Abstract
Mating system transitions dramatically alter the evolutionary trajectories of genomes that can be revealed by contrasts of species with disparate modes of reproduction. For such transitions in Caenorhabditis nematodes, some major causes of genome variation in selfing species have been discerned. And yet, we have only limited understanding of species-wide population genetic processes for their outcrossing relatives, which represent the reproductive state of the progenitors of selfing species. Multilocus–multipopulation sequence polymorphism data provide a powerful means to uncover the historical demography and evolutionary processes that shape genomes. Here we survey nucleotide polymorphism across the X chromosome for three populations of the outcrossing nematode Caenorhabditis remanei and demonstrate its divergence from a fourth population describing a closely related new species from China, C. sp. 23. We find high genetic variation globally and within each local population sample. Despite geographic barriers and moderate genetic differentiation between Europe and North America, considerable gene flow connects C. remanei populations. We discovered C. sp. 23 while investigating C. remanei, observing strong genetic differentiation characteristic of reproductive isolation that was confirmed by substantial F2 hybrid breakdown in interspecific crosses. That C. sp. 23 represents a distinct biological species provides a cautionary example of how standard practice can fail for mating tests of species identity in this group. This species pair permits full application of divergence population genetic methods to obligately outcrossing species of Caenorhabditis and also presents a new focus for interrogation of the genetics and evolution of speciation with the Caenorhabditis model system.
Keywords: Caenorhabditis remanei, speciation, molecular population genetics, population structure
EVOLUTIONARY transitions in mating systems can drastically alter population genetic processes, shifting patterns of genetic variability and the efficacy of natural selection (Charlesworth and Wright 2001; Wright et al. 2008). Potentially complicating inferences about the effects of such transitions in nature, past and present demographic histories also shape the population genetic structure and variation of a species. Several plant taxa, in particular, provide some of the most well-described population genetics systems that are composed of both selfing and outcrossing species in the same genus [e.g., Arabidopsis (Ross-Ibarra et al. 2008), Capsella (St. Onge et al. 2011), Eichornia (Ness et al. 2010), Mimulus (Sweigart and Willis 2003), and Solanum (Arunyawat et al. 2007)]. Animal population genetics models of such transitions are rare, with the notable exception of Caenorhabditis nematodes (Cutter et al. 2009). But even in Caenorhabditis, multipopulation and global population genetic studies have been limited primarily to selfing species. A thorough understanding of the effect of the mating system on species-wide and local population diversity for species like Caenorhabditis elegans requires evaluation of many populations for obligately outbreeding relatives. To fill this gap, here we investigate patterns of genetic variation for the outcrossing species Caenorhabditis remanei and its divergence from outcrossing C. sp. 23 in a multilocus, multipopulation framework.
Owing to its experimental tractability and extensive molecular genetic resources, C. elegans remains one of the most widely studied biological model organisms, especially in the realms of developmental biology, neurobiology, and molecular genetics. It now also serves as a model to study evolutionary transitions in the mode of reproduction (Braendle and Felix 2006; Haag 2009), with several species having evolved self-fertile hermaphroditism (androdioecy) from male–female (gonochoristic) ancestors, including C. elegans (reviewed in Kiontke et al. 2004, 2011; Hill et al. 2006; Denver et al. 2011). Although it is uncertain when selfing arose within each lineage, current evidence suggests that the origin of selfing might be relatively recent (Cutter et al. 2008, 2010; Rane et al. 2010), implying that the majority of the time separating extant selfing species from outcrossing relatives must have occurred in the ancestral, gonochoristic state. Population genetic variation for C. remanei has so far focused on only a single population or on small pooled samples (Graustein et al. 2002; Jovelin et al. 2003, 2009; Haag and Ackerman 2005; Cutter et al. 2006b, 2008; Jovelin 2009) and only recently has the related outcrossing species C. sp. 5 been considered in a multipopulation context (Wang et al. 2010; Cutter et al. 2012). C. sp. 5 is found only in eastern Asia, with a much smaller geographic range than the circumglobal north temperate distribution of C. remanei (Sudhaus and Kiontke 2007; Kiontke et al. 2011). Consequently, we still have a narrow view of how evolutionary processes have molded the species-wide genomic landscape for gonochoristic Caenorhabditis species. Understanding the progenitors of self-fertile species like C. elegans requires a solid understanding of extant representatives that share the ancestral reproductive state.
Individual wild strains of C. remanei have been collected previously from across the Northern Hemisphere, but the only large population sample collected so far, in which worms were found in phoretic association with several species of wood lice, derives from Ohio (Baird 1999). Previous work showed that genetic diversity in this population is ∼20-fold greater than in global samples of self-fertilizing relatives and that it has not been subject to strong demographic perturbations (Graustein et al. 2002; Jovelin et al. 2003; Cutter et al. 2006a; Cutter 2008; Jovelin 2009). And yet, it is unclear how much genetic variation differs among populations of C. remanei, to what extent they are interconnected by gene flow, and whether they have been afflicted by complex demographic changes and/or locally restricted natural selection. Because it is critical to analyze DNA sequence variation at multiple loci across multiple populations to address these issues, here we investigate diversity at 20 nuclear loci from three populations of C. remanei (Ohio, Ontario, Germany), representing much of the known geographic range for this species. In the process of exploring C. remanei population variation, we discovered a fourth population representing a new species, C. sp. 23, that is partially reproductively isolated from C. remanei. Because of high divergence between previously characterized relatives, cases of young species pairs have been virtually nonexistent in Caenorhabditis. One recent exception is the discovery that Caenorhabditis briggsae and C. sp. 9 are incompletely reproductively isolated from each other and capable of producing fertile hybrids (Woodruff et al. 2010; Kozlowska et al. 2012). Our finding of partial reproductive isolation between C. remanei and C. sp. 23 is distinctive in that it includes two species of Caenorhabditis that must outcross obligately and for which we have population samples of each. This provides a powerful new model for research in divergence population genetics and speciation.
Materials and Methods
Nematode isolation from natural populations
We isolated 17 isofemale lines of C. remanei from the Koffler Scientific Reserve located in King City, Ontario, Canada, referred in this study as the “Ontario” strains, during fall 2007 (Supporting Information, Table S1). These strains were associated with several different species of terrestrial isopods (pill bugs/wood lice) in rotten logs and old wood. We also included 19 isofemale strains from near Kiel, referred to as the “German” strains gifted to us by Hinrich Schulenburg; one strain each from Tennessee and Maine, courtesy of Erik Andersen; one strain from Jiangsu, China (JU724); and two strains from Japan, courtesy of Marie-Anne Felix. Finally, we collected 9 additional isofemale lines from Wuhan City, China, referred to as the “Wuhan” or “Chinese” strains in combination with JU724. These strains were isolated from either pill bugs or rotten vegetative matter in the soil (Table S1). Mating tests of samples from both Ontario and Wuhan with C. remanei strain SB146 produced viable progeny, leading us to initially infer species identity as C. remanei. However, our identification of high genetic differentiation of the Wuhan strains with those from Ontario, Ohio, and Germany during the course of this study led us to test species cohesion further. Subsequent crosses yielded considerable F2 hybrid breakdown between Ohio and Wuhan populations (reported in Results), leading us to conclude that the Wuhan samples represent a distinct species closely related to C. remanei. The Wuhan and JU724 strains were then designated C. sp. 23 (K. Kiontke, personal communication).
Molecular methods
Genomic DNA was amplified from a single male worm from each isofemale line by directly adding the single male to Qiagen Repli-g Mini kit reactions as described (Cutter 2008). Diluted aliquots of the amplified samples were then used as template for locus-specific amplification by PCR, using 20 primers used previously (Cutter 2008) (Table S2). These loci represent an arbitrary set of genes with long exons, distributed across the putative X chromosome of C. remanei. The focus on the X chromosome aimed to ease analysis by avoiding heterozygous base calls in the sequence reads from male DNA samples, as males are hemizygous for the X chromosome. Both the forward and the reverse strands were sequenced by the University of Arizona Genetics Core sequencing facility. For the complete data set of 20 loci, the number of isofemale strains for which we obtained sequence data are indicated in Table S2. We were unable to obtain sequence information for loci of some strains because of failed PCR and sequencing reactions. Strains with missing data were excluded from some analyses. The new data were combined in analysis with published nucleotide polymorphism data for the Ohio population and reference strains PB4641 and SB146 (Cutter 2008). New sequences have been deposited in GenBank under accessions JX077956–JX078938.
