Abstract
Overexpression of the multidrug efflux pump MDR1 confers resistance to the antifungal drug fluconazole on Candida albicans. It has been reported that two types of MDR1 promoters exist in C. albicans and that homozygosity for the allele with higher activity may promote fluconazole resistance. We found that the two MDR1 promoter alleles in strain SC5314 were equally well activated by inducing chemicals or hyperactive forms of the transcription factors Mrr1 and Cap1, which control MDR1 expression. In addition, no loss of heterozygosity at the MDR1 locus was observed in MDR1-overexpressing clinical C. albicans strains that developed fluconazole resistance during therapy.
TEXT
Infections by the fungal pathogen Candida albicans are commonly treated with fluconazole, an antifungal drug that inhibits ergosterol biosynthesis. C. albicans can develop resistance to fluconazole during therapy by various mechanisms, including mutations in the ERG11 gene, which encodes the drug target enzyme; overexpression of ERG11; or upregulation of multidrug efflux pumps that transport fluconazole out of the cell (16). The constitutive overexpression of efflux pumps in fluconazole-resistant strains is caused by gain-of-function mutations in the transcriptional regulators Mrr1 and Tac1, which control the expression of the major facilitator MDR1 and the ABC transporters CDR1 and CDR2, respectively (17). As C. albicans is a diploid organism, resistance mutations first occur in one of the two alleles of a gene. This is frequently followed by loss of heterozygosity, which further increases the drug resistance of the resulting homozygous strains. Indeed, loss of heterozygosity has been observed in many fluconazole-resistant clinical isolates containing mutations in ERG11, MRR1, or TAC1 (3–6, 14, 18, 29). Similarly, loss of heterozygosity has also been found in a fluconazole-resistant strain with a gain-of-function mutation in Upc2, the transcriptional regulator of ERG11 and other ergosterol biosynthesis genes (9).
The diploid genome of C. albicans exhibits a high degree of heterozygosity (12). In a given strain, the two alleles of a gene usually are not identical but differ from one another to various degrees. It has been recently reported that two types of MDR1 alleles exist in C. albicans which can be distinguished by specific polymorphic nucleotides in the promoter region (2). One of these promoter alleles was found to confer higher MDR1 expression than the other allele. Many clinical isolates contained two alleles of the higher-activity type, whereas strains containing only the less active allele were rare, and it was suggested that the higher-activity alleles of the MDR1 promoter could promote the development of drug resistance (2). These observations indicated that in strains containing both types of MDR1 alleles, loss of heterozygosity would be an additional mechanism of increased drug resistance.
In most C. albicans strains, including reference strain SC5314, MDR1 is not significantly expressed under standard growth conditions, but it is induced in the presence of certain chemicals, like benomyl or H2O2 (7, 8, 13, 21). Consequently, deletion of MDR1 in such strains does not result in hypersusceptibility of the mutants to fluconazole (19, 23, 25). In contrast, in fluconazole-resistant strains that have acquired activating mutations in the transcriptional regulator Mrr1 and overexpress the efflux pump, deletion of MDR1 causes a partial loss of drug resistance, demonstrating that MDR1 and other Mrr1 target genes contribute to the increased fluconazole resistance of these strains (25, 32). In our laboratory, we have used reporter gene fusions to unravel the role of cis-acting sequences and trans-regulatory factors in inducible and constitutive MDR1 expression (5, 11, 15, 18, 24–27, 31). For this purpose, the GFP reporter gene, which encodes green fluorescent protein, was placed under the control of the MDR1 promoter from fluconazole-susceptible strain SC5314. As SC5314 is heterozygous for MDR1 and contains both types of MDR1 promoters, it seemed possible that some conclusions about the regulation of MDR1 expression might be valid only for the cloned promoter and not for the other MDR1 promoter allele of this strain. Therefore, in the present study, we directly compared the inducibility of the two MDR1 promoter alleles by chemicals that are known to stimulate MDR1 expression and their constitutive activation by hyperactive transcription factors that control MDR1 expression. In addition, we investigated if loss of heterozygosity at the MDR1 locus is associated with the development of fluconazole resistance in MDR1-overexpressing clinical C. albicans isolates.
