Abstract
We have isolated a dominant, auxin-insensitive mutant of Arabidopsis thaliana, massugu2 (msg2), that displays neither hypocotyl gravitropism nor phototropism, fails to maintain an apical hook as an etiolated seedling, and is defective in lateral root formation. Yet other aspects of growth and development of msg2 plants are almost normal. These characteristics of msg2 are similar to those of another auxin-insensitive mutant, non-phototropic hypocotyl4 (nph4), which is a loss-of-function mutant of AUXIN RESPONSE FACTOR7 (ARF7) (Harper et al., 2000). Map-based cloning of the MSG2 locus reveals that all four mutant alleles result in amino acid substitutions in the conserved domain II of an Auxin/Indole-3-Acetic Acid protein, IAA19. Interestingly, auxin inducibility of MSG2/IAA19 gene expression is reduced by 65% in nph4/arf7. Moreover, MSG2/IAA19 protein binds to the C-terminal domain of NPH4/ARF7 in a Saccharomyces cerevisiae (yeast) two-hybrid assay and to the whole latter protein in vitro by pull-down assay. These results suggest that MSG2/IAA19 and NPH4/ARF7 may constitute a negative feedback loop to regulate differential growth responses of hypocotyls and lateral root formation.
INTRODUCTION
The plant hormone auxin acts in diverse processes during the course of plant development. For example, it functions as positional signal in vascular (Hardtke and Berleth, 1998; Mattsson et al., 1999; Berleth et al., 2000) and floral development (Sessions et al., 1997; Nemhauser et al., 2000). In addition, it has long been postulated that auxins mediate tropic responses to abiotic stimuli such as light and gravity (Went and Thimann, 1937). The molecular basis of auxin's role in the regulation of a wide range of developmental processes had remained largely elusive. However, recent studies have led to the proposal that products of the AUXIN RESPONSE FACTOR (ARF) and Auxin/Indole-3-Acetic Acid (Aux/IAA) gene families function in a negative (auto)feedback loop in which particular combinations of ARF and Aux/IAA proteins control specific auxin-mediated responses via regulation of gene expression (Liscum and Reed, 2002).
ARF proteins were initially identified by their ability to bind to auxin-responsive promoter elements (AuxREs) (Ulmasov et al., 1997a) and were subsequently shown to bind to such elements via an N-terminal DNA binding domain (DBD), thus regulating the expression of genes containing such promoter elements in an auxin-dependent fashion (Ulmasov et al., 1999a, 1999b). Twenty-three ARF genes are discernable in the Arabidopsis thaliana nuclear genome (Guilfoyle and Hagen, 2001; Liscum and Reed, 2002). ARF proteins can act as either transcriptional activators or repressors, a property determined by the sequence and corresponding structure of the middle region (MR) of the proteins (Ulmasov et al., 1999a). Auxin appears not to regulate the binding of ARFs to AuxREs but rather the transcriptional activity of ARFs bound to AuxREs via a modulation of stability of the Aux/IAA proteins that can heterodimerize, through sharedC-terminal domains (CTDs), with the ARF proteins to repress their activity (Hagen and Guilfoyle, 2002; Hellmann and Estelle, 2002; Tiwari et al., 2003).
To date, loss-of-function mutations have been described for three ARF genes: ETTIN (ETT)/ARF3, MONOPTEROS (MP)/ARF5, and NON-PHOTOTROPIC HYPOCOTYL4 (NPH4)/ARF7. The wild-type ETT/ARF3 encodes one of two ARFs lacking a CTD (Sessions et al., 1997; Ulmasov et al., 1999b; Guilfoyle and Hagen, 2001), and disruption of this gene results in aberrant auxin-dependent pattern formation of the gynoecium (Sessions, 1997; Sessions et al., 1997; Nemhauser et al., 2000). MP/ARF5 mutants are defective in vascular development, both patterning and differentiation (Berleth and Jürgens, 1993; Przemeck et al., 1996; Hardtke and Berleth, 1998). We have shown previously that mutations in NPH4/ARF7 (Harper et al., 2000) cause defects in differential growth responses of hypocotyls of etiolated seedlings, namely phototropism, gravitropism, and apical hook maintenance (Liscum and Briggs, 1995, 1996; Watahiki and Yamamoto, 1997; Stowe-Evans et al., 1998). Although ETT/ARF3 acts as a transcriptional repressor in transient expression assays (Tiwari et al., 2003), both MP/ARF5 and NPH4/ARF7 show promoting activity, consistent with their Q-rich MR (Ulmasov et al., 1999a). Significantly, nph4 null mutants exhibit dramatically impaired auxin-induced expression of a number of genes (Stowe-Evans et al., 1998). Thus, at least in the cases of MP/ARF5 (Mattsson et al., 2003) and NPH4/ARF7, loss-of-function mutant phenotypes likely result from impaired auxin-induced expression of particular sets of genes.
Aux/IAA genes were first identified as genes whose transcripts were rapidly (within several minutes) induced by auxin (Walker and Key, 1982; Theologis et al., 1985; Ainley et al., 1988; Yamamoto et al., 1992b). There are as many as 29 Aux/IAA genes present in the A. thaliana nuclear genome (Liscum and Reed, 2002). Most of the encoded Aux/IAA proteins share four conserved domains: domains I, II, III, and IV (Abel et al., 1995). Domains I and II are unique to Aux/IAA proteins, whereas domains III and IV are shared with the CTD of ARF proteins (Hagen and Guilfoyle, 2002; Liscum and Reed, 2002). Although little is known about the function of domain I, domain II appears to act as a regulatory domain that confers instability to the Aux/IAA proteins (Worley et al., 2000; Ouellet et al., 2001; Ramos et al., 2001; Tiwari et al., 2001). Auxin appears to enhance Aux/IAA instability by promoting their interaction with SCFTIR1 (Gray et al., 2001), an E3 ubiquitin ligase that tags proteins for degradation by the 26S proteasome (Hellmann and Estelle, 2002).
Domains III and IV of Aux/IAA proteins serve as protein–protein interaction domains that promote both homodimerization and heterodimerization between members of the Aux/IAA and ARF families (Ulmasov et al., 1997a; Kim et al., 1997). Aux/IAA proteins are generally thought to function as transcriptional repressors (Abel et al., 1994; Ulmasov et al., 1997b; Gray et al., 2001; Tiwari et al., 2001; Tian et al., 2002) and because ARFs appear to act as potent transcriptional regulators (Ulmasov et al., 1999b), formation of Aux/IAA-ARF heterodimers is thought to repress ARF activity (Ulmasov et al., 1997b; Tiwari et al., 2001, 2003). Thus, in cells containing low levels of auxin, Aux/IAA-ARF heterodimers would be predicted to persist, resulting in inhibition of ARF-dependent changes in transcription. By contrast, cells experiencing high levels of endogenous auxin would be expected to maintain little Aux/IAA protein, having ARFs dissociated from Aux/IAA proteins. This allows expression changes in typically auxin-regulated genes. ARF-dependent auxin induction of Aux/IAA gene expression would provide a means to rapidly dampen the auxin signal, as Aux/IAA-ARF complexes could replace ARFs on AuxREs (Liscum and Reed, 2002).
Dominant mutations have been identified in several Aux/IAA genes to date, and the collective physiological and molecular data strongly support the aforementioned hypotheses. In all cases these dominant mutations result in a single amino acid substitution at one of five consecutive conserved residues in domain II (Reed, 2001), resulting in stabilization of the proteins and their subsequent accumulation in the cell (Worley et al., 2000; Ramos et al., 2001; Tiwari et al., 2001). Therefore, according to the above model, auxin-mediated responses should be repressed in the dominant mutants. In fact, growth and gravitropism of root, hypocotyl, and/or inflorescence stem and formation of primary root, lateral roots, and/or root hairs are inhibited in most of the mutants. The bodenlos (bdl)/iaa12 dominant mutants represent a particularly informative example relative to a model of ARF-Aux/IAA regulation of auxin responses. First, the root meristem defects of the bdl/iaa12 mutants are very similar to those observed in the loss-of-function mp/arf5 mutants (Hamann et al., 1999). Second, BDL/IAA12 and MP/ARF5 exhibit overlapping expression patterns (Hardtke and Berleth, 1998; Hamann et al., 2002). Third and lastly, the BDL/IAA12 and MP/ARF5 proteins have been shown to interact in Saccharomyces cerevisiae two-hybrid assay. Together, these results suggest that auxin regulates initiation of root meristem through MP/ARF5-dependent changes in transcription, a process requiring the correct spatial and temporal interaction between BDL/IAA12 and MP/ARF5 (Hamann et al., 2002).
In an attempt to dissect the signaling pathway of auxin-mediated differential growth responses, a screen for A. thaliana mutants that do not exhibit growth curvature response after unilateral application of auxin-containing lanolin paste to hypocotyl was performed (Watahiki and Yamamoto, 1997). Mutants in two loci were identified, with all the mutations in one locus being recessive and all the mutations in the other being dominant. The former mutant locus was named massugu1 (msg1) (Watahiki and Yamamoto, 1997) and turned out to be allelic to nph4 (Liscum and Briggs, 1995) and transport inhibitor resistant5 (tir5) (Ruegger et al., 1997). Hence, all are loss-of-function alleles of ARF7 (Harper et al., 2000). As discussed in this study, the latter mutant locus, designated msg2, confers phenotypes similar to those of nph4 mutants, and its defects are essentially restricted to differential growth responses of the hypocotyl and formation of lateral roots. We show that the msg2 alleles represent dominant mutations in the Aux/IAA19 gene and that the auxin-dependent expression of MSG2/IAA19 is coupled to the activity of NPH4/ARF7. We also show that NPH4/ARF7 and MSG2/IAA19 can physically interact. These findings suggest that tropic responses of the hypocotyl may be determined by the dissociation-association state of NPH4/ARF7, whether it is present alone or as a complex with MSG2/IAA19, and that an intricate negative feedback loop likely exists between these two proteins to provide rapid and precise control over the tropic response, as light and gravity stimuli continually change in their intensity and direction relative to the growth axis during the development of the curvature response.
