACACAACAATCCA-3′) and DDB422 (5′-TCTTCAAAGCATCCGTCAACT-3′) for CG6650, and DDB137 (5′-CAGTCGGATCGATATGCTAAGC-3′) and DDB138 (5′-AATCTCCTTGCGCTTCTTGG-3′) for Rp49.
Western blotting
Late third-instar larvae, adult ovaries, or S2 tissue culture cells were homogenized, electrophoresed, and transferred to membranes as described (Von Stetina et al. 2008). Membranes were blocked and probed with 1:1000 rabbit polyclonal anti-Endos (c302) (Von Stetina et al. 2008), 1:1000 purified rabbit polyclonal anti-Gwl (Yu et al. 2004), 1:1000 mouse monoclonal anti-α-tubulin (DM1A; Sigma-Aldrich, St. Louis, MO), 1:50 mouse monoclonal anti-actin (JLA20, Developmental Studies Hybridoma Bank), or 1:5000 horseradish peroxidase (HRP)-conjugated mouse monoclonal anti-β-actin (C4; Santa Cruz Biotechnology, Santa Cruz, CA). HRP-conjugated goat anti-rabbit secondary antibodies (Jackson ImmunoResearch, West Grove, PA) were used at 1:4000 dilution for Endos, HRP-conjugated goat-anti-mouse secondaries (Bio-Rad, Hercules, CA) were used at 1:10,000 dilution for Gwl, and no secondary antibodies were used for the HRP-conjugated anti-β-actin blot. Immunoblotting of Xenopus egg extracts for the mitotic markers Gwl, Cdc25, phosphorylated MAPK, and tyrosine 15-phosphorylated Cdk1 has been described (Yu et al. 2006; Zhao et al. 2008). Signals were detected with the Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE) or by enhanced chemiluminescence (GE Healthcare, Piscataway, NJ).
C. elegans culture and genetics
Nematodes were cultured as described (Brenner 1974). N2 (Bristol) was used as wild type. The homozygous viable strain FX02810, carrying the tm2810 deletion allele in the gene K10C3.2, was obtained from the S. Mitani laboratory at Tokyo Women’s University Medical School (Tokyo, Japan). We confirmed by DNA sequence analysis that tm2810 is a 243-bp deletion beginning in the first exon and ending in the second exon, and results in an in-frame deletion of 34 amino acids (Q54–P87) from the predicted protein product of K10C3.2. We outcrossed this strain six times to N2 using PCR to follow the mutation; the outcrossed strain is designated MLG1 ensa-1(tm2810) and was used for all phenotypic characterization. Strain JK1553ces-1(n703) qDf9/unc-29(e1072) lin-11(n566) was obtained from the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources (NCRR). For brood size and embryo viability counts, worms were plated individually and transferred each day, and unhatched eggs and hatched larvae were counted. For larval growth studies, embryos were isolated by hypochlorite treatment and hatched into M9 buffer (42 mM Na2HPO4, 22 mM KH2PO4, 85 mM NaCl, 19 mM NH4Cl) to obtain semisynchronized L1 populations. L1 larvae were rocked in M9 for 48 hr prior to transfer to NGM plates with OP50 bacteria.
RNAi in C. elegans
We used the RNAi feeding method (Timmons et al. 2001) at 20°, with RNAi clones from the MRC geneservice RNAi library (Kamath et al. 2003) or from the library of Rual et al. (2004). RNAi with HT115(DE3) carrying the empty vector L4440 was used as a negative control. Embryos were hatched onto RNAi feeding plates and transferred to fresh plates with RNAi bacteria during larval and adult stages for RNAi knockdown over the life of the animal.
RNA was isolated from worms using Trizol reagent and reverse transcribed using an iScript cDNA synthesis kit (Bio-Rad). Quantitative PCR (qPCR) analysis of cDNAs used the SYBR Green system (Bio-Rad) on a Bio-Rad MyiQ real-time PCR detection system and was quantified relative to a standard curve. act-1 (actin) was used as an internal control. Primers used were: act-1F, CCAGGAATTGCTGATCGTATGCAGAA; act-1R, TGGAGAGGGAAGCGAGGATAGA; ensa-1F, CGCAAACCAATCCAATCATA; ensa-1R, GGTTGGGAAGGTGTTGAGAG.
Time-lapse microscopy of C. elegans embryos
Images of C. elegans embryos were obtained using a Leica DM RA microscope with a 63× HCX PL APO oil immersion lens, a Hamamatsu Orca-ER camera (Hamamatsu, Bridgewater, NJ) and OpenLab software (Perkin-Elmer, Waltham, MA). Embryos were mounted on 2% agarose pads on slides. The temperature of the microscope room was controlled to 20°.
Xenopus egg and C. elegans extracts
Gwl-depleted CSF (cytostatic factor) extracts from Xenopus eggs were prepared as described (Yu et al. 2006; Zhao et al. 2008). Xenopus interphase extracts were made by treating CSF extracts with CaCl2 (0.5 mM final concentration) and incubating at 23° for 40 min.
For C. elegans extracts, mixed-stage populations of embryos and worms were collected from plates in 130 mM NaCl, 25mM Tris pH 7.5 and pelleted by mild centrifugation. The pellet was mixed with an equal volume of M-PER protein extraction reagent (Thermo Fisher Scientific, Waltham, MA) and frozen. Just before use, these worm concentrates were thawed on ice, supplemented with another 0.2× volume of extraction reagent, and then lysed by a single pass through a French press (Sim-Aminco, Rochester, NY) at 15,000 psi.
Expression of Gwl and Endos proteins
Mutant Gwl proteins (point mutations and deletion mutations) were generated from a Gwl cDNA clone by using the QuikChange II site directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). All proteins were expressed in Sf9 insect cells using the Bac-to-Bac system (Invitrogen, Carlsbad, CA) and purified as previously described (Yu et al. 2006). Active Gwl was produced by treating the infected cells with OA to a final concentration of 100 nM for 12 hr before harvesting. The kinase dead (KD) allele of Xenopus Gwl used was G41S, previously characterized in (Yu et al. 2004; Yu et al. 2006).
His-tagged wild-type and S67A mutant Xenopus Endosulfine proteins were expressed in Escherichea coli and purified on nickel beads as described (Mochida et al. 2010); the constructs were the kind gift of S. Mochida (Kumamoto University, Kumamoto, Japan). S68A, S68D, and S68E point mutations of Drosophila Endos were introduced into pGEX4T2-endos (Drummond-Barbosa and Spradling 2004) using the QuikChange site-directed mutagenesis kit (Agilent), and proteins were expressed in BL21(DE3)pLysS (Agilent). Coding regions for C. elegans Endosulfine proteins were PCR amplified from cDNA libraries and cloned into pGEX6P-1 (GE Healthcare) using EcoRI and SalI. Phosphomimetic and nonphosphorylatable mutants in C. elegans glutathione S-transferase (GST)–Endos were made using the QuikChange II site-directed mutagenesis kit (Agilent). GST fusion proteins were purified on glutathione–Sepharose beads (GE Healthcare).
Drosophila in vitro expression cloning (DIVEC) interaction test
In vitro interaction tests were performed as described (Von Stetina et al. 2008), with slight modifications. cDNAs encoding Twins (PP2A–B regulatory subunit), Microtubule star (PP2A catalytic subunit), PP2A–B′ regulatory subunit, and Elgi (Von Stetina et al. 2008) were in vitro transcribed/translated in presence of [35S]methionine using the TNT system (Promega). GST fusion proteins with wild-type or mutant forms of Endos or GST alone (negative control) were bound to glutathione-Sepharose beads (GE Healthcare). The beads bound with protein were mixed with 3 μl of each 35S-radiolabeled protein in 150 μl of buffer A (50 mM Tris pH 8.0, 200 mM NaCl, 0.1% Tween-20, 1 mM PMSF, 1 mM DTT, 10 μg/ml protease inhibitor cocktail tables, EDTA free [Roche, Nutley, NJ]). Reactions were incubated at 4° for 2 hr on a rotating platform and then transferred to Wizard minicolumns (Promega) attached to a vacuum manifold. The bead-containing minicolumns were washed three times with 2.5 ml of buffer A and once with 2.5 ml of buffer B (50 mM Tris, pH 8.0, 50 mM NaCl, 1 mM PMSF). After the final wash, excess buffer was removed by centrifugation for 1 min at 10,000 rpm. Preheated (95°) 1× sample buffer was added to protein-bound beads, and proteins were eluted by another centrifugation of the columns. Samples of the eluates were separated by polyacrylamide gel electrophoresis and detected by autoradiography.
Kinase and phosphatase assays
To assess the relative abilities of Xenopus or Drosophila Gwl to phosphorylate Endosulfine from frogs, flies, or worms, kinase assays were performed for 10 min at 30° in 10 μl of kinase buffer (20 mM HEPES, pH 7.5; 10 mM MgCl2; 0.1 mg/ml BSA; and 3 mM β-mercaptoethanol) supplemented with 100 μM cold ATP and 1 μCi [γ-32P]ATP (3000 Ci/mmol) and with 50 ng of Gwl proteins (diluted into 1 μl of PBS + 50% glycerol) prepared from Sf9 insect cells infected with baculovirus constructs and treated with the phosphatase inhibitor okadaic acid to allow Gwl activation (Yu et al. 2006). A total of 1.5 μg of recombinant wild-type or mutant Endos protein (see below) was added to the reaction mix as substrate. Kinase reactions were terminated by addition of SDS sample buffer, and the samples were fractionated by SDS polyacrylamide gel electrophoresis. Radioactive Endos was identified by autoradiography.
The assay for the dephosphorylation of CDK phosphosites in Xenopus and C. elegans extracts was performed as described (Mochida and Hunt 2007). Briefly, the substrate consisted of a 25-amino acid fragment of the γ-isoform of human PP1 containing the CDK target site Thr311 fused to maltose binding protein. Cdk2/cyclin A (made as described in Blake-Hodek et al. 2012) was used to label this fusion protein in vitro to high specific activity with [γ-32P]ATP. The radiolabeled substrate was added to extracts, and the phosphate released was quantified as the percentage of the total input counts of substrate added to the assay. It has previously been shown that this substrate is specifically dephosphorylated in Xenopus extracts by PP2A associated with a B-type regulatory subunit (Mochida et al. 2009). In some experiments, recombinant Endos proteins were added at the indicated concentrations to inhibit PP2A/B55.