The two types of MDR1 promoter alleles in C. albicans can be distinguished by the presence or absence of an AseI restriction site (depending on the presence of an A or a G at position −306 upstream of the MDR1 coding region) and four linked single-nucleotide polymorphisms at positions −343, −154, −152, and −137 (2). Alleles with the AseI site have been termed A-type promoters, and alleles without the AseI site have been termed G-type promoters. Analysis of the sequence of the 1.1-kb MDR1 promoter fragment in our previously used PMDR1-GFP reporter construct showed that it contained all five polymorphic nucleotides that would classify it as a G-type promoter, which, according to the study by Bruzual and Kumamoto, are the lower-activity promoters (2). Strain SC5314, from which our cloned MDR1 promoter was derived, also contains an A-type promoter (2) and is therefore heterozygous at the MDR1 locus. To directly compare the activities of the two MDR1 promoter alleles of this model strain, we amplified the A-type MDR1 promoter from SC5314 with the same primers as before and inserted it instead of the G-type promoter into our reporter construct. A comparison of the sequences of the two MDR1 promoter alleles showed that they differed at 23 positions within the cloned region (see Fig. S1 in the supplemental material). We refer to the A-type promoter of strain SC5314 as allele 1 (MDR1-1) and to the G-type promoter as allele 2 (MDR1-2).
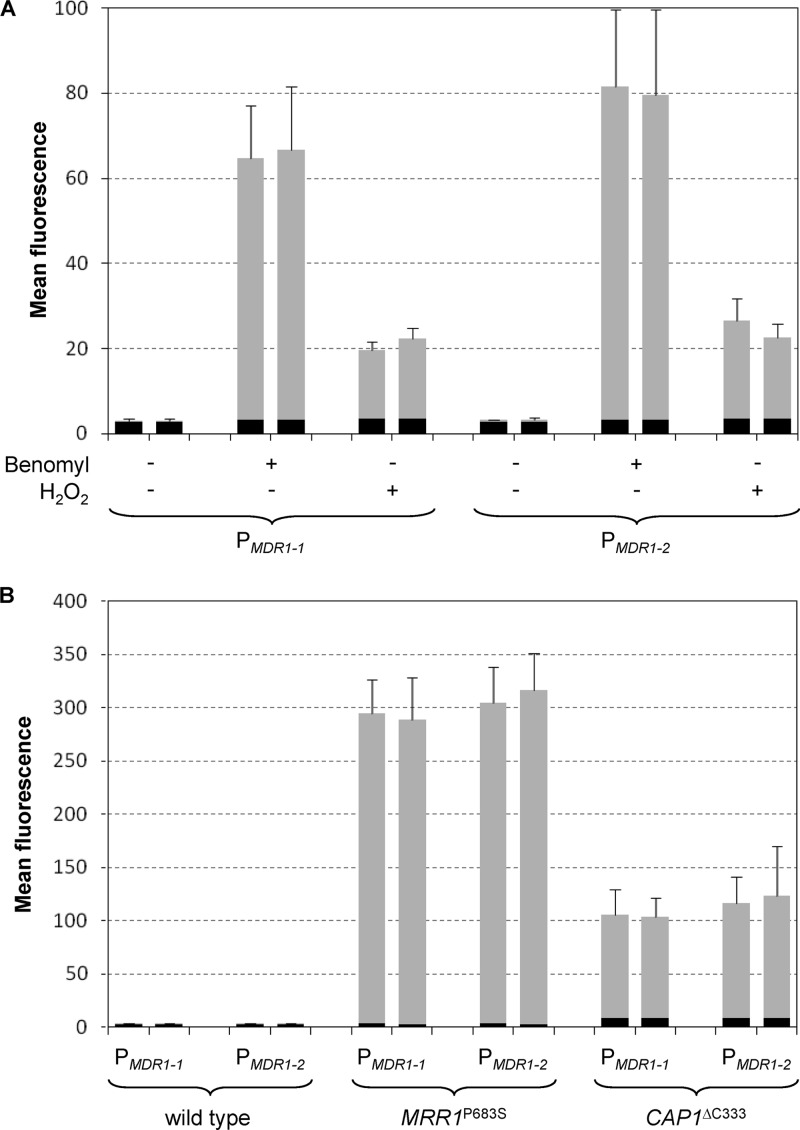
In order to test whether the MDR1 promoter alleles of strain SC5314 differ in their inducibility by chemicals that are commonly used to stimulate MDR1 expression, we integrated the PMDR1-1-GFP reporter fusion at an ectopic genomic locus (the ACT1 locus) of parental strain SC5314. Two independent transformants containing a single copy of the reporter fusion were kept for further analysis and compared with strains in which the GFP reporter gene had been integrated in an identical fashion under the control of the MDR1-2 promoter. The MDR1 promoters were induced by growing the cells in the presence of benomyl or H2O2 and GFP expression in the reporter strains was quantified by flow cytometry as described previously (18). Figure 1A shows that GFP was not detectably expressed in any of the reporter strains in the absence of inducers, demonstrating that the basal activities of both MDR1 promoter alleles are very low in strain SC5314. This is in line with previous observations that MDR1 mRNA or Mdr1 protein could not be detected under noninducing conditions in this strain background by Northern hybridization and Western blotting, respectively (1, 10, 13, 20). The two MDR1 promoters were similarly induced by benomyl and, at a lower level, by H2O2. Therefore, no differences in inducibility between the two MDR1 promoter alleles were detected in these experiments.