RESULTS
Isolation of msg2 Mutants That Are Defective in Auxin-Induced Growth-Curvature Response of Hypocotyls
We tested 74,000 M2 seeds (progeny of 44,000 M1 seeds) of mutagenized A. thaliana Columbia ecotype for the auxin-induced growth curvature response using lanolin containing 100 μM indole-3-acetic acid (IAA). This screen resulted in isolation of seven mutants. Genetic characterization indicated that four of the mutants carried a recessive mutation in the NPH4 locus, which were named nph4-104 to nph4-107 (Harper et al., 2000). The other three mutants represent independent dominant alleles at a different locus. We designated these mutants msg2-1 to msg2-3. We identified another dominant mutant from screening of 66,000 M2 seeds (progeny of 27,000 M1 seeds) by assaying the growth resistance of hypocotyls to 1-naphthaleneacetic acid. This mutant was another allele of msg2, which we designated msg2-4. We found no significant differences in phenotype in the four msg2 alleles.
msg2 Is Defective in Four Differential Growth Responses Observed in Hypocotyls
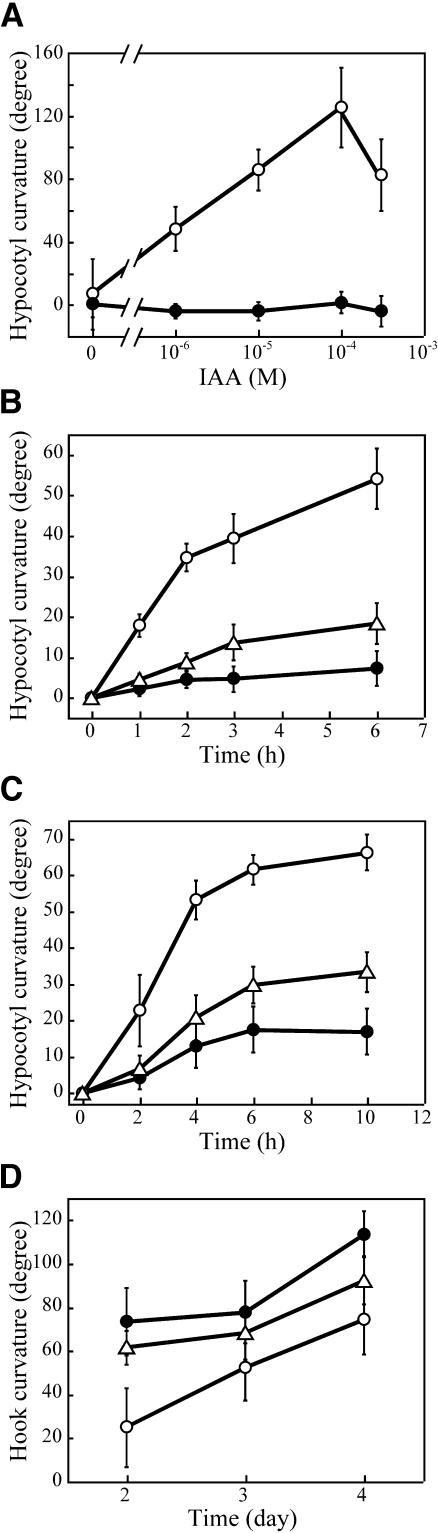
We first examined the growth curvature response of hypocotyls of the wild type and msg2-1 mutants induced by unilateral application of lanolin paste containing various concentrations of IAA. As shown in Figure 1A, growth curvature of the wild-type hypocotyls was increased in proportion to the concentration of IAA from 1 to 100 μM, then reduced at 300 μM IAA as reported previously (Watahiki and Yamamoto, 1997). By contrast, hypocotyls of msg2-1 were unable to bend in response to any concentration of IAA from 1 to 300 μM. The unresponsiveness of msg2-1 to IAA is essentially the same as that observed in nph4-102 (Watahiki and Yamamoto, 1997).
Figure 1.
Differential Growth Responses Observed in Hypocotyls of the Wild Type, msg2-1, and nph4-1.
(A) IAA-induced growth curvature observed 16 h after unilateral application of lanolin paste containing the indicated concentrations of IAA to hypocotyls. Values shown represent the mean ± sd of at least seven seedlings.
(B) Time course of gravitropic reorientation. Seedlings grown on vertically held plates for 3 d in darkness were turned 90° to a horizontal position, and the angle of hypocotyl curvature was measured at the indicated times thereafter; 90° represents complete reorientation upward. Values shown represent the mean ± se of four independent experiments, in which growth curvature of 11 seedlings was measured.
(C) Time course of second-positive phototropism. Seedlings grown on vertically held plates for 3 d in darkness were subjected to unilateral blue light (0.1 μmol·m−2·s−1), and the angle of hypocotyl curvature was measured. Values shown represent the mean ± se of six or seven independent experiments, in which growth curvature of 13 to 30 seedlings was measured.
(D) Hook curvature observed in dark-grown seedlings after induction of germination by incubating at 4°C for 3 d and then at 23°C for 1 d under continuous white light condition. Values shown represent the mean ± se of three independent experiments, in which hook curvature of 18 to 25 seedlings was measured.
Open circle, wild type; closed circle, msg2-1; open triangle, nph4-1.
We next examined the time course of hypocotyl gravitropism of etiolated seedlings grown initially on vertically oriented plates for 3 d and then reoriented 90° (Figure 1B). Rapid upward bending was observed in the wild-type hypocotyls for the initial 2 h after the start of gravistimulation; slower upward bending followed thereafter. Hypocotyls of nph4-1, which is a null allele of NPH4/ARF7 (Harper et al., 2000), responded to gravity much slower than the wild type, displaying a curvature only a third as intense as the wild type at 6 h. msg2-1 hypocotyls responded even more weakly to gravity, exhibiting only a slight upward bending at 6 h.
As shown in Figure 1C, we also examined hypocotyl phototropism in etiolated seedlings in response to long-term irradiation. Development of the phototropic curvature was slower in nph4-1 hypocotyls than the wild type, and the steady state curvature reached at 6 h after the start of unilateral irradiation with blue light was approximately half of that of the wild type. Hypocotyls of msg2-1 displayed a response approximately half as intense as nph4-1.
Lastly, we examined the maintenance of the apical hook in etiolated seedlings over time (Figure 1D). In the wild type a tightly closed hook was observed 2 d postgermination (DPG), after which gradual opening occurred from 2 to 4 DPG. Hooks of nph4-1 seedlings were more opened than those of the wild type at 2 DPG, and they remained more open than the wild type thereafter. msg2-1 hypocotyls exhibited a little more open hook structure than nph4-1 during the entire time period examined.
These results showed that msg2-1 was defective in the same four differential growth responses of the hypocotyl previously described for the nph4 mutants and that msg2-1 defects were as severe, or more severe, than those of nph4.
msg2 Hypocotyls Are Specifically Resistant to Auxin
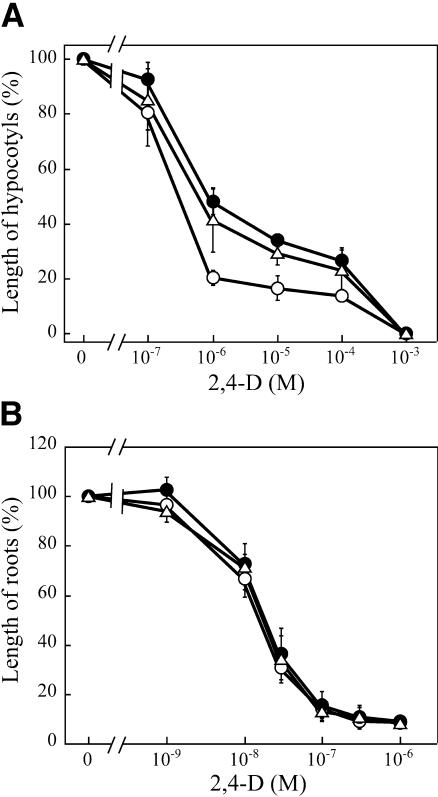
Effects of 2,4-D on hypocotyl and root growth were determined in an aqueous medium (Figure 2). Growth of hypocotyls was inhibited by 2,4-D in a dose-dependent manner in the wild type. msg2-1 and nph4-1 hypocotyls were more resistant to 2,4-D than those of the wild type, with msg2-1 possibly being slightly more resistant than nph4-1 (Figure 2A). Sensitivity of msg2-3 hypocotyls to the ethylene precursor 1-aminocyclopropane-1-carboxylic acid, the synthetic cytokinin 6-benzylaminopurine, and abscisic acid was also examined. msg2-3 was found to be as sensitive as the wild type to these compounds (data not shown), which is also the case for nph4-102 (Watahiki and Yamamoto, 1997). By contrast, neither msg2-1 nor nph4-1 roots showed any resistance to 2,4-D (Figure 2B). These results indicate that hypocotyls of msg2 were specifically resistant to auxin and that roots of msg2 are as sensitive as the wild type to auxin, as is the case for nph4 mutants.
Figure 2.
Effects of 2,4-D on Hypocotyl or Root Growth of the Wild Type, msg2-1, and nph4-1.
For measurement of hypocotyl growth (A), the seedlings were grown hydroponically for 5 d in the presence of 2,4-D at 23°C in darkness after germination was induced in the absence of 2,4-D. 2,4-D was added to the medium before germination when root growth (B) was examined. Organ length is expressed relative to the mean organ length of the same genotype in medium without 2,4-D. Each value represents the mean ± se of three independent experiments, in which ∼20 seedlings were used.
Open circle, wild type; closed circle, msg2-1; open triangle, nph4-1.
msg2 Develops Similar to the Wild Type with the Exception of Slightly Reduced Fecundity and Defects in Lateral Root Formation
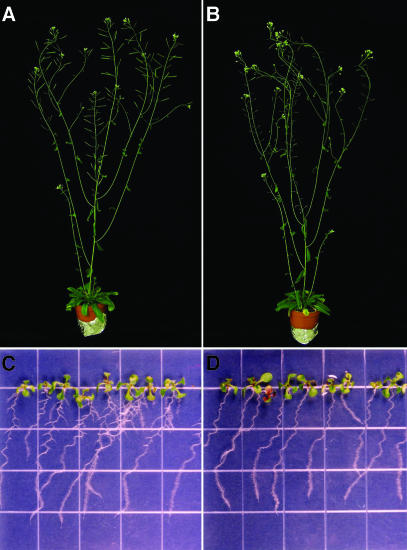
Figure 3 shows the wild type (Figures 3A and 3C) and msg2-1 (Figures 3B and 3D) as adult flowering plants (Figures 3A and 3B) and light-grown seedlings (Figures 3C and 3D). The msg2 plants are similar in size and morphology to the wild type, the only exception being slightly lowered fecundity as a result of a delay in mature fruit production (data not shown). Rosette leaves of msg2 were similar to those of the wild type, in contrast to what is observed in many auxin-insensitive mutants, in which hyponasty (nph4-103 [Watahiki and Yamamoto, 1997], suppressor of long hypocotyl2 [shy2] [Kim et al., 1996; Tian and Reed, 1999], and bdl [Hamann et al., 2002]), epinasty (nph4-1 [Stowe-Evans et al., 1998] and nph4-102 [Watahiki and Yamamoto, 1997]), or wrinkled leaves have been observed (auxin-resistant1 [axr1]; Lincoln et al., 1990). Hypocotyls and roots of msg2-1 seedlings grown under continuous white light and in darkness were as long as those of the wild type (data not shown); they also displayed normal root hair proliferation (Figures 3C and 3D).
Figure 3.
Morphology of msg2-1 Mutants.
(A) and (B) Six-week-old wild-type (A) and msg2-1 (B) plants grown under continuous white light at 23°C. Note the short infertile siliques in (B). Diameter of the pots was 5.5 cm.
(C) and (D) Eight-day-old wild-type (C) and msg2-1 (D) plants grown under continuous white light at 23°C on vertically held agar plates. Grid is 0.5 inches wide.
