Abstract
Bacteria consume dissolved organic matter (DOM) through hydrolysis, transport and intracellular metabolism, and these activities occur in distinct subcellular localizations. Bacterial protein subcellular localizations for several major marine bacterial groups were predicted using genomic, metagenomic and metatranscriptomic data sets following modification of MetaP software for use with partial gene sequences. The most distinct pattern of subcellular localization was found for Bacteroidetes, whose genomes were substantially enriched with outer membrane and extracellular proteins but depleted of inner membrane proteins compared with five other taxa (SAR11, Roseobacter, Synechococcus, Prochlorococcus, oligotrophic marine Gammaproteobacteria). When subcellular localization patterns were compared between genes and transcripts, three taxa had expression biased toward proteins localized to cell locations outside of the cytosol (SAR11, Roseobacter, and Synechococcus), as expected based on the importance of carbon and nutrient acquisition in an oligotrophic ocean, but two taxa did not (oligotrophic marine Gammaproteobacteria and Bacteroidetes). Diel variations in the fraction and putative gene functions of transcripts encoding inner membrane and periplasmic proteins compared to cytoplasmic proteins suggest a close coupling of photosynthetic extracellular release and bacterial consumption, providing insights into interactions between phytoplankton, bacteria, and DOM.
INTRODUCTION
Heterotrophic bacteria are major consumers of marine dissolved organic matter (DOM) (6). It has been estimated that as much as 50% of the photosynthetically fixed organic matter eventually passes through the DOM pool (5, 20, 50), formed via exudation or excretion from phytoplankton and the trophodynamic processes of sloppy feeding and viral lysis (17). It is known that 9 out of 10 marine bacterial cells are Gram negative (54), and their outer membrane is permeable to small DOM molecules of <600 Da (58). This indicates that the utilization of low molecular weight (LMW) DOM may not require the participation of extracellular and outer membrane-associated proteins.
A substantial amount of DOM in seawater has a molecular mass of >1,000 Da, however (e.g., 23 to 33% of DOM in surface oceans) (4, 10, 11, 46). The importance of this high molecular weight (HMW) DOM to bacterial growth has been demonstrated in a number of studies, including studies conducted in the northern Gulf of Mexico (4) and surface water of the North Pacific Subtropical Gyre (45). A data assimilative modeling approach used in a previous study (42) led to the estimation that HMW DOM can support up to 40% of heterotrophic bacterial carbon demand. To make use of these large organic compounds, marine bacterioplankton must mobilize extracellular and outer membrane-bound hydrolytic enzymes that break them down into smaller compounds capable of being transported across bacterial membranes.
The relevance of the size of DOM to bacterial utilization and DOM cycles was best illustrated by the size-reactivity continuum model (3), in which the bulk HMW DOM is more reactive than the bulk LMW DOM. Although LMW DOM supports higher bacterial growth efficiencies than HMW DOM, the latter fuels higher rates of bacterial growth and respiration (3, 4). Since utilization of different size classes of DOM requires proteins localized in different subcellular compartments, studying protein subcellular localization may be a valuable approach to understanding how bacteria interact with the chemical matrix of seawater.
In a Gram-negative bacterium, there are five possible subcellular compartments where proteins can be localized: cytoplasm, inner membrane, periplasm, outer membrane, and extracellular space. The subcellular localization of marine bacterial proteins can be explored with molecular sequence data from marine bacterial genomes (65), metagenomes (66), and metatranscriptomes (26, 51, 52, 57). Since different algorithms frequently make different predictions, we developed the meta-algorithm MetaP (41). Unlike an early meta-algorithm in which different subcellular localizations were treated as independent classes (39), in MetaP the neighborhood relations of different subcellular localizations are considered and predictions are made by summarizing weighted optimal and suboptimal prediction scores on neighboring compartments (41). In the present study using modified MetaP software with an improved performance for short read prediction, differences in protein subcellular localization were found for expressed genes compared with their genomes for several major marine bacterial lineages. Diel differences in protein subcellular localization and gene functions that provided insights into day/night shifts in bacterial activities were also observed.
MATERIALS AND METHODS
Marine bacterial genomic, metagenomic and metatranscriptomic data sets.
Four criteria were used to select marine bacterial lineages for subcellular localization prediction. First, all selected lineages had to be a dominant group in the ocean; they were members of the five most abundant lineages in surface oceans as identified by the Global Ocean Survey (GOS) (54). Second, multiple sequenced genomes (n > 5) within the lineage had to be available. Third, the lineages had to represent a wide phylogenetic diversity, including members from different phyla. Lastly, the lineages had to represent different life strategies, including heterotrophy versus autotrophy, and r versus K strategies. According to these considerations, six lineages were selected (see Table S1 in the supplemental material): Roseobacter, SAR11, Prochlorococcus, Synechococcus, oligotrophic marine gammaproteobacteria (OMG), and Bacteroidetes. These marine bacterial lineages span a wide range of genome sizes (see Table S1), including the highly reduced SAR11 and Prochlorococcus genomes (1 million to 2 million bp), the intermediate Synechococcus and OMG genomes (2 million to 4 million bp), the large Roseobacter genomes (3.5 million to 5 million bp), and the highly variable Bacteroidetes genomes (1.5 million to 9.7 million bp).
