Despite their identification >30 years ago, the mechanism of how chloroplast transit peptides direct precursor import remains enigmatic. This is more confounding in light of emerging genomic data that indicate ∼20% of plant genomes encode chloroplast-targeted proteins. This article develops a heuristic approach that may provide universal criteria for what makes a functional transit peptide.
Abstract
Despite the availability of thousands of transit peptide (TP) primary sequences, the structural and/or physicochemical properties that determine TP recognition by components of the chloroplast translocon are not well understood. By combining a series of in vitro and in vivo experiments, we reveal that TP recognition is determined by sequence-independent interactions and vectorial-specific recognition domains. Using both native and reversed TPs for two well-studied precursors, small subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase, and ferredoxin, we exposed these two modes of recognition. Toc34 receptor (34-kD subunit of the translocon of the outer envelope) recognition in vitro, preprotein binding in organellar, precursor binding in vivo, and the recognition of TPs by the major stromal molecular motor Hsp70 are specific for the physicochemical properties of the TP. However, translocation in organellar and in vivo demonstrates strong specificity to recognition domain organization. This organization specificity correlates with the N-terminal placement of a strong Hsp70 recognition element. These results are discussed in light of how individual translocon components sequentially interact with the precursor during binding and translocation and helps explain the apparent lack of sequence conservation in chloroplast TPs.
INTRODUCTION
The ability of plastids to import precursor proteins posttranslationally from the cytosol has been known for over 30 years (Dobberstein et al., 1977; Chua and Schmidt, 1979). Key to this process is the role of an N-terminal extension, known as the transit peptide (TP), which directs the precursor to the plastid membrane and through the translocons at the outer and inner chloroplast envelope membranes (TOC/TIC) (Bruce, 2000, 2001). Recent analysis of multiple plant and algal genomes using various TP identification tools indicates that the number of nuclear-encoded precursors ranges from ∼2100 in Arabidopsis thaliana to as high as ∼4800 in rice (Oryza sativa; Richly and Leister, 2004). Despite this large number of sequences available, fundamental understanding of how TPs function is still lacking.
Early analysis suggested that TPs may be composed of distinct homology blocks that share limited sequence similarity (Karlin-Neumann and Tobin, 1986). However, this hypothesis was challenged and replaced by a loose structural organization with three identifiable regions (von Heijne et al., 1989). Multiple efforts using mutagenesis (Pilon et al., 1995; Lee et al., 2002), deletion (Kindle, 1998; Kindle and Lawrence, 1998; Rensink et al., 1998, 2000), Ala scanning (Lee et al., 2006), domain swapping (Smeekens et al., 1986; de Castro Silva Filho et al., 1996), and the use of synthetic peptides (Perry et al., 1991; Schnell et al., 1991; Pinnaduwage and Bruce, 1996) have investigated the structure and function of only a few TPs in detail. However, these results are not extendable to other TPs based on sequence analysis, and the elucidation of common TP functional domains remains enigmatic. Although earlier attempts to identify homology blocks failed due to the high degree of sequence variation, it is still possible that TPs may contain a conserved motif or nonlinear peptide pattern that may provide some common mode of recognition (Lee et al., 2006). Moreover, a systematic approach involving in vivo targeting analysis indicates that individual amino acids do not contain specific targeting information, but the overall context of the amino acid sequence is critical for targeting to the chloroplast (Lee et al., 2002). Recent efforts to identify any universal signature motif in 208 experimentally confirmed TPs have not been fruitful, and it was concluded that these TPs are highly dissimilar (Lee et al., 2008). However, when these authors used a bioinformatics-based approach to pregroup TPs into seven subgroups, one or more conserved motifs were identified within a given subgroup but were not universal (Lee et al., 2008).
This suggests that TPs do not share any consensus motifs, yet each TP may contain different functional motifs that facilitate targeting and import. In light of the conserved nature of translocon protein composition and the high fidelity of protein targeting in vitro and in vivo, it is difficult to reconcile how individual TPs can engage a common set of translocon components, without some unifying information encoded within the TP. It is possible that sequence information does not define a TP, but rather the physicochemical properties of the TP determine its targeting activity. This may explain why TP prediction algorithms function in the absence of detectable sequence similarity. These physicochemical properties may be environmentally sensitive and/or context specific, behaving differently as a function of pH, in a membrane-like environment, or upon receptor binding. One example of this is the tendency of TPs to convert from a random coil in an aqueous inherent environment (von Heijne and Nishikawa, 1991) to an α-helix in the presence of membranes or membrane-mimetic environments (Bruce, 1998; Krimm et al., 1999; Wienk et al., 2000). Finally, it is possible that TP interaction with different components of the TOC and TIC as well as the stromal-localized components, such as stromal processing peptidase (SPP) and molecular chaperones, uses multiple mechanisms of recognition ranging from general physicochemical properties to specific sequence recognition.
Here, we attempted to differentiate the role of TP sequence-specific contributions from the physicochemical properties using TP sequences that have been inverted with respect to their N- to C- sequence, termed retro-peptides. These retro-TPs share no more similarity to their parent sequences than any random sequence (Haack et al., 1997); however, they share with their parents many identical properties, including (1) ratio of hydrophobic/hydrophilic amino acids, (2) a global amino acid composition, (3) chirality, (4) spacing of their constituent amino acid, (5) placement of secondary structures, and (6) potential mirroring of three-dimensional structure (Guptasarma, 1992; Battistutta et al., 1994; Lacroix et al., 1998). Thus, they contain the identical amino acid composition and its associated physicochemical properties yet are sequence divergent. This inherent property has attracted considerable interest in using retro-peptides to study various structure function relationships of peptides/proteins, including antimicrobial peptides (Pellegrini and von Fellenberg, 1999) and Leu zippers (Holtzer et al., 2000). They have also been used to examine protein folding (Olszewski et al., 1996; Lacroix et al., 1998) and antibody recognition (Guichard et al., 1994; Benkirane et al., 1995).
We developed a series of assays to evaluate how the well-studied TPs of the small subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) and ferredoxin and their retro-peptides interact with different components of the chloroplast import machinery. These assays include in vitro analyses of the interaction of pro- and retro-peptides with the isolated cytosolic receptor domain of the GTPase Toc34, the stromal molecular motor ATPase Hsp70, and the SPP. We perform in organellar analyses of the ability of isolated chloroplasts to bind and import pro- and retro-peptides and carry out in vivo analyses of onion (Allium cepa), Arabidopsis, and tobacco (Nicotiana tabacum) cells’ ability to sort, target, bind, and deliver pro- and retro-peptides to and into the chloroplast. Interestingly, we see that certain steps in the import process can recognize both the TP and the retro-peptide. Other steps are highly selective, such as in vitro and in vivo translocations. To reconcile these results, we further tested the requirements for the N-terminal sequence to be uncharged and largely nonpolar. This requirement seems to be a key determinant of the ability of a given sequence (pro or retro) to mediate translocation. These results are discussed in light of a possible general mechanism of TP recognition given the lack of sequence similarity that is so pervasive in chloroplast targeting sequences.
RESULTS
Generation and Analysis of Forward and Reverse Transit Sequences
To characterize the biophysical, biochemical, and targeting activities of the pro- and retro-TPs, we used an Escherichia coli expression system that allowed the production of these peptides without an attached epitope tag. This system used the self-cleavage activity of intein. Due to the heterologous nature of this system, we also were required to codon optimize these gene sequences. The DNA sequences of forward (pro) and reverse (retro) small subunit of Rubisco TPs (SSF and SSR) and ferredoxin TPs (FDF and FDR) are shown together with relative adaptiveness plots in Supplemental Figures 1A and 1B online. The optimized sequences have higher codon adaptive indices (CAI), the geometric mean of adaptiveness, at ∼0.8, whereas the native DNA sequences have an average CAI of 0.36 (see Supplemental Table 1 online).
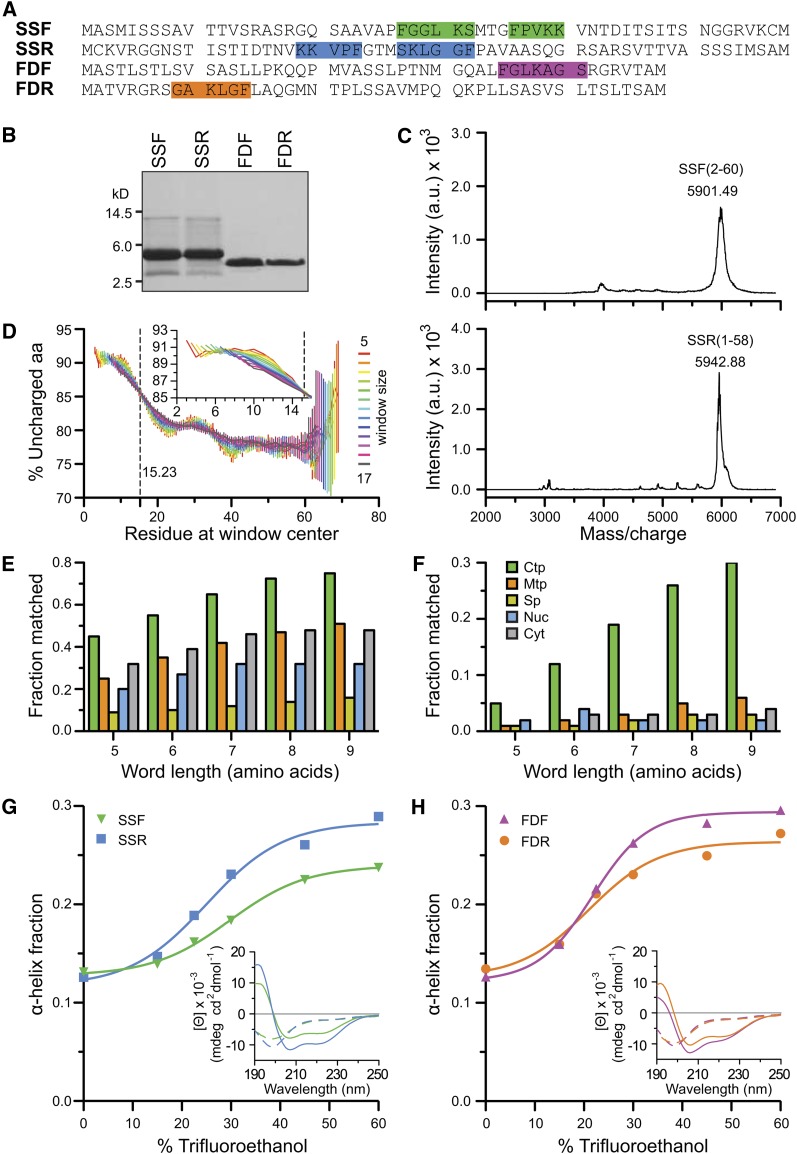
The codon-optimized synthetic DNAs were cloned into pTYB2 vector and sequence verified. The amino acid sequences of the peptides are presented in Figure 1A. The peptides were expressed in E. coli ER2566 cells, purified by chitin-affinity chromatography, intein-cleaved, and lyophilized. The yield and purity of the four peptides were confirmed by SDS-PAGE and matrix-assisted laser-desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Figures 1B and 1C). As is shown for SSF, there was a major species at 5901.49 mass-to-charge ratio that corresponds to the mass of SSF from amino acids 2 to 60 (Figure 1C). The theoretical average mass of SSF2-60 is 5903.81 D. The peak represents the +1 charge state that corresponds to a processed peptide missing the start Met, presumably due to the E. coli Met aminopeptidase. In addition, there were multiple peaks that correspond to different levels of Met oxidation of SSF2-60 following the major peak. As shown in Figure 1C for SSR, the major species at 5942.88 mass-to-charge ratio corresponds to the +1 charge state of SSR from amino acids 1 to 58 with four Met oxidations. The theoretical average mass is 5944.83 D. Similar analysis of the other peptides also confirmed their purity and sequence.
Figure 1.
Purification of Forward and Retro-TPs and Bioinformatic and Secondary Structure Analyses of TPs.
(A) The sequences of SSF, SSR, FDF, and FDR. FGLK motifs are highlighted.
(B) SDS-PAGE gel of the four peptides.
(C) MALDI-TOF spectra of SSF and SSR peptides. a.u., arbitrary units.
(D) Analysis of N-terminal uncharged region in TP data set (n = 912). Means ± se are shown. Inset shows zoom-in without error bars. aa, amino acids.
(E) and (F) The percentage of sequences matched to the TargetP results using a flexible rule 35 and a strict rule 22, respectively. The rules are defined in Supplemental Table 3 online. Ctp, predicted chloroplast localization; Cyt, predicted cytoplasm; Mtp, predicted mitochondrial localization; Nuc, predicted nuclear localization; Sp, predicted secretory pathway.
(G) and (H) Both forward and reverse peptides share similar TFE-induced random coil to α-helix transitions. Insets report the representative CD spectra. Dashed and solid lines generated from 0 and 60% TFE, respectively.
Bioinformatic Analysis of Functional TP Domains
There has been a proliferation of software to predict subcellular localization of proteins. Most of these tools are based on N-terminal sequence analysis. We analyzed the four sequences by seven online prediction tools. The results of these are shown in Supplemental Table 2 online. All of the programs identified SSF and FDF as chloroplast TPs. It is interesting that while SSR was recognized by ChloroP, PredSL, and ProtComp as a TP, FDR was predicted to be a mitochondrial presequence by iPSORT, Predotar, PredSL, and ProteinProwler.