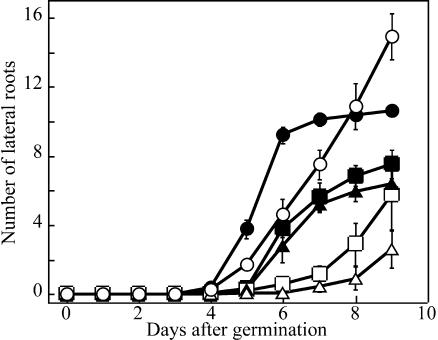
On the other hand, the formation of lateral roots was strongly inhibited in msg2-1 (Figures 3C, 3D, and 4). It was also inhibited in nph4-1 to a lesser extent than in msg2-1 (Figure 4; E.L. Stowe-Evans and E. Liscum, unpublished data). msg2-1 did not initiate normal numbers of lateral roots in response to exogenous IAA. However, msg2-1 was not completely insensitive to IAA because it produced more lateral roots upon auxin treatment. By comparison, nph4-1 showed similar reduced sensitivity to exogenous IAA with respect to lateral root formation (Figure 4).
Figure 4.
Time Course of Lateral Root Formation in the Wild Type, msg2-1, and nph4-1, and Its Induction by Exogenous Auxin.
After induction of germination, seedlings were grown on vertically held agar plates under continuous white light condition. For examination of auxin-induced lateral root formation, 3-d-old seedlings were transferred to the medium containing 40 nM IAA. Data represent the mean ± se of three independent experiments, in which 8 to 13 seedlings were examined.
Open circle, wild type; open triangle, msg2-1; open square, nph4-1; closed symbols, induction by exogenous auxin.
MSG2 Encodes One of the Aux/IAA Proteins, IAA19
The MSG2 locus was mapped to the upper arm of chromosome 3 near nga162 using cleaved amplified polymorphic sequence (CAPS) molecular markers. More precise mapping with polymorphic markers derived from P1 clone ends located MSG2 between two P1 clones, MJK13 and MSJ11. In 752 chromosomes examined, five and two recombinants were found at the proximal (MJK13LB) and distal (MSJ11RB) markers, respectively. No recombinants were found with the closest molecular marker, MJK13RB.
The Arabidopsis Genome Initiative (2000) revealed that a member of the Aux/IAA gene family, IAA19 (Kim et al., 1997), is located on MJK13, ∼3 kb from the MJK13RB marker. Using oligonucleotide primers synthesized according to the genomic DNA sequence of the Columbia ecotype, IAA19 was amplified from the four msg2 alleles and sequenced. The results showed that msg2-1 contains a single nucleotide change predicted to cause an amino acid substitution from Pro-76 to Ser (Figure 5), which had been reported to occur in another dominant Aux/IAA mutation, solitary-root/iaa14 (Fukaki et al., 2002). Substitutions from Pro-76 and Pro-75 to Leu were observed in msg2-3 and msg2-4, respectively, which had been also observed for axr3-1/iaa17 (Rouse et al., 1998) and iaa28-1 (Rogg et al., 2001), respectively. Overexpression of iaa1 with the Pro-75 to Leu substitution has been shown to cause aberrant phenotypes similar to these mutations (Park et al., 2002). For msg2-2, a substitution from Gly-73 to Arg was found. An analogous Gly to Glu substitution has been found in shy2-3/iaa3 (Tian and Reed, 1999). Together, all of the msg2 alleles had mutations in the five amino acid residue–long region of domain II, where all the dominant Aux/IAA gene mutations have been found previously (Figure 5). These results indicate that MSG2 gene encodes IAA19.
Figure 5.
The IAA19 Protein and msg2 Mutations.
Dominant mutations in the conserved domain II of Aux/IAA proteins reported to date are also shown for comparison. Amino acid residues almost perfectly conserved in the 24 domain II–containing Aux/IAA proteins are indicated by asterisks.
Auxin-Induced Gene Expression of MSG2/IAA19 Depends on NPH4/ARF7
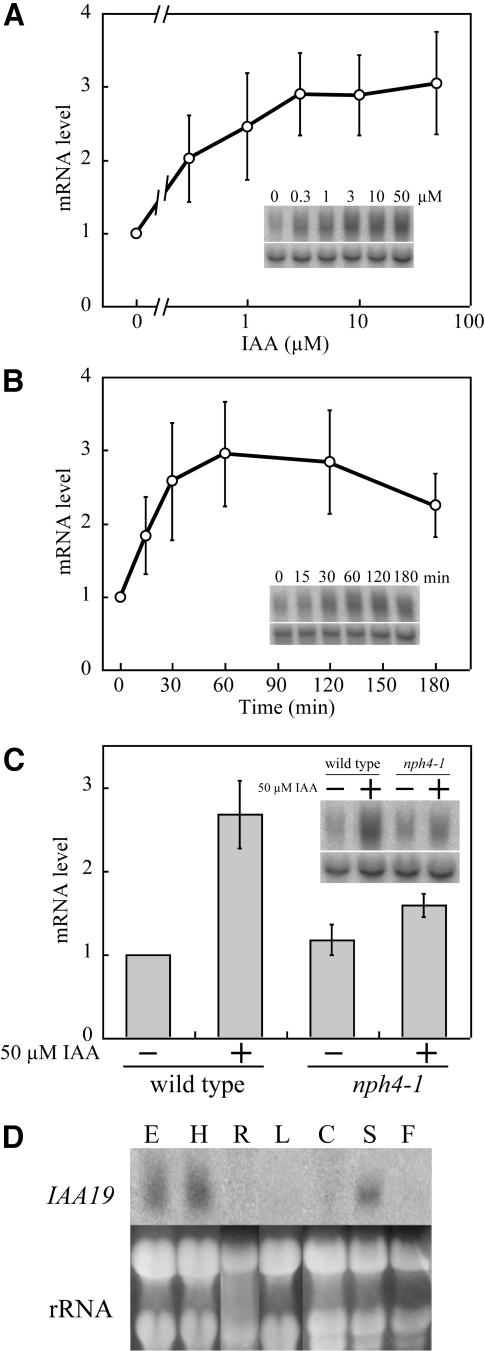
A full-length cDNA of MSG2/IAA19 gene was isolated by the use of 5′ rapid amplification of cDNA ends (RACE). The cDNA obtained started 76 bp upstream of the first ATG codon. A TATAA box–like sequence was found 34 bp upstream of the transcription start site, and three copies of the canonical AuxRE, TGTCTC (Ulmasov et al., 1995), were found 151, 168, and 207 bp upstream of the start site, respectively. Because MSG2/IAA19 has been shown auxin inducible (Tian et al., 2002; Mattsson et al., 2003; Nakamura et al., 2003; Zhao et al., 2003), we examined dose–response curve and time course of auxin induction of MSG2/IAA19 in dark-grown seedlings with RNA gel blot analysis. mRNA levels of MSG2/IAA19 started to increase at 0.3 μM IAA and reached a plateau at 3 μM IAA, where approximately threefold induction was observed (Figure 6A). Auxin induction of MSG2/IAA19 was rapid; accumulation of MSG2/IAA19 mRNA was obvious 15 min after the start of IAA treatment and reached a maximal level after the treatment for 60 min (Figure 6B).
Figure 6.
RNA Gel Blot Analysis of Expression of MSG2/IAA19 Gene.
(A) and (B) Dose–response curve (A) and time course (B) of auxin induction of MSG2/IAA19 mRNA in 3-d-old etiolated seedlings of the wild type. Seedlings were grown hydroponically at 23°C in the dark and treated with IAA for 1 h in (A) and with 50 μM IAA in (B).
(C) Quantitative estimation of MSG2/IAA19 mRNA in 3-d-old etiolated seedlings of the wild type and nph4-1 treated with 50 μM IAA for 1 h. Seedlings were grown hydroponically at 23°C.
(D) Tissue specificity of MSG2/IAA19 expression. Etiolated seedlings (E) were grown hydroponically for 3 d. Etiolated hypocotyls (H) were obtained from 3-d-old seedlings grown on agar plates. Roots (R) were obtained from 9-d-old light grown seedlings grown on agar plates. Rosette (L) and cauline leaves (C), inflorescence stems (S), and flowers (F) were prepared from 5-week-old plants grown on soil under continuous light condition.
Twenty micrograms of total RNA were electrophoresed in each lane. In (A) to (C), values represent the mean ± sd of three independent experiments. Signals of actin8 mRNA (bottom panel, inset of [A] to [C]) or rRNA bands stained with ethidium bromide (D) were shown as a loading control.
Auxin induction of MSG2/IAA19, together with the similarity of the msg2 and nph4 phenotypes, prompted us to ask whether the auxin inducibility of MSG2/IAA19 is dependent on NPH4/ARF7 function. As shown in Figure 6C, RNA gel blot analysis indicates that the steady state mRNA level of MSG2/IAA19 increases 2.7-fold after treatment with 50 μM IAA for 1 h. The MSG2/IAA19 mRNA level was not affected significantly by nph4-1 mutation when seedlings were not treated with IAA, however the auxin-induced increase in MSG2/IAA19 abundance is ∼65% reduced in the nph4-1 background relative to the wild type. These results confirmed that MSG2/IAA19 is an auxin-inducible gene and demonstrate that NPH4/ARF7 is a major factor responsible for auxin inducibility of MSG2/IAA19 gene expression in etiolated seedlings.
Expression of MSG2/IAA19 Is Strongest in Hypocotyls of Etiolated Seedlings
When MSG2/IAA19 transcript expression in different organs was examined by RNA gel blot analysis, the highest steady state level of mRNA was observed in hypocotyls of etiolated seedlings (Figure 6D). No transcript was detected in roots, leaves, or flowers, whereas moderate signal was observed in inflorescence stems. These observations are consistent with phenotypes observed for msg2 mutants, namely that most defects are associated with etiolated hypocotyls.