Fig 1.
(A) Inducibility of the MDR1 promoter alleles of strain SC5314 by benomyl and H2O2. Reporter strains expressing GFP under the control of the indicated MDR1 promoter allele were grown in the absence (−) or presence (+) of benomyl or H2O2, and the mean fluorescence of the cells (arbitrary units) was determined by flow cytometry. The results obtained with two independently generated reporter strains are shown in each case (means and standard deviations of three independent cultures). (B) Constitutive activation of the MDR1 promoter alleles of strain SC5314 by hyperactive forms of Mrr1 and Cap1. Strains were grown to log phase in YPD medium and analyzed by flow cytometry. The results obtained with two independently generated reporter strains are shown in each case (means and standard deviations of four or five independent cultures). The strains used (see Table S1 in the supplemental material) were SCMPG2S2A and -B (wild type, PMDR1-1), SCMPG2A and -B (wild type, PMDR1-2), SCMRR1R34MPG2S2A and -B (MRR1P683S, PMDR1-1), SCMRR1R34MPG2A and -B (MRR1P683S, PMDR1-2), SCCAP1R14MPG2S2A and -B (CAP1ΔC333, PMDR1-1), and SCCAP1R14MPG2A and -B (CAP1ΔC333, PMDR1-2). The background fluorescence of the parental strains, which do not contain the GFP gene, is indicated by the black part of each column.
As explained above, the low basal expression levels of MDR1 do not contribute to the wild-type tolerance of subinhibitory fluconazole concentrations in drug-susceptible strains. Bruzual and Kumamoto used a fluconazole-resistant strain containing a gain-of-function mutation in Mrr1 to compare the activities of four A-type and four G-type MDR1 promoters in an isogenic background (2). They found that the A-type promoters were, on average, 5-fold more active than the G-type promoters. To compare the constitutive activities of the two MDR1 promoter alleles of strain SC5314 in the presence of hyperactive Mrr1, we introduced the reporter fusions into a derivative of this strain in which the endogenous MRR1 alleles had been replaced with the MRR1P683S allele from a fluconazole-resistant clinical isolate. Figure 1B shows that both MDR1 promoters were constitutively activated at comparable levels in the presence of a hyperactive Mrr1.
