Abstract
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved self-degradative process, which involves the regular turnover of cellular components via sequestering damaged macromolecules and transporting them for lysosomal degradation. In the past few years, the scientific community has produced remarkable advances in our understanding of the genes that are involved in autophagy and of their profound effects on various diseases. Recently, a new class of noncoding RNAs, known as microRNAs (miRNAs), has been demonstrated to play crucial roles in diverse biological processes including development, cell differentiation and apoptosis. Here, we review the current understanding about miRNAs focusing on their involvement in the autophagy process. Intriguingly, several confirmed targets of these autophagy-miRNAs are also important regulators in the crosstalk between autophagy and apoptosis. Furthermore, transcripts involved in autophagy and apoptosis may indirectly modulate each other by competing for common miRNA binding sites. Thus, miRNAs potentially work as molecular switches between these two intimately connected processes and contribute to the cell fate decision.
Keywords: miRNAs, autophagy, apoptosis, cell signaling, cancers
Introduction
Macroautophagy is an evolutionarily conserved cellular catabolic process in which proteins and organelles are eliminated through delivery to lysosomes.1 Although in a few cases autophagy may play a role in the execution of cell death, it is usually a cytoprotective mechanism in maintaining homeostasis and protecting cells from nutrient stress.2 Deregulation in autophagy has been implicated in numerous human disease, including developmental disorders, neurodegenerative disease and cancers.1 Autophagy is an intrinsic cellular process, which needs to be tightly controlled. In addition, the autophagic pathway should actively exchange information with other cellular processes such as apoptosis. Recently, one group of endogenous noncoding RNAs, miRNAs, have been found to be involved in the regulation of autophagy, and their roles in modulating the crosstalk between autophagy and apoptosis are emerging.
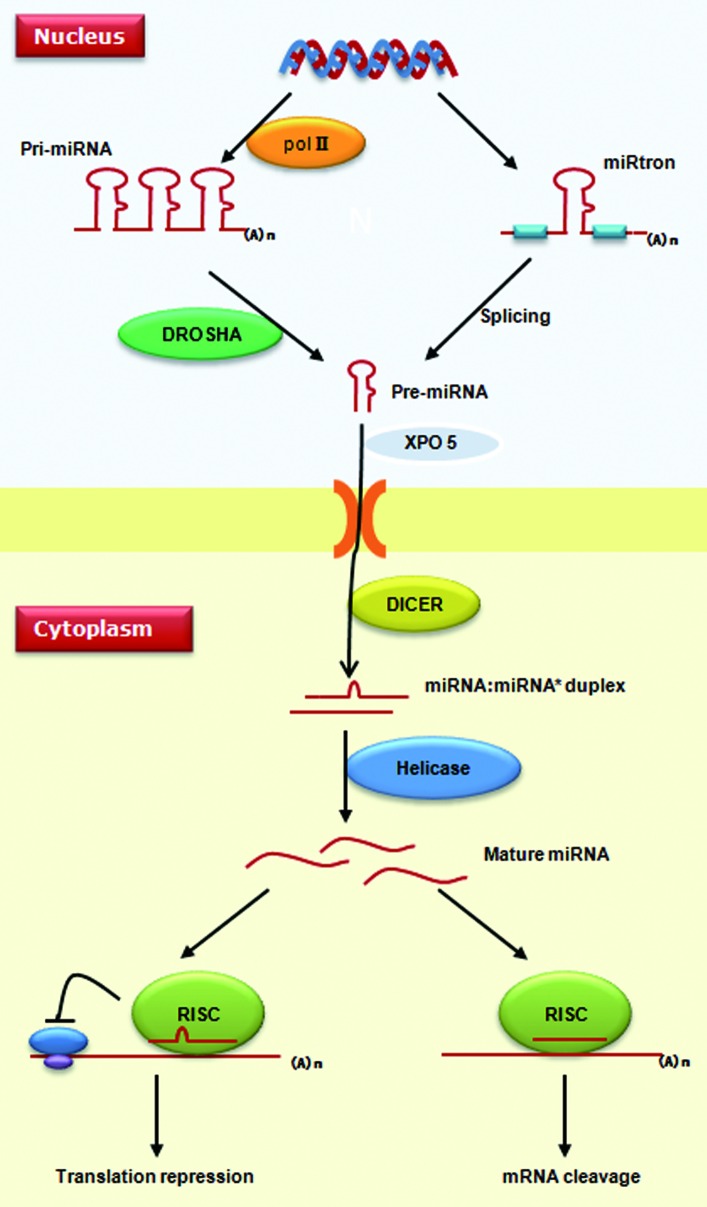
miRNAs are ~22-nucleotide long regulatory molecules, which are usually phylogenetically conserved.3 The biogenesis and action mechanism of metazoan miRNAs are summarized in Figure 1. miRNAs reside in protein-coding, intronic or intergenic regions throughout the genome. They can be produced from their own promoters or transcribed together with their host genes (in the cases of miRNAs located in introns). Mammalian miRNAs tend to cluster along the genome, and this clustering property plays an important role in guaranteeing the coordinate expression of different miRNAs.4,5 Metazoan miRNAs are mainly transcribed by RNA polymerase II, which produce hundreds or thousands of nucleotide-long products called primary miRNAs (pri-miRNAs).6 In the nucleus, the DROSHA nuclease complex cleaves metazoan pri-miRNAs into 70-nucleotide hairpins, known as precursor-miRNAs (pre-miRNAs). After being transported to the cytosol by XPO5 (exportin 5), pre-miRNAs are further processed into mature miRNAs by another RNase III DICER1.3 The metazoan pre-miRNAs can also be produced from spliced introns. This “miRtron” pathway is conserved among diverse mammals as well as in drosophilids and nematodes.7-9
Figure 1. Biogenesis and action mechanism of miRNAs. Metazoan miRNAs are transcribed into pri-miRNA by RNA polymerase II. In the nucleus, precursor-miRNA can be produced either by processing of pri-miRNAs via the DROSHA complex or by the miRtron pathway without RNase. Pre-miRNAs are translocated into the cytoplasm by XPO5, where another RNase, DICER1, cleaves pre-miRNAs to generate a miRNA:miRNA* duplex. Single strands of mature miRNA enter into the miRISC complex to recognize target genes. miRNAs can silence a target gene via either repressing protein translation or enhancing mRNA degradation.
