Abstract
Lower global DNA methylation is associated with genomic instability and it is one of the epigenetic mechanisms relevant to carcinogenesis. Emerging evidence for several cancers suggests that lower overall levels of global DNA methylation in blood are associated with different cancer types, although less is known about breast cancer. We examined global DNA methylation levels using a sibling design in 273 sisters affected with breast cancer and 335 unaffected sisters from the New York site of the Breast Cancer Family Registry. We measured global DNA methylation in total white blood cell (WBC) and granulocyte DNA by two different methods, the [3H]-methyl acceptance assay and the luminometric methylation assay (LUMA). Global methylation levels were only modestly correlated between sisters discordant for breast cancer (Spearman correlation coefficients ranged from -0.08 to 0.24 depending on assay and DNA source). Using conditional logistic regression models, women in the quartile with the lowest DNA methylation levels (as measured by the [3H]-methyl acceptance assay) had a 1.8-fold (95% CI = 1.0–3.3) higher relative association with breast cancer than women in the quartile with the highest DNA methylation levels. When we examined the association on a continuous scale, we also observed a positive association (odds ratio, OR = 1.3, 95% CI = 1.0–1.7, for a one unit change in the natural logarithm of the DPM/μg of DNA). We observed no association between measures by the LUMA assay and breast cancer risk. If replicated in prospective studies, this study suggests that global DNA methylation levels measured in WBC may be a potential biomarker of breast cancer risk even within families at higher risk of cancer.
Keywords: blood, breast cancer, epigenetics, global DNA methylation, global methylation, LUMA, luminometric methylation assay, methyl acceptance assay, white blood cell, [3H]-methyl acceptance assay
Introduction
DNA methylation is mitotically inheritable and, while stable, greater differences in DNA methylation patterns have been observed in older compared with younger monozygotic twins, suggesting that endogenous and exogenous changes across the life course may influence these changes.1 DNA methylation levels have also been found to be associated with selected environmental exposures.2 Global DNA methylation, also referred to as genomic DNA methylation, is the overall content of 5-methyl cytosine (5-mC) in the genome. Accumulating evidence suggests that global levels of DNA methylation are lower in tumor tissue when compared with adjacent tissue, indicating a significant role of DNA hypomethylation in cancer progression.3-6
There are two main mechanisms through which changes in DNA methylation relate to cancer initiation and progression.7 One is the silencing of expression of tumor suppressor genes by increased methylation of their promoter regions. The other one is general hypomethylation of the genome that leads to genomic instability by activation of transposable elements, viral sequences and genes. Genomic instability is an established hallmark of cancer progression and is characterized by increased mutation rates due to specific events such as the mutation of important gatekeeper genes and genome-wide events such as large chromosomal rearrangements.8
Increasingly, epidemiologic studies are measuring global levels of DNA methylation in peripheral tissue, such as blood. The literature is constantly evolving, but the results of many early studies suggest that lower global DNA methylation in white blood cells (WBC) is associated with many different cancers including colorectal, bladder, gastric, head and neck, lung and breast cancer.9 Most epidemiologic studies of global methylation have focused on measuring overall 5-mC content or DNA methylation levels in retrotransposable elements such as LINE-1 and Alu.10-17 Some of these early studies considered LINE-1 and Alu as surrogate epigenetic measures of global DNA methylation levels because these elements are the most abundant retrotransposons in the human genome and their silencing is regulated by epigenetic mechanisms.18 However, further research has indicated that DNA methylation levels at these sequences are not correlated with global 5-mC content11,19 and that these elements may be differentially associated with disease outcomes and lifestyle factors.9,20 There are currently several different methods used to measure global DNA methylation in epidemiologic studies, and new assays for large samples are under development.9,20 To be relevant for human population studies, the specified assay needs to yield consistent results and be high-throughput in addition to providing an accurate measurement of genomic 5-mC content. Based on these criteria, it is not clear which of the available assays are most appropriate for population research.
