Abstract
Gram-negative bacteria expel various toxic chemicals via tripartite efflux pumps belonging to the resistance-nodulation-cell division (RND) superfamily. These pumps span both the inner and outer membranes of the cell. The three components of these tripartite systems are an inner membrane, substrate-binding transporter (or pump); a periplasmic membrane fusion protein (or adaptor); and an outer membrane-anchored channel. These three efflux proteins interact in the periplasmic space to form the three-part complexes. We previously presented the crystal structures of both the inner membrane transporter CusA and membrane fusion protein CusB of the CusCBA tripartite efflux system from Escherichia coli. We also described the co-crystal structure of the CusBA adaptor-transporter, revealing the trimeric CusA efflux pump assembles with six CusB protein molecules to form the complex CusB6-CusA3. We here report three different conformers of the crystal structures of CusBA-Cu(I), suggesting a mechanism on how Cu(I) binding initiates a sequence of conformational transitions in the transport cycle. Genetic analysis and transport assays indicate that charged residues, in addition to the methionine pairs and clusters, are essential for extruding metal ions out of the cell.
INTRODUCTION
Efflux pumps of the RND superfamily are integral membrane proteins that derive energy from the proton motive force. In Gram-negative bacteria, these pumps play major roles in the intrinsic and acquired tolerance of antibiotics and toxic compounds.1,2 The E. coli CusA heavy-metal efflux pump is a large inner membrane RND protein that is responsible for extruding Cu(I) and Ag(I) ions, which are biocidal.3,4 CusA operates with a periplasmic membrane fusion protein, CusB, and an outer membrane channel, CusC, to form a functional protein complex. The resulting CusCBA tripartite efflux system spans the entire cell envelope and confers resistance to Cu(I) and Ag(I) by exporting these metal ions directly out of the cell.3,4
The crystal structures of individual components of this three-part complex system have been determined. The structure of CusA suggests that this RND pump exists as a homotrimer.5 Each subunit of CusA consists of 12 transmembrane helices (TM1–TM12) and a large periplasmic domain formed by two periplasmic loops between TM1 and TM2, and TM7 and TM8, respectively. The periplasmic domain of CusA can be divided into a pore domain (comprising sub-domains PN1, PN2, PC1 and PC2) and a CusC docking domain (containing sub-domains DN and DC). The structures indicate that this transporter utilizes methionine pairs and clusters to bind and export Cu(I) and Ag(I) ions.5,6
Overall, the structure of CusB demonstrates that this adaptor protein is folded into a four-domain elongated structure, ~120 Å long and ~40 Å wide.7 The first three domains (domains 1–3) of the protein are mostly β-strands. However, the fourth domain (domain 4) is all α-helices and is folded into a three-helix bundle structure. Interestingly, the co-crystal structure of the CusBA adaptor-transporter reveals that the trimeric CusA pump associates with six CusB molecules to form the CusB6-CusA3 complex.8 Thus, the entire tripartite efflux assembly is expected to be in the form of CusC3-CusB6-CusA3, which span both the inner and outer membranes of E. coli to export Cu(I) and Ag(I) ions. This assemblage is indeed in good agreement with the predicted 3:6:3 polypeptide ratios of these three-part complexes.9,10
Recently, the crystal structure of the CusC channel has also been resolved,11 suggesting that this protein resembles the architectures of TolC12 and OprM.13 The trimeric CusC channel consists of a membrane-anchoring β-barrel domain and an elongated periplasmic α-helical tunnel.11 The periplasmic tunnel is ~100 Å long with an outermost diameter of ~35 Å at the tip of the tunnel.
