Abstract
G protein–coupled receptor (GPCR) kinases (GRKs) instigate the desensitization of activated GPCRs via phosphorylation that promotes interaction with arrestins, thereby preventing the interaction of GPCRs with heterotrimeric G proteins. A current proposed model of GRK1 activation involves the binding of activated rhodopsin (Rho*) to the N–terminal region of GRK1. Perhaps concomitantly, this N–terminal region also stabilizes a closed, active conformation of the kinase domain. To further probe this model, we mapped changes in the backbone flexibility of GRK1 as it binds to its two substrates, adenosine triphosphate (Mg2+·ATP) and Rho*. We found that the conformational flexibility of GRK1 was reduced in the presence of either Mg2+·ATP and/or Rho*, with Mg2+·ATP having the greatest effect. In a truncated form of GRK1 lacking the N–terminal region (ΔN–GRK1), peptides that directly interact with ATP were not as dramatically stabilized by adding Mg2+·ATP, and dynamics were greater in the interface between the large lobe of the kinase domain and the regulator of G protein signaling homology domain. In the presence of Mg2+·ATP, the influence of Rho* versus Rho was negligible on GRK1 dynamics.
Normal visual function relies on the ability of rhodopsin (Rho) to respond to light stimuli and then quickly return to the dark adapted state 1. Recovery of the dark state is in part orchestrated by G protein–coupled Rho receptor (GPCR) kinase 1 (GRK1) which desensitizes activated rhodopsin (Rho*) 2, 3 by docking to and phosphorylating residues in the C–terminal tail of the receptor 4–7. This phosphorylation in turn promotes an interaction with the capping protein, arrestin, that blocks subsequent interactions with transducin 4. Humans lacking GRK1 suffer from so–called Oguchi disease 8, experiencing normal light adaptation at low levels of ambient light but abnormally slow dark–adaptation at high levels of illumination 9. We hypothesized that without a functional GRK1 in vivo, decay of Rho* to opsin and regeneration with 11–cis–retinal probably would constitute the sole pathway for recovering rod sensitivity 9. In mice, inactivation of the GRK1 gene eliminates light–dependent phosphorylation of Rho*, resulting in a larger and longer lasting single–photon response than in wild–type animals 10.
The currently proposed mechanism of GRK1 activation is believed to consist of two distinct steps. Rho* first binds to the N–terminal ~19 residues of the kinase, inducing the formation of an α–helix (Fig. 1). This helix then interacts with the kinase domain in a manner that bridges the large and small lobes and stabilizes a more closed, active conformational state. The importance of the N–terminal 19 amino acid residues for activation of GRK1 has already been described 11, 12. Truncation of this N–terminal region in GRK1, GRK2 or GRK5 abolished receptor phosphorylation 12–14. Moreover, mutation of residues in the small lobe of the GRK kinase domain that interact directly with the N–terminal helix, as observed in the structure of GRK6 in a relatively closed conformation 15, greatly compromised GPCR phosphorylation 16.
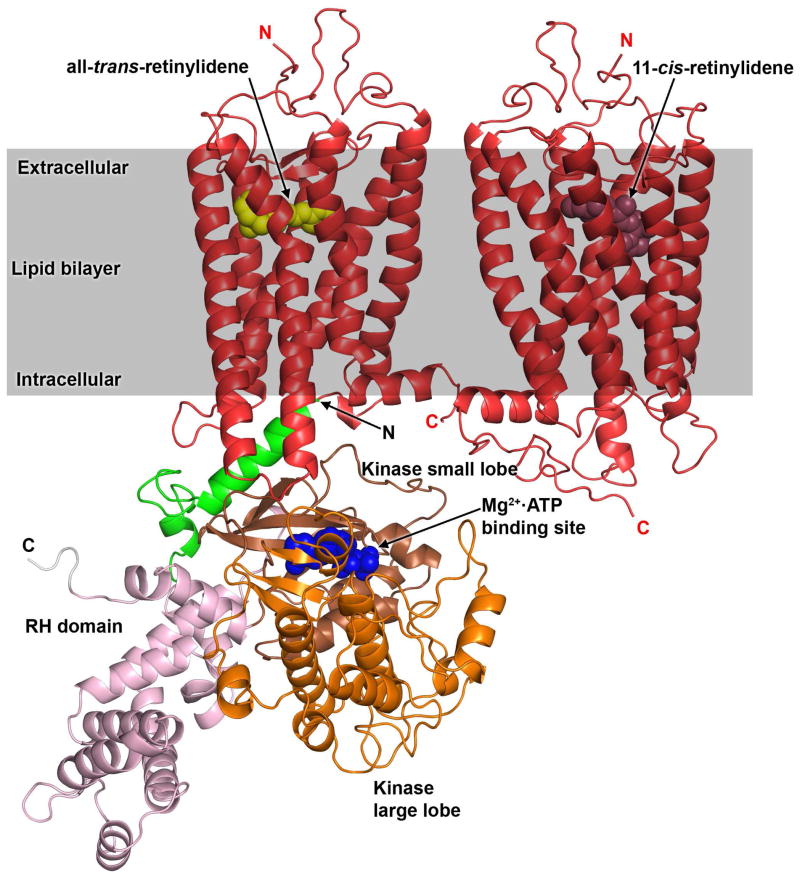
Figure 1. Model of the Rho–GRK1 complex.
GRK1 modeled in a closed conformation with an ordered N–terminal helix is docked to Rho* using the C–terminal helix of Gαs present in the β2AR–Gs crystal structure as a guide 16, 33. The coordinates for the model, in Protein Data Bank format, are available upon request. Positions of the ordered N– and C–termini are labeled with N and C, respectively. Domains of GRK1 are represented as follows: N–terminus region (Ser1–Pro42) in green, the RH domain in pink (Pro43 to Glu184 and Trp515 to Arg532) and the kinase domain small lobe in brown (Asp185 to Asn268) and large lobe in orange (Gly269 to Pro514). The C–terminal region of GRK1 is shown in gray. The GRK1 model was constructed using a homology modeling strategy based on the X–ray of the GRK6 structure 15. The expected position of the lipid bilayer relative to the Rho*–GRK1 complex is illustrated by the grey rectangle. In this cartoon, a theoretical Rho*–Rho heterodimer is shown in red, based on the recently modeled H8–H8 dimer orientation, although monomeric Rho is also an efficient substrate for GRK1 in vitro 34–36. All–trans–retinylidene and 11–cis–retinylidene are depicted using yellow and brown spheres, respectively.