Results
Endos is required for the transition between mitotic prometaphase and metaphase in Drosophila
We previously described female sterile alleles of Drosophila endos associated with P-element insertions into its 5′-UTR (Drummond-Barbosa and Spradling 2004; Von Stetina et al. 2008). Oocytes produced by endos00003 mutant females fail to progress from prophase into metaphase of meiosis I. These oocytes exhibit delayed and abnormal nuclear envelope breakdown, do not form/maintain proper meiotic spindles, and have much reduced levels of phosphorylated Cdk1 targets (Von Stetina et al. 2008). The few embryos from endos00003 females that initiate development show defects in early maternally controlled mitoses, suggesting that endos might play a role in mitosis (Von Stetina et al. 2008).
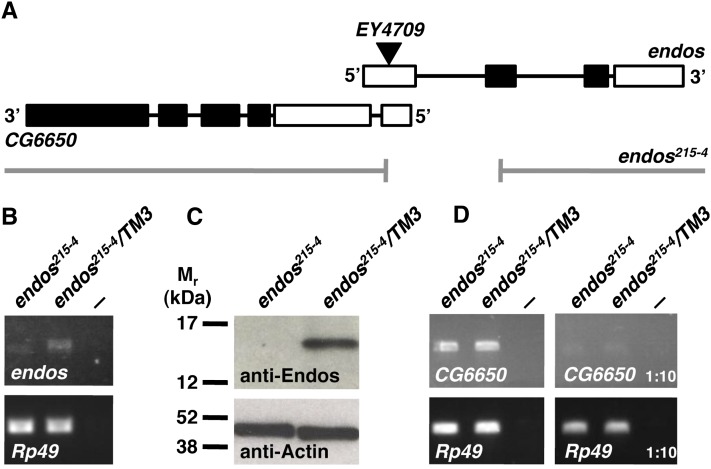
The endos00003 allele strongly disrupts but does not completely eliminate Endos expression (Drummond-Barbosa and Spradling 2004). To test more rigorously the requirements for Endos in cell divisions, we generated the null allele endos215-4 by imprecise P-element excision (Figure 1). The endos215-4 allele contains a ∼580-bp deletion, including the first 71 bp of the open reading frame (Figure 1A). RT–PCR shows an endos215-4 mRNA species of reduced size and amount (Figure 1B); further, we did not detect any Endos protein in endos215-4 homozygous larvae (Figure 1C). The endos215-4 deletion also may eliminate a small part of the neighboring gene CG6650 (see Figure 1A). However, endos215-4 homozygous larvae have normal levels of CG6650 mRNA (Figure 1D), so CG6650 expression is not grossly disrupted. The endos215-4 deletion therefore behaves as a specific null allele of endos.
Figure 1 .
Isolation of an endos null allele. (A) The endos215-4 allele generated by imprecise excision of EY4709 (triangle). Thick bars represent exons (coding region in black) and black lines represent introns. The neighboring gene CG6650 encodes two mRNA isoforms and one coding region; only RA–CG6650 overlaps with endos, as shown here. The gap indicates the deleted region in endos215-4. (B) RT–PCR analysis showing reduced levels of endos mRNA in endos215-4 homozygous third-instar larvae. Rp49 is the control transcript. endos215-4/TM3 are heterozygous controls. The negative control (—) is RT–PCR without a template. (C) Western blot showing that Endos protein is completely lacking in endos215-4 homozygous larvae. Actin was used as a loading control. (D) RT–PCR analysis showing that expression of CG6650 is unaffected in endos215-4 homozygous mutants. Controls are as in B. Right: RT–PCR using 1:10 dilutions of RT reactions.
Although endos215-4 homozygous late third-instar larvae and pupae survive, no homozygous mutant adults eclosed. This was also true of endos215-4 hemizygotes (endos215-4 /deletion), indicating that the lethality is caused by the endos215-4 excision and not by a distant second site mutation (Table 1). Late larval/pupal lethality is characteristic of many mitotic mutations in Drosophila: Animals survive to these stages using maternal gene products that become depleted in larval tissues such as the brain (Gatti and Goldberg 1991). We thus examined endos null mutant larval brains for cell-cycle progression defects. These animals exhibit clear evidence of delayed/arrested cell-cycle transit. For example, the mitotic index is more than twofold higher in endos mutant brains than in controls (Table 1).
Table 1. Characteristics of endos, gwl, and twins mutant combinations.
| Genotype | Viability/fertility | Brain size | MI | Early (%) | Late regular (%) | Late irregular (%) |
|---|---|---|---|---|---|---|
| Canton S (control) | V, MF, FF | N | 5.55 | 67.1 | 32.9 | 0 |
| endos215-4 homozygote | LL/P | N | 11.79 | 78.9 | 21.1 | 0 |
| endos215-4/Df(3L)fzGF3b | LL/P | N | 12.34 | 76.2 | 23.8 | 0 |
| gwl716 homozygote | LL/P | N | 12.42 | 78.7 | 21.3 | 0 |
| gwl180 homozygote | EL | — | — | — | — | — |
| gwl716/gwl180 | Esc, MS, FSN | N | 12.02 | 81.8 | 18.2 | 0 |
| gwl716/gwl1080 | Esc, MS, FSN | — | — | — | — | — |
| gwl716/gwl2790 | Esc, MS, FSN | — | — | — | — | — |
| endos215-4 gwl716 homozygote | LL/P | N | 8.65 | 86.4 | 13.6 | 0 |
| gwl180 twinsP/gwl716 | V, MF, FSE | N | 6.67 | 77.5 | 20.9 | 1.6 |
| gwl180 twinsP/gwl1028 | V, MF, FSE | — | — | — | — | — |
| gwl180 twinsP/gwl2790 | V, MF, FSE | — | — | — | — | — |
| endos215-4 twinsP/endos215-4 | V, MF, FSE | N | 7.00 | 67.3 | 32.7 | 0 |
| twinsP/twins196 | LL/P | S | 7.30 | 72.5 | 17.1 | 10.4 |
| endos215-4 twinsP/endos twinsP | LL/P | S | 7.51 | 82.4 | 6.6 | 11.0 |
| gwl180 twinsP/ twins196 | LL/P | S | 8.92 | 82.2 | 8.7 | 9.1 |
| gwl180 twinsP/gwl180 twinsP | LL/P | S | 6.94 | 78.7 | 9.6 | 11.7 |
| gwl180 twinsP/ twinsP | LL/P | — | — | — | — | — |
| gwl180 twinsP/ twins430 | LL/P | — | — | — | — | — |
For mitotic parameters, n = 500 mitotic figures (5 larval brains) for each genotype. —, not determined. V, viable, MF, Male fertile, FF, Female fertile. FSN, Female sterile (females do not lay eggs). FSE, female sterile (females lay eggs that do not develop). LL/P, die as late larvae or pupae. EL, die as early (first or second instar) larvae. Esc, escapers that eclose several days late and have rough eyes and wing defects. N, normal sized larval brains. S, small larval brains with few dividing cells. MI, mitotic index. Early represents the percentage of dividing cells in prophase/prometaphase. Late regular represents the percentage of dividing cells with normal metaphase/anaphase/telophase. Late irregular represents the percentage of dividing cells with lagging chromosomes or other irregularities during anaphase/telophase.
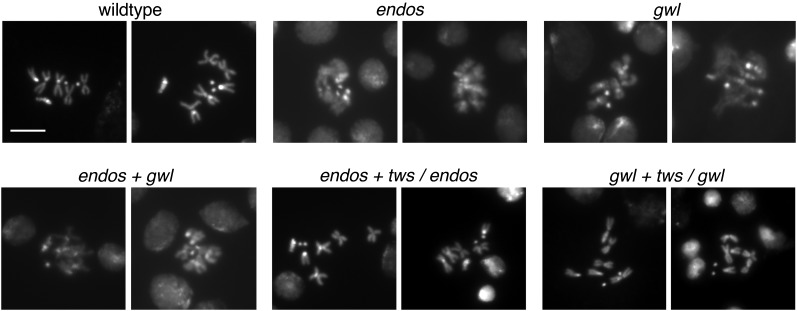
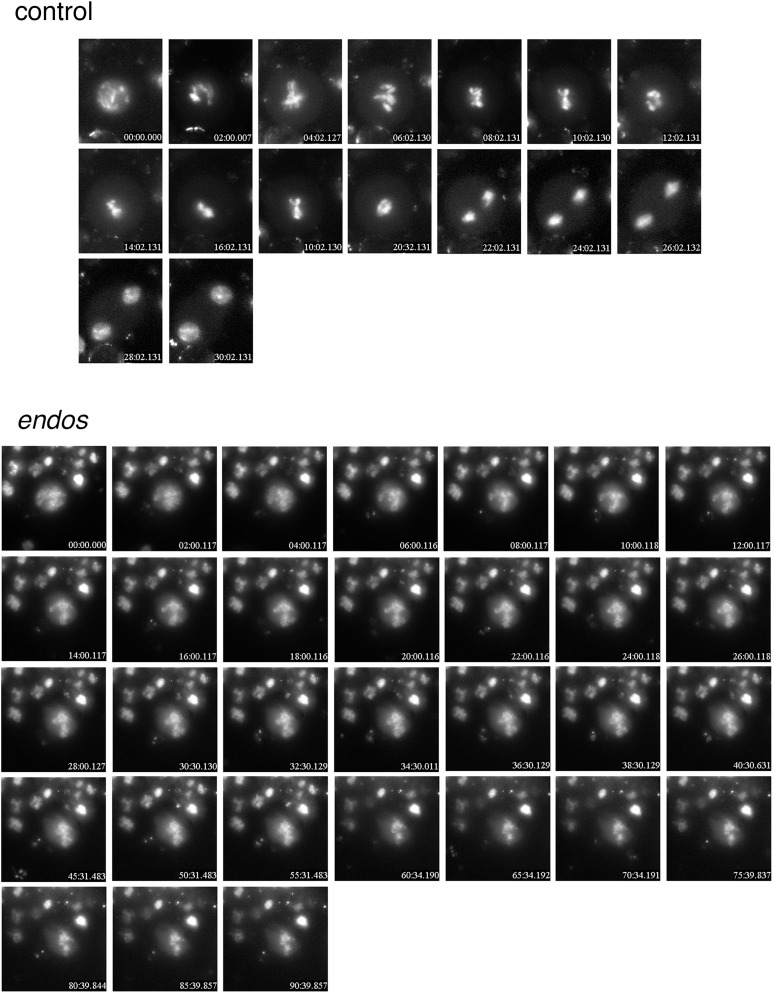
The major roadblock to mitosis in endos mutant brains occurs prior to metaphase, as the proportion of cells in early mitotic stages (prophase and prometaphase) is increased in mutants, while the proportion in late stages (metaphase, anaphase, and telophase) is decreased (Table 1). In karyotypes made from larval brains, endos null mutant chromosomes are strikingly undercondensed relative to wild type (Figure 2), consistent with cell-cycle delay disrupting the chromosome condensation that normally accompanies the prometaphase-to-metaphase transition. Time-lapse movies of mitotic divisions in living larval neuroblasts verify this conclusion (Figure 3; Supporting Information, File S1 and File S2). Cells chosen for filming already exhibited chromosome condensation typical of prophase/prometaphase. In wild type, these neuroblasts (n = 8) form a metaphase plate and then enter anaphase within 30 min. However, of 10 endos null mutant neuroblasts, none formed a metaphase plate by 60 min, and several cells failed to achieve metaphase even after 2 hr. The nuclear envelope did not break down, and chromosome condensation remained incomplete.