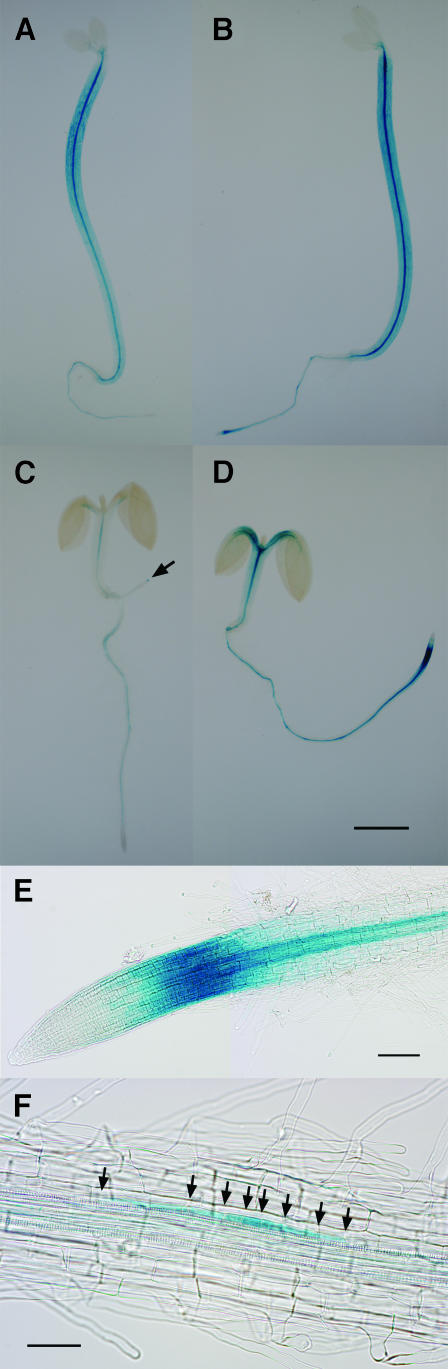
To gain a more precise picture of MSG2/IAA19 expression patterns, an IAA19 promoter:β-glucuronidase (GUS) fusion was introduced into the wild-type A. thaliana, and expression of the gene was examined histochemically. As shown in Figure 7A, transgenic seedlings grown in darkness in the absence of exogenous auxin exhibited intense GUS staining within the central stele of the hypocotyl, although epidermis, cortex, and endodermis of hypocotyls were also stained, especially in the apical region. Faint staining was observed in vascular tissues in the cotyledons and within the central stele of roots. Though root tips were not stained, columella root caps were sometimes stained weakly (data not shown). When treated with 50 μM IAA for 3 h, the GUS staining was increased in the entire region of the hypocotyl and root, with strongest staining in hypocotyl vasculature and the elongation zone of root tips (Figure 7B).
Figure 7.
GUS Staining of Wild-Type Seedlings Transgenic for the IAA19 Promoter:GUS Fusion.
Seedlings were grown hydroponically in the dark ([A] and [B]) or under continuous white light condition ([C] and [D]) for 3 d at 23°C. They were then treated with ([B] and [D]) or without ([A] and [C]) 50 μM IAA for 3 h. A lateral root is indicated by arrow in (C). Staining in primary root tips (E) and lateral root primordia (F) after the IAA treatment was also shown. Arrows in (F) point to cell walls indicating anticlinal cell divisions. Bars = 2 mm in (D) (the same magnification from [A] to [D]) and 0.1 mm in (E) and (F).
Figure 7C demonstrates that GUS staining was generally weaker in light-grown seedlings as compared with etiolated seedlings, and that staining is almost entirely limited to vasculature in the hypocotyls, petioles of cotyledons, and the stele of roots. Faint staining was also observed at the very tip of young lateral roots (Figure 7C, arrowhead). Almost no staining was observed in cortical tissues. Upon treatment with IAA, the staining in the vasculature of aerial organs and the stele of roots increased with strong signals in root tips and petioles of cotyledons (Figure 7D). Cortical cells in the hypocotyl also showed staining. IAA-treated roots showed intense GUS staining in elongation zone and a cell layer around the vasculature (Figure 7E). In the basal part of IAA-treated primary roots, early primordia of lateral roots, which are judged as stage I primordia (Malamy and Benfey, 1997) from anticlinal cell divisions, exhibited the staining (Figure 7F). These observations suggest that MSG2/IAA19 expression occurs in pericycle cells of roots after auxin treatment.
Loss-of-Function Mutants of MSG2/IAA19 Exhibit No Obvious Phenotypic Defects
In an attempt to determine more specifically the function of MSG2/IAA19, we performed screen for loss-of-function mutants in T-DNA (Biotechnology Center, University of Wisconsin, Madison, WI; Krysan et al., 1999) and transposon (Sainsbury Laboratory, John Innes Centre, Norwich, UK; Tissier et al., 1999) insertion lines of A. thaliana. Mutant lines obtained from the former and the latter populations were named msg2-21 and msg2-22, respectively. In msg2-21, a T-DNA was found to be inserted 165 bp upstream of the transcription start site, in the center of one of the three AuxREs in the IAA19 promoter. No MSG2/IAA19 mRNA signals were detected by RNA gel blot analysis of etiolated msg2-21 seedlings in the absence of auxin, and only a slight signal was observed after treatment with 50 μM IAA for 3 h (data not shown). In msg2-22, a transposon is inserted in the second exon of MSG2/IAA19, which changes the amino acid sequence of IAA19 after Gly-111 in domain IV. RNA gel blot analysis of msg2-22 showed that a longer, less abundant mRNA is produced in etiolated msg2-22 seedlings not treated with auxin. As observed with the msg2-21 mutant, the abundance of the altered msg2-22 transcript increased upon auxin treatment (data not shown). These results suggest that msg2-21 and msg2-22 likely represent loss-of-function mutants of MSG2, although neither may be null alleles. Yet neither mutant exhibited any significant changes in the four differential growth responses for which the alterations were observed in the dominant msg2/iaa19 mutants (data not shown). In the case of the loss-of-function mutations of SHY2/IAA3, a subtle phenotypic change was observed, such as increased lateral root formation, increased wavy root curvature, and accelerated root reorientation (Tian and Reed, 1999). msg2-21, however, displayed normal root growth in a similar condition (data not shown). These results may suggest that other Aux/IAA proteins function redundantly with MSG2/IAA19, as has also been hypothesized for other Aux/IAA proteins, including SHY2/IAA3 (Tian and Reed, 1999).
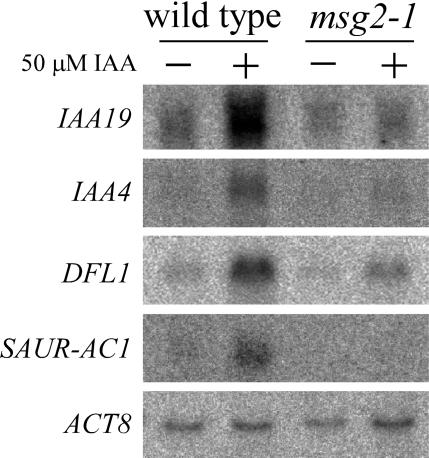
The Dominant msg2-1 Mutation Represses Expression of Auxin Early Genes IAA4, DFL1, and SAUR-AC1
We examined expression of IAA4 (AtAux2-11) (Conner et al., 1990; Abel et al., 1995), DFL1 (Nakazawa et al., 2001), and SAUR-AC1 genes (Gil et al., 1994) in msg2-1 background by RNA gel blot analysis (Figure 8). These genes belong to auxin early gene families Aux/IAA, GH3, and SAUR, respectively. In the wild-type seedlings, mRNA level of each gene was increased by treatment with 50 μM IAA for 1 h as reported previously. However, in msg2-1 seedlings induction by auxin treatments was reduced for each gene. These results indicated that the msg2-1 mutation partially blocked expression of the auxin early genes tested. Reduced SAUR-AC1 and IAA4 expression has also been observed previously in nph4 null mutants by RNA gel blot analysis (Stowe-Evans et al., 1998).
Figure 8.
RNA Gel Blot Analysis of Expression of Auxin Early Genes IAA4, DFL1, SAUR-AC1, and MSG2/IAA19 in msg2-1 Background.
Seedlings were grown hydroponically at 23°C in the dark and treated with 50 μM IAA for 1 h. Twenty micrograms of total RNA were electrophoresed in each lane, and actin8 (ACT8) mRNA bands were shown as a loading control.
We also checked expression of msg2/iaa19 gene in its dominant mutant background and obtained similar results, namely msg2/iaa19 negatively autoregulated its own gene (Figure 8). Essentially the same results have been reported for SHY2/IAA3 (Oono et al., 2002; Tian et al., 2002) and AXR2/IAA7 (Nagpal et al., 2000).
MSG2/IAA19 Interacts with the NPH4/ARF7-CTD in a S. cerevisiae Two-Hybrid Assay, and MSG2/IAA19 and NPH4/ARF7 Interact Physically in Vitro
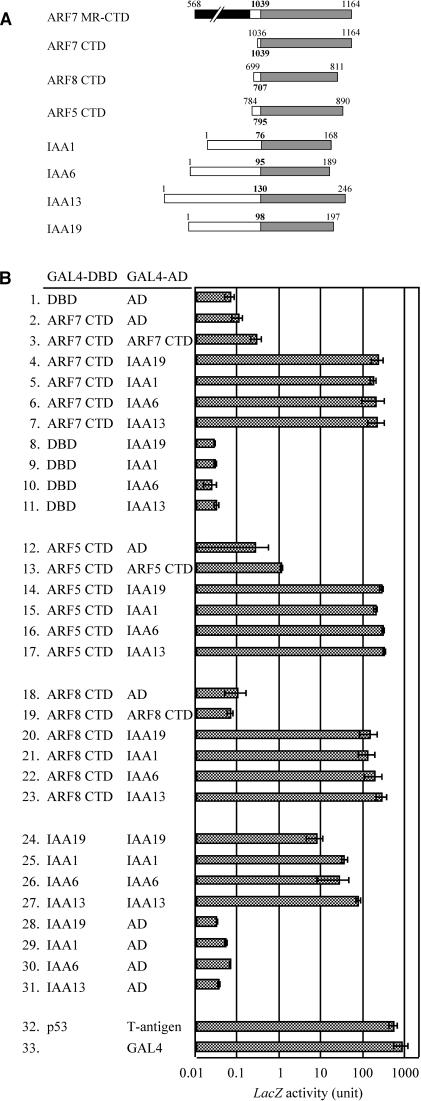
The phenotypes of the dominant msg2/iaa19 mutants have been found to be very similar to those of loss-of-function nph4/arf7 mutants, with defects generally being restricted to differential growth responses of the hypocotyl and the formation of lateral roots. These findings, together with the knowledge that domain II mutations increase the abundance of Aux/IAA proteins (Colón-Carmona et al., 2000), raise the possibility that the msg2 defects may arise from suppression of ARF7 function by formation of a more stable ARF7-iaa19 heterodimer. In an attempt to address this question, we used an S. cerevisiae two-hybrid assay to determine if IAA19 and ARF7 could physically interact. IAA1, IAA6, and IAA13 were also used as potential interacting proteins. IAA6 is most similar to IAA19 of the 29 Aux/IAA proteins with respect to their primary structure of domains III and IV, the ARF-interacting domain, whereas IAA1 and IAA13 are rather remotely related to IAA19 (Liscum and Reed, 2002). ARF5/MP and ARF8 were also used in the assay, as they are closely related to ARF7 with respect to amino acid sequence of their CTDs.
ARF7 is composed of an N-terminal DBD, transcriptional activator MR, and protein-interacting CTD (Ulmasov et al., 1997a; Hagen and Guilfoyle, 2002). We first checked whether a truncated ARF7 protein consisting of just the MR and CTD (ARF7 MR-CTD) could activate transcription of β-galactosidase (LacZ) reporter gene when fused to GAL4 DBD. The GAL4 DBD-ARF7 MR-CTD activated the reporter gene expression approximately half as strongly as did GAL4 protein (data not shown). When only CTD of ARF7 was fused to GAL4 DBD, expression of the reporter gene was not induced significantly (Figure 9B, bars 1 and 2). These results are consistent with the previous observation that ARF7 promotes transcription of reporter gene in transient assay of Daucus carota (carrot) protoplasts (Ulmasov et al., 1999a). The strong activation capacity of ARF7 MR-CTD ruled out the use of the construct in the S. cerevisiae two-hybrid assay, and the fusion protein between ARF7 CTD and GAL4 DBD was used for further experiments to estimate interaction between ARF7 and Aux/IAA proteins.