The marine metagenomic data set used in this study was a subset of GOS samples from open ocean and coastal waters combined according to geographical information (see Table S2 in the supplemental material). The marine metatranscriptomic data sets were from previously published studies of four marine surface waters: the Hawaiian Ocean Time-Series (HOT) station (51), the southeastern U.S. coast (26), the Bermuda Atlantic Time-Series Study (BATS) enriched with dimethylsulfoniopropionate (DMSP) (57), and southeastern U.S. coastal water enriched with compounds derived from phytoplankton or vascular plants (52). The GOS peptide database and the metatranscriptomic DNA short read data sets were downloaded from the CAMERA metagenome collections (http://camera.calit2.net/). Since the genes relevant to phototrophy were highly expressed in the transcriptomes (e.g., cyanobacterial photosystem genes, bacteriochlorophyll-related genes, proteorhodopsin; see Data Set S1 in the supplemental material), were expressed mainly during the day and rarely during the night, and most were found to be associated with cell membranes, inclusion of these gene transcripts might have biased the true signal of predicted patterns of subcellular localization, e.g., in comparison between day and night metatranscriptomic data sets at the HOT station. Therefore, data sets were analyzed before and after removal of these sequences for all metatranscriptomic data sets. Since the conclusions reached were identical, only the data after removal of phototrophy-related sequences were shown.
To determine the amino acid sequences of the metatranscriptomic data sets, cDNA sequences were translated in silico using six frames. Peptide fragments with at least 50 amino acids were analyzed by BLAST against the NCBI microbial RefSeq (reference sequence) database (53), and the fragments with significant hits (bit score of >40 and E value of <10−6) were retained and used for further analyses. The metagenomic and metatranscriptomic sequences were binned to the closest bacterial genome by identifying the top hit in BLAST searches against a custom bacterial genome database with a cutoff E value of 0.1. The custom database consists of all prokaryotic genomes downloaded from NCBI in 2010, and marine prokaryotic genomes that were not found in NCBI but were found in the Gordon and Betty Moore Foundation Marine Microbial Genome Sequencing Project (http://moore.jcvi.org/moore/). The latter includes several members in the Roseobacter clade, the SAR11 clade, the OMG group, the marine Bacteroidetes, and the genera Prochlorococcus and Synechococcus. Some metagenomic and metatranscriptomic data sets were obtained from coastal regions where Prochlorococcus cells are rarely found, and it was not possible to analyze protein subcellular localization for this lineage in those data sets.
MetaP consensus algorithm for subcellular localization prediction of fragmentary sequences.
The MetaP program for predicting protein subcellular localization for metagenomic sequences (41) is a consensus algorithm, and its accuracy is dependent on that of the multiple predictors incorporated into the final algorithm. In its original version (41), all of the three base predictors (CELLO, SUBLOC, LOCTREE) in MetaP used amino acid compositional bias or a variant of this property to construct a support vector machine (SVM) for predicting localization (30, 48, 67). The LOCTREE software (48), however, additionally employs the Signal-P program, which makes predictions based on the presence of signal peptides (21). Since signal peptides are usually located at the N-terminal parts, which are likely to be missed in metagenomic and metatranscriptomic sequences, this property made LOCTREE less reliable for metagenomic and metatranscriptomic sequences and subsequently reduced the accuracy of MetaP.
In the present new version of MetaP (v 2.0), LOCTREE was replaced with PSLDOC. PSLDOC software is an application of SVM utilizing gapped dipeptide compositional bias (16), making it potentially useful in predicting fragmentary peptides. Using training data sets (see Data Set S2 in the supplemental material), which consisted of protein sequences whose subcellular localization had been experimentally verified (41), the new version of MetaP was found to have better performance (see Tables S3 and S4 in the supplemental material) and therefore was used in the present study.
Among the three base predictors, CELLO and PSLDOC are able to predict membrane proteins, whereas SUBLOC can predict only nonmembrane proteins. These results indicate that the current version of MetaP is suitable for predicting nonmembrane proteins, and the membrane proteins must be identified by CELLO or PSLDOC and left out before MetaP is employed. Since PSLDOC (see Table S5 in the supplemental material) was found to perform better than CELLO (see Table S6 in the supplemental material) in membrane protein prediction, the former was therefore used to identify membrane proteins as a first step, and the remaining sequences were then predicted by all three base predictors (CELLO, PSLDOC, SUBLOC) and MetaP. This procedure was used to process all possible protein sequences in the selected marine bacterial genomes, metagenomes, and metatranscriptomes (see Tables S7, S8, and S9 at http://www.marsci.uga.edu/facultypages/moran/publications/supplements.html).
Statistical analysis.
To visualize whether the subcellular localization is taxon-specific and whether genes and gene transcripts differ with regard to subcellular localization in major marine bacterial lineages, the dimensionality of the subcellular localization was reduced and plotted using High-Throughput Multi-Dimensional Scaling (HiT-MDS) software (22, 55). Pearson's correlation coefficients were derived between coordinates of each of the two dimensions and data on the subcellular localization (see Table S10 in the supplemental material).
Differences in the subcellular localization patterns between gene transcripts (from metatranscriptomes) and genes (from genomes and metagenomes) were analyzed by the method of Baggerly et al. (7), a statistical method for count data which considers both within-library (library size effect) and between-library (biological replicate effect) variations. The within-library variation captures the sampling variability, and the between-library variation accounts for variations between individuals within the sample group (7). The same approach was used to test for subcellular localization differences in gene expression between day and night genes in surface ocean microbial assemblages at the HOT station.
To see what functions drive the differences between day and night gene expression, the determined peptide sequences were searched against the Clusters of Orthologous Groups (COG) (56) located in the NCBI Conserved Domain Database (44) using the RPSBLAST program (2) with a cutoff E value of 0.1 (see Table S11 in the supplemental material).
