Abstract
Human milk contains antimicrobial factors such as lysozyme and lactoferrin that are thought to contribute to the development of an intestinal microbiota beneficial to host health. However, these factors are lacking in the milk of dairy animals. Here we report the establishment of an animal model to allow the dissection of the role of milk components in gut microbiota modulation and subsequent changes in overall and intestinal health. Using milk from transgenic goats expressing human lysozyme at 68%, the level found in human milk and young pigs as feeding subjects, the fecal microbiota was analyzed over time using 16S rRNA gene sequencing and the G2 Phylochip. The two methods yielded similar results, with the G2 Phylochip giving more comprehensive information by detecting more OTUs. Total community populations remained similar within the feeding groups, and community member diversity was changed significantly upon consumption of lysozyme milk. Levels of Firmicutes (Clostridia) declined whereas those of Bacteroidetes increased over time in response to the consumption of lysozyme-rich milk. The proportions of these major phyla were significantly different (P < 0.05) from the proportions seen with control-fed animals after 14 days of feeding. Within phyla, the abundance of bacteria associated with gut health (Bifidobacteriaceae and Lactobacillaceae) increased and the abundance of those associated with disease (Mycobacteriaceae, Streptococcaceae, Campylobacterales) decreased with consumption of lysozyme milk. This study demonstrated that a single component of the diet with bioactivity changed the gut microbiome composition. Additionally, this model enabled the direct examination of the impact of lysozyme on beneficial microbe enrichment versus detrimental microbe reduction in the gut microbiome community.
INTRODUCTION
Human milk not only provides the newborn with all the nutrition it needs to grow and develop but also provides factors that promote health and combat infection. One of the main nonspecific host defense factors in human milk is lysozyme. Lysozyme is a naturally occurring antimicrobial enzyme found in the tears, saliva, and milk of all mammals that lyses a specific link in the peptidoglycan layer of bacterial cell walls, resulting in cell lysis (33). Along with lactoferrin and secretory IgA, lysozyme contributes to the nonspecific immunity associated with milk consumption. A number of epidemiological studies have documented the benefits of human milk, including advantages in general health, growth, and development and protection against a number of acute and chronic diseases in infancy and beyond (16, 22). In addition to its role in host defense, human milk also helps with the establishment of a beneficial gut microbiota. Breast-fed infants have been shown to have a more healthy and simple gut microbiota consisting mainly of bifidobacteria, lactobacilli, and staphylococci, while formula-fed infants have a more complex microbiota, with coliforms, clostridia, enterococci, streptococci, and bacteroides all being prevalent in the gut (1, 15, 34). In addition to human milk oligosaccharides and other bioactive compounds, another reason for the growth of fewer facultative anaerobes in breast-fed infants is thought to be the presence of antimicrobial factors such as lysozyme in human milk (18, 34).
The composition and function of the intestinal microbiota are just beginning to be defined and ascertained, but it is widely accepted that it plays a role in both health and disease (41). Due to the purported role of human milk in gut microbiota community formation, a source of milk rich in lysozyme may shift the microbial population ratios in the gut during milk consumption toward those microbes associated with health for the host. While cow and goat milk is readily available, it is low in key health-promoting antimicrobial components for humans. Human milk contains 400 μg/ml lysozyme, while cow milk and goat milk contain 3,000 (0.13 μg/ml) and 1,600 (0.25 μg/ml) times less lysozyme, respectively (8). We have developed transgenic dairy goats that express human lysozyme (HLZ) in their milk at 68% the level normally found in human milk (29) with the goal of incorporating the beneficial protective properties of human milk into readily available livestock milk in order to promote the intestinal and overall health of people of all ages. We previously demonstrated that consumption of milk from HLZ-transgenic goats by animal models results in the modulation of Escherichia coli and total coliform levels in the small intestine as determined using standard culture techniques (30), resistance to intestinal colonization by pathogenic E. coli (5), and histological and cytokine changes coupled with increases in serum metabolite makers indicative of improved gastrointestinal (GI) health (6, 10).
In order to determine if HLZ-rich milk can modulate gut microbiota composition, this study conducted feeding trials in young pigs followed by a more in-depth fecal microbiota assessment using 16S rRNA clone libraries and the G2 Phylochip. The pig was selected as a model organism since it represents a monogastric animal with GI anatomy, function, and metabolic regulation similar to those of humans (2). The use of pigs as a relevant human medical model is well documented (27). Furthermore, pigs and humans share similarities in GI microbial diversity (20) and pig studies allow more invasive sampling, induction of disease states, and a variety of nutritional intervention approaches. This study found that HLZ milk modulated GI microbiota by increasing the ratio of beneficial bacteria and decreasing disease-causing microbe numbers.
MATERIALS AND METHODS
Milk and milk analysis.
Production and characterization of the HLZ-transgenic line has been previously described (28, 29). This line of transgenic goats produces active HLZ in their milk at an average level of 270 μg/ml without disrupting the gross composition of the milk in terms of total fat and protein content (29). For the feeding to pigs, milk was collected by machine at the morning and evening milkings from HLZ-transgenic and nontransgenic control goats from the University of California (UC) Davis dairy goat herd. Milk was pooled into respective containers, pasteurized to 74°C, and stored at 4°C prior to feeding to animals. Milk was used for 1 week before being discarded and replaced with a new batch. Each batch of milk was analyzed by Western blotting to confirm the presence and quantity of HLZ. Milk was also collected from a total of six individual HLZ-transgenic and equal numbers of age- and breed-matched nontransgenic control animals for a more in-depth analysis using two-dimensional (2-D) gel analysis coupled with mass spectrometry. Milk was collected at peak lactation (2 months) from two transgenic animals each in their 1st, 2nd, and 3rd lactations and equal numbers of breed- and parity-matched nontransgenic controls. A total of 200 μg of protein from each milk sample was subjected to 2-D gel analysis followed by matrix-assisted laser desorption ionization–time of flight/time of flight (MALDI-TOF/TOF) mass spectrometry as previously described (32). Briefly, separation in the first dimension by isoelectric point was carried out using Immobiline DryStrips (GE Healthcare, Piscataway, NJ) (pH 3 to 5.6; 11 cm in length) followed by separation in the second dimension by size on 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (Protean Xi; Bio-Rad, Hercules, CA). Gels were then stained in Coomassie blue and scanned, and proteins were quantified using the All-to-One warping strategy with Delta 2D gel analysis software (Decodon GmbH, Greifswald, Germany). A total of 29 individual spots were then extracted from each of three gels containing milk from transgenic goats in their 2nd and 3rd lactation and their three corresponding control gels and processed using a Montage In-Gel Digest Zip kit (Millipore, Billerica, MA) followed by MALDI-TOF/TOF mass spectrometry. The same 29 spots were chosen on each of the six gels. Protein identification and annotation were carried out using GPS Explorer software with the Mascot search algorithm and DeNovo Explorer modules included in the 4700 Explorer software (Applied Biosystems, Foster City, CA). All animals were housed and cared for under AAALAC-approved conditions.