For this study, we selected two different measures of global DNA methylation, the overall incorporation of labeled methyl groups by the [3H]-methyl acceptance assay and the use of methylation specific enzymes to differentiate between methylated and non-methylated DNA by the luminometric methylation assay (LUMA).21-23 Both assays have been used in studies investigating associations of global DNA methylation and environmental exposures or disease outcomes.24-32 These assays differ by targeted genomic regions and have different methodological advantages and limitations. The [3H]-methyl acceptance assay is variable and requires much time and sample manipulation; however, it covers the whole genome. The LUMA assay is a surrogate measure of overall 5-mC content. It provides information on DNA methylation at 5′-CCGG-3′ sequences throughout the genome. Although the coverage represents a small fraction of all possible CpG sites, the assay is easy to perform, which makes it amenable to be used in epidemiologic studies. Here, we applied both methodologies to investigate the association between biomarkers of DNA methylation and the risk of breast cancer using a sibling-based design in women participating in the New York site of the Breast Cancer Family Registry (BCFR). This is a study population of high-risk breast cancer families, a large portion of which have no known genetic risk mutations in BRCA1 or BRCA2.
Results
We report the overall demographic characteristics of the study participants in Table 1. Measurements of DNA methylation levels by the [3H]-methyl acceptance assay showed that affected sisters had a mean DPM level that was higher than unaffected sisters by 9081 DPM/μg, indicating lower overall levels of DNA methylation. LUMA DNA methylation levels were similar between affected and unaffected sisters, 67.1% vs. 67.5% respectively.
Table 1. Demographics, mutation status and DNA methylation levels of the study population, New York site of the BCFR.
| Affected Sisters |
Unaffected Sisters |
|||
|---|---|---|---|---|
| n = 273 | n = 335 | |||
| |
N |
Mean (SD) or % |
N |
Mean (SD) or % |
|
Age |
|
|
|
|
| Continuous |
273 |
49.6 (11.4) |
335 |
48.2 (11.2) |
| Race‡ |
|
|
|
|
| White |
169 |
62.6% |
189 |
56.4% |
| Hispanic |
67 |
24.8% |
102 |
30.4% |
| Other |
34 |
12.6% |
44 |
13.1% |
| Smoking status |
|
|
|
|
| Current |
15 |
5.5% |
41 |
12.2% |
| Former |
96 |
35.2% |
101 |
30.1% |
| Never |
162 |
59.3% |
193 |
57.6% |
|
BRCA1 Mutation |
|
|
|
|
| Positive |
22 |
8.1% |
13 |
3.9% |
| Negative |
251 |
91.9% |
322 |
96.1% |
|
BRCA2 Mutation |
|
|
|
|
| Positive |
12 |
4.4% |
9 |
2.7% |
| Negative |
261 |
95.6% |
326 |
97.3% |
| [3H]-methyl acceptance assay (DPM/μg) |
|
|
|
|
| Continuous |
233 |
97111 (76348) |
295 |
88030 (70841) |
| LUMA (% DNA meth) |
|
|
|
|
| Continuous | 263 | 67.1 (7.6) | 321 | 67.5 (7.3) |
Race information was not available for three unaffected subjects
We observed modest correlations between sisters in global DNA methylation levels for both assays (range -0.08–0.24) (Table 2). For the [3H]-methyl acceptance assay, we observed statistically significant Spearman Correlation Coefficients for WBC and pooled sources of DNA (0.24, p = 0.006; 0.19, p = 0.006, respectively), but not for granulocytes (-0.08, p = 0.51). For the LUMA assay, we observed statistically significant Spearman Correlation Coefficients for the pooled source (0.16, p = 0.01).
Table 2. Correlation of the global DNA methylation measures between sisters discordant for breast cancer by blood cell type, New York site of the BCFR.
| Granulocytes | Total White Blood Cells | Pooled Source | |||||||
|---|---|---|---|---|---|---|---|---|---|
| |
N |
r |
p value |
N |
r |
p value |
N |
r |
p value |
|
[3H]-methyl acceptance assay (DPM/μg) |
64 |
-0.08 |
0.51 |
134 |
0.24 |
0.006 |
198 |
0.19 |
0.006 |
| LUMA (% DNA methylation) | 80 | 0.21 | 0.06 | 155 | 0.13 | 0.10 | 235 | 0.16 | 0.01 |
Table 3 reports the associations between DNA methylation and breast cancer (by assay and by quartile and continuous measures). As measured by the [3H]-methyl acceptance assay, women who had lower DNA methylation had higher odds of breast cancer than women who had higher levels of methylation (OR = 1.81, 95%CI = 0.99–3.32 for the quartile with the lowest level of methylation compared with the highest for the pooled sources). We also found an elevated, but not statistically significant, association with breast cancer for lower DNA methylation in granulocytes (OR = 2.42, 95%CI = 0.86–6.83) and WBC (OR = 1.51, 95%CI = 0.68–3.38). When we examined DPM/μg as a continuous variable, we observed a 50% increased relative association with increasing DPM/μg in total WBC DNA (OR per 1 unit increase in the natural logarithm of DPM/μg = 1.49, 95%CI = 1.03–2.16). When we pooled the two sources of DNA, a 1 unit increase in the log of DPM/μg was associated with a 34% increase in the relative association with breast cancer (95%CI = 1.03–1.74).