Sequence alignment and analysis
We confirmed sequence quality and aligned and manually edited sequences with the programs SEQUENCHER 4.10 and BioEdit 7.0.9. For each sampling locality, we used DnaSP 5.10 (Librado and Rozas 2009) to calculate standard population genetics summary statistics. Polymorphism was calculated from pairwise differences (πsyn-JC, πrep) and segregating sites (θsyn, θrep) separately for synonymous sites and replacement sites, using the Jukes–Cantor multiple hits correction (Jukes and Cantor 1969). A further correction for codon usage bias (FOP) was applied to the estimates of πsyn-JC to remove correlation between synonymous diversity and codon bias because translational selection induces codon bias in highly expressed genes in C. remanei (Cutter and Charlesworth 2006; Cutter et al. 2006b). We consider these adjusted values (πneu, θneu) to be our most accurate estimates of neutral polymorphism [πneu = πsyn-JC – (b – a × FOP) + (b – a × 0.36)], where b and a represent the intercept and slope of a linear regression of πsyn-JC on FOP, calculated separately for each population of C. remanei. For populations from Ohio, Ontario, and Germany, respectively, our correction used values for b of 0.0988007, 0.0932738, and 0.0564747; values for a were 0.0997853, 0874978, and 0.0436242. The value FOP = 0.36 represents unbiased codon usage, given the C. elegans optimal codons (Stenico et al. 1994), which are conserved among species of Caenorhabditis (Cutter et al. 2008). This correction for codon bias was applied to the data set of synonymous polymorphisms in C. remanei, but not C. sp. 23 because FOP was found not to be correlated to πsyn-JC in C. sp. 23. The numbers of synonymous and nonsynonymous segregating sites, including numbers of biallelic and triallelic sites, were tallied for each population. Additionally, each population was assessed for the number of shared and unique polymorphisms. Tests of neutrality were conducted for each population, using C. sp. 23 as outgroup for C. remanei populations and using C. remanei genome reference strain PB4641 as outgroup for the C. sp. 23 Wuhan population [Tajima’s D for synonymous site and nonsynonymous sites (Tajima 1989), Fu and Li’s D, F, D*, and F* (Fu and Li 1993), Fay and Wu’s H (Fay and Wu 2000)]. Furthermore, we calculated several population differentiation statistics: FST, NST (Jukes–Cantor-corrected FST), GST, average number of nucleotide substitutions per site (Dxy), and the net nucleotide substitutions per site (Da) between pairs of populations, after defining the populations in DnaSP. We computed pairwise divergence between species at synonymous sites using the Nei and Gojobori method implemented in DnaSP (Nei and Gojobori 1986). Neighbor-network trees were created using SPLITSTREE 4.10 from concatenated sequences (Huson and Bryant 2006), which explicitly displays recombination in the ancestry of the sample.
Population structure analysis
We used the Bayesian clustering algorithm implemented in the program STRUCTURE 2.3 (Pritchard et al. 2000; Falush et al. 2003) to further investigate population genetics structure among populations of C. remanei and C. sp. 23. A subset of 17 loci was used in the analysis of C. remanei populations alone and also for the combined data set containing both C. remanei and C. sp. 23. We used these 17 loci for which we had the least missing data (n = 39 and n = 46 haploid individuals when C. sp. 23 was excluded or included, respectively). The STRUCTURE 2.3 algorithm assumes independence of loci, with no background linkage disequilibrium (LD) within populations. We considered each polymorphic site to represent a distinct locus [n = 508 and n = 897 single nucleotide polymorphisms (SNPs) when C. sp. 23 was excluded or included, respectively], which formally violates the background LD assumption for sites within a gene. However, construction of haplotypes per gene yields unique haplotypes for each individual, which precludes further haplotype analysis by STRUCTURE. Moreover, LD decays rapidly in C. remanei (Figure S1), suggesting that complications due to linkage within genes might not be too problematic. Furthermore, our analysis incorporates sites from 17 independent genes that will mitigate among-site LD, so we proceeded, presuming that signals of population structure will dominate over any background levels of LD.
We explored several model options in STRUCTURE, but settled on the “linkage” model with correlated allele frequencies, which can account for non-independence between the SNPs within genes as a function of the base-pair distances between SNPs that we defined. We initially ran four independent runs of K = 1 genetic clusters to estimate the allele frequency prior parameter, λ. We then fixed the estimated value λ = 0.62 in all further runs. We ran STRUCTURE for clusters ranging from K = 1 to K = 8, with 10 independent runs for each K and a burn-in period of 100,000 steps followed by 500,000 iterations. We determined the K with highest median probability of the data across runs and also used the ΔK method for inferring the most appropriate K (Evanno et al. 2005). Then, for a chosen K value, we used the run that had the highest likelihood estimate to assign cluster proportions to individuals. We carried out runs on two data partitions: (i) the combined data set for strains of both C. remanei and C. sp. 23 and (ii) the C. remanei strains alone. The STRUCTURE output was then visualized with the help of the program Structure Harvester (Earl and vonHoldt 2011).
Hybrid crosses between C. remanei and C. sp. 23
As an initial investigation into reproductive isolation between C. remanei and C. sp. 23, we performed interspecific F1 hybrid crosses between the two species and compared them with intraspecific crosses of each species. We used three isofemale strains from the Ohio population of C. remanei (PB213, PB214, PB219) and three isofemale strains of C. sp. 23 (VX0081, VX0084, and VX0088) for performing the crosses. Intraspecific crosses were performed between distinct strains to avoid any confounding of inbreeding depression (Dolgin et al. 2007). A total of 10 replicates of intraspecific C. remanei crosses, 11 replicates of intraspecific C. sp. 23 crosses, and 17 replicates of interspecific crosses were performed between different strain combinations of the two species. The 17 interspecific crosses included 9 replicates with C. remanei as the female parent, with the remaining 8 replicates involving the reciprocal with C. sp. 23 as the female parent. Each cross allowed one virgin female to mate with four to six males for 24 hr, after which the males were removed. The females were picked as fourth stage larvae (L4) to ensure that they were virgin. F1 progeny grew until adulthood, after which they were placed at 4° to stop their development for counting. We counted the total lifetime progeny produced from each cross. Subsequently, as the first batch of F1 hybrids grew into adults, we placed one L4 female with four to six males to perform the F1 × F1 sibling crosses. The F2 progeny, as for the F1’s, were allowed to grow into adults and then counted. We counted total lifetime numbers of F2 progeny from each of the crosses. For the F2 data set, we obtained a total of 9 replicates of each intraspecific cross type and 15 replicates for the interspecific crosses. We cultured and performed all crosses on NGM-lite agar petri plates seeded with Escherichia coli strain OP50 and maintained the crosses at 25°.