It had been thought that metazoan miRNAs lead to translation attenuation by targeting the 3′UTRs; however, novel findings challenged this notion because miRNA-mediated metazoan mRNA cleavage could also be observed in many cases.10,11 The first 2–8 bases of a mature miRNA sequence usually play a pivotal role in target recognition in metazoans and are routinely used in most bioinformatics algorithms to search for target mRNAs.12 Therefore, different miRNAs can share common target mRNAs if they possess similar “seed” regions.
miRNAs play a critical role in a broad range of biological processes such as developmental timing, differentiation and tissue morphogenesis.3,13 Deregulation of miRNAs is also involved in a wide spectrum of diseases. For example, more than half of miRNA genes are located at fragile sites or cancer-associated genomic regions and are often aberrantly expressed in human cancers.14,15
miRNAs Regulate Autophagy and Contribute to Disease Progression
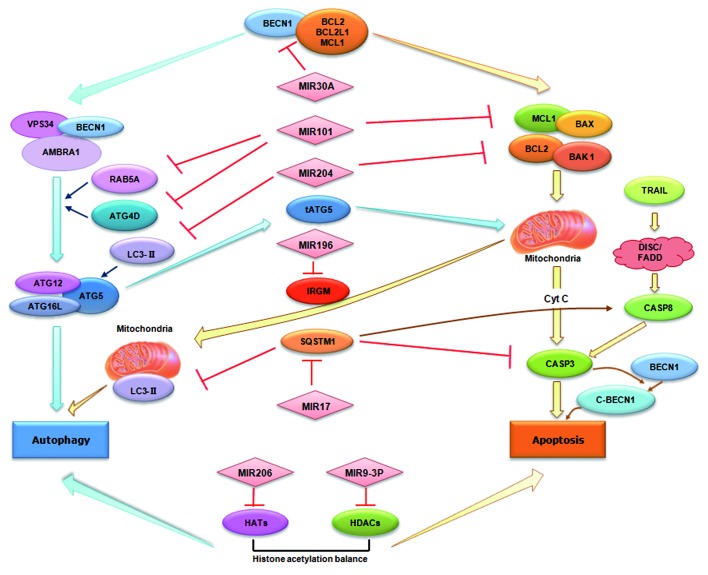
The regulatory role of miRNAs in autophagy was first uncovered in 2009 when BECN1, an important autophagy-promoting gene, was shown to be post-transcriptionally modulated by MIR30A.16 Soon after this report, a number of miRNAs participating in autophagy have been associated with certain diseases including cancers, cardiac pathologies and Crohn disease. In the following, we will discuss these miRNAs and their relevance to autophagy (summarized in Table 1 and Fig. 2).
Table 1. miRNAs with relevance in autophagy.
| miRNAs | Chromosome location | Disease relevance | Targets relevant to autophagy | Function | References |
|---|---|---|---|---|---|
| MIR30A |
6q13 |
Targets B-Myb oncogene during cellular senescence |
BECN1 |
Regulation of autophagic response to rapamycin in cancer cells |
16, 19 |
| MIR206 |
6p12.2 |
Blocks the growth of human rhabdomyosarcoma and breast cancer cells |
KAT6A (MYST3) |
Modulation of histone acetylation |
23, 27, 28, 35 |
| miR-9-3p |
1q22, 5q14.3, 15q26.1 |
Downregulated in the prefrontal cortex of schizophrenic subjects; contributes to tumor metastasis |
HDAC4 and HDAC5 |
Modulation of histone acetylation |
29, 33, 35 |
| MIR101 |
1p31.3, 9p24.1 |
Locus is lost in clinically localized prostate cancers or metastatic prostate cancers |
RAB5A, ATG4D and STMN1 |
Sensitizes breast cancer cells to 4-OHT-mediated cell death. |
43, 45 |
| MIR17,20,93 and 106 |
13q31.3, 7q22 |
Elevated in B-cell lymphoma and modulate tumor formation |
SQSTM1 (p62) |
Promote hematopoietic cell expansion |
56, 58 |
| MIR204 |
9q21.12 |
Downregulated in the NCI60 tumor cell lines; Involved in pulmonary arterial hypertension |
MAP1LC3 |
Regulation of cardiomyocyte autophagy induced by hypoxia-reoxygenation |
62–64 |
| MIR196 | 17q21.32, 12q13.13, 7p15.2 |
Potential roles in melanoma and acute leukemia | IRGM | Deregulation of IRGM-dependent xenophagy | 67, 68, 73 |
Figure 2. miRNAs in autophagy and their emerging roles in crosstalk with apoptosis. miRNAs can directly target autophagy-associated proteins such as BECN1 and RAB5A. Alternatively, miRNAs may indirectly modulate autophagy regulators such as HDACs and HATs. As shown in the figure, miRNAs can also mediate the crosstalk between autophagy and apoptosis via either targeting the common regulators for both pathways such as BECN1 and SQSTM1, or via regulating multiple targets involved in the two pathways.
MIR30A
The MIR30 subfamily contains five paralogs: MIR30A, B, C, D and E, which map at different genomic positions. MIR30A and other members of the MIR30 family are abundantly expressed in the adult prefrontal cortex.17 Previous studies have found that MIR30A can bind to a conserved site at the 3′UTR of BDNF (brain-derived neurotrophic factor), a key regulator during cortical development and maturation.17 Alterations in BDNF expression have been reported in a plethora of neuropsychiatric diseases.18 A recent investigation suggested MIR30 family members are also upregulated during cellular senescence. The MYBL2 (B-Myb) oncogene, an important regulator of the cell cycle, was identified as a bona fide target of MIR30A during senescence.19 Furthermore, blocking the activity of MIR30A inhibits cellular senescence.19 All of the above lines of evidence indicate MIR30A is a potential tumor suppressor.