Although this study investigates two of the best-characterized TPs, from the small subunit of Rubisco and ferredoxin, which are both localized in the stroma, highly abundant and associated with photosynthesis, they have very limited sequence similarity. In fact, SSF shares 21.2, 12.0, and 2.2% identity and 42.4, 20.0, and 10.9% similarity with SSR, FDF, and FDR, respectively. Likewise, FDF shares 14.3, 12.0, and 2.2% identity and 42.9, 20.0, and 10.9% similarity with FDR, SSF, and SSR, respectively. Although the pro- and retro-peptides have identical physicochemical properties, they share very little sequence similarity; therefore, any similarity in activity of two TPs (SSF and FDF) must be based on properties beyond simple sequence similarity.
Despite the failure of bioinformatic algorithms to identify universally conserved motifs within TPs, there have been two short domains identified as highly characteristic of TPs. One is the short uncharged N-terminal segment that has been observed in most chloroplast TPs (von Heijne et al., 1989). This domain has also been suggested to be capable of functioning as a strong Hsp70 binding domain and is possibly key to the formation of translocation intermediates via its recognition by intermembrane space (IMS) or stromal Hsp70s (Blond-Elguindi et al., 1993; Fourie et al., 1994; Ivey et al., 2000). To verify this N-terminal property of TPs, we performed an analysis of this tendency on a data set of the 912 most confidently predicted TPs from the Arabidopsis genome (Figure 1D). The percentage of uncharged amino acids within a specific residue length window was calculated along the length of TPs. The values shown were averaged across the data set. Regardless of window size, the percentage of uncharged amino acids showed the transition from highly uncharged at almost 91% at the N terminus to moderately uncharged at ∼78% at the C terminus, confirming the uncharged bias of the N terminus. When data within the first 30 amino acids were fitted to sigmoidal curve, the half transition point was determined to be 15.23, indicating the border of the N-terminal uncharged region is within the first 15 amino acids; the transition actually starts at around residue 10 (Figure 1D, inset).
A second semiconserved TP motif was first suggested by Pilon and coworkers to be involved in the chloroplast recognition of TPs (Karlin-Neumann and Tobin, 1986; Pilon et al., 1995). This group identified a loose FGLK motif that was found at least once in 27 characterized TPs. Taking this analysis further, we performed a rigorous analysis of the statistical occurrence of this motif in TPs, other targeting sequences, and N-terminal sequences of cytosolic proteins. This analysis used the curated 940 targeting sequences previously used as a training set for the TargetP program (Emanuelsson et al., 2007). We applied a rule-based or heuristic approach using the neural net program MEME (Bailey and Elkan, 1994) to look for the FGLK motif. The rules employed are listed in Supplemental Table 3 online. Most of the rules require the residues FGLK, and all rules exclude acidic residues Asp and Glu. Of the applied rules, rule 35 resulted in the highest correlation with TargetP results and yielded >70% agreement when the word length was >8. The discrimination of this rule compared with the training set of mitochondrial, secreted, nuclear, and cytosolic proteins is shown in Figure 1E. However, when the stricter rule 22 was applied, it provided the most discriminatory prediction of chloroplast TPs (Figure 1F). These heuristics confirm the TP specificity of the FGLK motif, which loosely can be generalized as a motif containing (in no particular order): (1) an aromatic amino acid, (2) a turn-inducing or helix-breaking amino acid, (3) small nonpolar amino acid, (4) a basic amino acid, and (5) absence of Asp and Glu. SSF has two of these motifs, whereas FDF has only one, and the positions of the FGLK motifs are highlighted in Figure 1A. Interestingly, it is this region in SSF that shows the most sequence similarity with its retro version (53.3% identity and 66.6% similarity), suggesting that evolution provided a motif with some targeting activity independent of the orientation of binding.
Structure of the Forward and Retro-TPs
It has been well documented that TPs lack secondary structure in an aqueous environment. This unusual property has led them to be proposed as the perfect random coil (von Heijne and Nishikawa, 1991) and also suggest that they may belong to an interesting group of proteins known as intrinsically disordered proteins (IDPs) (Dyson and Wright, 2005). An inversion of the amino acid sequence could feasibly alter the propensity of these peptides to form secondary structures in solution and thus alter and potentially limit their recognition properties. Using circular dichroism (CD) spectroscopy, we investigated the secondary structure content of the four peptides in both aqueous buffers and in the helix-inducing solvent trifluoroethanol (TFE) (Bruce, 1998; Wienk et al., 1999). The α-helical content of SSF and SSR (Figure 1G), and FDF and FDR (Figure 1H) as a function of %TFE is shown. The actual CD spectra are shown in Supplemental Figure 1C online. In aqueous buffer, all four peptides had only a very limited α-helical content (∼12%). However, as TFE was increased, all of the peptides showed a TFE-induced increase in secondary structure. The spectra and deconvolution indicate that despite the opposite orientation of amino acid, the forward and reverse peptides adopted similar secondary structure. The only noticeable difference was that SSR seemed to have a slightly higher tendency to form α-helices than its forward counterpart, SSF.
Reverse Peptides Stimulate GTP Hydrolysis
One specific and potentially mechanistic role of TPs identified to date has been their ability to function as GTPase activating proteins (GAPs) and stimulate the GTP hydrolysis of the receptor GTPases found in the TOC (Jelic et al., 2002; Reddick et al., 2008). As previously reported by our laboratory, SSF stimulates GTP hydrolysis of pea (Pisum sativum) Toc34 in a GAP-like manner and does not function as a guanine nucleotide exchange factor to modulate the rate of nucleotide exchange (Reddick et al., 2007, 2008). Although this activity has also been observed for several peptides, the mapping of this activity has only been refined to the C-terminal 26 amino acid of SSF (Schleiff et al., 2002).
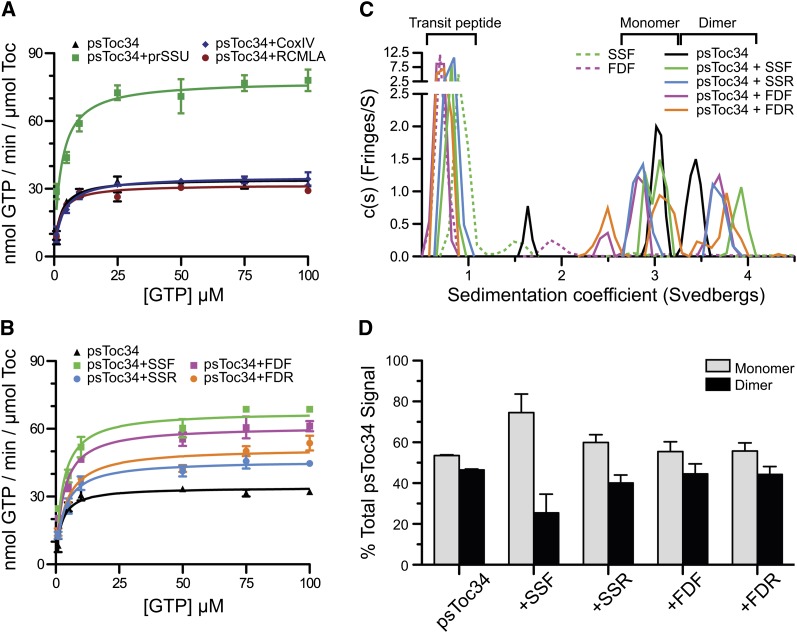
Because of the resolved nature and mechanistic significance of this TP activity, we designed experiments to see if the retro-TPs could also stimulate the GTPase activity of ps-Toc34 in vitro. Neither of the negative controls, a mitochondrial presequence CoxIV or a model unstructured polypeptide RCMLA, increased the maximal velocity (Vmax) nor did they alter the Michaelis constant (Km) (Figure 2A) as we and others have demonstrated for TPs (Jelic et al., 2002; Reddick et al., 2007). We analyzed the effects of adding 25 molar excess of each of the four TPs and determined the resultant Vmax and Km (Figure 2B). As expected, both of the forward TPs stimulated the hydrolytic rate by approximately twofold. The retro-TPs elicited GTPase activity of Toc34, however, at a much lower level than that of the pro-peptides, but still greater than that of negative controls. The observation that the retro-TPs still stimulated Toc34 GTPase activity with a slightly lower level indicates that physicochemical properties of TP alone are sufficient for GAP activity. Whether Toc34 GTPase activity alone is sufficient for full precursor import will be analyzed in the subsequent sections.
Figure 2.
Effect of TPs on Toc34 Hydrolysis Rate and Monomer:Dimer Ratio.
(A) The Vmax and Km of ps-Toc34 alone or in the presence of a 25 molar excess of the unstructured peptide RCMLA, the mitochondrial presequence CoxIV or prSSU.
(B) Hydrolysis rate of Toc34 alone or in the presence of 25 molar excess of SSF, FDF, SSR, or FDR. Means ± se are shown in (A) and (B).
(C) Analytical ultracentrifugation analysis shows ps-Toc34 exists in a monomer-dimer equilibrium with a slow transition rate (black line). Addition of SSF biases the equilibrium of Toc34 toward monomer.
(D) Quantitation of the monomeric and dimeric Toc34 species with and without various TPs. Means ± sd are shown.
TP-Induced Monomerization of Toc34
A second specific interaction of TPs has been their ability to disrupt the stability of the Toc34 homodimer. The concentration-dependent monomer-dimer equilibrium of ps-Toc34 has been well documented (Sun et al., 2002; Weibel et al., 2003; Reddick et al., 2007; Yeh et al., 2007; Koenig et al., 2008). Although the specific mode of binding is not known, it is clear that TP interaction with Toc34 dimer shifts the equilibrium toward a monomer. A dynamic equilibrium existed between the 3.0 S monomeric and 3.4 S dimeric species of ps-Toc34 in solution when analyzed by analytical ultracentrifugation (Figures 2C, black line, and 2D). Binding of TP increased the sedimentation coefficient of Toc34 homodimer to 3.9 S. Analysis of the areas under the curves of monomer and dimer peaks revealed that addition of SSF disrupted the 3.9 S dimer and increased the relative amount of 3.0 S monomer Toc34 (Figures 2C, green versus black lines, and 2D). While quantitatively not the same as that induced by SSF, SSR also stimulated the dimer-to-monomer transition of ps-Toc34 (Figures 2C, blue line, and 2D). SSF stimulated what was effectively a 50% monomer-dimer equilibrium to be biased toward essentially a 75/25% monomer-dimer distribution, whereas the addition of SSR resulted in only a 60/40% monomer-dimer distribution (Figure 2D). Here, we have shown that reversing SSF to SSR only slightly impaired the wild type in vitro activity. This indicates that the isolated TOC component ps-Toc34 is able to bind and be stimulated (either in its hydrolytic activity or in its monomer-dimer equilibrium) by both forward and reverse TPs. Interestingly, neither FDF nor FDR biased the monomer-dimer equilibrium of Toc34 (Figure 2D). The relationship between how TPs stimulate GTP hydrolysis and how this activity correlates with the disruption of Toc34 homodimer is unclear; however, the fact that the ferredoxin TPs contains only one FGLK motif may suggest that a very high local concentration of two FDF/FDR peptides would be required to be functionally equivalent to the apparent concentration of the two tethered motifs found within a single SSF/SSR peptide.
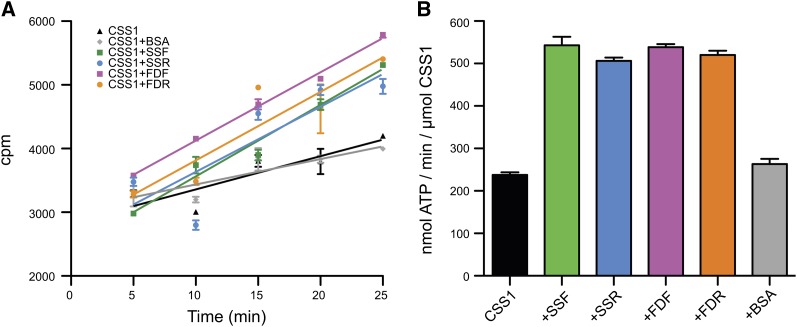
TP Stimulation of CCS1 ATP Hydrolysis
CSS1 is the Hsp70 homolog in the chloroplast stroma and is responsible for binding to TPs as they emerge from the TIC during chloroplast import (Ivey et al., 2000). CSS1 uses the energy of ATP hydrolysis to actively pull the TP through the TOC and TIC, thus completing import. We wanted to test if forward and reverse peptides stimulated the ATPase activity of CSS1 equally. Spinach (Spinacia oleracea) CSS1 exhibited a basal ATPase rate of ∼230 nmol ATP/min/µmol CSS1, and this rate was not stimulated further by addition of BSA (Figure 3). The addition of SSF, SSR, FDF, or FDR stimulated the rate of CSS1 ATP hydrolysis to very similar levels. Taken with the CD data of the unfolded peptides (Figures 1H and 1I), this suggested that unfolded peptides stimulate CSS1 and sequence information was not a factor.
Figure 3.
Effect of TPs on CSS1 Hydrolysis Rate.
(A) Time course of ATP hydrolysis rate of spinach CCS1 upon the addition of the different peptides. cpm, counts per minute.
(B) A bar graph of the calculated rate of ATP hydrolysis derived from the time course in (A) (n = 3, and the error bars indicate the mean ± se).