RESULTS
Lineage-specific subcellular localization.
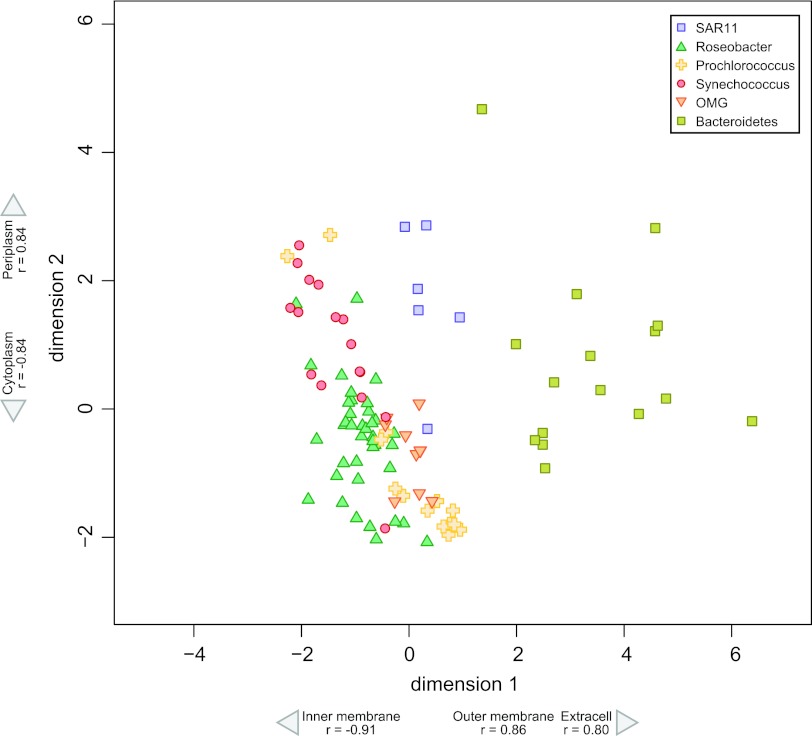
In most cases, the subcellular distribution of proteins was observed to be more similar among genomes within a lineage than across different lineages, suggesting lineage-specific localization patterns. There was a clear separation of Bacteroidetes from the other lineages attributable to a greater fraction of outer membrane and extracellular proteins and a lower fraction of inner membrane proteins (P < 0.001 in all cases) (Fig. 1). The remaining five lineages were separated to a lesser extent. Roseobacter and SAR11 genomes had distinct patterns, with the former having a larger fraction of inner membrane proteins and a lower fraction of outer membrane and extracellular proteins (P < 0.001 in all cases) (Fig. 1). Prochlorococcus was also distinguishable from Synechococcus with the exception of a few strains, based on a greater proportion of cytoplasmic proteins and a lower proportion of periplasmic proteins in the former (P < 0.001 in both cases) (Fig. 1).
Fig 1.
High-throughput multidimensional scaling (HiT-MDS) plot of MetaP using whole-genome-sequence-predicted protein subcellular localization among major marine bacterial lineages.
Differences in subcellular localization between genes and expressed genes.
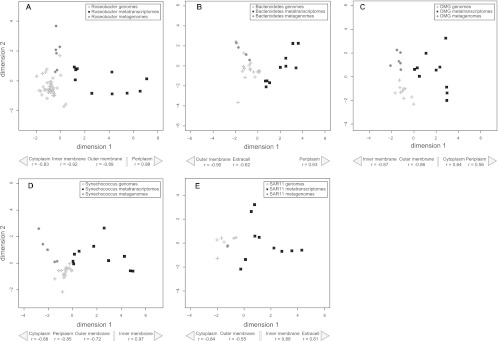
For each major marine bacterial lineage, a multidimensional scaling (MDS) figure was plotted that incorporated subcellular localization assignments for genomes in the lineage together with metagenomic and metatranscriptomic reads attributed to the lineage. The transcription pattern was distinguishable from the genomic potential pattern in all lineages, suggesting that genes which encode proteins localized to certain subcellular compartments were preferentially expressed. In all lineages, genes encoding outer membrane proteins represented a lower fraction of the metatranscriptomes than of the genomes and metagenomes (P < 0.001 in all cases) (Fig. 2). In addition, the Roseobacter, Bacteroidetes, and OMG clades had a greater fraction of genes encoding periplasmic proteins and a lower fraction of genes encoding inner membrane proteins in their metatranscriptomes than in their genomes and metagenomes (P < 0.001 in all cases) (Fig. 2A, 2B, 2C). In contrast, Synechococcus had the reverse trend, with genes encoding inner membrane proteins preferentially expressed (P < 0.001) and genes encoding periplasmic proteins less expressed (P < 0.01) (Fig. 2D). The SAR11 clade also had a greater fraction of genes encoding inner membrane proteins in the metatranscriptomes than in the genomes and metagenomes (P < 0.001) (Fig. 2E). Genes encoding extracellular proteins showed no differences between metagenomes and metatranscriptomes in the Roseobacter, OMG, and Synechococcus groups. In SAR11, genes encoding extracellular proteins represented a greater fraction in the metatranscriptomes than in the genomes and metagenomes (Fig. 2E), whereas in Bacteroidetes (Fig. 2B) these genes had lower expression (P < 0.001 in both cases). Lastly, genes encoding cytoplasmic proteins had a lower representation in transcriptomes in Roseobacter, Synechococcus and SAR11 (P < 0.001 in all cases) (Fig. 2A, 2D, 2E), while a reverse pattern was observed in the OMG clade (P < 0.001) (Fig. 2C).