Recently, Zhu et al. demonstrated that MIR30A expression is inhibited when cells are subjected to nutrient depletion or rapamycin treatment, respectively.16 MIR30A negatively regulates BECN1 both at the mRNA and protein level in human breast, lung and glioma cancer cell lines. Overexpression of MIR30A leads to the BECN1-dependent suppression of autophagic activity in cancer cells. BECN1 is identified as a potent inducer of autophagy and plays a key role in tumorigenesis and neurodegenerative diseases.20,21 Previous studies have documented the decreased expression of BECN1 in human breast, ovarian and brain cancers.20,22 This finding elucidated a novel regulation mechanism for BECN1 and further suggested that blocking of BECN1 by MIR30A may contribute to cancer progression.
MIR206 and miR-9-3p
MIR206 is transcribed together with MIR133B as a miRNA cluster. Expression of MIR133B and MIR206 increase during muscle cell differentiation and human fetus development.23,24 A master myogenic transcriptional factor, MYOD1, induces MIR206 transcription in muscle.25MIR206 can block human rhabdomyosarcoma growth by targeting the c-Met 3′UTR both in rhabdomyosarcoma cell lines and in xenotransplanted mice.23,24 In addition, MIR206 is reported to markedly decrease in estrogen receptor α-positive human breast cancer tissues.26 In MCF-7 breast cancer cells, overexpression of MIR206 results in reduced cell proliferation and enhanced apoptosis via downregulating endogenous estrogen receptor α and other estrogen receptor-associated coregulatory proteins.27,28 The human MIR9 subfamily contains three genes, MIR9-1, MIR9-2 and MIR9-3, which are separately located at 1q22, 5q14.3 and 15q26.1. miR-9-3p and miR-9-5p denote mature miRNAs originating from 3′ and 5′ arms of the same hairpin structure, respectively. miR-9-3p is significantly downregulated in the prefrontal cortex of subjects with schizophrenia compared with healthy controls.29 Recently, it has been demonstrated that MIR9 exerts its tumor suppressor role in various cancers including ovarian tumor,30 gastric cancer,31 cervical cancer32 and breast cancer.33 Expression of MIR9 is activated by MYC (v-myc myelocytomatosis viral oncogene homolog) and MYCN (v-myc myelocytomatosis viral related oncogene, neuroblastoma derived) in breast cancer cells, which further contributes to metastasis.33
A recent investigation indicated increased expression of MIR206 and reduced expression of miR-9-3p in primary waldenstrom macroglobulinemia (WM) cells.34 WM is a rare, nonHodgkin lymphoma characterized by an arrest of B lymphocytes after somatic hypermutation and before isotype class switching. The abnormal expression of these two miRNAs leads to alteration of balances between autophagy and apoptosis via modulating histone acetylation in WM cells.35
Histone deacetylases (HDACs) and histone acetyl transferases (HATs) control the chromatin structure status, and the balance of these two families of enzymes is crucial to gene transcription.36 In many malignancies including WM, this balance is disrupted, which is characterized by significantly increased expression of HDACs and by significantly decreased expression of HATs.34 Since HDAC4, HDAC5 and KAT6A (MYST3) were identified as targets for miR-9-3p and MIR206, respectively, the increased expression of MIR206 and reduced expression of miR-9-3p deregulate histone acetylation and lead to autophagy-dependent cell toxicity.35 It should be noted that although the modulation of autophagy by HDAC suppression has been implicated in the pathogenesis of human diseases,37,38 how histone acetylation affects autophagy remains largely unknown. For example, Shao et al. found that suberoylanilide hydroxamic acid, a potent inhibitor of HDAC, can induce cancer cell death, which had unambiguous morphological features of autophagosome formation.37 However, Cao et al. recently reported that suppressing of HDAC attenuates cardiac hypertrophy via autophagy inhibition.38 How can this discrepancy be explained? An obvious explanation is cell-type specificity. Alternatively, cells may choose different routes to link HDAC with autophagy. Therefore, further investigations are needed to delineate the underlying molecular mechanism.
MIR101
MIR101 is an established tumor suppressor across many cancer types.39-44 It has two genomic loci, which are on chromosome 1 (MIR101-1) and chromosome 9 (MIR101-2). Based on genomic PCR, one or both of the two genomic loci are found to be somatically lost in clinically localized prostate cancers or metastatic prostate cancers.43EZH2 (Enhancer of zeste homolog 2), which encodes a mammalian histone methyltransferase that epigenetically regulates cancer cell survival and metastasis, was identified as a key target of MIR101 in prostate cancer cells and in non-small cell lung cancers.43,44 Other confirmed targets for MIR101 include MCL1 (myeloid cell leukemia sequence 1) and FOS (FBJ murine osteosarcoma viral oncogene homolog) oncogene in hepatocellular carcinoma,40,42MAGI2 (membrane-associated guanylate kinase, WW and PDZ domain containing 2) in breast cancer cell lines41 and the MYCN gene in neuroblastoma cell lines.39 Therefore, MIR101 promotes apoptosis and suppresses tumorigenesis.