In Vitro Chloroplast Binding Assays
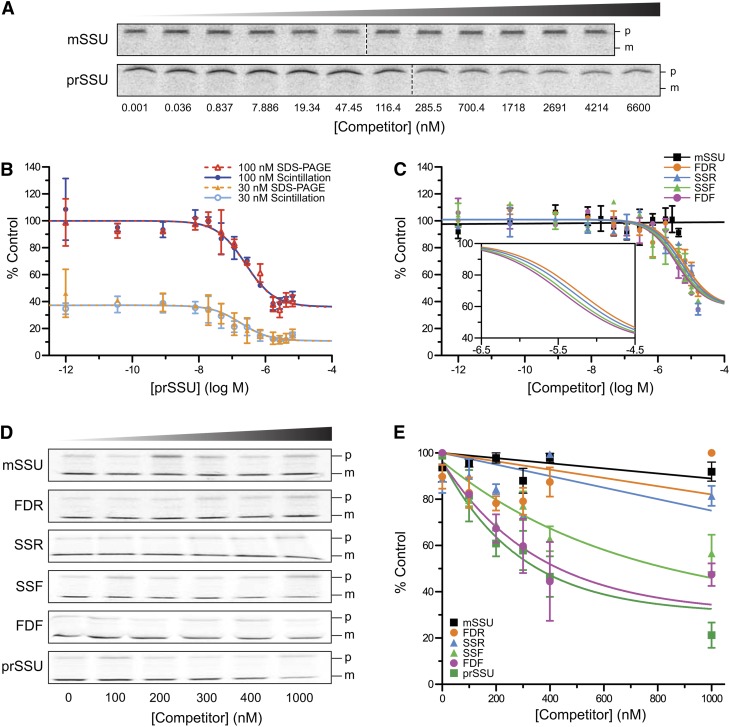
During chloroplast protein import, it is possible to trap an intermediate state specifically associated with the chloroplast, but not yet internalized (Olsen and Keegstra, 1992). This intermediate can be observed using radioactivity or by various fluorescence assays, such as flow cytometry or confocal microscopy, that permit the quantification and imaging of the bound precursor (Subramanian et al., 2001). In the past, we employed quantitative import competition assays that used competitive inhibitors to determine specific inhibitory values of the import process, such as the inhibitor constant (Ki) and the half maximal inhibitory concentration (IC50) (Dabney-Smith et al., 1999). Here, we used the competition assays to evaluate the binding of the radiolabeled precursor of small subunit of Rubisco (35S-prSSU). Binding was abrogated and eventually prevented by the addition of nonradiolabeled competitors. These competitors bound to one or more of the components of the translocon and blocked binding of the 35S-prSSU (Figures 4A to 4C). Since there was only one bound species (i.e., no processed form in Figure 4A), we developed a liquid scintillation counting (LSC) assay that would be more rapid and robust than the SDS-PAGE autoradiography assay that is traditionally used. For comparison, the LSC and autoradiograph were used to determine the equilibrium dissociation constant (Kd) of prSSU (Figure 4B). The LSC yielded nearly the same Kd as the SDS-PAGE autoradiography-based assay at 153.8 and 153.1 nM, respectively (see Supplemental Table 4 online). Thus, the separation by SDS-PAGE is not required, and we report that LSC can be used to determine the results of this assay in a quantitative manner precisely and quickly. Using this more rapid assay, we then tested the ability of SSF, SSR, FDF, and FDR to function as competitive inhibitors in binding. As expected, the mature domain of the small subunit of Rubisco (mSSU) did not compete with 35S-prSSU for chloroplast binding (Figure 4C, black). Both forward peptides, SSF and FDF, competed for binding (Figure 4C, green and magenta); surprisingly, both reverse peptides, SSR and FDR, also competed (Figure 4C, blue and orange circles). The Ki and IC50 of the forward peptides were slightly lower than those of the reverse peptides (see Supplemental Table 5 online).
Figure 4.
Both Forward and Retro-TPs Compete for Chloroplast Binding, but Retro-TPs Do Not Compete for Chloroplast Import.
(A) Representative autoradiograms of SDS-PAGE gels from binding assays using 100 nM 35S-prSSU in the presence of mSSU or prSSU as indicated. Only the bound precursors (p) were detected. The import-processed mature proteins (m) are undetected. The conditions of the assays prevented the import of 35S-prSSU.
(B) Homologous binding assays of prSSU measured by scintillation counting (solid lines) or SDS-PAGE autoradiography (dashed lines). To improve fitting confidence, two concentrations of 35S-prSSU, 30 nM (bottom traces) and 100 nM (top traces), were globally fitted together. Fittings with data collected from autoradiography of SDS-PAGE gels were comparable to those collected using scintillation counting (n = 2, and means ± sd are shown).
(C) Competitive binding assays of the competitors using scintillation counting. Inset shows close-up fitting curves of the peptides (n = 3, and means ± sd are shown). The data of mSSU were fitted to a straight line.
(D) Representative autoradiograms of SDS-PAGE gels from the import assays in the presence of 100 nM 35S-prSSU. Competitors are indicated on the left. Concentrations are shown at the bottom. p and m, precursor and mature protein sizes.
(E) Quantification of imported 35S-prSSU from the import assay partially represented in (D). In (D) and (E), means ± sd are shown (n = 3). The data from mSSU, SSR, and FDR were fitted to straight lines.
In Vitro Chloroplast Import Assays
When incubated with precursor proteins, isolated chloroplasts import and process precursors to mature proteins, as evidenced by a shift in size from a larger precursor to a smaller TP-devoid mature protein (Figure 4D) (Friedman and Keegstra, 1989; Dabney-Smith et al., 1999). The import competitions can be performed by titrating competitors to a constant concentration of 35S-prSSU. The amount of imported 35S-prSSU determines the competitiveness of the competitors. We performed assays with the forward and reverse TPs, as well as mSSU and prSSU as negative and positive controls, respectively (Figure 4D). As expected, prSSU competed against 35S-prSSU (Figure 4E, dark green) and mSSU did not (Figure 4E, black). Both forward peptides SSF and FDF competed for import (Figure 4E, green and magenta). Unlike the earlier in vitro hydrolysis, monomerization, and binding assays, these import assays reveal that SSR and FDR do not compete (Figure 4E, blue and orange), suggesting that the chloroplast translocon was able to distinguish between forward and reverse sequences effectively. The inhibition curves were fitted to one-phase exponential decay. The IC50 of prSSU, FDF, and SSF was determined to be 202.7, 247.8, and 480.3 nM, respectively.
In Vivo Imaging of Pro- and Retro-Peptide Fusion Proteins
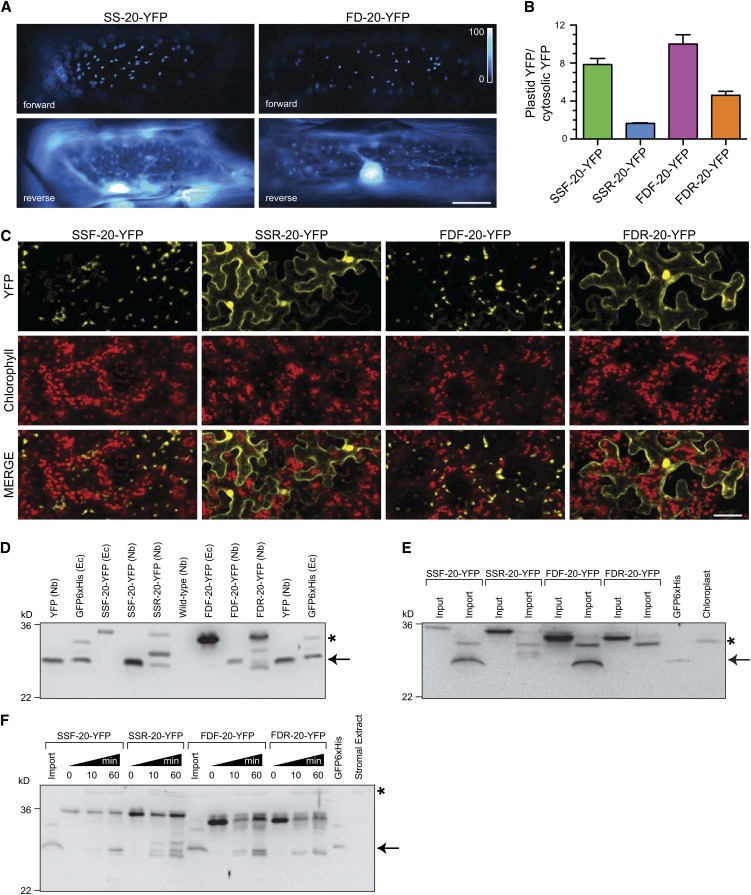
In addition to in vitro competition assays, we directly tested the efficiency of the forward and reverse TPs in directing the import of yellow fluorescent protein (YFP) into plastids/chloroplasts. The fusion proteins are chimeric proteins containing a TP fused to the first 20 amino acids of tobacco mSSU followed by YFP. The localization of transiently expressed TP-YFP fusion proteins were observed in onion epidermis peels, tobacco leaves, and Arabidopsis seedlings, as shown in Figures 5A and 5C and Supplemental Figure 2A online, respectively. In agreement with in vitro import assays, the forward TPs, SSF and FDF, directed YFP to the plastids/chloroplasts as observed by the punctate pattern in onion cells (Figure 5A), colocalization with chlorophyll in tobacco (Figure 5C), and colocalization with plastid cyan fluorescent protein marker in Arabidopsis (see Supplemental Figure 2A online). Using the retro-TP fusion proteins, both SSR and FDR showed low efficiency in directing YFP to the plastids/chloroplasts. Most of the YFP signals were observed outside of plastids/chloroplasts (Figures 5A and 5C; see Supplemental Figure 2A online).
Figure 5.
Plastid Targeting of Fluorescence Proteins Directed by Forward and Retro-TPs.
In vivo plastid targeting functions of TPs were observed using N terminus fusions of TPs linked to the first 20–amino acid sequence of mSSU from tobacco followed by YFP.
(A) Localization patterns of transiently expressed TP-YFP fusion proteins in onion epidermis cells observed under ×20 objective. SS and FD, TPs of small subunit of Rubisco and ferredoxin, respectively. Bar = 50 μm.
(B) Quantitative analysis of plastid targeting represented in (A) (n = 20). Error bars indicate the mean ± se.
(C) Localization patterns of transiently expressed YFP fusion proteins in N. benthamiana leaves. Autofluorescence of the chlorophylls is used as a chloroplast marker. Only some YFP signals overlap with chlorophyll signals because plastids in epidermis cells do not contain chlorophylls. Top labels indicate TPs. Left labels indicate fluorescent signals. Bar = 50 μm.
(D) Immunoblot analysis of YFP targeting in total protein extracts from tobacco leaves (similar to the leaves in [C]) with anti-GFP antibody. Top labels the expressed proteins. Nb and Ec indicate tobacco and E. coli extracts, respectively. The mature forms (arrow) with sizes similar to YFP were observed in all fusion proteins expressed in tobacco, indicating the import and the cleavage site occur before YFP domain. In addition, precursor and intermediate forms were observed in retro-peptide fusion proteins compared with full-length precursor from E. coli. The mature form of SSR-20-YFP showed further degradation. Asterisk indicates nonspecific bands from E. coli extract.
(E) Immunoblot analysis of in vitro imported fusion proteins. Equal protein amounts of reisolated chloroplasts were loaded. The nonspecific species from chloroplast is indicated with an asterisk.
(F) Immunoblot analysis of in vitro stromal processing assay with anti-GFP antibody. The processed forms (arrow) are comparable to that of in vivo import and in vitro import assay ([D] and [E]). All of the fusion proteins can be cleaved in stromal extract. The nonspecific species from stromal extract is indicated with an asterisk.
One of the challenges in analyzing results from these in vivo assays is the lack of a quantitative measure similar to the Ki and IC50 generated from in vitro assays. Here, we developed a method to quantify the efficiency of TP in directing the import of YFP into the plastids in onion cells. The efficiency is expressed as the relative intensity ratio between the plastid YFP and cytosolic YFP signals. The ratio measurements were performed on the transiently expressed onion cells similar to that of Figure 5A, and the results are shown in Figure 5B. As expected, the average ratios for SSF and FDF were high at 7.85 and 10.0, respectively, because most of the YFP was targeted to the plastids. SSR and FDR showed lower average ratios at 1.65 and 4.61, respectively, indicating lower efficiency in directing the import.
Immunoblotting of Import and Processing of Pro- and Retro-TP Fusion Proteins
To investigate the ability of chloroplast SPP to process the chimeric fusion proteins and to confirm the in vivo import of the proteins, immunoblotting was used to detect YFP at the C terminus of fusion proteins. Immunoblotting of total protein extract from tobacco leaves transiently expressing fusion proteins showed processed forms that are indicative of protein targeting into the chloroplasts (Figure 5D). Retro-TP fusion proteins resulted in a lower amount of processed forms, indicating the lower efficiency of import similar to the in vivo imaging results. Note that the fusion protein expression levels were not equal. Different amounts of protein extracts were loaded to visualize approximately equal signals. In vitro import of the fusion protein was performed and is shown in Figure 5E. The reisolated chloroplasts showed processed forms similar to total protein extracts from tobacco for forward TPs, confirming that the processing occurred in the chloroplasts. Because the same amount of reisolated chloroplast protein was loaded here, we failed to detect the processed forms of retro-TP fusion proteins, indicating that they were imported with less efficiency. In vitro stromal processing assays were performed using the stromal extract (Figure 5F). Here, the SPP was able to cleave all fusion proteins into the processed species similar to in vivo and in vitro import of the fusion proteins.