Feeding trials.
Eight crossbred, specific-pathogen-free Yorkshire full-sibling male pigs from the UC Davis Swine Facility were weaned at 21 days of age and housed together, receiving the same treatment and diet until 8 weeks of age, at which time they were placed into groups of two in connected environmental chambers. All pigs had ad libitum access to a standard ration of dry feed (UC Davis Pig Starter Diet; Associated Feed, Turlock, CA) and water throughout the trial. In addition to the standard diet, each pen received 500 ml of pasteurized nontransgenic control goat milk twice daily delivered through a lixit container for a period of 2 days in order for the animals to become accustomed to consuming milk. Each animal was then placed into an individual pen, with four animals receiving pasteurized milk from nontransgenic control goats and four receiving pasteurized milk from HLZ-transgenic goats for a period of 14 days. Pigs that were cohoused for the adjustment period were given the same type of milk. Each animal was dosed with 250 ml of its respective milk allotment twice daily for 4 days, 300 ml twice daily for 3 days, and then 350 ml twice daily for the remaining 6 days. Fresh feces samples were collected from each pair of animals before any milk was given (day 1) and then from individual animals at the end of the adjustment period and before the milk type was switched (day 3), 24 h after the start of HLZ milk administration (day 4), and then every other day for the remainder of the trial. After 14 days of milk dosing, the animals were subjected to necropsy and sections of the duodenum and ileum taken for total coliform and E. coli counts using Petrifilm count plates (3M, St. Paul, MN) as previously described (5).
16S rRNA gene sequencing.
Fecal samples from days 1 (no milk), 3 (end of adjustment period), 6 (72 h on HLZ milk), and 17 (endpoint after 14 days of HLZ milk) were processed for sequence analysis. All feces samples were stored at 4°C for no longer than 4 days prior to bacterial DNA extraction using a QIAmp DNA stool kit with the protocol for pathogen detection (Qiagen, Valencia, CA). Primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1392R (5′-GACGGGCGGTGTGTAC-3′) were used to amplify the 16S rRNA gene sequence (21). PCRs were performed as recommended by Polz and Cavanaugh (39) to reduce bias in amplification. Briefly, PCR was performed in 50-μl reaction volumes containing 50 ng DNA and 1 μM each primer under the following conditions: 1 cycle of 95°C for 5 min, 20 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1.5 min, and 1 cycle of 10 min at 72°C. Resulting PCR products were ligated into StrataClone PCR cloning kit vector (Agilent Technologies, Santa Clara, CA) to create a library of clones for sequencing. A total of 192 clones per sample were grown in 96-well plates in LB freezing media, and DNA templates were prepared by rolling-circle amplification using a TempliPhi HT amplification kit (GE Healthcare, Piscataway, NJ) and then sequenced using primer 1392R and a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA).
Resulting DNA sequences were subjected to base-calling by PHRED and quality trimming and vector removal by LUCY and used for comparison to 16S rRNA gene sequences from the Ribosomal Database Project (release 10; http://rdp.cme.msu.edu/). Only sequences with unambiguous reads of greater than 400 bp were used. Clones with more than 97% sequence identity were defined as operational taxonomic units (OTUs) and assigned to a phylum using the Classifier software available at http://rdp.cme.msu.edu/classifier/classifier.jsp (49), which assigns an OTU sequence to a phylum using a naïve Bayesian rRNA classifier trained on the known type strain 16S sequences. Complete 16S rRNA libraries for each feeding group were compared to each other using the Student t test and the Library Compare software available at http://rdp.cme.msu.edu/comparison/comp.jsp (49), which estimates the likelihood that the frequency of membership in a given taxon is the same for the two libraries. The percentage of a phylum in one library was considered significantly different (P < 0.05) from the percentage in another library if the two statistical methods (Student's t test and Compare) were in agreement. As one control-fed pig was found to be an outlier via G2 Phylochip analysis, this pig was not used in the analysis of the 16S rRNA data.
G2 Phylochip.
Analysis of fecal microbial diversity was also conducted on the endpoint samples (day 17) using the G2 Phylochip as described by Parnell et al. (37, 38). Briefly, the ribosomal 16S gene was amplified by PCR utilizing bacterium-specific primers (F [5′-AGAGTTTGATCCTGGCTCAG-3′] and R [5′-ACGGCTACCTTGTTAGCACTT-3′]). Amplified and labeled DNA (4 μg) was hybridized to the G2 Phylochip for 16 h at 48°C, with rotation at 60 rpm. The chip was washed, stained, and scanned following the standard Affymetrix protocol at the Center for Integrated BioSystems, Utah State University (Logan, UT). Raw .cel files were normalized using RMA (17). The presence or absence of each OTU was determined using Phylotrac (www.phylotrac.org). An OTU was considered present if 90% of the probes in the OTU probe set had a ratio of PM/MM > 1.3. OTUs called present in at least one of the samples were included in further analysis. Hierarchical clustering of data was done using HCE 3.5 (42). Principal component analysis was done using TMEV 4.6.1 (40). SAM (46) was used to identify OTUs statistically significantly different between the control and treatment groups. Phylogenetic groups were created based on the phylum, class, order, and family of each OTU present on the Phylochip. Statistical overrepresentation of the OTUs due to the treatment within specific phylogenetic groups was estimated using GSEA (44). 16S sequences for OTUs and family-wide consensus sequences were obtained from Greengenes (12). The 16S sequences were aligned using MUSCLE (14), maximum-likelihood trees were built using RAxML (43), and iTOL ver. 2.1.1 (24) was used to project log2 ratios of data from the treatment versus control groups onto the 16S trees.
