Abstract
We report retrospective analysis of the clinical and antimicrobial susceptibility data of 140 Streptococcus pseudopneumoniae isolates. Strains were isolated mostly from respiratory tract samples from patients with underlying diseases. In the case of infection, pneumonia, mainly aspiration pneumonia, was the most frequent (27.1% of the patients). We documented high rates of decreased susceptibilities and resistance to erythromycin and tetracycline (57% and 43% of the isolates, respectively), as well as reduced susceptibility to penicillin in 21% of the isolates.
TEXT
Streptococcus pseudopneumoniae is a recently designated species included in the S. oralis-S. mitis group and closely related to Streptococcus pneumoniae that may be identified using appropriate phenotypic and/or molecular assays (1, 8, 11, 12). Pathogenic significance of S. pseudopneumoniae in respiratory tract diseases is still unclear in spite of its association with both a history of chronic obstructive pulmonary disease (COPD) and exacerbation of COPD (4, 6). However, comparative genomic hybridization detected pneumococcal virulence factors in S. pseudopneumoniae (5, 10), while total genome sequencing revealed virulence gene content intermediate between those of S. pneumoniae and S. mitis, beside acquired antibiotic resistance genes (9). Antimicrobial susceptibility studies are scarce. Indeed, a unique study was conducted in New Zealand, and it may not be representative of the resistance of the species in other parts of the world (7).
Here we retrospectively analyzed the clinical circumstances of isolation and the antimicrobial susceptibility profiles of 140 S. pseudopneumoniae isolates recovered in 140 patients admitted to the University Hospital of Montpellier, France, over a 3-year period.
The isolates were recovered between January 2009 and December 2011 from respiratory tract specimens, i.e., bronchoalveolar lavage fluid samples (n = 22 [15.7%]), sputum and endotracheal aspirates (n = 116 [82.9%]), sinus (n = 1), and conjunctiva (n = 1) samples. The patients were hospitalized in intensive care units (n = 67), in medicine (n = 48), or surgery (n = 5) departments, or attended at the Cystic Fibrosis (CF) center (n = 20). Species identification was based on optochin (Oxoid Ltd., Basingstoke, England) susceptibility in an ambient atmosphere and resistance or intermediate susceptibility in a 5% CO2 atmosphere, insolubility in 1% sodium deoxycholate (BD Diagnostics, Heidelberg, Germany), and a positive reaction with the AccuProbe Pneumococcus test (bioMérieux, Marcy l'Etoile, France) (1, 6). Despite recent reports recommending molecular methods, such as recA gene sequencing or real-time PCR assays targeting the Spn9802 fragment and the autolysin gene lytA, for accurate identification of S. pseudopneumoniae (8, 12), the association of one molecular and two phenotypic assays we used here allowed confident identification, because optochin-resistant or deoxycholate-insoluble S. pneumoniae isolates are rarely observed (as reviewed in reference 10).
Reviewing the clinical data associated with S. pseudopneumoniae isolation revealed that pulmonary exacerbation was observed in 6 out of the 20 CF patients. For the 120 other patients, clinical diagnoses were as follows: pneumonia (n = 38), bronchitis (n = 22, including 16 patients with history of COPD, asthma, bronchiectasis, or cancer), chronic sinusitis (n = 1), respiratory tract colonization without a sign of infection (n = 49), and unknown diagnosis (n = 10). Clinical features and bacteriological data are summarized in Table 1 for the 38 patients with pneumonia. Most of them presented aspiration pneumonia and were hospitalized in intensive care units (n = 20). S. pseudopneumoniae was solely isolated in 5 cases, was isolated together with other potential bacterial pathogens, mainly Staphylococcus aureus or Haemophilus influenzae, in 16 cases, and was recovered in other mixed microflora in the 17 remaining cases (Table 1). When quantitative culture data were available, S. pseudopneumoniae was mostly found at bacterial counts above cutoffs used to define the presence of infection (14 out of 17 cases). The outcome was mostly favorable regardless of the administered antimicrobial treatment, but 3 patients died from causes that were not related to pneumonia (Table 1). For 18 patients with subsequent bacteriological analysis, S. pseudopneumoniae was not cultured from respiratory tract samples.