Figure 2 .
Condensation state of larval neuroblast chromosomes in gwl–endos–PP2A/B55 module mutants. Brains were dissected from late third-instar larvae, treated with colchicine, and squashed; chromosomes were visualized with Hoechst 33258. Genotypes: wild type (Canton S); endos215-4/endos215-4; gwl716/gwl716 ; endos215-4 gwl716/endos215-4 gwl716 ; endos215-4 twinsP/endos215-4 ; and gwl716 twinsP/gwl716. Chromosomes are undercondensed in endos and gwl mutants, but heterozygosity for twinsP suppresses this phenotype.
Figure 3 .
Prophase/prometaphase arrest of endos215-4 mutant neuroblasts. Brains were dissected from late larvae of either w1118; P{H2AvD-RFP}/+ (controls expressing red fluorescent protein-labeled histone H2AvD) or w1118; P{H2AvD-RFP}/+; endos215-4/endos215-4. Living neuroblasts chosen for filming had already begun chromosome condensation and were thus in prophase. Frames are shown at 2-min intervals. Wild-type cells (n = 8) all progressed through anaphase by 30 min. None of the endos mutant neuroblasts formed a clearcut metaphase plate or displayed anaphase chromosome separation after 60 min or more (n = 10).
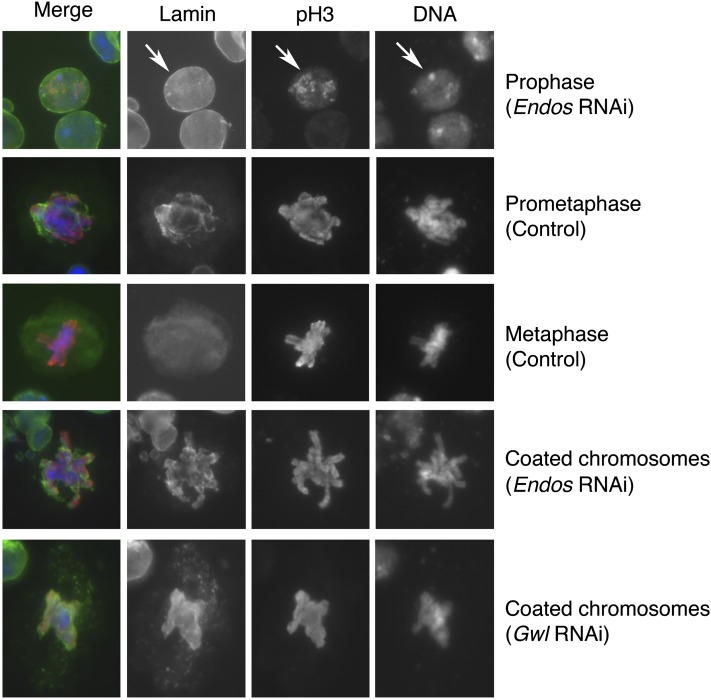
We also examined the requirement for Endos in Drosophila S2 tissue culture cells depleted for Endos by RNA interference (RNAi). Treating the cells with double-strand endos RNA efficiently removed Endos (Figure S1A). Chromosomes prepared from these cells are markedly undercondensed (Figure S1B and Table 2), similar to endos null neuroblasts (see Figure 2). By staining fixed cells for phosphoHistone H3 (pH3; a marker for M phase) and for nuclear lamins, we found that nuclear envelope breakdown was severely compromised in endos RNAi cells (Figure 4). The proportion of pH3-positive cells showing diffuse, metaphase-like lamin staining is thus considerably lower in Endos RNAi cells (13.6%) than in controls (28.4%) (Table 3). Interestingly, endos RNAi cells displayed a unique phenotype in which remnants of the partially broken-down nuclear lamin coated the chromosomes (Figure 4 and Table 3).
Table 2. Effects of RNAi on chromosome condensation (in S2 cells treated with colchicine).
| Genotype | Normal condensation (%) | Undercondensed (%) | Hypercondensed (%) | Hypercondensed +PSCSa (%) |
|---|---|---|---|---|
| Control | 80.3 | 19.7 | 0 | 0 |
| Endos RNAi | 42.0 | 58.0 | 0 | 0 |
| Gwl RNAi | 32.1 | 67.9 | 0 | 0 |
| Endos+Gwl RNAi | 33.2 | 66.8 | 0 | 0 |
| Twins RNAi | 26.0 | 0 | 48.0 | 16.0 |
| Twins+Endos RNAi | 38.1 | 9.5 | 52.4 | 0 |
| Twins+Gwl RNAi | 25.9 | 7.4 | 66.7 | 0 |
n = 500 for each genotype.
Precocious sister chromatid separation.
Figure 4 .
Incomplete nuclear envelope breakdown upon gwl or endos RNAi. S2 cells treated with control dsRNA or with endos or gwl dsRNAs were fixed and stained for DNA, phospho-histone H3 (pH3), and lamin. In prophase cells (top row; arrows), lamin stains the nuclear envelope; cells that do not stain with anti-pH3 are in interphase. In prometaphase cells, the chromosomes are condensed and the nuclear envelope is breaking down. In metaphase cells, the chromosomes have congressed to the metaphase plate, and lamin staining is diffuse throughout the cell. The bottom two rows illustrate phenotypes seen only in gwl or endos RNAi cells, in which lamin closely associates with condensed chromosomes.
Table 3. Nuclear lamin phenotypes in pH 3-positive S2 cells.
| Genotype | Prophase (%) | Prometaphase partial NEB (%) | Prometaphase lamin around chromosomes (%) | Metaphase diffuse lamin staining (%) |
|---|---|---|---|---|
| Control | 19.6 | 52.0 | 0 | 28.4 |
| Endos RNAi | 36.5 | 20.4 | 29.5 | 13.6 |
| Gwl RNAi | 29.5 | 32.0 | 33.6 | 4.7 |
| Endos+Gwl RNAi | 26.8 | 30.9 | 32.6 | 9.7 |
| Twins RNAi | 18.4 | 46.9 | 0 | 34.7 |
| Twins+Endos RNAi | 17.9 | 69.0 | 0 | 13.1 |
| Twins+Gwl RNAi | 19.0 | 62.0 | 0 | 19.0 |
n = 500 for each genotype.
Endos and Gwl function in a common pathway
The effects on cell-cycle progression just described are essentially identical to those caused by depletion of Gwl (Yu et al. 2004; Archambault et al. 2007). Indeed, cells lacking Gwl similarly exhibit delay/arrest prior to metaphase, resulting in chromosome undercondensation and impaired nuclear envelope breakdown (Figure 2, Figure 4, Figure S1, Table 1, Table 2, and Table 3).
To test the idea that endos and gwl operate in a common pathway, we constructed double mutants homozygous for both endos215-4 (null) and gwl716 (a strong hypomorph). These animals show the same neuroblast phenotypes with similar severity as either mutant alone (Figure 2; Table 1). In addition, we found that tissue culture cells simultaneously depleted of endos and gwl by RNAi share the same chromosome condensation and nuclear envelope breakdown defects seen in cells treated with either dsRNA alone (Figure S1; Tables 2 and 3). The close similarities of the double- and single-mutant phenotypes, whether in larval neuroblasts or S2 cells, strongly suggest that endos and gwl function together within a single pathway. However, because some mitotic parameters differ slightly between larval brains of double and single mutants (Table 1), it remains possible that Endos and/or Gwl might have additional functions independent of the other protein. Indeed, we present evidence below suggesting that Endos may have Gwl-independent roles during female meiosis.
Gwl phosphorylates Endos Ser68, enhancing its association with the B55 regulatory subunit of PP2A
In vertebrates, Gwl phosphorylates a single well-conserved site corresponding to Ser68 in Drosophila Endos (see Figure S2). Xenopus and Drosophila Gwl kinases and Endos proteins are interchangeable in this reaction (Figure 5A). Alanine mutations at Ser68 in fly Endos or its equivalent in frog Endos prevent the phosphorylation (Figure 5A). This phosphorylation of Endos by Gwl also seems to occur in vivo. Treatment of Drosophila brains with the phosphatase inhibitor okadaic acid produces phosphorylations that reduce Endos protein’s electrophoretic mobility (Figure S3), consistent with the existence of two mobility forms of Endos in ovaries (Drummond-Barbosa and Spradling 2004). Most of this phosphorylation is due to Gwl kinase, because levels of slow-migrating Endos are much reduced in gwl mutant brains (Figure S3).