The bZip transcription factor Cap1 is also involved in the regulation of MDR1 expression (1, 21, 25). No gain-of-function mutations in Cap1 have been found so far in fluconazole-resistant clinical C. albicans isolates, but C-terminally truncated Cap1 is hyperactive and constitutively activates the MDR1 promoter, albeit less efficiently than does hyperactive Mrr1 (1, 25). To assess whether this hyperactive Cap1 protein might differentially activate the two types of MDR1 promoter alleles, the reporter fusions were also introduced into a derivative of strain SC5314 in which the endogenous CAP1 alleles had been replaced with the CAP1ΔC333 allele. As shown in Fig. 1B, both MDR1 promoter alleles were activated to similar levels by the hyperactive Cap1 protein. These results demonstrate that the two MDR1 alleles of strain SC5314 are equally well activated by two different hyperactive transcription factors that control MDR1 expression and would therefore be expected to contribute equally to the increased drug resistance of such strains.
The findings that A-type MDR1 promoters exhibited higher activity than G-type promoters and that homozygosity for A-type promoters was common in clinical C. albicans isolates, whereas homozygosity for G-type promoters was rarely observed, have led to the hypothesis that homozygosity for A-type MDR1 promoters is another mechanism that contributes to the development of fluconazole resistance in C. albicans (2). The mechanisms of fluconazole resistance in clinical C. albicans isolates can be best studied in serial isolates from individual patients that developed drug resistance over time (30). To investigate if loss of heterozygosity at the MDR1 locus is associated with MDR1 overexpression and increased fluconazole resistance in clinical isolates, we first reexamined the MDR1 promoter sequences of two well-studied matched pairs of susceptible and resistant isolates from AIDS patients (6). Isolates F2 and G2 are fluconazole-susceptible isolates that do not detectably (by Northern hybridization) express MDR1, and isolates F5 and G5 are matched fluconazole-resistant isolates from the same patients that overexpress MDR1 due to the acquisition of gain-of-function mutations in the transcriptional regulator Mrr1 and homozygosity for the mutated MRR1 allele (18). Previous work in our lab demonstrated that susceptible isolates F2 and G2 contained two polymorphic MDR1 promoter alleles that differed at various positions and that both alleles were retained in the corresponding resistant isolates, F5 and G5 (31). Inspection of the MDR1 promoter sequences demonstrated that both alleles of these isolates were of the G type, although one of the two alleles had a T at position −343 instead of the A that usually occurs at this position in G-type promoters (Table 1). This result demonstrated that the A at position −343 is not invariably linked with the other four diagnostic nucleotides in G-type alleles. In addition, and contrary to expectations, in these two cases, MDR1 overexpression and fluconazole resistance developed in strains that contained only G-type alleles.
Table 1.
Polymorphisms in the MDR1 promoters of clinical C. albicans isolates
| Isolate | Allele(s) | Nucleotidea at position: |
||||
|---|---|---|---|---|---|---|
| −343 | −306 | −154 | −152 | −137 | ||
| F2 (Flus)b | 1 | T | G | A | T | A |
| 2 | A | G | A | T | A | |
| F5 (Flur)b | 1 | T | G | A | T | A |
| 2 | A | G | A | T | A | |
| G2 (Flus)b | 1 | T | G | A | T | A |
| 2 | A | G | A | T | A | |
| G5 (Flur)b | 1 | T | G | A | T | A |
| 2 | A | G | A | T | A | |
| B3 (Flus)c | 1, 2 | T | A | G | A | C |
| B4 (Flur)c | 1, 2 | T | A | G | A | C |
| 5044 (Flus)c | 1, 2 | A | G | A | T | A |
| 5052 (Flur)c | 1, 2 | A | G | A | T | A |
| 1442 (Flus)c | 1, 2 | A | A/G | A/G | A/T | A/C |
| 2271 (Flur)c | 1, 2 | A | A/G | A/G | A/T | A/C |
| 5833 (Flus)c | 1, 2 | A | A/G | A/G | A/T | A/C |
| 6692 (Flur)c | 1, 2 | A | A/G | A/G | A/T | A/C |
| 1490 (Flus)c | 1, 2 | T | A/G | A/G | A/T | A/C |
| 1587 (Flur)c | 1, 2 | T | A/G | A/G | A/T | A/C |
| DSY291 (Flus)c | 1, 2 | T | A | A/G | A/T | A/C |
| DSY292 (Flur)c | 1, 2 | T | A | A/G | A/T | A/C |
| DSY2285 (Flus)c | 1, 2 | T | G | A/G | A/T | A/C |
| DSY2286 (Flur)c | 1, 2 | T | G | A/G | A/T | A/C |
| Signature | A type | T | A | G | A | C |
| Signature | G type | A | G | A | T | A |
Only the diagnostic nucleotides that define A- and G-type alleles, named after the nucleotide at position −306 (gray column) are shown. The signature sequences of A-type and G-type alleles are shown at the bottom.
