Abstract
A haloalkane dehalogenase, DpcA, from Psychrobacter cryohalolentis K5, representing a novel psychrophilic member of the haloalkane dehalogenase family, was identified and biochemically characterized. DpcA exhibited a unique temperature profile with exceptionally high activities at low temperatures. The psychrophilic properties of DpcA make this enzyme promising for various environmental applications.
TEXT
Haloalkane dehalogenases (HLDs) (EC 3.8.1.5) are enzymes that catalyze the hydrolysis of a carbon-halogen bond in halogenated aliphatic hydrocarbons, releasing a corresponding alcohol, a halide ion, and a proton as the reaction products (17). HLDs catalyze reactions of great environmental and biotechnological significance. They have potential applications in bioremediation (33), biosensing (4, 6), decontamination of warfare agents (30), synthesis of optically pure compounds (29, 35), cellular imaging, and protein analysis (25, 27). The effective use of biocatalysts in these applications requires availability of enzymes with specific properties under process conditions (5). Isolation of novel enzymes from extremophilic microorganisms represents an effective approach for obtaining biocatalysts with desired properties (34). The requirement of low temperature for selected practical applications of HLDs prompted us to explore the domain of psychrophilic proteins (7).
To identify HLDs with potentially psychrophilic properties, we performed sequence database searches. We used six HLD family members—LinB (26), DhaA (23), DhlA (21), DmbB (19), DrbA (20), and DmbC (20)—as queries for the PSI-BLAST (1) search against the nr database of NCBI (February 2009 version) (32). PSI-BLAST was conducted with the E-value thresholds of 10−10 for the initial BLAST search and the threshold of 10−15 for inclusion of the sequence in a position-specific matrix. A set of 11,208 sequences collected after three iterations of PSI-BLAST were clustered with the software program CLANS (13) to distinguish HLDs from other related sequences. The best resolution of clusters was observed at the P value of 10−35. The HLD cluster was composed of 308 nonunique sequences originating from 268 different sources, including unclassified organisms and artificial constructs. Information about source organisms of all putative HLDs was collected from the Genomes OnLine Database, v2.0 (24), and the Entrez Genome Project database of NCBI (32). Collected sequences were aligned using the MUSCLE software program (10). Putative HLDs were identified in the genomes of 16 psychrophilic or psychrotolerant organisms (see Table S1 in the supplemental material), including Psychrobacter cryohalolentis K5. P. cryohalolentis K5 was originally isolated from saline-water lenses obtained from 40,000-year-old Siberian permafrost with an in situ temperature from −9 to −11°C (2). The bacterium formed small nonmotile coccoid rods and could be cultured on marine agar in smooth and nonpigmented colonies. The strain was selected for genome sequencing for its ability to reproduce at −10°C with a generation time of 39 days and rapid growth at low temperature (2). Additional studies of P. cryohalolentis K5 examined critical temperature, growth efficiency, and protein expression at subzero temperatures (3).
The putative HLD from P. cryohalolentis K5 (NCBI accession no. YP_580518; hereafter referred to as DpcA), belonging to the HLD-I subfamily (9), was prepared and characterized as follows. An optimized recombinant gene, dpcA-His6, was artificially synthesized (Mr. Gene, Regensburg, Germany) and subcloned into the expression vector pET21b (Novagen, Madison, WI). His-tagged DpcA was expressed under the control of a T7 promoter in Escherichia coli ArcticExpress (DE3) (Stratagene, La Jolla, CA) and purified to homogeneity by metallo-affinity chromatography with Ni-nitrilotriacetic acid resin (Qiagen, Hilden, Germany). The protein yield obtained from purification was 30 mg of protein from a 1-liter cell culture.