Table 1.
Clinical and bacteriological data for the 38 patients with clinical diagnosis of pneumonia reported in this studya
| Patient | Age (yr)/Sex | Bacteriological data |
Clinical data |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Bacterial countb | Associated organism(s) (count)b | VAP | IP | Background | Smokingc | Antibiotic treatmentd | Clinical outcomee | ||
| 1 | 62/M | BAL | 105 | Escherichia coli (105), H. influenzae (105) | + | − | COPD, alcoholism, hypertension | + | CTX | I |
| 2 | 55/M | BAL | 5 × 104 | Mixed salivary microflora | − | − | COPD | + | AMC | I |
| 3 | 66/M | BAL | 104 | H. influenzae (>106), S. aureus (10) | − | − | COPD, alcoholism, hypertension, diabetes | + | AMC | I |
| 4 | 55/M | BAL | 104 | H. influenzae (105) | − | − | Tuberculosis | NS | CTX | I |
| 5 | 75/F | BAL | 102 | H. influenzae (102) | − | − | Pulmonary fibrosis | NS | NS | I |
| 6 | 64/M | BAL | 104 | None | − | − | Scleroderma | NS | NS | I |
| 7 | 48/M | ETA | 3+ | Escherichia coli (1+), H. influenzae (2+) | − | + | Alcoholism, diabetes, stroke | + | CTX | I |
| 8 | 68/M | ETA | 2+ | S. aureus (3+) | − | + | Hypertension, bleeding brain | NS | FEP | De |
| 9 | 35/M | ETA | 3+ | S. aureus (3+) | − | + | VI | + | NS | I |
| 10 | 48/F | ETA | 2+ | Klebsiella oxytoca (1+) | − | + | Asthma, alcoholism, VI | + | AMC | I |
| 11 | 48/F | ETA | 3+ | Mixed salivary microflora | − | − | Meningeal bleeding | NS | CRO-GEN | I |
| 12 | 20/M | ETA | 1+ | Neisseria meningitidis | − | + | Polytrauma | + | CTX | I |
| 13 | 68/F | ETA | 2+ | Mixed salivary microflora | − | + | Auricular fibrillation, stroke | NS | CTX-MTZ | I |
| 14 | 45/M | ETA | 3+ | Neisseria meningitidis (2+) | − | + | Polytrauma | NS | AMC | I |
| 15 | 55/F | ETA | ND | Mixed salivary microflora | − | + | Alcoholism, hypertension, VI | NS | AMC | I |
| 16 | 56/F | ETA | 3+ | Klebsiella pneumoniae (2+) | − | + | Hypertension, diabetes, cardiogenic shock | + | CTX-MTZ | I |
| 17 | 65/F | ETA | ND | Moraxella catarrhalis | − | + | Auricular fibrillation | NS | CTX | I |
| 18 | 56/M | ETA | 3+ | Mixed salivary microflora | − | + | Alcoholism, cardiac arrest | + | AMC | Df |
| 19 | 52/M | ETA | ND | Mixed salivary microflora | − | + | Alcoholism, hanging | + | AMC | I |
| 20 | 63/M | ETA | 1+ | S. aureus (3+) | − | + | Hypertension, auricular fibrillation, stroke | NS | NS | I |
| 21 | 50/F | ETA | 3+ | Mixed salivary microflora | − | + | Alcoholism, hypertension, chronic renal failure | + | CRO-MTZ | I |
| 22 | 60/M | ETA | 3+ | Mixed salivary microflora | − | + | Hypertension, chronic renal failure, stroke | − | AMC | I |
| 23 | 74/M | ETA | 1+ | S. aureus (1+) | − | + | Huntington's disease, cranial trauma | NS | AMC-GEN | I |
| 24 | 85/F | ETA | 3+ | Mixed salivary microflora | − | + | Hypertension, auricular fibrillation | NS | AMC | I |
| 25 | 27/M | ETA | 2+ | Yeast (1+) | − | + | COPD, emphysema, VI | + | CFM | I |
| 26 | 27/M | ETA | 2+ | Yeast (1+) | + | − | Renal transplantation, measles | NS | AMX | I |