Results
Levels of nucleotide polymorphism
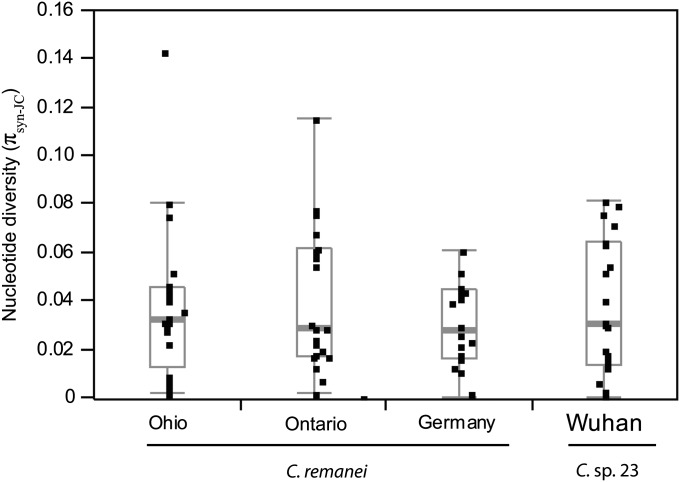
We quantified nucleotide polymorphism for 20 loci from 49 isofemale strains of C. remanei, representing three collection localities, as well as 9 isofemale strains of its putative sister species, C. sp. 23. Our estimates of polymorphism derive from 14.6 kbp of coding sequences for each strain. Local diversity in the three populations of C. remanei averaged πneu = 5.5% (mean πsyn-JC = 3.6% uncorrected for codon bias), with polymorphism in a pooled sample of these populations being somewhat higher (pooled πneu = 6.2%; pooled πsyn-JC = 4.3%); note the strong influence of correcting for codon bias on estimates of neutral polymorphism (Figure S2). The three populations of C. remanei (Ohio, Ontario, and Germany) did not differ significantly from each other in overall levels of synonymous-site nucleotide diversity (population mean πneu across loci ranged from 4 to 6.3%) (Figure 1, Table 1). The German population of C. remanei, however, had nominally lower genetic diversity than both of the North American populations (Figure 1, Table 1). The Wuhan population sample of C. sp. 23 had comparable levels of synonymous-site diversity that were not significantly different from those of the C. remanei populations (average πsyn-JC = 3.7%; θsyn = 3.2%) (Figure 1, Table 1, and Table S3). As expected, nucleotide diversity at nonsynonymous sites was much lower than that at synonymous sites, with πrep averaging ∼0.08% across populations of C. remanei and 0.097% for C. sp. 23 (Table S3). The ratio of replacement to synonymous polymorphism (πrep/πsyn-JC) averaged just 0.023 in C. remanei. Despite the similarity in mean diversity across populations, there is considerable heterogeneity in the amount of nucleotide polymorphism among the 20 loci examined here (Figure 1). At the upper extreme, the locus Cre-D1005.1, which was previously reported to be particularly polymorphic (πsyn = 12.8%) (Cutter 2008), was estimated to have similarly high levels of nucleotide polymorphism for all the populations studied (average πneu = 11.8%).
Figure 1 .
Synonymous-site nucleotide diversity (πsyn-JC, Jukes–Cantor corrected for multiple hits) in local population samples of C. remanei and C. sp. 23. Boxplots indicate the median (thick gray bar) and interquartile range of values. Points beyond the whiskers represent potential outliers.
Table 1. Summary of nucleotide diversity for each locus and population sample.
| πsyn-JC |
πneu |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
C. remanei |
C. sp. 23 |
C. remanei |
C. sp. 23 | |||||||
| Locus | Ohio | Ontario | Germany | Pooled | Wuhan | Ohio | Ontario | Germany | Pooled | Wuhan |
| Cre-dpy-8 | 0.02970 | 0.01688 | 0.02108 | 0.03760 | 0.06357 | 0.06063 | 0.04400 | 0.03460 | 0.06110 | 0.06357 |
| Cre-vit-2 | 0.02806 | 0.02877 | 0.02903 | 0.03906 | 0.02899 | 0.08105 | 0.07523 | 0.05219 | 0.07932 | 0.02899 |
| Cre-lam-2 | 0.03138 | 0.01687 | 0.02325 | 0.01563 | 0.04049 | 0.05563 | 0.03813 | 0.03385 | 0.03405 | 0.04049 |
| Cre-ncr-1 | 0.00886 | 0.03053 | 0.01097 | 0.02013 | 0.01272 | 0.02014 | 0.04042 | 0.01590 | 0.02870 | 0.01272 |
| Cre-pcca-1 | 0.03297 | 0.06802 | 0.03884 | 0.05633 | 0.06401 | 0.06460 | 0.09576 | 0.05267 | 0.08036 | 0.06401 |
| Cre-nmy-1 | 0.02192 | 0.06107 | 0.04104 | 0.04767 | 0.03128 | 0.03938 | 0.07638 | 0.04867 | 0.06094 | 0.03128 |
| Cre-glit-1 | 0.04397 | 0.05827 | 0.04538 | 0.05948 | 0.00228 | 0.04896 | 0.06264 | 0.04756 | 0.06327 | 0.00228 |
| Cre-spc-1 | 0.00779 | 0.01222 | 0.01242 | 0.01132 | 0.05211 | 0.04192 | 0.04214 | 0.02734 | 0.03725 | 0.05211 |
| Cre-myo-2exon8 | 0.00333 | 0.00161 | 0.05160 | 0.03216 | 0.02996 | 0.04394 | 0.03722 | 0.06936 | 0.06302 | 0.02996 |
| Cre-lfi-1 | 0.07481 | 0.07799 | 0.00000 | 0.06617 | 0.07619 | 0.09447 | 0.09523 | 0.00000 | 0.08111 | 0.07619 |
| Cre-cht-1 | 0.04188 | 0.02193 | 0.02561 | 0.02290 | 0.07951 | 0.06523 | 0.04240 | 0.03582 | 0.04064 | 0.07951 |
| Cre-myo-2exon7 | 0.00611 | 0.01744 | 0.01792 | 0.02335 | 0.01542 | 0.04263 | 0.04946 | 0.03389 | 0.05110 | 0.01542 |
| Cre-pgp-14 | 0.04635 | 0.05480 | 0.04349 | 0.05390 | 0.07121 | 0.06740 | 0.07326 | 0.05269 | 0.06990 | 0.07121 |
| Cre-Y102A11A.8 | 0.08009 | 0.06205 | 0.06049 | 0.07572 | 0.05422 | 0.08169 | 0.06345 | 0.06119 | 0.07693 | 0.05422 |
| Cre-ifa-1 | 0.03084 | 0.02853 | 0.01556 | 0.03106 | 0.01735 | 0.06706 | 0.06029 | 0.03140 | 0.05858 | 0.01735 |
| Cre-D1005.1 | 0.14298 | 0.11562 | 0.04504 | 0.12308 | 0.08109 | 0.16463 | 0.13461 | 0.05451 | 0.13953 | 0.08109 |
| Cre-F47A4.5 | 0.04043 | 0.02375 | 0.02588 | 0.03313 | 0.00000 | 0.04582 | 0.02847 | 0.02824 | 0.03722 | 0.00000 |
| Cre-let-2 | 0.00204 | 0.00743 | 0.00171 | 0.00378 | 0.01930 | 0.04694 | 0.04680 | 0.02134 | 0.03790 | 0.01930 |
| Cre-alg-1 | 0.05156 | 0.07616 | 0.04557 | 0.07887 | 0.00000 | 0.07401 | 0.09585 | 0.05539 | 0.09593 | 0.00000 |
| Cre-E01G6.1 | 0.03526 | 0.01955 | 0.04314 | 0.03859 | 0.00597 | 0.05143 | 0.03372 | 0.05021 | 0.05087 | 0.00597 |
| Mean | 0.03802 | 0.03997 | 0.02990 | 0.04350 | 0.03728 | 0.06288 | 0.06177 | 0.04034 | 0.06239 | 0.03728 |
πsyn-JC, synonymous-site diversity corrected for multiple hits; πneu, neutral synonymous-site diversity corrected for codon bias.
Given the relatively high diversity in C. remanei and C. sp. 23, we also scored the number of sites that were hit multiply that gave rise to more than two variants segregating per site. However, populations did not differ notably in the number of triallelic segregating sites (with the exception of locus Cre-D1005.1); we observed no tetra-allelic polymorphisms in any of the populations. In Cre-D1005.1, we found six triallelic sites segregating in the Ohio sample of C. remanei, compared to zero triallelic sites for this locus in the Ontario and German samples.