The sequences of both MDR1 promoter alleles of these isolates were determined in a previous study (31).
The MDR1 promoters of these isolates were amplified by PCR, followed by direct sequencing of the PCR products. Polymorphic nucleotides could therefore not be assigned to one or the other allele.
To investigate if loss of heterozygosity at the MDR1 locus occurred in other MDR1-overexpressing, fluconazole-resistant clinical C. albicans isolates, we determined the MDR1 promoter sequences in seven additional isolate pairs in which the resistant isolate had acquired a gain-of-function mutation in Mrr1 and overexpressed MDR1 (5). For this purpose, the MDR1 promoters of these isolates were amplified by PCR and the PCR products were directly sequenced (Table 1). Two of the seven isolate pairs contained only one type of MDR1 promoter; isolates B3 and B4 contained a typical A-type promoter, whereas isolates 5044 and 5052 contained a canonical G-type promoter. The remaining isolates were heterozygous. Isolate pairs 1442–2271, 5833–6692, and 1490–1587 contained both an A-type and a G-type promoter, but the A-type promoters in the former two pairs contained an A instead of a T at position −343, and the G-type promoter of the latter pair contained a T instead of an A at this position. The situation in the last two isolate pairs was even more unusual. Both MDR1 promoter alleles of isolates DSY291 and DSY292 contained the AseI site with the A at position −306, but the nucleotides at positions −154, −152, and −137 did not correspond to the signature sequence of an A-type allele in one of the alleles. Conversely, both MDR1 alleles of isolates DSY2285 and DSY2286 lacked the AseI site and contained a G at position −306, but the nucleotides at the other four positions did not match the G-type signature in one of the two alleles. The results obtained by direct sequencing of the PCR-amplified MDR1 promoters were fully confirmed by Southern hybridization analysis of AseI-digested genomic DNA of all isolates, which verified the presence or absence of the AseI site in one or both MDR1 promoter alleles (data not shown). These results demonstrate that in the nine isolate pairs studied in which fluconazole resistance was associated with MDR1 overexpression, A-type MDR1 promoters were not more frequent than G-type promoters. In addition, all of the strains that contained two different MDR1 promoter alleles retained both of them after the development of fluconazole resistance; i.e., no loss of heterozygosity was observed in any of the strains. Finally, the proposed signature sequence of A- and G-type MDR1 promoters is not well conserved; the nucleotide at position −306, which determines the presence or absence of the AseI site, is not invariably linked with any of the other four nucleotides of the consensus sequence.