| 27 | 68/M | ETA | 107 | None | + | − | COPD, alcoholism, pulmonary cancer | + | TZP | I |
| 28 | 75/M | Sputum | 107 | Mixed salivary microflora | − | − | Ischemic heart disease | NS | CTX-LVX | I |
| 29 | 6/M | Sputum | 106 | S. aureus (106) | − | − | Encephalopathy | − | AMC | I |
| 30 | 47/M | Sputum | 107 | Yeast (106) | − | − | Alcoholism, hepatitis C, pancreatitis | + | IPM-LVX | I |
| 31 | 82/F | ETA | 106 | S. aureus (108) | − | + | Hypertension, stroke | NS | NS | I |
| 32 | 44/M | ETA | 5 × 106 | None | − | − | Thrombophilia | + | AMC | I |
| 33 | 60/M | Sputum | 108 | Mixed salivary microflora | − | − | COPD | + | NS | I |
| 34 | 51/M | ETA | 106 | None | − | − | Pulmonary cancer | + | AMC | I |
| 35 | 49/F | Sputum | ND | None | − | − | Emphysema | + | AMC | I |
| 36 | 8/M | Sputum | 106 | Mixed salivary microflora | − | − | Chronic bronchitis | − | Josamycin | I |
| 37 | 87/M | Sputum | 108 | S. aureus (108) | − | − | COPD, hypertension, asthma | NS | AMC | I |
| 38 | 85/F | Sputum | 5 × 107 | S. aureus (107) | − | + | Stroke | NS | AMC | Df |
M, male; F, female; BAL, bronchoalveolar lavage fluid; ETA, endotracheal aspirate; ND, not determined; VAP, ventilator-associated pneumonia; IP, inhalation/aspiration pneumonia; COPD, chronic obstructive pulmonary disease; VI, voluntary intoxication; NS, not specified; I, improvement; D, death; AMX, amoxicillin; AMC, amoxicillin-clavulanic acid; CFM, cefixime; CRO, ceftriaxone; CTX, cefotaxime; FEP, cefepime; GEN, gentamicin; IPM, imipenem; LVX, levofloxacin; MTZ, metronidazole; TZP, piperacillin-tazobactam.
When quantitative culture was performed, bacterial count for S. pseudopneumoniae is expressed as CFU/ml and significant counts for infection appear in bold type; when qualitative culture was performed, microbial growth per plate is reported semiquantitatively according to the method of Washington (13) as many (3+), moderate (2+), or few (1+), depending on how far out from the inoculum site colonies appeared, with an organism growing in all streaked areas being reported as 3+, with the aim of estimating the relative loads for the different species cultured from the sample. Yeast strains were not identified.
Past or present.
Antimicrobial treatments that were adjusted according to the antibiogram results are indicated in bold type. For patient 25, amoxicillin-clavulanic acid treatment was switched to cefixime, the strain being resistant to penicillin G and showing decreased susceptibility to amoxicillin; for patient 29, treatment associating cefotaxime and gentamicin was switched to amoxicillin-clavulanic acid, the strain being resistant to cefotaxime, showing decreased susceptibility to penicillin G but being fully susceptible to amoxicillin.
Pneumonia resolution.
Not related to S. pseudopneumoniae infection. Strains recovered from patients 8 and 18 were fully susceptible to the 3 β-lactam agents tested, while the strain recovered from patient 38 showed decreased susceptibility to penicillin G but full susceptibility to amoxicillin and cefotaxime.