Tests of neutrality and deviations from population equilibrium
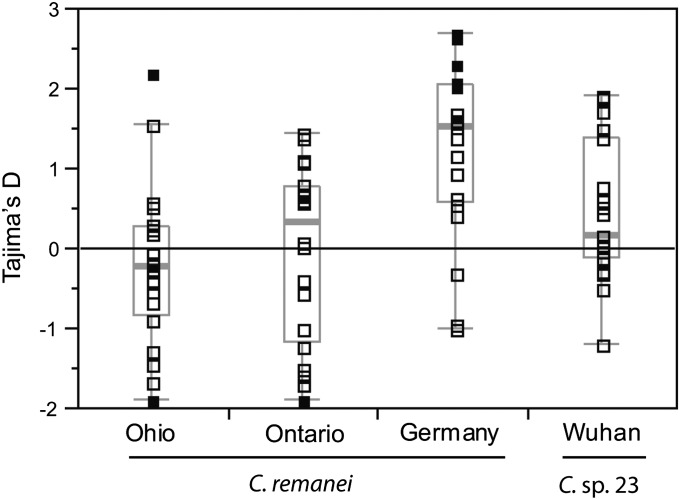
Demographic processes like population-size changes, structure, and migration can skew the spectrum of variant site frequencies all across the genome relative to neutral equilibrium conditions. We used tests of neutrality to detect and infer gross demographic perturbations from widespread deviations across the 20 loci on the X chromosome. Overall, the two North American (Ohio and Ontario) populations3 of C. remanei do not deviate substantially from neutral expectations for the site-frequency spectrum. As shown previously (Cutter 2008), there is a slight trend toward a negative skew in the variant frequencies in the Ohio population (mean synonymous-site Tajima’s D = −0.203) (Figure 2; Table S4). This negative skew is also reflected in Fu and Li’s D and F statistics and in Fu and Li’s D* and F* statistics (Table S4), which focus on derived allele frequencies with respect to the inferred states relative to outgroup C. sp. 23. Tajima’s D (DTaj) for the Ontario population has very little skew, on average (mean synonymous-site DTaj = −0.037; median DTaj = 0.32), although the distribution of DTaj values across loci appears somewhat bimodal (Figure 2). The German sample, on the other hand, has an overall positive skew in the variant frequency spectrum (mean synonymous-site DTaj = +1.3; mean Fu and Li’s D and F are 1.40 and 1.66, respectively). We found 6 of 20 loci for the German sample to have significantly positive values of DTaj and observed that 10 of the 20 loci have significantly positive values for Fu and Li’s D and F statistics (Table S4). Given these dramatic multilocus deficits of rare variants in the German sample, we hypothesize that demographic events specific to the German population sample of C. remanei have drastically affected its patterns of polymorphism. C. sp. 23 has a slightly positive mean Tajima’s D across loci (+0.45), but no loci yielded individually significant deviations from zero. Fu and Li’s D and F statistics in C. sp. 23 also support this slight positive skew, and four loci deviate significantly for D and F from the expectations of neutral equilibrium (Cre-dpy-8, Cre-vit-2, Cre-lam-2, Cre-lfi-1; Table S4), but this test might be anti-conservative for these four loci, which have zero singletons segregating.
Figure 2 .
Tajima’s D measure of the site-frequency spectrum for synonymous sites in populations of C. remanei and C. sp. 23. Boxplots show the median (thick gray bar) and interquartile range of values, with points beyond the whiskers indicating potential outliers. Solid boxes indicate values of Tajima’s D that differ significantly from standard neutral expectations; open boxes represent nonsignificant values.
Even though there is no overall deviation from neutrality in the Ohio and Ontario population samples, two of the loci have significantly negative values of Tajima’s D (both Cre-spc-1 in the Ohio population and Cre-E01G6.1 in the Ontario population have DTaj = −1.88, P < 0.05). Both of these loci also differ significantly from neutrality for Fu and Li’s D* and F*, although McDonald–Kreitman tests (McDonald and Kreitman 1991), using C. sp. 23 as the outgroup, failed to detect any signature of positive selection on the partial coding sequence available to us. We noted strong negative values for Fay and Wu’s H at Cre-spc-1 in all three populations of C. remanei (H = −4.04, −4.15, and −3.35 for Ohio, Ontario, and Germany, respectively), further indicating that some portion of this gene, or a closely linked locus, could be a possible target of a selective sweep.
Population differentiation
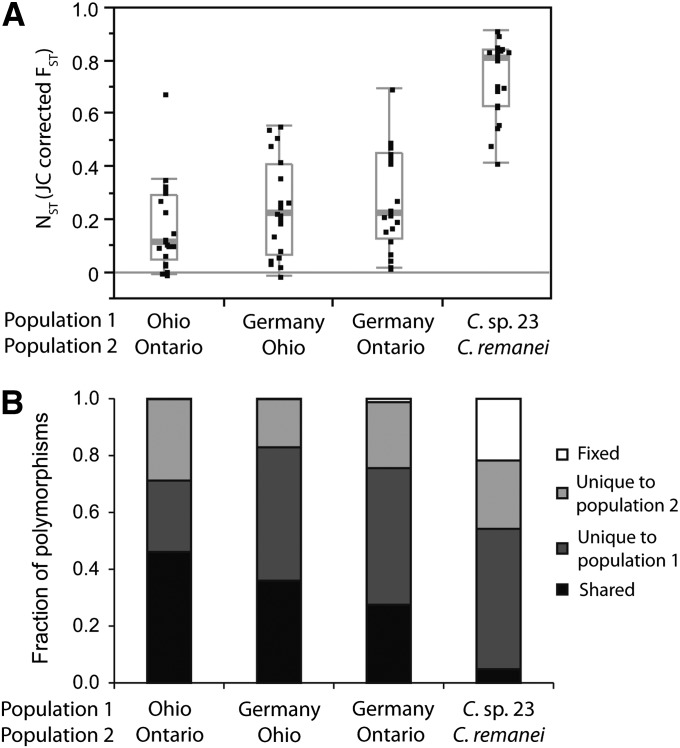
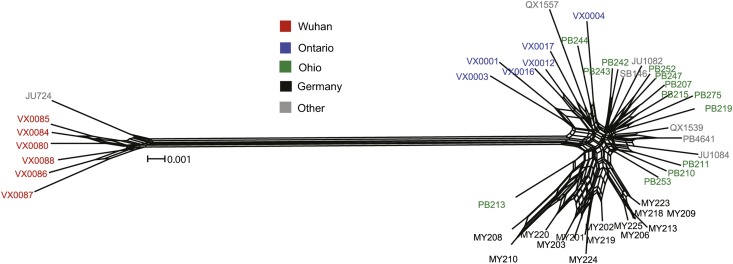
We sought to determine how much of C. remanei’s genetic diversity, species-wide, could be explained by differences among populations. In this context, we note that, at the onset of this study, we presumed strains from Wuhan, China (later designated C. sp. 23) to be simply another population of C. remanei, owing to the presence of fertile F1 progeny in mating tests to a standard isofemale strain of C. remanei. After observing extremely high population genetic differentiation, we hypothesized and subsequently confirmed that C. sp. 23 represents a species closely related to C. remanei that is partially reproductively isolated from it. Specifically, we calculated standard metrics of population differentiation between pairwise populations of C. remanei and between C. remanei and C. sp. 23, finding substantial differentiation for each of the following: FST, NST, GST, DXY, and Da (Table S5). FST between species averaged 0.73 across loci, being significantly > 0 for every locus in pairwise comparisons of the Wuhan sample of C. sp. 23 strains to each of the three C. remanei populations (Figure 3A). Similarly, measures of average and net nucleotide divergence also are high between species (mean DXY = 0.036, mean Da = 0.027), and synonymous-site pairwise divergence averages KS-JC = 0.168. The differentiation between C. sp. 23 and populations of C. remanei is visually clear in the neighbor-network tree derived from the concatenated data set of all strains of C. remanei and C. sp. 23 (Figure 4). Finally, we gained some further insight into the population differentiation between the two species by analyzing the number of unique and shared variants between them. C. sp. 23 shares only 4.8% of variants with C. remanei, indicating that 95.2% of the variants are unique to one species or the other, with 21.8% representing fixed differences (Figure 3B).
Figure 3 .
Population differentiation within C. remanei and between C. remanei and C. sp. 23. (A) Boxplots of NST (Jukes–Cantor corrected FST) for pairs of populations or species showing median (thick gray bar) and interquartile range of values, with values beyond the whiskers indicating potential outliers. (B) Cumulative proportions of fixed, unique, and shared polymorphisms indicate the disparity of population pairs within C. remanei relative to C. sp. 23.
Figure 4 .
Neighbor network for globally sampled C. remanei and C. sp. 23 based on concatenated sequence for 17 nuclear loci. Population samples of C. remanei (Ohio, Ontario, and Germany) and C. sp. 23 (Wuhan) are color coded. Strains labeled in gray represent geographically isolated samples. Nucleotide distances (Jukes–Cantor corrected) exclude gaps; reticulation indicates potential recombination in the ancestry of the strains.
