Abstract
The obligate predator Bdellovibrio bacteriovorus HD100 shows a large set of proteases and other hydrolases as part of its hydrolytic arsenal needed for its predatory life cycle. We present genetic and biochemical evidence that open reading frame (ORF) Bd3709 of B. bacteriovorus HD100 encodes a novel medium-chain-length polyhydroxyalkanoate (mcl-PHA) depolymerase (PhaZBd). The primary structure of PhaZBd suggests that this enzyme belongs to the α/β-hydrolase fold family and has a typical serine hydrolase catalytic triad (serine-histidine-aspartic acid) in agreement with other PHA depolymerases and lipases. PhaZBd has been extracellularly produced using different hypersecretor Tol-pal mutants of Escherichia coli and Pseudomonas putida as recombinant hosts. The recombinant PhaZBd has been characterized, and its biochemical properties have been compared to those of other PHA depolymerases. The enzyme behaves as a serine hydrolase that is inhibited by phenylmethylsulfonyl fluoride. It is also affected by the reducing agent dithiothreitol and nonionic detergents like Tween 80. PhaZBd is an endoexohydrolase that cleaves both large and small PHA molecules, producing mainly dimers but also monomers and trimers. The enzyme specifically degrades mcl-PHA and is inactive toward short-chain-length polyhydroxyalkanoates (scl-PHA) like polyhydroxybutyrate (PHB). These studies shed light on the potentiality of these predators as sources of new biocatalysts, such as an mcl-PHA depolymerase, for the production of enantiopure hydroxyalkanoic acids and oligomers as building blocks for the synthesis of biobased polymers.
INTRODUCTION
Polyhydroxyalkanoates (PHAs) are optically active biopolyoxoesters composed of R-3-hydroxy fatty acids, which represent a complex class of storage polyesters. A wide variety of taxonomically different groups of microorganisms (domains Bacteria and Archaea) produce intracellular homopolymers or copolymers containing different alkyl groups at the beta position in aerobic and anaerobic environments (36, 37, 42). At present, PHAs are classified into two major classes, short-chain-length PHAs (scl-PHA) with C4 and C5 monomers and medium-chain-length PHAs (mcl-PHA) with C6 to C14 monomers (37). PHAs are accumulated as inclusions in the bacterial cytoplasm in response to inorganic nutrient limitations and play a role as a sink for carbon and reducing equivalents and in synchronizing global metabolism to the availability of resources in PHA-producing microorganisms (16). It is now evident that intracellular accumulation of PHAs enhances the survival of several bacteria under environmental stress conditions imposed in water or soil (6, 27, 35).
PHAs can be catabolized by many microorganisms through extracellular or intracellular processes depending on the PHA localization (11, 12, 26). Intracellular PHA can be hydrolyzed by intracellular depolymerases, which are permanently associated to the PHA granule (11, 49). Their hydrolytic activity is controlled by the carbon demand of the PHA producer cells (10, 11, 18, 19, 26). The study of the physiological role of intracellular depolymerases in the mcl-PHA metabolism in Pseudomonas putida was recently addressed, demonstrating that the intracellular depolymerase PhaZ plays a fundamental role in the bacterial carbon metabolism by maintaining the PHA turnover, where synthesis and mobilization constitute a continuous and simultaneous cycle (10, 11, 45, 61).
Extracellular PHA can be utilized as a carbon and energy source by PHA producer or nonproducer microorganisms. This PHA is released to the medium by producer microorganisms after death, and the granules spread into the environment can be hydrolyzed by secreted PHA depolymerases of microbial origin into water-soluble oligomers and monomers that can be used as a carbon source (26). The ability to degrade extracellular scl-PHA is more widespread among bacteria than the ability to degrade mcl-PHA. Thus, many extracellular scl-PHA depolymerases have been characterized in depth over the last decade, and a considerable number of genes have been identified (1, 3, 4, 24, 26, 29, 40). The prototype of extracellular mcl-PHA depolymerases is that of Pseudomonas fluorescens GK13 (here PhaZGK13) (21, 26, 52).
Based on the analysis of the PHA depolymerase database (29), we identified a potential extracellular mcl-PHA depolymerase-coding sequence from the complete genome of Bdellovibrio bacteriovorus HD100 (open reading frame [ORF] Bd3709). Bdellovibrio and related organisms known as BALOs (Bdellovibrio and like organisms) (8, 55) are obligate bacterial predators ubiquitously found in the environment. The predator B. bacteriovorus HD100 is a highly motile Gram-negative deltaproteobacterium that invades the periplasm of other Gram-negative bacteria. This predator undergoes a complex life cycle in the periplasm of the prey, which culminates in killing of the prey cell (58). The life cycle of B. bacteriovorus is biphasic, alternating a nonreplicative attack phase and a periplasmic growth phase (48). During the reproductive phase, Bdellovibrio replicates its DNA and grows using the prey as a source of nutrients, forming the bdelloplast (a rounded prey cell containing the predator due to cell wall modifications). Finally, Bdellovibrio septates into individual cells, synthesizes flagella, and releases the new motile predators into the environment (attack phase) to seek further preys (58). In spite of its very small size (about 0.2 to 0.5 μm wide and 0.5 to 2.5 μm long), it carries a quite large genome (3.8 Mb) predicted to encode 3,584 proteins (47). The genome of B. bacteriovorus HD100 encodes a large set of proteases and other hydrolases, which are used throughout its life cycle for prey entry, degradation of prey components, and exit from the bdelloplast (47).
In this work we demonstrate that the ORF Bd3709 certainly encodes a novel mcl-PHA depolymerase enzyme (here named PhaZBd). The depolymerase was exclusively produced in the extracellular medium in hypersecretor Tol-pal mutants of Escherichia coli and P. putida (33, 34), facilitating the PhaZBd purification and characterization.
MATERIALS AND METHODS
Chemicals.
Octanoic acid (1-14C labeled) (50 mCi/mmol) was from American Radiolabeled Chemicals. All other products were of analytical quality or high-performance liquid chromatography (HPLC) grade.
Bacterial strains, plasmids, and growth conditions.
The bacterial strains, plasmids, and oligonucleotides used are listed in Table 1. E. coli and P. putida strains were grown in LB medium, in nutrient broth (NB) medium (51), or in M63 minimal medium (38) with 0.2% glucose as the carbon source, with shaking (250 rpm) at 37°C and 30°C, respectively. The appropriate selection antibiotic, kanamycin (50 μg ml−1) or gentamicin (10 μg ml−1), was added when needed. Growth was monitored with a Shimadzu UV-260 spectrophotometer at 600 nm. Solid media were supplemented with 1.5% (wt/vol) agar. For poly-(hydroxyoctanoate-co-hydroxyhexanoate) [P(HO-co-HX), also named mcl-PHA] accumulation, P. putida KT2442 was cultured in 200 ml of minimal medium with 0.1 N M63 (10, 11), using 15 mM octanoic acid as a carbon source in a 500-ml flask. B. bacteriovorus HD100 was grown in two-membered cultures with P. putida as prey in HEPES buffer (25 mM HEPES amended with 2 mM CaCl2 · 2H2O and 3 mM MgCl2 · 3H2O [pH 7.8]) as previously described (32). To obtain isolated plaques of B. bacteriovorus HD100, serial dilutions from 10−1 to 10−7 were made in diluted nutrient broth (DNB) liquid medium. A 0.1-ml volume of the appropriate dilution was mixed with 0.5 ml of P. putida in HEPES buffer, and the mixture was vortexed and plated onto DNB solid medium by using the double-overlay method (32). Isolated plaques were observed after 48 h of incubation at 30°C (see Fig. S1A in the supplemental material).
Table 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant genotype, description, or sequence (5′ to 3′) | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH10B | F′ mcrA Δ(mrr hsdRMS-mcrBC) ϕ80dlacΔM15 Δ(lacX74-deoR) recA1 araD139 Δ(ara-leu)7697 galU galK λ rpsL endA1 nupG | Invitrogen |
| K1041 | Hfr(PO2A) garB10 fhuA22 phoA4(Am) tolQ7(Am) zbg-2225::Tn10 ompF627(T2R) fadL701(T2R) relA1 pitA10 spoT1 rrnB-2 mcrB1 creC510 | CGSCb no. 7788 |
| BL21(DE3) | F− ompT hsdS gal dcm λ(DE3) | Novagen |
| P. putida strains | ||
| KT2442 | P. putida mt-2 without TOL plasmid, hsdR Rfr | 20 |
| AΩ | KT2440 tolA::ΩKm (insertion after codon 180) | 33 |
| Plasmids | ||
| pIZ1016 | pBBR1MCS-5 derivative cloning plasmid, Gmr | 39 |
| pET29 | Expression vector for producing His-tagged proteins in E. coli BL21(DE3); Kmr | Novagen |
| pIZBd1 | pIZ1016 derivative containing 816-bp XbaI/HindIII-digested phaZBd gene from B. bacteriovorus HD100 | This study |
| pETBd1 | pET29 derivative containing the NdeI/XhoI-digested phaZBd gene from B. bacteriovorus HD100 expressing PhaZBd with a C-terminal six-His tag | This study |
| pIZPZ | pIZ1016 derivative with 914-bp SmaI/HindIII phaZ gene from P. fluorescens GK13 | 9 |
| Primersa | ||
| V01 | CCCTCTAGAGAATGATATCTATTTCAACTATTTATGGT | This study |
| V02 | CCCAAGCTTAAACTAAGGAGTCCCGGATGAAAAAAC | This study |
| V06 | GGGAATTCCATATGAAAAAACTTTTAGCAGGTGTCTTTGGTGTTGTGG | This study |
| V07 | CCGCTCGAGTTTATGATTGATGAACCAGTTGGTGATC | This study |
Engineered endonuclease sites on the oligonucleotides are underlined.