Fig 2.
High-throughput multidimensional scaling (HiT-MDS) plot of MetaP using genome, metagenome, and metatranscriptome sequence-predicted protein subcellular localization of marine bacteria. (A) Roseobacter, (B) Bacteroidetes, (C) oligotrophic marine Gammaproteobacteria (OMG), (D) Synechococcus, and (E) SAR11.
Day versus night gene expression patterns for subcellular localization.
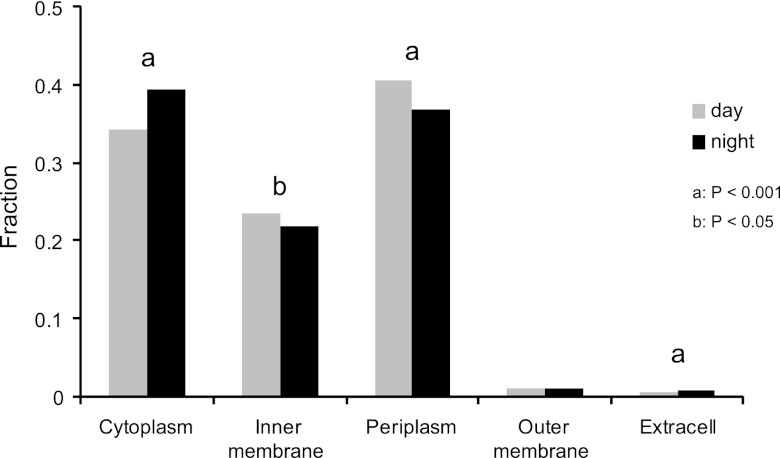
Day and night gene expression patterns for protein subcellular localization were compared for the bacterioplankton assemblage sampled from surface water at the HOT station ALOHA, defined by the 6-nautical-mile radius circle centered at 22°45′N, 158°W (51). The night sample was collected at 0300 on 11 November 2005 and the daytime sample was collected at 1300 on 13 November 2005 (51). Genes encoding the periplasmic (P < 0.001) and inner membrane (P < 0.05) proteins were enriched in the day versus the night. In contrast, genes encoding the cytoplasmic (P < 0.001) and extracellular (P < 0.001) proteins were enriched in the night versus the day. There was no statistically significant diel difference in expression for genes encoding outer membrane proteins (Fig. 3).
Fig 3.
Differential gene expression in protein subcellular localizations between day and night in surface waters of the North Pacific Subtropical Gyre. The letter above the bars indicates the significance level: a, P < 0.001; b, P < 0.05.
DISCUSSION
Members of Bacteroidetes are recognized for the ability to degrade biopolymers (34), a process considered the rate-limiting step in HMW DOM mineralization (29, 33). Bacteroidetes have been shown to comprise the largest fraction of bacteria consuming chitin, polysaccharides, and proteins, but the smallest fraction consuming amino acids (18), and they participate less in the uptake of dissolved free amino acids than do Alphaproteobacteria and Gammaproteobacteria (1). These observed ecological roles are backed up by comparative genomic analyses. Bacteroidetes members Gramella forsetii KT0803 and Polaribacter sp. MED152 have a greater representation of outer membrane receptors for polymeric compounds and a smaller fraction of inner membrane transporters and periplasmic substrate-binding proteins for monomeric compounds than Pelagibacter ubique (SAR11), Ruegeria pomeroyi (the Roseobacter clade), or marine Gammaproteobacteria (8, 27). Thus, this finding that the genomes and metagenomes of Bacteroidetes are the most distinctive with regard to outer/inner membrane protein ratios corroborates the significance of biopolymer degradation in driving the ecological differentiation of this lineage (Fig. 1).
Considering the importance of nutrient acquisition for bacteria to survive in the oligotrophic ocean environment, genes encoding proteins localized in the cytoplasm were expected to comprise a smaller fraction of the metatranscriptomes than the genomes and metagenomes. Among the five taxonomic groups studied, Roseobacter, SAR11 and Synechococcus were consistent with this expectation, while OMG showed the reverse trend and Bacteroidetes did not exhibit a differential pattern. One possible interpretation is that the first three bacterial groups allocate more resources to acquire substrates from the environment. Similarly, subcellular localization patterns between the single marine autotroph analyzed here and the heterotrophic taxa showed that Synechococcus expressed fewer periplasmic proteins and more inner membrane proteins relative to their abundance in genomes and metagenomes, while Roseobacter, Bacteroidetes, and OMG expressed more periplasmic and fewer inner membrane proteins than expected from their genomes. Since the periplasm is enriched with hydrolytic enzymes and nutrient-binding proteins (49, 61), and the inner membrane is known to harbor various nutrient transporters (19), this finding is consistent with different cell-nutrient interactions in which the heterotrophs play a larger role in organic matter degradation while Synechococcus transports a higher proportion of nutrients that do not need hydrolysis prior to transport, such as inorganic nutrients and small DOM compounds. In fact, a proteomics study of a freshwater cyanobacterium showed that identified transporter proteins were mainly for inorganic nutrient acquisition (31). It is interesting that SAR11 had a ratio of inner/outer membrane protein transcripts consistent with Synechococcus but differing from the other heterotrophic bacteria. A recent study showed that SAR11 is specialized for uptake of small organic molecules while Synechococcus has a high capacity for phosphate uptake (68). Thus, the coincident pattern of SAR11 and Synechococcus in expression of inner membrane proteins is likely driven by different ecological functions. The above discussion is based on an assumption that protein abundance and mRNA levels are positively correlated, which has been shown to be the case for populations of some model organisms under steady-state conditions (40). Currently, however, very little is known about coordination of mRNA and protein pools in marine bacteria under dynamic environmental conditions.