We also quantified differentiation among the three populations of C. remanei. Pairwise comparison of the German population with the Ohio and Ontario populations yielded a slightly stronger average level of differentiation (mean FST = 0.24 and 0.28, respectively) compared to pairwise differentiation between Ohio and Ontario (mean FST = 0.17) (Figure 3A). On a per-locus basis, a considerable number of loci yielded significant population differentiation, albeit generally of moderate magnitude: 17 of 20 for Ontario–Germany and 15 of 20 loci for the Ohio–Germany pairwise comparison. In contrast, only 9 of the 20 loci in the Ohio–Ontario pairwise comparison were found to have an FST significantly greater than zero. Whether this moderate disparity in the level of genetic structure for intracontinental vs. intercontinental populations of C. remanei reflects an effect of isolation by distance is unclear at this point. More population sampling from continental Europe and elsewhere is required to determine whether a strong signal of genetic isolation with geographic distance is present, as in C. sp. 5 (Wang et al. 2010). Hence, we presently conclude simply that our data indicate that the German population is moderately genetically differentiated from the North American ones. Among the three C. remanei populations analyzed pairwise, Ohio and Ontario share the most polymorphic variants (248) (Figure 3B), whereas either of them paired with the German sample share fewer variants (135 and 181, respectively, for Ontario and Ohio). This is consistent with the FST evidence that differentiation is lower for Ohio–Ontario than for Ohio–Germany and Ontario–Germany. Only moderate population structure, despite an oceanic divide, suggests a picture of ongoing, extensive migration among the collection localities of C. remanei.
Population structuring
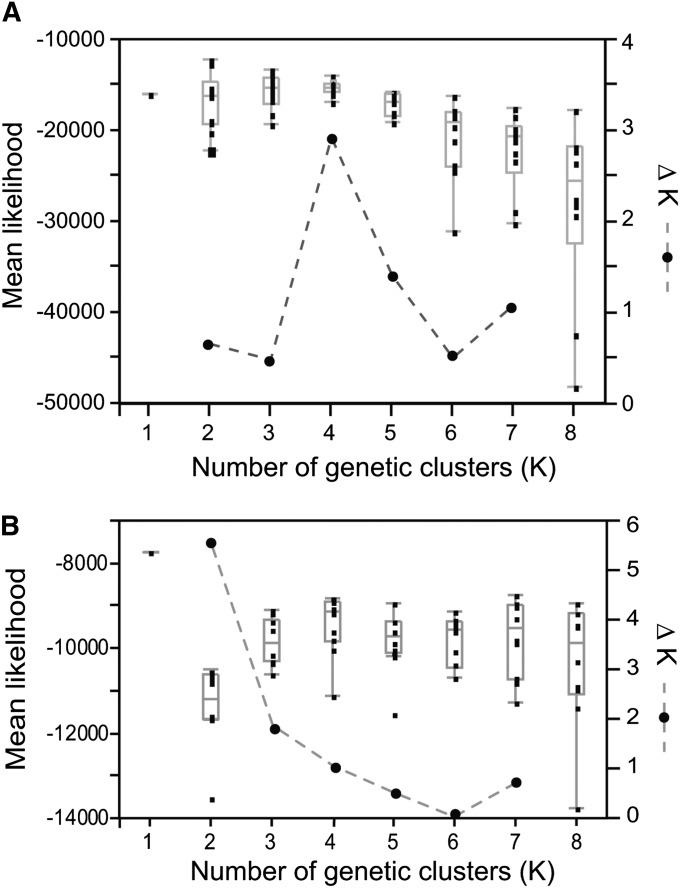
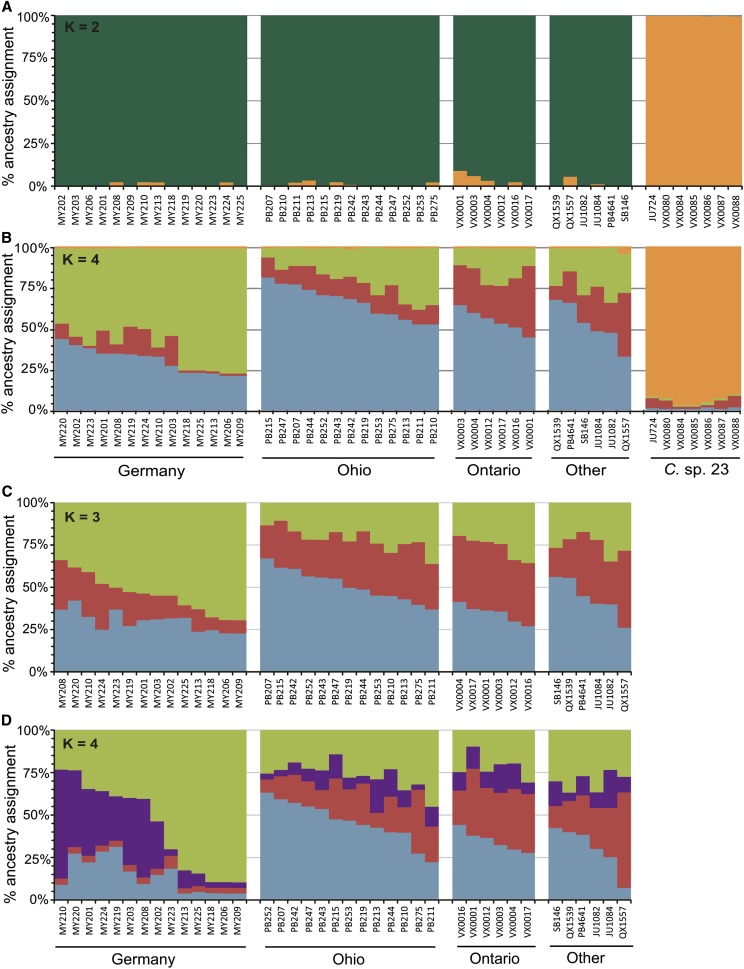
We used the STRUCTURE program to estimate population structuring (Pritchard et al. 2000; Falush et al. 2003; Hubisz et al. 2009). STRUCTURE assumes independence of loci without any background LD and hence is not typically applied to polymorphism data based on resequencing. However, because LD decays so rapidly in C. remanei (Cutter et al. 2006a; Cutter 2008), we employed STRUCTURE for our data set by treating each polymorphic site as a distinct locus. Using STRUCTURE’s “linkage” model for which we defined base-pair distances for sites within a gene fragment and free recombination between gene fragments, we first analyzed a combined data set that contained all populations of C. remanei and C. sp. 23. The clustering analysis identified the maximum likelihood of the data given the model for runs of K = 2 genetic clusters (Figure 5A). The mean maximum likelihood, however, was nominally highest for K = 4, although there was much overlap of likelihoods from K = 2 to K = 5 (Figure 5A). Falush et al. (2003) recommends choosing a biologically plausible K at the point where the mean maximum likelihood plateaus. By this criterion, K = 2 is most appropriate, separating C. remanei strains clearly from C. sp. 23. However, the ΔK method (Evanno et al. 2005) suggests that the modal value of K = 4 model best fits the data (Figure 5A). In this case, C. sp. 23 remains distinct, with additional substructure evident within C. remanei, which is qualitatively consistent with our analysis below using the data partition restricted to strains of C. remanei alone. Regardless of the choice of K > 1, the clustering analysis partitions each species into separate clusters with very little admixture between them. In this regard, when using K = 2 and K = 4, there are no differences in the assignment of C. remanei and C. sp. 23 (Figure 6, A and B).
Figure 5 .
Mean likelihood and ΔK plot for STRUCTURE runs based on (A) the combined data set of C. remanei and C. sp. 23 and (B) the data set containing only C. remanei strains. Boxplots of the estimated mean likelihood of the data (left axis), given K genetic clusters, indicating the median and interquartile range. Solid circles connected by dashed lines indicate ΔK (right axis).
Figure 6 .
Clustering analysis from STRUCTURE for the combined data set including strains of C. remanei and C. sp. 23 (A and B) and for the data set restricted to C. remanei strains only (C and D). Cumulative bar plots indicate the percentage ancestry of each strain from each inferred genetic cluster for a given value of K genetic clusters. Each genetic cluster is distinguished by a distinct color; colors are not necessarily equivalent among panels. The panels depict the STRUCTURE run with the highest likelihood for the given K value in the corresponding data partition.