Folding and secondary structure of DpcA were studied by circular dichroism (CD) spectroscopy. Far-UV CD spectra were recorded for DpcA and other related HLDs at room temperature using a Jasco J-810 spectropolarimeter (Jasco, Tokyo, Japan). All tested enzymes exhibited CD spectra with one positive peak at 195 nm and two negative maxima at 208 and 220 to 222 nm, characteristic of α-helical content (11), suggesting proper folding of DpcA. Thermally induced denaturation of DpcA was tested by monitoring ellipticity at 221 nm with increasing temperature. The melting temperature (Tm) calculated from the curve was 34.7 ± 0.6°C, revealing that DpcA possesses the lowest thermal stability of all related enzymes, whose melting temperatures lie within the range of 39.2 to 58.5°C (15, 18–21, 26, 31).
The native structure of DpcA was examined by gel filtration chromatography using the elution buffer consisting of 50 mM Tris-HCl at pH 7.5 with the presence of 150 mM NaCl. A Superdex 200 10/300 GL column (GE Healthcare, Waukesha, WI) was calibrated with aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (44 kDa), carbonic anhydratase (29 kDa), and RNase A (13,7 kDa). DpcA was determined to be a monomer under tested conditions. The most biochemically characterized HLDs occur as monomers (8, 15, 21, 26); some of them are dimers or form large oligomeric complexes (8, 15, 20).
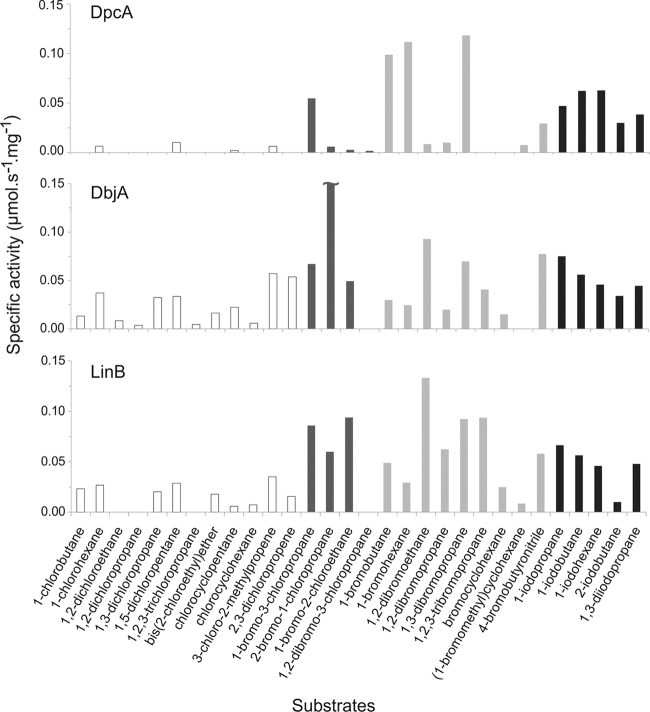
The substrate specificity of DpcA was assayed with 30 different halogenated substrates (see Table S2 in the supplemental material) at 25°C by a colorimetric Iwasaki method (16). The released halide ions were measured after adding mercuric thiocyanate and ferric ammonium sulfate spectrophotometrically at 460 nm. DpcA generally exhibited better activities toward longer substrates (number of carbon atoms in the chain ≥ 3) with the following substituent preferences: brominated > iodinated ≫ chlorinated. In order to compare the activity and substrate specificity of DpcA with those of other HLDs, principal component analysis (PCA) was carried out as previously described by Koudelakova et al. (22). Compared to those of other HLDs, the overall level of activity of DpcA was moderate (see Fig. S1 in the supplemental material). DpcA has one of the narrowest substrate specificity profiles of all biochemically characterized HLDs. It has exceptionally high activity toward 1-bromobutane (0.099 μmol s−1 mg−1), 1-bromohexane (0.112 μmol s−1 mg−1), and 1,3-dibromopropane (0.118 μmol s−1 mg−1), while it has zero activity toward short chlorinated substrates (Fig. 1). PCA classified DpcA into substrate specificity group IV (SSG-IV) together with the haloalkane dehalogenases DatA, DbeA, and DmbC (see Fig. S1 in the supplemental material).
Fig 1.