The elevated daytime expression of proteins in the periplasm and inner membrane in the North Pacific Subtropical Gyre metatranscriptome suggests a greater flux of organic matter and inorganic nutrients into the bacterioplankton assemblage during the day than during the night. This is consistent with previous studies showing that the turnover rate of dissolved free amino acid can be much greater in the day (15, 23). Several phototrophy-relevant genes (e.g., proteorhodopsin, pufM, pufL, cyanobacterial photosystems) were highly expressed during the day, and these are localized in either the cytoplasmic membrane or the thylakoid membrane. However, the finding of differential diel localization patterns for expressed genes remained the same whether or not these gene transcripts directly linked to solar energy capture were included.
The sources of the bioreactive DOM being processed by bacterioplankton include phytoplankton exudation, zooplankton grazing (through “sloppy feeding” mechanisms), and viral lysis (14), as well as potentially through photodegradation of refractory DOM (47). Zooplankton grazing has not been found to have a diel pattern (36, 60), or if so, the rates are higher at night (59). Evidence of diel patterns of viral lysis is controversial (32, 62, 63), but biopolymer-enriched viral lysis products would likely require mobilization of hydrolytic enzymes, which was not evident in the subcellular localization data. Finally, solar radiation has been shown to both increase bioreactivity of refractory DOM and decrease bioreactivity of labile DOM (9). Thus, the subcellular localization signal indicative of higher diurnal bacterial DOM flux is hypothesized to represent a response to increases in exudates from actively photosynthesizing phytoplankton (43, 64).
Tight coupling of autotrophic production and heterotrophic consumption of organic matter has been observed previously. Fuhrman et al. (24) found significant diel covariations of chlorophyll, bicarbonate incorporation, bacterial abundance and thymidine incorporation, while Gasol et al. (25) showed diel changes in both DNA and protein synthesis by bacteria, and Kuipers et al. (35) observed a diel rhythm in frequency of dividing cells, cell volume, and bacterial abundance. These studies provide physiological evidence of daily rhythms of bacterial activities consistent with the subcellular localization predictions of gene expression observed here.
The finding of elevated daytime expression of genes encoding proteins localized in the inner membrane and periplasm but not in the outer membrane and extracellular space provides clues as to the chemical composition of bioreactive DOM. Since small compounds freely diffuse across the bacterial outer membrane without the involvement of extracellular hydrolytic enzymes and transporters located there, the subcellular localization pattern points to a predominant daytime flux of low molecular weight (LMW) DOM. This is consistent with previous studies showing phytoplankton exudates consisting mainly of compounds 300 to 600 Da in size (38) representing a wide range of LMW DOM which is rapidly consumed by bacteria (12, 13), though exudates from natural diatom populations in the North Sea dominated by macromolecules have also been reported (37).
The finding of elevated nighttime expression of genes encoding extracellular proteins suggests the transport of higher molecular weight DOM at night. Microbial cell ruptures due to zooplankton grazing and viral lysis are likely to release HMW compounds in the form of membrane and other biopolymeric materials, and HMW DOM accounts for a greater fraction of phytoplankton release in the dark than in the light (28). Bacteria may therefore bias gene expression patterns toward the extracellularly localized proteins necessary to utilize a HMW-enriched bioreactive nighttime DOM pool.
The differential day versus night gene expression pattern in regard to protein subcellular localization was backed up by observations of relative abundance of gene transcripts with different functional categories. Some examples were summarized in Table 1 (see also Table S12 in the supplemental material). Cytoplasmic protein families showing preferential transcription during the night included those involved in cellular synthesis and maintenance (e.g., DNA polymerase, helicases, topoisomerases, replication-related ATPases, DNA repair proteins). In contrast, families driving the differential pattern of periplasmic and inner membrane proteins included genes involved in nutrient hydrolysis and transport (e.g., various permeases and hydrolases). Families involved in energy production and detoxification of reactive oxygen species were preferentially transcribed during the daytime, and they were found in multiple subcellular compartments (Table 1; see also Table S12 in the supplemental material). These gene families appear to be contributing to the differential patterns of day and night expression in different subcellular compartments.
Table 1.