Figure 9.
Molecular Interaction between ARF CTDs and Aux/IAA Proteins in a S. cerevisiae Two-Hybrid Assay.
SFY526 host cells were transformed with plasmids derived from pGBT9 and pGAD424, and the transformants were then assayed for LacZ activity. Interaction between the fusion of the murine p53 protein and the GAL4 DBD (p53) and the fusion of the SV40 large T-antigen and the GAL4 AD (T-antigen) were measured as a positive control. Effects of the wild-type GAL4 protein were also checked. Data represent the mean ± sd of three to nine independent colonies.
(A) Schematic diagram showing the intact and truncated ARF and Aux/IAA proteins used in S. cerevisiae two-hybrid analysis. Shaded boxes indicate CTD. A hatched box indicates the MR of ARF7. The bold numbers indicate the first amino acid residue of CTDs. The other numbers indicate the positions of deletions with regard to each full-length protein.
(B) Molecular interaction between four Aux/IAA proteins and CTD of three ARF proteins.
Although the GAL4 DBD-ARF7 CTD fusion proteins alone did not stimulate reporter gene expression (Figure 9B, bar 2), cotransformation of S. cerevisiae cells with GAL4 DBD-ARF7 CTD and GAL4 activation domain (AD)-ARF7 CTD constructs increased LacZ activity significantly (Figure 9B, bar 3), indicating homotypic interaction between ARF7 CTD. However, when GAL4 AD-IAA19 proteins were introduced into S. cerevisiae instead of GAL4 AD-ARF7 CTD, much stronger activation of the reporter genes was observed (Figure 9B, bar 4). Because transformation of GAL4 AD-IAA19 alone did not induce expression of the reporter genes (Figure 9B, bar 8), this result demonstrates heterodimer formation between ARF7 CTD and IAA19. Furthermore, heterodimerizations of ARF7 CTD with the other Aux/IAA proteins, IAA1, IAA6 or IAA13, were also detected to a similar extent (Figure 9B, bars 5 to 7). We also examined whether a dominant mutation of MSG2/IAA19 affected its heterotypic interaction with ARF7 CTD. S. cerevisiae cells harboring both msg2-1/iaa19 and ARF7 CTD constructs exhibited LacZ activity as strong as that observed with the wild-type IAA19 and ARF7 CTD constructs (data not shown), indicating that the amino acid substitution occurring in msg2-1 protein did not affect molecular interaction between MSG2/IAA19 and ARF7 CTD. Essentially the same conclusion has been made on interaction between axr3-1/iaa17 and ARF1 or ARF5/MP (Ouellet et al., 2001).
The same two-hybrid experiments were performed for ARF5 and ARF8 CTDs. ARF5 CTD interacted with IAA19 as strongly as did ARF7 CTD (Figure 9B, bars 12 to 14). ARF5 CTD also showed similar capacity to heterodimerize with the other Aux/IAA proteins (Figure 9B, bars 15 to 17). In the case of ARF8 CTD no significant homotypic interaction was detected (Figure 9B, bars 18 and 19), yet strong heterodimerization between ARF8 CTD and IAA19 was observed as indicated by the significant reporter activity (Figure 9B, bar 20). Similar heterodimer formation was observed between ARF8 CTD and the other Aux/IAA proteins (Figure 9B, bars 21 to 23).
Finally, we examined the capacity of the Aux/IAA proteins to homodimerize because strong interactions were observed for heterodimerization between the Aux/IAA proteins and the ARF CTDs. The results presented in Figure 9B (bars 24 to 31) show that the capacity of each Aux/IAA protein to form homodimers was similar to their capacity to form heterodimers with the ARF CTDs.
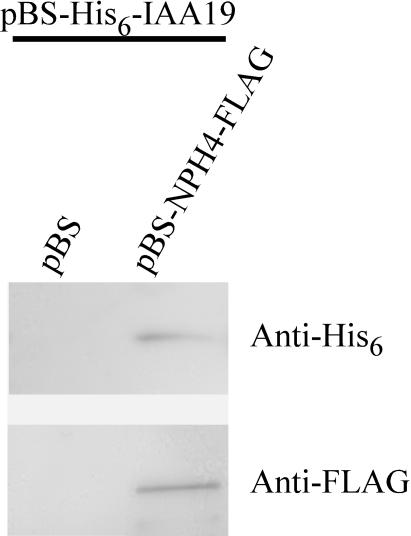
To confirm interaction between MSG2/IAA19 and NPH4/ARF7 CTD observed above, pull-down assays were performed using His-tagged MSG2/IAA19 and FLAG-tagged NPH4/ARF7 expressed in Escherichia coli (Figure 10). Crude extracts of E. coli expressing each protein were mixed, and protein complexes were precipitated with anti-FLAG antibody. When crude extracts of E. coli harboring an empty plasmid were mixed with those expressing recombinant His6-MSG2, no MSG2 was detected by anti-His6 antibody in the immunoprecipitates. However, both MSG2 and NPH4 were observed in them when crude extracts of E. coli cells expressing a NPH4 fusion protein were mixed with those expressing MSG2 fusion protein. These results showed that MSG2 proteins could physically interact with NPH4 proteins in vitro.
Figure 10.
Pull-Down Assays with His-Tagged MSG2/IAA19 and FLAG-Tagged NPH4/ARF7.
Crude extracts of E. coli cells expressing either fusion protein were mixed and immunoprecipitated with immobilized anti-FLAG antibody. Samples eluted with FLAG peptide were subjected to protein gel blot analysis. A plasmid, pBluescript II KS+ containing no inserts (pBS), was used as a negative control.
DISCUSSION
The Loss-of-Function nph4/arf7 and Dominant msg2/iaa19 Mutants Exhibit Similar Defects in Differential Growth Responses of Hypocotyls and the Formation of Lateral Roots
Screens for seedlings that failed to exhibit hypocotyl curvature in response to unilaterally applied auxin have to date yielded mutations in two loci: (1) loss-of-function alleles of the NPH4/ARF7 gene (Liscum and Briggs, 1995; Ruegger et al., 1997; Watahiki and Yamamoto, 1997; Harper et al., 2000) and (2) dominant mutations in the MSG2/IAA19 gene (this study). Both classes of mutants also dramatically reduce three endogenous differential growth responses observed in hypocotyls, namely gravitropism, phototropism, and apical hook structure maintenance in etiolated seedlings (Figure 1; Liscum and Briggs, 1995, 1996; Watahiki and Yamamoto, 1997; Watahiki et al., 1999). Furthermore, these mutants are insensitive to auxin with respect to growth inhibition of the hypocotyl by supraoptimal concentrations of auxin (Figure 2; Watahiki and Yamamoto, 1997; Stowe-Evans et al., 1998). These observations provide strong genetic support for the Cholodny-Went theory that proposes that such differential growth responses arise because of a lateral redistribution of and response to auxin (Went and Thimann, 1937).
In this study, MSG2 has been shown to encode an Aux/IAA protein, IAA19. Most of Aux/IAA proteins of A. thaliana contain four conserved domains, domains I to IV (Abel et al., 1995). One of the remarkable characteristics of the protein family is its fast degradation rate (Abel et al., 1994), which is enhanced by auxin (Gray et al., 2001; Zenser et al., 2001). Domain II has been shown to serve as a dominant transferable degradation signal when translationally fused to a reporter gene (Worley et al., 2000). In fact, all that is essential to confer instability is the pentapeptide domain II core sequence, GWPPV/I/L, which is nearly completely conserved among 24 domain II–containing Aux/IAA proteins in A. thaliana (Ramos et al., 2001). All four dominant msg2 mutant alleles result from amino acid substitution in the pentapeptide segment of domain II, as is the case for the dominant mutations of six other Aux/IAA genes reported so far (Figure 5). The mutations in the domain II core sequence reduce the degradation rate of Aux/IAA proteins, leading to accumulation of the mutated proteins in the cell (Colón-Carmona et al., 2000; Worley et al., 2000; Ramos et al., 2001; Tiwari et al., 2001). The dominant nature of the domain II–aux/iaa mutations, ability of these mutations when introduced into an otherwise wild-type background to confer similar mutant phenotypes, and ability to identify intragenic suppressors have led to the conclusion that the stable domain II mutant aux/iaa represents gain-of-function proteins (Liscum and Reed, 2002). We therefore concluded that the dominant msg2 mutations also represent gain-of-function genotypes.
NPH4 encodes the auxin-regulated transcriptional activator ARF7 (Harper et al., 2000). Most of the ARF proteins containC-terminal regions that are structurally similar to domains III and IV of the Aux/IAA proteins (Ulmasov et al., 1997a; Hagen and Guilfoyle, 2002). These CTDs function as protein–protein interaction domains that allow the ARF and Aux/IAA proteins to form both homodimers and heterodimers (Kim et al., 1997; Ulmasov et al., 1997a). Aux/IAA proteins have been shown to repress the transcriptional activity of ARF proteins by heterodimerizing with the ARF (Ulmasov et al., 1997b; Tiwari et al., 2001, 2003). Considering these findings, it is especially noteworthy that phenotypic defects of the dominant gain-of-function msg2/iaa19 mutants are very similar to those of the loss-of-function nph4/arf7 mutants. As mentioned earlier, the phenotypes of these mutants are, for the most part, restricted to differential growth responses of the hypocotyl and the formation of lateral roots. The msg2/iaa19 and nph4/arf7 mutants are different from each other in only two respects: hyponastic or epinastic leaves are observed in nph4 (Watahiki and Yamamoto, 1997; Stowe-Evans et al., 1998) and are not seen in msg2, and the slightly lower fecundity of msg2 is not observed in nph4. Thus, except for these two properties, the defects in msg2 are similar to, or more severe than, those of nph4, suggesting that the dominant gain-of-function msg2 phenotypes likely result from repression of NPH4/ARF7 activity by msg2/iaa19 proteins because of accumulation of the latter mutant proteins. In cases in which the defects of msg2 are more severe than those of nph4, it is possible that the mutant msg2/iaa19 protein interacts with another ARF protein(s) that is, at least partially, redundant in function with NPH4/ARF7.
Unique Expression Patterns of MSG2/IAA19 May in Part Determine the Specificity of Defects Observed in the Dominant Gain-of-Function msg2/iaa19 Mutants
Compared with the six dominant mutations of Aux/IAA genes previously described in the literature, msg2/iaa19 is distinct in its specific defects in differential growth responses of the hypocotyl and formation of lateral roots. The other dominant gain-of-function aux/iaa mutants are smaller in size to various extents than the wild type and show more pleiotropic defects than msg2/iaa19 mutants. For example, msg2/iaa19 is specifically resistant to auxin, whereas shy2/iaa3 (Tian and Reed, 1999), axr2/iaa7 (Wilson et al., 1990), axr3/iaa17 (Leyser et al., 1996), iaa28 (Rogg et al., 2001), and solitary-root/iaa14 (Fukaki et al., 2002) are additionally resistant to abscisic acid, cytokinin, and/or ethylene. In contrast to the axr2/iaa7, axr3/iaa17 (Nagpal et al., 2000), and shy2/iaa3 (Kim et al., 1998; Tian and Reed, 1999; Nagpal et al., 2000) mutants, which exhibit a partial constitutively photomorphogenetic (cop/det) phenotype as etiolated seedlings, no obvious photomorphogenetic responses are observed in dark-grown msg2/iaa19 seedlings.