CGSC, Coli Genetic Stock Center, Yale University.
DNA manipulations and plasmid constructions.
DNA manipulations and other molecular biology techniques were essentially performed as described previously (51). Transformation of E. coli and P. putida cells was carried out by electroporation (Gene Pulser; Bio-Rad) (15). DNA fragments were purified by standard procedures using Gene Clean (BIO 101, Inc.). To construct pIZBd1, the 816-bp DNA fragment coding for the putative depolymerase of B. bacteriovorus HD100 was PCR amplified by using the oligonucleotides V01 and V02 (Table 1), and 1 to 2 mg of an agar slide containing several preying plaques of B. bacteriovorus HD100 growing on P. putida was used as the template (see Fig. S1A in the supplemental material). For the rest of the PCR amplifications, 0.05 μg of DNA template was used. The amplification mixture also contained 2 U of AmpliTaq DNA polymerase (Perkin-Elmer Applied Biosystems, Norwalk, CT), 0.5 μg of each deoxynucleotide triphosphate, and 2.5 mM MgCl2 in the buffer recommended by the manufacturer. Conditions for amplification were chosen according to the GC content of the corresponding oligonucleotides. The PCR product was digested with endonucleases XbaI and HindIII (Table 1) and cloned into the broad-host-range vector pIZ1016 (Table 1). Similar procedures were performed to generate the expression vector pETBd1, a pET29-derived vector expressing the C-terminal six-His-tagged PhaZBd (Table 1). In this case, oligonucleotides V06 and V07 were used as well as NdeI and XhoI endonucleases (Table 1). All these constructions were confirmed by sequencing using an ABI Prism 3730 DNA sequencer.
Expression of the phaZBd gene product in E. coli and P. putida strains.
A preculture of E. coli DH10B(pIZBd1) cells was incubated overnight in LB medium plus gentamicin. The culture was diluted with LB medium until an optical density at 600 nm (OD600) of 0.2 was reached. When OD600 reached 0.5, the culture was induced with 0.2 mM isopropyl 1-thio-β-d-galactopyranoside (IPTG), and cells were further incubated for 16 h. One hundred milliliters of culture was centrifuged and resuspended in 10 ml of 50 mM phosphate buffer (pH 8). Cells were broken by a 4-fold passage through a French press (1,000 lb/in2) (Aminco) and centrifuged at 27,000 × g. The resulting supernatant (soluble crude extract fraction) and pellet (insoluble fraction) were stored for further analysis. To analyze the culture supernatants, the strains were grown in M63 minimal medium with 0.2% glucose. After 16 h, cultures were centrifuged at 4,500 × g for 10 min. Five hundred milliliters of the culture supernatants was filtered through 0.22-μm filters (Millipore Corp) and then lyophilized, resuspended in 50 ml of 50 mM phosphate buffer (pH 8), and dialyzed for 15 h at 4°C against the same buffer. Similar procedures were performed for P. putida strains, inducing gene expression with 3 mM IPTG.
Electrophoretic techniques and determination of N-terminal amino acid sequence.
SDS-polyacrylamide gel electrophoresis was performed routinely as described before (31). The protein bands were stained with Coomassie brilliant blue G-250, and quantification was carried out by densitometric scanning and calibrating with broad-range molecular mass markers from Bio-Rad. The protein content of unstained crude extracts was calculated as previously described (5). The protein fractions containing PhaZBd were separated in native 7.5% (wt/vol) polyacrylamide gels (without SDS and 2-mercaptoethanol) and subjected to PHA depolymerase activity assay (see below). To determine the N-terminal sequence, the protein bands were transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad) after the SDS-PAGE (57) using the Transblot SD cell (Bio-Rad, Hercules, CA) at 25 V for 3 h. The membrane filter was stained with 0.1% amido black and destained with 5% acetic acid. The protein band on the filter was excised and subjected to N-terminal amino acid sequence analysis by automatic Edman degradation using a gas phase protein sequencer (Procise Protein Sequencing System) connected to online model 610A (Applied Biosystems, Foster, CA).
Preparation of periplasmic fraction.
E. coli DH10B(pIZBd1) cells were fractionated into periplasmic and spheroplastic protein fractions. To this aim, 25 ml of E. coli DH10B (pIZBd1) cells expressing PhaZBd depolymerase as described above was centrifuged and resuspended in 2.4 ml of lysis solution (100 mM Tris-HCl [pH 8], 0.5 mM EDTA, and 20% sucrose [wt/vol]) plus 0.1 mg/ml lysozyme (Sigma). After 20 min of incubation at 4°C, the solution was heated at 37°C for 10 min and the formation of spheroplasts was confirmed by phase-contrast microscopy. Spheroplasts were stabilized by adding 20 mM MgCl2 and centrifuged at 8,000 × g for 20 min to obtain the periplasmic fraction (supernatant) and the spheroplastic fraction (sediment). In order to compare activities, the spheroplastic fraction was resuspended in the same volume of buffer (2.4 ml).
Similar procedures were performed for cellular fractionation of P. putida (pIZBd1) cells, with minor modifications: 1 mg/ml lysozyme and heating for 2 h at 37°C after the incubation at 4°C.
ICDH assay.
Isocitrate dehydrogenase (ICDH), an enzyme involved in the tricarboxylic acid pathway, is localized in the cytoplasm exclusively. Therefore, the amount of ICDH found in the E. coli DH10B(pIZBd1) cellular fractions (see above) is a measure of the degree of cell lysis. ICDH activity was measured according to a procedure described previously (54) with minor modifications. Briefly, 600 μl of assay mixtures containing 50 mM phosphate buffer (pH 8), 5 mM MgCl2, 2 mM NADP, 1 mM isocitrate (pH 7.0), and 100 μg of the cellular fractions was incubated at 30°C, and the absorbance at 340 nm was recorded. The enzyme activity was calculated from the rate of absorbance decrease at 340 nm and the extinction coefficient of NADH, 6,220 M−1 cm−1.
Purification of His-tagged PhaZBd protein.
A preculture of E. coli BL21(DE3)(pETBd1) cells was incubated in LB medium plus kanamycin with shaking (250 rpm) at 30°C. When OD600 reached 1, the culture was diluted to an optical density of 0.05 and incubated at 30°C. When OD600 reached 0.4, the culture was incubated at lower temperature and agitation (16°C and 150 rpm) until OD600 reached 0.6 to 0.7. At this point, 0.1 mM IPTG was added, and the cultures were further incubated at 16°C and 150 rpm for 16 h. Fifty milliliters of culture was centrifuged at 4,500 × g and resuspended in 3 ml of 50 mM sodium phosphate (pH 8.0). Cells were broken by sonication treatment and centrifuged at 15,000 × g. Enzyme purification was performed with the Ni-nitrilotriacetic acid (Ni-NTA) silica spin kit (Qiagen) according to the protocol described by the manufacturer. Briefly, the soluble crude extract fraction supplemented with 10 mM imidazole was loaded onto the Ni-NTA agarose column provided in the kit, equilibrated with 50 mM sodium phosphate buffer (pH 8.0) and 10 mM imidazole, and centrifuged at 400 × g. After loading, the column was washed with 5 ml of the same buffer, and protein was eluted with 50 mM sodium phosphate (pH 8.0) and 500 mM imidazole. All the purification steps were carried out at 4°C.
Purification of PhaZBd produced by P. putida AΩ(pIZBd1).
P. putida AΩ (pIZBd1) cells were grown, and cell-free supernatant broth was filtered, lyophilized, and dialyzed as described above. Subsequently, sodium chloride was added to the concentrated supernatant at a final concentration of 0.5 M. Then, the solution was centrifuged at 10,000 × g for 10 min and loaded onto an Octyl FF/fast flow Sepharose column (GE Healthcare, Sweden) equilibrated with 50 mM sodium phosphate buffer (pH 8.0) and 0.5 M NaCl using a BioLogic LP chromatographic system (Bio-Rad). The column was extensively washed with 5 ml of the same buffer and finally eluted with a linear decreasing gradient of 0.5 to 0 M NaCl. Protein was monitored at 280 nm, and PHA depolymerase activity of the purification fractions was detected as described below.