RESULTS
Milk analysis.
The overall protein composition of milk from HLZ-transgenic goats was not statistically different from that of milk from nontransgenic control goats. A total of 136 protein spots were analyzed on 2-D gels, and the numbers of proteins and their locations and intensities were not significantly different between transgenic and control samples (Table 1; see also Fig. S1 in the supplemental material). Furthermore, 23 of the 29 spots chosen for mass spectrometry analysis were identified and all corresponded to proteins normally found in milk (Table 1). Pigs consuming HLZ milk for 14 days had significantly lower numbers of coliforms in the duodenum than did control-fed pigs (P = 0.019; Table 2). The numbers of coliforms and E. coli in the ileum were lower but not significantly different in HLZ-fed pigs (P = 0.487 and P = 0.332, respectively).
Table 1.
Identification and quantification of proteins in HLZ and control milk
| Gel | Protein identification | Spot density (% vol) |
SwissProt accession no. | Mascot score | Peptide count | ||
|---|---|---|---|---|---|---|---|
| Control (n = 6) | HLZ (n = 6) | P | |||||
| ID32847 | Serum albumin precursor | 0.681 ± 0.133 | 0.728 ± 0.135 | 0.811 | P14639 | 81 | 28 |
| ID32983 | β-Casein precursor | 1.492 ± 0.367 | 1.357 ± 0.277 | 0.774 | P11839 | 196 | 8 |
| ID34457_N | β-Casein precursor | 17.278 ± 0.979 | 17.192 ± 0.950 | 0.951 | Q9TSI0 | 66 | 3 |
| ID34461_N | β-Casein precursor | 19.646 ± 1.461 | 20.338 ± 1.345 | 0.735 | P11839 | 136 | 8 |
| ID34468 | αs1-Casein precursor | 4.001 ± 0.822 | 4.476 ± 0.897 | 0.704 | P18626 | 161 | 8 |
| ID34477 | β-Casein precursor | 5.829 ± 0.216 | 5.655 ± 0.509 | 0.759 | Q9TSI0 | 176 | 5 |
| ID34478 | αs1-Casein A short form | 1.600 ± 0.426 | 1.342 ± 0.434 | 0.680 | gi|999049 | 76 | 3 |
| ID34484 | β-Casein precursor | 5.208 ± 0.504 | 5.152 ± 0.463 | 0.936 | P33048 | 166 | 2 |
| ID34485_N | β-Casein precursor | 2.317 ± 0.539 | 2.423 ± 0.232 | 0.859 | P11839 | 61 | 4 |
| ID34488_N | β-Casein precursor | 2.904 ± 0.205 | 2.975 ± 0.220 | 0.817 | P33048 | 191 | 6 |
| ID34504_N | κ-Casein precursor | 1.642 ± 0.263 | 1.363 ± 0.165 | 0.390 | P02670 | 211 | 4 |
| ID34505 | κ-Casein precursor | 8.761 ± 0.464 | 9.148 ± 0.294 | 0.497 | P02670 | 171 | 6 |
| ID34516 | αs1-Casein precursor | 0.715 ± 0.274 | 0.587 ± 0.228 | 0.727 | P04653 | 151 | 10 |
| ID34517 | αs1-Casein precursor | 0.871 ± 0.277 | 1.083 ± 0.325 | 0.630 | P18626 | 146 | 7 |
| ID34530 | α-Lactalbumin | 12.180 ± 0.719 | 12.066 ± 0.692 | 0.911 | P00712 | 86 | 2 |
| ID46338_N | αs2-Casein precursor | 14.202 ± 0.753 | 14.198 ± 1.007 | 0.997 | P04654 | 126 | 10 |
| ID46339_N | αs2-Casein precursor | 9.162 ± 0.543 | 9.306 ± 0.660 | 0.870 | P33049 | 111 | 14 |
| ID46344_N | αs2-Casein precursor | 7.574 ± 0.413 | 7.393 ± 0.321 | 0.737 | P04654 | 103 | 7 |
| ID46345_N | αs2-Casein precursor | 2.099 ± 0.192 | 2.158 ± 0.283 | 0.866 | P04654 | 91 | 13 |
| ID46347_N | αs2-Casein precursor | 4.080 ± 0.388 | 4.110 ± 0.495 | 0.963 | P04654 | 96 | 16 |
| ID46351 | αs2-Casein precursor | 4.507 ± 0.583 | 4.671 ± 0.426 | 0.825 | P33049 | 121 | 8 |
| ID46387 | β-Lactoglobulin precursor | 17.566 ± 1.001 | 18.033 ± 2.158 | 0.848 | P02756 | 181 | 10 |
| ID7607 | αs2-Casein precursor | 1.422 ± 0.236 | 1.809 ± 0.285 | 0.320 | P04654 | 116 | 12 |
Table 2.
Total coliform and E. coli counts in intestinal segments at day 17
| Segment and organism(s) | log CFU/g ± SD for indicated pig group |
|
|---|---|---|
| Control fed (n = 3) | HLZ fed (n = 4) | |
| Duodenum | ||
| Coliforms | 1.93 ± 0.81 | 0.25 ± 0.5a |
| E. coli | 0 | 0 |
| Ileum | ||
| Coliforms | 3.66 ± 1.46 | 3.01 ± 0.82 |
| E. coli | 3.66 ± 1.46 | 2.36 ± 1.65 |
HLZ-fed pigs significantly different from controls (P = 0.019).
16S rRNA clone sequencing analysis.