To probe this model for GRK activation further, we used deuterium exchange mass spectrometry (DXMS) to characterize the structural flexibility of GRK1 in the presence or absence of its substrates. We hypothesized that in the presence of Mg2+·ATP we should observe changes in the overall flexibility consistent with multiple crystal structures of GRK1 in complex with ATP and/or ADP. Moreover in the presence of Rho*, we should observe changes in the stability of the N–terminal 19 amino acids of GRK1 and perhaps in regions of the protein kinase that this region interacts with 15. As a control, we used a GRK1 molecule lacking the first 19 amino acid residues (ΔN–GRK1). We found that both Mg2+·ATP and Rho* induced marked reductions in the flexibility of GRK1 whereas deletion of the N–terminal 19 amino acids resulted in increased flexibility in the active site and interdomain contacts of this enzyme.
EXPERIMENTAL PROCEDURES
Expression and purification of bovine GRK1 and ΔN–GRK1
High–Five cells were infected with viruses encoding residues 1–535 of GRK1 (hence lacking the C–terminal farnesylation site) or ΔN–GRK1 (residues 20–535) for 48 h prior to harvesting and purified as described previously 16, 17. Both proteins were engineered to have C–terminal hexa–histidine tags. Briefly, cell pellets were resuspended in lysis buffer (20 mM HEPES, pH 7.5, containing 150 mM NaCl and 10 mM β–mercaptoethanol) supplemented with phenylmethylsulfonyl fluoride and leupeptin. Cells were homogenized, lysed sonication, and debris was pelleted by ultracentrifugation. The soluble fraction was loaded onto a Ni–NTA column, washed with lysis buffer supplemented with 10 mM imidazole, and eluted with lysis buffer containing 200 mM imidazole at pH 7.5. Protein–containing fractions were pooled, diluted to reduce the NaCl concentration below 50 mM and loaded onto a Source15S ion exchange column (GE Healthcare). The column was eluted with an increasing NaCl gradient (0 to 500 mM) and the protein eluted between 100 to 200 mM NaCl. Fractions containing the protein were pooled, concentrated and loaded onto a tandem S200 size exclusion column (GE Healthcare) pre–equilibrated with 20 mM HEPES, pH 7.5, containing 50 mM NaCl and 2 mM DTT. All purification steps were performed at 4 °C. Purified protein was then aliquoted, flash frozen in liquid nitrogen and stored at −80 °C for future use.
Preparation of rod outer segment (ROS) membranes
ROS membranes were prepared from 100 bovine retinas under dim red light by using discontinuous sucrose gradients as described 18. Membranes were diluted in 40 ml of hypotonic buffer (5 mM Bis–Tris–propane (BTP) pH 7.5, containing 0.1 mM EDTA and 1 mM DTT) and proteins were released by gentle homogenization followed by centrifugation at 25,000g at 4 °C for 30 min. This extraction procedure was repeated three times. Combined membranes were suspended in 3 ml of 10 mM BTP, pH 7.5, 100 mM NaCl and either used immediately or frozen at −80 °C. Rho concentrations were determined from their absorption at 500 nm by using the extinction coefficient ε = 40,600 M−1cm−1 18.
Amide hydrogen–deuterium exchange of GRK1 and ΔN–GRK1
Hydrogen–deuterium exchange experiments were performed as follows: 10 μg of GRK1 in 10 mM HEPES, pH 7.5, containing 0.15 M NaCl and 1 mM DTT were incubated in 80 μl D2O on ice for 10, 20, 30, 60, 300, 600 and 1800 s. After each incubation, deuterium exchange was terminated by adding 10 μl of quench solution (D2O with pH adjusted to 2.4 with HCl). Immediately thereafter, 10 μl of pepsin (1.7 mg ml−1; Worthington, Lakewood, NJ) was added to the solution, The final pH was 2.5. Then the sample was digested for 8 min on ice before injecting the resulting peptides on a Luna 20 × 2.00 mm C18 column (Phenomenex, Torrance, CA) with a temperature controlled autosampler set to 4 °C attached to a Hewlett–Packard 1100 HPLC system (Agilent Technologies, Santa Clara, CA). Peptides were eluted with the following gradients: 0–4 min, 98% H2O in 0.1% (v/v) formic acid (A) and 2% isopropanol in 0.1% (v/v) formic acid (B); 4–12 min, 98% to 2% A. Peptides were directed into a Finnigan LXQ (Thermo Finnigan, Waltham, MA) equipped with a electrospray ionization source operating at 300 °C. Tandem MS2 were collected after collision–induced dissociation of ions with a normalized collision energy set to 35 eV. Deuterium exchange experiments conducted in the presence of Mg2+·ATP were performed as described for apo–GRK1. Concentrations of ATP and Mg2+ were 1 and 2 mM, respectively.
Amide hydrogen–deuterium exchange of GRK1 and ΔN–GRK1 in the presence of Rho or Rho*
These experiments were initiated after mixing either GRK1 or ΔN–GRK1 with either ROS or photoactivated–ROS membranes (GRK1:Rho=1:2, w/w). ROS membranes were activated with light (150 W) delivered by fiber light through a 480–520 nm band pass filter for 30 s. Hydrogen–deuterium exchange was performed in the dark. Exposure to D2O was carried out in a manner similar to that described for GRK1 in the absence of Rho. Before digestion with pepsin, samples were terminated with quenching solution (pH 2.4) supplemented with 1 mM Mg2+ and 1 mM Ca2+ chloride salts. All digestions were carried out in tubes wrapped with aluminum foil. Then they were centrifuged at 10,000×g for 30 s. This additional step, as compared to the procedure described above for apo–GRK1, was included to pellet ROS membranes. Supernatants were collected and processed as described above for apo–GRK1 samples.
Before starting production–run experiments, we first evaluated the effects adding 1 mM Mg2+ and 1 mM Ca2+ to the quench solution for Rho and Rho* samples. The effect of this combination was first investigated on the deuterium uptake of GRK1. This control was critical because we had observed that addition of Mg2+ in D2O used to dilute samples caused significant conformational changes in apo–GRK1 (vide infra). In all the present experiments, the pH was maintained at 2.5. Addition of 1 mM Mg2+ and 1 mM Ca2+ to the pH 2.4 quench solution had no detectable effect on the deuterium uptake of apo–GRK1 (data not shown), allowing us to compare the deuterium uptake profile of GRK1 with profiles obtained in the presence of Rho or Rho*.