In the absence of the CusB adaptor, two distinct structures of CusA were obtained by x-ray crystallography.5 These structures probably capture two different conformational states of the pump in the transport cycle. One key feature of the CusA pump is that its external periplasmic cleft, formed by subdomains PC1 and PC2, can be open and closed. In the absence of Cu(I) or Ag(I), the cleft is closed. However, in the presence of Cu(I) or Ag(I), this cleft becomes open. The bound Cu(I) or Ag(I) is found to coordinate at the center of a three-methionine binding site, formed by M573, M623 and M672.5 This methionine triad is probably responsible for the selectivity of the pump.
The apo-CusA conformation should represent the “resting” state where the external periplasmic cleft is closed. However, the Cu(I) and Ag(I)-bound CusA structures should correspond to the “binding” state where the periplasmic cleft is open. Upon metal ion transport, the pump must go through other transient states to actively remove the metal ions. Here, we present new crystal structures of the CusBA-Cu(I) efflux complexes, which represent different intermediates transitioning between the “binding” and “resting” states in the transport cycle. The structures also indicate that the conserved charged residues located at the periplasmic domain of CusA are essential for the transport of metal ions.
RESULTS
Crystal structures of the CusBA-Cu(I) complexes
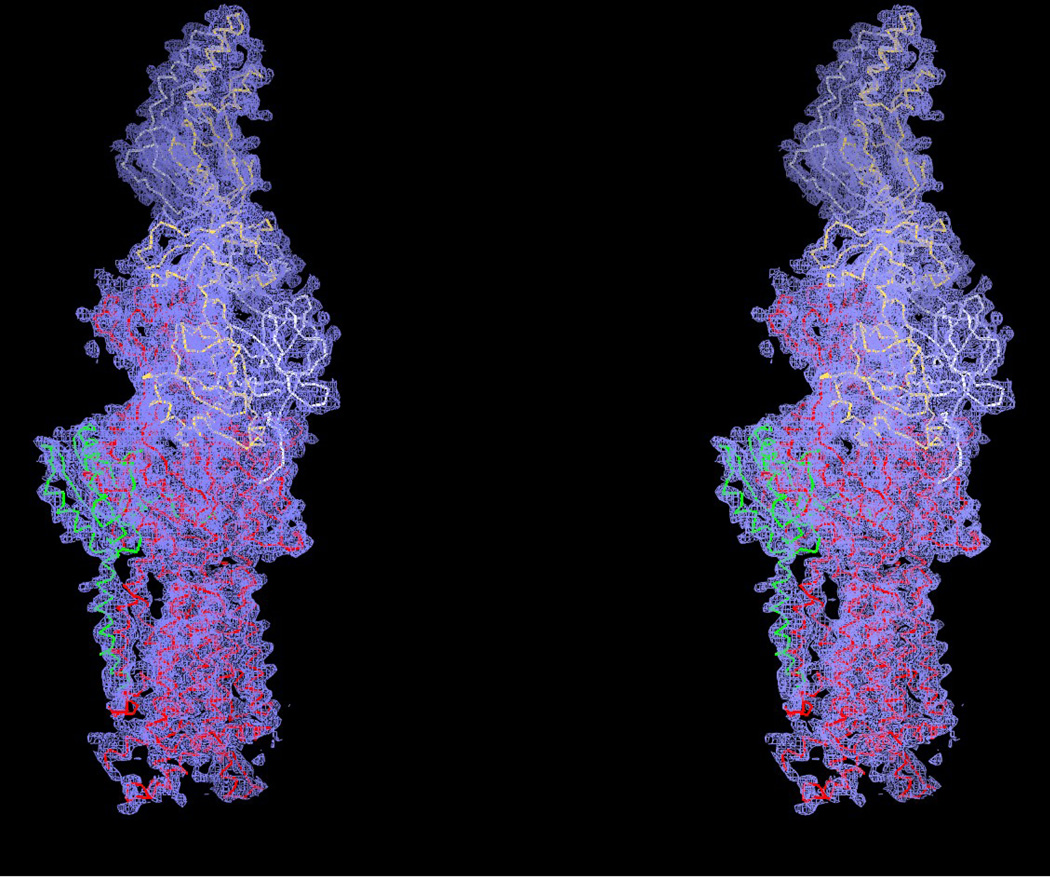
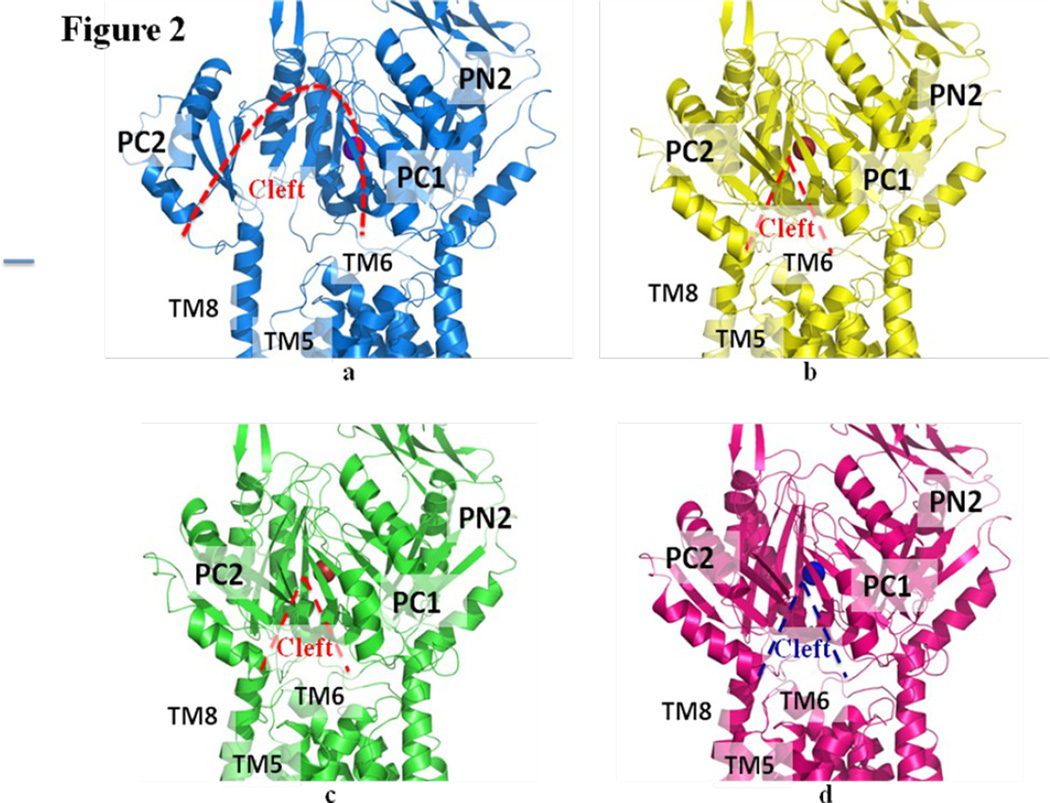
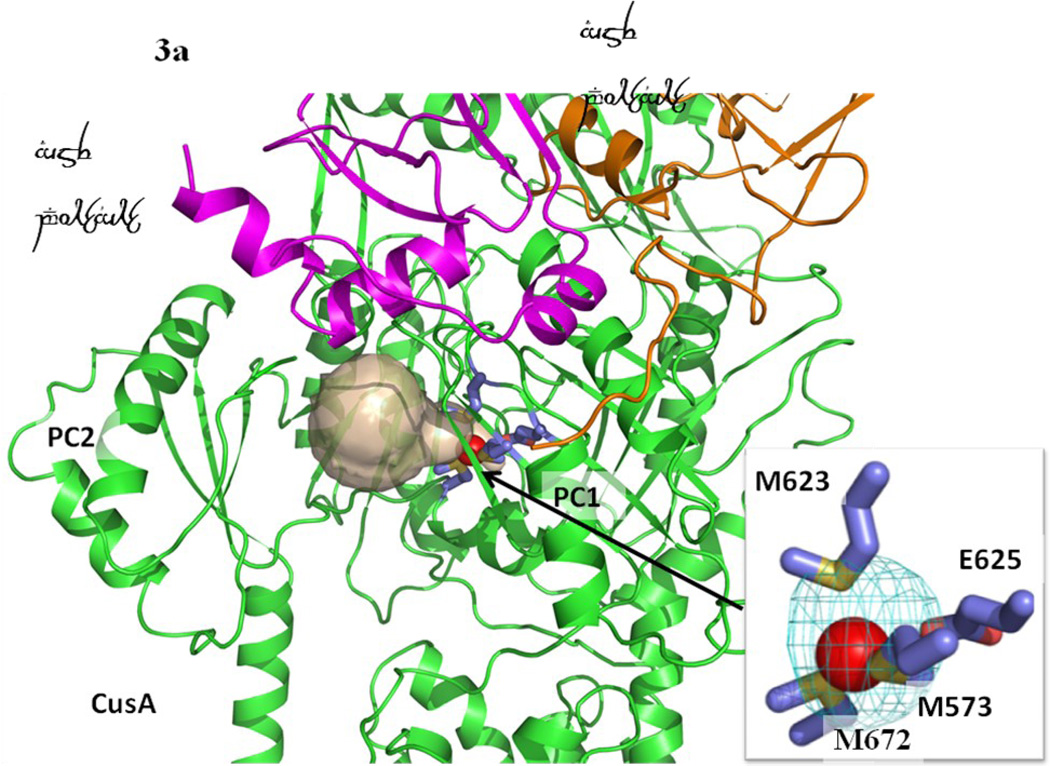
There are three distinct conformations of CusBA-Cu(I) based on the crystal structures, and all these structures contain one CusA and two CusB protomers in the asymmetric unit (Table 1). One of the structures, designed as form I, constitutes two mixed conformations within a single Cα chain of CusA. The structure reveals that the Cα chain of residues 664–717 and 814–888, which contribute to form the horizontal helix, subdomain PC2 and TM8 of CusA, displays two distinct conformations (forms Ia and Ib) (Fig. 1). The occupancy distribution of forms Ia and Ib are in the ratio of 0.22:0.78. The conformation of the CusA molecule in form Ia appears to be similar to but is also different from the CusA-Cu(I) structure. Like the “binding” conformation, the periplasmic cleft is open in the form Ia structure (Figs. 1 and 2). The conformation of CusA in form Ib seems to be closer to but is distinct from the apo-CusA structure of the pump. In this conformation, the periplasmic cleft is closed, similar to that of the “resting” state. In both forms Ia and Ib, a single bound Cu(I) is found to coordinate residues M573, M623 and M672, which forms the distinct three-methionine ion binding site inside the cleft of the periplasmic domain of CusA. Surprisingly, the nearby conserved charged residue E625 seems to be involved in the binding (Fig. 3). In comparison with the structure of the “binding” state, the horizontal helix, formed by residues 665–675, inside the cleft of the structure of form Ib is significantly lower in position (Fig. 4). The change can be interpreted as a 10° downward tilting motion of the C-terminal end of the horizontal helix in this structure when compared with the CusA-Cu(I) form. This motion also shifts M672 away from M573 and M623, seemingly to disassemble the three-sulfur coordination site and to release the bound Cu(I) ion from this site. The change of the horizontal helix is less obvious in the form Ia structure, but it can be interpreted as a 3° downward tilt at the C-terminal end in comparison with that of CusA-Cu(I). In addition, the transmembrane helices 5 and 6 (residues 447–495) are found to shift downward by 5 Å with respect to the membrane plane in both forms Ia and Ib, mimicking the change in position of the horizontal helix, when compared with the conformation of the “binding” state (Fig. 4). In contrast to the “binding” and “resting” structures, no continuous channel is found in the CusA protomer of forms Ia and Ib as indicated by the program CAVER (http://loschmidt.chemi.muni.cz/caver) (Fig. 3a and b). Because the conformations of forms Ia and Ib are different from those of the “binding” and “resting” forms, these conformations are designated as “pre-extrusion 1” and “pre-extrusion 2” states, respectively.