Interestingly, in a functional screen for miRNAs that regulate the autophagic flux in breast cancer cells, MIR101 was found to be a potent inhibitor of autophagy.45 MIR101 targets the genes encoding the autophagy-related proteins RAB5A, ATG4D and STMN1. RAB5A is a member of the RAS oncogene family, which is conserved from yeast to humans. The RAB5A protein can exhibit GTPase activities and is identified as a key regulator of endocytosis.46 However, a recent investigation suggested RAB5 is involved in autophagosome formation and regulates ATG5–ATG12 conjugation.47 In addition, siRNA inhibition of RAB5A blocks basal and rapamycin-induced autophagy.45
ATG4D belongs to the ATG4 family of cysteine-type endopeptidases, which is a homolog of yeast Atg4.48 In mammals, ATG4 cleaves the C terminus of LC3 (yeast Atg8) to form cytosolic LC3-I, which is covalently conjugated to the lipid phosphatidylethanolamine (PE) on autophagosomal membranes. Atg4-Atg8 conjunction is a crucial step in the autophagosome biogenesis pathway. Currently, four mammalian ATG4 paralogs [ATG4A-ATG4D] and six ATG8 paralogs with varied substrate specificity have been cloned. Previous studies suggested that although ATG4B is the main regulator of LC3 in mammalian cells, other members of the ATG4 family may be specific for other individual Atg8 orthologs.48,49
STMN1 is a gene coding for a ubiquitous cytosolic phosphoprotein. Previous studies suggested STMN1 modulates depolymerization of interphase and mitotic microtubules based on the transition of unphosphorylated and phosphorylated forms.50STMN1 is ectopically expressed in a variety of cancer types.51 Silencing of STMN1 inhibits tumor growth in breast cancer cells, primary melanomas and osteosarcomas.52,53 Excessive expression of STMN1 causes a partial block of MIR101-mediated inhibition of autophagy, indicating its importance as a MIR101 target.45
MIR17, 20, 93 and 106
The MIR17, 20, 93 and 106 genes, are highly conserved between species and share the same AAGUGC ‘seed’ region and target specificity. MIR17 and MIR20 belong to the MIR17-92 cluster. The human MIR17-92 cluster is mapped to 13q31.3, a region amplified in diffuse B-cell lymphomas (DLBCLs), follicular lymphomas, Burkitt’s lymphomas and lung carcinoma.54 The MIR17-92 cluster is markedly elevated in B-cell lymphoma and lung cancers.55,56 MIR106B, MIR205 and MIR93 consist of a paralog of the MIR17-92 cluster. This miRNA cluster locates at chromosome 7q22, a region also amplified in several cancers. In recent years, the oncogenic properties of the MIR17-92 and MIR106-25 clusters have been extensively investigated.55,56 These two clusters are induced via MYC and E2F1 signals. Subsequent overexpression of these miRNAs downregulates p21 which is required for cell cycle arrest.57 MIR106A, located in the MIR106A-92 cluster, is the second paralog of MIR17-92. Although the function of this cluster remains obscure, it may also be associated with oncogenic development at least in cancer cells.
Recently, these AAAGUGC seed-containing miRNAs were found highly expressed in myeloid progenitors and blocked in mature neutrophils.58 SQSTM1 (sequestosome 1/p62), a multiple domain protein that acts as a signaling hub, was identified as a key target for these miRNAs. SQSTM1 can interfere with autophagy via binding to the autophagic regulator Atg8/LC3.59 SQSTM1 has also been implicated in a variety of cellular events such as the NFκB and proteasome pathways.60 During ligand-induced neutrophil differentiation, SQSTM1 regulates colony-stimulating factor 3 receptor stability and mitogen-activated protein kinase signaling. Therefore, these AAAGUGC seed-containing miRNAs enhance the expansion of myeloid 32D cells and primary hematopoietic progenitors by modulating SQSTM1-regulated cellular events.58 Notably, in autophagy-defective and apoptosis-incompetent tumor cells, metabolic stress results in SQSTM1 aggregation, which further triggers a positive feedback loop for the generation of reactive oxygen species, enhanced genomic instability and tumorigenesis.61 Thus, elimination of SQSTM1 through upregulation of AAAGUGC seed-containing miRNAs may potentially inhibit the proliferation of these tumor cells.
MIR204
MIR204 is located within the intron of the TRPM3 gene at 9q21.12. Its expression is downregulated in pulmonary artery smooth muscle cells and in clinical samples, suggesting that MIR204 plays a critical role in the etiology of human pulmonary arterial hypertension.62 In addition, compared with normal tissues, MIR204 is significantly lower in the NCI60 tumor cell line panel.63 In a recent study, Xiao et al. found that hypoxia-reoxygenation induces cellular autophagy in cardiomyocytes of neonatal rats.64 Concurrently, MIR204 is significantly decreased and the ratio of LC3-II/LC3-I is increased. However, it remains to be determined whether MIR204 inhibits autophagy by directly targeting MAP1LC3.
MIR196
MIR196 is an evolutionarily conserved miRNA in vertebrate species. This family consists of three members including two MIR196A genes (MIR196A1 and MIR196A2) and one MIR196B gene. These miRNAs appear to be expressed from intergenic regions in HOX gene clusters and mediate the posttranscriptional restriction of HOX gene expression during development.65,66 Recent studies have extended the function of MIR196 to tumorgenesis. For example, MIR196 can repress several transcription factors and play a regulatory role in melanoma and acute leukemia.67,68 Indeed, a number of genetic polymorphisms in the precursor or mature MIR196 have been associated with susceptibility and risk in different cancer types.69-71
Crohn disease is a complex inflammatory bowel disease, which can affect any area of the gastrointestinal tract, from the mouth to the anus.72 MIR196 is overexpressed in the inflammatory intestinal epithelia of individuals with Crohn disease.73 Bioinformatics analysis revealed the potential recognition sequence of MIR196 on the coding region of the IRGM (immunity-related GTPase family, M gene). Interestingly, a synonymous variant polymorphism of IRGM is located within the ‘seed’ region. Subsequent functional analysis indicated that MIR196 downregulates the IRGM protective variant (c.313B) but not the risk-associated allele (c.313T).73 The involvement of IRGM in the innate immune response is mediated via regulating autophagy in response to intracellular pathogens, or xenophagy through mitochondria.74 Therefore this is the first example suggesting a miRNA-associated synonymous polymorphism influencing autophagy and disease risk.
Roles of miRNAs in Crosstalk between Autophagy and Apoptosis
Apoptosis is a controlled process of cell death occurring when cells face irreversible stress. By apoptosis, cells eliminate the damaged cells that may be harmful to the organism and maintain normal cell homeostasis. Apoptosis can be induced either by various cellular insults mediated through the mitochondria (intrinsic pathway) or cell surface death receptors (extrinsic pathway). Members of the BCL2 family play crucial roles in regulating apoptosis.75 Upon induction, upstream BH3-only proteins (such as BCL2L11/BIM, BAD and BBC3/PUMA) inactive anti-apoptosis BCL2 family members (BCL2, MCL1, BCL2L1). Then these anti-apoptotic family members relieve pro-apoptotic BAX and BAK1, which are translocated to the mitochondrial membrane and result in cytochrome c release and mitochondrial fission. In the extrinsic apoptosis pathway, components of the death-inducing signaling complex (DISC) including surface receptors of the death receptor, adaptor proteins (FADD and TRADD) and CASP8 and CASP10 are activated upon stimuli. Both intrinsic and extrinsic apoptotic pathways converge on the level of CASP3 activation, which in turn cleaves various intracellular substrates and cause the morphological changes observed in apoptotic cells.75
Both autophagy and apoptosis play important roles in the development, cellular homeostasis and oncogenesis of mammals. They may be triggered by common upstream signals, resulting in combined autophagy and apoptosis, or be mutually exclusive.76,77 Intriguingly, several confirmed targets of autophagy-miRNAs are also important mediators in the crossregulation between autophagy and apoptosis (Fig. 2).