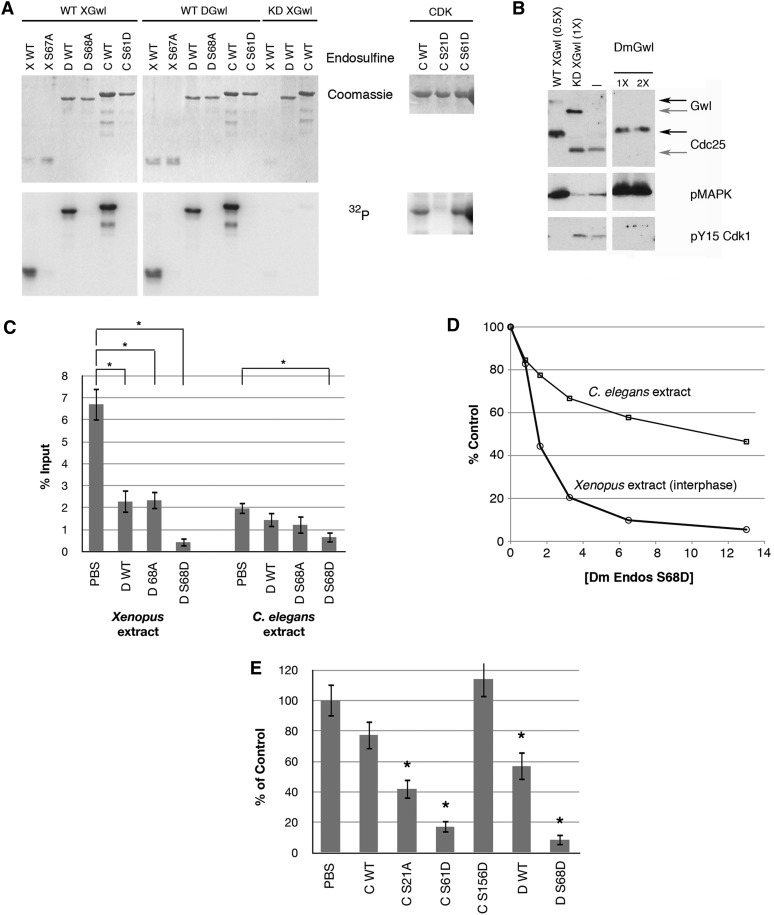
Figure 5 .
Conservation of Gwl and Endos function. (A) In vitro phosphorylation of Endos by Gwl. Recombinant Gwl proteins [wild type (WT) from Xenopus (X) or Drosophila (D); and kinase dead (KD, G41S) from Xenopus] were incubated with recombinant fusion proteins containing the Gwl target site of frog, fly, or worm Endos proteins (X WT, D WT, and C WT, respectively), or mutants in which the target site S was changed to A (X S67A or D S68A) or D (C S61D). WT frog and fly enzymes efficiently phosphorylate WT but not mutant Endos proteins from all three species. Right: in vitro phosphorylation reaction using Cdk2/cyclin A and C. elegans Endos (either WT, or with S21D or S61D mutations). CDK phosphorylation of worm Endos is mostly directed to the S21 site. (B) Fly Gwl functions in frog egg extracts. Endogenous Gwl was depleted from CSF extracts (Yu et al. 2006). The extracts exit M phase within 20 min due to activation of PP2A/B55, as shown by the loss of Cdc25 and MAPK phosphorylations, and the appearance of the Y15 inhibitory phosphorylation of Cdk1. The extracts remain in M phase if supplemented immediately by active WT, but not KD, frog Gwl. WT Drosophila Gwl (DmGwl) also prevents M-phase exit. Black arrows at right indicate phosphorylated M-phase Gwl and Cdc25; gray arrows the unphosphorylated interphase forms. Anti-frog Gwl does not detect fly Gwl on Western blots. 0.5×, 1×, and 2× indicate levels of exogenous Gwl relative to the endogenous levels in extracts. (C) Drosophila Endos works in heterologous systems. Fly Endos (WT, S68A, or S68D) or phosphate buffered saline (PBS) was added at 15 μM to interphase extracts from frog eggs or C. elegans adults whose total protein concentrations were adjusted to be the same. Phosphatase activity directed against a model proline-directed site was assayed as described in Materials and Methods. As previously shown for Xenopus Endos (Gharbi-Ayachi et al. 2010; Mochida et al. 2010), fly Endos suppresses PP2A/B55 phosphatase activity against proline-directed substrates. The S68D phosphomimetic mutant of fly Endos enhances this effect. Fly Endos also inhibits phosphatase activity against the same substrate in C. elegans extracts. (D) Dose–response curve for fly Endos S68D (micromolar concentrations) on dephosphorylation of the same substrate. (E) The S61D phosphomimetic mutant of C. elegans Endos strongly suppresses PP2A/B55 phosphatase. Indicated proteins were added at a final concentration of 15 μM to Xenopus interphase extracts, and the dephosphorylation of the model proline-directed substrate was measured. For parts C and E, N = 5 and asterisks indicate P < 0.05 relative to control.
Given the interchangeability in vitro, we were curious to determine whether fly Endos and/or Gwl proteins can function in Xenopus egg extracts. We first asked if fly Gwl could replace frog Gwl in M-phase-arrested CSF extracts. Immunodepletion of Xenopus Gwl causes rapid M phase exit that can be prevented equally efficiently by adding either frog or fly recombinant Gwl (Figure 5B). This is surprising because Gwl proteins from flies and frogs have insertions of hundreds of unrelated amino acids into the kinase domains (Yu et al. 2004). We next asked whether Drosophila Endos could suppress PP2A/B55 phosphatase in interphase extracts (made by treating CSF extracts with calcium to induce M-phase exit). Addition of fly Endos to these extracts can indeed reduce PP2A/B55 activity against a model CDK-phosphorylated substrate (Figures 5, C–E). Nonphosphorylated Drosophila Endos suppresses PP2A/B55 to some extent, but this effect is strongly enhanced by the S68D phosphomimetic Endos mutant (Figures 5, D and E).
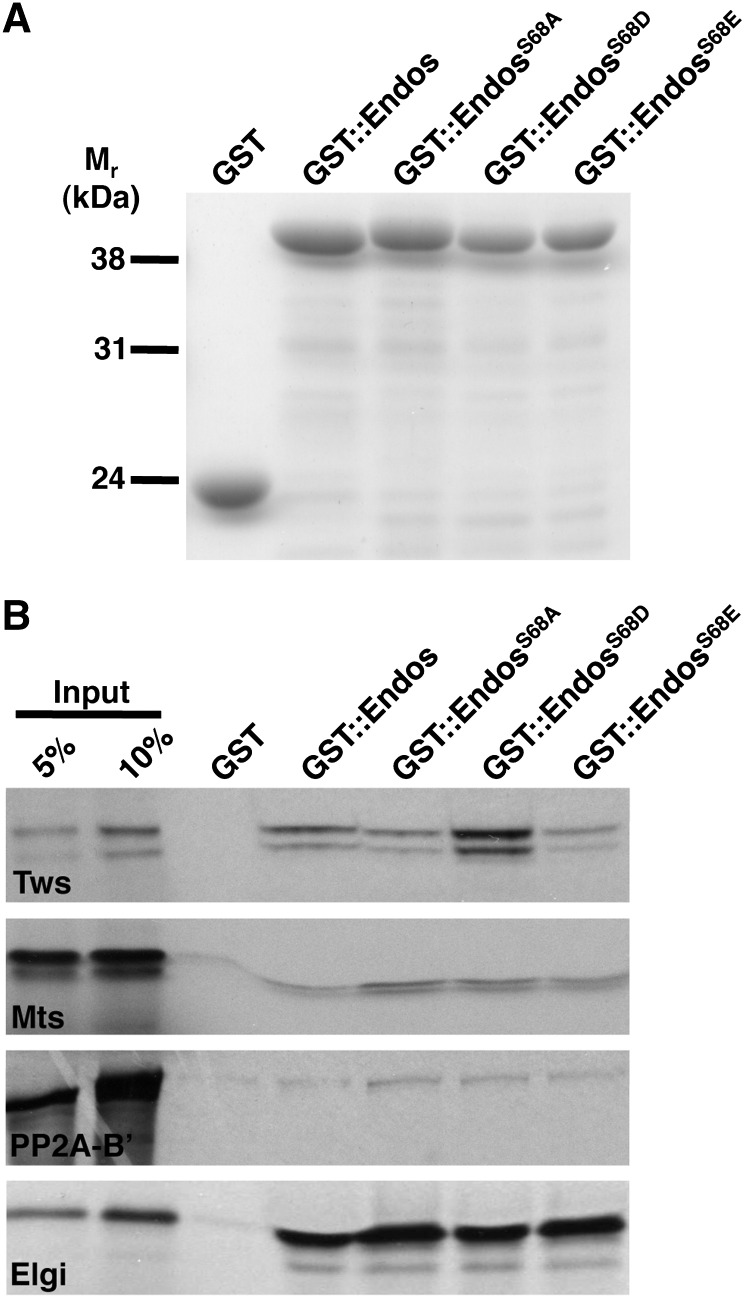
In vertebrate systems, the suppressive effects of Gwl and Endos on PP2A/B55 are thought to be mediated by physical interactions between Gwl-phosphorylated Endos and the phosphatase (Gharbi-Ayachi et al. 2010; Mochida et al. 2010). We have corroborated this idea using Drosophila proteins (Figure 6 and Figure S4). In a pull-down assay, approximately 10% of GST-tagged wild-type fly Endos associates with in vitro transcribed/translated, 35S-labeled Twins, the single Drosophila B55-type protein. This interaction is strengthened when fly Endos is altered with the S to D, but not the S to E, phosphomimetic mutation of Ser68 (Figure 6 and Figure S4), as anticipated by the model.
Figure 6 .