Examples of biological functions driving differences between day and night gene transcription in different subcellular compartments
| General function | COG | Function | Subcellular localization | Change (%)a |
|---|---|---|---|---|
| Cellular synthesis and maintenance | COG1202 | Superfamily II helicase, archaea-specific | Cytoplasmic | 100 |
| COG1112 | Superfamily I DNA and RNA helicases and helicase subunits | Cytoplasmic | 100 | |
| COG1241 | Predicted ATPase involved in replication control, Cdc46/Mcm family | Cytoplasmic | 100 | |
| COG1697 | DNA topoisomerase VI, subunit A | Cytoplasmic | 100 | |
| COG3569 | Topoisomerase IB | Cytoplasmic | 100 | |
| COG2177 | Cell division protein | Cytoplasmic | 100 | |
| COG5214 | DNA polymerase alpha-primase complex, polymerase-associated subunit B | Cytoplasmic | 100 | |
| COG1591 | Holliday junction resolvase, archaeal type | Cytoplasmic | 100 | |
| COG1471 | Ribosomal protein S4E | Cytoplasmic | 100 | |
| COG5241 | Nucleotide excision repair endonuclease NEF1, RAD10 subunit | Cytoplasmic | 100 | |
| Various nutrient permeases and hydrolases | COG3687 | Predicted metal-dependent hydrolase | Inner membrane | 100 |
| COG0672 | High-affinity Fe2+/Pb2+ permease | Inner membrane | 100 | |
| COG4139 | ABC-type cobalamin transport system, permease component | Inner membrane | 100 | |
| COG0737 | 5′-nucleotidase/2′,3′-cyclic phosphodiesterase | Inner membrane | 100 | |
| COG1113 | Gamma-aminobutyrate permease and related permeases | Inner membrane | 100 | |
| COG1984 | Allophanate hydrolase subunit 2 | Inner membrane | 100 | |
| COG4160 | ABC-type arginine/histidine transport system, permease component | Inner membrane | 100 | |
| COG2998 | ABC-type tungstate transport system, permease component | Inner membrane | 100 | |
| COG4778 | ABC-type phosphonate transport system, ATPase component | Periplasmic | 100 | |
| COG1001 | Adenine deaminase | Periplasmic | 100 | |
| COG5266 | ABC-type Co2+ transport system, periplasmic component | Periplasmic | 100 | |
| COG0832 | Urea amidohydrolase (urease) beta subunit | Periplasmic | 100 | |
| COG0221 | Inorganic pyrophosphatase | Periplasmic | 100 | |
| Detoxification of reactive oxygen species | COG1773 | Rubredoxin | Cytoplasmic | −636 |
| COG0278 | Glutaredoxin-related protein | Cytoplasmic | −665 | |
| COG1331 | Highly conserved protein containing a thioredoxin domain | Inner membrane | 100 | |
| COG3634 | Alkyl hydroperoxide reductase, large subunit | Inner membrane | 100 | |
| COG2032 | Cu/Zn superoxide dismutase | Periplasmic | 100 | |
| COG0694 | Thioredoxin-like proteins and domains | Periplasmic | 100 | |
| Energy production | COG2920 | Dissimilatory sulfite reductase (desulfoviridin), gamma subunit | Inner membrane | 100 |
| COG2221 | Dissimilatory sulfite reductase (desulfoviridin), alpha and beta subunits | Cytoplasmic | −1,934 |
For a given cytoplasmic protein family, change (%) is the relative change in gene transcript abundance from night to day. For a given inner membrane or periplasmic protein family, change (%) is the relative change in gene transcript abundance from day to night. Gene transcript abundance was normalized by the library size. A value of change (%) greater than 0 indicates an increase, while a value smaller than 0 indicates a decrease.
This predictive framework for subcellular localization of proteins in ocean bacteria therefore contributes to a model of diel cellular activities in which photosynthetic exudates dominated by labile LMW DOM diffuse freely across bacterial outer membrane and are hydrolyzed in the periplasm and/or transported through inner membrane permeases and transporters during the day. These acquired nutrients provide the raw materials for greater cytoplasm-dominated synthesis activities during the night, along with a relatively greater investment in proteins mediating HMW DOM uptake.
Supplementary Material
ACKNOWLEDGMENTS
I thank R. Benner and M. A. Moran for discussions, A. Rivers for advice on data analysis, M. Strickert for assistance with HiT-MDS software, K. A. Baggerly for discussion of statistical methods for count data, J. Hu for advice on using PSLDoc, J. Tang for installing PSLDoc, C. English for assistance with graphics, and staff in the Georgia Advanced Computing Resource Center at the University of Georgia for computing assistance.
This research was funded by grants from the Gordon and Betty Moore Foundation and the National Science Foundation (MCB-0702125).
Footnotes
Published ahead of print 6 July 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alonso-Saez L, Gasol JM. 2007. Seasonal variations in the contributions of different bacterial groups to the uptake of low-molecular-weight compounds in northwestern Mediterranean coastal waters. Appl. Environ. Microbiol. 73:3528–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amon RMW, Benner R. 1996. Bacterial utilization of different size classes of dissolved organic matter. Limnol. Oceanogr. 41:41–51 [Google Scholar]
- 4. Amon RMW, Benner R. 1994. Rapid cycling of high-molecular-weight dissolved organic matter in the ocean. Nature 369:549–552 [Google Scholar]
- 5. Azam F, et al. 1983. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10:257–263 [Google Scholar]
- 6. Azam F, Hodson RE. 1977. Size distribution and activity of marine microheterotrophs. Limnol. Oceanogr. 22:492–501 [Google Scholar]
- 7. Baggerly KA, Deng L, Morris JS, Aldaz CM. 2003. Differential expression in SAGE: accounting for normal between-library variation. Bioinformatics 19:1477–1483 [DOI] [PubMed] [Google Scholar]