miRNAs can inhibit the common regulators of these two pathways. For example, previous investigations have suggested the physical interaction between BECN1 and proteins in the anti-apoptotic family (BCL2, MCL1, BCL2L1) is pivotal for the conversation between the two pathways.78-80 Under normal conditions, BECN1 and anti-apoptotic BCL2 family members can bind to each other to maintain cellular homeostasis. When cells face stress conditions, BECN1 and BCL2 family members disassociate, thereby promoting autophagy and inhibiting apoptosis, respectively.78-80 In addition to MIR30A, which can reduce the cytoplasmic level of BECN1, several other miRNAs have been demonstrated to reduce the expression level of the anti-apoptotic family members such as BCL2 and MCL1.81,82 Thus, miRNAs may be actively involved in the regulation of both autophagy and apoptosis signals based on modulation of the protein-protein interactions. SQSTM1 is another common mediator under the control of miRNAs. Recent data suggest SQSTM1 can modulate the polyubiquitination and aggregation of CASP8, which is essential for the extrinsic apoptotic pathway.83 On the other hand, SQSTM1 can negatively regulate the degradation of the autophagic protein LC3 by the 20S proteasome.84 Therefore, collective evidence implicates a potential mechanism of SQSTM1 underlying the interplay between the apoptosis and autophagic pathways.
In fact, autophagy and apoptosis share many essential genes, ranging from common players such as TP53 (p53) and ATG5 to signal transduction mediators such as DAPK1 (death-associated protein kinase 1) and EEF2 (eukaryotic translation elongation factor 2).76,85,86 It would be interesting to identify whether or not other miRNAs could coordinately regulate both apoptosis and autophagy signals based on modulating these proteins. Alternatively, as we summarized in Figure 2, some miRNAs can simultaneously modulate multiple targets, which function either in autophagy or in apoptosis. For example, MIR101 potently targets the genes encoding the autophagy-associated proteins RAB5A, ATG4D and STMN1; and also the anti-apoptotic protein MCL1.42,45 During hypoxia-reoxygenation, MIR204 blocks autophagy by modulating the LC3-II protein whereas in cholangiocarcinoma cells the exogenous expression of MIR204 negatively regulates BCL2 and facilitate chemotherapeutic drug-triggered apoptosis.87
Working Models for the Mechanisms of miRNAs in Autophagy and the Crosstalk with Apoptosis
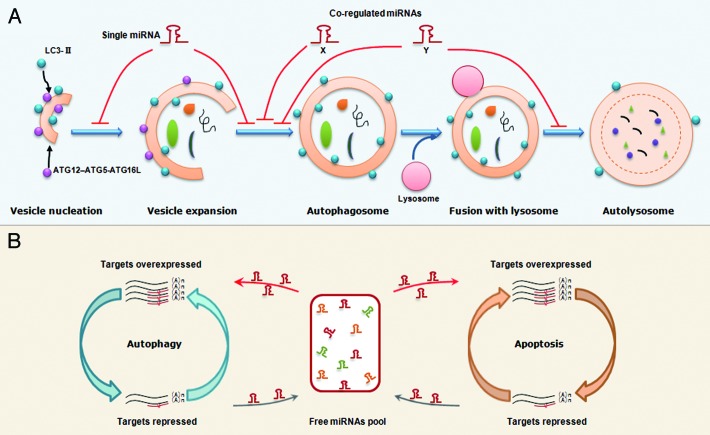
Previous proteolysis analyses have suggested most miRNAs only downregulate their individual target moderately.88,89 However, some miRNAs can function as molecular on-off switches to completely shut down a cellular process. The trick perhaps lies in the multitarget characteristics of miRNAs. Since one single miRNA can concurrently target multiple genes, the multiple autophagy-related proteins that control different steps of autophagy may be regulated by the same miRNAs (Fig. 3A). Indeed, in the above MIR101 example, this miRNA represses three important autophagy-associated genes. Importantly, overexpression of STMN1 can only partially rescue MIR101-mediated autophagic inhibition.45 This result indicates that other MIR101 targets including RAB5A and ATG4D still act in autophagy, and further strengthens the point that a single miRNA can take many routes to modulate autophagy.
Figure 3. Proposed models that miRNAs regulate the autophagy and the balance between autophagy and apoptosis. (A) An illustration of miRNA regulation of autophagy. A single miRNA can modulate multiple targets in different steps of autophagy. Alternatively, several co-regulated miRNAs can modulate the different steps in a cooperative manner although each miRNA regulates its specific target. (B) A working model for miRNA-mediated crosstalk between autophagy and apoptosis. Transcripts involved in different cellular processes but with common miRNA binding sites can modulate each other by competing for miRNA binding. For example, overexpression of autophagy-related genes will result in binding of more miRNA molecules,thereby leading to fewer miRNA molecules free to bind to apoptosis-related transcripts, which share similar miRNA binding sites. Thus, the miRNA-medicated conversation between autophagy and apoptosis is regular and intensive.
Alternatively, several coregulated miRNAs can modulate the same or different autophagic steps in a cooperative manner, although each miRNA regulates its specific target (Fig. 3A). This situation is illustrated in the MIR17 seed family example. MIR17, 20, 93 and 106, which all contain an AAAGUGC seed region, are expressed in hematopoietic cells at different stages of myeloid development.58 Experiments demonstrate that they all regulate the same target, SQSTM1.58 In fact, this cooperation may not be restricted to a set of miRNAs with a similar seed. For example, previously we and others independently found a functional link between members within the same miRNA cluster.4,90 Since mammalian miRNAs with a proximal locus tend to share the same transcriptional unit, upon stimulation, different members of the same miRNA cluster can be concurrently induced or repressed, thus controlling a biological process in a cooperative manner.