The relatively specific defects observed in msg2/iaa19 gain-of-function mutants are shared with those observed in nph4/arf7 loss-of-function mutants. In the context of a model in which Aux/IAA and ARF proteins can form a regulatory feedback loop (Liscum and Reed, 2002; see below), these phenotypes predict that there should be a specific interaction between MSG2/IAA19 and NPH4/ARF7, as determined either by inherent properties of the two proteins or by their spatial and temporal expression patterns relative to other Aux/IAAs and ARFs. Results from our S. cerevisiae two-hybrid studies suggest that little structural specificity exists with respect to Aux/IAA–ARF interactions. In particular, we have found that although IAA19 can certainly interact strongly with ARF7 CTD, it also interacted with the other two ARFs we tested, MP/ARF5 and ARF8 (Figure 9). Moreover, ARF7 CTD interacted strongly with all three of the additional Aux/IAAs we tested: IAA1, SHY1/IAA6 and IAA13 (Figure 9). Similar promiscuous Aux/IAA–ARF interactions have been observed by others (Ouellet et al., 2001).
Two of the Aux/IAA–ARF interactions we observed, namely MSG2/IAA19-MP/ARF5 and SHY1/IAA6-NPH4/ARF7, represent important findings relative to the development of a model of phenotypic specificity for Aux/IAA and ARF protein actions. Because there is almost no overlap between the phenotypes of msg2 and mp mutants, or shy1 and nph4 mutants (Liscum and Reed, 2002), the interactions of the wild-type protein pairs in S. cerevisiae argue that either structural specificity is determined by additional protein factors as part of a bigger transcriptional complex or that these protein pairs do not usually have a chance to interact because they are expressed in different cells. Although the first possibility has yet to be addressed for any Aux/IAA-ARF system, the altered phenotypes observed in each of the previously identified dominant aux/iaa mutants have been found to more or less coincide with expression pattern of the corresponding gene. For example, bdl/iaa12 mutants display embryonic defects, and IAA12 gene is expressed in embryo (Hamann et al., 2002). Defects in lateral root initiation and apical dominance observed in the iaa28 mutants are reflected by strongest expression of IAA28 in roots and inflorescence stems (Rogg et al., 2001). SHY2/IAA3, whose dominant mutants exhibit short, agravitropic hypocotyls (Kim et al., 1996, 1998; Tian and Reed, 1999), is expressed mainly in hypocotyls of dark-grown seedlings (Tian et al., 2002).
Although NPH4/ARF7 appears, at least grossly at the level of RNA gel blot analysis, to be constitutively expressed and is not sensitive to changes in auxin concentration (Ulmasov et al., 1999b), MSG2/IAA19 does exhibit specificity in expression. Similar to what has been observed for other Aux/IAA genes, the patterns of expression of MSG2/IAA19 are predictable based on the phenotypes of the dominant msg2 mutants. Furthermore, the expression patterns of MSG2/IAA19 (Figures 6 and 7) are distinct in many ways from those observed for other Aux/IAA genes. First, almost no expression of MSG2/IAA19 was observed in leaves or floral organs (Figure 6D), in stark contrast to the expression patterns observed for most of the Aux/IAA genes (Abel et al., 1995; Rogg et al., 2001). The lack of MSG2/IAA19 expression in green tissues likely results from its dramatic downregulation in light (Figure 7), which is at least partly dependent upon phytochrome A (Tepperman et al., 2001). Second, although our IAA19 promoter:GUS fusion studies have demonstrated that MSG2/IAA19 is expressed in the root, its expression is more restricted relative to other Aux/IAAs, in which similar studies have been done (Rogg et al., 2001; Fukaki et al., 2002; Tian et al., 2002). Third, although SHY2/IAA3 is expressed in the hypocotyl, the organ in which MSG2/IAA19 exhibits its strongest expression (Figures 6D and 7A), the two genes do exhibit different regulation within the hypocotyl. In particular, SHY2/IAA3 shows both downregulation and upregulation by light being dependent upon the presence or absence of sucrose (Tian et al., 2002), whereas MSG2/IAA19 expression is downregulated by light independent of sugars (Figures 7C and 7D; R.M. Harper and E. Liscum, unpublished data). Thus, it appears that MSG2/IAA19 may have specific patterns of expression distinct from that of other Aux/IAA genes and that these differences likely contribute to its phenotypic specificity that overlaps with NPH4/ARF7 function. Our analysis of loss-of-function msg2/iaa19 mutants, however, suggests that some functional redundancy does exist between MSG2/IAA19 and some other member(s) of the Aux/IAA family.
MSG2/IAA19 and NPH4/ARF7 Likely Form a Negative Feedback Loop to Control Differential Growth Responses in the Hypocotyl
MSG2/IAA19 is an auxin-inducible gene (Figure 6; Tian et al., 2002; Zhao et al., 2003), and its promoter contains three copies of a canonical AuxRE (TGTCTC) (Ulmasov et al., 1995; Nakamura et al., 2003) to which ARF proteins have been shown to selectively bind to modulate auxin-responsive transcription (Ulmasov et al., 1997a; 1999b). Interestingly, we have found that 65% of the auxin inducibility of MSG2/IAA19 expression is dependent on NPH4/ARF7 activity (Figure 6C). This finding suggests the presence of a feedback loop consisting of NPH4/ARF7 and MSG2/IAA19; MSG2/IAA19 expression is upregulated by auxin through NPH4/ARF7, but the resultant MSG2/IAA19 protein represses NPH4/ARF7 activity. A feedback circuit such as this would allow for precise spatial and temporal control of tropic responses, which result from transient localized changes in cell expansion in response to tropic stimuli, which change in intensity and potentially direction, where differential growth curvature develops.
It has recently been shown that the level of Aux/IAA proteins is reduced in response to auxin treatment using Aux/IAA:luciferase fusions expressed in D. carota protoplasts (Tiwari et al., 2001) and A. thaliana plants (Zenser et al., 2001, 2003). Based on these findings, the level of MSG2/IAA19 protein would be expected to decrease in response to increased auxin concentrations. In a tropic context, this would lead to derepression of NPH4/ARF7, resulting in activation of downstream genes whose products promote cell elongation. The Cholodny-Went theory (Went and Thimann, 1937) predicts that a lateral gradient of auxin is produced in response to tropic stimulation, with auxin concentrations being greatest on the side away from the stimulus. Thus, induction of NPH4/ARF7-dependent cell-elongation genes would occur differentially across the hypocotyl, being highest where auxin is highest and MSG2/IAA19 is lowest. Now comes into play the negative feedback loop: As NPH4/ARF7 activity increases, the level of MSG2/IAA19 would also be expected to increase as a result of NPH4/ARF7-dependent transcriptional activation of MSG2/IAA19. This would in turn result in the rerepression of NPH4/ARF7, such that cell-elongation gene targets would no longer be more actively transcribed in the flank of the hypocotyl away from the tropic stimulation, and thus differential growth would cease. The data presented here are entirely consistent with this model, yet additional studies will be required to decipher the details by which this ARF-Aux/IAA pair cooperates so precisely to properly regulate differential growth responses although not apparently affecting growth in general.
METHODS
Plant Materials and Growth Conditions
M2 seeds of A. thaliana ecotype Columbia, mutagenized with ethyl methanesulfonate, were obtained from Lehle Seeds (Round Rock, TX). Seeds were first imbibed in water in the dark at 4°C for 2 d. They were surface-sterilized as described by Watahiki et al. (1995) and sown on nutrient agar plates that contained half-strength MS salts, 1% (w/v) sucrose, half-strength B5 vitamin (Gamborg et al., 1968), 1% (w/v) agar, and 2.3 mM Mes, pH 5.8. Plants were grown at 23°C under continuous illumination at a fluence rate of 8.2 W m−2 obtained from three 40-W white fluorescent tubes (FL40SW; Mitsubishi-Osram, Yokohama, Japan). In some experiments, plants were grown on a 1:1 (v/v) mixture of vermiculite:Metromix 350 (Scotts-Sierra, Marysville, OH). All msg2 mutants were backcrossed twice to Columbia wild type before further analysis.
Differential Growth Responses
Hypocotyl curvature tests were performed as described by Watahiki and Yamamoto (1997). In brief, after induction of germination, seedlings were grown in a row on the nutrient agar plate under dim white light at 0.24 W m−2 for 30 to 36 h to promote hook opening and elongation of hypocotyls. After unilateral application of the lanolin containing IAA to hypocotyl, the seedlings were grown for 12 to 15 h under dim red light. They were then laid on agar plate, and an image of them was taken by an image scanner (GT-7600U; Epson, Suwa, Japan). The angle was determined digitally from the image using appropriate software (NIH Image or Image-Pro Plus; Media Cybernetics, Silver Spring, MD).
To determine gravitropism of hypocotyls, seedlings were grown on vertically oriented plates for 3 d in the dark and then turned 90° to a horizontal position. For second-positive phototropism, 3-d-old etiolated seedlings grown as above were irradiated with unilateral blue light at a fluence rate of 0.1 μmol·m−2·s−1 obtained by blue light–emitting diodes (λmax = 470 ± 30 nm; Stick-B16, Tokyo Rikakikai, Tokyo, Japan). An image of the seedlings was taken with a digital camera (C-4040 Zoom; Olympus, Tokyo, Japan) at different times under dim green light. For determination of growth curvature maintained in hook structure, seedlings were grown in the dark on horizontally held agar plates. They were then laid on agar plate, and an image was captured with a digital camera.
Growth Resistance to 2,4-D
Seedlings were grown hydroponically in the above-mentioned nutrient medium without agar. For determination of 2,4-D resistance of hypocotyls, seeds, after the cold treatment and surface-sterilization, were placed in the medium under continuous white light at 8.2 W m−2 at 23°C for 24 h to induce germination. After the medium was exchanged for a medium supplemented with various concentrations of 2,4-D, seedlings were further grown in darkness for 5 d. 2,4-D was added to the medium before germination when growth of roots was examined. Seedlings were fixed with 5% formaldehyde and 10% acetic acid before measurement of length of the hypocotyls and the roots.
Genetic Characterization
The genetic location of msg2 was established by determining the linkage between the mutant gene and codominant CAPS (Konieczny and Ausubel, 1993), derived CAPS (Michaels and Amasino, 1998), or simple sequence length polymorphisms (Bell and Ecker, 1994). A homozygous msg2-1 plant (ecotype Columbia) was crossed to a wild-type plant (ecotype Landsberg), and the resulting F1 plants were allowed to self-fertilize to generate an F2 population. The F2 population was scored for segregation by the hypocotyl curvature response using lanolin containing 100 μM IAA.