Comparison of substrate specificity profiles of DpcA and two other biochemically characterized haloalkane dehalogenases, DbjA and LinB. Substrate specificities toward a set of 30 chlorinated (white), chlorobrominated (dark gray), brominated (light gray), and iodinated (black) substrates were determined. DbjA activity toward 2-bromo-1-chloropropane (0.42 μmol s−1 mg−1) is trimmed due to scaling limitations of the figure.
1-Bromobutane, identified as one of the best substrates for DpcA, was used for the determination of temperature and pH profiles. The temperature profile of DpcA was measured in the range of temperatures from 5°C to 35°C. The highest activity of the enzyme was observed at 25°C (Fig. 2). This property is unique and has not been observed for any other HLD so far. The highest activities of other related enzymes lay in the temperature interval between 40 and 50°C (15, 18–21, 26). Another important finding is that the DpcA enzyme retained almost 27% of its maximal activity at 5°C. Both observations were consistent with the fact that DpcA originates from the psychrophilic organism.
Fig 2.
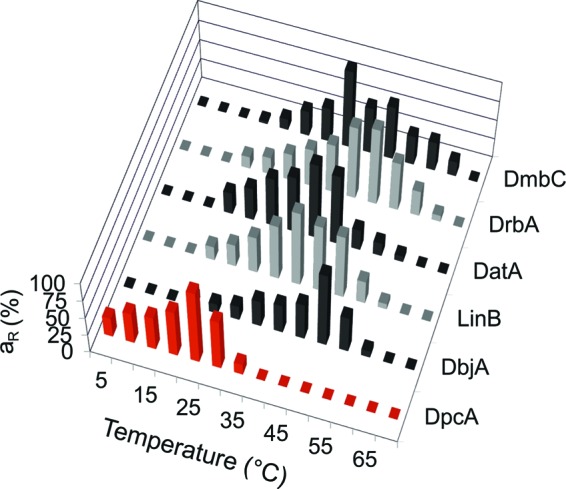
Comparison of the temperature profiles of DpcA with those of five other biochemically characterized haloalkane dehalogenases, DbjA, LinB, DatA, DrbA, and DmbC. The data are expressed as relative activities (aR).
The effect of pH on DpcA activity was measured over the pH range of 5.2 to 10.1. The maximal activity of DpcA was detected at pH 8.7, which was similar to findings with other HLDs (15, 18–21, 26). The protein loses all activity below pH 6.4 and above pH 9.6.
A steady-state kinetic analysis was performed with two substrates, 1-bromobutane and 1,3-dibromopropane, for which the enzyme exhibited the highest activity. The kinetics of DpcA both with 1-bromobutane (K0.5 [the substrate concentration at which half-maximal velocity is achieved according to the cooperativity mode] = 3.83 ± 0.66 mM; kcat = 1.10 ± 0.07 s−1) and with 1,3-dibromopropane (K0.5 = 5.77 ± 1.26 mM; kcat = 2.72 ± 0.24 s−1) follow a mechanism involving positive cooperative substrate binding with Hill coefficients n = 1.60 ± 0.14 and 1.50 ± 0.37, respectively (see Fig. S2 in the supplemental material).
The enantioselectivity of DpcA was assessed by determination of the kinetic resolution of racemic β-brominated alkanes (2-bromopentane and 2-bromoheptane) and α-brominated esters (methyl 2-bromobutyrate and ethyl 2-bromobutyrate). The DpcA exhibited excellent enantioselectivity toward α-brominated esters, with a preference for the R over the S enantiomer (E value = 105 for methyl 2-bromobutyrate; E value > 200 for ethyl 2-bromobutyrate), but no enantioselectivity was observed toward β-brominated alkanes.