PHA depolymerase assays.
A quick and simple qualitative procedure for estimating the mcl-PHA depolymerase activity was performed by spot test on indicator plates. A homogeneous latex suspension was obtained as previously described (52, 53). PHA agar plates were prepared by adding 1.5% (wt/vol) agar to the PHA latex suspension (6 mg/ml) in 50 mM phosphate buffer (pH 8) and pouring this onto prewarmed (37°C) glass slides or petri plates. After solidification, an enzyme solution was dropped onto 5-mm-diameter holes made in the PHA agar plates, and the plates were incubated at 37°C for 8 h. The diameters of the resulting clearing zones correlated with the depolymerase activity. In the same way, PHB agar plates were prepared with a homogeneous sonicated PHB suspension (1 mg/ml) in 50 mM phosphate buffer (pH 8) and 1.5% (wt/vol) agar. PhaZBd activity toward PHB was compared to that of the depolymerase from Streptomyces exfoliatus purified as previously described (22).
The PHA depolymerase activity of the enzyme was determined by measuring the turbidity decrease of the PHA suspension. The turbidimetric assay method was performed as described elsewhere (24). The reaction mixture of 500 μl contained 0.6 mg/ml PHA latex, 0.002 mg/ml lysozyme (to avoid unspecific binding of the PhaZBd to the polymer substrate), 0.2 M Tris-HCl (pH 8), and 0.5 M NaCl. After preincubation at 37°C for 10 min, the reaction was started by the addition of 240 μl of solution containing 50 to 100 μg of total protein, and the turbidity decrease at OD650 was monitored with a Beckman DU250 spectrophotometer during 60 min. The turbidity decrease was linear during the assay period, and controls without the enzyme were carried out in parallel. All the activity assays were performed in triplicate, and the maximum error was below 5%.
To prepare the substrate for the radioactive assay, P. putida strain KT2442 was cultured as described above for mcl-PHA production, in the presence of 10 μCi of [14C]octanoic acid, as previously described (11). The labeled polymer, here named PHA*, was purified and used to prepare a polymer-water suspension (PHA* latex), which was used as the substrate to assay the depolymerase activity of the PhaZBd enzyme. The assay mixture of 250 μl contained 0.6 mg/ml lysozyme-preincubated PHA* latex (7,000 cpm), 0.2 M Tris-HCl (pH 8.0), 0.5 M NaCl, and 155 μl solution containing 50 to 100 μg of protein. The mixture was incubated at 37°C for 3 h, and the reaction was stopped by adding 10 μl of formaldehyde. The samples were filtrated (PVDF membrane; Millipore), and the radioactivity present in 200-μl aliquots of the eluted fractions was determined in a scintillation counter. This assay was used to biochemically characterize PhaZBd as well as thermal stability. For the latter determination, PhaZBd solutions were incubated from 25 to 70°C for 5 h. To investigate the effects of various chemical reagents on the mcl-PHA hydrolyzing activity of PhaZBd, the chemical reagent was initially preincubated for 20 min at room temperature with the reaction mixture containing PhaZBd. Then, the enzymatic reaction was started by adding 10 μl of the lysozyme-preincubated PHA* latex suspension containing 150 μg of PHA*.
Enzymatic hydrolysis of PHA polymer and identification of the PhaZBd reaction products.
To identify the PhaZBd hydrolysis products, two reaction mixtures were subjected to enzymatic hydrolysis in parallel with 150 μg of mcl-PHA for different reaction times (60 min and 24 h). The mixtures were developed in the presence of 15 μg of semipurified PhaZBd as a standard turbidimetric assay. The degradation products were identified by analyzing the supernatant of the reaction mixture by HPLC-mass spectrometry (HPLC-MS). The HPLC-MS experiments were carried out on a Finnigan Surveyor (Thermo Electron) pump coupled with a Finnigan LXQ TM (Thermo Electron) ion trap mass spectrometer. The separation was performed at 40°C on a 2.1- by 150-mm (3.5-μm particle size) XTerra MS C18 column (Waters) at a flow rate of 100 μl/min and an injection volume of 25 μl. The mobile phase was 0.1% ammonium hydroxide in water (A) and 0.1% ammonium hydroxide in methanol (B). The following elution program was used: at the start, 95% A and 5% B; after 3 min, the percentage of B was linearly increased to 95% in 7 min, kept constant for 20 min, ramped to the original composition in 5 min, and then equilibrated for 10 min. The detection was monitored by MS-electrospray ionization (MS-ESI) spectrometry at a source voltage of 4.5 kV and at a capillary heat of 200°C. All spectra were recorded in full-scan mode (m/z = 50 to 1,500).
RESULTS
Identification and functional characterization of a putative extracellular mcl-PHA depolymerase in the predator bacterium B. bacteriovorus HD100.
A B. bacteriovorus HD100 gene coding for a putative extracellular PHA depolymerase was identified by inspection of the PHA depolymerase engineering database (29). The sequence is classified within the class of extracellular mcl-PHA depolymerases (www.ded.uni-stuttgart.de) (29). The encoded polypeptide of 271 amino acids (molecular mass, 29.8 kDa) revealed significant amino acid sequence similarity (60.8%) to the extracellular mcl-PHA depolymerase from P. fluorescens GK13 (see Fig. S1B in the supplemental material) (53). Analysis of the N terminus of the amino acid sequence of B. bacteriovorus depolymerase using the algorithm of the signal P 3.0 server predicted the presence of a cleavage site for peptidase I located between positions 20 and 21 of the preprotein (see Fig. S1B in the supplemental material), suggesting the secretion of the protein across the cytoplasmic membrane. The proposed signal peptide consists of 20 amino acids with a positively charged N region, a hydrophobic H region, and a C region with the predicted cleavage site, matching the criteria given for Gram-negative bacteria type II secretion system (41, 43). The most confirmatory feature of the PhaZBd primary structure was the occurrence of the lipase consensus sequence (G-I-S-S-G) (see Fig. S1B in the supplemental material) (2). Moreover, the high content of aromatic (11%) and uncharged aliphatic (40%) amino acids in the mature protein predicted a highly hydrophobic polypeptide. All these characteristics suggest that the ORF Bd3709 codes for an extracellular mcl-PHA depolymerase.
To verify this hypothesis, a recombinant E. coli strain producing the PhaZBd depolymerase was constructed. The phaZBd gene was directly amplified from isolated plaques of B. bacteriovorus HD100 growing on P. putida KT2442 as prey (see Fig. S1A in the supplemental material) (see Materials and Methods for details). E. coli DH10B(pIZBd1) grown in LB medium produced a soluble protein of 30 kDa (see Fig. S2A in the supplemental material), which correlates with the predicted molecular mass of the mature protein deduced from its amino acid sequence (27.8 kDa). Subsequently, mcl-PHA depolymerase activity was analyzed in the soluble crude extract by spot test in PHA agar plates (Fig. 1A). In a comparison with the control strain E. coli DH10B(pIZ1016) carrying the empty vector, only the crude extract of E. coli DH10B (pIZBd1) was able to hydrolyze mcl-PHA latex as shown by the formation of a clearing zone around the spot well. Moreover, PhaZBd specifically hydrolyzes mcl-PHA because no activity was detected in the drop test analysis on denatured-PHB agar plates (data not shown).
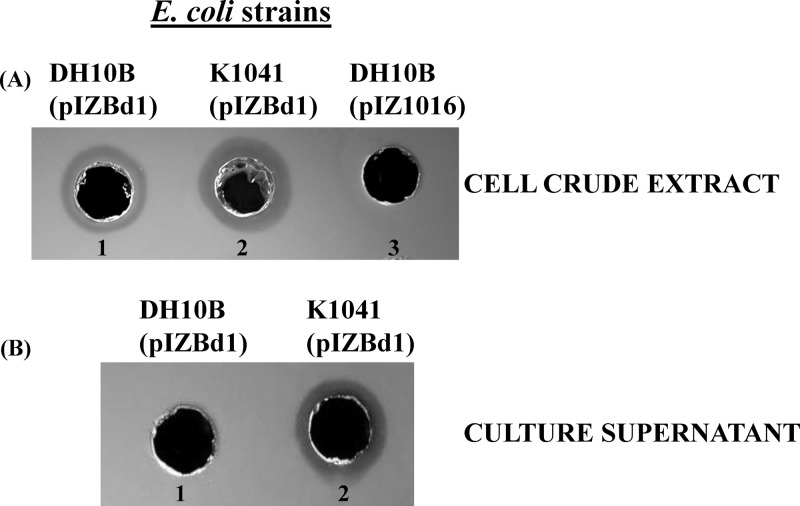
Fig 1.