Genomic alterations are now recognized as an important mechanism of C. albicans adaptation to changes in its environment (22, 28). The presence of antifungal drugs selects for adaptive mutations in the drug target or in transcription factors that control the expression of genes whose upregulation confers increased drug resistance. The appearance of such mutations in one allele of a gene is often followed by loss of heterozygosity, which further increases the resistance of strains that became homozygous for the mutated allele (3–6, 9, 14, 18, 22, 25, 28–29). The high degree of heterozygosity in its diploid genome expands the genetic repertoire of C. albicans, and homozygosity for an advantageous allele may allow better survival under selective conditions, even in the absence of new mutations. In this context, the recent proposal that homozygosity for an MDR1 promoter allele with higher activity may promote fluconazole resistance is relevant for our understanding of the evolution of drug resistance in C. albicans (2). This hypothesis also implied that conclusions drawn from experiments in which the regulation of the MDR1 promoter was studied by using reporter gene fusions might be valid only for the MDR1 promoter driving the expression of the reporter gene and not necessarily for the other type of MDR1 promoter. The results of our present study demonstrate that in C. albicans reference strain SC5314, the two promoters are equally well activated by chemicals that induce MDR1 expression and by hyperactive forms of transcription factors Mrr1 and Cap1, which confer constitutive MDR1 overexpression. Therefore, previous conclusions about the regulation of MDR1 expression were not biased by the analysis of only one of the two promoter alleles of this strain.
Bruzual and Kumamoto compared the abilities of four A-type and four G-type MDR1 promoters from different C. albicans strains to drive the expression of the GFP reporter gene in a host strain containing hyperactive Mrr1 and found that all A-type promoters produced larger amounts of GFP mRNA than did the G-type promoters (2). One of the A-type promoters was from strain CAI4, a derivative of strain SC5314, but the G-type allele from this strain was not included for comparison in that study. Our results indicate that no differences between the activities of these two promoter alleles would have been observed. The different activities of the various MDR1 promoters that were reported in that study must therefore be due to sequence differences other than those found in the alleles of strain SC5314 (see Fig. S1 in the supplemental material).
Our results imply that in strain SC5314, loss of heterozygosity for the MDR1 promoter would not increase MDR1 expression and therefore not contribute to the development of fluconazole resistance. In this respect, it is interesting that fluconazole-resistant derivatives of strain SC5314, which were selected in vitro during growth in the presence of fluconazole, retained both polymorphic MDR1 alleles, demonstrating that loss of heterozygosity at the MDR1 locus did not play a role in fluconazole resistance development in any of these strains (2). Five of the fluconazole-resistant strains overexpressed MDR1 and contained gain-of-function mutations in the transcriptional regulator Mrr1 (5, 20). Therefore, the presence of the drug did not select for MDR1 homozygosity either before or after the acquisition of the MRR1 mutation. In contrast, four of the five MDR1-overexpressing strains had become homozygous for the mutated MRR1 allele (5), demonstrating that loss of heterozygosity was a common event under the experimental conditions as soon as one of two polymorphic alleles of a gene conferred a significant advantage.
The results obtained with strain SC5314 and its derivatives do not rule out the possibility that loss of heterozygosity at the MDR1 locus may contribute to fluconazole resistance development in other C. albicans strains. We therefore compared the MDR1 promoter sequences in nine previously characterized matched pairs of fluconazole-susceptible and MDR1-overexpressing, fluconazole-resistant clinical isolates. Seven of the susceptible isolates contained two different MDR1 promoter alleles, which were retained in the corresponding resistant isolates; i.e., in none of these cases did we observe loss of heterozygosity (Table 1). Two isolate pairs (B3-B4 and 5044–5052) were homozygous within the sequenced region of the MDR1 promoter. In these strains, loss of heterozygosity may have occurred at an earlier stage during fluconazole therapy. However, one of the isolate pairs (5044–5052) was homozygous for a G-type promoter, which is contrary to expectations if A-type alleles confer higher MDR1 expression.