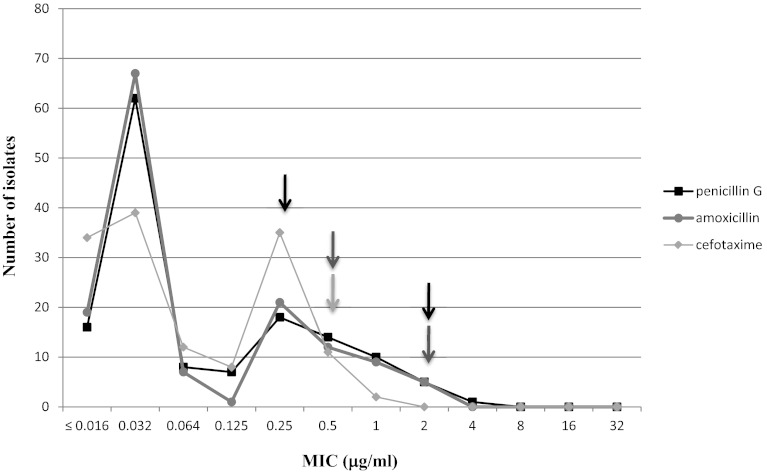
According to the routine procedures in our laboratory, the MICs of penicillin, amoxicillin, and cefotaxime were determined by using the Etest method (AB Biodisk, Solna, Sweden), while the in vitro activities of other drugs were determined by the disk (Bio-Rad, Marne-la-Coquette, France) diffusion method according to guidelines edited by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (3). MIC and zone diameter results were interpreted based on breakpoints established for viridans group streptococci by the EUCAST (3) and the Antibiogram Committee of the French Society of Microbiology (CASFM) (2), respectively, and are given in Fig. 1 and in Table 2. MIC50s of penicillin G, amoxicillin, and cefotaxime were 0.03 μg/ml, while MIC90s were 1, 0.5, and 0.25 μg/ml, respectively. Decreased susceptibility to penicillin G and amoxicillin was observed for 29 and 14 isolates, respectively, while resistance to β-lactams was observed for three isolates (Table 2). Empirical antibiotic treatment was adjusted to the antibiogram results in these three cases (patients 25 and 29 in Table 1 and one CF patient with pulmonary exacerbation). Resistance to erythromycin was the most frequent, followed by resistance to tetracycline and trimethoprim-sulfamethoxazole. A majority of the isolates showing decreased susceptibility or resistance to erythromycin (60.3%) exhibited an efflux phenotype; the macrolide-lincosamide-streptogramin B (MLS) phenotype observed for the other 18 isolates was mainly an inducible phenotype (n = 13) as deduced from the antagonism observed between erythromycin and lincomycin disks placed 30 mm apart. No isolate displayed a high level of resistance to aminoglycosides, and all isolates were susceptible to pristinamycin, levofloxacin, moxifloxacin, and glycopeptides. Resistance to erythromycin, tetracycline, and trimethoprim-sulfamethoxazole was more frequently observed among penicillin-intermediate and -resistant isolates than among penicillin-susceptible isolates (Table 2), but the rate of efflux phenotype was higher among penicillin-susceptible isolates (76.4% versus 21.7%). Similarly, higher rates of tetracycline-resistant isolates and of isolates with decreased susceptibility to penicillin were observed among erythromycin-resistant isolates than among erythromycin-susceptible isolates (Table 2). It is noteworthy that using the corresponding breakpoints for S. pneumoniae would have increased the rate of strains displaying decreased susceptibility to penicillin to 39.3% (n = 49 strains) instead of 21.4% (30 strains), while the 2 strains resistant to cefotaxime would have been categorized as intermediate. Categorization to other agents remained unchanged.
Fig 1.
Penicillin G, amoxicillin, and cefotaxime MIC distributions. Black, dark-gray, and light-gray arrows indicate breakpoints established by the EUCAST for penicillin G, amoxicillin, and cefotaxime, respectively.
Table 2.