Production of PhaZBd in E. coli and qualitative mcl-PHA depolymerase activity determination. (A) Enzyme activity measured in mcl-PHA agar plates of 20 μg of protein of the soluble crude extract fractions of E. coli DH10B (pIZBd1) (spot 1), E. coli TolQ mutant K1041(pIZBd1) (spot 2), and negative control E. coli DH10B(pIZ1016) (spot 3). (B) Enzyme activity measured in mcl-PHA agar plates of 10 μg of protein of the culture supernatants of E. coli DH10B(pIZBd1) (spot 1) and E. coli TolQ mutant K1041(pIZBd1) (spot 2).
These results indicated that the cloned phaZBd gene codes in fact for an mcl-PHA depolymerase. PHA depolymerase assays of cell-free culture supernatant from E. coli DH10B(pIZBd1) cells grown in minimal medium did not show extracellular depolymerase activity, even if the culture supernatant was concentrated 30-fold by lyophilization (Fig. 1B, spot 1). Similar results were obtained when the heterologous host strain P. putida KT2442 transformed with pIZBd1 (Table 1) was used to express the phaZBd gene. As described for the E. coli host, depolymerase activity was exclusively detected in the soluble crude extract of P. putida KT2442(pIZBd1) (Fig. 2A).
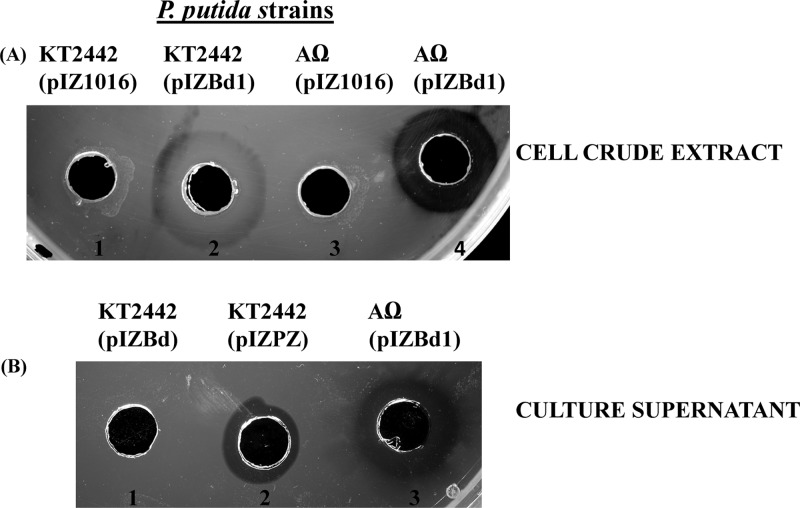
Fig 2.
Production of PhaZBd in P. putida and qualitative mcl-PHA depolymerase activity determination. (A) Enzyme activity measured in mcl-PHA agar plates of 20 μg of protein of the soluble crude extract fraction of negative control P. putida KT2442(pIZ1016) (spot 1), P. putida KT2442(pIZBd1) (spot 2), P. putida TolA mutant AΩ negative control (pIZ1016) (spot 3), and P. putida TolA mutant AΩ(pIZBd1) (spot 4). (B) Enzyme activity measured in mcl-PHA agar plates of 20 μg of protein of the culture supernatants of P. putida KT2442(pIZBd1) (spot 1), positive control P. putida KT2442(pIZPZ) (spot 2), and P. putida TolA mutant AΩ(pIZBd1) (spot 3).
The above results demonstrated that the phaZBd gene certainly encodes an mcl-PHA depolymerase that is located inside the cell when expressed in E. coli DH10B and P. putida KT2442. To ascribe depolymerase activity to the 30-kDa band, the protein was subjected to N-terminal sequencing. The N-terminal amino acid sequence of PhaZBd present in the soluble extracts of E. coli DH10B(pIZBd1) and P. putida KT2442(pIZBd1) strains was determined to be AKKASNC (see Fig. S2 in the supplemental material). This result indicates that the PhaZBd precursor is processed to the mature form, suggesting a periplasmic location of the depolymerase, probably through a type II secretion system. To confirm the mcl-PHA depolymerase activity of the phaZBd gene product, we aimed to purify the PhaZBd depolymerase by metal affinity chromatography. For this purpose, a C-terminal His-tagged PhaZBd was overproduced in E. coli strain BL21(DE3)(pETBd1) (Table 1). Although most of the fused PhaZBd protein sedimented as insoluble inclusion bodies, we were able to detect PHA hydrolysis in the soluble crude extracts, suggesting that a significant fraction of the protein was produced in a soluble form. Thus, the soluble His-PhaZBd enzyme could be semipurified by metal chelate chromatography. A spot test assay showed that His-PhaZBd was unable to spread and disseminate along the PHA agar plate, and efficient hydrolysis was obtained only when the protein extract was directly applied onto the PHA agar plate (data not shown). This result indicates that His-PhaZBd is in an active but aggregated form. In addition, we have determined the PHA depolymerase activity of semipurified His-PhaZBd by using a radioactive assay with 14C-labeled PHA (PHA*) (see Materials and Methods for details). Despite the high sensitivity of the assay, His-PhaZBd activity was almost undetectable after 3 h of incubation at 37°C, and only when shaking the reaction mixture every 10 min was PHA* hydrolysis detected. Under these conditions, His-PhaZBd showed a specific activity of 13.7 ± 0.6 μg PHA* min−1mg−1.
Subcellular location of PhaZBd in E. coli and P. putida host strains.
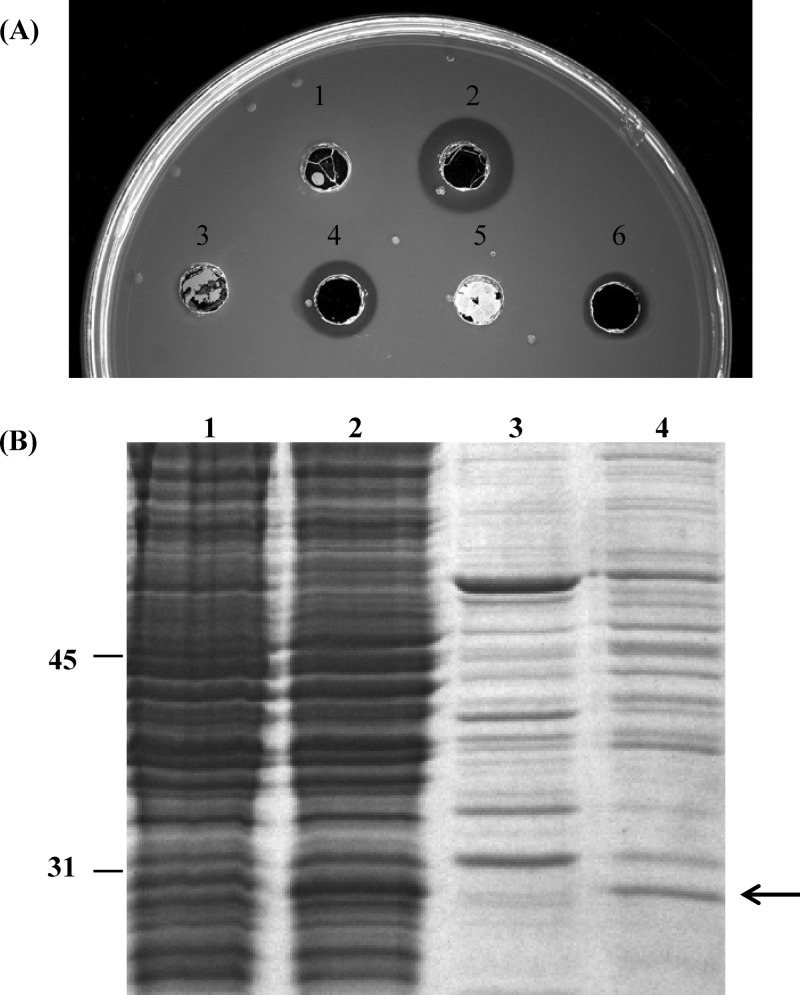
The results presented in the previous section strongly suggested that the processed mature form of PhaZBd was accumulated in the periplasm of E. coli and P. putida strains, since no depolymerase activity was detected in the extracellular medium (Fig. 1 and 2). To confirm the cellular location of PhaZBd in E. coli DH10B(pIZBd1), we isolated the periplasmic fraction of the cultures and analyzed the depolymerase activity using isocitrate dehydrogenase (ICDH) as a control cytoplasmic marker. As expected, most of the ICDH activity was associated with the soluble extract of the spheroplasts (previously broken by ultrasonic treatment) showing an activity of 0.26 μmol min−1 mg−1, whereas in the periplasmic fraction the ICDH activity was 0.04 μmol min−1 mg−1, which accounts for 15% of total ICDH activity. The depolymerase activity found in a spot test assay of the isolated fractions of E. coli DH10B(pIZBd1) is shown in Fig. 3A. PHA hydrolysis was observed when the nonsonicated spheroplast fraction was added (68 μg of total protein content) (Fig. 3A, spot 2). This finding suggests that in the spheroplasts, the PhaZBd depolymerase might be located on the external face of the membrane. Besides, depolymerase activity was also found in the supernatant of the ultracentrifuged periplasmic fraction (6 μg of total protein content) (Fig. 3A, spot 4) and of the resulting sediment (0.15 μg of total protein content) (Fig. 3A, spot 6). As expected, the isolated fractions of the control sample [E. coli DH10B(pIZ1016)] did not hydrolyze the polymer (Fig. 3A, spots 1, 3, and 5). These results are in agreement with those obtained by SDS-PAGE analyses, showing the presence of PhaZBd in both spheroplastic and ultracentrifuged periplasmic fractions (Fig. 3B). The N-terminal amino acid sequence of the PhaZBd present in the periplasm showed a processed protein. Similar results were obtained for P. putida KT2442(pIZBd1) (data not shown). These cellular location studies confirmed the PhaZBd depolymerase accumulation inside the cell, mainly in the periplasmic space in an active, soluble, and processed form.