A variety of structural features responsible for psychrophilic properties of enzymes have been described in the scientific literature, including an increased flexibility of molecular structure, a higher abundance of glycine residues, a lower abundance of proline and arginine residues, a reduced number of ion pairs, aromatic interactions, hydrogen bonds, helix dipoles, or buried nonpolar residues, an exposure of a higher proportion of nonpolar residues to the solvent, and increased accessibility of the active site cavity (12, 14, 28). Analysis of amino acid composition showed no significant differences between DpcA and biochemically characterized HLDs from mesophilic organisms (see Table S3 in the supplemental material). Neither multiple sequence alignment revealed any sequence features specific for putative HLDs from cold-adapted bacteria. We are therefore working on determination of the tertiary structure of DpcA to investigate the structural features responsible for DpcA psychrophilicity.
The biochemical characterization of the novel HLD DpcA from P. cryohalolentis K5 described in this article revealed several unique characteristics of this enzyme, including its narrow substrate specificity and high activity at low temperatures. Psychrophilic properties of DpcA make this biocatalyst highly attractive for environmental applications, e.g., biosensing and bioremediation in subsurface environments.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the European Regional Development Fund (CZ.1.05/2.1.00/01.0001), the Grant Agency of the Czech Republic (P207/12/0775), and the Grant Agency of the Czech Academy of Sciences (IAA401630901). MetaCentrum is acknowledged for providing access to computing facilities, supported by the Ministry of Education of the Czech Republic (LM2010005).
Footnotes
Published ahead of print 28 April 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakermans C, Tsapin AI, Souza-Egipsy V, Gilichinsky DA, Nealson KH. 2003. Reproduction and metabolism at −10°C of bacteria isolated from Siberian permafrost. Environ. Microbiol. 5:321–326 [DOI] [PubMed] [Google Scholar]
- 3.Bakermans C, Nealson KH. 2004. Relationship of critical temperature to macromolecular synthesis and growth yield in Psychrobacter cryopegella. J. Bacteriol. 186:2340–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bidmanova S, Chaloupkova R, Damborsky J, Prokop Z. 2010. Development of an enzymatic fiber-optic biosensor for detection of halogenated hydrocarbons. Anal. Bioanal. Chem. 398:1891–1898 [DOI] [PubMed] [Google Scholar]
- 5.Bornscheuer UT, Pohl M. 2001. Improved biocatalysts by directed evolution and rational protein design. Curr. Opin. Chem. Biol. 5:137–142 [DOI] [PubMed] [Google Scholar]
- 6.Campbell DW, Muller C, Reardon KF. 2006. Development of a fiber optic enzymatic biosensor for 1,2-dichloroethane. Biotechnol. Lett. 28:883–887 [DOI] [PubMed] [Google Scholar]
- 7.Cavicchioli R, Siddiqui KS, Andrews D, Sowers KR. 2002. Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 13:253–261 [DOI] [PubMed] [Google Scholar]
- 8.Chaloupkova R, Prokop Z, Sato Y, Nagata Y, Damborsky J. 2011. Stereoselectivity and conformational stability of haloalkane dehalogenase DbjA from Bradyrhizobium japonicum USDA110: the effect of pH and temperature. FEBS J. 278:2728–2738 [DOI] [PubMed] [Google Scholar]
- 9.Chovancova E, Kosinski J, Bujnicki JM, Damborsky J. 2007. Phylogenetic analysis of haloalkane dehalogenases. Proteins 67:305–316 [DOI] [PubMed] [Google Scholar]
- 10.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fasman GD. 1996. Circular dichroism and the conformational analysis of biomolecules, p 740. Plenum Press, New York, NY [Google Scholar]
- 12.Feller G, Gerday C. 2003. Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. Microbiol. 1:200–208 [DOI] [PubMed] [Google Scholar]
- 13.Frickey T, Lupas A. 2004. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20:3702–3704 [DOI] [PubMed] [Google Scholar]
- 14.Georlette D, et al. 2004. Some like it cold: biocatalysis at low temperatures. FEMS Microbiol. Rev. 28:25–42 [DOI] [PubMed] [Google Scholar]