Table 1.
Data collection and structural refinement statistics of the CusBA-Cu(I) complexes.
| Form 1 “Pre-extrusions 1 and 2 mixed states” |
Form 2 “Pre-extrusion 2 state” |
Form 3 “Extrusion state” |
CusB-D405A- Cu(I) |
CusB-R669A- Cu(I) |
|
|---|---|---|---|---|---|
| Data Collection | |||||
| Space group | R32 | R32 | R32 | R32 | R32 |
| Cell dimensions | |||||
| a, b, c (Å) | 160.34, 160.34, 685.07 |
159.64, 159.64, 679.83 |
159.79, 139.79, 683.09 |
159.64, 159.64, 681.10 |
159.79, 159.79, 683.09 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Wavelength (Å) | 1.37 | 0.979 | 0.979 | 0.979 | 0.979 |
| Resolution (Å) | 50-3.42 (3.51-3.45) |
50-3.90 (4.00-3.90) |
50-3.30 (3.39-3.30) |
50-3.10 (3.18-3.10) |
50-4.20 (4.31-4.20) |
| Rpim (%)a | 8.8 (45.6) | 10.2 (46.7) | 6.4 (47.2) | 6.6 (41.0) | 12.6 (31.9) |
| Average I/σI | 8.9 (2.0) | 6.8 (2.0) | 11.0 (1.9) | 10.3 (2.5) | 6.4 (2.8) |
| Completeness (%) | 99.9 (100) | 93.8 (95.1) | 100 (100) | 100 (100) | 99.9 (100) |
| Redundancy | 5.3 (5.4) | 5.7 (5.8) | 12.4 (12.4) | 5.3 (5.4) | 5.2 (5.4) |
| Total reflections | 239,469 | 164,514 | 638,674 | 326,934 | 131,349 |
| Unique reflections | 45,301 | 28,809 | 51,324 | 61,292 | 25,268 |
| Refinement | |||||
| PDB code | 3T56 | 3T51 | 3T53 | 4DNT | 4DOP |
| Resolution (Å) | 50-3.45 | 50-3.90 | 50-3.30 | 50-3.10 | 50-4.20 |
| No. reflections | 41,030 | 26,937 | 42,148 | 59,909 | 23,337 |
| Rwork/Rfree | 25.9/29.4 | 25.8/32.4 | 24.7/28.5 | 21.9/24.4 | 22.1/26.8 |
| R.m.s deviations | |||||
| Bond lengths (Å) | 0.003 | 0.004 | 0.003 | 0.003 | 0.003 |
| Bond angles (°) | 0.747 | 0.724 | 0.619 | 0.675 | 0.617 |
| Ramachandran | |||||
| most favored | 84.4 | 78.5 | 86.1 | 87.9 | 85.8 |
| additional allowed | 14.7 | 20.5 | 13.6 | 11.8 | 13.7 |
| generously allowed | 0.9 | 0.9 | 0.3 | 0.3 | 0.4 |
| disallowed | 0.1 | 0.1 | 0 | 0 | 0.1 |
Because of the high multiplicity, Rpim (precision indicating merging R-factor)22 was used to obtain the high-resolution cut-off.
Figure 1.
Stereo view of the electron density map at a resolution of 3.45 Å. This is a 2Fo – Fc electron density map (blue mesh) contoured at 1.0 σ. The Cα traces of residues 664-717 and 814–888, showing a distinct conformation of the structure of form Ia are in green. The Cα traces of form Ib of CusA, and molecules 1 and 2 of CusB are in red, white and yellow, respectively.
Figure 2.