Endos and B55/Twins physically interact in vitro. (A) Coomassie blue-stained gel showing that loading of GST::Endos on beads was similar. (B) Autoradiogram showing that GST::EndosS68D beads bind in vitro transcribed/translated 35S-labeled B55/Twins (Tws) with higher affinity than GST::Endos, GST::EndosS68A, or GST::EndosS68E beads. 35S-labeled microtubule star (Mts, the catalytic subunit of PP2A) and PP2A-B’ (the B56 regulatory subunit of PP2A) associate with the beads at background levels. 35S-labeled Elgi, a strong Endos interactor (Von Stetina et al. 2008), was used as a positive control and GST beads as a negative control.
Gwl phosphorylation of Endos Ser68 is required for meiotic maturation in Drosophila
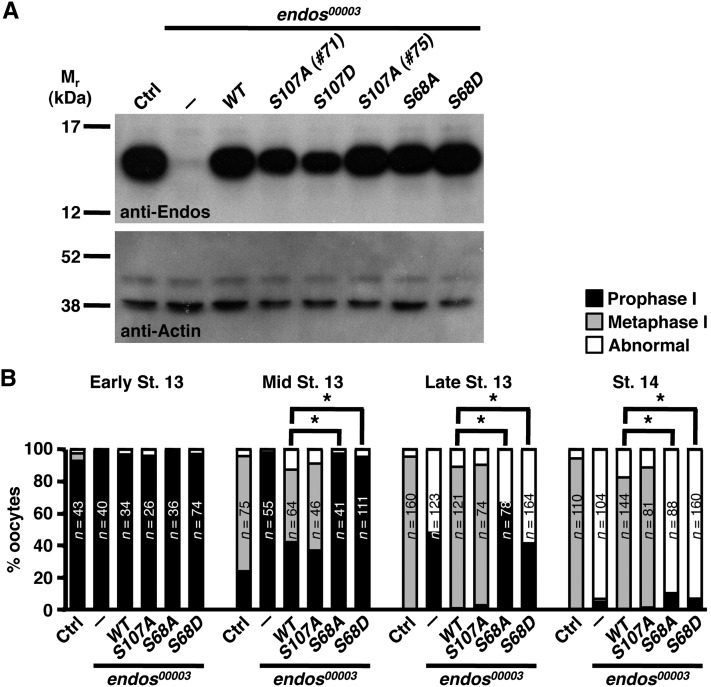
We next explored the in vivo functional significance of Endos phosphorylation by Gwl. We have previously shown that hypomorphic endos00003 homozygous oocytes have impaired progression from prophase I to metaphase I. Germline-specific expression of wild-type Drosophila or human Endos rescues this phenotype and restores fertility to endos00003 females (Von Stetina et al. 2008) (Figure 7). To test if the Gwl phosphorylation site is critical for meiotic maturation, we generated transgenes encoding nonphosphorylatable and phosphomimetic forms of Endos Ser68. These transgenic proteins were stable and expressed at wild-type levels (Figure 7A). Germline expression of unphosphorylatable EndosS68A did not rescue endos00003, consistent with an in vivo requirement for Endos phosphorylation by Gwl (Figure 7B). Expression of phosphomimetic EndosS68D also failed to rescue oocyte meiotic maturation (Figure 7B). These findings suggest that Endos Ser68 phosphorylation by Gwl is critical for meiotic progression and that this phosphorylation must be temporally regulated for proper meiotic maturation. Of interest, Endos with a nonphosphorylatable mutation of the conserved predicted PKA site at Ser107 still rescues the endos mutant phenotype (Figure 7), indicating that Endos activity is unlikely to depend upon PKA phosphorylation.
Figure 7 .
The Gwl target site is specifically required for Endos function in meiotic maturation. (A) Western blots of ovary extracts from wild-type control (Ctrl) or endos00003 females expressing wild-type, phosphomimetic, or nonphosphorylatable Endos proteins. Germline-specific expression of the UAS-endos transgenes was driven by nanos-Gal4::VP16. Two different lines (#71 and #75) of the S107A mutant were analyzed. Actin was the loading control. (B) Percentage of oocytes at the indicated developmental stages in prophase I, metaphase I, or with abnormal DNA morphology, showing that S68 (the Gwl site), but not S107 (the putative PKA site), is required for meiotic maturation. The number of oocytes examined is written inside the bars. Asterisks: P < 0.001.
Twins mutations dominantly suppress endos and gwl loss-of-function in mitosis and male meiosis
The model that Gwl and Endos inhibit the activity of the antimitotic phosphatase PP2A/B55 during M phase suggests that reduction in dosage of B55 might suppress the cell-cycle effects of gwl or endos mutations. Indeed, we observed this to be the case in tissue culture cells. Chromosomes in cells subjected to simultaneous RNAi for gwl and twins, or for endos and twins, were not undercondensed, as was the case upon RNAi for gwl or endos alone. Instead, chromosomes in the double RNAi cells were actually overcondensed. Loss of twins is thus epistatic to loss of gwl or endos, as would be expected from the model (Figure S1 and Table 2). The nuclear envelope in double RNAi cells broke down normally, in contrast with gwl or endos RNAi cells (Table 3).
We also tested this model in vivo by examining the effects on gwl or endos mutants of heterozygosity for the severe loss-of-function allele twinsP. Heterozygosity for twinsP could in fact completely or partially revert many phenotypes associated with gwl or endos loss-of-function. Several combinations of gwl loss-of-function alleles cause aberrant development in which most mutant animals die as late larvae or pupae, although some adult escapers emerge. These escapers eclose several days late, and they show obvious signs of incorrect cell divisions including high mitotic indices in brains; small, rough eyes; and wing margin defects (Table 1). Remarkably, when the twinsP allele halves the twins dosage, these gwl mutants now eclose as adults at the normal time with normal eye and wing morphologies. The mitotic index in the larval brains of these animals is near normal, and the chromosome undercondensation phenotype is reversed (Figure 2). Strikingly, animals completely lacking endos but heterozygous for twinsP also develop into normal adults whose larval brains display mitotic parameters identical with controls (Table 1) and no chromosome undercondensation (Figure 2). Males carrying semilethal or lethal alleles for gwl or endos but heterozygous for twinsP are fertile, but their sisters are not (Table 1), indicating that rescue of female meiosis and/or early embryonic mitoses does not occur.
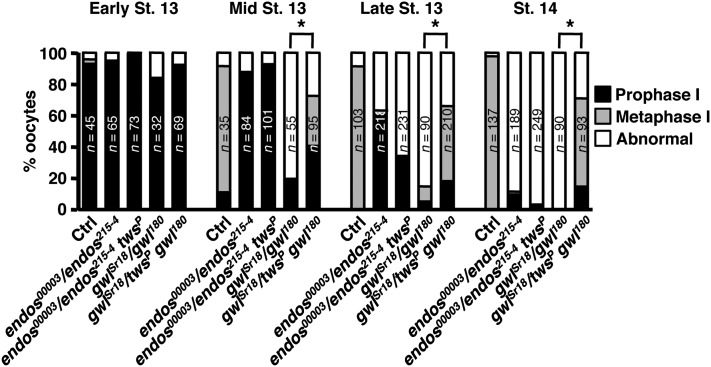
Heterozygosity for twins suppresses ovarian phenotypes associated with female sterile alleles of gwl but not endos
The results just described suggest that female meiotic maturation is more sensitive to the Gwl–Endos system than other types of cell division. Indeed, female sterile alleles of both genes with less severe loss-of-function are known (Archambault et al. 2007; Von Stetina et al. 2008). We directly compared in further detail the meiotic maturation phenotypes of oocytes in viable, essentially hemizygous endos00003/endos215-4 and gwlSr18/gwl180 females. As previously described for endos00003 homozygotes (Von Stetina et al. 2008), endos00003/endos215-4 oocytes show prolonged prophase I, failure to initiate/maintain metaphase I, and highly dispersed or undetectable DNA at stage 14. gwlSr18/gwl180 mutant females also fail to maintain metaphase I; however, they do not show prolonged prophase I (see Figure 8) and have a sister chromatid cohesion defect that results in fewer dispersed DNA masses than seen in endos mutant oocytes (Archambault et al. 2007; Von Stetina et al. 2008).
Figure 8 .
Heterozygosity for twins rescues the meiotic maturation defects of gwl but not endos mutants. Percentage of oocytes at the indicated developmental stages in prophase I, metaphase I, or with abnormal DNA morphology, showing that reducing twins dosage significantly rescues the meiotic maturation defect of gwlSr18/gwl180, but not of endos00003/endos215-4. The number of oocytes examined is written inside the bars. Asterisks: P < 0.001.
To establish whether the Gwl–Endos system also works through PP2A/B55 in female meiosis, we tested whether reduction of twins dosage affects the ovarian defects of gwl or endos viable, female sterile genotypes. Removing one copy of twins clearly rescues a subset of gwlSr18/gwl180 defects (early oogenesis and meiotic maturation; see below), although gwlSr18/twinsP gwl180 females remain sterile, presumably due to later failure during meiosis II or early embryonic mitoses. Almost all gwlSr18/gwl180 females have one or two rudimentary ovaries (likely due to defects in early germ cell mitoses), in agreement with the reduced number of eggs they lay (Figure S5, A, B, and D). In contrast, in gwlSr18/twinsP gwl180, defects in ovarian morphology and egg laying are significantly alleviated (Figure S5, A, C, and D). The twinsP mutation also dominantly suppressed the meiotic maturation defects of gwl mutants. All gwlSr18/gwl180 females fail to progress into (or maintain) metaphase I, while in gwlSr18/twinsP gwl180 approximately 50% of stage 14 oocytes had normal metaphase I DNA morphology (Figure 8). Gwl’s roles in female gonad development and meiosis thus ultimately involve repression of PP2A/B55.
The situation in endos00003/endos215-4 females is different. These females do not have rudimentary ovaries (Drummond-Barbosa and Spradling 2004), perhaps due to higher residual levels of Endos expression in early oogenesis. Nevertheless, the twinsP mutation did not suppress meiotic maturation defects of endos00003/endos215-4 females (Figure 8).