In vitro susceptibilities of 140 isolates of Streptococcus pseudopneumoniae to 15 antimicrobial agentsa
| Antimicrobial agent (disk charge, μg) | Breakpointsb | ZD50c or MIC50 | ZD90d or MIC90 | ZD range or MIC range | Susceptibility of group (ne)f |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
|
S. pseudopneumoniae (140) |
% I + R (n) for subgroup (n) |
|||||||||
| % I + R (n) | % R (n) | SPSP (110) | SPDSP (30) | SPSE (62) | SPDSE (78) | |||||
| Penicillin G | ≤0.25–>2 | 0.03 | 1 | ≤0.016–4 | 21.4 (30) | 0.7 (1) | NA | NA | 11.3 (7) | 29.5 (37) |
| Amoxicillin | ≤0.5–>2 | 0.03 | 0.5 | ≤0.016–2 | 10 (14) | 0 | 0 | 46 (14) | 12.9 (8) | 15.4 (12) |
| Cefotaxime | ≤0.5–>0.5 | 0.03 | 0.25 | ≤0.016–1 | NA | 1.4 (2) | 0 | 3,3 (1) | 3.2 (2) | 1.3 (1) |
| Kanamycin (1,000) | ≥14–<10 | 27 | 22 | 17–39 | 0 | 0 | 0 | 0 | 0 | 0 |
| Gentamicin (500) | ≥17–<11 | 29 | 24 | 19–39 | 0 | 0 | 0 | 0 | 0 | 0 |
| Tetracycline (30) | ≥23–<21 | 27 | 12 | 6–39 | 42.9 (60) | 42.1 (59) | 33.6 (37) | 67 (20) | 8 (5) | 66.7 (52) |
| Chloramphenicol (30) | ≥23–<23 | 28 | 24 | 13–39 | NA | 1.4 (2) | 0.9 (1) | 3.4 (1) | 3.2 (2) | 0 |
| Erythromycin (15) | ≥26–<24 | 17 | 6 | 6–39 | 57.1 (80) | 55.7 (78) | 50 (55) | 73 (22) | NA | NA |
| Pristinamycin (15) | ≥22–<19 | 30 | 26 | 24–39 | 0 | 0 | 0 | 0 | 0 | 0 |
| SXT (1.25/23.75) | ≥19–<16 | 24 | 11 | 6–39 | 18.6 (26) | 12.9 (18) | 11.8 (13) | 36.7 (11) | 16.1 (10) | 17.9 (14) |
| Levofloxacin (5) | ≥20–<17 | 25 | 21 | 20–39 | 0 | 0 | 0 | 0 | 0 | 0 |
| Moxifloxacin (5) | ≥24–<21 | 29 | 25 | 24–40 | 0 | 0 | 0 | 0 | 0 | 0 |
| Rifampin (30) | ≥29–<24 | 32 | 29 | 19–40 | 7.1 (10) | 2.1 (3) | 1.82 (2) | 0 | 1.6 (1) | 1.3 (1) |
| Vancomycin (30) | ≥17 | 26 | 24 | 22–35 | 0 | 0 | 0 | 0 | 0 | 0 |
| Teicoplanin (30) | ≥17 | 24 | 22 | 20–38 | 0 | 0 | 0 | 0 | 0 | 0 |
Determined by Etest (penicillin G, amoxicillin, and cefotaxime) or disk diffusion method according to European Committee on Antimicrobial Susceptibility Testing guidelines (3). ZD, zone diameter (in mm); SP, Streptococcus pseudopneumoniae; SPSP, S. pseudopneumoniae susceptible to penicillin; SPDSP, S. pseudopneumoniae with decreased susceptibility and resistance to penicillin; SPSE, S. pseudopneumoniae susceptible to erythromycin; SPDSE, S. pseudopneumoniae with decreased susceptibility and resistance to erythromycin; SXT, trimethoprim-sulfamethoxazole; NA, not applicable.
Breakpoints were given according to the EUCAST guidelines for viridans group streptococci for penicillin G, amoxicillin, and cefotaxime MICs and according to the CASFM guidelines for zone diameters for other antimicrobial agents (2).
ZD50, mean zone diameter (in mm) over which 50% of the isolates were inhibited.
ZD90, mean zone diameter (in mm) over which 90% of the isolates were inhibited.
n, no. of isolates.
I, intermediate; R, resistant.