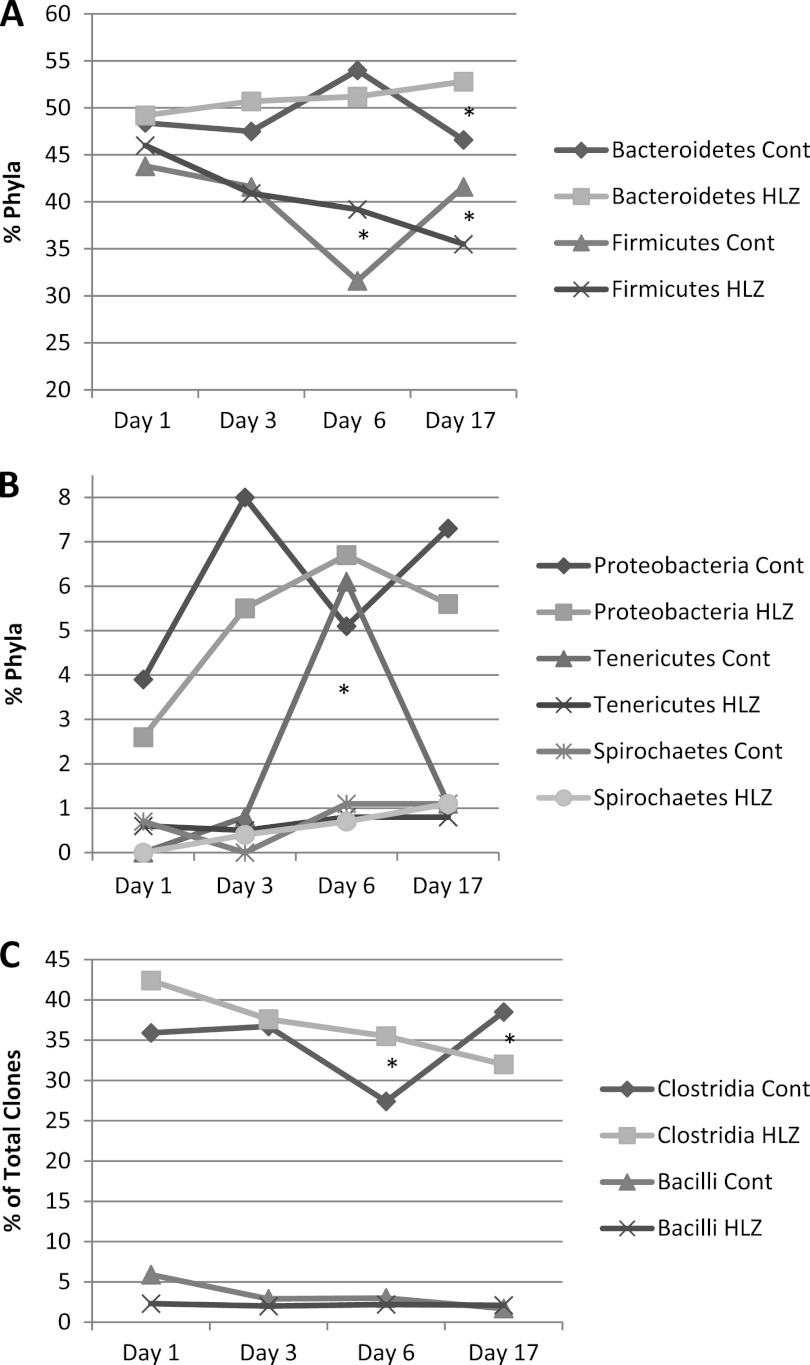
The fecal microbial profile determined using 16S rRNA gene sequencing showed that all animals started with similar fecal microbial populations, as no significant differences in phyla among animals at day 1 and day 3 were observed (Fig. 1). The fecal microbial profile changed significantly after 14 days of HLZ milk consumption (Table 3). Animals receiving HLZ milk had an overrepresentation of Bacteroidetes approaching significance (P = 0.07) and significant underrepresentation of Firmicutes (P = 0.038) compared to animals receiving control goat milk. Within the Bacteroidetes, members of the order Bacteroidales were present in a significantly greater proportion in HLZ-fed animals (P = 0.035), with a significantly higher representation from the genera Paraprevotella and Parabacteroides (P = 0.033; Table 4).
Fig 1.
Changes in phyla over time. Percentages of clones assigned to (A) major phyla, (B) minor phyla, and (C) Firmicutes determined by 16S rRNA sequencing are shown. HLZ, pigs fed milk from HLZ-transgenic animals; Cont, pigs fed milk from nontransgenic control animals. *, significantly different (P < 0.05).
Table 3.
Fecal microbial profile after 14 days of consuming HLZ or control milk
| Phylum | % of clones in indicated pig groupa |
||
|---|---|---|---|
| Control fed (n = 3; 356 clones) | HLZ fed (n = 4; 532 clones) | P | |
| Bacteroidetes | 46.6 | 52.8 | 0.070 |
| Firmicutes | 41.6 | 35.5b | 0.038 |
| Proteobacteria | 7.3 | 5.6 | 0.317 |
| Tenericutes | 1.1 | 0.8 | 0.537 |
| Spirochaetes | 1.1 | 1.1 | 1 |
| Actinobacteria | 0.3 | 0 | 0.321 |
| Lentisphaerae | 0 | 0.4 | 0.430 |
| Fusobacteria | 0 | 0.6 | 0.258 |
| Unclassified | 2.0 | 3.2 | |
Data represent the percentages of clones assigned to a phylum using 16S rRNA gene sequencing. Clone values in column headings represent the number of sequences in each library.
HLZ-fed pigs significantly different from controls (P < 0.05).
Table 4.