Since autophagy and apoptosis share some essential mediators, the direct targeting of the common proteins by miRNAs will ultimately affect signal transduction in both pathways (Fig. 2). In addition, we speculate that transcripts involved in autophagy and apoptosis can indirectly modulate each other by competing for common miRNA binding sites. As indicated in the schema shown in Figure 3B, when autophagy-related proteins are repressed, more miRNA molecules are liberated into the free miRNA pool and further enter into the apoptotic pathway to target apoptosis-related proteins. By contrast, the accumulation of genes encoding autophagy-related proteins will result in the binding of more miRNA molecules, thereby leading to fewer miRNAs being free to bind apoptosis-related transcripts with similar binding sites. Thus, the miRNAs maintain regular and intensive crosstalk between autophagy and apoptosis.
This miRNA-mediated mutual regulation between transcripts with similar binding sites has great implications in cancer biology. Poliseno et al. found that PTENP1, the pseudogene of PTEN, possesses well-conserved miRNA binding sites and is biologically active since it can compete with PTEN for miRNA binding. Therefore PTENP1 modulates the cellular levels of PTEN and exerts a tumor suppressor role.91 Since this mechanism depends only on the competition between the 3′UTRs, it can be expected that it is not limited to this gene and its pseudogene. Indeed, the same group of researchers discovered that there are a network of competing endogenous mRNAs that in a mutually reciprocal manner center on PTEN.92,93 Recently, a set of investigations has extend this concept to long-coding RNAs and to all protein-coding mRNAs.94,95 Based on the shared miRNA binding sites, protein-coding genes within the autophagic and apoptotic pathways can maintain cellular homeostasis and collectively decide cell fate. Deregulation of this miRNA-based “housekeeping” mechanism may contribute to disease progression.
Conclusions and Perspectives
Over the past few decades, tremendous interest has focused on understanding the regulatory role of miRNAs under various physiological and disease conditions. In this review, we summarized the recent findings concerning miRNAs involved in autophagy. Ever since the first autophagy-associated miRNA, MIR30A, was discovered in 2009, our knowledge of miRNAs in autophagy has accumulated rapidly. However, the understanding of this field is still in its infancy. For example, direct targeting of autophagy-related genes has only been experimentally shown in a few cases at present.16,45,58,73 Since the identification of gene products that participate in autophagy is continually expanding,96 it is likely that other autophagy-associated miRNAs will be found in the near future. Besides, it is important to develop efficient research tools for manipulating autophagy. We propose that miRNAs may be used to block autophagy-associated genes at both the mRNA and protein levels. As seen in the MIR101 example, one single miRNA is enough to block both basal and rapamycin-induced autophagy via targeting multiple autophagy-associated genes.
miRNAs also represent an additional layer in the intricate interconnection between autophagy and apoptosis. As we discussed above, miRNAs may target the common regulators for both processes or simultaneously modulate multiple targets essential to each pathway. Recent developments have further indicated that miRNAs may work as a general mediator between protein-coding genes based on shared binding sites.97 Therefore, multiple miRNAs together with their multiple downstream genes form a complicated regulation network. Use of systems level analysis, such as the efforts made in analyzing transcriptional and miRNA-based post-transcriptional regulation in the autophagy-lysosomal pathway, will help to elucidate the underlying connection across cellular processes.98 In addition, a detailed deciphering of the crucial role of miRNAs in the interplay between autophagy and apoptosis have profound clinical implications since the evasion of cell death underlies tumorigenesis and represents a major obstacle to successful therapies.99 Therefore, such efforts are imperative to improve our understanding of miRNAs in tumorigenesis and facilitate the design of appropriate therapies targeting this novel group of molecules.
Acknowledgments
We are grateful to Dr. Daniel J. Klionsky and two anonymous reviewers for their constructive comments and help. We would like to thank Mr. Zhenkun Zhang and Jinjiang Lu for collection of supporting data. We are in debt to Mr. Junfeng Wang for assistance on graphic preparation. The research is supported by the science foundation of the education department of Henan province (Grant No. 2011A180009) and a grant from Henan University of Technology (#2009BS040).
Glossary
Abbreviations:
- BECN1
Beclin 1
- BCL2
B-cell CLL/lymphoma 2
- MCL1
myeloid cell leukemia sequence 1
- BCL2L1
BCL2-like 1
- VPS34
class III phosphatidylinositol 3-kinase
- AMBRA1
autophagy/Beclin 1 regulator 1
- RAB5A
RAS-associated protein RAB5A
- LC3
microtubule-associated protein 1 light chain 3 alpha
- ATG
AuTophaGy-related
- tATG5
truncated form of autophagy-related protein 5
- IRGM
immunity-related GTPase family, M
- SQSTM1
p62/sequestosome 1
- HATs
histone acetyltransferases
- TRAIL
TNF- related apoptosis-inducing ligand
- BAK1
BCL2-antagonist/killer 1
- HDACs
histone deacetylases
- BAX
BCL2-associated X protein
- FADD
Fas-associated via death domain
- DISC
death-inducing signaling complex
- CASP
apoptosis-related cysteine peptidase
- C-BECN1
C terminal fragments of BECN1
- 3′UTR
3′ untranslated region
Note
After this manuscript was revised, the role of MIR376B on autophagy was reported in a recent paper.100
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/19629
References
- 1.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–10. doi: 10.1038/nrm2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu J, Wong C. A computational screen for mouse signaling pathways targeted by microRNA clusters. RNA. 2008;14:1276–83. doi: 10.1261/rna.997708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Megraw M, Sethupathy P, Corda B, Hatzigeorgiou AG. miRGen: a database for the study of animal microRNA genomic organization and function. Nucleic Acids Res. 2007;35(Database issue):D149–55. doi: 10.1093/nar/gkl904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–36. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–6. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–90. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 11.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L, Xu J, Yang D, Tan X, Wang H. Computational approaches for microRNA studies: a review. Mamm Genome. 2010;21:1–12. doi: 10.1007/s00335-009-9241-2. [DOI] [PubMed] [Google Scholar]