Mass spectrometry of pepsin digests
Peptides resulting from digestion with pepsin were identified based on their MS2 spectra compared to their theoretical y and b ions. First, the raw files format (Thermo Finnigan, Waltham, MA) was converted to a mzXML file format through instrument–associated software. Then the resulting files were searched against the primary sequences of GRK1 and ΔN–GRK1 (Fig. 2) with the MassMatrix search engine 19. The quality of each peptide search was evaluated by estimating three statistical scores, namely pp, pp2, and ptag described in detail elsewhere 20. Each deuterated peptide was identified by using both its retention time (min) and the charge state of its undeuterated control counterpart.
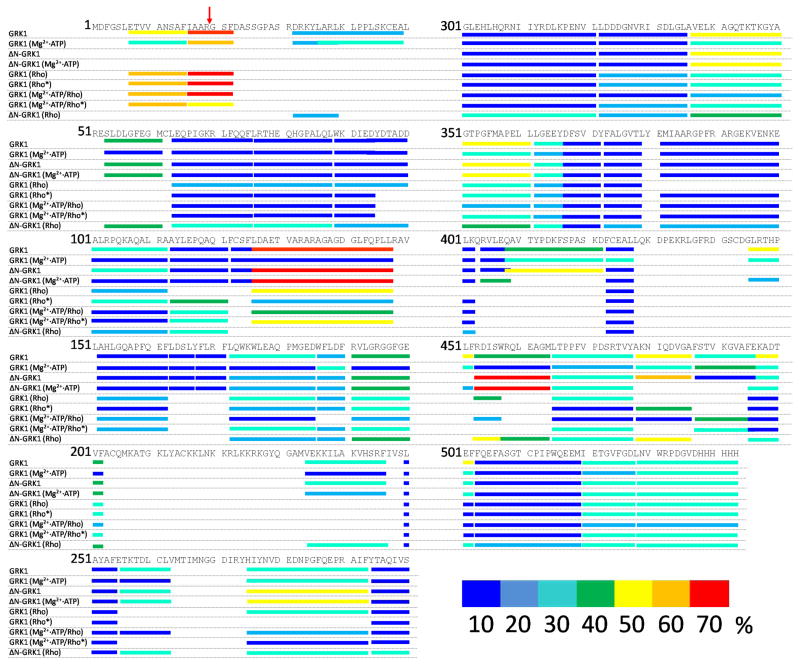
Figure 2. Proteolytic cleavage of GRK1/ΔN–GRK1 by pepsin and deuterium uptake mapped on the primary sequence.
Peptide coverage after proteolytic digestion of bovine GRK1 by pepsin is mapped on the primary sequence of the kinase. Peptide fragments identified by MS2 following an 8 min digestion resulted in 96% coverage. Normalized deuterium uptake was evaluated according to the flowchart presented in Fig. 3 and described in Experimental Procedures. Uptake is color–coded as follows: 0–10%, dark blue; 10–20% blue; 20–30%, cyan; 30–40%, green; 40–50%, yellow; 50–60% orange, 60–70% red. The red arrow indicates the position of the N–terminus of ΔN–GRK1.
Analysis of deuterium exchange products
Undeuterated signals were identified by MS2 as emanating from bona fide GRK1 peptic fragments by comparing the theoretical peptide product ions to ions obtained after collision–induced dissociation of the specific ion. Deuterated peptides were additionally identified by their retention times as described above. Eluted peaks were processed in a semi–automatic fashion whereby the peak envelope and the consequent centroid were evaluated with HDExpress software 21. For each peptic peptide, we calculated the maximum theoretical number of exchangeable protons. For Pro–containing peptides, the maximum exchangeable number was decreased by one for each Pro residue. Protons of amino acid residue side chains have very rapid exchange rates and thus were not included. Whereas deuterium exchange at peptide bond sites is expected during the dilution procedure, peptide deuteriums located on amino acid side–chains also are exchanged with protons during the liquid chromatography step. The total percent of deuterium uptake was evaluated for each peptide and represented with Origin 8 SR0 software (OriginLab Corporation, ver. 8.0725, Northampton, MA) (Fig. 3C) as a function of D2O incubation time as described in detail elsewhere 22. Error bars represent standard errors of the means (SEM) with statistical significance assessed by the Student’s t test. Data from triplicate experiments were averaged. Deuterium exchange was color–coded based on the total percent of the theoretical maximum deuterium uptake defined for each peptide. The code was as follows: dark blue, 0–10%; light blue, 10–20%; cyan, 20–30%; green, 30–40%; yellow, 40–50%; orange, 50–60%; and red, 60–70%. Color–coded molecular models illustrating deuterium exchange profiles were created with Pymol and Chimera 23.
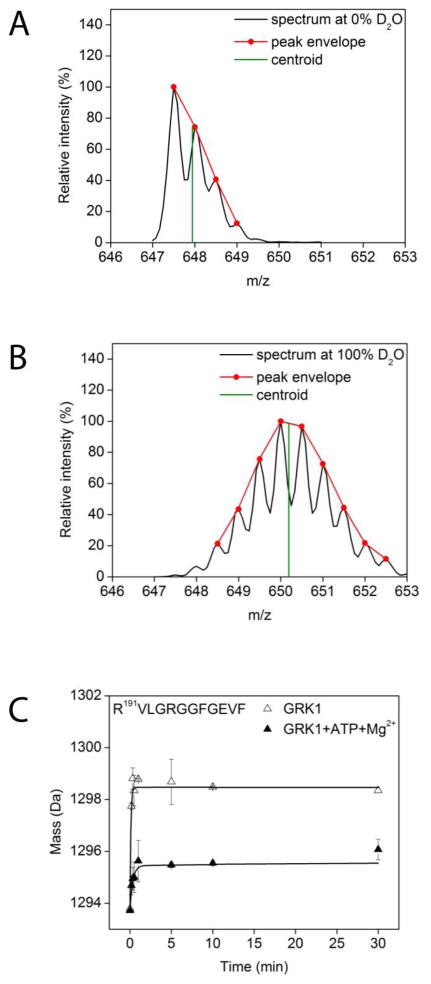
Figure 3. Typical workflow for evaluation of hydrogen–deuterium exchange.
(A) The spectrum of a doubly charged peptide with the sequence R191VLGRGGFGEVF. For illustration purposes, the peak envelope is shown in red whereas the centroid calculated by using the algorithm described in Experimental Procedures is shown in green. (B) The same peak at the same retention time as the undeuterated species in (A). (C) Typical deuterium uptake of the R191VLGRGGFGEVF peptide under two different experimental conditions. The absolute mass of the R191VLGRGGFGEVF peptide as a function of incubation time is shown by the open triangles for apo–GRK1 and closed triangles for Mg2+·ATP loaded GRK1.