Structures of the CusBA-Cu(I) efflux complexes. (a) Ribbon diagram of the structure of form Ia (blue) viewed in the membrane plane. The bound copper is shown as a purple sphere. This structure represents the “pre-extrusion 1” state of the pump. (b) Ribbon diagram of the structure of form Ib (yellow) viewed in the membrane plane. The bound copper is shown as a red sphere. This structure represents the “pre-extrusion 2” state of the pump. (c) Ribbon diagram of the structure of form II (green) viewed in the membrane plane. The bound copper is shown as a red sphere. This structure represents the “pre-extrusion 2” state of the pump. (d) Ribbon diagram of the structure of form III (magenta) viewed in the membrane plane. The bound copper is shown as a blue sphere. This structure represents the “extrusion” state of the pump. For clarity, only the periplasmic domain (subdomains PN2, PC1 and PC2) and part of the transmembrane region of CusA are shown in (a)–(d). The molecules of CusB in each complex are not included.
Figure 3.
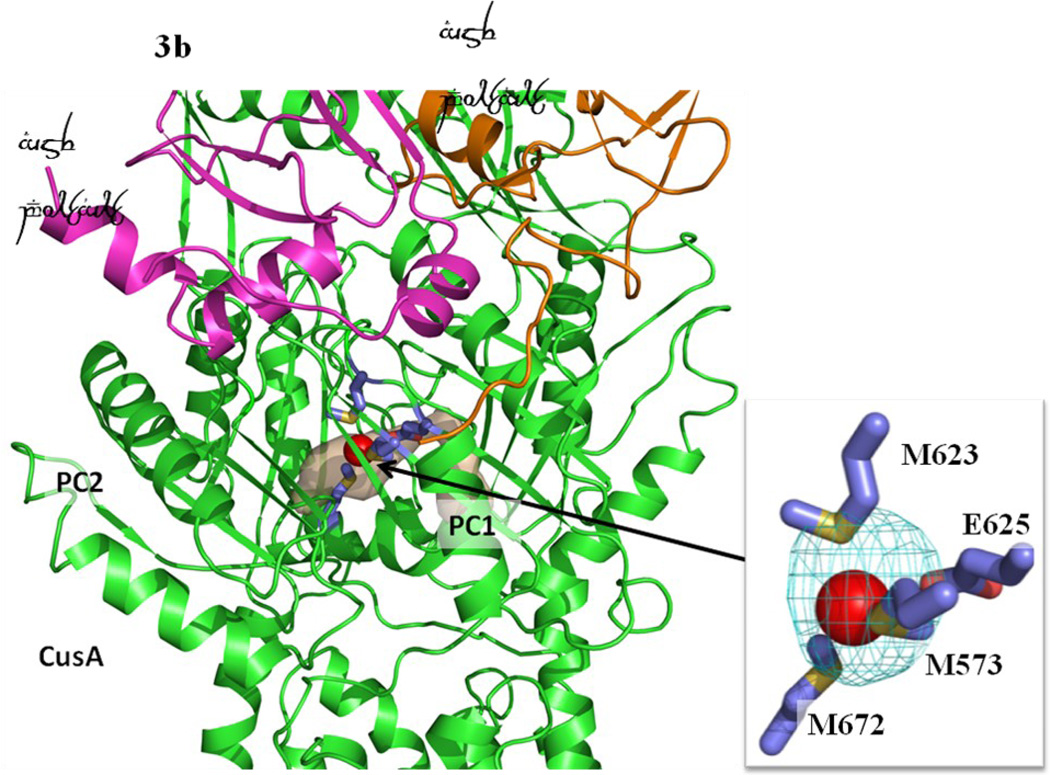
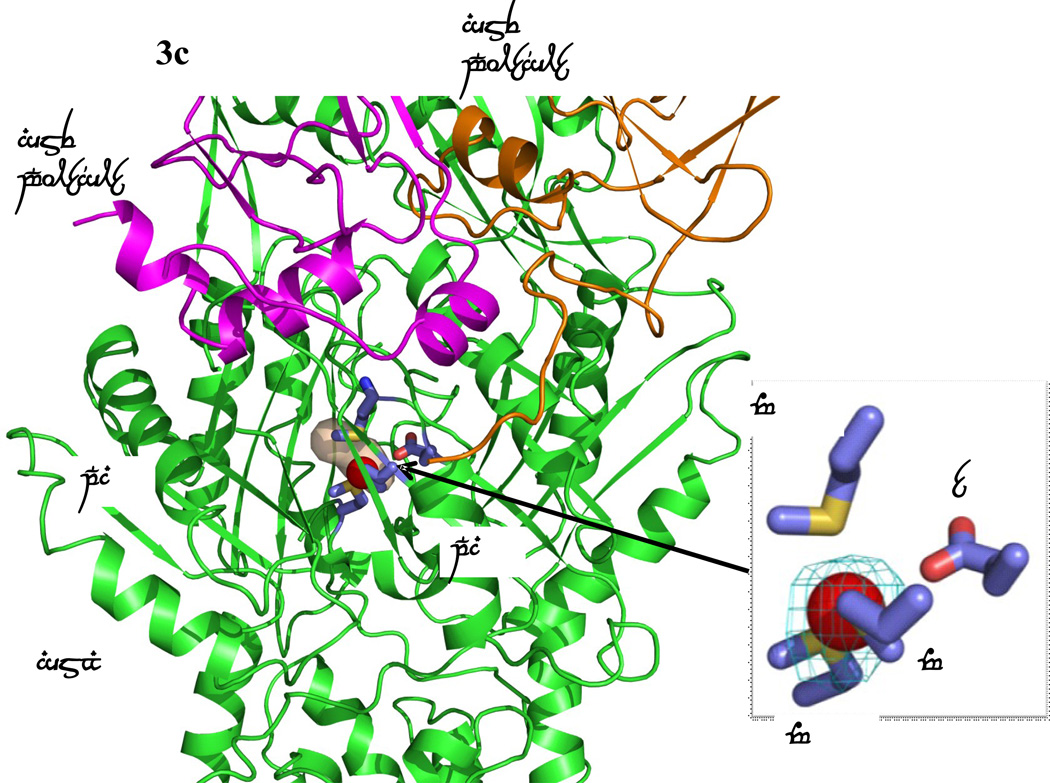
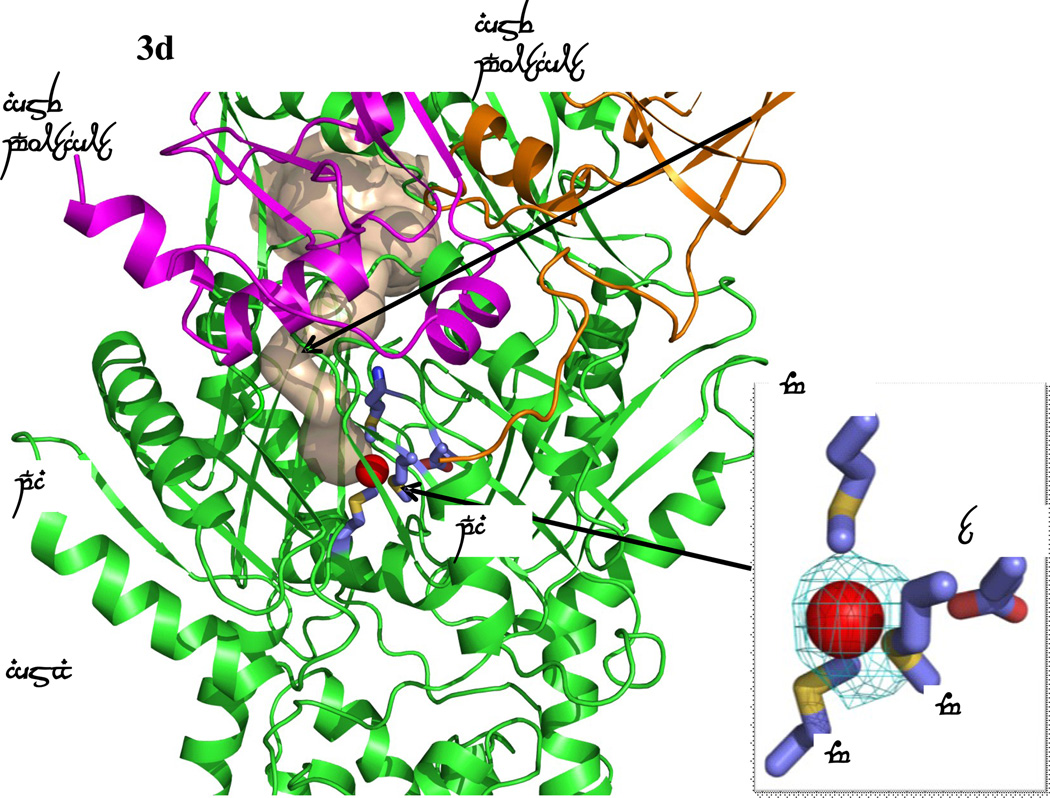
Formation of channels at different conformational states. (a) The channel formed by a protomer of the form Ia structure of CusBA at the periplasmic domain is in brown color. The bound Cu(I) is shown as a red sphere. M573, M623, M672 and E625, which form a Cu(I) binding site, are shown as sticks (slate, carbon; blue, nitrogen; red, oxygen). The CusA and two CusB protomers are in green, orange and magenta ribbons, respectively. M573, M623, M672 and E625, which form a Cu(I) binding site of the form Ia conformation, are shown as sticks (slate, carbon; blue, nitrogen; red, oxygen). (b) The channel formed by a protomer of the form Ib structure of CusBA at the periplasmic domain is in brown color. The bound Cu(I) is shown as a red sphere. Anomalous map of the bound Cu(I), contoured at 3σ, is in cyan. M573, M623, M672 and E625, which form a Cu(I) binding site of the form Ib conformation, are shown as sticks (slate, carbon; blue, nitrogen; red, oxygen). The CusA and two CusB protomers are in green, orange and pink ribbons, respectively. (c) The channel formed by a protomer of the form II structure of CusBA at the periplasmic domain is in brown color. The bound Cu(I) ions are shown as a red sphere. Anomalous map of bound Cu(I) ion, contoured at 3σ, is in cyan. Residues forming the Cu(I) binding sites M573, M623, E625 and M672 are shown as sticks (slate, carbon; blue, nitrogen; red, oxygen). The CusA and two CusB protomers are in green, orange and magenta ribbons, respectively. (d) The channel formed by a protomer of the form III structure of CusBA at the periplasmic domain is in brown color. The bound Cu(I) is shown as a red sphere. Anomalous map of the bound Cu(I), contoured at 3σ, is in cyan. M573, M623, E625 and M672, which form the Cu(I) binding site, are shown as sticks (slate, carbon; blue, nitrogen; red, oxygen). The CusA and two CusB protomers are in green, orange and magenta ribbons, respectively.
Figure 4.