Fig 3.
Cellular location of PhaZBd in E. coli. (A) Enzyme activity measured in mcl-PHA agar plates of cellular fractions: DH10B (pIZ1016) spheroplasts (spot 1); DH10B (pIZBd1) spheroplasts (spot 2); DH10B (pIZ1016) supernatant of the ultracentrifuged periplasmic fraction (spot 3); DH10B (pIZBd1) supernatant of the ultracentrifuged periplasmic fraction (spot 4); DH10B (pIZ1016) pellet of the ultracentrifuged periplasmic fraction (spot 5); and DH10B (pIZBd1) pellet of the ultracentrifuged periplasmic fraction (spot 6). (B) SDS-PAGE analysis of the cellular fractions: lane 1, DH10B (pIZ1016) spheroplasts; lane 2, DH10B (pIZBd1) spheroplasts; lane 3, DH10B (pIZ1016) supernatant of the ultracentrifuged periplasmic fraction; lane 4, DH10B (pIZBd1) supernatant of the ultracentrifuged periplasmic fraction. The arrow shows the position of PhaZBd.
Extracellular production of PhaZBd in E. coli and P. putida host strains.
To produce the depolymerase extracellularly, we tested a different strategy by using hypersecretor Tol-pal mutants of E. coli and P. putida as hosts. These mutants exhibit severe alterations in outer membrane integrity that allow the release of periplasmic proteins to the culture medium (33, 34). To this aim, the pIZBd1 plasmid was introduced into a variety of Tol-pal mutants of E. coli and P. putida. Remarkably, PHA depolymerase activity was detected in the cell-free culture supernatants of mineral medium fermentations of these mutants. Among them, an E. coli TolQ mutant carrying the pIZBd1 plasmid [E. coli K1041 (pIZBd1)] (Table 1) was selected as the best candidate for depolymerase production (Fig. 1B, spot 2). The extracellular depolymerase activity was consistent with the presence of a 30-kDa protein band visualized by SDS-PAGE of the cell-free culture supernatant (see Fig. S2B in the supplemental material). The other selected strain was a P. putida TolA mutant carrying the pIZBd1 plasmid [P. putida AΩ(pIZBd1)] (Table 1). Its activity was compared to that of the extracellular mcl-PHA depolymerase from P. fluorescens GK13 (PhaZGK13) using the clearing halo system. To this aim, the cell-free culture supernatant of the recombinant P. putida KT2442(pIZPZ) harboring the phaZGK13 depolymerase gene (Table 1) was assayed (Fig. 2B, spot 2). These results demonstrated that Tol-pal mutants of E. coli and P. putida favor the release of the PhaZBd depolymerase into the extracellular medium, suggesting that it is certainly secreted and accumulated in the periplasm. However, it is worth mentioning that the PhaZGK13 depolymerase was secreted into the extracellular medium by the wild-type P. putida strain, evincing differences in the mechanisms applied by this strain to secrete both depolymerases.
As an alternative approach to avoid the problems caused by the aggregation of the His-tagged depolymerase, we produced and purified the native PhaZBd from the culture supernatant of P. putida AΩ(pIZBd1). This strain was selected as the best producer because it renders a higher activity in the spot test assay of the culture supernatant than that of the recombinant E. coli K1041 (pIZBd1) (Fig. 2B and 1B, respectively). The proteins from the cell-free supernatant of P. putida AΩ(pIZBd1) were separated by hydrophobic interaction chromatography on Octyl-Sepharose. The PhaZBd protein band could not be detected even after concentrating the culture supernatant, but it was bound to the column, becoming visible after its elution (see Fig. S3 in the supplemental material). Subsequently, N-terminal amino acid sequencing of this band confirmed its identity as a PhaZBd depolymerase and also that the signal peptide was processed to render the mature form. This partially purified extract of PhaZBd (depolymerase represents about 10% of total protein) (see Fig. S3 in the supplemental material) enabled us to determine the biochemical properties of the enzyme (see below).
Biochemical properties of PhaZBd.
The specific activity of the PhaZBd eluted from Octyl-Sepharose was calculated by turbidimetry as 560 ± 62 μg PHA min−1 mg−1, considering an apparent extinction coefficient (ε) for PHA of 2.1 μl μg−1 cm−1. By the radioactive assay (see Materials and Methods), PhaZBd showed a specific activity of 55 ± 2 μg PHA* min−1 mg−1. As expected, the control sample obtained from the recombinant P. putida AΩ (pIZ1016) did not release radioactive soluble products. It is worth mentioning that the activity determined by the radioactive method cannot be directly compared to that of the nonradioactive method, since in the first one the high-molecular-weight radioactive products released by the enzyme are eliminated by filtration for the final counting. Nevertheless, due to the high sensitivity and accuracy of the radioactive method, it was used in the following steps to determine the optimal pH and temperature of the enzyme. PhaZBd was active in a pH range of 5 to 11 with an optimal activity at pH 10, but at pH 12 the enzyme retained only 3% of its activity. PhaZBd was very active between 4 and 45°C, showing its maximal activity at 37°C, but the activity decreased drastically to 29% when the reaction was carried out at 50°C. In addition, thermal inactivation experiments showed that the enzyme was stable from 25 to 37°C for at least 24 h of incubation, whereas it was slightly deactivated after 5 h at 50°C, since it lost 11% of its activity. It was completely deactivated after 5 h at 70°C. The enzyme retained 100% of its activity for at least 2 months at 4°C.
In addition, when the effect of several ions and other compounds was studied (Table 2), we observed that it was important to maintain a high ionic strength (0.5 M NaCl) in the reaction mixture for optimal activity. Particularly, 1 mM CaCl2, 1 mM MgSO4, or 10 mM EDTA did not affect enzyme activity, suggesting that magnesium and calcium ions are not required as cofactors for the enzyme activity. Ionic and nonionic detergents, except sodium cholate, inhibited PhaZBd activity even at low concentrations (Table 2).
Table 2.
Effects of various chemicals on PhaZBd depolymerase activity
| Reagent (unit) | Final concn | Relative activity (%) |
|---|---|---|
| None (NaCl, 0.5 M; Tris-HCl, 0.2 M) | 100 | |
| NaCl (M) | 0.2 | 95 |
| 0 | 81 | |
| MgSO4 (mM) | 1 | 97 |
| CaCl2 (mM) | 1 | 106 |
| EDTA (mM) | 10 | 110 |
| KCl (mM) | 25 | 100 |
| Triton X-100 (%, vol/vol) | 0.01 | 10 |
| 0.001 | 96 | |
| Tween 80 (%, vol/vol) | 0.05 | 8 |
| 0.005 | 21 | |
| 0.0005 | 81 | |
| SDS (%, vol/vol) | 0.05 | 53 |
| 0.01 | 108 | |
| DOCa (%, vol/vol) | 0.05 | 16 |
| 0.005 | 88 | |
| CTABb (%, vol/vol) | 0.05 | 12 |
| 0.005 | 88 | |
| Cyclodextrin (mM) | 1 | 99 |
| 0.6 | 108 | |
| Sodium cholate (mM) | 0.02 | 130 |
| 0.2 | 118 | |
| 2 | 43 | |
| PMSFc (mM) | 1 | 90 |
| 10 | 40 | |
| EDC (mM) | 10 | 105 |
| DTT (mM) | 1 | 103 |
| 10 | 55 |
DOC, deoxycholate.
CTAB, cetyltrimethylammonium bromide.
Dissolved in isopropanol; value has been corrected for inhibition by the solvent.