CAPS markers, MJK13RB and MSJ11RB, were generated in the southern ends of P1 clones MJK13 and MSJ11, respectively. For MJK13RB marker, PCR was performed using a forward primer, 5′-ACGTACTTTTGGATTCGTTCAAGCC-3′, and a reverse primer, 5′-AAACAACTGCAGCTGAAGCAAGCCC-3′. Digestion of the 1973 bp-long PCR product with DdeI yielded a 666 bp-long fragment in Columbia, whereas the fragment was digested into two smaller fragments in ecotype Landsberg. For MSJ11RB marker, PCR was performed with a forward primer, 5′-TGGTAAGCTATGCAATTGG-3′, and a reverse primer, 5′-GATCTGTCTTTCCTTTTTTG-3′. Digestion of the 2012 bp-long PCR product with MboII resulted in a ∼990 bp-long fragment in Landsberg. By contrast, the fragment was divided into 529 bp- and 462 bp-long fragments in Columbia. A derived CAPS marker, MJK13LB, was created in the northern end of MJK13. PCR was performed using a forward primer, 5′-CCCTAAACCCTTCCCTCTCTTTATATTAGC-3′, and a reverse primer, 5′-ATTTTTGATCGCAGTGTACTGGGACTGATC-3′. The 74 bp-long PCR product was digested into two smaller fragments with TaqαI only in Landsberg.
Cloning and Sequencing of IAA19 Gene in msg2 Mutants
To sequence msg2 mutant alleles, IAA19 gene was amplified from msg2 mutants in three independent PCRs. The primers were 5′-CATAATTGTATCAAATTGTGAGAGG-3′ and 5′-AAACCATAACATGAAATTTTGTTGC-3′; they were designed based on the complete DNA sequence of the IAA19 gene from Columbia (The Arabidopsis Genome Initiative, 2000). PCRs were performed using Expand High Fidelity PCR system (Boehringer Mannheim, Basel, Switzerland). The amplified genomic DNA fragments were cloned into pT7Blue vector (Novagen, Darmstadt, Germany) using DNA ligation kit, version 2 (Takara, Kusatsu, Japan). Sequencing reactions were performed using Thermo Sequenase primer cycle sequencing kit 7-deaza dGTP (Amersham, Buckinghamshire, UK), and denaturing gel electrophoresis was run by a DNA sequencer (4000L; LI-COR, Lincoln, NE).
RNA Preparation and RNA Gel Blot Analysis
Total RNA was extracted from A. thaliana tissues using the SDS/phenol methods as described by Ausubel et al. (1990) or RNeasy Plant Mini kit (QIAGEN, Hilden, Germany). Total RNA (20 μg) was subjected to electrophoresis in a 1.2% agarose gel that contained 2.2 M formaldehyde (Ausubel et al., 1990), and RNAs were vacuum transferred to nylon membrane (Hybond-N+; Amersham) with 1 M ammonium acetate for 1 h. The membrane was treated with 0.05 M NaOH for 5 min to fix RNA and washed twice for 5 min with 2 × SSPE buffer. 32P-labeled probes for cDNA were synthesized by random-primer method with Megaprime DNA-labeling kit (Amersham). The membrane was hybridized with the probe, and RNA signal detected as described previously (Yamamoto et al., 1992a). RNA gel blot analysis was performed with at least three independent RNA samples.
Rapid Amplification of cDNA Ends
The 5′ end of IAA19 cDNA was determined by 5′ RACE by the use of5′-Full RACE Core set (Takara) according to the manufacturer's directions. First-strand synthesis was primed using a 5′ phosphorylated gene-specific primer, 5′-TTGGCTCGAACCAAGATCCATCTT-3′, and total RNA as a template. After ligation with T4 RNA ligase, nested PCR was performed. Primers for the first PCR were 5′-CGGTTTGTTCTTACCGGAAGAAAA-3′ and 5′-CGACGCCGCTTTCACATTGATCAC-3′; those for the second PCR were 5′-GTAAGGAAGCTTCGACCACGAAAG-3′ and5′-TACCCGACGACGTCATATTCATCT-3′. The PCR products were cloned into pT7Blue and sequenced as described above.
Transgenic Lines and GUS Staining
To construct the IAA19 promoter:GUS reporter plasmid, a 2051-bp fragment of genomic DNA including the upstream region of IAA19 was amplified from Columbia genomic DNA using the IAA19 promoter forward primer, 5′-CCATCTAGATAACTAACCGAAAACATAAGC-3′, and the IAA19 promoter reverse primer, 5′-CCGTCTAGATTCTTGAACTTCTTTTTTTCC-3′ (XbaI sites are underlined), and cloned into pBI-H1 (Kimura et al., 1993). The amplified sequence and junctions of the construct were confirmed by sequencing. The plasmids were introduced into Agrobacterium tumefaciens strain pGV2260 by electroporation, which was then used to inoculate the wild-type Columbia plants by flower dip method (Clough and Bent, 1998). Three dozen T1 plants were screened on medium containing 20 μg/mL hygromycin.
GUS expression was examined by incubating seedlings in 100 mM sodium phosphate, pH 7.0, containing 1 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronide, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 1 mM EDTA, and 0.1% Tween-20 at 25°C for 24 h. Seedlings were dehydrated and rehydrated by passing through a graded ethanol series to remove chlorophyll. After immersion in clearing solution (chloral hydrate:glycerol:water = 100 g:10 g:25 mL), they were observed with a microscope (Zeiss Axioplan [Zeiss, Jena, Germany] or Leica MZ12 [Leica Microsystems, Wetzlar, Germany]) equipped with a digital camera (DXM1200; Nikon, Tokyo, Japan).
Screening for T-DNA or Transposon Insertion Lines
We screened gene-disruption lines from the collection in Sainsbury Laboratory, John Innes Centre, Norwich, United Kingdom (Tissier et al., 1999), and Biotechnology Center, University of Wisconsin (Krysan et al., 1999). In the former collection, a pool of 50 lines had been identified by Sainsbury Laboratory, which contained a line carrying a defective-Spm (dSpm) element in IAA19 gene. The IAA19 insertion line was screened by PCR using two IAA19-specific primers, 5′-CATGAATTCATGGAGAAGGAAGGACTCGGG-3′ and 5′-CCCGAATTCTCTTTCTGAAGATAATTATGC-3′ (EcoRI sites are underlined), and two dSpm-specific primers, dSpm1 and dSpm11 (Tissier et al., 1999). The position of the dSpm insertion was determined by sequencing the PCR products amplified between dSpm11 and the former IAA19-specific primer. PCR screening of the collection of University of Wisconsin was performed by the use of a IAA19-specific primer, 5′-TTAAATTAATGAACCAGCTCCTTGCTTCT-3′, and a T-DNA specific JL-202 primer, 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′. We measured nucleotide sequence of the PCR products amplified using the IAA19 promoter reverse primer described above and JL-202 primer to determine a position of T-DNA insertion.
S. cerevisiae Two-Hybrid Assay
The molecular interactions between Aux/IAA and ARF proteins were analyzed with the MATCHMAKER two-hybrid system (Clontech, Palo Alto, CA). DNA fragments for the proteins were amplified by PCR methods, and the PCR products were cloned into pGBT9 and pGAD424 vectors. The amplified sequences and the junctions of all constructs were confirmed by sequencing. S. cerevisiae SFY526 was used as the host strain for the S. cerevisiae two-hybrid system (Harper et al., 1993). All vectors were cotransformed into S. cerevisiae cells according to the manufacturer's instruction (the lithium acetate method). The transformants were selected on the synthetic dropout minimal medium plates, free of Trp and Leu. The liquid LacZ assay was performed withO-nitrophenyl β-D-galactopyranoside as substrate, according to the procedure provided by Clontech.
Pull-Down Assay
A full-length predicted open reading frame for ARF7 fused in frame with the FLAG tag at 3′ terminus was subcloned into vector pBluescript II KS+ (pBS-ARF7-FLAG; Stratagene, La Jolla, CA). A full-length predicted open reading frame for IAA19 was cloned in frame into the 6× His-tagged expression vector pET15b (Novagen, Madison, WI). Fifty milliliters of E. coli BL21 cells carrying each plasmid were grown at 30°C to an OD of ∼0.6 and then induced with 0.4 mM isopropyl-β-D-thiogalactopyranoside for 3 h. Crude extracts of BL21 cells were prepared by disruption of cells by freezing and thawing, microcentrifugation for 15 min, and passage through a Sephadex G-25 column (Amersham Pharmacia Biotech, Uppsala, Sweden) equilibrated with 50 mM Tris-HCl buffer, pH 7.5, containing 150 mM NaCl, 0.5% Nonidet NP-40, 1 mM phenylmethylsulfonyl fluoride, 5 mM benzamidine, 5 μg/mL leupeptin, 5 μg/mL pepstatin, and 5 μg/mL aprotinin. Crude extracts containing each of the ARF7-FLAG and His6-IAA19 proteins were mixed and incubated at 4°C with gentle agitation for 13 h. Volume and total protein contents of the mixture were 1.0 mL and 2.4 mg, respectively. Then, 10 μL of anti-FLAG M2 agarose beads (Sigma, St.Louis, MO) were added and further incubated for 6 h in the same condition. The beads were collected by brief centrifugation, washed three times in the above buffer, and eluted with 50 μL of the above buffer containing 100 μg/mL FLAG peptide. The eluate was subjected to SDS-PAGE and immunoblotting with anti-FLAG M2 antibody (Sigma) or anti-poly His antibody (Sigma) and alkaline phosphatase conjugated with anti-mouse IgG goat antibody (Jackson Immunoresearch, West Grove, PA).
Acknowledgments
We thank Kenzo Nakamura for pBI-H1, Athanasios Theologis for IAA1, IAA4, IAA6, and IAA13 cDNA clones, Miki Nakazawa for advice on transformation of A. thaliana, Satoshi Tabata for sequence information of A. thaliana genome, and Satoko Ichikawa for technical assistance. Screening for IAA19 disruption lines was conducted in part by Biotechnology Center, University of Wisconsin, and Sainsbury Laboratory, John Innes Centre. This work was carried out in part in Laboratory of Genetic Research, Center for Advanced Science and Technology, Hokkaido University, Japan, and was supported in part by Grants-in-Aid for Scientific Research in Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology to K.T.Y. and National Science Foundation Grant MCB-0077312 to E.L.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Kotaro T. Yamamoto (kty@sci.hokudai.ac.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.018630.
References
- Abel, S., Nguyen, M.D., and Theologis, A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251, 533–549. [DOI] [PubMed] [Google Scholar]
- Abel, S., Oeller, P.W., and Theologis, A. (1994). Early auxin-induced genes encode short-lived nuclear proteins. Proc. Natl. Acad. Sci. USA 91, 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainley, W.M., Walker, J.C., Nagao, R.T., and Key, J.L. (1988). Sequence and characterization of two auxin-regulated genes from soybean. J. Biol. Chem. 263, 10658–10666. [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1990). Current Protocols in Molecular Biology. (New York: Wiley InterScience).