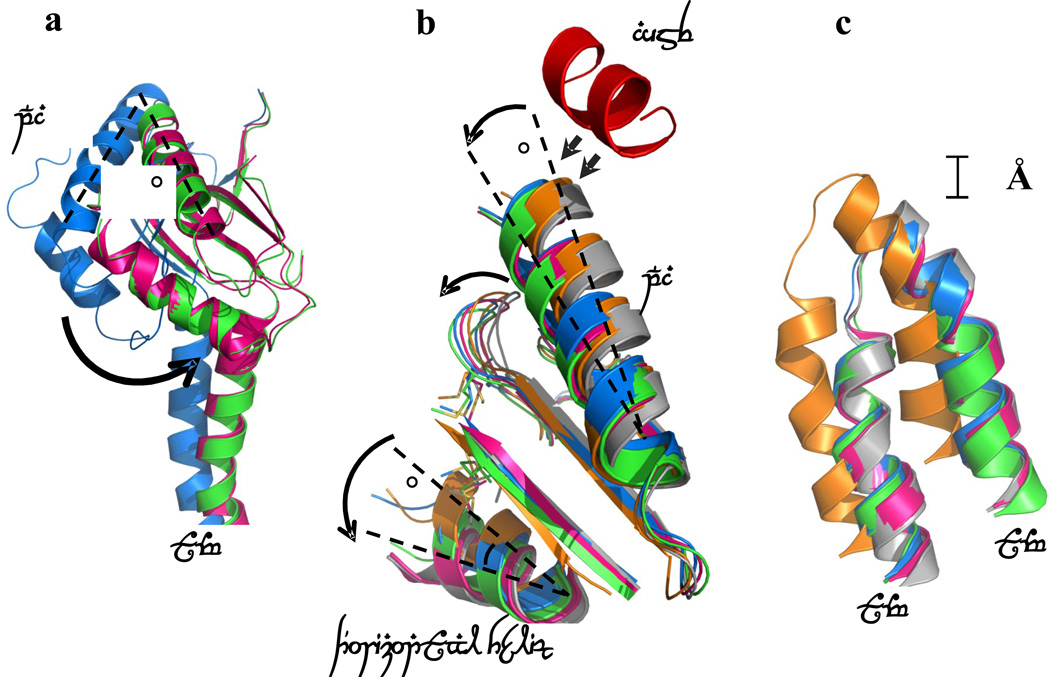
Different conformational states of CusBA-Cu(I). (a) Superimposition of subdomain PC2 and TM8 of each state of the pump. The arrow indicates a 30° swing of PC2 upon conformational transition from the “pre-extrusion 1” (cyan) to “pre-extrusion 2” (green) and “extrusion” (magenta) states. The structures of forms Ia, II and III are used to represent the “pre-extrusion 1”, “pre-extrusion 2” and “extrusion” states, respectively. (b) Superimposition of subdomain PC1 and the horizontal helix of each state of the pump. The figure illustrates the change in conformations of the PC1 helix (residues 582–589) and the flexible loop (residues 609–626) upon CusB binding. For clarity, only the short C-terminal helix (residues 391–400) of molecule 1 of CusB (red) is included. The change in conformation of the horizontal helix at different states of the pump is also shown in this superimposition (apo-CusBA, gray; CusA-Cu(I), orange; form Ia, cyan; form II, green; form III, magenta). (c) Superimposition of the transmembrane helices 5 (TM5) and 6 (TM6) of each state of the pump. The figure illustrates the change in positions of TM5 and TM6 (residues 447–495) within the transport cycle (apo-CusBA, gray; CusA-Cu(I), orange; form Ia, cyan; form II, green; form III, magenta). As both forms Ib and II are in the same state, the conformation of form Ib is not included in (a)–(c) for clarity.
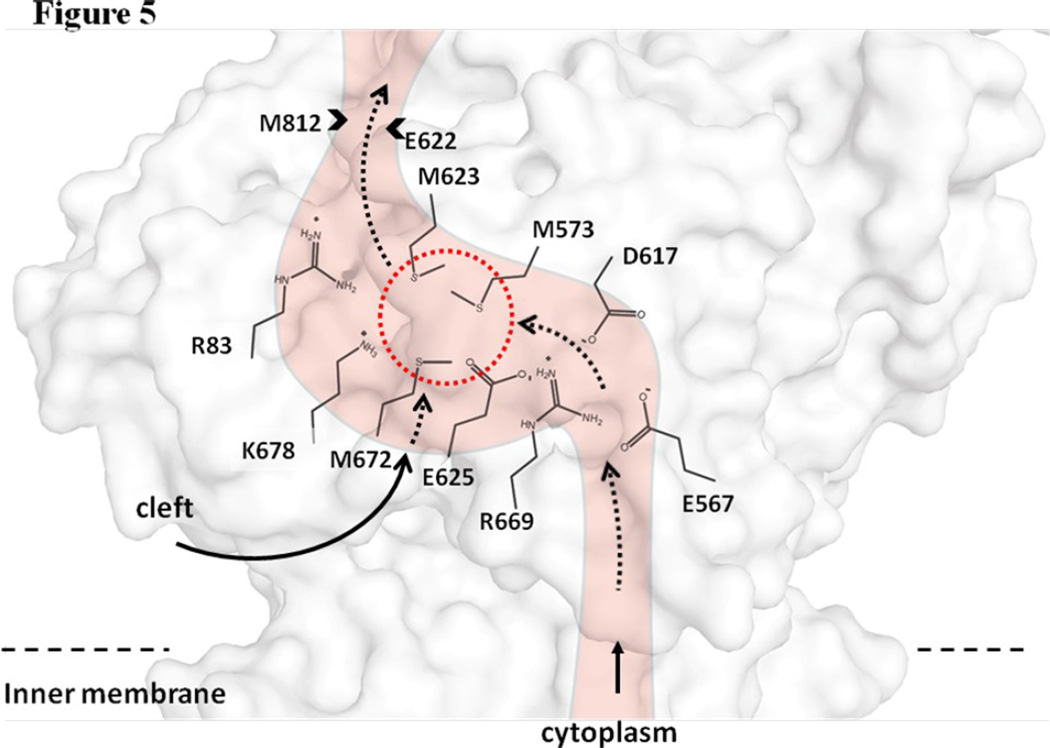
The overall conformation of the second CusBA-Cu(I) structure (form II) is almost identical to the form Ib structure, indicating that this structure should also represent the “pre-extrusion 2” state. A strong peak at the copper edge was observed at the periplasmic domain of CusA in this CusBA-Cu(I) complex (Fig. 3c). The program CAVER illustrates that the channel formed by the methionine relay network of each protomer of CusA has been closed up in this conformation. The only cavity that can be identified is the space nearby the copper signal (Fig. 3c). This copper signal is found in the familiar three-methionine binding site formed by M573, M623 and M672. However, its binding mode is quite distinct from that found in the “binding” state. First, the C-terminal end of the horizontal helix (residues 665–675) significantly tilts downward by ~10° in this “pre-extrusion 2” state in comparison with the CusA-Cu(I) form. This motion shifts the position of M672 away from M573 and M623, probably weakening the binding for Cu(I) within the methionine triad. Secondly, coupled with this movement, the side chain of a conserved anionic charged residue E625, located deep inside the periplasmic cleft, is found to flip towards the bound Cu(I) and interact with this ion by electrostatic interaction. In addition, a cluster of conserved charged residues, including R83, E567, D617, E625, R669 and K678, is found nearby this copper signal. Interestingly, these conserved charged residues line along the metal-ion transport channel formed by the methionine relay network (Figs. 5 and S1), suggesting that these charged residues may be important for the transporter’s functioning.
Figure 5.
Cu(I) binding site and conserved charged residues. This is a schematic representation of the CusA channel. The conserved residues R83, E567, D617, E625, R669 and K678, lining the channel at the periplasmic domain are indicated. The dotted circle marks the location of the Cu(I) binding site formed by the methionine triad M573, M623 and M672. The paths for metal transport through the periplasmic cleft and transmembrane region are illustrated with black curves.
The third CusBA-Cu(I) structure (form III) is nearly identical to that of apo-CusBA, and one bound Cu(I) is found to coordinate M573, M623, M672 and E625 (Fig. 3d). The conformation of CusA in this complex structure is also very similar to that of apo-CusA at the “resting” state. However, a closer inspection of these structures suggests that the CusA protomers in the two CusBA complexes are in a different conformational state when compared with the apo-CusA form. Binding of the CusB adaptor triggers a subtler but significant conformational change at the upper portion of subdomain PC1 of the CusA transporter. It is observed that a short C-terminal helix (residues 391–400) of molecule 1 of CusB directly interacts with and pushes a helix (residues 582–589) located at the upper half of PC1 of CusA (Fig. 4). The consequence is that the N-terminal end of this PC1 helix is found to tilt downwards by 8° in the CusBA form in comparison with the apo-CusA structure. This tilting motion in turn further pushes a loop (residues 609–626) located right below the PC1 helix downward. On the basis of these structures, CusB may be involved in tuning the width of the channel formed by the methionine metal-ion relay network to its optimal level through the above mechanism. The structure of form III is thus designated as an “extrusion” state of the pump.