There are two potential explanations for differences in how twins interacts with gwl or endos during meiotic maturation. Either these differences arise from varying strengths of gwl vs. endos female sterile mutant combinations, or they reflect potential gwl- and twins-independent roles of endos during meiosis. The fact that endos strongly interacts with other factors such as the E3 ubiquitin ligase Elgi (Von Stetina et al. 2008) (also see Figure 6) favors the latter possibility.
In C. elegans, deletion of the conserved Gwl target site does not disrupt cell-cycle progression
Given the importance of the Gwl–Endos–PP2A/B55 pathway to cell division in insects and vertebrates, we are intrigued by puzzling aspects of the evolution of this system, particularly with respect to nematodes. Worm genomes do not have obvious orthologs of Gwl, but paradoxically they do encode a single Endos protein that retains the highly conserved sequence motif targeted by Gwl in flies and vertebrates, and by the related kinase Rim15 in yeast (Figure S2). Indeed, fly or frog Gwl can efficiently phosphorylate this conserved site (S61) in C. elegans Endos in vitro (Figure 5A). Moreover, recombinant worm Endos with the S61D phosphomimetic mutant suppresses PP2A/B55 activity in frog extracts (Figure 5E).
To test the importance of this conserved site to the worm, we examined possible phenotypic effects of the mutation ensa-1(tm2810), which deletes 23 amino acids of C. elegans Endos including the conserved residues surrounding the Gwl target site (see Figure S2). We found that animals homozygous or hemizygous for ensa-1(tm2810) are perfectly viable and fecund (Table 4). Strikingly, embryonic development of ensa-1(tm2810) homozygotes occurs in near-perfect synchrony with controls (Figure 9 and File S3). Careful measurements suggest that the first two mitotic divisions of ensa-1(tm2810) embryos may be slightly delayed (∼10% longer) relative to controls (Table 5 and Figure S6), but these minor effects are not characteristic of later cell cycles because mutants hatch at the same time as wild type (Figure 9). We conclude that the conserved Gwl site in Endos is dispensable for somatic and germline cell divisions in C. elegans. We do not presently know whether the remainder of Endos performs an essential function in worms. RNAi of Endos is without obvious effect (Table 4), except perhaps again during the first two embryonic divisions (Table 5). However, the knockdown is incomplete, as 7–10% of the normal amount of ensa-1 mRNA persists (Figure S6C). We therefore cannot exclude the possibility that a null mutation for ensa-1 might affect worm viability or fecundity even if removal of the Gwl site in this protein does not.
Table 4. The C. elegans ensa-1 gene is not essential for viability or germline proliferation.
| Genotype (n) | % embryonic lethality (n) | Average brood ± SD |
|---|---|---|
| 20° | ||
| Wild type (N2) | 0.8 (2257) | 251 ± 30 (9) |
| ensa-1(tm2810) | 0.6 (1673) | 239 ± 24 (7) |
| ensa-1(tm2810)/qDf9a | 26.5 (3556) | 274 ± 43 (12) |
| +/qDf9 | 26.1 (1233) | 206 ± 33 (6) |
| 25° | ||
| Wild type(N2) | 2.1 (2092) | 209 ± 34 (10) |
| ensa-1(tm2810) | 2.3 (1552) | 172 ± 31 (9) |
| ensa-1(tm2810)/qDf9 | 26.4 (2058) | 172 ± 39 (12) |
| +/qDf9 | 28.7 (663) | 174 ± 66 (4) |
| 25° | ||
| L4440b RNAi | 0.8 (2638) | 240 ± 39 (11) |
| ensa-1(tm2810); L4440 RNAi | 0.6 (2369) | 237 ± 41 (10) |
| ensa-1(RNAi) | 1.1 (1816) | 259 ± 32 (8) |
| ensa-1(tm2810); ensa-1(RNAi) | 0.5 (2148) | 239 ± 73 (9) |
| ensa-1(tm2810); glb-30(RNAi)c | 0.7 (1416) | 202 ± 60 (7) |
| cdc-14(RNAi) | 1.2 (1240) | 248 ± 30 (5) |
| ensa-1(tm2810); cdc-14(RNAi) | 2.0 (1424) | 237 ± 72 (5) |
| sur-6(RNAi) | 98.5 (1228) | 136 ± 41 (9) |
| ensa-1(tm2810); sur-6(RNAi) | 99.0 (848) | 106 ± 43 (8) |
qDf9 is a deletion encompassing ensa-1 that also removes sur-6 (endoding the B55 regulatory subunit of PP2A).
Empty vector control.
glb-30 encodes a protein of unknown function with very slight homology to ensa-1.
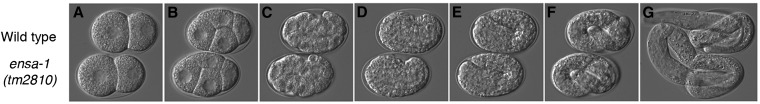
Figure 9 .
ensa-1(tm2810) embryos develop at wild-type rates. Images are taken from a time-lapse movie at 20° (File S3). Wild-type (top) and ensa-1(tm2810) embryos were mounted for viewing just prior to first cleavage. (A) Two-cell stage (time = 14 min after start of movie); (B) four-cell stage (30 min); (C) gastrulation (1 hr 40 min); (D) bean stage (6 hr 15 min); (E) comma stage (7 hr); (F) threefold stage (8 hr, 40 min); (G) hatch (13 hr, 40 min).
Table 5. Mutation or RNAi of the C. elegans ensa-1 gene causes slight delays in AB and P1 cell cycles.
| Genotype | AB cell-cycle time, min ± SD. | P1 cell-cycle time, min ± SD. | N |
|---|---|---|---|
| N2 (wild type) | 15.35 ± 0.24 | 17.30 ± 0.28 | 12 |
| N2, control RNAi | 15.07 ± 0.30 | 17.18 ± 0.38 | 11 |
| ensa-1(tm2810) | 16.31 ± 0.61* | 18.60 ± 0.83* | 17 |
| ensa-1(RNAi) | 16.10 ± 0.26* | 18.34 ± 0.26* | 9 |
| ensa-1(tm2810)/qDf9 | 17.62 ± 0.19* | 20.23 ± 0.30* | 10 |
AB and P1 cell-cycle times were measured from P0 NEBD to AB or P1 NEBD. (*) P < 2.0E-05; T-test to N2.
Discussion
Conserved role of the Gwl–Endos–PP2A/B55 module in Drosophila
The Gwl–Endos–PP2A/B55 module in flies works identically to that in vertebrates. Active fly Gwl phosphorylates Endos at a conserved site (Figure 5A); this phosphorylation enhances Endos’ binding to Twins (Figure 6 and Figure S4), inactivating PP2A/B55 activity against CDK-phosphorylated substates (Figure 5, B–D). Indeed, fly and vertebrate Gwl or Endos are interchangeable in assays for in vitro phosphorylation or suppression of PP2A/B55 in frog egg extracts (Figure 5, A and B). Furthermore, human Endos can, just as wild-type fly Endos, rescue endos00003 female sterility (Von Stetina et al. 2008). This highly conserved module is critical to virtually all types of Drosophila cell cycles, including those in mitotic early embryos; larval neuroblasts; primordial testes; imaginal tissues for the eyes, wings, and abdominal tergites; and tissue culture cells; as well as meiosis in oocytes (Figures 2–4, Figure S1, and Tables 1–3; Yu et al. 2004; Archambault et al. 2007). The major effects of depleting either Endos or Gwl are exerted through the same pathway converging on PP2A/B55. However, either protein might play other roles. For example, Endos interacts genetically and physically with the E3 ubiquitin ligase Elgi (Von Stetina et al. 2008).
During the preparation of this manuscript, two partially overlapping studies reached similar conclusions about the general conservation of this system in flies (Rangone et al. 2011; Wang et al. 2011). One difference is that the null endos215-4 mutants we describe die late in late larval/pupal stages of development, whereas the null endos mutants in the other reports survived as adults. These adult escapers showed clear cell-cycle defects similar to those of flies with trans-heterozygote combinations of gwl mutations (see Table 1), including much delayed eclosion, rough eyes, defective wings, male sterility, and female sterility associated with a failure to lay eggs. The lethality of endos215-4 cannot be due to a second-site lethal mutation on the same chromosome because trans-heterozygotes between endos215-4 and a deletion of this region also die as late larvae/pupae. We can also exclude that the endos215-4 P-element excision grossly disrupts expression of the neighboring gene CG6650 (Figure 1), consistent with the existence of a second transcription initiation site for CG6650 that is intact in endos215-4 (http://www.flybase.org). Of special note, the nonlethal endos1 null allele (Rangone et al. 2011) removes the same portion of CG6650’s 5′-UTR, yet all endos1 phenotypes are rescued by a genomic construct that includes endos but excludes CG6650 (Rangone et al. 2011). Finally, the fact that the lethality of endos215-4 is reversed by removal of one copy of twins argues strongly that this lethality is indeed due to loss of endos function. Most likely, phenotypic differences between endos null alleles reflect varying genetic backgrounds; indeed, we have described the existence of many endos enhancers throughout the genome (Von Stetina et al. 2011). Regardless of the cause, these differences in the gross effects of endos null mutations do not affect our major conclusion that most types of cell division in flies occur essentially normally in the absence of Endos if the Twins dose is lowered.
Suppression of endos or gwl mutations by heterozygosity for twins
The model that Gwl and Endos collaborate to inactivate PP2A/B55 during M phase suggests that loss-of-function mutations in twins (encoding B55) might compensate for loss-of-function of gwl or endos. This prediction was fulfilled in experiments involving both animals and tissue culture cells. The strong twinsP allele dominantly suppresses many effects of mutations in either gwl or endos; suppression is also seen in gwl or endos RNAi S2 cells simultaneously treated with dsRNA for twins (Figures 2, 4, and 8; Figure S1 and Figure S5; and Tables 1 and 2). Most remarkable is the finding that animals homozygous for the endos215-4 null mutation and without any detectable Endos protein (Figure 1C), but heterozygous for twinsP, survive to adulthood with no mitotic difficulties. In contrast with the “escaper” phenotype described above, these adults eclose at the same time as balancer-bearing siblings and have wild-type eyes and wings. Males are completely fertile. Larval brains reveal wild-type values for all mitotic parameters examined (Table 1). Thus, when the dosage of twins is halved, most kinds of cell division are insensitive to the loss of the Gwl/Endos pathway.