- 8. Bauer M, et al. 2006. Whole genome analysis of the marine Bacteroidetes ‘Gramella forsetii’ reveals adaptations to degradation of polymeric organic matter. Environ. Microbiol. 8:2201–2213 [DOI] [PubMed] [Google Scholar]
- 9. Benner R, Biddanda B. 1998. Photochemical transformations of surface and deep marine dissolved organic matter: effects on bacterial growth. Limnol. Oceanogr. 43:1373–1378 [Google Scholar]
- 10. Benner R, Biddanda B, Black B, McCarthy M. 1997. Abundance, size distribution, and stable carbon and nitrogen isotopic compositions of marine organic matter isolated by tangential-flow ultrafiltration. Mar. Chem. 57:243–263 [Google Scholar]
- 11. Benner R, Pakulski JD, McCarthy M, Hedges JI, Hatcher PG. 1992. Bulk chemical characteristics of dissolved organic matter in the ocean. Science 255:1561–1564 [DOI] [PubMed] [Google Scholar]
- 12. Biddanda B, Benner R. 1997. Carbon, nitrogen, and carbohydrate fluxes during the production of particulate and dissolved organic matter by marine phytoplankton. Limnol. Oceanogr. 42:506–518 [Google Scholar]
- 13. Bjornsen PK. 1988. Phytoplankton exudation of organic matter: why do healthy cells do it? Limnol. Oceanogr. 33:151–154 [Google Scholar]
- 14. Carlson CA. 2002. Production and removal processes, p 91–151 In Hansell DA, Carlson CA. (ed), Biogeochemistry of marine dissolved organic matter. Elsevier Science, Philadelphia, PA: [Google Scholar]
- 15. Carlucci AF, Craven DB, Henrichs SM. 1984. Diel production and microheterotrophic utilization of dissolved free amino acids in waters off southern California. Appl. Environ. Microbiol. 48:165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang J-M, et al. 2008. PSLDoc: protein subcellular localization prediction based on gapped-dipeptides and probabilistic latent semantic analysis. Proteins 72:693–710 [DOI] [PubMed] [Google Scholar]
- 17. Church MJ. 2008. Resource control of bacterial dynamics in the sea, p 335–382 In Kirchman DL. (ed), Microbial ecology of the oceans. John Wiley & Sons, Hoboken, NJ: [Google Scholar]
- 18. Cottrell MT, Kirchman DL. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Daley DO, et al. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308:1321–1323 [DOI] [PubMed] [Google Scholar]
- 20. Ducklow HW. 1999. The bacterial component of the oceanic euphotic zone. FEMS Microbiol. Ecol. 30:1–10 [Google Scholar]
- 21. Dyrlov Bendtsen J, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 22. Fester T, Schreiber F, Strickert M. 2009. CUDA-based multi-core implementation of MDS-based bioinformatics algorithms, p 67–79 In Proceedings of the German Conference on Bioinformatics, Halle, Germany http://www2.ipk-gatersleben.de/∼schreibe/publications/FesterSS09.pdf [Google Scholar]
- 23. Fuhrman JA. 1990. Dissolved free amino acid cycling in an estuarine outflow plume. Mar. Ecol. Prog. Ser. 66:197–203 [Google Scholar]
- 24. Fuhrman JA, Eppley RW, Hagstrom A, Azam F. 1985. Diel variations in bacterioplankton, phytoplankton, and related parameters in the Southern California Bight. Mar. Ecol. Prog. Ser. 27:9–20 [Google Scholar]
- 25. Gasol JM, et al. 1998. Diel variations in bacterial heterotrophic activity and growth in the northwestern Mediterranean Sea. Mar. Ecol. Prog. Ser. 164:107–124 [Google Scholar]
- 26. Gifford SM, Sharma S, Rinta-Kanto JM, Moran MA. 2011. Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. ISME J. 5:461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. González JM, et al. 2008. Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria). Proc. Natl. Acad. Sci. U. S. A. 105:8724–8729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hama T, Yanagi K, Hama J. 2004. Decrease in molecular weight of photosynthetic products of marine phytoplankton during early diagenesis. Limnol. Oceanogr. 49:471–481 [Google Scholar]
- 29. Hoppe H-G, Arnosti C, Herndl GJ. 2002. Ecological significance of bacterial enzymes in the marine environment, p 73–108 In Burns RG, Dick RP. (ed), Enzymes in the environment: activity, ecology, and applications. Marcel Dekker, New York [Google Scholar]
- 30. Hua S, Sun Z. 2001. Support vector machine approach for protein subcellular localization prediction. Bioinformatics 17:721–728 [DOI] [PubMed] [Google Scholar]
- 31. Huang F, et al. 2002. Proteomics of Synechocystis sp. strain PCC 6803. Mol. Cell. Proteomics 1:956–966 [DOI] [PubMed] [Google Scholar]
- 32. Jacquet S, et al. 2002. Flow cytometric analysis of an Emiliana huxleyi bloom terminated by viral infection. Aquat. Microb. Ecol. 27:111–124 [Google Scholar]
- 33. Karl DM, Bjorkman KM. 2002. Dynamics of DOP, p 249–366 In Hansell DA, Carlson CA. (ed), Biogeochemistry of marine dissolved organic matter. Academic Press, Cleveland, OH [Google Scholar]
- 34. Kirchman DL. 2002. The ecology of Cytophaga–Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91–100 [DOI] [PubMed] [Google Scholar]
- 35. Kuipers B, van Noort GV, Vosjan J, Herndl GJ. 2000. Diel periodicity of bacterioplankton in the euphotic zone of the subtropical Atlantic Ocean. Mar. Ecol. Prog. Ser. 201:13–25 [Google Scholar]
- 36. Lampert W, Taylor BE. 1985. Zooplankton grazing in a eutrophic lake: implications of diel vertical migration. Ecology 66:68–82 [Google Scholar]
- 37. Lancelot C. 1984. Extracellular release of small and large molecules by phytoplankton in the Southern Bight of the North Sea. Estuar. Coast. Shelf. Sci. 18:65–77 [Google Scholar]