- 15.Hasan K, et al. 2011. Biochemical characteristics of the novel haloalkane dehalogenase DatA isolated from the plant pathogen Agrobacterium tumefaciens C58. Appl. Environ. Microbiol. 77:1881–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwasaki I, Utsumi S, Ozawa T. 1952. New colorimetric determination of chloride using mercuric thiocyanate and ferric ion. Bull. Chem. Soc. Jpn. 25:226 [Google Scholar]
- 17.Janssen DB. 2004. Evolving haloalkane dehalogenase. Curr. Opin. Chem. Biol. 8:150–159 [DOI] [PubMed] [Google Scholar]
- 18.Jesenska A, et al. 2002. Cloning and expression of the haloalkane dehalogenase gene dhmA from Mycobacterium avium N85 and preliminary characterization of DhmA. Appl. Environ. Microbiol. 68:3724–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jesenska A, et al. 2005. Cloning, biochemical properties, and distribution of mycobacterial haloalkane dehalogenases. Appl. Environ. Microbiol. 71:6736–6745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jesenska A, et al. 2009. Biochemical characterization of haloalkane dehalogenase DrbA and DmbC, representatives of a novel subfamily. Appl. Environ. Microbiol. 75:5157–5160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keuning S, Janssen DB, Witholt B. 1985. Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. J. Bacteriol. 163:635–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koudelakova T, et al. 2011. Substrate specificity of haloalkane dehalogenases. Biochem. J. 435:345–354 [DOI] [PubMed] [Google Scholar]
- 23.Kulakova AN, Larkin MJ, Kulakov LA. 1997. The plasmid-located haloalkane dehalogenase gene from Rhodococcus rhodochrous NCIMB 13064. Microbiology 143:109–115 [DOI] [PubMed] [Google Scholar]
- 24.Liolios K, et al. 2010. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 38:D346–D354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Los GV, Wood K. 2007. The HaloTag: a novel technology for cell imaging and protein analysis. Methods Mol. Biol. 356:195–208 [DOI] [PubMed] [Google Scholar]
- 26.Nagata Y, et al. 1997. Purification and characterization of haloalkane dehalogenase of a new substrate class from a hexachlorohexane-degrading bacterium, Sphingomonas paucimobilis UT26. Appl. Environ. Microbiol. 63:3707–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohana RF, et al. 2009. HaloTag7: a genetically engineered tag that enhances bacterial expression of soluble proteins and improves protein purification. Protein Expr. Purif. 68:110–120 [DOI] [PubMed] [Google Scholar]
- 28.Paredes DI, Watters K, Pitman DJ, Bystroff C, Dordick JS. 2011. Comparative void-volume analysis of psychrophilic and mesophilic enzymes: structural bioinformatics of psychrophilic enzymes reveals sources of core flexibility. BMC Struct. Biol. 11:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pieters RJ, Spelberg LJH, Kellogg RM, Janssen DB. 2001. The enantioselectivity of haloalkane dehalogenases. Tetrahedron Lett. 42:469–471 [Google Scholar]
- 30.Prokop Z, Oplustil F, DeFrank J, Damborsky J. 2006. Enzymes fight chemical weapons. Biotechnol. J. 1:1370–1380 [DOI] [PubMed] [Google Scholar]
- 31.Sato Y, et al. 2005. Two rhizobial strains, Mesorhizobium loti MAFF303099 and Bradyrhizobium japonicum USDA110, encode haloalkane dehalogenases with novel structures and substrate specificity. Appl. Environ. Microbiol. 71:4372–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sayers EW, et al. 2010. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 38:D5–D16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stucki G, Thuer M. 1995. Experiences of a large-scale application of 1,2-dichloroethane degrading microorganisms for groundwater treatment. Environ. Sci. Technol. 29:2339–2345 [DOI] [PubMed] [Google Scholar]
- 34.van den Burg B. 2003. Extremophiles as a source for novel enzymes. Curr. Opin. Microbiol. 6:213–218 [DOI] [PubMed] [Google Scholar]
- 35.Westerbeek A, Szymanski W, Feringa BL, Janssen DB. 2011. Dynamic kinetic resolution process employing haloalkane dehalogenase. ACS Catal. 1:1654–1660 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.