Significant differences in bacterial genera by 16S rRNA gene sequencing after 2 and 14 days of HLZ milk consumption
| Phylum | Class | Order | Family | Genus | P | % of total clones in indicated pig group |
|
|---|---|---|---|---|---|---|---|
| Control fed | HLZ fed | ||||||
| Day 6 | |||||||
| Tenericutes | Mollicutes | Anaeroplasmatales | Anaeroplasmataceae | Anaeroplasma | 0.000103 | 6.1 | 0.8 |
| Firmicutes | 0.0114 | 31.4 | 38.9 | ||||
| Clostridia | Clostridiales | 0.0048 | 27.4 | 35.5 | |||
| Ruminococcaceae | 0.105 | 11.0 | 14.3 | ||||
| Oscillibacter | 0.00932 | 3.8 | 7.6 | ||||
| Veillonellaceae | 0.174 | 5.5 | 7.6 | ||||
| Anaerovibrio | 0.029 | 0.8 | 2.7 | ||||
| Lachnospiraceae | 0.881 | 7.2 | 7.4 | ||||
| Roseburia | 0.00012 | 3.4 | 0.3 | ||||
| Bacteroidetes | Bacteroidia | Bacteroidales | 0.358 | 54.0 | 51.2 | ||
| Prevotellaceae | 0.0183 | 50.0 | 42.8 | ||||
| Prevotella | 0.0008 | 50.0 | 35.8 | ||||
| Day 17 | |||||||
| Firmicutes | 0.038 | 42.1 | 35.5 | ||||
| Clostridia | Clostridiales | 0.045 | 38.5 | 31.9 | |||
| Ruminococcaceae | 0.317 | 14.3 | 12.0 | ||||
| Subdoligranulum | 0.038 | 1.4 | 0.2 | ||||
| Veillonellaceae | 0.992 | 5.6 | 5.6 | ||||
| Anaerovibrio | 0.012 | 2.2 | 0.6 | ||||
| Bacilli | Lactobacillales | 0.682 | 1.7 | 2.1 | |||
| Lactobacillaceae | Lactobacillus | 0.671 | 0.3 | 0.6 | |||
| Streptococcaceae | Streptococcus | 0.97 | 1.4 | 1.5 | |||
| Bacteroidetes | 0.070 | 46.6 | 52.8 | ||||
| Bacteroidia | Bacteroidales | 0.035 | 44.7 | 52.1 | |||
| Prevotellaceae | 0.215 | 39.0 | 43.2 | ||||
| Paraprevotella | 0.033 | 0 | 1.3 | ||||
| Porphyromonadaceae | 0.097 | 2.8 | 5.1 | ||||
| Parabacteroides | 0.033 | 0 | 1.3 | ||||
The consumption of control milk during the adjustment period promoted the growth of members of Proteobacteria (P = 0.0063), specifically, of the genus Succinivibrio (P < 0.0048). Over time, levels of Bacteroidetes steadily increased and were accompanied by a steady decrease in levels of Firmicutes in HLZ-fed pigs whereas these populations fluctuated in control-fed pigs (Fig. 1A). There were significant differences in the levels of Firmicutes in control and HLZ-fed pigs by day 6 (P = 0.013), and they remained significantly different at day 17 (P = 0.038). On day 6, HLZ-fed pigs also had significantly fewer Tenericutes (genus Anaeroplasma) than did control-fed pigs (P < 0.001, Table 4). Within the Firmicutes, levels of the class Clostridia gradually decreased over time in HLZ-fed pigs and were significantly different from those in control-fed pigs at both day 6 (P = 0.005) and day 17 (P = 0.045), with more fluctuation in the control-fed animals (Fig. 1B and Table 4). There were significantly fewer bacteria from the Subdoligranulum and Anaerovibrio genera in HLZ-fed animals at day 17 (Table 4). Levels of Bacilli were steadily maintained in HLZ-fed animals and decreased over time in control-fed animals, although these differences were not statistically different (P = 0.43 at day 6 and P = 0.682 at day 17) (Fig. 1C).
G2 Phylochip analysis.
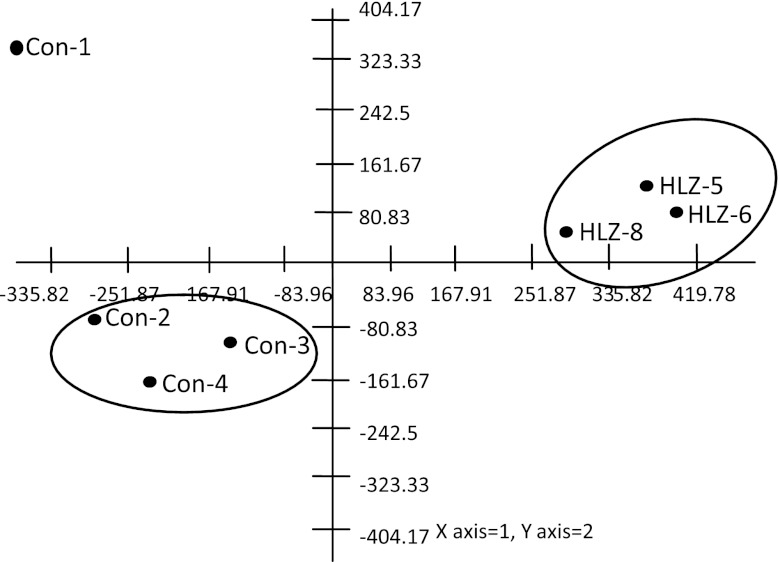
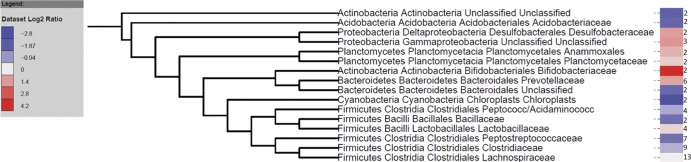
All microbes and trends found in the clone library of the endpoint (day 17) were also found to be present using the G2 Phylochip; plus, many additional OTUs were observed. The Phylochip analysis identified a total of 500 OTUs across seven samples (18 phyla, 38 classes, 64 orders, and 91 families) compared to 13 phyla, 18 classes, 17 orders and 32 families detected with the clone libraries. The total fecal community populations of all animals consuming HLZ milk for 14 days were similar to each other, as were the populations of bacteria in the feces of three of the four control-fed animals (Fig. 2). Using principal component analysis (PCA), the six samples were segregated into two distinct clusters. Based on the PCA, one of the control samples (Con-1 in Fig. 2) was a clear outlier and hence was removed from further statistical analysis. A total of 113 OTUs were significantly different (q [adjusted P value taking into consideration false-discovery rate] ≤ 0.05) across the control and HLZ-fed groups (see Fig. S2 in the supplemental material). Figure 3 summarizes the differences at the family level, which had at least two OTUs that were significantly different between the treatment and control groups (total of 64 significant OTUs). Among all groups, Clostridales were statistically underrepresented in HLZ-fed pigs (q = 0.069), while Bacteroidetes were overrepresented in HLZ-fed pigs (q = 0.1) compared to pigs fed control milk. Interestingly, beneficial microbes such as Bifidobacteriaceae (2 OTUs) and Lactobacillaceae (2 OTUs) were also enriched upon consumption of HLZ milk whereas detrimental microbes such as Streptococcaceae (1 OTU) and Campylobacterales (2 OTUs) were depleted with consumption of HLZ milk (see Table S1 in the supplemental material).