During the study period, 1,139 S. pneumoniae isolates were collected from respiratory specimens, highlighting an incidence of about 12 S. pseudopneumoniae for 100 S. pneumoniae isolates, higher than that reported previously (4). No mixed infection due to S. pneumoniae and S. pseudopneumoniae was observed. Based on clinical and laboratory findings of this study, S. pseudopneumoniae appeared as a respiratory tract colonizer or as a mild opportunistic pathogen involved in pneumonia and bronchitis, most usually isolated together with other potential pathogens and recovered mainly in patients with various underlying chronic diseases. Among the latter, COPD, which was previously reported as a condition associated with S. pseudopneumoniae isolation, was rarely observed in this study (15 out of the 120 non-CF patients, including 5 patients with bronchitis and 7 with pneumonia) (4, 6). We documented high rates of resistance to erythromycin and tetracycline, as previously reported (6, 7). Of note, erythromycin-resistant S. pseudopneumoniae isolates mainly exhibited an efflux phenotype, contrasting with the MLS resistance phenotype observed in 87.3% of the S. pneumoniae isolated in our institution during the study period. One-fifth of the isolates showed reduced susceptibility to penicillin, with frequent associated resistances. Despite the limitation of being retrospective, our study gave antimicrobial susceptibility results roughly similar to those of previous work from a different continent (7) and contributes to a better knowledge of the antimicrobial susceptibility of S. pseudopneumoniae. However, regarding the clinical implication currently reported for S. pseudopneumoniae, whether strain categorization should be based on breakpoints established for S. pneumoniae or for viridans group streptococci must still be clarified by studies with various antimicrobials used in treatment.
Footnotes
Published ahead of print 29 May 2012
REFERENCES
- 1. Arbique JC, et al. 2004. Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and a description of Streptococcus pseudopneumoniae sp. nov. J. Clin. Microbiol. 42:4686–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Comité de l'Antibiogramme de la Société Française de Microbiologie (CASFM) 2011. Recommandations 2011. Société Française de Microbiologie, Paris, France: http://www.sfm-microbiologie.org Accessed 3 April 2011 [Google Scholar]
- 3. European Committee on Antimicrobial Susceptibility Testing (EUCAST) 2011. Antimicrobial susceptibility testing and clinical breakpoints. http://www.eucast.org Accessed 17 November 2011
- 4. Harf-Monteil C, Granello C, Le Brun C, Monteil H, Riegel P. 2006. Incidence and pathogenic effect of Streptococcus pseudopneumoniae. J. Clin. Microbiol. 44:2240–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnston C, et al. 2010. Detection of large numbers of pneumococcal virulence genes in streptococci of the mitis group. J. Clin. Microbiol. 48:2762–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Keith ER, Podmore RG, Anderson TP, Murdoch DR. 2006. Characteristics of Streptococcus pseudopneumoniae isolated from purulent sputum samples. J. Clin. Microbiol. 44:923–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keith ER, Murdoch DR. 2008. Antimicrobial susceptibility profile of Streptococcus pseudopneumoniae isolated from sputum. Antimicrob. Agents Chemother. 52:2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leung M, et al. 2012. Streptococcus pseudopneumoniae identification by pherotype: a method to assist understanding of a potentially emerging or overlooked pathogen. J. Clin. Microbiol. 50:1684–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shahinas D, et al. 2011. Whole-genome sequence of Streptococcus pseudopneumoniae isolate IS7493. J. Bacteriol. 193:6102–6103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simões AS, et al. 2010. Highly penicillin-resistant multidrug-resistant pneumococcus-like strains colonizing children in Oeiras, Portugal: genomic characteristics and implications for surveillance. J. Clin. Microbiol. 48:238–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sistek V, et al. 2011. Development of a real-time PCR assay for the specific detection and identification of Streptococcus pseudopneumoniae using the recA gene. Clin. Microbiol. Infect. doi:10.1111/j.1469-0691.2011.03684.x [DOI] [PubMed] [Google Scholar]
- 12. Wessels E, Schelfaut JJG, Bernards AT, Claas ECJ. 2012. Evaluation of several biochemical and molecular techniques for identification of Streptococcus pneumoniae and Streptococcus pseudopneumoniae and their detection in respiratory samples. J. Clin. Microbiol. 50:1171–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Washington JA. 1996. Principles of diagnosis, chapter 10. In Baron S. (ed), Medical microbiology, 4th ed University of Texas Medical Branch at Galveston, Galveston, TX: http://www.ncbi.nlm.nih.gov/books/NBK8014/ [PubMed] [Google Scholar]