- 13.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–87. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 14.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X, et al. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy. 2009;5:816–23. doi: 10.4161/auto.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. 2008;17:3030–42. doi: 10.1093/hmg/ddn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Angelucci F, Brenè S, Mathé AA. BDNF in schizophrenia, depression and corresponding animal models. Mol Psychiatry. 2005;10:345–52. doi: 10.1038/sj.mp.4001637. [DOI] [PubMed] [Google Scholar]
- 19.Martinez I, Cazalla D, Almstead LL, Steitz JA, DiMaio D. miR-29 and miR-30 regulate B-Myb expression during cellular senescence. Proc Natl Acad Sci U S A. 2011;108:522–7. doi: 10.1073/pnas.1017346108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 21.Jaeger PA, Wyss-Coray T. Beclin 1 complex in autophagy and Alzheimer disease. Arch Neurol. 2010;67:1181–4. doi: 10.1001/archneurol.2010.258. [DOI] [PubMed] [Google Scholar]
- 22.Miracco C, Cosci E, Oliveri G, Luzi P, Pacenti L, Monciatti I, et al. Protein and mRNA expression of autophagy gene Beclin 1 in human brain tumours. Int J Oncol. 2007;30:429–36. [PubMed] [Google Scholar]
- 23.Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, et al. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–78. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan D, Dong XdaE, Chen X, Wang L, Lu C, Wang J, et al. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem. 2009;284:29596–604. doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H. miR-206 Expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res. 2008;68:5004–8. doi: 10.1158/0008-5472.CAN-08-0180. [DOI] [PubMed] [Google Scholar]
- 27.Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21:1132–47. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 28.Adams BD, Cowee DM, White BA. The role of miR-206 in the epidermal growth factor (EGF) induced repression of estrogen receptor-alpha (ERalpha) signaling and a luminal phenotype in MCF-7 breast cancer cells. Mol Endocrinol. 2009;23:1215–30. doi: 10.1210/me.2009-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo LM, Pu Y, Han Z, Liu T, Li YX, Liu M, et al. MicroRNA-9 inhibits ovarian cancer cell growth through regulation of NF-kappaB1. FEBS J. 2009;276:5537–46. doi: 10.1111/j.1742-4658.2009.07237.x. [DOI] [PubMed] [Google Scholar]
- 31.Luo H, Zhang H, Zhang Z, Zhang X, Ning B, Guo J, et al. Down-regulated miR-9 and miR-433 in human gastric carcinoma. J Exp Clin Cancer Res. 2009;28:82. doi: 10.1186/1756-9966-28-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu X, Schwarz JK, Lewis JS, Jr., Huettner PC, Rader JS, Deasy JO, et al. A microRNA expression signature for cervical cancer prognosis. Cancer Res. 2010;70:1441–8. doi: 10.1158/0008-5472.CAN-09-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–56. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roccaro AM, Sacco A, Chen C, Runnels J, Leleu X, Azab F, et al. microRNA expression in the biology, prognosis, and therapy of Waldenström macroglobulinemia. Blood. 2009;113:4391–402. doi: 10.1182/blood-2008-09-178228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roccaro AM, Sacco A, Jia X, Azab AK, Maiso P, Ngo HT, et al. microRNA-dependent modulation of histone acetylation in Waldenstrom macroglobulinemia. Blood. 2010;116:1506–14. doi: 10.1182/blood-2010-01-265686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 37.Shao Y, Gao Z, Marks PA, Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci U S A. 2004;101:18030–5. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao DJ, Wang ZV, Battiprolu PK, Jiang N, Morales CR, Kong Y, et al. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci U S A. 2011;108:4123–8. doi: 10.1073/pnas.1015081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buechner J, Tømte E, Haug BH, Henriksen JR, Løkke C, Flægstad T, et al. Tumour-suppressor microRNAs let-7 and mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br J Cancer. 2011;105:296–303. doi: 10.1038/bjc.2011.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li S, Fu H, Wang Y, Tie Y, Xing R, Zhu J, et al. MicroRNA-101 regulates expression of the v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) oncogene in human hepatocellular carcinoma. Hepatology. 2009;49:1194–202. doi: 10.1002/hep.22757. [DOI] [PubMed] [Google Scholar]
- 41.Sachdeva M, Wu H, Ru P, Hwang L, Trieu V, Mo YY. MicroRNA-101-mediated Akt activation and estrogen-independent growth. Oncogene. 2011;30:822–31. doi: 10.1038/onc.2010.463. [DOI] [PubMed] [Google Scholar]
- 42.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, et al. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–42. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 43.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–9. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang JG, Guo JF, Liu DL, Liu Q, Wang JJ. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J Thorac Oncol. 2011;6:671–8. doi: 10.1097/JTO.0b013e318208eb35. [DOI] [PubMed] [Google Scholar]
- 45.Frankel LB, Wen J, Lees M, Høyer-Hansen M, Farkas T, Krogh A, et al. microRNA-101 is a potent inhibitor of autophagy. EMBO J. 2011;30:4628–41. doi: 10.1038/emboj.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, et al. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–28. doi: 10.1016/0092-8674(92)90306-W. [DOI] [PubMed] [Google Scholar]
- 47.Ravikumar B, Imarisio S, Sarkar S, O’Kane CJ, Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci. 2008;121:1649–60. doi: 10.1242/jcs.025726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mariño G, Uría JA, Puente XS, Quesada V, Bordallo J, López-Otín C. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278:3671–8. doi: 10.1074/jbc.M208247200. [DOI] [PubMed] [Google Scholar]
- 49.Li M, Hou Y, Wang J, Chen X, Shao ZM, Yin XM. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem. 2011;286:7327–38. doi: 10.1074/jbc.M110.199059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gavet O, Ozon S, Manceau V, Lawler S, Curmi P, Sobel A. The stathmin phosphoprotein family: intracellular localization and effects on the microtubule network. J Cell Sci. 1998;111:3333–46. doi: 10.1242/jcs.111.22.3333. [DOI] [PubMed] [Google Scholar]
- 51.Rana S, Maples PB, Senzer N, Nemunaitis J. Stathmin 1: a novel therapeutic target for anticancer activity. Expert Rev Anticancer Ther. 2008;8:1461–70. doi: 10.1586/14737140.8.9.1461. [DOI] [PubMed] [Google Scholar]
- 52.Phadke AP, Jay CM, Wang Z, Chen S, Liu S, Haddock C, et al. In vivo safety and antitumor efficacy of bifunctional small hairpin RNAs specific for the human Stathmin 1 oncoprotein. DNA Cell Biol. 2011;30:715–26. doi: 10.1089/dna.2011.1240. [DOI] [PubMed] [Google Scholar]