Fig 2.
Projection of samples on the first two principal component axes based on the PCA of OTU abundance from the Phylochip analysis. HLZ, pigs fed milk from HLZ-transgenic animals; Con, pigs fed milk from nontransgenic control animals.
Fig 3.
Summarized log2 ratios for families that had at least two significantly different (q ≤ 0.05) OTUs between the control and HLZ-fed treatment groups at day 17 as determined by Phylochip analysis. The numbers beside the heat map represent the OTUs that were significantly different in each family.
DISCUSSION
Human milk contains numerous bioactive components that can modulate the infant gut microbiome and thus benefit the host. We used a model system of pigs and transgenic goat milk containing HLZ to examine the effects of consumption of this antimicrobial milk component on beneficial microbe enrichment versus detrimental microbe reduction in the gut microbial community. We demonstrated that consumption of lysozyme-rich milk indeed altered gut microbiota populations and shifted the microbial population to those microbes associated with activities beneficial for the host. The changes in microbial populations seen can be attributed to the presence of HLZ in milk. Fine analysis of milk composition using 2-D gels indicated that there were no off-target proteins being produced and no endogenous proteins whose production was being diminished by the expression of HLZ and therefore that all effects of the milk were a result of the presence of HLZ. Moreover, human milk oligosaccharides which are known modulators of intestinal microbiota are not found in the same diversity or quantity in goat milk (9), demonstrating that HLZ alone could contribute this important function to the milk of dairy animals.
Microbial abundance increases along the length of the digestive tract (35), with the largest diversity and most bacteria found in the colon, and therefore the microbiota of feces is distinct from that of the rest of the GI tract in both pigs (7) and humans (4, 41). Using standard culturing techniques, total coliform and E. coli counts in the duodenum and ileum of HLZ-fed pigs tended to be lower than in control-fed pigs, similar to results seen when starting with 14-day-old pigs (5, 30). These results indicate the reproducibility of the effect of HLZ milk in differently aged animals at this level of detail. As shown using molecular techniques and fecal samples, the overall level of Proteobacteria was not significantly lower in HLZ-fed animals than in controls (5.6% versus 7.3%) and there were no differences in E. coli levels in the feces of HLZ-fed and control-fed animals, likely a difference in sample location (small intestine versus feces). Proteobacteria are a common component of the distal gut, and in vitro work has demonstrated that HLZ milk acts in a bactericidal fashion toward E. coli (31).
The predominant phyla present in the feces of both feeding groups of pigs at all time points were the Firmicutes and Bacteroidetes, similar to previous studies with both pig and human subjects (7, 13, 23), suggesting that the pig is a good animal model for studying the GI microbiome and the consequences of microbiota manipulation. The microbial populations were not statistically different between individuals at the start of the trial (day 1, no milk) at the phylum level, indicating that all pigs were starting with similar fecal microbiotas. It should be noted that while samples were prepared after storage at 4°C, which has been shown to have an impact on detected microbial diversity (36), all samples were subjected to similar storage conditions, thus allowing direct comparison between samples. After the 3-day period of adjustment to consumption of milk, there were significantly more Proteobacteria (namely, Succinivibrio) present than on day 1, but there were no significant differences between animals. While milk alone altered the fecal microbiota, all animals started with similar microbiotas before the administration of HLZ milk and thus all subsequent changes seen were due to direct effects of HLZ milk and not natural variation. The elevated level of Succinivibrio seen after consumption of milk commenced was maintained over all later time points in both feeding groups. Succinivibrio commonly inhabit the rumen of dairy animals to aid in digestion, and thus, consumption of goat milk in general increased levels of this microbe either by stimulating the growth of the low levels of Succinivibrio already present or by reducing the levels of other microbes, thereby allowing more growth of Succinivibrio. Using 16S rRNA gene sequencing of clone libraries, no Succinivibrio bacteria were found to be present in the milk itself (data not shown).
Previous work has demonstrated that a majority (95%) of the Firmicutes in the human gut were Clostridia (13). Similar findings were seen here, as after 14 days of milk consumption, 91% of the Firmicutes in control-fed animals were Clostridia. In HLZ-fed animals, levels of Clostridia were reduced from 92.4% (day 2) to 89.5% of total Firmicutes at day 17. While Firmicutes are important for supplying energy in the form of short-chain fatty acids, a skewed ratio of Firmicutes to Bacteroidetes has been implicated in obese mice and humans (25, 26), with elevated levels of Firmicutes and decreased levels of Bacteroidetes in obese subjects. An increase in Firmicutes has been associated with an increased ability to extract energy from the diet and/or promote the deposition of this energy in the form of fat (19, 45). After 14 days of HLZ milk consumption, the proportion of Firmicutes in the fecal microbiota was underrepresented and that of Bacteroidetes overrepresented compared to animals consuming control goat milk. The gut surface area of the animals used in this study was investigated previously and was found to be related to an improved absorptive capacity of the intestinal epithelium, as HLZ-fed pigs tended to have longer villi and had a significantly thinner lamina propria in the duodenum (10). A 20% increase in the proportional representation of Bacteroidetes in fecal microbiota in humans has been correlated with a decrease in nutrient absorption (19). Here we observed a more modest increase in fecal Bacteroidetes content (6.2%) upon consumption of HLZ milk that was related to changes in cellular ultrastructure in the small intestine. While the energy content of the feces was not determined in this study, the resulting histological changes represent an intestinal epithelium better able to absorb what nutrients were present if less energy was indeed available due to the underrepresentation of the energy supplying Firmicutes. Combined, these results suggest that microbiota composition can influence the physical state of the intestine. It remains to be determined what other metabolic and functional changes occur in response to HLZ-induced modulation of gut microbiota.