The one exception to the normality of endos215-4 twinsP/endos215-4 (or gwl twinsP/gwl) adults is the sterility of females (Table 1). This phenomenon fits both with the existence of gwl and endos female sterile alleles and with the facts that removal of one copy of twins does not rescue meiotic maturation defects in endos00003/endos215-4 females and only rescues a subset of sterility-causing defects in gwlSr18/gwl180 females (Figure 8 and Figure S5). Why are the requirements for Gwl and particularly for Endos stronger during female meiosis (and perhaps early embryonic mitoses) than in other cell divisions, including meiosis in males? One possibility is that Endos and Gwl may have Twins-independent roles specifically during female meiotic maturation, consistent with our identification of the Endos interactor Elgi (an E3 ubiquitin ligase), whose disruption induces premature meiotic maturation (Von Stetina et al. 2008). A second, nonexclusive possibility relates to the fact that oocytes and early embryos display very high levels of Gwl, Endos, and Twins expression (http://flybase.org; Drummond-Barbosa and Spradling 2004), presumably due to the large size of the egg that must eventually support many syncytial embryonic mitoses. Gwl and Endos might be particularly important in eggs and early embryos to counteract the high levels of PP2A/B55 present at these developmental stages.
Role of the Gwl-Endos–PP2A/B55 module in C. elegans
Although the Gwl–Endos–PP2A/B55 pathway is critical to all types of cell division analyzed in Drosophila and vertebrates, it is dispensable for cell divisions in C. elegans (Figure 9, Table 4, and File S3). We presume that elements of this pathway still operate in nematodes, even if cell division can occur normally in their absence. First, it is likely that proline-directed M-phase phosphorylations are removed at anaphase onset in worms by PP2A/B55 as in other multicellular eukaryotes: (a) PP2A and B55 (SUR-6) are both required for mitotic progression in C. elegans embryos (Kao et al. 2004), (b) the dephosphorylation of proline-directed substrates in C. elegans extracts is suppressed by Drosophila S68D Endos (Figure 5C), and (c) Cdc14, the phosphatase that dephosphorylates such sites in yeast (Visintin et al. 1998; Mocciaro and Schiebel 2010; and see below), is unlikely to substitute for PP2A/B55, because loss of Cdc14 in worms does not cause mitotic defects (Table 4; Roy et al. 2011). Second, nematode PP2A/B55 could be regulated through Endos, because worm Endos with a phosphomimetic mutation of the conserved Gwl target site (S61D) suppresses PP2A/B55 activity in frog extracts (Figure 5E). In addition, homozygosity for the ensa-1(tm2810) mutation removing S61 is associated with statistically significant, although minor, delays in progression through the first two embryonic divisions (Table 5 and Figure S6). These delays are slightly more pronounced in ensa-1(tm2810) hemizygotes, suggesting that the mutant allele has residual function (Table 5). Such slight effects could explain the conservation of this site (and indeed of Endos itself) in nematodes, particularly if Endos regulation of PP2A/B55 has a special role when worms are stressed in nature. Given that nematode genomes do not have obvious Gwl homologs (Figure S7A), this scenario would require an as-yet-unknown worm kinase to phosphorylate the Gwl target site in Endos.
The most definitive finding from our investigations in C. elegans is the near-complete absence of phenotypic effects in homozygotes for a deletion of the Gwl target site or in RNAi animals with reduced Endos expression (<10% of wild-type levels) (Figure 9, Table 4, and File S3). These results contrast starkly with the requirements for Gwl and Endos in Drosophila and vertebrate cell cycles. One possible explanation is that worm Endos could be regulated by phosphorylations at sites other than the S61 Gwl site. For example, nematode Endos could be directly regulated by MPF during M phase, without the mediation of Gwl. Only a single proline-directed site in Endos potentially targeted by MPF is conserved throughout the nematode lineage; this is S156 in C. elegans Endos (Figure S2). However, S156D mutant Endos does not inhibit PP2A/B55 in frog egg extracts (Figure 5E). Another candidate proline-directed site is S21, the major target for CDK phosphorylation of C. elegans Endos in vitro (Figure 5A). S21D mutant Endos has modest ability to inhibit PP2A/B55 in frog extracts (Figure 5E), but S21 is not conserved in all nematodes (Figure S2B).
Another interesting possibility for explaining the normality of animals with the ensa-1(tm2810) mutation or subjected to ensa-1 RNAi is the existence of a redundant mechanism for downregulating PP2A/B55 during M phase. This putative system would need to be newly evolved in the nematode lineage or it might exist in other metazoans yet not suffice to compensate for the loss of Gwl/Endos.
The simplest explanation for the normal cell divisions of ensa-1 worms is that the levels of PP2A/B55 in C. elegans cells are so low that its M-phase downregulation assumes less importance. Indeed, C. elegans extracts display less than one-third the dephosphorylation activity against a CDK-phosphorylated substrate than Xenopus egg extracts display at similar protein concentrations (Figure 5C). This hypothesis also fits our findings that mitosis and male meiosis in flies can occur relatively normally in the complete absence of Endos and near absence of Gwl if the dosage of PP2A’s B55 regulatory subunit is cut in half (Figure 2 and Figure S1; Tables 1 and 2).
Evolution and function of the Gwl-Endos–PP2A/B55 module
Gwl and Endos are found only in certain eukaryotic lineages. Genes encoding kinases somewhat related to Gwl appear in all major branches of the Eukaryota (Figure S7A). However, evidence suggests that functional orthologs of vertebrate and insect Gwl are characterized by certain amino acid sequences (residues 180–222, 708–739, and 864–878 in the human enzyme) containing sites whose phosphorylation ensures Gwl’s activation at M phase (Blake-Hodek et al. 2012). By this criterion, authentic Gwl first evolved subsequent to the separation between fungi and metazoans (Figure S7A). Authentic Gwl is widespread throughout all metazoans except the Nematoda, indicating that it was lost specifically in the nematode lineage. In contrast, database searches reveal likely Endosulfine family members in all Unikonts including fungi and nematodes (Figure S7B). An outstanding feature of all these Endos proteins is the remarkable conservation of the Gwl target site (Figure S2), even in nematode species lacking Gwl.
Fungal genomes encode Endosulfine proteins, as well as kinases related to Gwl that lack the sequences required for their M-phase activation. In S. cerevisae, the Gwl-like kinase Rim15 phosphorylates the Igo1 and Igo2 Endos-like proteins at a site homologous to that targeted by Gwl in animal Endos (Talarek et al. 2010). The phosphorylation of Igo1/2 by Rim15 does not influence M-phase entry; instead, this event helps initiate the reversible quiescent state (G0) caused by nutrient limitation. Phosphorylated Igo proteins associate with the mRNA decapping activator Dhh1 to protect specific mRNAs from degradation during the G0 program (Talarek et al. 2010; Luo et al. 2011).
These considerations suggest a possible scenario for Gwl–Endos–PP2A/B55 module evolution. The most primitive Unikonts are likely to have had an Endos-like protein that could be targeted by an ancestral Gwl-like kinase. The phosphorylated Endos-like protein probably influenced pathways other than M-phase control (for example, the mRNA protection pathway during G0 in yeast). Only with the later appearance of authentic Gwl that can be activated specifically during M phase could this module play a decisive role in the interphase-to-M phase switch. This scenario is consistent with the phylogenies in Figure S7, but limited aspects of the pathway’s cell-cycle function might have evolved earlier. The Cek1 Gwl-like kinase in S. pombe acts as a multicopy suppressor of mitotic blockage associated with certain mutations (Samejima and Yanagida 1994; Samuel et al. 2000), although the mechanisms involved are obscure and mutations in cek1 do not obviously affect cell proliferation in otherwise wild-type backgrounds.
One perplexing aspect of the apparent change in the module’s end function concerns the opposite roles played by PP2A/B55 in S. cerevisiae and higher eukaryotes. In metazoans, the primary phosphatase removing CDK-driven phosphorylations during M phase entry is PP2A/B55 (Castilho et al. 2009; Lindqvist et al. 2009; Mochida et al. 2009). PP2A/B55 thus needs to be turned off during M-phase entry and M-phase proper by Gwl and Endos, but this phosphatase must be turned on during M-phase exit and interphase. However, in budding yeast, PP2A heterotrimers with the Cdc55 B55-type regulatory subunit must be turned on during M phase to help disable networks that would otherwise sequester Cdc14 (the major anti-CDK phosphatase in yeast) in the nucleolus (Wang and Ng 2006; Yellman and Burke 2006; Bizzari and Marston 2011; Kerr et al. 2011). Conversely, M-phase exit in yeast requires inactivation of Cdc55 by Separase (Queralt et al. 2006; Queralt and Uhlmann 2008).
These disparate underpinnings to the logic of PP2A/B55 activation and inactivation in yeast and metazoans suggest that implementation of the Gwl-Endos pathway to inactivate PP2A/B55 during M phase is likely to have accompanied PP2A/B55’s acquisition of Cdc14’s role in yeast as the major phosphatase targeting CDK-driven phosphorylations. One interesting speculation is that Gwl-Endos would allow metazoans to accumulate higher levels of PP2A/B55 than yeasts, particularly in large cells such as oocytes. This idea fits recent models suggesting that the Gwl-Endos mechanism for silencing PP2A/B55 allows sharper M phase–interphase transitions (Domingo-Sananes et al. 2011; Krasinska et al. 2011). This hypothesis is also consistent with our findings in flies and worms of cases in which the functions of Gwl and Endos can be bypassed, particularly when the levels of PP2A/B55 are low.