Table 3. Conditional Logistic Regression analysis of global DNA methylation biomarkers and breast cancer risk (pooled and by source), New York site of the BCFR.
| DNA Methylation | Odds Ratios and 95% Confidence Intervals for Overall Pooled sample and Stratified by Sourcea |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
[3H]-methyl acceptance assayb |
|
Pooled (n = 458) |
|
Granulocytes (n = 156) |
|
Total WBC (n = 302) |
||||
| OR |
95% CI |
OR |
95% CI |
OR |
95% CI |
|||||
| Quartile (DPM/μg DNA) |
Q4 (> 115215); lower overall methylation |
1.81 |
0.99–3.32 |
2.42 |
0.86–6.83 |
1.51 |
0.68–3.38 |
|||
| Q3 (68940 – 115215) |
1.81 |
1.02–3.20 |
1.34 |
0.46–3.87 |
1.76 |
0.84–3.67 |
||||
| Q2 (41692 – 68940) |
0.82 |
0.46–1.46 |
0.91 |
0.42–2.00 |
0.67 |
0.28–1.61 |
||||
| Q1 (≤ 41692); higher overall methylation |
1 |
|
1 |
|
1 |
|
||||
| Continuous (log DPM/μg DNA) |
1 unit change, less methylation |
1.34 |
1.03–1.74 |
1.18 |
0.82–1.70 |
1.49 |
1.03–2.16 |
|||
| | ||||||||||
| LUMAc | |
Pooled (n = 551) |
Granulocytes (n = 201) |
Total WBC (n = 350) |
||||||
| OR |
95% CI |
OR |
95% CI |
OR |
95% CI |
|||||
| Quartile (DNA meth %) |
Q1 (≤ 62.9), lower overall methylation |
0.92 |
0.53–1.59 |
1.18 |
0.43–3.24 |
0.8 |
0.41–1.55 |
|||
| Q2 (62.9 - 68.0) |
0.71 |
0.40–1.24 |
1.12 |
0.39–3.21 |
0.58 |
0.29–1.14 |
||||
| Q3 (68.0 - 73.0) |
1.26 |
0.75–2.12 |
1.9 |
0.79–4.55 |
0.98 |
0.51–1.91 |
||||
| Q4 (> 73.0); higher overall methylation |
1 |
|
1 |
|
1 |
|
||||
| Continuous | 1 unit change, less methylation | 1.00 | 0.97–1.02 | 1.02 | 0.97–1.07 | 0.99 | 0.96–1.02 | |||
a OR = odds ratio, 95% CI = 95% confidence intervals; adjusted for age at blood draw and smoking status. bHigher values on assay mean lower DNA methylation. cLower values on assay mean lower DNA methylation
Table 3 also reports our results for the LUMA assay. As Table 3 summarizes, we did not find any consistent pattern with the levels of DNA methylation measured by LUMA in granulocytes, total WBC or when both sources are pooled together.