One important issue regarding gut microbiota communities is their stability over time (3). Animals fed control milk were more prone to fluctuation in the relative proportions of phyla present after the 3-day adjustment period. The significant increase in Tenericutes at day 6 in control-fed animals was related to one individual; however, the proportions of rest of the phyla were more consistent between individuals of the same feeding group. Gut microbiota populations have been reported to have minimal variation over time within individuals but are highly variable between people (11). The greater fecal community member stability seen with HLZ milk consumption may have been due to the presence of HLZ acting to quiet challenges to the gut, the short interval between sample analysis, or a brief environmental perturbation, although all animals were housed together and exposed to the same environment. Changes in diet have been shown to rapidly induce changes in fecal microbiota (within 3 to 4 days), after which the microbiota was maintained for several weeks until the diet was altered again (48). Here, the introduction of milk altered fecal microbiota in a similarly short time frame; however, the changes were not static. The changes being seen in both feeding groups over time may reflect natural variation in microbiota stability over a shorter time period and point to HLZ being able to continuously modulate community member diversity in a similar pattern over time.
Overall, the G2 Phylochip analysis supported our findings obtained with 16S rRNA gene sequencing and offered more details. Both methods identified Bacteroidetes and Firmicutes as the main phyla and the presence of fewer Firmicutes and more Bacteroidetes in HLZ-fed animals compared to control-fed animals. Within phyla and families, levels of community members were both increasing and decreasing in response to HLZ milk consumption, with the exception of Bacteroidetes, where levels of all members were elevated compared to the results from control-fed animals. Greater variability within phyla has been reported in previous studies (13); however, HLZ milk was also able to change the bacterial abundance at the phylum level.
Lysozyme-rich milk was capable of significantly altering community member diversity in a fashion that allowed identification of the feeding group based on the resulting fecal microbiota. Whether these changes can influence overall health and resistance to disease remains to be determined, but the types of changes seen are associated with improved gut health. The more comprehensive data generated with the Phylochip demonstrated that, compared to the microbiota of control-fed pigs, the microbiota of pigs fed HLZ-rich milk more closely resembled that of human infants being breast fed, with the enrichment of Bifidobacteriacea and Lactobacillacea, both biomarkers of increased gut and host health (47). These beneficial changes were accompanied by the reduction of the numbers clostridia and Streptococcaceae, which are components of the fecal microbiota of infants fed formula that lacks lysozyme, as well as by decreased levels of disease-related bacteria such as Mycobacteriaceae and Campylobacterales. It is likely that the antimicrobial action and/or cationic properties of HLZ were contributing to the selection of these beneficial bacteria by preventing the growth of others. Taken as a whole, the data suggest that addition of HLZ alone to milk is able to modulate intestinal microbiota in a fashion that is similar to the modulation seen with human milk.
Untangling the complex interactions of nutrient, host, and bacteria is a substantial challenge. The results of this study support the hypothesis that milk components actively modified the gut microbiome, resulting in community membership changes to increase levels of beneficial microbes and reduce levels of undesirable community members. They also support the use of pigs as a model animal for gut microbiota research and, along with the use of HLZ-rich milk, offer an approach not only for the study of the mechanism of action of lysozyme at the level of the gut but also for the direct manipulation of gut microbiota with the potential to improve GI disorders, including diarrhea and inflammatory bowel diseases such as Crohn's disease and colitis.
Supplementary Material
ACKNOWLEDGMENTS
We thank the UC Davis College of Agricultural and Environmental Sciences Genomics Facility for carrying out template preparation and sequencing reactions for the 16S rRNA gene sequencing. We kindly thank Jan Carlson and Kent Parker of the UC Davis Goat and Swine Facilities, respectively, for animal assistance and care. We also thank Caitlin Cooper for animal feeding and sample collection.
This project was supported in part by the Biotechnology Risk Assessment Program competitive grant 2008-33522-04842 from the USDA National Institute of Food and Agriculture (NIFA).
Footnotes
Published ahead of print 29 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Adlerberth I, Wold AE. 2009. Establishment of the gut microbiota in Western infants. Acta Paediatr. 98: 229–238 doi:10.1111/j.1651-2227.2008.01060.x [DOI] [PubMed] [Google Scholar]
- 2. Aigner B, et al. 2010. Transgenic pigs as models for translational biomedical research. J. Mol. Med. 88: 653–664 [DOI] [PubMed] [Google Scholar]
- 3. Bik EM. 2009. Composition and function of the human-associated microbiota. Nutr. Rev. 67: S164–S171 [DOI] [PubMed] [Google Scholar]
- 4. Booijink CC, et al. 2010. High temporal and inter-individual variation detected in the human ileal microbiota. Environ. Microbiol. 12: 3213–3227 [DOI] [PubMed] [Google Scholar]
- 5. Brundige DR, Maga EA, Klasing KC, Murray JD. 2008. Lysozyme transgenic goats' milk influences gastrointestinal morphology in young pigs. J. Nutr. 138: 921–926 [DOI] [PubMed] [Google Scholar]
- 6. Brundige DR, Maga EA, Klasing KC, Murray JD. 2010. Consumption of pasteurized human lysozyme transgenic goats' milk alters serum metabolite profile in young pigs. Transgenic Res. 19: 563–574 doi:10.1007/s11248-009-9334-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Castillo M, et al. 2007. Application of 16S rRNA gene-targetted fluorescence in situ hybridization and restriction fragment length polymorphism to study porcine microbiota along the gastrointestinal tract in response to different sources of dietary fibre. FEMS Microbiol. Ecol. 59: 138–146 [DOI] [PubMed] [Google Scholar]
- 8. Chandan RC, Parry RM, Shahani KM. 1968. Lysozyme, lipase, and ribonuclease in milk of various species. J. Dairy Sci. 51: 606–607 doi:10.3168/jds.S0022-0302(68)87036-5 [Google Scholar]