As stated above, we suggested the presence of a catalytic triad in PhaZBd, in agreement with most PHA depolymerases and lipases (26), and thus, the effect of serine esterase inhibitors, such as phenylmethylsulfonyl fluoride (PMSF) was studied. As expected, 10 mM PMSF inhibited the PhaZBd activity up to 60%, suggesting that the enzyme behaves as a typical serine hydrolase. Besides, the reducing agent dithiothreitol (DTT) also inhibited the PhaZBd enzymatic activity (50% inhibition at 10 mM DTT), pointing to the existence of essential disulfide bonds for the enzyme activity. Furthermore, the enzyme retained its activity after being treated with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC).
Analysis of the products released after PHA hydrolysis catalyzed by PhaZBd.
Enzymatic hydrolysis of P(HO-co-HX) latex catalyzed by PhaZBd depolymerase was assessed by HPLC, and the identities of the resulting peaks were determined by MS (see Fig. S4 in the supplemental material). This analysis revealed the existence of five chromatographic peaks, corresponding to the molecular masses of the deprotonated HO monomer (m/z 159) (peak 1), the deprotonated HX-HO diester (m/z 273) and the dimer adduct of HX-HO diester (m/z 547) (peak 2), the deprotonated HO diester (m/z 301) and the dimer adduct of HO diester (m/z 603) (peak 3), the deprotonated HO-HX-HO triester (m/z, 415) and the dimer adduct of HO-HX-HO triester (m/z 830) (peak 4), and the deprotonated HO triester (m/z 443) and the dimer adduct of HO triester (m/z 836) (peak 5).
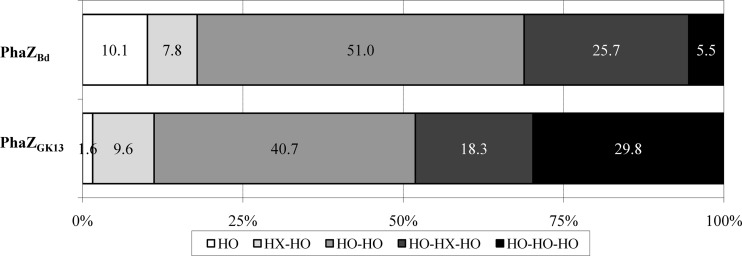
The mcl-PHA hydrolysis products catalyzed by PhaZBd after 1 h of hydrolysis showed that the amounts of dimers released (58.8% of total hydrolyzed products) were higher than those of monomers or trimers (Fig. 4). Remarkably, these results confirmed that PhaZBd is a true depolymerase that behaves as an endoexohydrolase, which hydrolyzes both large and small polyester molecules. Like the depolymerase of P. fluorescens GK13, dimers were identified as the main products of PHA hydrolysis. However, after 1 h of reaction, PhaZGK13 (Fig. 4) showed a trimer accumulation greater than that of PhaZBd, which conversely generates greater quantities of dimers and monomers. Further analyses at 24 h of reaction time confirmed that, independently of the reaction time used, the dimer 3-HO-HO was the main product of the PHA hydrolysis, i.e., 51.7% of total hydrolysis products detected at 1 h (Fig. 4) and 54.1% at 24 h (data not shown).
Fig 4.
Comparison of the P(HO-co-HX) degradation products identified by HPLC-MS after 1 h of enzymatic hydrolysis of PhaZBd versus PhaZGK13. HO, hydroxyoctanoate monomer; HX-HO, dimer of hydroxyhexanoate and hydroxyoctanoate; HO-HO, dimer of hydroxyoctanoate; HO-HX-HO, trimer of hydroxyhexanoate and hydroxyoctanoate; HO-HO-HO, trimer of hydroxyoctanoate.
DISCUSSION
The great hydrolytic arsenal of the predator B. bacteriovorus HD100 plays an important role in the lysis and destruction of prey cell structures throughout different stages of the predator's life cycle, specifically, for prey entry, degradation of biopolymers, and final exit from the bdelloplast (47). The predatosome of Bdellovibrio is composed of at least 150 genes encoding proteases and peptidases, 10 genes encoding glycanases, 15 genes encoding lipases, and 89 genes encoding other hydrolytic enzymes. This density of hydrolytic enzyme-coding genes is the highest in bacterial genomes sequenced so far, after the small genome of Buchnera aphidicola (47). The enzymatic activities of these gene products must be well timed and organized, and the transcriptional control of these genes is likely to be carried out by precise mechanisms (reviewed in reference 56). The large array of predicted hydrolytic enzymes revealed by the B. bacteriovorus HD100 genome sequence provides a valuable reservoir of new enzymes that could be used for biotechnological purposes, with a great potential for environmental industrial applications.
In this work, we report genetic and biochemical evidence that B. bacteriovorus HD100 encodes a novel mcl-PHA depolymerase as part of its predatosome. Besides the possibility of discerning its role on predator physiology, the interest in the study of PHA hydrolytic enzymes lies in its putative industrial use for the production of chiral R-hydroxyalkanoic acids (RHAs) as building blocks for the synthesis of biobased polymers. These biobased monomers are important chiral starting scaffolds in material, fine chemical, pharmaceutical, and medical industries (7, 46).
Several bacterial extracellular PHA depolymerases have been identified and characterized so far (4, 26, 28, 29). PHA depolymerases belong to the α/β-hydrolase fold family and have a catalytic triad (serine-histidine-aspartic acid) as the active site, with the exception of the intracellular scl-PHA depolymerase superfamily (29). The serine is part of a typical pentapeptide lipase box (Gly-X1-Ser-X2-Gly), found in almost all known serine hydrolases. Additionally, a second noncatalytic histidine residue is also conserved near the oxyanion hole (26). The organization of the new PhaZBd depolymerase fully agrees with these general characteristics (see Fig. S1 in the supplemental material).
In contrast to a large variety of well-characterized scl-PHA depolymerases, only a few mcl-PHA depolymerases have been described so far. The mcl-PHA depolymerase of P. fluorescens GK13 is the only one that has been purified and studied at the molecular level (21, 52, 53). The substrate binding domain of PhaZGK13 is located at the N-terminal half of the enzyme, but the residues involved in the binding site have not been determined yet (26). The extracellular mcl-PHA depolymerase from B. bacteriovorus HD100 reported in this study displays a high sequence similarity to that of P. fluorescens GK13 (see Fig. S1 in the supplemental material), except for the signal peptide region. The sequences of both signal peptides suggested the implication of the general secretion pathway (GSP), also called type II secretion system. GSP is a two-step process in which the substrates are first translocated over the cytoplasmic membrane by the Sec apparatus (13, 17, 50). In the periplasm, the exoproteins fold into their (near-)native conformation and are then recognized by proteins of the GSP machinery via the xcp genes (13), which translocates the substrates across the outer membrane. The two-step mechanism involves a stopover of the secreted proteins into the periplasm, where the protein is trapped only when the secretion machine is faulty. According to our findings, the PhaZBd might be secreted by the predator but not released to the extracellular medium by the prey bacteria. This issue opens new questions regarding the natural cellular localization of the enzyme, since the extracellular medium for the predator is either the periplasm of the prey or the culture medium itself, depending on the life cycle stage.
The depolymerases have been classified as intra- or extracellular enzymes, with the exception of the PHB depolymerase from Rhodospirillum rubrum (PhaZ1Rru), which is located in the periplasm (25, 59). The physiological function of R. rubrum PHB depolymerase remains to be elucidated, since PHB granules are located intracellularly. PhaZ1Rru is close to extracellular PHB depolymerases, according to its sequence, its putative catalytic amino acid triad, and the presence of a functional signal peptide, although it differs by its inability to hydrolyze denatured PHB, very likely due to the absence of a substrate-binding domain (25, 59). As expected, the mcl-PHA depolymerase from B. bacteriovorus HD100 shares no amino acid homology with the PHB depolymerase of R. rubrum. However, as occurs in R. rubrum, despite the existence of a signal peptide that is processed during transport across the cytoplasmic membrane, we failed to detect extracellular PhaZBd depolymerase activity after its expression in heterologous hosts (E. coli and P. putida). Moreover, we demonstrate that the processed depolymerase is accumulated in the periplasm. Since B. bacteriovorus HD100 is an obligate predator that develops in the periplasm of other bacteria, a periplasm-located depolymerase makes sense physiologically, as it supports the host PHA degradation during the predator developmental cycle.