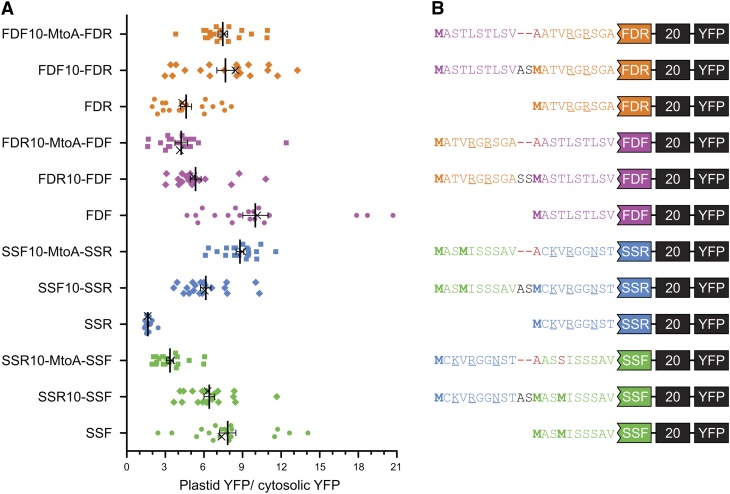
Role of the First 10 N-terminal Residues on in Vivo Targeting
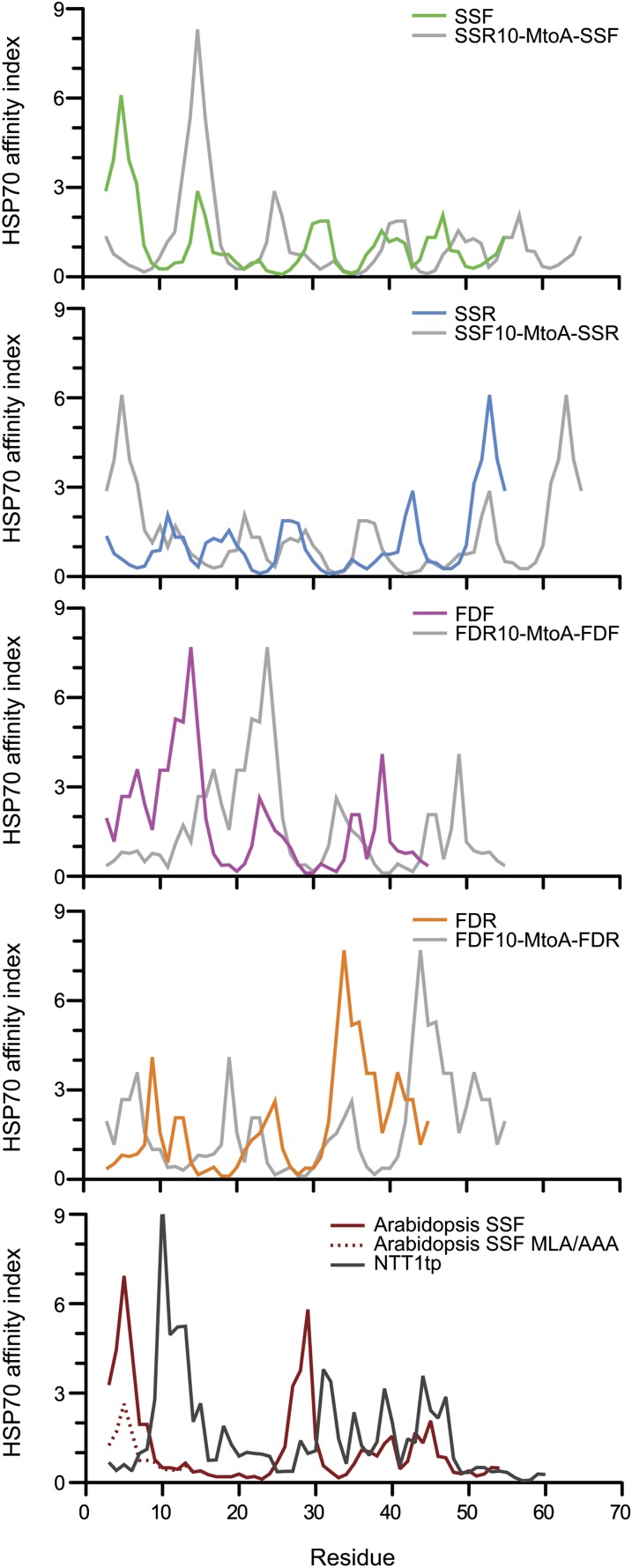
Based on previous observations that TPs harbor an uncharged N terminus (von Heijne et al., 1989) and our analysis in Figure 1D, it is possible that the weak import efficiency of retro-TPs is due to charged amino acid present within the N terminus. To investigate the role of an uncharged N terminus, the charged N termini retro-TP YFP fusion proteins were altered by appending an extra 10 amino acids corresponding to the N terminus of the opposite pair TP (i.e., the extra SSR fusion protein contains the N-terminal 10 amino acid of SSF at the N-terminal of former SSR fusion protein) as shown in Figure 6B. The alteration of the uncharged N termini of pro-TP fusion proteins to charged N termini was generated in the same manner. In vivo targeting using onion epidermis cells was performed (see Supplemental Figure 2B online), and the plastid/cytosolic YFP ratio was calculated (Figure 6A). The extra residues seem to invert the import efficiencies of the former TPs. The fusion proteins containing Met following the extra sequence show moderate change compared with the fusion proteins with an Ala substitution of Met. This result indicates the possibility of two translation start sites at the first and second Met residues, which potentially leads to the production of a protein mixture containing both low and high import efficient proteins. In addition to the ratios obtained from 12 h after transformation, in some cases, we calculated the ratio using the images captured 24 h after transformation (see Supplemental Figure 2C online). The results showed that the ratios obtained from 24 h are always slightly higher than those from 12 h. Finally, using two algorithms to predict the Hsp70 binding site, we observed that all import-efficient peptides harbor a strong Hsp70 binding site at the N terminus, whereas the sites are not present in the import-deficient peptides (Figure 7; see Supplemental Figure 3 online).
Figure 6.
Uncharged N Terminus of TP Determines Chloroplast Import Efficiency.
Extra amino acid sequences representing the first 10–amino acid sequence from the opposite TP were added to the N terminus of each TP-YFP fusion construct. In vivo plastid targeting was observed via transient expression in onion epidermis.
(A) Plastid targeting efficiencies of TP-YFP fusion constructs. Left labels indicate TPs in the constructs. The extra 10 residues were named based on the sourced TP and indicated with suffix 10. MtoA indicates the substitution of the internal Met with Ala. Ratio intensities were measured from 20 cells. The images of cells marked with X are shown in Supplemental Figure 2B online.
(B) The partial N terminus sequences of the constructs used in (A). Met residues are in bold. Substitutions are in red. Additional amino acid residues from restriction sites are in black. Charged amino acids are underlined.
Figure 7.
Hsp70 Binding Site Prediction.
Two Hsp70 binding site prediction algorithms developed from E. coli Hsp70, DnaK, and binding assays were used in the analysis. Prediction results using algorithms by Ivey et al. (2000) derived from Gragerov et al. (1994) are shown here, and those of Rüdiger et al. (1997) are shown in Supplemental Figure 3 online. The import-efficient peptides seem to harbor a strong predicted Hsp70 binding site at the N terminus (area under curve within the first 10 amino acids) where it is lacking in the import-deficient peptides. The results generated from the algorithm derived from Gragerov et al. show strong agreement in every prediction. However, the algorithm developed by Rüdiger et al. show some disagreement in the predictions of SSF and SSR where the calculated energy contribution within the first 10 residues shows no difference. NTT1tp is the TP of Arabidopsis nucleotide transporter 1.
DISCUSSION
Flexible Recognition of TPs by Toc34
Although previous studies have reported that cytosolic factors initially recognize TPs/precursors, kinetic arguments suggest that in vitro import can attain import rates without cytosolic factors to support organelle biogenesis (Pilon et al., 1992; May and Soll, 2000). This very rapid binding and translocation occurs despite a very low density distribution of TOC complexes on the plastid outer membrane (≤1 TOC/13,600 nm2) (Friedman and Keegstra, 1989; Schleiff et al., 2003). One potential strategy to increase the kinetics of productive TP binding with the TOC complex is to relax the structural constraints of the recognition mechanism.
Although the structural basis of initial binding is unclear, it is probably mediated by one of the TOC GTPases. While the interaction between TP and Toc34 clearly has been shown (Sveshnikova et al., 2000; Jelic et al., 2002; Schleiff et al., 2002; Reddick et al., 2008), it is unknown how and where Toc34 binds to the TP. The shortest peptide directly interacting with Toc34 is the B1 peptide corresponding to amino acids 22 to 47 of tobacco SSF (Schleiff et al., 2002). These residues in pea and Arabidopsis SSFs contain two FGLK motifs (Pilon et al., 1995), suggesting that the FGLK motif is required for Toc34 recognition. Since our heuristic analysis of targeting sequences showed the best discrimination of chloroplast TPs when the FGLK motif was required, this motif may be specifically recognized by Toc34 during chloroplast protein import. We have shown that both pro- and retro-TPs can stimulate Toc34 GTP hydrolysis in vitro. This ability to interact productively with the TP independent of collision orientation is supported by the observation that both SSF and SSR can shift the monomer-dimer equilibrium of Toc34 toward the monomer form. Disruption of Toc34 dimerization by TPs may or may not be required for effective preprotein import but does appear to be a consequence of either TP binding and/or structural changes arising from GTP hydrolysis.
TPs are largely unstructured in solution, forming a perfect random coil (von Heijne and Nishikawa, 1991), suggesting that TPs may be a new example of IDPs or at least contain significant segments that are predicted to be IDPs. Mechanistically, the ability of IDPs to assume different conformations when interacting with different binding partners (Kriwacki et al., 1996; Tompa et al., 2005; Uversky et al., 2008; Narayan et al., 2011) may explain how multiple interactions can be accommodated by the relatively short TPs during binding and translocation. The kinetics of favorable recognition between two components may be accelerated by increasing the encounter productivity using the fly-casting model (Shoemaker et al., 2000), which proposes that TPs are rapidly recognized regardless of whether the peptide binds to the receptor with one topological orientation or the opposite. This model of binding would accommodate both the forward and retro-TPs being recognized by one or more of the TOC GTPases. The ability to successfully recognize TPs with either an N- or C-terminal orientation would greatly accelerate preprotein binding and processing. This may explain how kinetics of posttranslational preprotein import in vitro may be able to match the maximum rates predicted in vivo during greening and chloroplast development (Pilon et al., 1992) without the need for cytosolic factors or the topological facilitation provided during the cotranslational processes of protein transport.
Chaperone Interactions with TPs
Our work has clearly implicated the N-terminal region of two model TPs as being important for translocation. Prior work has shown multiple interactions of TP N termini with lipids (Pilon et al., 1995; Pinnaduwage and Bruce, 1996) and the import receptor Toc159 (Lee et al., 2009). However, our study demonstrates that retro-TPs were able to compete with prSSU for binding to the chloroplasts, yet are unable to direct fusion protein import into the plastids, suggesting that the discrimination of TPs during translocation is mediated by components trans-relative to the outer envelope components. Based on prior work in our lab (Ivey et al., 2000) and others (Rial et al., 2000; Zhang and Glaser, 2002), there is a clear precedent for N-terminal sequences of TPs to interact with the Hsp70 class of molecular chaperones that may be located in either the IMS or the stroma. Although TP interactions with chaperones localized in the IMS could not be ruled out, only two (Hsp70-6 and Hsp70-7) of the 14 Arabidopsis Hsp70s are predicted to be chloroplast localized and both are localized in the stroma (Ratnayake et al., 2008). Moreover, there is no evidence of any Hsp93 homolog to be localized to the IMS (Constan et al., 2004). Thus, it appears that the critical role of the N-terminal residues is in fact mediated by stromal chaperones.
Although Hsp70s were initially proposed to mediate multiple steps in chloroplast protein import (Marshall et al., 1990), similar to what has been observed in mitochondria (Tomkiewicz et al., 2007), it is a member of the Hsp100 family, stromal Hsp93, that may drive import (Cline and Dabney-Smith, 2008; Jarvis, 2008). The essentiality of this chaperone family has been shown in Arabidopsis where double knockout of the two homologs HSP93-V (ClpC1) and HSP93-III (ClpC2) proved lethal (Kovacheva et al., 2007). Although Hsp93 interacts with translocon components as well as precursors (Nielsen et al., 1997), interaction with TPs remains unclear. Furthermore, recent in vivo work in both Arabidopsis (Su and Li, 2010) and Physcomitrella patens (Shi and Theg, 2010) has clearly implicated Hsp70s as having a role in chloroplast protein import. Recent evidence suggests that chloroplasts may have two separate chaperone systems facilitating protein translocation into the stroma: the cpHsc70 system and the Hsp93/Tic40 system (Su and Li, 2010). Furthermore, protein import in chloroplasts from the cphsc70-1 hsp93-V double mutant had a more severe import defect than either of the single mutants, suggesting that the two proteins function independently, possibly interacting with a discreet subset of substrates.
Although Hsp93 and Hsp70 may both play vital roles in chloroplast protein import, considerably more is known about TP recognition by Hsp70 (Ivey et al., 2000; Rial et al., 2000; Zhang and Glaser, 2002). Bioinformatic and experimental approaches demonstrate that the N-terminal region of most TPs (>75%) has the highest affinity for Hsp70. Our work confirms this observation, as only the import-efficient TPs contained a strong Hsp70 binding site at the N terminus. Interestingly, the fact that all four of our TPs were able to interact productively with CSS1 in solution, yet placement of a non-Hsp70 binding segment in front of an existing binding domain reduced the translocation of a precursor in vivo, evokes a specific placement requirement for functional Hsp70-interacting sequences. Although we observed in vitro ability of forward and reverse TPs to interact with two individual translocon components, Toc34 and CSS1, the sequence determinants for actual translocation of the precursors through the TOC are more complex and strongly influenced by the N-terminal 10 amino acids. The importance of the highly uncharged N terminus of TPs (von Heijne et al., 1989) in chloroplast import has been shown repeatedly both in vitro (Pilon et al., 1995; Rensink et al., 1998) and in vivo (Lee et al., 2002; Lee et al., 2006, 2008, 2009). Collectively, it appears that the N terminus of the TP is involved only in the binding step.