- 9. Chichlowski M, German JB, Lebrilla CB, Mills DA. 2011. The influence of milk oligosaccharides on microbiota of infants: opportunities for formulas. Annu. Rev. Food Sci. Technol. 2: 331–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cooper CA, Brundige DR, Reh WA, Maga EA, Murray JD. 2011. Lysozyme transgenic goats' milk positively impacts intestinal cytokine expression and morphology. Transgenic Res. 20: 1235–1243 doi:10.1007/s11248-011-9489-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Costello EK, et al. 2009. Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697 doi:10.1126/science.1177486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DeSantis TZ, et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72: 5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eckburg PB, et al. 2005. Diversity of the human intestinal microbial flora. Science 308: 1635–1638 doi:10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 doi:10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fallani M, et al. 2010. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J. Pediatr. Gastroenterol. Nutr. 51: 77–84 [DOI] [PubMed] [Google Scholar]
- 16. Gartner LM, et al. 2005. Breastfeeding and the use of human milk. Pediatrics 115: 496–506 doi:10.1542/peds.2004-2491 [DOI] [PubMed] [Google Scholar]
- 17. Irizarry RA, et al. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31: e15 doi:10.1093/nar/gng015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Isaacs CE. 2005. Human milk inactivates pathogens individually, additively, and synergistically. J. Nutr. 135: 1286–1288 [DOI] [PubMed] [Google Scholar]
- 19. Jumpertz R, et al. 2011. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am. J. Clin. Nutr. 94: 58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lamendella R, Domingo JW, Ghosh S, Martinson J, Oerther DB. 2011. Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol. 11: 103 doi:10.1186/1471-2180-11-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–147 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 22. Le Huërou-Luron I, Blat S, Boudry G. 2010. Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr. Res. Rev. 23: 23–36 [DOI] [PubMed] [Google Scholar]
- 23. Leser TD, et al. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68: 673–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Letunic I, Bork P. 2011. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39: W475–W478 doi:10.1093/nar/gkr201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ley RE, et al. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 102: 11070–11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444: 1022–1023 doi:10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- 27. Lunney JK. 2007. Advances in swine biomedical model genomics. Int. J. Biol. Sci. 3: 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maga EA, et al. 2003. Increased efficiency of transgenic livestock production. Transgenic Res. 12: 485–496 doi:10.1023/A:1024257906647 [DOI] [PubMed] [Google Scholar]
- 29. Maga EA, et al. 2006. Production and processing of milk from transgenic goats expressing human lysozyme in the mammary gland. J. Dairy Sci. 89: 518–524 doi:10.3168/jds.S0022-0302(06)72114-2 [DOI] [PubMed] [Google Scholar]
- 30. Maga EA, Walker RL, Anderson GB, Murray JD. 2006. Consumption of milk from transgenic goats expressing human lysozyme in the mammary gland results in the modulation of intestinal microflora. Transgenic Res. 15: 515–519 doi:10.1007/s11248-006-0014-3 [DOI] [PubMed] [Google Scholar]
- 31. Maga EA, Cullor JS, Smith W, Anderson GB, Murray JD. 2006. Human lysozyme expressed in the mammary gland of transgenic dairy goats can inhibit the growth of bacteria that cause mastitis and the cold-spoilage of milk. Foodborne Pathog. Dis. 3: 384–392 [DOI] [PubMed] [Google Scholar]
- 32. Maga EA, Daftari P, Kültz D, Penedo MCT. 2009. Prevalence of αs1-casein genotypes in American dairy goats. J. Anim. Sci. 87: 3464–3469 [DOI] [PubMed] [Google Scholar]
- 33. Masschalck B, Michiels CW. 2003. Antimicrobial properties of lysozyme in relation to foodborne vegetative bacteria. Crit. Rev. Microbiol. 29: 191–214 [DOI] [PubMed] [Google Scholar]
- 34. Mountzouris KC, McCartney AL, Gibson GR. 2002. Intestinal microflora of human infants and current trends for its nutritional modulation. Br. J. Nutr. 87: 405–420 doi:10.1079/BJNBJN2002563 [DOI] [PubMed] [Google Scholar]
- 35. O'Hara AM, Shanahan F. 2006. The gut flora as a forgotten organ. EMBO Rep. 7: 688–693 doi:10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ott SJ, et al. 2004. In vitro alterations of intestinal bacterial microbiota in fecal samples during storage. Diagn. Microbiol. Infect. Dis. 50: 237–245 [DOI] [PubMed] [Google Scholar]
- 37. Parnell JJ, et al. 2010. Functional biogeography as evidence of gene transfer in hypersaline microbial communities. PLoS One 5: e12919 doi:10.1371/journal.pone.0012919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Parnell JJ, Rompato G, Crowl TA, Weimer BC, Pfrender ME. 2011. The effect of disturbance on phylogenetic diversity in microbial communities. Aquat. Microb. Ecol. 64: 267–273 [Google Scholar]
- 39. Polz MF, Cavanaugh CM. 1998. Bias in template to product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64: 3724–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saeed AI, et al. 2006. TM4 microarray software suite. Methods Enzymol. 411: 134–193 doi:10.1016/S0076-6879(06)11009-5 [DOI] [PubMed] [Google Scholar]
- 41. Sekirov I, Russell SL, Antunes LC, Finlay BB. 2010. Gut microbiota in health and disease. Physiol. Rev. 90: 859–904 [DOI] [PubMed] [Google Scholar]
- 42. Seo J, Gordish-Dressman H, Hoffman EP. 2006. An interactive power analysis tool for microarray hypothesis testing and generation. Bioinformatics 22: 808–814 doi:10.1093/bioinformatics/btk052 [DOI] [PubMed] [Google Scholar]
- 43. Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57: 758–771 [DOI] [PubMed] [Google Scholar]
- 44. Subramanian A, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Turnbaugh PJ, Gordon JI. 2009. The core gut microbiome, energy balance and obesity. J. Physiol. 587: 4153–4158 doi:10.1113/jphysiol.2009.174136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ventura M, et al. 2009. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat. Rev. Microbiol. 7: 61–71 [DOI] [PubMed] [Google Scholar]
- 48. Walker AW, et al. 2011. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 5: 220–230 doi:10.1038/ismej.2010.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang QG, Garrity M, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73: 5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.