The PhaZBd shows high similarity to other extracellular mcl-PHA depolymerases: it is 48% identical to that of Pseudomonas alcaligenes (AAO73963.1), 45% identical to that of Rhodococcus equi (ABN70847.1), 46% identical to that of P. alcaligenes (AAQ72538.1), and 42% identical to that of Pseudomonas luteola (AAV51817.1). PhaZBd is the most distant member in the phylogenic tree of the extracellular mcl-PHA depolymerase family (http://www.ded.uni-stuttgart.de/CONTENT/trees/tree8.html), suggesting the existence of significant phylogenic differences between PhaZBd and the mcl-PHA depolymerases reported so far. From an evolutionary perspective, the high similarity of PhaZBd and PhaZGK13 suggests acquisition of the encoding gene from the prey bacteria by lateral gene transfer (LGT). However, comparative analysis of the GC content of the PhaZBd depolymerase-coding region to the whole genome of B. bacteriovorus HD100, showing 52.2% versus an average of 50.64%, suggests an ancient acquisition of the gene, in agreement with previously published LGT studies of the predator bacterium reporting that no cases of recent prey-derived LGT were detected (23, 47).
The genome of B. bacteriovorus HD100 codes for a second putative depolymerase (ORF Bd2637), classified as a member of superfamily 2 of extracellular scl-PHA depolymerases (type 2) homologous family 6 in the PHA depolymerase database (29). Whether this enzyme degrades prey PHA or plays other roles in Bdellovibrio has been previously discussed by members of the Jurkevitch lab (14). These authors detected the expression of a protein similar to PHB granule-associated proteins, suggesting that the predator might degrade the PHB produced by the prey. This finding is in fact intriguing because B. bacteriovorus was not described as a PHA producer strain. In this sense, we could not find any other gene related specifically with PHA metabolism, including genes for the prototype PHA synthases of classes I to IV (44). Furthermore, the analysis of the flanking regions of the two depolymerase-coding genes, ORF Bd3709 (localized at the coordinates 3591244 to 3590429 of the B. bacteriovorus HD100 genome) and ORF Bd2637 (localized at the coordinates 2555631 to 2556614 of the B. bacteriovorus HD100 genome) did not show the presence of related PHA metabolism-coding genes.
Another striking conclusion reached from this work is the use of Tol-pal mutants for expressing heterologous enzymes, not only in E. coli hosts but also in the generally recognized as safe (GRAS)-certified strain P. putida KT2440 (60). In fact, we were able to produce the PhaZBd depolymerase as an extracellular active form in E. coli and P. putida hypersecretor Tol-pal mutants. The extracellular production of the enzyme facilitates its biochemical characterization. The enzyme specifically degrades mcl-PHA, being inactive toward scl-PHA like PHB. The 3-HO-HO dimer was identified as the main product of enzymatic hydrolysis by HPLC-MS analysis, although the enzyme can also produce monomers. The comparison between PhaZBd and PhaZGK13 reveals several similarities with respect to the main hydrolytic products (HO-HO dimers) or the inability to hydrolyze scl-PHAs (21, 52, 53). Regarding the biochemical characteristics, PhaZBd and PhaZGK13 show several similarities like the PMSF inhibitory effect, indicating that both enzymes behave as typical serine hydrolases (21), but also clear differences; for example, the activity of PhaZBd is not affected by the presence of the chelating agent EDTA, but it is affected by DTT, which indicates the existence of essential disulfide bonds for enzyme activity. PhaZBd differs from PhaZGK13 by its sensitivity against nonionic detergents such as Tween 80, which suggests a hydrophobic region in the catalytic site.
PHA hydrolytic extracellular enzymes have always been considered part of the carbon source assimilation machinery (26). We report genetic and biochemical evidence that B. bacteriovorus HD100 encodes a novel mcl-PHA depolymerase as part of its potential predatosome. Although the physiological role of this hydrolytic enzyme in terms of predator fitness is still an open question, very likely it provides fatty acids as carbon sources to the predator bacterium, which has a full complement of metabolic enzymes needed for the beta-oxidation pathway of fatty acids (47).
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Jurkevitch and P. García for helpful discussions. The technical work of A. Valencia is greatly appreciated.
This work was supported by the Ministry of Economía y Competitividad (BIO2007-67304, BIO2010-21049, CSD2007-00005; CISC 201120E050) and by European Union Grants (NMP2-CT-2007-026515).
Footnotes
Published ahead of print 15 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abe H, Doi Y. 1999. Structural effects on enzymatic degradabilities for poly[(R)-3-hydroxybutyric acid] and its copolymers. Int. J. Biol. Macromol. 25: 185–192 [DOI] [PubMed] [Google Scholar]
- 2. Arpigny JL, Jaeger KE. 1999. Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343: 177–183 [PMC free article] [PubMed] [Google Scholar]
- 3. Behrends A, Klingbeil B, Jendrossek D. 1996. Poly(3-hydroxybutyrate) depolymerases bind to their substrate by a C-terminal located substrate binding site. FEMS Microbiol. Lett. 143: 191–194 [DOI] [PubMed] [Google Scholar]
- 4. Braaz R, Handrick R, Jendrossek D. 2003. Identification and characterization of the catalytic triad of the alkaliphilic thermotolerant PHA depolymerase PhaZ7 of Paucimonas lemoignei. FEMS Microbiol. Lett. 224: 107–112 [DOI] [PubMed] [Google Scholar]
- 5. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- 6. Castro-Sowinski S, Burdman S, Matan O, Okon Y. 2010. Natural functions of bacterial polyhydroxyalkanoates, p 39–61 In Chen GQ. (ed), Plastics from bacteria: natural functions and applications (Microbiology monographs, vol 14). Springer-Verlag, Berlin, Germany [Google Scholar]
- 7. Chen GQ. 2011. Biofunctionalization of polymers and their applications. Adv. Biochem. Eng. Biotechnol. 125: 29–45 [DOI] [PubMed] [Google Scholar]
- 8. Chen H, Athar R, Zheng G, Williams HN. 2011. Prey bacteria shape the community structure of their predators. ISME J. 5: 1314–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Eugenio LI. 2009. Ph.D. thesis The Complutense University, Madrid, Spain [Google Scholar]
- 10. de Eugenio LI, et al. 2010. The turnover of medium-chain-length polyhydroxyalkanoates in Pseudomonas putida KT2442 and the fundamental role of PhaZ depolymerase for the metabolic balance. Environ. Microbiol. 12: 207–221 [DOI] [PubMed] [Google Scholar]
- 11. de Eugenio LI, et al. 2007. Biochemical evidence that phaZ gene encodes a specific intracellular medium chain length polyhydroxyalkanoate depolymerase in Pseudomonas putida KT2442. J. Biol. Chem. 282: 4951–4962 [DOI] [PubMed] [Google Scholar]
- 12. de Eugenio LI, García JL, García P, Prieto MA, Sanz JM. 2008. Comparative analysis of the physiological and structural properties of a medium chain length polyhydroxyalkanoate depolymerase from Pseudomonas putida KT2442. Eng. Life Sci. 8: 260–267 [Google Scholar]
- 13. de Groot A, Gerritse G, Tommassen J, Lazdunski A, Filloux A. 1999. Molecular organization of the xcp gene cluster in Pseudomonas putida: absence of an xcpX (gspK) homologue. Gene 226: 35–40 [DOI] [PubMed] [Google Scholar]
- 14. Dori-Bachash M, Dassa B, Pietrokovski S, Jurkevitch E. 2008. Proteome-based comparative analyses of growth stages reveal new cell cycle-dependent functions in the predatory bacterium Bdellovibrio bacteriovorus. Appl. Environ. Microbiol. 74: 7152–7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dower WJ, Miller JF, Ragsdale CW. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16: 6127–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Escapa IF, García JL, Bühler B, Blank LM, Prieto MA. 2012. The polyhydroxyalkanoate metabolism controls carbon and energy spillage in Pseudomonas putida. Environ. Microbiol. 14: 1049–1063 [DOI] [PubMed] [Google Scholar]
- 17. Filloux A. 2011. Protein secretion systems in Pseudomonas aeruginosa: an essay on diversity, evolution, and function. Front. Microbiol. 2: 155 doi:10.3389/fmicb.2011.00155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Foster LJ, Stuart ES, Tehrani A, Lenz RW, Fuller RC. 1996. Intracellular depolymerase and polyhydroxyoctanoate granule integrity in Pseudomonas oleovorans. Int. J. Biol. Macromol. 19: 177–183 [DOI] [PubMed] [Google Scholar]
- 19. Foster LJ, Lenz RW, Fuller RC. 1999. Intracellular depolymerase activity in isolated inclusion bodies containing polyhydroxyalkanoates with long alkyl and functional substituents in the side chain. Int. J. Biol. Macromol. 26: 187–192 [DOI] [PubMed] [Google Scholar]
- 20. Franklin FC, Bagdasarian M, Bagdasarian MM, Timmis KN. 1981. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc. Natl. Acad. Sci. U. S. A. 78: 7458–7462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gangoiti J, Santos M, Llama MJ, Serra JL. 2010. Production of chiral (R)-3-hydroxyoctanoic acid monomers, catalyzed by Pseudomonas fluorescens GK13 poly(3-hydroxyoctanoic acid) depolymerase. Appl. Environ. Microbiol. 76: 3554–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. García-Hidalgo J, Hormigo D, Prieto MA, Arroyo M, de la Mata I. 2012. Extracellular production of Streptomyces exfoliatus poly(3-hydroxybutyrate) depolymerase in Rhodococcus sp. T104: determination of optimal biocatalyst conditions. Appl. Microbiol. Biotechnol. 93: 1975–1988 [DOI] [PubMed] [Google Scholar]
- 23. Gophna U, Charlebois RL, Doolittle WF. 2006. Ancient lateral gene transfer in the evolution of Bdellovibrio bacteriovorus. Trends Microbiol. 14: 64–69 [DOI] [PubMed] [Google Scholar]
- 24. Handrick R, et al. 2001. A new type of thermoalkalophilic hydrolase of Paucimonas lemoignei with high specificity for amorphous polyesters of short chain-length hydroxyalkanoic acids. J. Biol. Chem. 276: 36215–36224 [DOI] [PubMed] [Google Scholar]
- 25. Handrick R, Reinhardt S, Kimmig P, Jendrossek D. 2004. The “intracellular” poly(3-hydroxybutyrate) (PHB) depolymerase of Rhodospirillum rubrum is a periplasm-located protein with specificity for native PHB and with structural similarity to extracellular PHB depolymerases. J. Bacteriol. 186: 7243–7253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jendrossek D, Handrick R. 2002. Microbial degradation of polyhydroxyalkanoates. Annu. Rev. Microbiol. 56: 403–432 [DOI] [PubMed] [Google Scholar]
- 27. Kadouri D, Jurkevitch EY, Okon Castro-Sowinski S. 2005. Ecological and agricultural significance of bacterial polyhydroxyalkanoates. Crit. Rev. Microbiol. 31: 55–67 [DOI] [PubMed] [Google Scholar]
- 28. Kim DY, Kim HW, Chung MG, Rhee YH. 2007. Biosynthesis, modification and biodegradation of bacterial medium-chain-length polyhydroxyalkanoates. J. Microbiol. 45: 87–97 [PubMed] [Google Scholar]
- 29. Knoll M, Hamm TM, Wagner F, Martínez V, Pleiss J. 2009. The PHA depolymerase engineering database: a systematic analysis tool for the diverse family of polyhydroxyalkanoate (PHA) depolymerases. BMC Bioinform. 10: 89 doi:10.1186/1471-2105-10-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reference deleted.