Our findings provide examples of the separate recognition requirements during binding and translocation. The retro peptides behave differently from other N-terminal mutated TPs (Pilon et al., 1995; Rensink et al., 1998; Lee et al., 2002; Lee et al., 2006, 2009) such that they are undistinguishable from pro-TPs in preprotein binding (Figures 2 and 4C) yet were unable to direct translocation in vivo (Figures 5A to 5C) or in vitro (Figures 4E and 5E). These retro-peptides will permit us to elucidate further the role of the N-terminal region of TP as a stromal-protein interacting site.
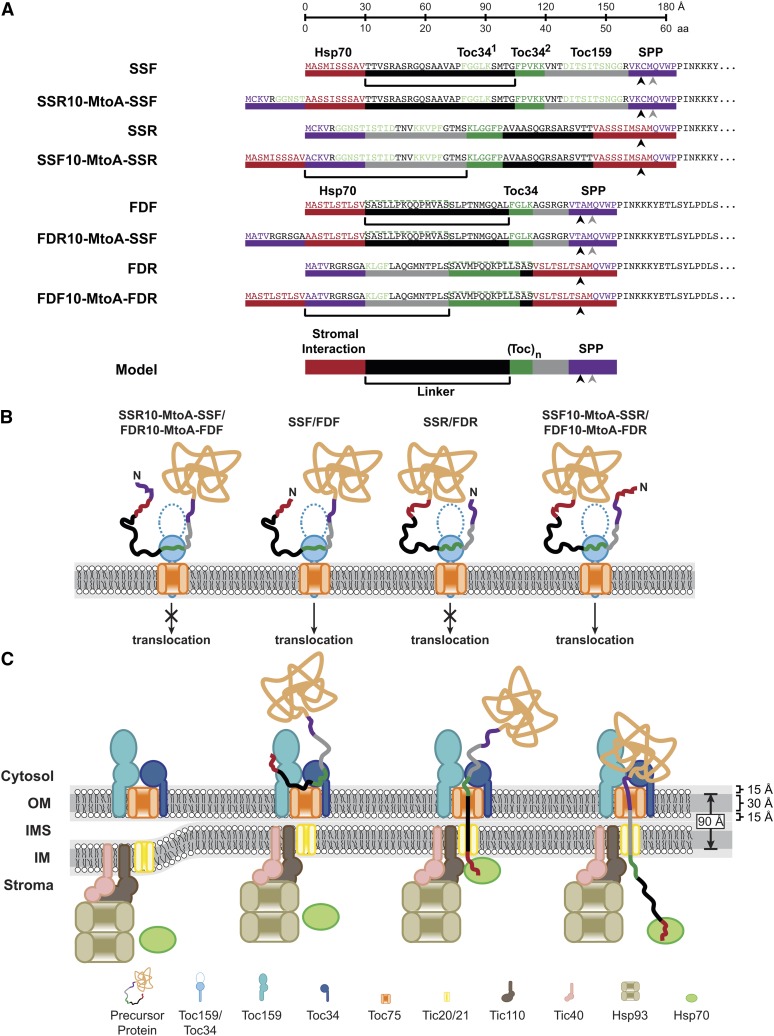
Bimodal Model of TP Design
Based on previous studies and our observation that the N-terminal Hsp70 binding site of the TP is the major determinant for translocation, we propose a model describing a bimodal TP architecture (shown at the bottom of Figure 8A), which allows a TP to engage TOC receptors and stromal chaperones concurrently. We suggest the TP contains an N-terminal stromal protein recognition site linked to a TOC receptor recognition site via a linker region along with a SPP recognition site at the C terminus (Figure 8A). We demonstrate that the N-terminal Hsp70 binding site of SSF and FDF determines the translocation of preproteins into chloroplasts (Figures 6, 7A, and 7B). Evidence also suggests that stromal Hsp70 is a chloroplast translocation motor (Theg et al., 1989; Marshall et al., 1990; Schnell et al., 1994; Shi and Theg, 2010; Su and Li, 2010). Based on the trapping and pulling model in endoplasmic reticulum and mitochondria protein import (Tomkiewicz et al., 2007), and the proposed unfolding and pulling model in chloroplast import (Keegstra and Cline, 1999), it is tempting to predict that interaction of the TP with the stromal chaperones is required to trap or pull the precursor into the chloroplasts, which is supported by our observation of a requirement for an N-terminal Hsp70 binding site for preprotein import. Apart from stromal Hsp70, Hsp93 is another example of stromal protein involved in chloroplast import, although the Hsp93 recognition sequence on the TP is unclear (Nielsen et al., 1997; Chou et al., 2006; Kovacheva et al., 2007). We propose that TPs harbor a stromal protein recognition site at their N termini, which allow the stromal proteins to trap and/or pull TP during translocation process (rule 1).
Figure 8.
Bimodal Model of TP Design.
(A) The recognition elements in pro-, retro-peptide, and the MtoA constructs are shown. The sequence was highlighted and marked with color bars to indicate different elements. Predicted Hsp70 binding sites are colored red. Proposed Toc34 (FGLK) and experimentally determined Toc159 binding sites are colored green. Predicted SPP recognition sites are colored in purple, with the black and gray arrowheads indicating TargetP predicted and actual cleavage sites, respectively. Top ruler bar shows the amino acid length and the length of experimentally determined unfolded protein (∼3.0 Å/amino acid [aa]). The black bar indicates the linker region. The general model of a TP is shown in the bottom.
(B) Depiction of translocation competent and incompetent fusion protein interactions with Toc34 receptor. As unstructured proteins, pro- and retro-peptides can engage Toc34 in opposite orientations. However, only the proteins containing an N-terminal Hsp70 site are able to translocate across the membrane.
(C) Depiction of concurrent TP recognition by Toc34 and stromal Hsp70. As detailed in the Discussion, an import-efficient TP is proposed to harbor an N-terminal stromal interacting site and TOC receptor binding site separated by the linker with a preferred length that allows the concurrent engagement of a TOC receptor and stromal motors through the double membrane. The hydrophobic core of the double membrane at the contact site is assumed to be 90 Å. IM, inner membrane of the chloroplast envelope; IMS, intermembrane space; OM, outer membrane of the chloroplast envelope.
Other portions of TPs have been shown to interact with lipids and TOC components to be targeted to the chloroplast surface (Bruce, 2000). Manipulation of the Toc34 recognition site by deletion of FGLK motif in FDF (Rensink et al., 1998) or Ala scanning of the second FGLK motif in Arabidopsis SSF (Lee et al., 2006) was shown to inhibit translocation but still permit binding. Thus, TP-mediated precursor translocation is determined by both the N-terminal Hsp70 binding site and the Toc34 binding site. Nevertheless, the N-terminal Hsp70 binding site seems to be the major determinant for translocation, as it seems that it can overrule Toc34 recognition. As evidence for this, both pro-TPs and retro-TPs contain FGLK motifs, but only those harboring an N-terminal Hsp70 binding site are efficiently translocated (Figures 1A, 4D, 4E, 5, and 6; see Supplemental Figures 2 and 3 online). Upon deletion of the N-terminal Hsp70 binding site, Δ1-14 (Pilon et al., 1995) and Δ6-14 (Rensink et al., 1998) of FDF, and ΔT1 (amino acids 2 to 12) of E1α-subunit of pyruvate dehydrogenase TP (Lee et al., 2009), translocation was abrogated. By contrast, a variant of N-terminal Hsp70 binding site mutant of Arabidopsis SSF, the MLM/AAA mutant (Met-5, Lue-6, and Met-11 were substituted with Ala), has decreased import efficiency (Lee et al., 2009), suggesting that the MLM/AAA mutant has lower Hsp70 affinity than that of the wild type as predicted in Figure 7. Interestingly, translocation was restored when the N terminus of MLM/AAA mutant was fused to the C terminus of E1α TP in wild-type Arabidopsis (Lee et al., 2009), suggesting that this new C terminus may provide a higher affinity binding site and compensate for the low affinity Hsp70 binding site in MLM/AAA. In fact, the translocation of MLM/AAA chimera TP was not restored when expressed in the Toc159 knockout mutant ppi2, suggesting that Toc159 interaction can substitute for reduced Hsp70 affinity (Lee et al., 2009). Thus, there is a direct connection between binding and translocation steps in chloroplast import. While the stromal protein interaction at the N terminus could possibly provide the trapping and pulling mechanism, the initial interaction with TOC components may capture or trap the TP at the surface. The TOC GTPases may provide a mechanical pushing force through domain movements in response to nucleotide status.
Spatial Requirements for Concurrent TP Recognition
The interconnection between stromal and surface interactions can occur efficiently only if both are occurring concurrently or sequentially within the rapid timeframe of protein import. Thus, for a given unfolded preprotein to engage two binding elements concurrently during translocation, the relative spatial distance between interaction domains may be a critical feature of TP design. We observed that the FGLK motif location of FDF and SSF is relatively conserved, suggesting the existence of a preferred TOC interaction site. Assuming an N-terminal Hsp70 binding site within the first 10 amino acids of a TP, the linker length between the stromal interaction site and the TOC interaction site is 22, 24, and 25 amino acids in Arabidopsis SSF, FDF, and pea SSF, respectively. We proposed that there is a linker with preferred length of ≥22 amino acids that connects the N-terminal stromal interaction site with the TOC receptor interaction site (rule 2). Using the TP of Arabidopsis nucleotide transporter 1, the overall TP length requirement has been shown to be ∼60 amino acids to translocate titin protein (Bionda et al., 2010). However, based on the cleavage site predictions, the length of this TP is only 21 amino acids, which is much shorter than most of the TPs. In relation to our model, we predicted that this TP also contains an N-terminal Hsp70 binding site (Figure 7). According to the cleavage site prediction, this TP could not accommodate a preferred linker size. However, when analyzed further, we observed an FGLK motif at amino acids 35 to 39, allowing a linker of 24 amino acids. Thus, it is possible for the two rules to be accommodated, even if the TOC interacting domain falls within the mature domain C-terminal to the SPP cleavage site.
Prior work suggests that protein import takes place at contact sites between the chloroplast outer and inner envelope membranes (Schnell et al., 1990), which are rich in galactolipids (Bruce, 1998). The thickness of mono- and digalactosyldiacylglycerol bilayers has been measured to be 55 and 60 Å, respectively (Marra, 1985; Bottier et al., 2007). Within this distance, we assume an organization similar to a model bilayer containing a 30-Å hydrophobic core with two 15-Å polar regions on the two surfaces (White and von Heijne, 2008). For two tightly pressed membranes, the thickness of the two hydrophobic cores of outer and inner membranes would be ∼90 Å (Figure 8C).
The conformation of a TP during translocation is unknown, so we looked at other systems where extended peptide lengths are known for fully unstructured peptides. Using atomic force microscopy, the extendibility of peptides under different forces has been measured and suggests an average length of ∼3.0 Å/amino acid (Rief et al., 1997; Chyan et al., 2004). Using the thickness of the chloroplast envelope and this value for the extended TP, the shortest linker of 22 amino acids would have a length of 66 Å. Although this observation suggests that the linker could not span 90-Å double membranes, the N terminus of TP could reach the stromal side with the length of 96 Å through the addition of 10 amino acids of Hsp70 binding site. Thus, the translocation in our model can occur only where the TP is bound within the TOC complex or released from TOC receptor prior to interaction with stromal protein (Figure 8C). In this model, the TOC receptor functions both to target TP from the cytosol to chloroplast and also functions to prevent the TP from escaping the translocon prior to stromal capture. Evidence for this dual trapping model was observed previously using a modified TP with an N-terminal epitope tag (His-S), which introduces multiple charged residues at the N terminus (Subramanian et al., 2001). This TP is able to compete for binding but is unable to be imported into the chloroplast. We now suspect that the His-S tag prevents the natural N terminus from interacting with the stromal chaperones. In support of this, binding of the TP was not influenced by the addition of ATP; however, when GTP was added, binding was disrupted, suggesting that GTP promotes preprotein release/transfer, which if not captured by the stromal chaperone, may escape from the translocon.
Conclusion
In this work, we combine both quantitative in vitro and in vivo analyses of the efficacy of chloroplast TPs with a large-scale bioinformatics analysis of TP sequences. Our results converge on a new model of modular design TP organization and function. This design requires the placement of a specific N-terminal domain of the TP that must be able to interact productively with one or more Hsp70 class of molecular chaperones. This domain is followed by a second element that interacts with one or more components of the TOC apparatus, which can promote binding, yet alone cannot support translocation. This supports the prior evidence that translocation is driven by a stromal ATP-dependent process that may include (but not be limited to) Hsp70-mediated recognition. Although these sequences are degenerate in nature, we observe a key spacing requirement that may reflect the coordinated translocation of the preprotein across both membranes at contact sites where the TOC and TIC complexes are tightly appressed. With this advance, we are now in position to design TP variants that may allow these spatial and energetic requirements to be tested directly. Finally, this advance may provide insight into the evolution of chloroplast preproteins and partially explain the high variability of TPs length, composition, and sequence.