In the “pre-extrusion 2” structures, the channel formed by the methionine metal-ion relay network is occluded (Fig. 3b and c). It is observed that the region nearby E622 and M812 form the narrowest region immediately after the three-methionine binding site. Interestingly, M812 was found to be significant in relieving copper sensitivity of E. coli.4 Thus, an empirical measurement defined as the Cα–Cα distance between E622 and M812 was used to identify the degree of opening of the channel. These distances are 13 Å, 12 Å, 10 Å, 10 Å and 11 Å for the “resting”, “binding”, “pre-extrusion 1”, “pre-extrusion 2” and “extrusion” states, respectively. In the “resting” state, the channel is fully open. This channel is also open and is believed to be at its optimal degree of opening in the “extrusion” form.
It should be noted that the conformation of the CusB hexamer does not change much at various states of the efflux complex. Superimpositions of molecules 1 and 2 of CusB onto their corresponding molecules at different states give rmsd between 0.2–0.5 Å.
It is also worth noting that, unlike the CusA structures, no continuous channel is found spanning the transmembrane region of these three CusBA-Cu(I) complexes. The only tunnel-like feature observed by CAVER is the periplasmic channel observed in the “extrusion” state (form III) of the pump (Fig. 3d).
In vivo and vitro functional studies
The structures of CusBA-Cu(I) highlight the importance of charged residues for the transporter’s functioning. These conserved charged residues (R83, E567, D617, E625, R669 and K678) seem to form a network and line along the wall of the methionine relay tunnel in the periplasmic domain of CusA. These residues were mutated into alanines (R83A, E567A, D617A, E625A, R669A, K678A) or aspartate (E625D), and the corresponding mutant transporters were expressed in BL21(DE3)ΔcueOΔcusA that lacks both the cueO and cusA genes (Fig. S2). All these mutant proteins were expressed in the bacterial membrane. In addition, the secondary structures of these mutant transporters were the same as that of the wild-type in detergent solution as suggested by CD spectroscopy (Fig. S3). The ability of these mutant transporters to confer copper resistance was then tested in vivo using susceptibility assay. The results indicate that all these CusA mutants are unable to relieve the copper sensitivity of strain BL21(DE3)ΔcueOΔcusA (Table 2), suggesting that these residues are critical for the function of the pump.
Table 2.
MICs of copper for different CusA mutants expressed in E. coliBL21(DE3)ΔcueOΔcusA
| Gene inBL21(DE3)ΔcueOΔcus A |
MIC (mM) of CuSO4 |
|---|---|
| Empty vector | 0.50 |
| cusA (wild-type) | 2.25 |
| cusA (R83A) | 0.50 |
| cusA (E567A) | 0.50 |
| cusA (D617A) | 0.50 |
| cusA (E625A) | 0.50 |
| cusA (E625D) | 0.50 |
| cusA (R669A) | 0.50 |
| cusA (K678A) | 0.50 |
To investigate whether these charged residues are essential for metal ion transport, the purified R83A, E567A, D617A, E625A, R669A and K678A mutant proteins, which are shown to abolish the function of the pump, were reconstituted into liposomes containing the fluorescent indicator Phen Green SK (PGSK) in the intravesicular space, respectively. A stopped-flow transport assay was used to determine whether these proteoliposomes can capture metal ions from the extravesicular medium. When Ag+ ions were added into the extravesicular medium, no quenching of the fluorescence signal were detected. Unlike the wild-type CusA pump, the results suggest that these mutant transporters are incapable of taking up Ag+ from the extravesicular medium into the intravesicular space of the proteoliposomes (Fig. S4a). These collective experiments provide compelling evidence that these conserved charged residues are critical for the transporter’s functioning.
The stopped-flow assay was also used to study the transport activity of the reconstituted CusBA proteoliposomes. The results suggest that the CusBA proteoliposomes are more active than those liposomes containing CusA only for metal transport as indicated by the position of the 50% attenuation of the fluorescent signal (Fig. S4b). No transport activity has been observed in liposomes containing CusB only, indicating that these CusB protoliposomes cannot uptake silver ions (Fig. S4b).
Crystal structures of the CusB-mutant CusA-Cu(I) complexes
Previous study suggested that the charged residues, D405, E939 and K984, located at the transmembrane region form the proton-relay network of the pump. Replacements of these residues with alanines (D405A, E939A and K984A) disrupted the hydrogen-bonded network, which in turn abolished the function of the pump.5 Thus, the D405A mutant was chosen to produce the CusB-D405A-Cu(I) co-crystal (Table 1). Surprisingly, no copper signal was found in this co-crystal, indicating that the D405A mutant is not capable of binding Cu(I) (Fig. S5a). This result is indeed in good agreement with the functional studies in which a mutation on this residue abolishes the function of the pump. The overall structure of CusB-D405A-Cu(I) resembles the apo-CusBA conformation. Superimposition of these two structures gives an rmsd of 0.2 Å.
The CusA mutant R669A was also used to crystallize and produce the CusB-R669A-Cu(I) co-crystal. Again, no copper signal was found in this co-crystal (Fig. S5b). The overall structure of this mutant co-crystal is nearly identical to the apo-CusBA structure. A superimposition of these two co-crystals results in an rmsd of 0.3 Å. It is likely that the D405A and R669A mutants are trapped in one of the transient states of the pump and thus cannot change their conformation to go through the transport cycle.
DISCUSSION
Four lines of evidence suggest that the structures identified can be understood in terms of sequential transition of conformations leading to the extrusion of metal ions from the CusA pump. First, the sequential decrease in distance between subdomains PC1 and PC2 from the “binding” to “resting” states. This change highlights the sequential movements of CusA upon transiting from one state to the other. Secondly, a subtler shift in position of the horizontal helix leading to the departure of M672 from the methionine triad and a release of the bound Cu(I) from these methionines. In addition, transmembrane helices 5 and 6 are found to shift in position at different conformational states (Fig. 6), mimicking the change of the horizontal helix. Lastly, a smaller change in conformation of subdomain PC1 of CusA upon CusB binding, which in turn controls the degree of opening of the channel formed by the methionine relay network. These structures also suggest that the CusB adaptor may play an essential role in tuning the extent of channel opening upon conformational transition through different states.
Figure 6.
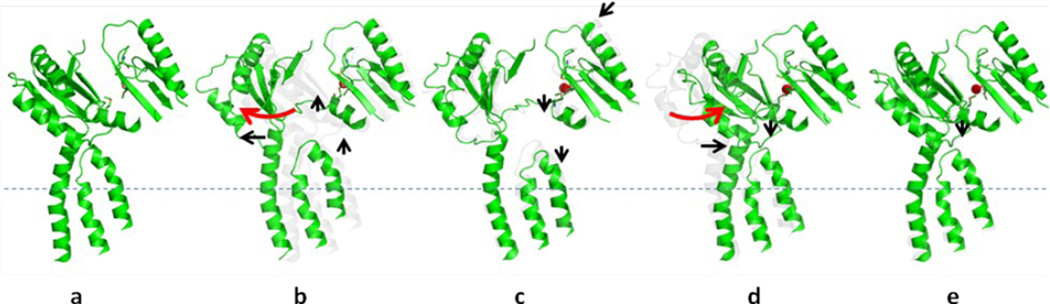
Sequential conformational transition of the CusBA pump. This figure depicts stepwise motions of CusA in the adaptor-transporter complex transitioning between the “resting” and “extrusion” states (a, “resting” state; b, “binding” state; c, “pre-extrusion 1” state; d, “pre-extrusion 2” state; e, “extrusion” state). For clarity, only subdomains PC1 and PC2 in the periplasmic domain and transmembrane helices 5, 6 and 8 in the transmembrane region of CusA are shown. The bound copper is in red sphere. The red arrows indicate the change in conformation of the protein within the transport cycle.