The sequence analysis of the MDR1 promoters of the clinical isolates also revealed that the linkage of the signature nucleotides of A-type and G-type promoters is not as tight as previously suggested (2). In our set of strains, there was no linkage of the nucleotide at position −343 (A or T) to the other four nucleotides of the signature sequence (Table 1). In two cases (isolate pairs DSY291-DSY292 and DSY2285-DSY2286), the presence or absence of the AseI site, which depends on the presence of an A or a G at position −306, respectively, was not even linked to the other consensus nucleotides of A- and G-type promoters at positions −154, −152, and −137 in one of the two alleles (note that we did not determine linkage between the latter nucleotides themselves in the two alleles of a strain). Therefore, A-type and G-type promoters cannot be easily defined in some cases and the presence or absence of the AseI site, which was the only criterion used for the identification of MDR1 promoter alleles in an extended set of clinical isolates (2), will not always indicate which type of promoter is present. Using the AseI site as the defining criterion, Bruzual and Kumamoto found significantly more A/A strains (37 out of 44) than G/G strains (7 out of 44) among the homozygous strains in their collection of clinical isolates (2). This was not the case in our more limited set of clinical isolate pairs that developed fluconazole resistance during therapy. Only two of these contained the AseI site in both MDR1 alleles, whereas four other isolate pairs possessed two MDR1 alleles without the AseI site.
It is conceivable that some polymorphisms in the MDR1 promoter region may affect the binding of transcriptional regulators and thereby MDR1 expression levels, and differences in the activities of various MDR1 promoter alleles have been clearly demonstrated (2). In an extreme case, if one allele is completely inactive, MDR1 expression levels would double when a heterozygous strain became homozygous for the active allele and this might provide a selective advantage when the cells were exposed to fluconazole, at least in strains containing a gain-of-function mutation in Mrr1 or in the presence of a stimulus that induces MDR1 expression. Whether loss of heterozygosity for MDR1 occurs under selective pressure could be tested with strains that are heterozygous for MDR1 promoters with different activities. However, all of the MDR1 promoter alleles that have been compared in an isogenic background were derived from different strains and the existence of strains containing both a high-activity and a low-activity MDR1 promoter allele, which could be used for that purpose, remains to be demonstrated. So far, there is no documented example of loss of heterozygosity at the MDR1 promoter as a mechanism of fluconazole resistance, and if it occurs at all, it seems to be rare in clinical C. albicans isolates.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Deutsche Forschungsgemeinschaft (DFG grants MO 846/6, SFB 630, and IRTG 1522).
Footnotes
Published ahead of print 21 May 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Alarco AM, Raymond M. 1999. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181:700–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bruzual I, Kumamoto CA. 2011. An MDR1 promoter allele with higher promoter activity is common in clinically isolated strains of Candida albicans. Mol. Genet. Genomics 286:347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coste A, et al. 2007. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot. Cell 6:1889–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coste A, et al. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172:2139–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dunkel N, Blaß J, Rogers PD, Morschhäuser J. 2008. Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol. Microbiol. 69:827–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Franz R, et al. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob. Agents Chemother. 42:3065–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gupta V, et al. 1998. Identification of polymorphic mutant alleles of CaMDR1, a major facilitator of Candida albicans which confers multidrug resistance, and its in vitro transcriptional activation. Curr. Genet. 34:192–199 [DOI] [PubMed] [Google Scholar]