Molecular modeling
Because there are no crystal structures of GRK1 in a closed state in which its N–terminal 19 amino acids is ordered, molecular models of GRK1 and ΔN–GRK1 were generated with the Modeller modeling package 24. Primary sequences of GRK1 and ΔN–GRK1 (Fig. 2) together with the X–ray structure of GRK6 in complex with sangivamycin 15 were used as input parameters for the modeling procedure. Models were visually interpreted using Pymol 25.
Statistical analyses
Graphical and statistical analyses were carried out with Origin 8 SR0 software. Error bars represent standard error of the mean (SEM). Data were derived from at least 3 independent experiments.
RESULTS
Dynamics of apo– and Mg2+·ATP–loaded GRK1
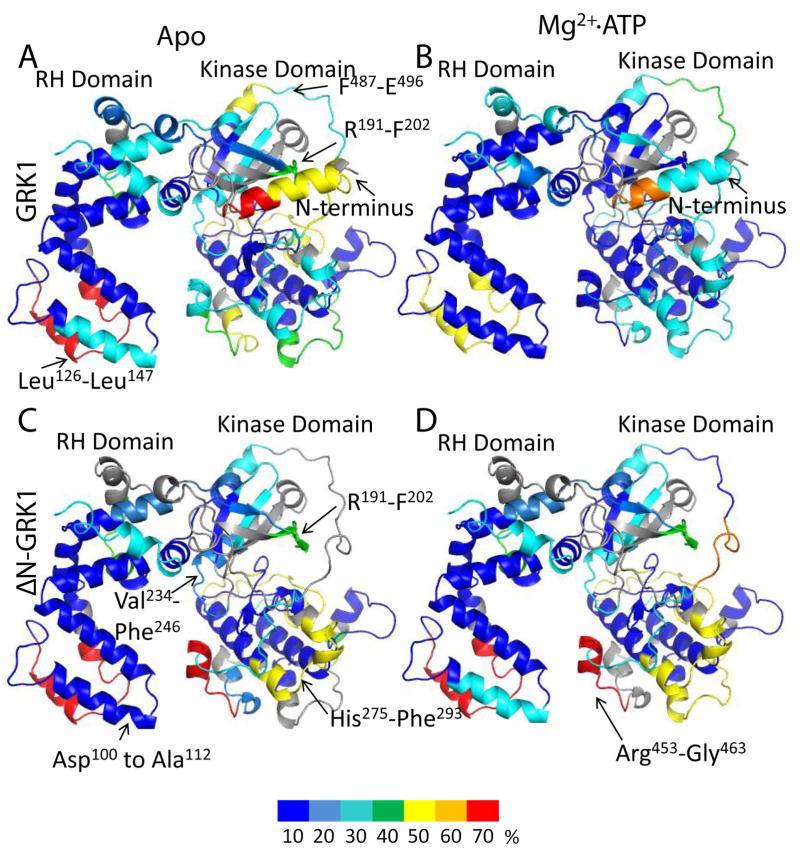
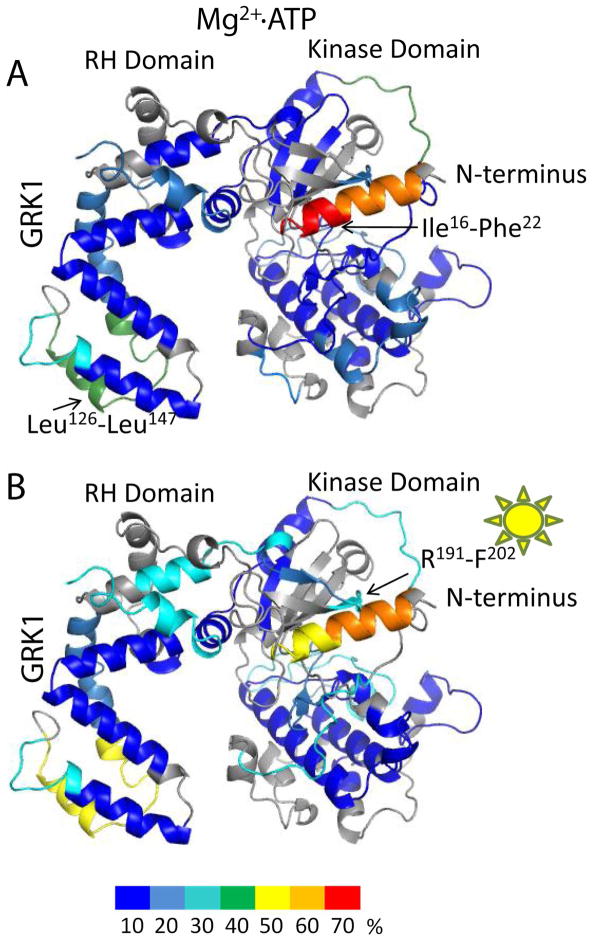
After incubation in D2O, regions buried in the hydrophobic core of apo–GRK1 had limited deuterium uptake (Fig 4 in blue), i.e., less than 10% of the theoretical maximum (see Analysis of deuterium exchange). Regions in the vicinity of the Mg2+·ATP binding site, the active site tether (AST) loop that passes adjacent to the active site, and the regulator of G protein signaling homology (RH) domain were found to have increased amide hydrogen exchange, i.e. >50% of the theoretical maximum, (Fig. 4A). The highly conserved Mg2+·ATP binding motif (P–loop) was fully characterized by the R191VLGRGGFGEVF peptide. As expected from previous studies, the N–terminus was found to be highly disordered (>40% uptake). Incubation of GRK1 with Mg2+·ATP significantly decreased deuterium uptake in all peptide fragments, consistent with the dramatic thermostabilization observed for GRK1 upon binding ATP 26. The highly dynamic regions that were observed in apo–GRK1 exhibited exchange rates of less than 10% in most locations in the presence of Mg2+·ATP. The one region that still had more than 50% exchange was located in the RH domain between residues Leu126 and Leu147 (Fig. 4B), corresponding to the α6 helix and α6–α7 loop of the RH domain. The high deuterium exchange rates of this region are consistent with its relatively high temperature factors in crystal structures of GRK1 in complex with ATP 17. GRK1 autophosphorylation sites are found at residues Ser488 and Thr489, which are included in the F487STVKGVAFE peptide. This peptide showed increased deuterium uptake in the Mg2+·ATP bound form as compared to the apo–form (Fig. 2; Fig. 4A,B). Interestingly, deuterium uptake by the N–terminal 22 amino acids was also reduced by the addition of Mg2+·ATP, suggesting that this region is folded against the kinase more of the time.