- 38. Lignell R. 1990. Excretion of organic carbon by phytoplankton: its relation to algal biomass, primary productivity and bacterial secondary productivity in the Baltic Sea. Mar. Ecol. Prog. Ser. 68:85–99 [Google Scholar]
- 39. Liu J, Kang S, Tang C, Ellis LBM, Li T. 2007. Meta-prediction of protein subcellular localization with reduced voting. Nucleic Acids Res. 35:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu P, Vogel C, Wang R, Yao X, Marcotte EM. 2007. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotech. 25:117–124 [DOI] [PubMed] [Google Scholar]
- 41. Luo H, Benner R, Long RA, Hu J. 2009. Subcellular localization of marine bacterial alkaline phosphatases. Proc. Natl. Acad. Sci. U. S. A. 106:21219–21223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Luo Y-W, Friedrichs MAM, Doney SC, Church MJ, Ducklow HW. 2010. Oceanic heterotrophic bacterial nutrition by semilabile DOM as revealed by data assimilative modeling. Aquat. Microb. Ecol. 60:273–287 [Google Scholar]
- 43. Mague TH, Friberg E, Hughes DJ, Morris I. 1980. Extracellular release of carbon by marine phytoplankton; a physiological approach. Limnol. Oceanogr. 25:262–279 [Google Scholar]
- 44. Marchler-Bauer A, et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McCarren J, et al. 2010. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc. Natl. Acad. Sci. U. S. A. 107:16420–16427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McCarthy M, Hedges J, Benner R. 1996. Major biochemical composition of dissolved high molecular weight organic matter in seawater. Mar. Chem. 55:281–297 [Google Scholar]
- 47. Moran MA, Zepp RG. 1997. Role of photoreactions in the formation of biologically labile compounds from dissolved organic matter. Limnol. Oceanogr. 42:1307–1316 [Google Scholar]
- 48. Nair R, Rost B. 2005. Mimicking cellular sorting improves prediction of subcellular localization. J. Mol. Biol. 348:85–100 [DOI] [PubMed] [Google Scholar]
- 49. Pieper R, et al. 2008. Characterizing the dynamic nature of the Yersinia pestis periplasmic proteome in response to nutrient exhaustion and temperature change. Proteomics 8:1442–1458 [DOI] [PubMed] [Google Scholar]
- 50. Pomeroy LR. 1974. The oceans food web, a changing paradigm. Bioscience 24:499–504 [Google Scholar]
- 51. Poretsky RS, et al. 2009. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ. Microbiol. 11:1358–1375 [DOI] [PubMed] [Google Scholar]
- 52. Poretsky RS, Sun S, Mou X, Moran MA. 2010. Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ. Microbiol. 12:616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pruitt KD, Tatusova T, Klimke W, Maglott DR. 2009. NCBI reference sequences: current status, policy and new initiatives. Nucleic Acids Res. 37:D32–D36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rusch DB, et al. 2007. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5:e77 doi:10.1371/journal.pbio.0050077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Strickert M, Teichmann S, Sreenivasulu N, Seiffert U. 2005. High-throughput multi-dimensional scaling (HiT-MDS) for cDNA-array expression data, p 625–633 In Duch W, Kacprzyk J, Oja E, Zadrozny S. (ed), Artificial neural networks: biological inspirations–ICANN 2005, vol 3696. Springer; Berlin/Heidelberg: [Google Scholar]
- 56. Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637 [DOI] [PubMed] [Google Scholar]
- 57. Vila-Costa M, et al. 2010. Transcriptomic analysis of a marine bacterial community enriched with dimethylsulfoniopropionate. ISME J. 4:1410–1420 [DOI] [PubMed] [Google Scholar]
- 58. Weiss M, et al. 1991. Molecular architecture and electrostatic properties of a bacterial porin. Science 254:1627–1630 [DOI] [PubMed] [Google Scholar]
- 59. Welschmeyer NA, Copping AE, Vernet M, Lorenzen CJ. 1984. Diel fluctuation in zooplankton grazing rate as determined from the downward vertical flux of pheopigments. Mar. Biol. 83:263–270 [Google Scholar]
- 60. White JR, Roman MR. 1991. Measurement of zooplankton grazing using particles labelled in light and dark with [methyl-3H] methylamine hydrochloride. Mar. Ecol. Prog. Ser. 71:45–52 [Google Scholar]
- 61. Wilmes B, et al. 2011. Cytoplasmic and periplasmic proteomic signatures of exponentially growing cells of the psychrophilic bacterium Pseudoalteromonas haloplanktis TAC125. Appl. Environ. Microbiol. 77:1276–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Winget DM, Wommack KE. 2009. Diel and daily fluctuations in virioplankton production in coastal ecosystems. Environ. Microbiol. 11:2904–2914 [DOI] [PubMed] [Google Scholar]
- 63. Winter C, Herndl GJ, Weinbauer MG. 2004. Diel cycles in viral infection of bacterioplankton in the North Sea. Aquat. Microb. Ecol. 35:207–216 [Google Scholar]
- 64. Wolter K. 1982. Bacterial incorporation of organic substances released by natural phytoplankton populations. Mar. Ecol. Prog. Ser. 7:287–295 [Google Scholar]
- 65. Yooseph S, et al. 2010. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature 468:60–66 [DOI] [PubMed] [Google Scholar]
- 66. Yooseph S, et al. 2007. The Sorcerer II Global Ocean Sampling Expedition: expanding the universe of protein families. PLoS Biol. 5:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yu CS, Chen YC, Lu CH, Hwang JK. 2006. Prediction of protein subcellular localization. Proteins 64:643–651 [DOI] [PubMed] [Google Scholar]
- 68. Zubkov MV, et al. 2007. Microbial control of phosphate in the nutrient-depleted North Atlantic subtropical gyre. Environ. Microbiol. 9:2079–2089 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.