- 31. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227: 680–685 [DOI] [PubMed] [Google Scholar]
- 32. Lambert C, Smith MCM, Sockett RE. 2003. A novel assay to monitor predator-prey interactions for Bdellovibrio bacteriovorus 109J reveals a role for methyl-accepting chemotaxis proteins in predation. Environ. Microbiol. 5: 127–132 [DOI] [PubMed] [Google Scholar]
- 33. Llamas MA, Ramos JL, Rodríguez-Herva JJ. 2000. Mutations in each of the tol genes of Pseudomonas putida reveal that they are critical for maintenance of outer membrane stability. J. Bacteriol. 182: 4764–4772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lloubès R, et al. 2001. The Tol-Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res. Microbiol. 152: 523–529 [DOI] [PubMed] [Google Scholar]
- 35. López NI, Haedo AS, Méndez BS. 1999. Evaluation of Xanthomonas campestris survival in a soil microcosm system. Int. Microbiol. 2: 111–114 [PubMed] [Google Scholar]
- 36. Luengo J, García B, Sandoval A, Naharroy G, Olivera ER. 2003. Bioplastics from microorganisms. Curr. Opin. Microbiol. 6: 251–260 [DOI] [PubMed] [Google Scholar]
- 37. Madison LL, Huisman GW. 1999. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 63: 21–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 39. Moreno-Ruiz E, Hernáez MJ, Martínez-Pére O, Santero E. 2003. Identification and functional characterization of Sphingomonas macrogolitabida strain TFA genes involved in the first two steps of the tetralin catabolic pathway. J. Bacteriol. 185: 2026–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Numata K, et al. 2006. Adsorption of biopolyester depolymerase on silicon wafer and poly[(R)-3-hydroxybutyric acid] single crystal revealed by real-time AFM. Macromol. Biosci. 6: 41–50 [DOI] [PubMed] [Google Scholar]
- 41. Paetzel M, Dalbey RE, Strynadka NC. 2000. The structure and mechanism of bacterial type I signal peptidases. A novel antibiotic target. Pharmacol. Ther. 87: 27–49 [DOI] [PubMed] [Google Scholar]
- 42. Prieto MA, de Eugenio LI, Galán B, Luengo JM, Witholt B. 2007. Synthesis and degradation of polyhydroxyalkanoates, p 397–428 In Ramos JL, Filloux A. (ed), Pseudomonas: a model system in biology. Pseudomonas, vol V Springer, Berlin, Germany [Google Scholar]
- 43. Pugsley AP. 1993. The complete general secretory pathway in Gram-negative bacteria. Microbiol. Rev. 57: 50–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rehm BHA. 2003. Polyester synthases: natural catalysts for plastics. Biochem. J. 376: 15–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ren Q, et al. 2009. Simultaneous accumulation and degradation of polyhydroxyalkanoates: futile cycle or clever regulation? Biomacromolecules 10: 916–922 [DOI] [PubMed] [Google Scholar]
- 46. Ren Q, Ruth K, Thöny-Meyer L, Zinn M. 2010. Enatiomerically pure hydroxycarboxylic acids: current approaches and future perspectives. Appl. Microbiol. Biotechnol. 87: 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rendulic S, et al. 2004. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303: 689–692 [DOI] [PubMed] [Google Scholar]
- 48. Ruby EG. 1992. The genus Bdellovibrio, p 3401–3412 In Balows A, Starr MP, Stolp H, Tr¨uper HG, Schlegel HG. (ed), The prokaryotes. Springer, Berlin, Germany [Google Scholar]
- 49. Saegusa H, Shiraki M, Kanai C, Saito T. 2001. Cloning of an intracellular poly[D(2)-3-hydroxybutyrate] depolymerase gene from Ralstonia eutropha H16 and characterization of the gene product. J. Bacteriol. 183: 94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Saier MH., Jr 2006. Protein secretion and membrane insertion systems in Gram-negative bacteria. J. Membr. Biol. 214: 75–90 [DOI] [PubMed] [Google Scholar]
- 51. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 52. Schirmer A, Jendrossek D. 1994. Molecular characterization of the extracellular poly(3-hydroxyoctanoic acid) [P(3HO)] depolymerase gene of Pseudomonas fluorescens GK13 and of its gene product. J. Bacteriol. 176: 7065–7073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schirmer A, Jendrossek D, Schlegel HG. 1993. Degradation of poly(3-hydroxyoctanoic acid) [P(3HO)] by bacteria: purification and properties of a [P(3HO)] depolymerase from Pseudomonas fluorescens GK13. Appl. Environ. Microbiol. 59: 1220–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Smith H, Bron S, Van Ee J, Venema G. 1987. Construction and use of signal sequence selection vectors in Escherichia coli and Bacillus subtilis. J. Bacteriol. 169: 3321–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sockett RE. 2009. Predatory lifestyle of Bdellovibrio bacteriovorus. Annu. Rev. Microbiol. 63: 523–539 [DOI] [PubMed] [Google Scholar]
- 56. Sockett RE, Lambert C. 2004. Bdellovibrio as therapeutic agents: a predatory renaissance? Nat. Rev. Microbiol. 2: 669–675 [DOI] [PubMed] [Google Scholar]
- 57. Speicher DW. 1994. Methods and strategies for the sequence analysis of proteins on PVDF membranes. Methods 6: 262–273 [Google Scholar]
- 58. Stolp H, Starr MP. 1963. Bdellovibrio bacteriovorus gen. et sp. n., a predatory, ectoparasitic and bacteriolytic microorganism. Antonie Van Leeuwenhoek 29: 217–248 [DOI] [PubMed] [Google Scholar]
- 59. Sznajder A, Jendrossek D. 2011. Biochemical characterization of a new type of intracellular PHB depolymerase from Rhodospirillum rubrum with high hydrolytic activity on native PHB granules. Appl. Microbiol. Biotechnol. 89: 1487–1495 [DOI] [PubMed] [Google Scholar]
- 60. Timmis KN. 2002. Pseudomonas putida, a cosmopolitan opportunist par excellence. Environ. Microbiol. 4: 779–781 [DOI] [PubMed] [Google Scholar]
- 61. Zinn M, et al. 2011. Growth and accumulation dynamics of poly(3-hydroxyalkanoate) (PHA) in Pseudomonas putida GPo1 cultivated in continuous culture under transient feed conditions. Biotechnol. J. 6: 1240–1252 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.