Figure 4. Normalized hydrogen–deuterium uptake by GRK1 and ΔN–GRK1.
Color coding based on the normalized deuterium uptake was mapped onto the three dimensional model of GRK1 as described in the “Molecular modeling” section. Color coding is as described in Fig. 2. For ΔN–GRK1, the N–terminal helix was removed from the model. The normalized deuterium uptake for (A) apo–GRK1, (B) GRK1 in the presence of Mg2+·ATP, (C) apo–ΔN–GRK1, and (D) ΔN–GRK1 in the presence of Mg2+·ATP. Regions are colored grey where uptake was not determined because it was either below the detection threshold or peptide signals overlapped.
Dynamics of apo– and Mg2+·ATP loaded–ΔN–GRK1
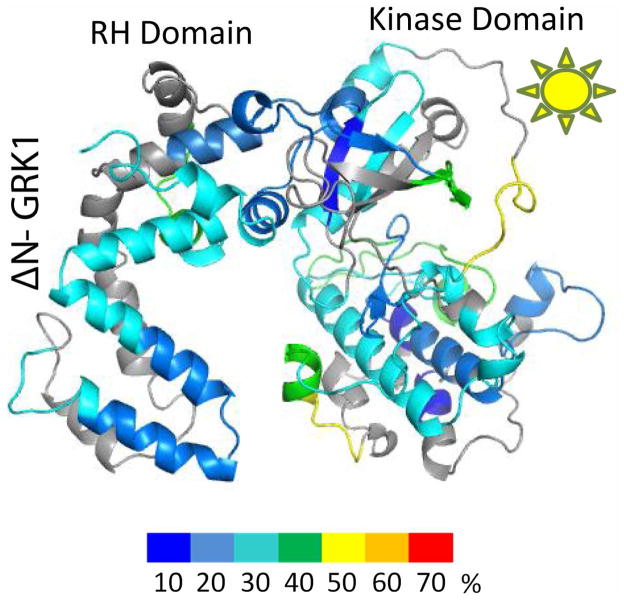
Apo–ΔN–GRK1 exhibited an deuterium uptake pattern similar to apo–GRK1 for most regions with the following exceptions. Arg453 to Gly463, which immediately precedes the AST loop, exhibited decreased flexibility in GRK1 after addition of Mg2+·ATP, but appeared unaffected by Mg2+·ATP binding in the ΔN–GRK1 molecule (Fig 4C, D). Arg191 to Phe202, the P–loop, remained highly dynamic even after Mg2+·ATP incubation with ΔN–GRK1. Two other peptides that were less affected in the ΔN–GRK1 molecule after exposure to Mg2+·ATP were Val234 to Phe246 within the active site, and His275 to Phe293, which docks with the AST region in the closed structure of GRK6. The proximity of these peptides to the active site suggests that the N–terminal region of GRK1 contributes to stabilization of the ATP–bound state of the enzyme. However, we were unable to measure a difference in thermostability between wild–type and ΔN–GRK1 either with or without Mg2+·ATP, nor were we able to show that ATP binds with a different affinity to these enzymes (Fig. S1 of the Supporting Information). Thus, the differences in deuterium exchange exhibited by wild–type and ΔN–GRK1 in the presence of Mg2+·ATP may reflect the ability of the kinase to sample the closed state (Fig. 1) rather than changes in affinity for nucleotide.
Remarkably, the peptide spanning the αJ helix, residues 452–464, exhibited much higher deuterium exchange in ΔN–GRK1 than GRK1, regardless of the presence of Mg2+·ATP, suggesting a more labile RH–kinase domain interface in ΔN–GRK1. Indeed, in one structure of GRK1 (PDB entry 3C50 17) the N–terminus, which is deleted in ΔN–GRK1, was observed to pack into the cavity between the RH and kinase domains. Thus, the N–terminus of GRK1 may help fix the RH–kinase domain interface when GRK1 is not engaged with a receptor or lipid membranes.
Dynamics of GRK1 in the presence of Rho and Rho*
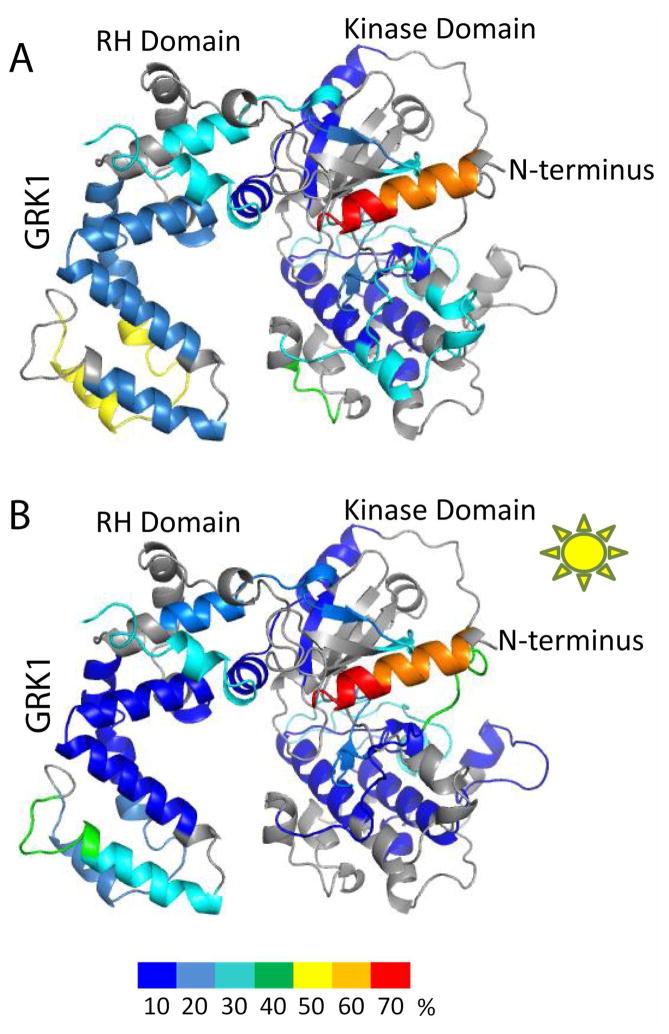
Addition of Rho to apo–GRK1 did not appear to induce dramatic changes in deuterium uptake, consistent with the fact that Rho is not in an activated state. Notably, no differences were observed for the N–terminal 22 amino acids (cf. Fig. 4A and Fig. 5A). Addition of Rho* to apo–GRK1 led to a generally decreased overall deuterium uptake compared to Rho, but not at the N–terminus (Fig. 5A, 5B). This result would be consistent with a requirement for adenine nucleotides or their analogs to locate in the active site pocket before the receptor docking competent state of GRK1 can be achieved. Interestingly, the N–terminal region (Asp7 to Phe22) exhibited higher deuterium exchange in either the presence of Rho or Rho* compared to apo–GRK1 (cf. Fig. 4A with Fig. 5A, B). The effects observed in this experiment could either be due to interactions of GRK1 with membranes, Rho, or Rho*. Thus, one interpretation is that the N–terminal region of GRK1 interacts non–specifically and transiently with Rho and/or lipids, as opposed to an intramolecular interaction, prior to activation by Rho*.