- 8. Harry JB, et al. 2005. Drug-induced regulation of the MDR1 promoter in Candida albicans. Antimicrob. Agents Chemother. 49:2785–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heilmann CJ, Schneider S, Barker KS, Rogers PD, Morschhäuser J. 2010. An A643T mutation in the transcription factor Upc2p causes constitutive ERG11 upregulation and increased fluconazole resistance in Candida albicans. Antimicrob. Agents Chemother. 54:353–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hiller D, Sanglard D, Morschhäuser J. 2006. Overexpression of the MDR1 gene is sufficient to confer increased resistance to toxic compounds in Candida albicans. Antimicrob. Agents Chemother. 50:1365–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hiller D, Stahl S, Morschhäuser J. 2006. Multiple cis-acting sequences mediate upregulation of the MDR1 efflux pump in a fluconazole-resistant clinical Candida albicans isolate. Antimicrob. Agents Chemother. 50:2300–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones T, et al. 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 101:7329–7334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karababa M, Coste AT, Rognon B, Bille J, Sanglard D. 2004. Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob. Agents Chemother. 48:3064–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. MacCallum DM, et al. 2010. Genetic dissection of azole resistance mechanisms in Candida albicans and their validation in a mouse model of disseminated infection. Antimicrob. Agents Chemother. 54:1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mogavero S, Tavanti A, Senesi S, Rogers PD, Morschhäuser J. 2011. Differential requirement of the transcription factor Mcm1 for activation of the Candida albicans multidrug efflux pump MDR1 by its regulators Mrr1 and Cap1. Antimicrob. Agents Chemother. 55:2061–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morschhäuser J. 2002. The genetic basis of fluconazole resistance development in Candida albicans. Biochim. Biophys. Acta 1587:240–248 [DOI] [PubMed] [Google Scholar]
- 17. Morschhäuser J. 2010. Regulation of multidrug resistance in pathogenic fungi. Fungal Genet. Biol. 47:94–106 [DOI] [PubMed] [Google Scholar]
- 18. Morschhäuser J, et al. 2007. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 3:e164 doi:10.1371/journal.ppat.0030164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morschhäuser J, Michel S, Staib P. 1999. Sequential gene disruption in Candida albicans by FLP-mediated site-specific recombination. Mol. Microbiol. 32:547–556 [DOI] [PubMed] [Google Scholar]
- 20. Riggle PJ, Kumamoto CA. 2006. Transcriptional regulation of MDR1, encoding a drug efflux determinant, in fluconazole-resistant Candida albicans strains through an Mcm1p binding site. Eukaryot. Cell 5:1957–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rognon B, Kozovska Z, Coste AT, Pardini G, Sanglard D. 2006. Identification of promoter elements responsible for the regulation of MDR1 from Candida albicans, a major facilitator transporter involved in azole resistance. Microbiology 152:3701–3722 [DOI] [PubMed] [Google Scholar]
- 22. Rustchenko E. 2007. Chromosome instability in Candida albicans. FEMS Yeast Res. 7:2–11 [DOI] [PubMed] [Google Scholar]
- 23. Sanglard D, Ischer F, Monod M, Bille J. 1996. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob. Agents Chemother. 40:2300–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sasse C, et al. 2011. The transcription factor Ndt80 does not contribute to Mrr1-, Tac1-, and Upc2-mediated fluconazole resistance in Candida albicans. PLoS One 6:e25623 doi:10.1371/journal.pone.0025623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schubert S, et al. 2011. Regulation of efflux pump expression and drug resistance by the transcription factors Mrr1, Upc2, and Cap1 in Candida albicans. Antimicrob. Agents Chemother. 55:2212–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schubert S, Popp C, Rogers PD, Morschhäuser J. 2011. Functional dissection of a Candida albicans zinc cluster transcription factor, the multidrug resistance regulator Mrr1. Eukaryot. Cell 10:1110–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schubert S, Rogers PD, Morschhäuser J. 2008. Gain-of-function mutations in the transcription factor MRR1 are responsible for overexpression of the MDR1 efflux pump in fluconazole-resistant Candida dubliniensis strains. Antimicrob. Agents Chemother. 52:4274–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Selmecki A, Forche A, Berman J. 2010. Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryot. Cell 9:991–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. White TC. 1997. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14α demethylase in Candida albicans. Antimicrob. Agents Chemother. 41:1488–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. White TC, Marr KA, Bowden RA. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wirsching S, Michel S, Köhler G, Morschhäuser J. 2000. Activation of the multiple drug resistance gene MDR1 in fluconazole-resistant, clinical Candida albicans strains is caused by mutations in a trans-regulatory factor. J. Bacteriol. 182:400–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wirsching S, Michel S, Morschhäuser J. 2000. Targeted gene disruption in Candida albicans wild-type strains: the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol. Microbiol. 36:856–865 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.