Supplementary Material
Acknowledgments
M.-Y.K., J.R.V.S., X.H., and D.D.-B. designed, performed, and interpreted the experiments shown in Figures 1, 6, 7, 8, and S5. E.B., C.P., M.P.S., and M.L.G. designed, performed, and interpreted the experiments displayed in Figures 2, 3, 4, and S1, and in Tables 1–3. B.C.W. and K.B.-H. designed, performed, and interpreted the experiments documented in Figures 5, S3, and S4. D.G.M. designed, performed, and interpreted the C. elegans work shown in Figures 9 and S6 and in Tables 4 and 5. D.D.-B. and M.L.G. wrote the manuscript. We are grateful to L. Zhang for technical support in generating the endos215-4 null allele and A. Palena for technical assistance in making dsRNAs and performing the cytological analysis of tissue culture cells. We also greatly appreciate the generosity of Professor Maurizio Gatti, in whose laboratory at the Universitá di Roma “La Sapienza” several of the experiments reported here were performed. This work was supported by National Institutes of Health grants R01 GM069875 to D.D.-B. and R01 GM048430 to M.L.G.
Footnotes
Communicating editor: D. I. Greenstein
Literature Cited
- Archambault V., Zhao X., White-Cooper H., Carpenter A. T., Glover D. M., 2007. Mutations in Drosophila Greatwall/Scant reveal its roles in mitosis and meiosis and interdependence with Polo kinase. PLoS Genet. 3: e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., Liao G., He Y., Carlson J. W., et al. , 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzari F., Marston A. L., 2011. Cdc55 coordinates spindle assembly and chromosome disjunction during meiosis. J. Cell Biol. 193: 1213–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake-Hodek K. A., Williams B. C., Zhao Y., Castilho P. V., Chen W., et al. , 2012. Determinants for activation of the atypical AGC kinase Greatwall during M phase entry. Mol. Cell. Biol. 32: 1337–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess A., Vigneron S., Brioudes E., Labbe J. C., Lorca T., et al. , 2010. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc. Natl. Acad. Sci. USA 107: 12564–12569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho P. V., Williams B. C., Mochida S., Zhao Y., Goldberg M. L., 2009. The M phase kinase Greatwall (Gwl) promotes inactivation of PP2A/B55delta, a phosphatase directed against CDK phosphosites. Mol. Biol. Cell 20: 4777–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N., Zhou C., Villen J., Beausoleil S. A., Bakalarski C. E., et al. , 2008. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 105: 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Sananes M. R., Kapuy O., Hunt T., Novak B., 2011. Switches and latches: a biochemical tug-of-war between the kinases and phosphatases that control mitosis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366: 3584–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond-Barbosa D., Spradling A. C., 2004. Alpha-endosulfine, a potential regulator of insulin secretion, is required for adult tissue growth control in Drosophila. Dev. Biol. 266: 310–321 [DOI] [PubMed] [Google Scholar]
- Gatti M., Goldberg M. L., 1991. Mutations affecting cell division in Drosophila. Methods Cell Biol. 35: 543–586 [DOI] [PubMed] [Google Scholar]
- Gatti M., Bonaccorsi S., Pimpinelli S., 1994. Looking at Drosophila mitotic chromosomes. Methods Cell Biol. 44: 371–391 [DOI] [PubMed] [Google Scholar]
- Gharbi-Ayachi A., Labbe J. C., Burgess A., Vigneron S., Strub J. M., et al. , 2010. The substrate of Greatwall kinase, Arpp19, controls mitosis by inhibiting protein phosphatase 2A. Science 330: 1673–1677 [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., et al. , 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Kao G., Tuck S., Baillie D., Sundaram M. V., 2004. C. elegans SUR-6/PR55 cooperates with LET-92/protein phosphatase 2A and promotes Raf activity independently of inhibitory Akt phosphorylation sites. Development 131: 755–765 [DOI] [PubMed] [Google Scholar]
- Kerr G. W., Sarkar S., Tibbles K. L., Petronczki M., Millar J. B., et al. , 2011. Meiotic nuclear divisions in budding yeast require PP2A(Cdc55)-mediated antagonism of Net1 phosphorylation by Cdk. J. Cell Biol. 193: 1157–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasinska L., Domingo-Sananes M. R., Kapuy O., Parisis N., Harker B., et al. , 2011. Protein phosphatase 2A controls the order and dynamics of cell-cycle transitions. Mol. Cell 44: 437–450 [DOI] [PubMed] [Google Scholar]
- Lindqvist A., Rodriguez-Bravo V., Medema R. H., 2009. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J. Cell Biol. 185: 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Talarek N., De Virgilio C., 2011. Initiation of the yeast G0 program requires Igo1 and Igo2, which antagonize activation of decapping of specific nutrient-regulated mRNAs. RNA Biol. 8: 14–17 [DOI] [PubMed] [Google Scholar]
- Mocciaro A., Schiebel E., 2010. Cdc14: A highly conserved family of phosphatases with non-conserved functions? J. Cell Sci. 123: 2867–2876 [DOI] [PubMed] [Google Scholar]
- Mochida S., Hunt T., 2007. Calcineurin is required to release Xenopus egg extracts from meiotic M phase. Nature 449: 336–340 [DOI] [PubMed] [Google Scholar]
- Mochida S., Ikeo S., Gannon J., Hunt T., 2009. Regulated activity of PP2A–B55 delta is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J. 28: 2777–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida S., Maslen S. L., Skehel M., Hunt T., 2010. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science 330: 1670–1673 [DOI] [PubMed] [Google Scholar]
- Queralt E., Uhlmann F., 2008. Separase cooperates with Zds1 and Zds2 to activate Cdc14 phosphatase in early anaphase. J. Cell Biol. 182: 873–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queralt E., Lehane C., Novak B., Uhlmann F., 2006. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell 125: 719–732 [DOI] [PubMed] [Google Scholar]
- Rahmani Z., Gagou M. E., Lefebvre C., Emre D., Karess R. E., 2009. Separating the spindle, checkpoint, and timer functions of BubR1. J. Cell Biol. 187: 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangone H., Wegel E., Gatt M. K., Yeung E., Flowers A., et al. , 2011. Suppression of scant identifies Endos as a substrate of Greatwall kinase and a negative regulator of protein phosphatase 2A in mitosis. PLoS Genet. 7: e1002225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S. H., Clayton J. E., Holmen J., Beltz E., Saito R. M., 2011. Control of Cdc14 activity coordinates cell cycle and development in Caenorhabditis elegans. Mech. Dev. 128: 317–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J. F., Ceron J., Koreth J., Hao T., Nicot A. S., et al. , 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14: 2162–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima I., Yanagida M., 1994. Identification of cut8+ and cek1+, a novel protein kinase gene, which complement a fission yeast mutation that blocks anaphase. Mol. Cell. Biol. 14: 6361–6371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel J. M., Fournier N., Simanis V., Millar J. B., 2000. spo12 is a multicopy suppressor of mcs3 that is periodically expressed in fission yeast mitosis. Mol. Gen. Genet. 264: 306–316 [DOI] [PubMed] [Google Scholar]
- Somma M. P., Ceprani F., Bucciarelli E., Naim V., De Arcangelis V., et al. , 2008. Identification of Drosophila mitotic genes by combining co-expression analysis and RNA interference. PLoS Genet. 4: e1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M., 1982. Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218: 341–347 [DOI] [PubMed] [Google Scholar]
- Talarek N., Cameroni E., Jaquenoud M., Luo X., Bontron S., et al. , 2010. Initiation of the TORC1-regulated G0 program requires Igo1/2, which license specific mRNAs to evade degradation via the 5′-3′ mRNA decay pathway. Mol. Cell 38: 345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Court D. L., Fire A., 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112 [DOI] [PubMed] [Google Scholar]
- Uemura T., Shiomi K., Togashi S., Takeichi M., 1993. Mutation of twins encoding a regulator of protein phosphatase 2A leads to pattern duplication in Drosophila imaginal discs. Genes Dev. 7: 429–440 [DOI] [PubMed] [Google Scholar]
- Vigneron S., Brioudes E., Burgess A., Labbe J. C., Lorca T., et al. , 2009. Greatwall maintains mitosis through regulation of PP2A. EMBO J. 28: 2786–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Craig K., Hwang E. S., Prinz S., Tyers M., et al. , 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2: 709–718 [DOI] [PubMed] [Google Scholar]
- Voets E., Wolthuis R. M., 2010. MASTL is the human orthologue of Greatwall kinase that facilitates mitotic entry, anaphase and cytokinesis. Cell Cycle 9: 3591–3601 [DOI] [PubMed] [Google Scholar]
- Von Stetina J. R., Tranguch S., Dey S. K., Lee L. A., Cha B., et al. , 2008. α-Endosulfine is a conserved protein required for oocyte meiotic maturation in Drosophila. Development 135: 3697–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Stetina J. R., LaFever K. S., Rubin M., Drummond-Barbosa D., 2011. A genetic screen for dominant enhancers of the cell cycle regulator α-Endosulfine identifies matrimony as a strong functional interactor in Drosophila. G3: Genes, Genomes, Genetics 1: 607–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Pinson X., Archambault V., 2011. PP2A-twins is antagonized by Greatwall and collaborates with polo for cell cycle progression and centrosome attachment to nuclei in Drosophila embryos. PLoS Genet. 7: e1002227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ng T. Y., 2006. Phosphatase 2A negatively regulates mitotic exit in Saccharomyces cerevisiae. Mol. Biol. Cell 17: 80–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellman C. M., Burke D. J., 2006. The role of Cdc55 in the spindle checkpoint is through regulation of mitotic exit in Saccharomyces cerevisiae. Mol. Biol. Cell 17: 658–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Fleming S. L., Williams B., Williams E. V., Li Z., et al. , 2004. Greatwall kinase: a nuclear protein required for proper chromosome condensation and mitotic progression in Drosophila. J. Cell Biol. 164: 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Zhao Y., Li Z., Galas S., Goldberg M. L., 2006. Greatwall kinase participates in the Cdc2 autoregulatory loop in Xenopus egg extracts. Mol. Cell 22: 83–91 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Haccard O., Wang R., Yu J., Kuang J., et al. , 2008. Roles of Greatwall kinase in the regulation of cdc25 phosphatase. Mol. Biol. Cell 19: 1317–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.