Figure 5. Normalized hydrogen–deuterium uptake by GRK1 in the presence of Rho and Rho*.
(A) Normalized deuterium uptake mapped on the model of GRK1 in the presence of Rho. (B) Normalized deuterium uptake in the presence of Rho*. Color coding is as described in Fig. 2.
Dynamics of Mg2+·ATP loaded GRK1 in the presence of Rho and Rho*
Here we observed reduced deuterium uptake by GRK1 in the presence of either Rho or Rho* (Fig. 6A, B). The differences between these states were negligible for most peptides studied. An exception was the region between Leu126 to Leu147, which is poorly ordered in most GRK1 crystal structures and exhibited many changes from experiment to experiment reported here. Thus it is difficult to assess whether the changes in this poorly conserved region are physiologically relevant. The presence of Rho* in Mg2+·ATP loaded GRK1 seemed to inhibit deuterium uptake in the N–terminal Ile16 to Phe22 peptide (Fig 6.A, 6.B). The difference in uptake was ~30% as compared to Mg2+·ATP–loaded GRK1 in the presence of Rho. In contrast, the uptake of the Asp7 to Phe15 peptide remained unaffected by light activation (Fig 6. A,B).
Figure 6. Normalized hydrogen–deuterium uptake of Mg2+·ATP loaded GRK1 in the presence of Rho and Rho*.
(A) Normalized deuterium uptake mapped on the model of GRK1 in the presence of Mg2+·ATP and Rho. (B) Normalized deuterium uptake in the presence of Mg2+·ATP and Rho*. Color coding is as described in Fig. 2.
Dynamics of ΔN–GRK1 in the presence of Rho*
As with apo–GRK1, addition of Rho* in ROS reduced the flexibility of ΔN–GRK1 (Fig. 7) but did not produce the same pattern of selective effects observed in GRK1. This could be attributed in part to the inability of the ΔN–GRK1 molecule to interact efficiently with Rho*. In the case of ΔN–GRK1, the missing N–terminus affected deuterium uptake, not only in regions located far from the N–terminus but also in the P–loop (residues Arg191 to Phe202), as observed for ΔN–GRK1 in other liganded states (Fig. 4C and D, Fig. 8A).
Figure 7. Normalized hydrogen–deuterium uptake by ΔN–GRK1 in the presence of Rho*.
Color coding is as described in Fig. 2.
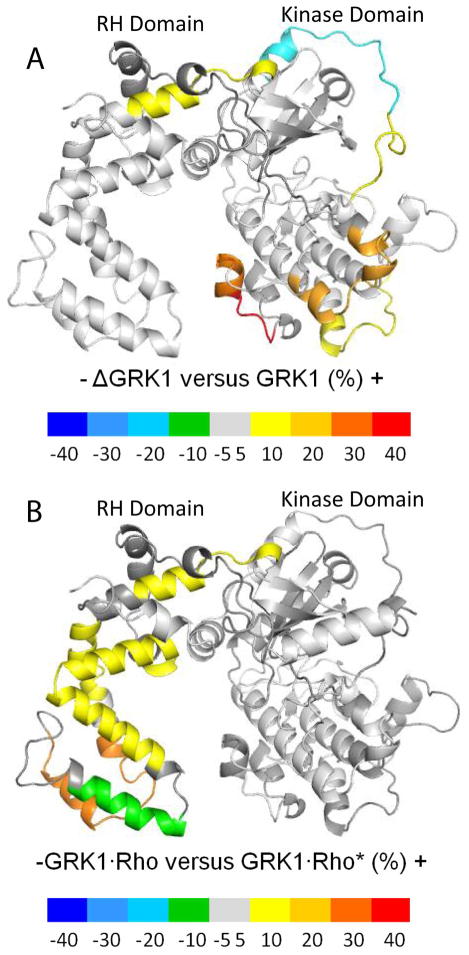
Figure 8. Differences in normalized hydrogen–deuterium exchange.
(A) Differences in normalized deuterium uptake between ΔN–GRK1 and apo–GRK1. (B) Differences in normalized deuterium uptake between GRK1 in the presence of either Rho or Rho*. Color coding was assigned based on the value of the difference between the normalized deuterium uptake values between the states shown. Negative differences (associated to blue, cyan, light blue, and green) denote higher uptake in the case of GRK1 (panel A) or when Rho* is present (panel B). Positive differences (associated to yellow, orange, and red) denote higher uptakes in ΔN–GRK1 (panel A) or when Rho is present (panel B). Regions that have absolute differences less than 10% or were not included in the analysis are depicted in grey.
DISCUSSION
In the current study, we found that residues 1–535 of GRK1 contain regions of considerable flexibility, as do many globular proteins, and that this flexibility was reduced in the presence of either of its substrates: the nucleotide complex Mg2+·ATP and/or the protein Rho*. Stability was further enhanced when both substrates were added. A truncated form of GRK1 missing the first 19 amino acid residues, ΔN–GRK1, failed to display most of these effects, implying that the first 19 amino acids help to dictate the overall dynamics of GRK1 in the presence of either Mg2+·ATP or Rho*.