METHODS
Codon Optimization and Cloning of Synthetic DNA
The wild-type amino acid sequences of Silene latifolia ferredoxin and pea (Pisum sativum) small subunit of Rubisco TPs were codon optimized using Gene Designer 1.0.7.3 (Villalobos et al., 2006) based on the codon usage of Escherichia coli highly expressed genes (Hénaut and Danchin, 1996). The adaptiveness and CAI values were calculated as previously described (Sharp and Li, 1987). For cloning, the NdeI and XmaI sites were added (see Supplemental Figure 1A online). The synthetic DNAs (Epoch Biolabs) were cloned into a pTYB2 vector (New England Biolabs) and confirmed by sequencing.
Protein Expression and Purification
Recombinant, ΔTM-His-tagged ps-Toc34 was prepared as previously published (Reddick et al., 2007). Forward and reverse peptides were expressed from pTYB2 constructs and purified as described previously (Reddick et al., 2008). The peptides were resolved by SDS-PAGE using a 19.2% Tris/Tricine gel and visualized by Coomassie Brilliant Blue staining and by MALDI-TOF MS. The prSSU and mSSU proteins were expressed from pET-11d constructs and purified as described previously (Reddick et al., 2008). The proteins were solubilized in buffer containing 8 M urea, 50 mM DTT, and 20 mM Tris-HCl, pH 8.0. Radiolabeled prSSU was generated as previously described (Reddick et al., 2008). YFP fusion proteins were expressed from pET-30a constructs and purified as described for prSSU.
MALDI-TOF MS
MALDI-TOF MS was performed on a Bruker Daltonics Microflex mass spectrometer. Positive ion mode mass analysis of TP sequences was performed as previously described (Reddick et al., 2007) with minor amendments. Briefly, the target plate was streaked with 50 mg/mL paraffin wax in chloroform and dried under vacuum prior to applying 2 μL peptide/matrix mixture (5 pmol/μL peptide in 10 mg/mL α-cyano-4-hydroxycinnaminic acid) to increase the signal-to-noise ratio (Jackson-Constan et al., 2001). The spots were desiccated for 15 min and were washed twice with 2 μL of 10 mM diammonium hydrogen citrate.
Bioinformatic Analysis of TP Motifs
Pairwise alignment of the TP sequences was performed with the Needleman-Wunsch algorithm using Needle program in EMBOSS package (Rice et al., 2000). BLOSUM45 to 90 were tested. When gap opening and extension penalties were set as 10 and 5, respectively, BLOSUM55 generated the highest scores. The %identity and %similarity computed from BLOSUM55 are reported.
For analysis of the N-terminal property of TPs, a data set of Arabidopsis thaliana TP sequences was generated. TargetP was used to predicted TPs from the Arabidopsis genome (Emanuelsson et al., 2007). All of predicted TPs assigned reliability classes 1 and 2 with lengths from 35 to 71 amino acids (within mean ± sd) were collected into a data set resulting in a total of 912 sequences. For analysis, percentage of uncharged amino acid (not Lys, Arg, Asp, Glu, or His) was calculated within a subsequence. A window of length L was defined and moved along the whole length of the sequence. The calculated percentage in each window was assigned to the amino acid position at the center. The average percentage of uncharged amino acids of each position is the average from all the sequence in the data set. The calculation was repeated from window length L of 5 to 17 amino acids. The percentage of uncharged amino acid data within the first 30 residues was fitted to the inverted Boltzmann sigmoidal model with nonlinear regression using GraphPad Prism 5.0 (GraphPad Software).
CD Spectroscopic Analysis of TPs
CD spectroscopy was performed on an Aviv Series 202 CD spectrophotometer (Aviv Biomedical). Lyophilized TP was resuspended in water and quantified using BCA assay (Pierce). The TP was diluted to a final concentration of 100 µM in 0, 15, 22.5, 30, 45, or 60% TFE in water. Spectra were collected at 25°C as 2-s averages at 1-nm intervals from 185 to 285 nm. Three spectra were averaged, corrected for buffer contributions, smoothed, and converted to molar ellipticity using the Aviv software, version 2.71. Deconvolution was performed using the CDPro software as described previously (Sreerama and Woody, 1999).
Phosphate Release Assay for GTP Hydrolysis
Measurements of phosphate release, scintillation counting, and fitting to the Michaelis-Menten equation for determination of Vmax and Km of ps-Toc34 were all performed as previously reported (Reddick et al., 2007, 2008).
CSS1 ATP Hydrolysis
CSS1 was purified from baby spinach (Spinacia oleracea) leaves obtained from a local market. The chloroplasts were isolated as previously described (Reddick et al., 2008), and the CSS1 was purified as previously reported (Ivey et al., 2000).
ATP hydrolysis activity measurements of CSS1 were performed using an activated charcoal, phosphate release assay (Reddick et al., 2008). In a flat-bottomed 96-well microplate, reactions were performed with 1 μM CSS1 in 10 mM HEPES-KOH, pH 7.5, 10 mM NaCl, 10 nM [γ-32P]ATP, and varying amounts of cold ATP (and 10 μM of TP) in a final volume of 100 μL. The addition of the requisite amount of CSS1 initiated the reaction, and at various time points, 12.5-μL aliquots of the reaction were removed to 200 μL of 10% (w/v) activated charcoal in 50 mM HCl and 5 mM H3PO4. This suspension was clarified via vacuum filtration, and 8-μL aliquots of the filtrate were mixed with MicroScint40 scintillation fluid and counted on a TopCount NXT Microplate Scintillation Counter (Perkin Elmer Life Sciences). At least three experiments were performed. Determination of enzymatic rate was performed as described previously (Reddick et al., 2007, 2008).
Sedimentation Velocity Experiments of ps-Toc34 and TPs
Sedimentation velocity analyses were performed using a Beckman Optima XL-I analytical ultracentrifuge using the interference mode. The ps-Toc34 and TPs were dialyzed extensively into GBS buffer (20 mM Tricine-KOH, pH 7.65, 1 mM MgCl2, 50 mM NaCl, and 1 mM β-mercaptoethanol [BME]) with 10,000 and 3500 molecular weight cutoffs, respectively. The analysis sample was generated by the gentle mixing of ps-Toc34, TPs, GTP, and dialysis buffer to a final concentration of 13.5 µM ps-Toc34, 135 µM TP, and 2 mM GTP. Analytical ultracentrifugation sample cells with sapphire windows and Epon charcoal-filled two sector 12-mm centerpieces were loaded with 400 μL of sample using the final dialysis buffer as reference. Interference scans were obtained at 50,000 rpm (∼200,000g) after temperature equilibration of the An-50 Ti rotor and centerpieces for at least 1 h at 25°C. A differential distribution of the sedimentation coefficients, c(s), of the samples was fit to the experimental data using the program SEDFIT (Schuck, 2000). The solvent viscosity, η, and density, ρ, were determined to be 0.00896 g cm−1 s−1 and 1.0003 g mL−1, respectively, at 25°C using an Anton Parr densitometer. The partial specific volume, vbar, of the protein mixture was estimated to be 0.7441 mL g−1 by inputting ps-Toc34 sequence into the program SEDNTERP (Lebowitz et al., 2002). The best-fit c(s) distribution was regularized as described previously (Dam and Schuck, 2004). The c(s) distribution was then transformed to a distribution of molecular mass. The fractions of ps-Toc34 monomer and dimer were calculated by integrating the area under the sedimentation distributions from 2.6 to 3.3 S and 3.3 to 4.4 S for monomer and dimer, respectively. Two separate experiments were performed, and the fraction values reported were the averages. For presentation, the distributions were exported and graphed using GraphPad Prism 5.0.
In Vitro Chloroplast Protein Binding Assays
The chloroplasts were isolated from 12- to 14-d-old dwarf pea seedlings (P. sativum Progress No. 9) as previously described (Reddick et al., 2008). The assays were done as previously described (Reddick et al., 2008). Briefly, 300 μL reaction contained 0.25 mg chlorophyll/mL chloroplasts, 100 nM 35S-prSSU, 10 mM DTT, 100 μM Na-ATP, 2 mM MgCl2, 1% BSA, 300 mM urea, 330 mM sorbitol, 50 mM HEPES-KOH, pH 8.0, and various amounts of competitor proteins/TPs. The assays were performed under dim light conditions to prevent ATP synthesis. The bindings were stopped after 25 min of equilibration at room temperature, and the chloroplasts were reisolated and analyzed by SDS-PAGE autoradiography and/or scintillation counting as previously described (Reddick et al., 2008). For the homologous binding of prSSU, two concentrations of 35S-prSSU at 30 and 100 nM were used. Two independent assays of each concentration were performed, and the data were globally fitted to a one-site homologous binding model using GraphPad Prism 5.0. Fitting parameters are shown in Supplemental Table 4 online. For other competitors, three independent assays were performed in the presence of 100 nM 35S-prSSU, and the data were fit to one-site competitive binding model with nonlinear regression using GraphPad Prism 5.0. The fitting parameters are shown in Supplemental Table 5 online.
In Vitro Chloroplast Protein Import Assays
The chloroplasts were prepared as described above. The import assays were performed using a modified method from Dabney-Smith et al. (1999). Briefly, the competition assays were performed in a 300-μL reaction containing 0.125 mg chlorophyll/mL chloroplasts, 100 nM 35S-prSSU, 1 mM BME, 2 mM Mg-ATP, 0.5% BSA, 250 mM urea, 330 mM sorbitol, 50 mM HEPES-KOH, pH 8.0, and various amounts of competitor proteins/TPs. The reactions were stopped after incubation for 15 min at room temperature. The chloroplasts were reisolated and separated by SDS-PAGE. Autoradiography was performed on a Storm 840 PhosphorImager (GE Healthcare) followed by quantification with ImageQuant software (GE Healthcare). At least three separate assays were performed. The values were normalized to the values from the reaction with no competitor controls, and the data were fitted to one-phase exponential decay model with nonlinear regression using GraphPad Prism 5.0.
For in vitro import of fluorescent fusion proteins, isolated baby spinach chloroplasts were used in the reaction containing 0.25 mg chlorophyll/mL chloroplasts, 400 mM fusion protein, 10 mM DTT, 2 mM Mg-ATP, 0.5% BSA, 300 mM urea, 330 mM sorbitol, and 50 mM HEPES-KOH, pH 8.0, in a total of 300 μL. The reactions were stopped after 20 min incubation at room temperature. The chloroplasts were re-isolated and separated on SDS-PAGE gel for immunoblot detection. At least two experiments were performed.
In Vitro Stromal Processing Assay
Spinach chloroplasts were prepared as described above. Stromal processing assay was modified from that previously described (Richter and Lamppa, 1998). Briefly, the chloroplasts were pelleted and resuspended in 5 mM HEPES-KOH, pH 7.5, at 0.8 mg/mL chlorophyll, incubated at 4°C for 30 min, and lysed using a Douce homogenizer (Wheaton). The lysate was centrifuged at 137,000g for 30 min, and the supernatant was used as the stromal extract. The assay was performed in 100-μL reaction volume containing 60 μL stromal extract, 250 nM YFP fusion proteins, 2 mM PMSF, and 20 mM HEPES-KOH, pH 7.5. The reactions were incubated at room temperature. Samples of 50 μL were taken at 0, 10, and 60 min after incubation. The samples were immediately mixed with equal volume of sample buffer and boiled for 3 min. The samples were used for immunoblotting. Two experiments were performed.
Construction of TP Fusion Protein Expression Plasmid
A plastid marker construct pAN187 (Nelson et al., 2007) based on pBluescript (Stratagene) was used as an expression plasmid backbone in particle bombardment transformation. The pAN187 contains tobacco (Nicotiana tabacum) small subunit of Rubisco TP and 20 amino acids of mature domain followed by YFP under the control of a double 35S promoter and a nos 3′ terminator. The TP in pAN187 was replaced by a new TP using NheI and MscI sites. The new TPs were amplified from the pTYB2 constructs. The new pAN197 were named pBS-SSF-YFP, pBS-SSR-YFP, pBS-FDF-YFP, and pBS-FDR-YFP.
The vectors containing extra sequence, the first 10 amino acids of the opposite TP, at the N terminus of preexisting TP, were generated by ligation of DNA fragments into the NheI site. The DNA fragment containing the first 10 amino acids of TPs were made by hybridization of two oligonucleotides. The new constructs were named pBS-SSF10-SSR-YFP, pBS-SSR10-SSF-YFP, and pBS-FDF10-FDR-YFP. The pBS-FDR10-FDF-YFP was generated differently. The DNA fragment containing double 35S promoter and the first 10 amino acids of FDR was amplified from pBS-FDR-YFP, digested with SacI and XbaI, and cloned into pBS-FDF-YFP using SacI and NheI sites.
The mutated constructs with Met at the N terminus of preexisting TP substituted by Ala/Ser were generated using overlap extension PCR technique (Ho et al., 1989). The final PCR products were cloned into the vectors using SacI and NotI sites to replace the former cassette. The mutated vectors were named pBS-pBS-SSF10-MtoA-SSR-YFP, pBS-SSR10-MtoA-SSF-YFP, pBS-FDF10-MtoA-FDR-YFP, and pBS-FDR10-MtoA-FDF-YFP. The mutated residues are shown in Figure 6B.
For Agrobacterium tumefaciens–mediated transformation, the expression cassettes from pBS-TP-YFPs were subcloned into the binary T-DNA vector pFGC19 using SacI and HindIII sites and named pFGC-SSF-YFP, pFGC-SSR-YFP, pFGC-FDF-YFP, and pFGC-FDR-YFP.
For expression in E. coli, the fusion protein coding regions were subcloned into pET-30a using NdeI and NotI sites. The NdeI site was introduced during the PCR. The new vectors were named pET-SSF-YFP, pET-SSR-YFP, pET-FDF-YFP, pET-FDR-YFP, and pET-NtSSF-YFP.