To further explore population structure within C. remanei, we analyzed the subset of the data containing only the C. remanei samples. Using the same linkage model, our analysis indicated that the maximum likelihood occurs for K = 1, indicative of a single population genetic cluster. Because background LD in our analysis might mask subtle structure, however, we also evaluated K > 1. The mean maximum likelihood for STRUCTURE runs increases from K = 2 to K = 4, but plateaus for K ≥ 4 (Figure 5B). Using the method of Evanno et al. (2005), the maximum rate of change of likelihood is found from K = 2 to K = 3 (Figure 5B). So we considered both K = 3 and K = 4 to perform cluster assignment of individual strains (Figure 6, C and D). We observed extensive admixture among all three populations of C. remanei, as evinced by intermediate cluster assignment probabilities across strains for both K = 3 and K = 4. The only disparity among locations appeared to distinguish the cluster assignment frequencies for German strains relative to their North American counterparts, with a greater skew in the assignment frequencies for Germany (Figure 6C). For the K = 4 model, we observed some subtle differentiation within the German strains (Figure 6D). The strains differentiated within Germany by STRUCTURE also appear as a slightly separated cluster of multi-locus genotypes in the neighbor-network diagram (Figure 4). Unless it simply reflects over-fitting, this individual assignment heterogeneity within the German sample could reflect recent admixture, as it does not correspond to any obvious details of sample collection. Notably, none of the geographic populations of C. remanei form a separate and distinct genetic cluster of their own, reinforcing the view of extensive gene flow across the species range (Figure 6, C and D).
Reproductive isolation between C. remanei and C. sp. 23
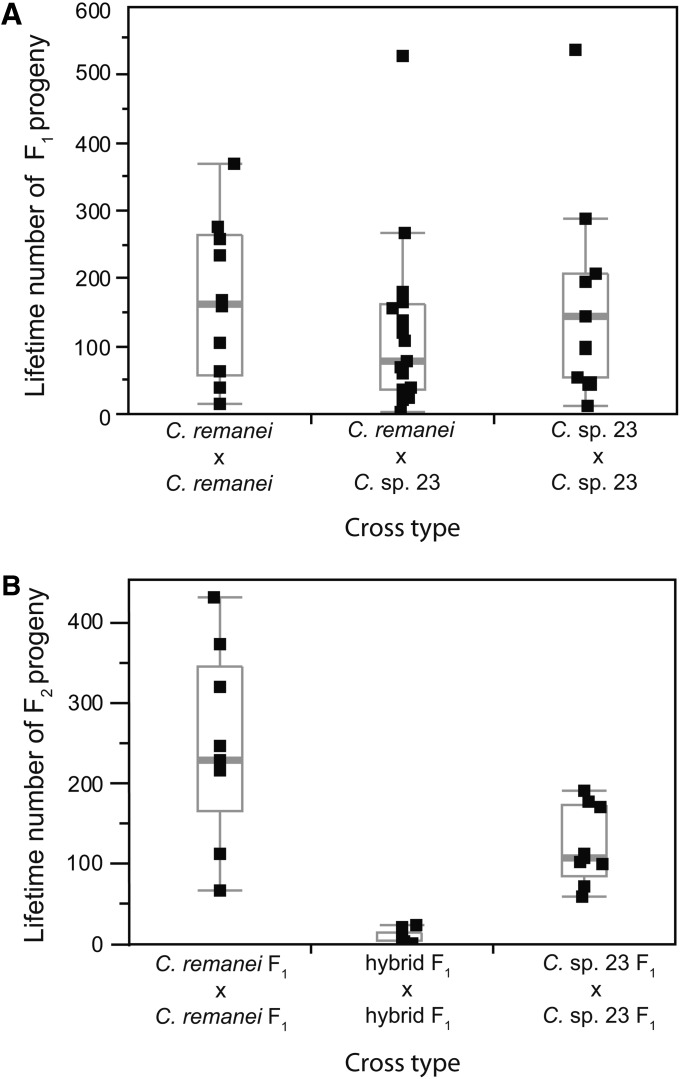
Because of the consistently high population differentiation indicated by distinct analyses for the European/North American populations relative to the Chinese strains, we hypothesized that the strains from China represent a distinct species. To test this hypothesis, we compared the viability of F1 and F2 hybrids derived from crosses within and between strains from Ohio and Wuhan. We did not detect a significant difference in the mean number of F1 progeny resulting from within- and between-population crosses, although the mean was nominally lower for the between-population crosses (Figure 7A). In many organisms, F1 hybrids between closely related species are nearly fully viable and fertile, and yet increased mortality and sterility occur in the second generation (F2) onward in a phenomenon known as “hybrid breakdown” (Coyne and Orr 2004). Therefore, we measured the total lifetime progeny production by F1 hybrids that were derived from either within- or between-population crosses. The F2’s were obtained following mating of one F1 female with four to six F1 males for 24 hr. Strikingly, we found significantly fewer F2’s from those animals derived from Ohio × Wuhan crosses as compared to either of the intrapopulation crosses (ANOVA, F2,30 = 39.73, P < 0.0001) (Figure 7B). Indeed, mean F2 progeny production was reduced up to 97% (Figure 7B). We therefore conclude that considerable F2 hybrid breakdown is present and that those genetically differentiated strains from China, now termed C. sp. 23, compose a distinct biological species from C. remanei with incomplete reproductive isolation.
Figure 7 .
Hybrid breakdown of F2 progeny between C. remanei and C. sp. 23. Boxplots indicate the median (thick gray bar) and interquartile range of (A) total lifetime F1 progeny and (B) total lifetime F2 progeny derived from intrapopulation crosses of C. remanei and C. sp. 23 and crosses between the two species. Points beyond the whiskers indicate potential outliers. Significantly fewer F2 progeny result from the hybrid cross between C. remanei and C. sp. 23 than from the intraspecies crosses (P < 0.001).
Discussion
Patterns of genetic variation and differentiation provide valuable insights into the evolutionary processes that cause divergence between closely related species. Numerous studies in diverse organisms have documented evidence of ongoing gene flow between recently separated species (Hey and Pinho 2012). In Caenorhabditis nematodes, however, only recently has a species pair been discovered that has close enough relatives to be partially reproductively isolated from one another (Cutter et al. 2010; Woodruff et al. 2010; Kozlowska et al. 2012). In addition to providing effective tools to study the genetic basis of reproductive isolation, these species also permit the answering of evolutionary questions related to divergence and gene flow on the path to speciation. Here, we discover another such species pair: C. remanei and C. sp. 23. These closely related taxa provide the first system of outcrossing species to which the methods of divergence population genetics can be applied in Caenorhabditis, as saturation of molecular divergence between most known species in the group is too great to infer ancestral states accurately (Cutter et al. 2009). This species pair also is intriguing because, initially, nematode strains isolated from China were identified through mating tests to simply represent a population of C. remanei. However, population genetics data uncovered that the Chinese nematodes were highly differentiated from C. remanei. Genetic crosses between the two now indicate that they are distinct biological species, partially reproductively isolated from one another owing to F2 hybrid breakdown. This case of morphologically highly similar, yet genetically very distinct, species opens the door to foundational investigations of speciation in the Caenorhabditis model system that are based on a close pair of species that both have obligately outbreeding reproduction.
Divergence of C. remanei with C. sp. 23
We investigated nucleotide polymorphism and divergence for 20 X-linked nuclear loci in C. remanei and C. sp. 23. Both species have an obligately outcrossing mode of reproduction with distinct male and female sexes representing extant examples of the reproductive state from which self-fertilization evolved in the related species C. elegans. We observed high genetic differentiation between strains collected from China (subsequently renamed C. sp. 23) and strains of C. remanei from North America and Europe (and Japan), a characteristic that is expected for populations that are at least partly reproductively isolated. The high differentiation is clear in neighbor-network diagrams (Figure 4) and was captured quantitatively (mean FST = 0.73, Figure 3A; STRUCTURE analysis, Figure 6, A and B). After further interrogation with crosses between the two genetic clusters, we conclude that C. remanei and C. sp. 23 do indeed represent distinct biological species that are partially reproductively isolated from one another. Strain JU724, isolated in Jiangsu, China, in 2005, was originally identified as conspecific to C. remanei on the basis of F1 mating tests (M. A. Felix, personal communication), but it clusters with the C. sp. 23 strains from Wuhan, China, in our population genetic analysis. Similarly, our crosses show that F1 progeny production is not strongly affected in crosses between the two species, but that F2 animals are substantially compromised, indicating extensive hybrid breakdown (Figure 7). We do not know whether F2 hybrid breakdown occurs owing to F1 hybrid male or female sterility, but future experiments will help elucidate this further.