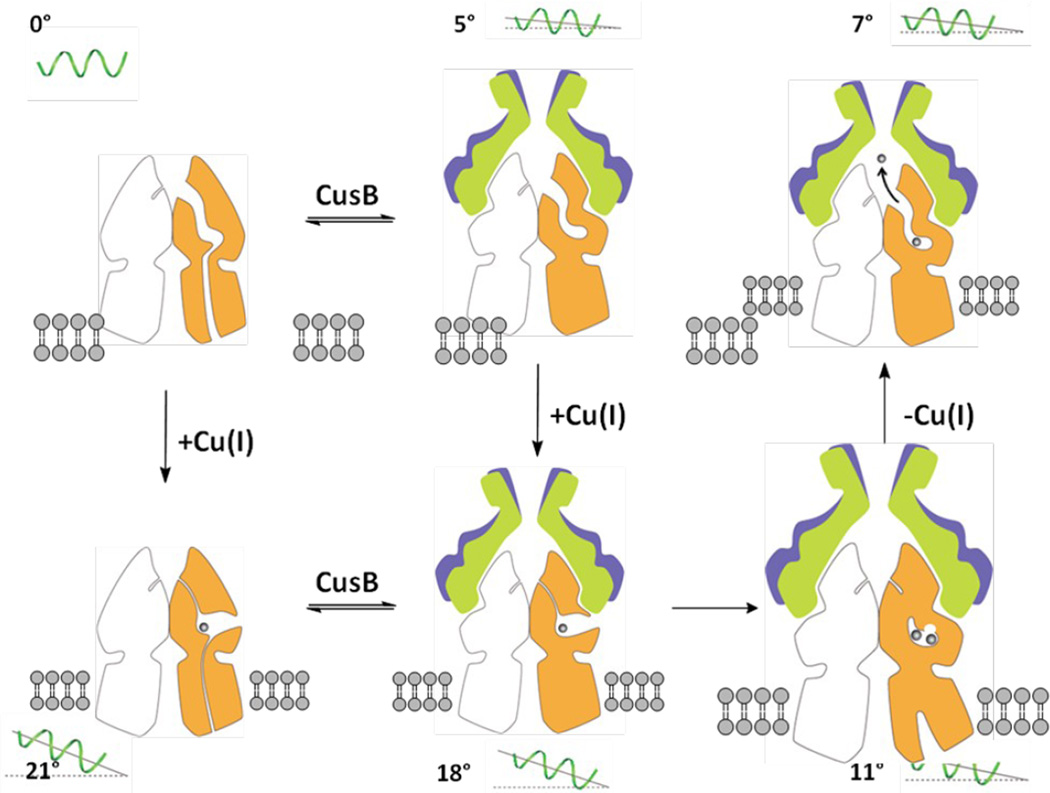
It should be noted that all these protein conformers were obtained via soaking, a procedure which may limit the degree of conformational transitions. Nonetheless, the co-crystal structures of CusBA-Cu(I) extend our knowledge on how successive motions in the CusBA efflux pump are coordinated and how CusBA extrudes the bound metal ions. This protein complex undergoes cycles of conformational changes that drive the passage of the metal ions. In the absence of both CusB and Cu(I) or Ag(I), the CusA pump prefers the “resting” conformation where the periplasmic cleft between subdomains PC1 and PC2 are closed. Cu(I) or Ag(I) binding triggers a large conformational change where subdomain PC2 performs a 30° swing to open this cleft. In the presence of CusB, the subsequent steps may be a transitions to the “pre-extrusion 1” and then “pre-extrusion 2” states in which PC2 is found to swing back to close the cleft (Fig. 7). The majority of the channel formed by the methionine relay network becomes occluded at the “pre-extrusion 2” state. Subsequently, the last step is a transition to the “extrusion” form where CusB is responsible for stabilizing the conformation of this state, and for tuning the channel at the optimal degree of opening to conduct metal ion export. It is very likely that the CusCBA pump operates through an alternating-access mechanism.14–16 Indeed, functional dynamics simulation has already suggested that the periplasmic cleft of CusA may alternately open and close upon metal transport.5 As the crystal structures of the CusBA adaptor-transporter complex depict that there are several intermediates participating in the transport cycle, it is difficult to correlate all these conformational states to those of AcrB. Nonetheless, the “binding” and “extrusion” states of the CusBA system should correspond to the “T” and “O” states of the AcrB pump,14–16 respectively.
Figure 7.
Proposed model of metal ion export within the transport cycle. The model is based on different structures obtained by x-ray crystallography. Molecules 1 and 2 of CusB and a protomer of CusA are colored blue, green and orange. The angle of inclination of the horizontal helix at each state is shown in the figure. For clarity, the front protomers (one CusA and two CusB molecules) are not included.
The metal ions can enter the CusA pump via the periplasmic cleft as well as the cytoplasm (Fig. 5). It is believed that CusA predominately carries out metal ion efflux through the periplasm.17 Thus, the major path for taking up metals should be across the cleft in the periplasmic space. During the transport cycle, the CusBA efflux complex must go through different transient states, including the “resting”, “binding”, “pre-extrusion 1”, “pre-extrusion 2” and “extrusion” conformations, to complete the transport cycle. Thus, in coordination with the motion of the periplasmic cleft, the transmembrane portion of the methionine relay channel should also be synchronized to open and close to accommodate for the export of metal ions. It is likely that the long N-terminal tail of CusB may be involved in the delivery of metal ions from the periplasmic space into the cleft of the CusA pump.8 Genetic data and transport assay in this study have shown that the conserved charged residues lining the methionine relay channel are essential for the transporter’s functioning. Therefore, it is possible that these conserved charged amino acids are responsible for passing the transported metal ion along from one residue to another in the methionine relay channel. Coupling with the methionine pairs and clusters, these conserved charged residues are deemed to be of importance in transferring the metal ions out of the cell.
Methods
Preparation of the CusBA-Cu(I) and CusB-mutant CusA-Cu(I) complexes
The procedures for cloning, expression and purification of the CusA and CusB proteins have been described previously.5,7 Co-crystals of the CusBA complex were obtained using sitting-drop vapor diffusion.8 Briefly, the CusBA crystals were grown at room temperature in 24-well plates with the following procedures. A 2 µl protein solution containing 0.1 mM CusA and 0.1 mM CusB in buffer solution containing 20 mM Na-HEPES (pH 7.5) and 0.05% (w/v) CYMAL-6 was mixed with a 2 µl of reservoir solution containing 10% PEG 6000, 0.1 M Na-HEPES (pH 7.5), 0.1 M ammonium acetate and 20% glycerol. The resultant mixture was equilibrated against 500 µl of the reservoir solution. Co-crystals of CusBA grew to a full size in the drops within two months. Typically, the dimensions of the crystals were 0.1 mm × 0.1 mm × 0.1 mm. The procedures for producing the CusB-CusA mutant crystals were the same as those of the wild-type crystals.
The CusBA-Cu(I) or CusB-mutant CusA-Cu(I) complex crystals were then prepared by incubating crystals of apo-CusBA or apo-CusB-mutant CusA in solution containing 10% PEG 6000, 0.1 M Na-HEPES (pH 7.5), 0.1 M ammonium acetate, 20% glycerol, 2 mM [Cu(CH3CN)4]PF6, 2 mM tris(2-carboxyethyl)phosphine (TCEP) and 0.05% (w/v) CYMAL-6 for 1 hour at 25°C. Cryoprotection was achieved by incubating the CusBA-Cu(I) or CusB-mutant CusA-Cu(I) crystals in cryoprotectant containing 10% PEG 6000, 0.1 M Na-HEPES (pH 7.5), 0.1 M ammonium acetate, 25% glycerol, 2 mM [Cu(CH3CN)4]PF6, 2 mM TCEP and 0.05% (w/v) CYMAL-6 for 5 min at 25°C.