All of the oligonucleotides used for cloning are listed in Supplemental Table 6 online. All of the constructs have been verified by sequencing.
Plant Transformation
Transient expression in onion (Allium cepa) epidermis peels and Arabidopsis seedlings using particle bombardment was done as previously described (Nelson et al., 2007). For each construct, 1 μg of plasmid DNA was used to coat the tungsten particles. Transient expression in tobacco (Nicotiana benthamiana) leaf was done as previously described (Sparkes et al., 2006). Agrobacterium strain GV3101 (pMP90) carrying the T-DNA expression constructs were used in the transformations.
Microscopy
For epifluorescence imaging, an Axiovert 200 M microscope (Zeiss) equipped with YFP/cyan fluorescent protein filters (filter set 52017; Chroma) was used. The images were captured with a ×63 (1.4 numerical aperture) plan-apo oil immersion objective unless stated otherwise. Image capture was done with a digital camera (Orca ER; Hamamatsu Photonics) controlled by OPENLAB software (Improvision). To remove the camera noise, two images were captured with the same exposure time: one with excitation light on and another one with light off. The camera noise from the dark image was subtracted from the fluorescence image pixel by pixel. For confocal imaging, a Leica SP2 laser scanning confocal microscope was used. YFP and chlorophyll were excited at 488 nm using an argon laser. Florescence emission signals from YFP and chlorophyll were recorded from 512 to 584 nm and 650 to 750 nm, respectively. The images were taken with an HC PL APO ×20 (0.7 numerical aperture) objective. All images were captured 12 h after transformation unless otherwise stated. Resizing and cropping of the images for presentation were done using Photoshop (Adobe).
Relative Intensity Ratio Measurement
Analysis of the camera noise subtracted images taken from epifluorescence microscopy was performed using ImageJ software (Abramoff et al., 2004). The intensity per pixel values from different areas in the images was calculated from the summation of intensity signals in the area divided by the total number of pixels in the area. A circle area was drawn to fit around the plastid to measure the plastid intensity per pixel. The same circle was then enlarged to threefold diameter. While sharing the same center, the enlarged circle area extended to incorporate intensity signals from cytosol. The cytosol intensity per pixel was calculated from the ring area located between the former and enlarged circles. A rectangular area outside the fluorescing cell in the same image was used to calculate the background intensity per pixel. The background intensity per pixel was subtracted from the plastid and cytosol intensity per pixel values. For each plastid, the ratio between the background removed plastid and cytosol intensity per pixel values was calculated. The ratio intensity of each cell is the average of all plastid ratio values. At least two plastids were used from each cell. The number of cells used in the measurement for each construct is stated in its figure legend.
Immunoblotting
In vivo import of YFP fusion proteins was analyzed based on transient expression of proteins in tobacco leaves. Total protein was extracted from leaf tissue expressing fusion proteins 2 d after transformation, as previously described (Isaacson et al., 2006). Briefly, the tissue was ground in liquid nitrogen in the presence of polyvinylpolypyrrolidone and homogenized in extraction buffer (10% trichloroacetic acid and 2% BME in acetone). The protein pellet was then resuspended in solubilization buffer (200 mM DTT, 20 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, and 8 M urea), boiled for 3 min, and stored in −80°C. Two separate transformations were performed.
The samples from in vivo import, in vitro import, and in vitro stromal processing assays were separated via 10 to 15% SDS-PAGE, transferred to polyvinylidene fluoride membrane, and detected with 1:5000 rabbit polyclonal anti–green fluorescent protein (GFP) (Gasper et al., 2009).
Hsp70 Binding Site Prediction
Perl scripts were generated based on previously described algorithms for Hsp70 binding site prediction (Gragerov et al., 1994; Rüdiger et al., 1997; Ivey et al., 2000). The TP sequences were analyzed by the scripts, and the scores based on each algorithm were generated. The scripts are available upon request.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: pea small subunit of Rubisco (CAA25390), pea Toc34 (Q41009), tobacco small subunit of Rubisco (P69249), and S. latifolia ferredoxin (CAA26281).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Codon Optimization and Far-UV CD Spectra of Forward and Retro Peptides.
Supplemental Figure 2. Plastid Targeting of Fluorescent Proteins by Forward and Retro-TPs.
Supplemental Figure 3. Hsp70 Binding Site Prediction.
Supplemental Table 1. Codon Adaptive Indices of the Competitors Based on Three Codon Usage Tables of E. coli.
Supplemental Table 2. Subcellular Targeting Predictions of Forward and Reverse Transit Peptides.
Supplemental Table 3. Rules Applied during Development of the Heuristic FGLK Motif Detection.
Supplemental Table 4. Curve Fitting Parameters for prSSU Homologous Binding Data.
Supplemental Table 5. Curve Fitting Parameters for Competitive Binding Data.
Supplemental Table 6. Sequences of Oligonucleotides Used in Cloning.
Supplementary Material
Acknowledgments
We thank Andreas Nebenführ for helpful discussions regarding microscopy and for providing pAN187 and pFGC19 constructs, John Dunlap for assistance in confocal microscopy, Rose Goodchild for anti-GFP antibody and GFP6xHis, Michael D. Vaughn for technical advice in the chloroplast binding assay, and Bijoyita Roy and Eusook Park for providing the seeds. This work was supported in part by a Science Alliance grant to L.E.R. and the National Science Foundation Program in Cell Biology to B.D.B. (MCB0628670 and MCB0344601).
AUTHOR CONTRIBUTIONS
P.C. and B.D.B. designed the research. P.C. and D.R.M. contributed computational tools. P.C., L.E.R, D.R.M., and I.M.C. performed the research and analyzed the data. P.C., L.E.R, I.M.C., and B.D.B wrote the article.
Glossary
- TP
transit peptide
- SPP
stromal processing peptidase
- Rubisco
ribulose-1,5-bis-phosphate carboxylase/oxygenase
- CAI
codon adaptive index
- MS
mass spectrometry
- MALDI-TOF
matrix-assisted laser-desorption ionization–time of flight mass spectrometry
- IMS
intermembrane space
- IDP
intrinsically disordered proteins
- CD
circular dichroism
- TFE
trifluoroethanol
- GAP
GTPase activating proteins
- LSC
liquid scintillation counting
- YFP
yellow fluorescent protein
- BME
β-mercaptoethanol
- GFP
green fluorescent protein
References
- Abramoff M.D., Magelhaes P.J., Ram S.J. (2004). Image processing with ImageJ. Biophotonics Int. 11: 36–42 [Google Scholar]
- Bailey T.L., Elkan C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36 [PubMed] [Google Scholar]
- Battistutta R., Bisello A., Mammi S., Peggion E. (1994). Conformation of retro-bombolitin I in aqueous solution containing surfactant micelles. Biopolymers 34: 1535–1541 [DOI] [PubMed] [Google Scholar]
- Benkirane N., Guichard G., Van Regenmortel M.H., Briand J.P., Muller S. (1995). Cross-reactivity of antibodies to retro-inverso peptidomimetics with the parent protein histone H3 and chromatin core particle. Specificity and kinetic rate-constant measurements. J. Biol. Chem. 270: 11921–11926 [DOI] [PubMed] [Google Scholar]
- Bionda T., Tillmann B., Simm S., Beilstein K., Ruprecht M., Schleiff E. (2010). Chloroplast import signals: The length requirement for translocation in vitro and in vivo. J. Mol. Biol. 402: 510–523 [DOI] [PubMed] [Google Scholar]
- Blond-Elguindi S., Cwirla S.E., Dower W.J., Lipshutz R.J., Sprang S.R., Sambrook J.F., Gething M.J. (1993). Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75: 717–728 [DOI] [PubMed] [Google Scholar]
- Bottier C., Géan J., Artzner F., Desbat B., Pézolet M., Renault A., Marion D., Vié V. (2007). Galactosyl headgroup interactions control the molecular packing of wheat lipids in Langmuir films and in hydrated liquid-crystalline mesophases. Biochim. Biophys. Acta 1768: 1526–1540 [DOI] [PubMed] [Google Scholar]
- Bruce B.D. (1998). The role of lipids in plastid protein transport. Plant Mol. Biol. 38: 223–246 [PubMed] [Google Scholar]
- Bruce B.D. (2000). Chloroplast transit peptides: Structure, function and evolution. Trends Cell Biol. 10: 440–447 [DOI] [PubMed] [Google Scholar]
- Bruce B.D. (2001). The paradox of plastid transit peptides: Conservation of function despite divergence in primary structure. Biochim. Biophys. Acta 1541: 2–21 [DOI] [PubMed] [Google Scholar]
- Chou M.L., Chu C.C., Chen L.J., Akita M., Li H.M. (2006). Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts. J. Cell Biol. 175: 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua N.H., Schmidt G.W. (1979). Transport of proteins into mitochondria and chloroplasts. J. Cell Biol. 81: 461–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyan C.L., Lin F.C., Peng H., Yuan J.M., Chang C.H., Lin S.H., Yang G. (2004). Reversible mechanical unfolding of single ubiquitin molecules. Biophys. J. 87: 3995–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K., Dabney-Smith C. (2008). Plastid protein import and sorting: Different paths to the same compartments. Curr. Opin. Plant Biol. 11: 585–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constan D., Patel R., Keegstra K., Jarvis P. (2004). An outer envelope membrane component of the plastid protein import apparatus plays an essential role in Arabidopsis. Plant J. 38: 93–106 [DOI] [PubMed] [Google Scholar]
- Dabney-Smith C., van Den Wijngaard P.W., Treece Y., Vredenberg W.J., Bruce B.D. (1999). The C terminus of a chloroplast precursor modulates its interaction with the translocation apparatus and PIRAC. J. Biol. Chem. 274: 32351–32359 [DOI] [PubMed] [Google Scholar]
- Dam J., Schuck P. (2004). Calculating sedimentation coefficient distributions by direct modeling of sedimentation velocity concentration profiles. Methods Enzymol. 384: 185–212 [DOI] [PubMed] [Google Scholar]
- de Castro Silva Filho M., Chaumont F., Leterme S., Boutry M. (1996). Mitochondrial and chloroplast targeting sequences in tandem modify protein import specificity in plant organelles. Plant Mol. Biol. 30: 769–780 [DOI] [PubMed] [Google Scholar]
- Dobberstein B., Blobel G., Chua N.H. (1977). In vitro synthesis and processing of a putative precursor for the small subunit of ribulose-1,5-bisphosphate carboxylase of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 74: 1082–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson H.J., Wright P.E. (2005). Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6: 197–208 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Fourie A.M., Sambrook J.F., Gething M.J. (1994). Common and divergent peptide binding specificities of hsp70 molecular chaperones. J. Biol. Chem. 269: 30470–30478 [PubMed] [Google Scholar]
- Friedman A.L., Keegstra K. (1989). Chloroplast protein import: Quantitative analysis of precursor binding. Plant Physiol. 89: 993–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasper R., Meyer S., Gotthardt K., Sirajuddin M., Wittinghofer A. (2009). It takes two to tango: regulation of G proteins by dimerization. Nat. Rev. Mol. Cell Biol. 10: 423–429 [DOI] [PubMed] [Google Scholar]
- Gragerov A., Zeng L., Zhao X., Burkholder W., Gottesman M.E. (1994). Specificity of DnaK-peptide binding. J. Mol. Biol. 235: 848–854 [DOI] [PubMed] [Google Scholar]
- Guichard G., Benkirane N., Zeder-Lutz G., van Regenmortel M.H., Briand J.P., Muller S. (1994). Antigenic mimicry of natural L-peptides with retro-inverso-peptidomimetics. Proc. Natl. Acad. Sci. USA 91: 9765–9769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guptasarma P. (1992). Reversal of peptide backbone direction may result in the mirroring of protein structure. FEBS Lett. 310: 205–210 [DOI] [PubMed] [Google Scholar]
- Haack T., Sánchez Y.M., González M.J., Giralt E. (1997). Structural comparison in solution of a native and retro peptide derived from the third helix of Staphylococcus aureus protein A, domain B: Retro peptides, a useful tool for the discrimination of helix stabilization factors dependent on the peptide chain orientation. J. Pept. Sci. 3: 299–313 [DOI] [PubMed] [Google Scholar]
- Hénaut A., Danchin A. (1996). Analysis and predictions from Escherichia coli sequences, or E. coli in silico. In Escherichia coli and Salmonella: Cellular and Molecular Biology, F. Neidhardt and R. Curtiss, eds (Washington, D.C.: ASM Press), pp. 2047–2066 [DOI] [PubMed]
- Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- Holtzer M.E., Braswell E., Angeletti R.H., Mints L., Zhu D., Holtzer A. (2000). Ultracentrifuge and circular dichroism studies of folding equilibria in a retro GCN4-like leucine zipper. Biophys. J. 78: 2037–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson T., Damasceno C.M., Saravanan R.S., He Y., Catalá C., Saladié M., Rose J.K. (2006). Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat. Protoc. 1: 769–774 [DOI] [PubMed] [Google Scholar]
- Ivey R.A., III, Subramanian C., Bruce B.D. (2000). Identification of a Hsp70 recognition domain within the rubisco small subunit transit peptide. Plant Physiol. 122: 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Constan D., Akita M., Keegstra K. (2001). Molecular chaperones involved in chloroplast protein import. Biochim. Biophys. Acta 1541: 102–113 [DOI] [PubMed] [Google Scholar]