C. sp. 23 has high genetic diversity, like other outcrossing species in the genus including C. remanei, C. brenneri, and C. sp. 5 (Cutter 2008; Jovelin 2009; Wang et al. 2010). The population sample from Wuhan, China, also shows no strong overall deviation from demographic equilibrium. C. sp. 23 now provides the newest example of an obligately outcrossing Caenorhabditis with sequence population polymorphism estimated to be >20 times that of selfing relatives, despite extensive morphological similarity within and between species (Graustein et al. 2002; Kiontke et al. 2011). Translational selection appears to strongly depress synonymous-site polymorphism in C. remanei such that corrected estimates of neutral diversity are ∼50% higher than corresponding values subject only to a Jukes–Cantor multiple-hits adjustment; we did not detect such an effect in C. sp. 23. Moreover, these outcrossing species have very high diversity in absolute terms, with few other eukaryotes known to harbor higher levels of polymorphism. Thus, a consistent portrait of highly genetically variable ancestral progenitors of C. elegans and C. briggsae is emerging, where evolutionary processes have drastically reduced the diversity in the derived selfing species in a complex interaction of selection, metapopulation dynamics, and recent origins of selfing (Cutter et al. 2009).
Demographic patterns in C. remanei
The three distinct populations of C. remanei that we sampled from North America and Europe (Ohio, Ontario, and Germany) each harbor considerable genetic diversity, with an average πneu= 5.5% consistent with previous estimates from a single population (Cutter et al. 2006a; Cutter 2008; Jovelin 2009). The North American populations also do not exhibit dramatic deviations from demographic equilibrium. By contrast, the German sample exhibits a deficit of rare variants across many loci, as well as nominally lower nucleotide diversity (Table 1 and Table S4), suggestive of recent admixture and/or bottleneck population-size changes. Indeed, a quarter to a half of the loci investigated manifest a significant positive skew in the site-frequency spectrum for the German sample, depending on the metric used. And yet, McDonald–Kreitman tests reveal no evidence of selection on any of the coding sequences investigated here. Therefore, we favor a demographic explanation for the peculiar patterns seen in the German sample of C. remanei. Because the German sample derives from rotting fruits in a private garden orchard near Kiel, it is possible that this locality was recently colonized from multiple sources. This contrasts with the forest habitat of the Ohio and Ontario samples, which we expect to represent a less disturbed habitat and to have been less subjected to anthropogenic influence. The genetic structure of European and North American plants and animals also has been affected by quaternary glacial cycles over the past 2.5 million years. Particularly since the last glacial maximum (∼20,000 years ago), massive changes have affected northern Europe. It is possible that the German sample could have been subject to genetic admixture following this natural process as well. In any case, our analysis indicates that the Ohio sample represents the population closest to equilibrium assumptions.
Genetic differentiation within C. remanei
C. remanei populations experience considerable gene flow and recombination that has mixed alleles across long distances, spanning oceans and continents. However, the German population appears to have greater genetic differentiation from the North American populations than is seen between the two North American localities. This suggests some degree of genetic isolation by distance, but sampling of additional populations of C. remanei will be essential to capture a more detailed portrait of genetic differentiation with respect to geography. The differentiation of the German strains also is notable in our STRUCTURE analysis of the C. remanei populations. Specifically, cluster assignments suggest a slightly distinct genetic makeup of the German sample relative to the Ohio and Ontario populations (Figure 6, C and D). Finally, we note that strains collected in Japan cluster genetically with other C. remanei strains, rather than with the geographically closer C. sp. 23 strains found in China (Figures 4 and 6). Thus, it does not appear that C. remanei is excluded in its range distribution from Asia by C. sp. 23.
Implications of newly discovered C. sp. 23
Discovery of a novel incipient species pair of obligately outcrossing species, the first such example in Caenorhabditis, emerged from our multilocus, multipopulation analysis of population genetic variation. Although studies of the genetic basis of post-zygotic isolation has a rich and long history in model organisms like Saccharomyces, Drosophila, Arabidopsis, and Mus, unfortunately Caenorhabditis has contributed little to this topic, primarily owing to the high genetic divergence between known species and the absence of fertile hybrids between them. Only recently has a species pair been discovered in which fertile F1 hybrid females result from interspecific crosses: the selfing C. briggsae and outcrossing C. sp. 9 (Woodruff et al. 2010; Kozlowska et al. 2012). While the distinct breeding systems of C. briggsae and C. sp. 9 are invaluable for understanding the origins of selfing in the genus Caenorhabditis, it has so far proven difficult to characterize genetically because of the absence of self-fertile hybrid progeny and the low incidence of hermaphrodites in subsequent generations (Woodruff et al. 2010). By contrast, both C. remanei and C. sp. 23 reproduce by outcrossing obligatorily, permitting exploration of the genetic basis of post-zygotic isolation without the complicating additional layering of breeding system evolution. The two species have so far been isolated in non-overlapping geographical locations, suggesting that genetic differences might have accumulated in allopatry, with limited-to-no gene flow between them. However, given the globally widespread distribution of many species in Caenorhabditis (Kiontke et al. 2011), this species pair could provide new insights if they can be found in sympatry.
The C. remanei–C. sp. 23 species pair also opens up opportunities to study this system in the context of divergence population genetics. Most species in the genus suffer from saturation of synonymous sites in pairwise divergence (Cutter et al. 2009), a problem that does not afflict C. remanei and C. sp. 23. We made use of this feature to compute unfolded metrics of the site-frequency spectrum (e.g., Fu and Li’s D and F and Fay and Wu’s H) and to conduct McDonald–Kreitman tests on the loci included in this study, which were inaccessible methods in previous work on this system. Future genome-scale analyses will be able to fully exploit these approaches.
Finally, the C. remanei–C. sp. 23 species pair serves to provide a word of caution about the routine laboratory practice of mating tests in Caenorhabditis species delimitation. Incipient species can form fertile F1 hybrids with strong breakdown occurring only in later generations (Coyne and Orr 2004). Hence, F1 mating tests provide a conservative test of the reproductive boundary for biological species, the details of which can benefit from examination of subsequent hybrid generations as well as population genetics analysis. C. remanei itself was the focus of historical taxonomic confusion (Baird et al. 1992, 1994; Sudhaus and Kiontke 1996; Baird and Yen 2000). With the increased intensity of Caenorhabditis collection from nature and the concomitant acceleration in the discovery of new species (Kiontke et al. 2011), it will become increasingly important to apply multi-tiered approaches to species identification.
Supplementary Material
Acknowledgments
We thank Erik Andersen, Marie-Anne Felix, and Hinrich Schulenburg for collecting and sharing strains and Huang Ai and Hui Liu for assistance with strain collection. We thank Richard Jovelin for insightful discussions on the project and the manuscript. G.-X.W. is supported by the National Natural Science Foundation of China (#31071998) and HuaZhong Normal University. A.D.C. is supported by the Natural Sciences and Engineering Research Council of Canada, the National Institutes of Health, and a Canada Research Chair.