Data collection, structural determination and refinement
All diffraction data were collected at 100K at beamline 24ID-C located at the Advanced Photon Source, using an ADSC Quantum 315 CCD-based detector. Diffraction data were processed using DENZO and scaled using SCALEPACK.18
The crystals belong to space group R32 (Table 1). The structures of the CusBA-Cu(I) and CusB-mutant CusA-Cu(I) complexes were phased using molecular replacement, utilizing the apo-CusBA structure (3NE5)8 determined earlier by our group as a search model. The models were refined using TLS refinement techniques adopting a single TLS body as implemented in PHENIX (20) leaving 5% of reflections in Free-R set. Iterations of refinement using PHENIX19 and CNS20 and model building in Coot21 lead to the current models (Table 1)
For the form I crystal data, the group occupancy of residues 664 through 888 in the two conformations (forms Ia and Ib) was refined under the constraint that individual occupancies added up to one. This increased one more parameter to the refinement process. The final refined occupancies were 0.22 and 0.78 for the conformations of forms Ia and Ib, respectively.
Susceptibility assays
The double knocked-out E. coli strain BL21(DE3)ΔcueOΔcusA was produced as described previously.5 The susceptibility to copper of E. coli BL21(DE3)ΔcueOΔcusA harboring pET15bΩcusA expressing the wild-type or mutant transporters, or the pET15b empty vector was tested on agar plates. Cells were grown in Luria Broth (LB) medium with 100 µg/ml ampicillin at 37 °C. When the OD600 reached 0.5, the cultures were induced with 0.5 mM IPTG and harvested in two hours after induction. The expression level of each mutant in BL21(DE3)ΔcueOΔcusA is similar to that of the wild-type transporter as indicated by Western blot analysis (Fig. S2). The minimum growth inhibitory concentrations (MICs) to copper of E. coli BL21(DE3)ΔcueOΔcusA(incoculum, 500 cells/ml) harboring these vectors were then determined using LB agar containing 50 µg/ml ampicillin, 0.1 mM IPTG and different concentrations of CuSO4 (0.25 to 2.75 mM in steps of 0.25 mM) (Table 2).
CD spectroscopy
CD spectra were measured at 25 °C using a Jasco J-710 spectropolarimeter with a cell path length of 1 mm. The concentrations of the apo-CusA and mutant-CusA proteins were 0.6 µM in buffer containing 10 mM sodium phosphate, pH 7.5, and 0.028% CYMAL-6. Spectra were recorded at wavelengths between 190 and 260nm. For each spectrum, 10 scans at a scanning speed of 10 nm/min and a data pitch of 0.1 nm were averaged.
Reconstitution and stop-flow transport assay
The Phen Green SK (PGSK) (Invitrogen) encapsulated proteoliposomes of the wild-type CusA, R83A, E567A, D617A, E625A, R669A or K678A was prepared as edescribed.5 In all cases, nearly 100% of these detergent solubilized CusA proteins were incorporated into the proteoliposomes through these procedures. The CusB and CusBA proteoliposomes were produced using the similar method except that the liposomes were made in the presence of 50 µg purified CusB in the reconstitution buffer containing 20 mM HEPES-KOH, pH 6.6. Transport experiments were performed at 25oC on a stopped-flow apparatus (Hi-Tech Scientific) connected to a spectrofluorometer (PerkinElmer LS55). Proteoliposomes and a transport assay buffer (20 mM HEPES-KOH pH 7.0 and 1 mM AgNO3) were loaded into a two separate syringes of equal volume. Transport reactions were initiated by pushing 400 µl fresh reactants at a 1:1 ratio through the 90 µl mixing cell at a flow rate of 2 ml/s. Stopped-flow traces were the cumulative average of three or four successive recordings at 530 nm with the excitation wavelength at 480 nm.
Data Deposition
Atomic coordinates and structure factors have been deposited with the Protein Data Bank under codes 3T56 (form I), 3T51 (form II), 3T53 (form III), 4DNT (CusB-D405A-Cu(I)) and 4DOP (CusB-R669A-Cu(I)).
Supplementary Material
Three different conformers of the crystal structures of CusBA-Cu(I) are reported.
These structures suggest a sequence of conformational transitions in the transport cycle.
Conserved charged residues of CusA are essential for extruding metal ions out of the cell.
Acknowledgements
This work is supported by an NIH Grant R01GM086431 (E.W.Y.). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MH., Jr The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development protein. J. Mol. Microbiol. Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- 2.Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003;27:313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 3.Franke S, Grass G, Nies DH. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiol. 2001;147:965–972. doi: 10.1099/00221287-147-4-965. [DOI] [PubMed] [Google Scholar]
- 4.Franke S, Grass G, Rensing C, Nies DH. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 2003;185:3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long F, Su C-C, Zimmermann MT, Boyken SE, Rajashankar KR, Jernigan RL, Yu EW. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature. 2010;467:484–488. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su C-C, Long F, Yu EW. The Cus efflux system removes toxic ions via a methionine shuttle. Prot. Sci. 2011;20:6–18. doi: 10.1002/pro.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su C-C, Yang F, Long F, Reyon D, Routh MD, Kuo DW, Mokhtari AK, Van Ornam JD, Rabe KL, Hoy JA, Lee YJ, Rajashankar KR, Yu EW. Crystal structure of the membrane fusion protein CusB from Escherichia coli. J. Mol. Biol. 2009;393:342–355. doi: 10.1016/j.jmb.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su C-C, Long F, Zimmermann MT, Rajashankar KR, Jernigan RL, Yu EW. Crystal structure of the CusBA heavy-metal efflux pump of Escherichia coli. Nature. 2011;467:484–488. doi: 10.1038/nature09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rensing C, Pribyl T, Nies DH. New functions for the three subunits of the CzcCBA cation-proton antiporter. J. Bacteriol. 1997;179:6871–6879. doi: 10.1128/jb.179.22.6871-6879.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stegmeier JF, Polleichtner G, Brandes N, Hotz C, Andersen C. Importance of the adaptor (membrane fusion) protein hairpin domain for the functionality of multidrug efflux pumps. Biochemistry. 2006;45:10303–10312. doi: 10.1021/bi060320g. [DOI] [PubMed] [Google Scholar]
- 11.Kulathila R, Kulathila R, Indic M, van den Berg B. Crystal structure of Escherichia coli CusC, the outer membrane component of a heavy-metal efflux pump. PLoS One. 2011;6:e15610. doi: 10.1371/journal.pone.0015610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane proteinTolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 13.Akama H, Kanemaki M, Yoshimura M, Tsukihara T, Kashiwag T, Yoneyama H, Narita S, Nakagawa A, Nakae T. Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end. J. Biol. Chem. 2004;279:52816–52819. doi: 10.1074/jbc.C400445200. [DOI] [PubMed] [Google Scholar]
- 14.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 15.Seeger MA, Schiefner A, Eicher T, Verrey F, Dietrichs K, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 16.Sennhauser G, Amstutz P, Briand C, Storchengegger O, Grütter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5:e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim E-H, Nies DH, McEvoy MM, Rensing C. Switch or funnel: how RND-type transport systems control periplasmic metal homeostasis. J. Bacteriol. 2011;193:2381–2387. doi: 10.1128/JB.01323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otwinowski Z, Minor M. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 19.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCroy AJ, Moriarty NW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 20.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Weiss MS, Hilgenfeld R. On the use of the merging R factor as a quality indicator for X-ray data. J. Appl. Crystallogr. 1997;30:203–205. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.