- Jarvis P. (2008). Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 179: 257–285 [DOI] [PubMed] [Google Scholar]
- Jelic M., Sveshnikova N., Motzkus M., Hörth P., Soll J., Schleiff E. (2002). The chloroplast import receptor Toc34 functions as preprotein-regulated GTPase. Biol. Chem. 383: 1875–1883 [DOI] [PubMed] [Google Scholar]
- Karlin-Neumann G.A., Tobin E.M. (1986). Transit peptides of nuclear-encoded chloroplast proteins share a common amino acid framework. EMBO J. 5: 9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K., Cline K. (1999). Protein import and routing systems of chloroplasts. Plant Cell 11: 557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K.L. (1998). Amino-terminal and hydrophobic regions of the Chlamydomonas reinhardtii plastocyanin transit peptide are required for efficient protein accumulation in vivo. Plant Mol. Biol. 38: 365–377 [DOI] [PubMed] [Google Scholar]
- Kindle K.L., Lawrence S.D. (1998). Transit peptide mutations that impair in vitro and in vivo chloroplast protein import do not affect accumulation of the gamma-subunit of chloroplast ATPase. Plant Physiol. 116: 1179–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig P., Oreb M., Rippe K., Muhle-Goll C., Sinning I., Schleiff E., Tews I. (2008). On the significance of Toc-GTPase homodimers. J. Biol. Chem. 283: 23104–23112 [DOI] [PubMed] [Google Scholar]
- Kovacheva S., Bédard J., Wardle A., Patel R., Jarvis P. (2007). Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. Plant J. 50: 364–379 [DOI] [PubMed] [Google Scholar]
- Krimm I., Gans P., Hernandez J.F., Arlaud G.J., Lancelin J.M. (1999). A coil-helix instead of a helix-coil motif can be induced in a chloroplast transit peptide from Chlamydomonas reinhardtii. Eur. J. Biochem. 265: 171–180 [DOI] [PubMed] [Google Scholar]
- Kriwacki R.W., Hengst L., Tennant L., Reed S.I., Wright P.E. (1996). Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc. Natl. Acad. Sci. USA 93: 11504–11509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix E., Viguera A.R., Serrano L. (1998). Reading protein sequences backwards. Fold. Des. 3: 79–85 [DOI] [PubMed] [Google Scholar]
- Lebowitz J., Lewis M.S., Schuck P. (2002). Modern analytical ultracentrifugation in protein science: A tutorial review. Protein Sci. 11: 2067–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Kim J.K., Lee S., Choi S., Kim S., Hwang I. (2008). Arabidopsis nuclear-encoded plastid transit peptides contain multiple sequence subgroups with distinctive chloroplast-targeting sequence motifs. Plant Cell 20: 1603–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Lee S., Lee G.J., Lee K.H., Kim S., Cheong G.W., Hwang I. (2006). Functional characterization of sequence motifs in the transit peptide of Arabidopsis small subunit of rubisco. Plant Physiol. 140: 466–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Lee S., Oh Y.J., Hwang I. (2009). Multiple sequence motifs in the rubisco small subunit transit peptide independently contribute to Toc159-dependent import of proteins into chloroplasts. Plant Physiol. 151: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.H., Kim D.H., Lee S.W., Kim Z.H., Hwang I. (2002). In vivo import experiments in protoplasts reveal the importance of the overall context but not specific amino acid residues of the transit peptide during import into chloroplasts. Mol. Cells 14: 388–397 [PubMed] [Google Scholar]
- Marra J. (1985). Controlled deposition of lipid monolayers and bilayers onto mica and direct force measurements between galactolipid bilayers in aqueous solutions. J. Colloid Interface Sci. 107: 446–458 [Google Scholar]
- Marshall J.S., DeRocher A.E., Keegstra K., Vierling E. (1990). Identification of heat shock protein hsp70 homologues in chloroplasts. Proc. Natl. Acad. Sci. USA 87: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May T., Soll J. (2000). 14-3-3 Proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell 12: 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan V., Halada P., Hernychova L., Chong Y.P., Zakova J., Hupp T.R., Vojtesek B., Ball K.L. (2011). A multiprotein binding interface in an intrinsically disordered region of the tumour suppressor protein interferon regulatory factor-1. J. Biol. Chem. 286: 14291–14303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nielsen E., Akita M., Davila-Aponte J., Keegstra K. (1997). Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 16: 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L.J., Keegstra K. (1992). The binding of precursor proteins to chloroplasts requires nucleoside triphosphates in the intermembrane space. J. Biol. Chem. 267: 433–439 [PubMed] [Google Scholar]
- Olszewski K.A., Kolinski A., Skolnick J. (1996). Does a backwardly read protein sequence have a unique native state? Protein Eng. 9: 5–14 [DOI] [PubMed] [Google Scholar]
- Pellegrini A., von Fellenberg R. (1999). Design of synthetic bactericidal peptides derived from the bactericidal domain P(18-39) of aprotinin. Biochim. Biophys. Acta 1433: 122–131 [DOI] [PubMed] [Google Scholar]
- Perry S.E., Buvinger W.E., Bennett J., Keegstra K. (1991). Synthetic analogues of a transit peptide inhibit binding or translocation of chloroplastic precursor proteins. J. Biol. Chem. 266: 11882–11889 [PubMed] [Google Scholar]
- Pilon M., Weisbeek P.J., de Kruijff B. (1992). Kinetic analysis of translocation into isolated chloroplasts of the purified ferredoxin precursor. FEBS Lett. 302: 65–68 [DOI] [PubMed] [Google Scholar]
- Pilon M., Wienk H., Sips W., de Swaaf M., Talboom I., van ’t Hof R., de Korte-Kool G., Demel R., Weisbeek P., de Kruijff B. (1995). Functional domains of the ferredoxin transit sequence involved in chloroplast import. J. Biol. Chem. 270: 3882–3893 [DOI] [PubMed] [Google Scholar]
- Pinnaduwage P., Bruce B.D. (1996). In vitro interaction between a chloroplast transit peptide and chloroplast outer envelope lipids is sequence-specific and lipid class-dependent. J. Biol. Chem. 271: 32907–32915 [DOI] [PubMed] [Google Scholar]
- Ratnayake R.M.U., Inoue H., Nonami H., Akita M. (2008). Alternative processing of Arabidopsis Hsp70 precursors during protein import into chloroplasts. Biosci. Biotechnol. Biochem. 72: 2926–2935 [DOI] [PubMed] [Google Scholar]
- Reddick L.E., Chotewutmontri P., Crenshaw W., Dave A., Vaughn M., Bruce B.D. (2008). Nano-scale characterization of the dynamics of the chloroplast Toc translocon. Methods Cell Biol. 90: 365–398 [DOI] [PubMed] [Google Scholar]
- Reddick L.E., Vaughn M.D., Wright S.J., Campbell I.M., Bruce B.D. (2007). In vitro comparative kinetic analysis of the chloroplast Toc GTPases. J. Biol. Chem. 282: 11410–11426 [DOI] [PubMed] [Google Scholar]
- Rensink W.A., Pilon M., Weisbeek P. (1998). Domains of a transit sequence required for in vivo import in Arabidopsis chloroplasts. Plant Physiol. 118: 691–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensink W.A., Schnell D.J., Weisbeek P.J. (2000). The transit sequence of ferredoxin contains different domains for translocation across the outer and inner membrane of the chloroplast envelope. J. Biol. Chem. 275: 10265–10271 [DOI] [PubMed] [Google Scholar]
- Rial D.V., Arakaki A.K., Ceccarelli E.A. (2000). Interaction of the targeting sequence of chloroplast precursors with Hsp70 molecular chaperones. Eur. J. Biochem. 267: 6239–6248 [DOI] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A. (2000). EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Richly E., Leister D. (2004). An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene 329: 11–16 [DOI] [PubMed] [Google Scholar]
- Richter S., Lamppa G.K. (1998). A chloroplast processing enzyme functions as the general stromal processing peptidase. Proc. Natl. Acad. Sci. USA 95: 7463–7468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rief M., Gautel M., Oesterhelt F., Fernandez J.M., Gaub H.E. (1997). Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 276: 1109–1112 [DOI] [PubMed] [Google Scholar]
- Rüdiger S., Germeroth L., Schneider-Mergener J., Bukau B. (1997). Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16: 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E., Soll J., Küchler M., Kühlbrandt W., Harrer R. (2003). Characterization of the translocon of the outer envelope of chloroplasts. J. Cell Biol. 160: 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E., Soll J., Sveshnikova N., Tien R., Wright S., Dabney-Smith C., Subramanian C., Bruce B.D. (2002). Structural and guanosine triphosphate/diphosphate requirements for transit peptide recognition by the cytosolic domain of the chloroplast outer envelope receptor, Toc34. Biochemistry 41: 1934–1946 [DOI] [PubMed] [Google Scholar]
- Schnell D.J., Blobel G., Pain D. (1990). The chloroplast import receptor is an integral membrane protein of chloroplast envelope contact sites. J. Cell Biol. 111: 1825–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell D.J., Blobel G., Pain D. (1991). Signal peptide analogs derived from two chloroplast precursors interact with the signal recognition system of the chloroplast envelope. J. Biol. Chem. 266: 3335–3342 [PubMed] [Google Scholar]
- Schnell D.J., Kessler F., Blobel G. (1994). Isolation of components of the chloroplast protein import machinery. Science 266: 1007–1012 [DOI] [PubMed] [Google Scholar]
- Schuck P. (2000). Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78: 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M., Li W.H. (1987). The codon Adaptation Index—A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15: 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.X., Theg S.M. (2010). A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell 22: 205–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker B.A., Portman J.J., Wolynes P.G. (2000). Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc. Natl. Acad. Sci. USA 97: 8868–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S., Bauerle C., Hageman J., Keegstra K., Weisbeek P. (1986). The role of the transit peptide in the routing of precursors toward different chloroplast compartments. Cell 46: 365–375 [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Sreerama N., Woody R.W. (1999). Molecular dynamics simulations of polypeptide conformations in water: A comparison of alpha, beta, and poly(pro)II conformations. Proteins 36: 400–406 [PubMed] [Google Scholar]
- Su P.H., Li H.M. (2010). Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian C., Ivey R., III, Bruce B.D. (2001). Cytometric analysis of an epitope-tagged transit peptide bound to the chloroplast translocation apparatus. Plant J. 25: 349–363 [DOI] [PubMed] [Google Scholar]
- Sun Y.J., Forouhar F., Li Hm H.M., Tu S.L., Yeh Y.H., Kao S., Shr H.L., Chou C.C., Chen C., Hsiao C.D. (2002). Crystal structure of pea Toc34, a novel GTPase of the chloroplast protein translocon. Nat. Struct. Biol. 9: 95–100 [DOI] [PubMed] [Google Scholar]
- Sveshnikova N., Soll J., Schleiff E. (2000). Toc34 is a preprotein receptor regulated by GTP and phosphorylation. Proc. Natl. Acad. Sci. USA 97: 4973–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theg S.M., Bauerle C., Olsen L.J., Selman B.R., Keegstra K. (1989). Internal ATP is the only energy requirement for the translocation of precursor proteins across chloroplastic membranes. J. Biol. Chem. 264: 6730–6736 [PubMed] [Google Scholar]
- Tomkiewicz D., Nouwen N., Driessen A.J. (2007). Pushing, pulling and trapping—Modes of motor protein supported protein translocation. FEBS Lett. 581: 2820–2828 [DOI] [PubMed] [Google Scholar]
- Tompa P., Szász C., Buday L. (2005). Structural disorder throws new light on moonlighting. Trends Biochem. Sci. 30: 484–489 [DOI] [PubMed] [Google Scholar]
- Uversky V.N., Oldfield C.J., Dunker A.K. (2008). Intrinsically disordered proteins in human diseases: Introducing the D2 concept. Annu. Rev. Biophys. 37: 215–246 [DOI] [PubMed] [Google Scholar]
- Villalobos A., Ness J.E., Gustafsson C., Minshull J., Govindarajan S. (2006). Gene Designer: A synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics 7: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G., Nishikawa K. (1991). Chloroplast transit peptides. The perfect random coil? FEBS Lett. 278: 1–3 [DOI] [PubMed] [Google Scholar]
- von Heijne G., Steppuhn J., Herrmann R.G. (1989). Domain structure of mitochondrial and chloroplast targeting peptides. Eur. J. Biochem. 180: 535–545 [DOI] [PubMed] [Google Scholar]
- Weibel P., Hiltbrunner A., Brand L., Kessler F. (2003). Dimerization of Toc-GTPases at the chloroplast protein import machinery. J. Biol. Chem. 278: 37321–37329 [DOI] [PubMed] [Google Scholar]
- White S.H., von Heijne G. (2008). How translocons select transmembrane helices. Annu. Rev. Biophys. 37: 23–42 [DOI] [PubMed] [Google Scholar]
- Wienk H.L., Czisch M., de Kruijff B. (1999). The structural flexibility of the preferredoxin transit peptide. FEBS Lett. 453: 318–326 [DOI] [PubMed] [Google Scholar]
- Wienk H.L., Wechselberger R.W., Czisch M., de Kruijff B. (2000). Structure, dynamics, and insertion of a chloroplast targeting peptide in mixed micelles. Biochemistry 39: 8219–8227 [DOI] [PubMed] [Google Scholar]
- Yeh Y.H., Kesavulu M.M., Li H.M., Wu S.Z., Sun Y.J., Konozy E.H., Hsiao C.D. (2007). Dimerization is important for the GTPase activity of chloroplast translocon components atToc33 and psToc159. J. Biol. Chem. 282: 13845–13853 [DOI] [PubMed] [Google Scholar]
- Zhang X.P., Glaser E. (2002). Interaction of plant mitochondrial and chloroplast signal peptides with the Hsp70 molecular chaperone. Trends Plant Sci. 7: 14–21 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.