Footnotes
Communicating editor: D. Begun
Literature Cited
- Arunyawat U., Stephan W., Stadler T., 2007. Using multilocus sequence data to assess population structure, natural selection, and linkage disequilibrium in wild tomatoes. Mol. Biol. Evol. 24: 2310–2322 [DOI] [PubMed] [Google Scholar]
- Baird S. E., 1999. Natural and experimental associations of Caeonrhabditis remanei with Trachelipus rathkii and other terrestrial isopods. Nematology 1: 471–475 [Google Scholar]
- Baird S. E., Yen W. C., 2000. Reproductive isolation in Caenorhabditis: terminal phenotypes of hybrid embryos. Evol. Dev. 2: 9–15 [DOI] [PubMed] [Google Scholar]
- Baird S. E., Sutherlin M. E., Emmons S. W., 1992. Reproductive isolation in Rhabditidae (Nematoda:Secernentea): mechanisms that isolate six species of three genera. Evolution 46: 585–594 [DOI] [PubMed] [Google Scholar]
- Baird S. E., Fitch D. H., Emmons S. W., 1994. Caenorhabditis vulgaris n. sp. (Secernentea: Rhabditidae): a necromenic associate of pill bugs and snails. Nematologica 40: 1–11 [Google Scholar]
- Braendle C., Felix M. A., 2006. Sex determination: ways to evolve a hermaphrodite. Curr. Biol. 16: R468–R471 [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Wright S. I., 2001. Breeding systems and genome evolution. Curr. Opin. Genet. Dev. 11: 685–690 [DOI] [PubMed] [Google Scholar]
- Coyne J., Orr H. A., 2004. Speciation. Sinauer Associates, Sunderland, MA [Google Scholar]
- Cutter A. D., 2008. Multilocus patterns of polymorphism and selection across the X chromosome of Caenorhabditis remanei. Genetics 178: 1661–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter A. D., Charlesworth B., 2006. Selection intensity on preferred codons correlates with overall codon usage bias in Caenorhabditis remanei. Curr. Biol. 16: 2053–2057 [DOI] [PubMed] [Google Scholar]
- Cutter A. D., Baird S. E., Charlesworth D., 2006a High nucleotide polymorphism and rapid decay of linkage disequilibrium in wild populations of Caenorhabditis remanei. Genetics 174: 901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter A. D., Felix M. A., Barriere A., Charlesworth D., 2006b Patterns of nucleotide polymorphism distinguish temperate and tropical wild isolates of Caenorhabditis briggsae. Genetics 173: 2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter A. D., Wasmuth J. D., Washington N. L., 2008. Patterns of molecular evolution in Caenorhabditis preclude ancient origins of selfing. Genetics 178: 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter A. D., Dey A., Murray R. L., 2009. Evolution of the Caenorhabditis elegans genome. Mol. Biol. Evol. 26: 1199–1234 [DOI] [PubMed] [Google Scholar]
- Cutter A. D., Yan W., Tsvetkov N., Sunil S., Felix M. A., 2010. Molecular population genetics and phenotypic sensitivity to ethanol for a globally diverse sample of the nematode Caenorhabditis briggsae. Mol. Ecol. 19: 798–809 [DOI] [PubMed] [Google Scholar]
- Cutter A. D., Wang G. X., Ai H., Peng Y., 2012. Influence of finite-sites mutation, population subdivision and sampling schemes on patterns of nucleotide polymorphism for species with molecular hyperdiversity. Mol. Ecol. 21: 1345–1359 [DOI] [PubMed] [Google Scholar]
- Denver D. R., Clark K. A., Raboin M. J., 2011. Reproductive mode evolution in nematodes: insights from molecular phylogenies and recently discovered species. Mol. Phylogenet. Evol. 61: 584–592 [DOI] [PubMed] [Google Scholar]
- Dolgin E. S., Charlesworth B., Baird S. E., Cutter A. D., 2007. Inbreeding and outbreeding depression in Caenorhabditis nematodes. Evolution 61: 1339–1352 [DOI] [PubMed] [Google Scholar]
- Earl D. A., vonHoldt B. M., 2011. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4: 359–361. [Google Scholar]
- Evanno G., Regnaut S., Goudet J., 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14: 2611–2620 [DOI] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J. K., 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J. C., Wu C. I., 2000. Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. X., Li W. H., 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graustein A., Gaspar J. M., Walters J. R., Palopoli M. F., 2002. Levels of DNA polymorphism vary with mating system in the nematode genus Caenorhabditis. Genetics 161: 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag E. S., 2009. Caenorhabditis nematodes as a model for the adaptive evolution of germ cells. Curr. Top. Dev. Biol. 86: 43–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag E. S., Ackerman A. D., 2005. Intraspecific variation in fem-3 and tra-2, two rapidly coevolving nematode sex-determining genes. Gene 349: 35–42 [DOI] [PubMed] [Google Scholar]
- Hey J., Pinho C., 2012. Population genetics and objectivity in species diagnosis. Evolution 66: 1413–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill R. C., de Carvalho C. E., Salogiannis J., Schlager B., Pilgrim D., et al. , 2006. Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev. Cell 10: 531–538 [DOI] [PubMed] [Google Scholar]
- Hubisz M. J., Falush D., Stephens M., Pritchard J. K., 2009. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 9: 1322–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H., Bryant D., 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23: 254–267 [DOI] [PubMed] [Google Scholar]
- Jovelin R., 2009. Rapid sequence evolution of transcription factors controlling neuron differentiation in Caenorhabditis. Mol. Biol. Evol. 26: 2373–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovelin R., Ajie B. C., Phillips P. C., 2003. Molecular evolution and quantitative variation for chemosensory behaviour in the nematode genus Caenorhabditis. Mol. Ecol. 12: 1325–1337 [DOI] [PubMed] [Google Scholar]
- Jovelin R., Dunham J. P., Sung F. S., Phillips P. C., 2009. High nucleotide divergence in developmental regulatory genes contrasts with the structural elements of olfactory pathways in Caenorhabditis. Genetics 181: 1387–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes T. H., Cantor C. R., 1969. Evolution of protein molecules, pp. 21–132 in Mammalian Protein Metabolism, edited by H. N. Munro. Academic Press, New York
- Kiontke K., Gavin N. P., Raynes Y., Roehrig C., Piano F., et al. , 2004. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc. Natl. Acad. Sci. USA 101: 9003–9008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K. C., Felix M. A., Ailion M., Rockman M. V., Braendle C., et al. , 2011. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol. Biol. 11: 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowska J. L., Ahmad A. R., Jahesh E., Cutter A. D., 2012. Genetic variation for postzygotic reproductive isolation between Caenorhabditis briggsae and Caenorhabditis sp. 9. Evolution 66: 1180–1195 [DOI] [PubMed] [Google Scholar]
- Librado P., Rozas J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 [DOI] [PubMed] [Google Scholar]
- McDonald J. H., Kreitman M., 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654 [DOI] [PubMed] [Google Scholar]
- Nei M., Gojobori T., 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426 [DOI] [PubMed] [Google Scholar]
- Ness R. W., Wright S. I., Barrett S. C., 2010. Mating-system variation, demographic history and patterns of nucleotide diversity in the tristylous plant Eichhornia paniculata. Genetics 184: 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane H. S., Smith J. M., Bergthorsson U., Katju V., 2010. Gene conversion and DNA sequence polymorphism in the sex-determination gene fog-2 and its paralog ftr-1 in Caenorhabditis elegans. Mol. Biol. Evol. 27: 1561–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Ibarra J., Wright S. I., Foxe J. P., Kawabe A., DeRose-Wilson L., et al. , 2008. Patterns of polymorphism and demographic history in natural populations of Arabidopsis lyrata. PLoS ONE 3: e2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenico M., Lloyd A. T., Sharp P. M., 1994. Codon usage in Caenorhabditis elegans: delineation of translational selection and mutational biases. Nucleic Acids Res. 22: 2437–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Onge K. R., Källman T., Slotte T., Lascoux M., Palmé A. E., 2011. Contrasting demographic history and population structure in Capsella rubella and Capsella grandiflora, two closely related species with different mating systems. Mol. Ecol. 20: 3306–3320 [DOI] [PubMed] [Google Scholar]
- Sudhaus W., Kiontke K., 1996. Phylogeny of Rhabditis subgenus Caenorhabditis (Rhabditidae, Nematoda). J. Zoolog. Syst. Evol. Res. 34: 217–233 [Google Scholar]
- Sudhaus W., Kiontke K., 2007. Comparison of the cryptic nematode species Caenorhabditis brenneri sp. n. and C. remanei (Nematoda: Rhabditidae) with the stem species pattern of the Caenorhabditis elegans group. Zootaxa 1456: 45–62 [Google Scholar]
- Sweigart A. L., Willis J. H., 2003. Patterns of nucleotide diversity in two species of Mimulus are affected by mating system and asymmetric introgression. Evolution 57: 2490–2506 [DOI] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. X., Ren S., Ren Y., Ai H., Cutter A. D., 2010. Extremely high molecular diversity within the East Asian nematode Caenorhabditis sp. 5. Mol. Ecol. 19: 5022–5029 [DOI] [PubMed] [Google Scholar]
- Woodruff G. C., Eke O., Baird S. E., Felix M. A., Haag E. S., 2010. Insights into species divergence and the evolution of hermaphroditism from fertile interspecies hybrids of Caenorhabditis nematodes. Genetics 186: 997–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. I., Ness R. W., Foxe J. P., Barrett S. C. H., 2008. Genomic consequences of outcrossing and selfing in plants. Int. J. Plant Sci. 169: 105–118 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.