Effects of Rho or Rho* on GRK1 dynamics
We noted that deuterium uptake by GRK1 (Fig. 4A) differed from the pattern observed when dark–adapted Rho purified from membranes was added to the mix (Fig 5A). However, GRK1 should selectively recognize only Rho*. Thus the observed differences could reflect a residual interaction of GRK1 with Rho or with the membrane. Upon exposure to light, however, we expected that GRK1 would interact with Rho* which in turn would either cause reduced flexibility of the GRK1 secondary structure or directly hinder deuterium update in segments involved in their interface. Indeed, we did find a decrease in deuterium uptake in GRK1 following addition of Rho* when compared to Rho (Fig. 8B). One N–terminal peptide (Ile16 to Phe22) showed decreased uptake (~30%) when GRK1 was incubated in the presence of Mg2+·ATP and Rho* when compared to GRK1 in the presence of Rho or Rho* alone. However, the other N–terminal peptide (Asp7 to Phe15) failed to show large differences expected to result from binding site protection. These results could be due to the high flexibility of this region, or to the fact that GRK1 is thought to rapidly dissociate from Rho*, such that the differences in deuterium exchange are too small to be detected under the conditions used for our experiments. Indeed, the interaction of GRK1 with Rho* is not as stable as that of transducin 27 and isolation of a stable GRK1–Rho* complex has not yet been reported. Moreover, GRK1 does not form so–called “extra–Meta II” wherein the Meta II state of Rho* is stabilized at the expense of other photoactivation products 27, 28. In contrast, a stable complex between Rho* and transducin has been isolated biochemically and visualized by electron microscopy using single particle reconstruction 29, 30. Therefore, consistent with our hydrogen deuterium exchange measurements, there seems to be only a small fraction of GRK1 bound to Rho* at any given time.
When the dynamics of GRK1 and Rho/Rho* were evaluated in the presence of Mg2+·ATP, the differences between those states were found to be negligible. Once Mg2+·ATP is bound, all other parameters that could affect deuterium uptake, such as the presence of lipid bilayers in ROS and/or Rho*, have minimal influence. However, there were a few regions where deuterium uptake did not follow this simple trend. For example, the Leu126 to Leu147 region is one of the most flexible regions in the GRK1 molecule, and even when Mg2+·ATP was bound this region exchanged up to 40–50% of the theoretical maximum (Fig. 4). This held true for all our preparations of GRK1 (>50% uptake), except when Rho* was present and Mg2+·ATP was absent (~20%) (Fig. 5B). The decrease in deuterium uptake following addition of Mg2+·ATP in peptides that encompass the active site, such as the P–loop, supports the view that ATP is binding as predicted by prior crystal structures. Furthermore, we could identify allosterically coupled regions that exhibit decreased uptake upon binding Mg2+·ATP such as that encompassed by Asp100 to Ala112 (Figs. 4 and 6).
When the N–terminal 19 amino acids of GRK1 were removed to create ΔN–GRK1, the most dramatic effect in the hydrogen deuterium uptake profile compared to GRK1 was localized to a single region between Arg453 and Met464 (Fig. 4C and D). This region is located at the interface between the kinase large lobe and the RH domain. It is possible that the increased deuterium uptake is induced by the rigid body movement of the kinase large lobe away from the RH domain, which is somehow favored in the absence of the N–terminal region. The presence of Rho* did reduce the flexibility in ΔN–GRK1 (Fig. 7) to the extent seen in the case of GRK1 when bound to Rho*, suggesting that the N–terminal region of GRK1 is not the only region that interacts with light–activated ROS. Although the binding of ATP seems to stabilize backbone atoms in the N–terminal region of GRK1 (Fig. 4A,B), this change is not associated with any measurable difference in thermostability or affinity of ΔN–GRK1 towards ATP (Fig. S1 of the Supporting Information). Indeed, multiple crystal structures have been determined for GRK1 in the presence of ATP or ADP, yet the N–terminal region has not been routinely observed nor has the kinase assumed what is considered to be a more active conformation 11, 17.
Before this study, GRK1 activation was proposed to involve three distinctive steps: docking of its N–terminal helix to the receptor, interaction of this helix with the kinase domain, and kinase domain closure. These events are likely highly coupled. Our deuterium exchange results suggest that, as expected, the GRK1 molecule becomes more rigid following binding of substrates such as Mg2+·ATP or Rho*. This agrees with the increased order and lower temperature factors in liganded states of GRK1 determined in prior crystallographic studies of GRK1 17. ΔN–GRK1 did not show the same deuterium exchange pattern as intact GRK1 for all its substrates, probably because ΔN–GRK1 is unable to carry out all of the required steps believed to result in sampling the closed, more active and presumably less dynamic conformation of this kinase. Thus, our results confirm that the N–terminal region in GRKs plays a multi–faceted role that could involve modulation of the interdomain contacts of GRK1 as well as interactions with membrane lipid interactions 31 and receptor binding 12, 32.
Supplementary Material
Acknowledgments
We would like to thank Dr. Marcin Golczak for help with the MS. We also thank Dr. L.T. Webster, Jr. and members of the Palczewski laboratory for critical comments on the manuscript.
ABBREVIATIONS
- AST
active site tether
- BTP
Bis–Tris–propane
- DTT
dithiothreitol
- DXMS
deuterium exchange mass spectrometry
- GRK1
G protein–coupled rhodopsin kinase 1
- ΔN–GRK1
GRK1 lacking the first 19 amino acids
- HEPES
4–(2–hydroxyethyl)–1–piperazineethanesulfonic acid
- MS
mass spectrometry
- MS2
tandem MS/MS
- RH
regulator of G protein signaling homology
- Rho
rhodopsin
- Rho*
photoactivated Rho
Footnotes
This research was supported in part by grants EY008061 and HL071818 from the National Institutes of Health. KP is John H. Hord Professor of Pharmacology.