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Berleth, T., and Jürgens, G. (1993). The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development 118, 575–587. [Google Scholar]
- Berleth, T., Mattsson, J., and Hardtke, C.S. (2000). Vascular continuity and auxin signals. Trends Plant Sci. 5, 387–393. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Colón-Carmona, A., Chen, D.L., Yeh, K.-C., and Abel, S. (2000). Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 124, 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner, T.W., Goekjian, V.H., LaFayette, P.R., and Key, J.L. (1990). Structure and expression of two auxin-inducible genes from Arabidopsis. Plant Mol. Biol. 15, 623–632. [DOI] [PubMed] [Google Scholar]
- Fukaki, H., Tameda, S., Masuda, H., and Tasaka, M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 genes of Arabidopsis. Plant J. 29, 153–168. [DOI] [PubMed] [Google Scholar]
- Gamborg, O.L., Miller, R.A., and Ojima, K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50, 151–158. [DOI] [PubMed] [Google Scholar]
- Gil, P., Liu, Y., Orbovic, V., Verkamp, E., Poff, K.L., and Green, P.J. (1994). Characterization of the auxin-inducible SAUR-AC1 gene for use as a molecular genetic tool in Arabidopsis. Plant Physiol. 104, 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Guilfoyle, T.J., and Hagen, G. (2001). Auxin response factors. J. Plant Growth Regul. 20, 281–291. [Google Scholar]
- Hagen, G., and Guilfoyle, T. (2002). Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385. [PubMed] [Google Scholar]
- Hamann, T., Benkova, E., Bäurle, I., Kientz, M., and Jürgens, G. (2002). The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 16, 1610–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann, T., Mayer, U., and Jürgens, G. (1999). The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126, 1387–1395. [DOI] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating axis formation and vascular development. EMBO J. 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J.W., Adami, G.R., Wei, N., Keyomarsi, K., and Elledge, S.J. (1993). The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816. [DOI] [PubMed] [Google Scholar]
- Harper, R.M., Stowe-Evans, E.L., Luesse, D.R., Muto, H., Tatematsu, K., Watahiki, M.K., Yamamoto, K., and Liscum, E. (2000). The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12, 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann, H., and Estelle, M. (2002). Plant development: Regulation by protein degradation. Science 297, 793–797. [DOI] [PubMed] [Google Scholar]
- Kim, J., Harter, K., and Theologis, A. (1997). Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94, 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, B.C., Soh, M.S., Hong, S.H., Furuya, M., and Nam, H.G. (1998). Photomorphogenic development of the Arabidopsis shy2–1D mutation and its interaction with phytochromes in darkness. Plant J. 15, 61–68. [DOI] [PubMed] [Google Scholar]
- Kim, B.C., Soh, M.S., Kang, B.J., Furuya, M., and Nam, H.G. (1996). Two dominant photomorphogenic mutations of Arabidopsis thaliana identified as suppressor mutations of hy2. Plant J. 9, 441–456. [DOI] [PubMed] [Google Scholar]
- Kimura, T., Takeda, S., Kyozuka, J., Asahi, T., Shimamoto, K., and Nakamura, K. (1993). The presequence of a precursor to the δ-subunit of sweet potato mitochondrial F1 ATPase is not sufficient for the transport of β-glucuronidase (GUS) into mitochondria of tobacco, rice and yeast cells. Plant Cell Physiol. 34, 345–355. [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser, H.M.O., Pickett, F.B., Dharmasiri, S., and Estelle, M. (1996). Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10, 403–413. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum, E., and Briggs, W. (1995). Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell 7, 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum, E., and Briggs, W.R. (1996). Mutations of Arabidopsis in potential transduction and response components of the phototropic signaling pathway. Plant Physiol. 112, 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum, E., and Reed, J.W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400. [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44. [DOI] [PubMed] [Google Scholar]
- Mattsson, J., Ckurshumova, W., and Berleth, T. (2003). Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 131, 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson, J., Sung, Z.R., and Berleth, T. (1999). Responses of plant vascular systems to auxin transport inhibition. Development 126, 2979–2991. [DOI] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1998). A robust method for detecting single-nucleotide changes as polymorphic markers by PCR. Plant J. 14, 381–385. [DOI] [PubMed] [Google Scholar]
- Nagpal, P., Walker, L.M., Young, J.C., Sonawala, A., Timpte, C., Estelle, M., and Reed, J.W. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, A., Higuchi, K., Goda, H., Fujiwara, M.T., Sawa, S., Koshiba, T., Shimada, Y., and Yoshida, S. (2003). Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol. 133, 1843–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa, M., Yabe, N., Ichikawa, T., Yamamoto, Y.Y., Yoshizumi, T., Hasunuma, K., and Matsui, M. (2001). DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 25, 213–221. [DOI] [PubMed] [Google Scholar]
- Nemhauser, J.L., Feldman, L.F., and Zambryski, P.C. (2000). Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127, 3877–3888. [DOI] [PubMed] [Google Scholar]
- Oono, Y., Ooura, C., and Uchimiya, H. (2002). Expression pattern of Aux/IAA genes in the iaa3/shy2–1D mutant of Arabidopsis thaliana (L.). Ann. Bot. 89, 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet, F., Overvoorde, P.J., and Theologis, A. (2001). IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell 13, 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.-Y., Kim, H.-J., and Kim, J. (2002). Mutation in domain II of IAA1 confers diverse auxin-related phenotypes and represses auxin-activated expression of Aux/IAA genes in steroid regulator-inducible system. Plant J. 32, 669–683. [DOI] [PubMed] [Google Scholar]
- Przemeck, G.K.H., Mattsson, J., Hardtke, C.S., Sung, R.Z., and Berleth, T. (1996). Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200, 229–237. [DOI] [PubMed] [Google Scholar]
- Ramos, J.A., Zenser, N., Leyser, O., and Callis, J. (2001). Rapid degradation of Auxin/Indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 12, 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W. (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6, 420–425. [DOI] [PubMed] [Google Scholar]
- Rogg, L.E., Lasswell, J., and Bartel, B. (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13, 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, D., Mackay, P., Stirnberg, P., Estelle, M., and Leyser, O. (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279, 1371–1373. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Hobbie, L., Brown, D., Bernasconi, P., Turner, J., Muday, G., and Estelle, M. (1997). Reduced naphthylphthalamic acid binding in the tir3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell 9, 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A. (1997). Arabidopsis (Brassicaceae) flower development and gynoecium patterning in wild-type and ettin mutants. Am. J. Bot. 84, 1179–1191. [PubMed] [Google Scholar]
- Sessions, A., Neuhauser, J.L., McColl, A., Roe, J.L., Feldmann, K.A., and Zambryski, P.C. (1997). ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124, 4481–4491. [DOI] [PubMed] [Google Scholar]
- Stowe-Evans, E.L., Harper, R.M., Motchoulski, A.V., and Liscum, E. (1998). NPH4, a conditional modulator of auxin-dependent differential growth responses in Arabidopsis. Plant Physiol. 118, 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman, J.M., Zhu, T., Chang, H.-S., Wang, X., and Quail, P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98, 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologis, A., Huynh, T.V., and Davis, R.W. (1985). Rapid induction of specific mRNAs by auxin in pea epicotyl tissue. J. Mol. Biol. 183, 53–68. [DOI] [PubMed] [Google Scholar]
- Tian, Q., and Reed, J.W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126, 711–721. [DOI] [PubMed] [Google Scholar]
- Tian, Q., Uhlir, N.J., and Reed, J.W. (2002). Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14, 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier, A.F., Marillonnet, S., Klimyuk, V., Patel, K., Torres, M.A., Murphy, G., and Jones, J.D.G. (1999). Multiple independent defective Suppressor-mutator transposon insertions in Arabidopsis: A tool for functional genomics. Plant Cell 11, 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, S.B., Hagen, G., and Guilfoyle, T.J. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, S.B., Wang, X.-J., Hagen, G., and Guilfoyle, T.J. (2001). AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13, 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1997. a). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. a). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96, 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. b). Dimerization and DNA binding of auxin response factors. Plant J. 19, 309–319. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Liu, Z.-B., Hagen, G., and Guilfoyle, T.J. (1995). Composite structure of auxin response elements. Plant Cell 7, 1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997. b). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J.C., and Key, J.L. (1982). Isolation of cloned cDNAs to auxin-responsive poly(A)+RNAs of elongating soybean hypocotyl. Proc. Natl. Acad. Sci. USA 79, 7185–7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watahiki, M.K., Mori, H., and Yamamoto, K.T. (1995). Inhibitory effects of auxin and related substances on the activity of an Arabidopsis glutathion S-transferase isozyme expressed in Escherichia coli. Physiol. Plant. 94, 566–574. [Google Scholar]
- Watahiki, M.K., Tatematsu, K., Fujihira, K., Yamamoto, M., and Yamamoto, K.T. (1999). The MSG1 and AXR1 genes of Arabidopsis are likely to act independently in growth-curvature responses of hypocotyls. Planta 207, 362–369. [DOI] [PubMed] [Google Scholar]
- Watahiki, M.K., and Yamamoto, K.T. (1997). The massugu1 mutation of Arabidopsis identified with failure of auxin-induced growth curvature of hypocotyl confers auxin insensitivity to hypocotyl and leaf. Plant Physiol. 115, 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Went, F.W., and Thimann, K.V. (1937). Phytohormones. (New York: MacMillan).
- Wilson, A.K., Pickett, F.B., Turner, J.C., and Estelle, M. (1990). A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol. Gen. Genet. 222, 377–383. [DOI] [PubMed] [Google Scholar]
- Worley, C.K., Zenser, N., Ramos, J., Rouse, D., Leyser, O., Theologis, A., and Callis, J. (2000). Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J. 21, 553–562. [DOI] [PubMed] [Google Scholar]
- Yamamoto, K.T., Mori, H., and Imaseki, H. (1992. a). Novel mRNA sequences induced by indole-3-acetic acid in sections of elongating hypocotyls of mung bean (Vigna radiata). Plant Cell Physiol. 33, 13–20. [Google Scholar]
- Yamamoto, K.T., Mori, H., and Imaseki, H. (1992. b). cDNA cloning of indole-3-acetic acid-regulated genes: Aux22 and SAUR from mung bean (Vigna radiata) hypocotyl tissue. Plant Cell Physiol. 33, 93–97.
- Zenser, N., Dreher, K.A., Edwards, S.R., and Callis, J. (2003). Acceleration of Aux/IAA proteolysis is specific for auxin and independent of AXR1. Plant J. 35, 285–294. [DOI] [PubMed] [Google Scholar]
- Zenser, N., Ellsmore, A., Leasure, C., and Callis, J. (2001). Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 98, 11795–11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Dai, X., Blackwell, H.E., Schreiber, S.L., and Chory, J. (2003). SIR1, an upstream component in auxin signaling identified by chemical genetics. Science 301, 1107–1110. [DOI] [PubMed] [Google Scholar]