- 53.Alli E, Yang JM, Hait WN. Silencing of stathmin induces tumor-suppressor function in breast cancer cell lines harboring mutant p53. Oncogene. 2007;26:1003–12. doi: 10.1038/sj.onc.1209864. [DOI] [PubMed] [Google Scholar]
- 54.Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–95. doi: 10.1158/0008-5472.CAN-03-3773. [DOI] [PubMed] [Google Scholar]
- 55.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–32. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 56.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191–4. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- 58.Meenhuis A, van Veelen PA, de Looper H, van Boxtel N, van den Berge IJ, Sun SM, et al. MiR-17/20/93/106 promote hematopoietic cell expansion by targeting sequestosome 1-regulated pathways in mice. Blood. 2011;118:916–25. doi: 10.1182/blood-2011-02-336487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 60.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–4. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Courboulin A, Paulin R, Giguère NJ, Saksouk N, Perreault T, Meloche J, et al. Role for miR-204 in human pulmonary arterial hypertension. J Exp Med. 2011;208:535–48. doi: 10.1084/jem.20101812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang FE, Zhang C, Maminishkis A, Dong L, Zhi C, Li R, et al. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010;24:1552–71. doi: 10.1096/fj.08-125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jian X, Xiao-yan Z, Bin H, Yu-feng Z, Bo K, Zhi-nong W, et al. MiR-204 regulate cardiomyocyte autophagy induced by hypoxia-reoxygenation through LC3-II. Int J Cardiol. 2011;148:110–2. doi: 10.1016/j.ijcard.2011.01.029. [DOI] [PubMed] [Google Scholar]
- 65.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 66.Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–4. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 67.Mueller DW, Bosserhoff AK. MicroRNA miR-196a controls melanoma-associated genes by regulating HOX-C8 expression. Int J Cancer. 2011;129:1064–74. doi: 10.1002/ijc.25768. [DOI] [PubMed] [Google Scholar]
- 68.Coskun E, von der Heide EK, Schlee C, Kühnl A, Gökbuget N, Hoelzer D, et al. The role of microRNA-196a and microRNA-196b as ERG regulators in acute myeloid leukemia and acute T-lymphoblastic leukemia. Leuk Res. 2011;35:208–13. doi: 10.1016/j.leukres.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 69.Hu Z, Shu Y, Chen Y, Chen J, Dong J, Liu Y, et al. Genetic polymorphisms in the precursor MicroRNA flanking region and non-small cell lung cancer survival. Am J Respir Crit Care Med. 2011;183:641–8. doi: 10.1164/rccm.201005-0717OC. [DOI] [PubMed] [Google Scholar]
- 70.Xu W, Xu J, Liu S, Chen B, Wang X, Li Y, et al. Effects of common polymorphisms rs11614913 in miR-196a2 and rs2910164 in miR-146a on cancer susceptibility: a meta-analysis. PLoS One. 2011;6:e20471. doi: 10.1371/journal.pone.0020471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Christensen BC, Avissar-Whiting M, Ouellet LG, Butler RA, Nelson HH, McClean MD, et al. Mature microRNA sequence polymorphism in MIR196A2 is associated with risk and prognosis of head and neck cancer. Clin Cancer Res. 2010;16:3713–20. doi: 10.1158/1078-0432.CCR-10-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brest P, Corcelle EA, Cesaro A, Chargui A, Belaïd A, Klionsky DJ, et al. Autophagy and Crohn’s disease: at the crossroads of infection, inflammation, immunity, and cancer. Curr Mol Med. 2010;10:486–502. doi: 10.2174/156652410791608252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet. 2011;43:242–5. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 74.Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E, et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol. 2010;12:1154–65. doi: 10.1038/ncb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 76.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 77.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–39. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Germain M, Nguyen AP, Le Grand JN, Arbour N, Vanderluit JL, Park DS, et al. MCL-1 is a stress sensor that regulates autophagy in a developmentally regulated manner. EMBO J. 2011;30:395–407. doi: 10.1038/emboj.2010.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luo S, Rubinsztein DC. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ. 2010;17:268–77. doi: 10.1038/cdd.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu J, Liao X, Wong C. Downregulations of B-cell lymphoma 2 and myeloid cell leukemia sequence 1 by microRNA 153 induce apoptosis in a glioblastoma cell line DBTRG-05MG. Int J Cancer. 2010;126:1029–35. doi: 10.1002/ijc.24823. [DOI] [PubMed] [Google Scholar]
- 83.Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–35. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 84.Gao Z, Gammoh N, Wong PM, Erdjument-Bromage H, Tempst P, Jiang X. Processing of autophagic protein LC3 by the 20S proteasome. Autophagy. 2010;6:126–37. doi: 10.4161/auto.6.1.10928. [DOI] [PubMed] [Google Scholar]
- 85.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–32. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 86.Cheng Y, Ren X, Zhang Y, Patel R, Sharma A, Wu H, et al. eEF-2 kinase dictates cross-talk between autophagy and apoptosis induced by Akt Inhibition, thereby modulating cytotoxicity of novel Akt inhibitor MK-2206. Cancer Res. 2011;71:2654–63. doi: 10.1158/0008-5472.CAN-10-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen L, Yan HX, Yang W, Hu L, Yu LX, Liu Q, et al. The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. J Hepatol. 2009;50:358–69. doi: 10.1016/j.jhep.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 88.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 90.Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, et al. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672–81. doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–95. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–57. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–81. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tian Y, Li Z, Hu W, Ren H, Tian E, Zhao Y, et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141:1042–55. doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 97.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jegga AG, Schneider L, Ouyang X, Zhang J. Systems biology of the autophagy-lysosomal pathway. Autophagy. 2011;7:477–89. doi: 10.4161/auto.7.5.14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 100.Korkmaz G, le Sage C, Tekirdag KA, Agami R, Gozuacik D. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy. 2012 doi: 10.4161/auto.8.2.18351. In press. [DOI] [PubMed] [Google Scholar]