SUPPORTING INFORMATION AVAILABLE
Supporting Information includes Figure S1, which compares the thermostability and ATP affinity of GRK1 and ΔN–GRK1. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Palczewski K. G protein–coupled receptor rhodopsin. Annu Rev Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maeda T, Imanishi Y, Palczewski K. Rhodopsin phosphorylation: 30 years later. Prog Retinal Eye Res. 2003;22:417–434. doi: 10.1016/s1350-9462(03)00017-x. [DOI] [PubMed] [Google Scholar]
- 3.Arshavsky VY, Lamb TD, Pugh EN., Jr G proteins and phototransduction. Annu Rev Physiol. 2002;64:153–187. doi: 10.1146/annurev.physiol.64.082701.102229. [DOI] [PubMed] [Google Scholar]
- 4.Wilden U, Hall SW, Kuhn H. Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48–kDa protein of rod outer segments. Proc Natl Acad Sci U S A. 1986;83:1174–1178. doi: 10.1073/pnas.83.5.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palczewski K, Buczylko J, Kaplan MW, Polans AS, Crabb JW. Mechanism of rhodopsin kinase activation. J Biol Chem. 1991;266:12949–12955. [PubMed] [Google Scholar]
- 6.Ohguro H, Rudnicka–Nawrot M, Buczylko J, Zhao X, Taylor JA, Walsh KA, Palczewski K. Structural and enzymatic aspects of rhodopsin phosphorylation. J Biol Chem. 1996;271:5215–5224. doi: 10.1074/jbc.271.9.5215. [DOI] [PubMed] [Google Scholar]
- 7.Ohguro H, Van Hooser JP, Milam AH, Palczewski K. Rhodopsin phosphorylation and dephosphorylation in vivo. J Biol Chem. 1995;270:14259–14262. doi: 10.1074/jbc.270.24.14259. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto S, Sippel KC, Berson EL, Dryja TP. Defects in the rhodopsin kinase gene in the Oguchi form of stationary night blindness. Nature Genet. 1997;15:175–178. doi: 10.1038/ng0297-175. [DOI] [PubMed] [Google Scholar]
- 9.Cideciyan AV, Zhao X, Nielsen L, Khani SC, Jacobson SG, Palczewski K. Null mutation in the rhodopsin kinase gene slows recovery kinetics of rod and cone phototransduction in man. Proc Natl Acad Sci U S A. 1998;95:328–333. doi: 10.1073/pnas.95.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen CK, Burns ME, Spencer M, Niemi GA, Chen J, Hurley JB, Baylor DA, Simon MI. Abnormal photoresponses and light–induced apoptosis in rods lacking rhodopsin kinase. Proc Natl Acad Sci U S A. 1999;96:3718–3722. doi: 10.1073/pnas.96.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang CC, Orban T, Jastrzebska B, Palczewski K, Tesmer JJ. Activation of G protein–coupled receptor kinase 1 involves interactions between its N–terminal region and its kinase domain. Biochemistry. 2011;50:1940–1949. doi: 10.1021/bi101606e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palczewski K, Buczylko J, Lebioda L, Crabb JW, Polans AS. Identification of the N–terminal region in rhodopsin kinase involved in its interaction with rhodopsin. J Biol Chem. 1993;268:6004–6013. [PubMed] [Google Scholar]
- 13.Yu QM, Cheng ZJ, Gan XQ, Bao GB, Li L, Pei G. The amino terminus with a conserved glutamic acid of G protein–coupled receptor kinases is indispensable for their ability to phosphorylate photoactivated rhodopsin. J Neurochem. 1999;73:1222–1227. doi: 10.1046/j.1471-4159.1999.0731222.x. [DOI] [PubMed] [Google Scholar]
- 14.Noble B, Kallal LA, Pausch MH, Benovic JL. Development of a yeast bioassay to characterize G protein–coupled receptor kinases. Identification of an NH2–terminal region essential for receptor phosphorylation. J Biol Chem. 2003;278:47466–47476. doi: 10.1074/jbc.M308257200. [DOI] [PubMed] [Google Scholar]
- 15.Boguth CA, Singh P, Huang CC, Tesmer JJ. Molecular basis for activation of G protein–coupled receptor kinases. EMBO J. 2010;29:3249–3259. doi: 10.1038/emboj.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang CC, Yoshino–Koh K, Tesmer JJ. A surface of the kinase domain critical for the allosteric activation of G protein–coupled receptor kinases. J Biol Chem. 2009;284:17206–17215. doi: 10.1074/jbc.M809544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh P, Wang B, Maeda T, Palczewski K, Tesmer JJ. Structures of rhodopsin kinase in different ligand states reveal key elements involved in G protein–coupled receptor kinase activation. J Biol Chem. 2008;283:14053–14062. doi: 10.1074/jbc.M708974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews RG, Hubbard R, Brown PK, Wald G. Tautomeric Forms of Metarhodopsin. J Gen Physiol. 1963;47:215–240. doi: 10.1085/jgp.47.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H, Freitas MA. MassMatrix: a database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics. 2009;9:1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu H, Freitas MA. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinform. 2007;8:133. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weis DD, Kass IJ, Engen JR. Semi–automated analysis of hydrogen exchange mass spectra using HX–Express. J Amer Soc Mass Spectrom. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 22.Orban T, Bereta G, Miyagi M, Wang B, Chance MR, Sousa MC, Palczewski K. Conformational changes in guanylate cyclase–activating protein 1 induced by Ca2+ and N–terminal fatty acid acylation. Structure. 2010;18:116–126. doi: 10.1016/j.str.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera––a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 24.Marti–Renom MA, Stuart A, Fiser A, Sánchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 25.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 26.Thal DM, Yeow RY, Schoenau C, Huber J, Tesmer JJ. Molecular mechanism of selectivity among G protein–coupled receptor kinase 2 inhibitors. Mol Pharmacol. 2011;80:294–303. doi: 10.1124/mol.111.071522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pulvermuller A, Palczewski K, Hofmann KP. Interaction between photoactivated rhodopsin and its kinase: stability and kinetics of complex formation. Biochemistry. 1993;32:14082–14088. doi: 10.1021/bi00214a002. [DOI] [PubMed] [Google Scholar]
- 28.Hofmann KP, Pulvermuller A, Buczylko J, Van Hooser P, Palczewski K. The role of arrestin and retinoids in the regeneration pathway of rhodopsin. J Biol Chem. 1992;267:15701–15706. [PubMed] [Google Scholar]
- 29.Jastrzebska B, Tsybovsky Y, Palczewski K. Complexes between photoactivated rhodopsin and transducin: progress and questions. Biochem J. 2010;428:1–10. doi: 10.1042/BJ20100270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jastrzebska B, Ringler P, Lodowski DT, Moiseenkova–Bell V, Golczak M, Muller SA, Palczewski K, Engel A. Rhodopsin–transducin heteropentamer: three–dimensional structure and biochemical characterization. J Struct Biol. 2011;176:387–394. doi: 10.1016/j.jsb.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pao CS, Barker BL, Benovic JL. Role of the amino terminus of G protein–coupled receptor kinase 2 in receptor phosphorylation. Biochemistry. 2009;48:7325–7333. doi: 10.1021/bi900408g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boguth CA, Singh P, Huang CC, Tesmer JJ. Molecular basis for activation of G protein–coupled receptor kinases. EMBO J. 2010;29:3249–3259. doi: 10.1038/emboj.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang CC, Tesmer JJ. Recognition in the face of diversity: interactions of heterotrimeric G proteins and G protein–coupled receptor (GPCR) kinases with activated GPCRs. J Biol Chem. 286:7715–7721. doi: 10.1074/jbc.R109.051847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knepp AM, Periole X, Marrink SJ, Sakmar TP, Huber T. Rhodopsin forms a dimer with cytoplasmic helix 8 contacts in native membranes. Biochemistry. 2012;51:1819–1821. doi: 10.1021/bi3001598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gurevich VV, Gurevich EV. GPCR monomers and oligomers: it takes all kinds. Trends Neurosci. 2008;31:74–81. doi: 10.1016/j.tins.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.