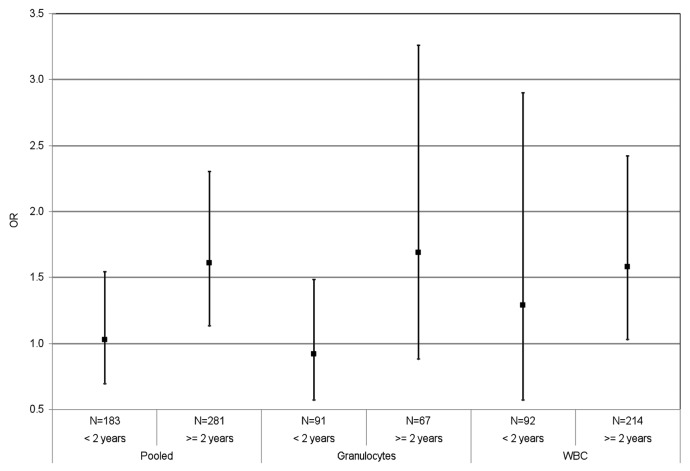
Figure 1 shows the results stratified by time since diagnosis for the [3H]-methyl acceptance assay. There was no overall association between DNA methylation level and breast cancer for those individuals diagnosed within two years of sample collection (OR = 1.29 for unit change in the log of DPM/μg, 95%CI = 0.57–2.90 for WBC; OR = 0.92, 95%CI = 0.57–1.48 for granulocytes; OR = 1.03, 95%CI = 0.69–1.54 for pooled sources). There were positive associations with breast cancer in the subgroup of sisters whose blood was collected more than two years after diagnosis (OR = 1.58 for unit change in the log of DPM/μg, 95%CI = 1.03–2.42 for WBC; and OR = 1.61, 95%CI = 1.13–2.30 for pooled sources).
Figure 1. Global DNA methylation measured by [3H]-methyl acceptance assay and breast cancer risk (pooled and by source), for specimens collected less than two years and more than two years after breast cancer diagnosis in the New York site of the BCFR. The OR is per unit change in log DPM/μg DNA
Discussion
We used two different global methylation assays that do not represent specific coding genomic regions to investigate directly the association between global DNA methylation levels and breast cancer in a high-risk population. We found that sisters with breast cancer had lower levels of DNA methylation measured by the [3H]-methyl acceptance assay than their unaffected sisters. A lower level of global DNA methylation might indicate increased genomic instability, which in turn could lead to increased cancer susceptibility. Similar results have been observed in previous studies of WBC with lower levels of global DNA methylation and at repetitive sequences observed in cancer cases compared with controls.10-17 Similarly, in a prospective study, Zhu et al. showed that lower DNA methylation of the repetitive element LINE-1 was indicative of increased overall cancer incidence and that lower DNA methylation levels at LINE-1 and Alu were related to increased cancer mortality.33 Our finding is consistent with the other study of breast cancer and DNA methylation levels, which found that unrelated cases had lower WBC DNA 5-mC content than controls.11
We observed some differences across DNA source for [3H]-methyl acceptance assay associations with breast cancer. However, both DNA sources, granulocytes and total WBC, had larger estimates for the higher quartiles, indicating stronger associations with lower overall global DNA methylation. We have previously reported differences in overall levels of global DNA methylation across cell type suggesting that association with disease may be assay- and cell type-specific.19 Epigenetic marks are tissue specific;34 however, granulocytes are a major fraction of WBC. It is not completely understood how much each blood cell type contributes to the epigenetic profile of WBC. Further work and consideration to cell type will be important in interpreting work from different epidemiologic studies.
We did not observe any consistent findings with the LUMA assay and breast cancer. The different results obtained by the two assays highlight the importance of considering coverage of the whole genome in determining relevant associations of DNA methylation level and breast carcinogenesis. The assays used in this study target different parts of the genome; we only found associations with the assay that has full genomic coverage, the [3H]-methyl acceptance assay. Previous results showed that measures from the same source of [3H]-methyl acceptance assay and LUMA do not significantly correlate for the same individual, supporting the conclusion that they are identifying different sets of genomic information.19
All the assays used to date to determine global DNA methylation levels have limitations when applied to epidemiologic studies. The main limitations of the [3H]-methyl acceptance assay are related to the reagents used, S-adenosyl methionine (SAM) and the bacterial methyltransferase SssI. The instability of SAM and the variable enzymatic activity of the SssI enzyme can affect the efficiency of the reaction. Additionally, this assay is very dependent on good genomic DNA quantification, which is hard to carry out in an easy and consistent manner.35 However, despite these limitations, our results suggest that overall coverage of the genome is important in understanding associations between global DNA methylation levels and cancer.
We have also observed a significant association between a decrease in WBC DNA methylation of the non-tandem repeat Sat2 and breast cancer.43 This finding and the results of the current study, might suggest that decreased global DNA methylation represents changes in Sat2 DNA methylation levels. Repeat sequence or element DNA hypomethylation might account largely for the contribution of the activation of these elements to overall genomic instability. Studies investigating surrogate markers of global DNA methylation, such as levels in sequences of transposon-derived elements that are abundantly distributed throughout the genome, have also found associations between cancer and lower levels of DNA methylation.10,12,13,17 We found no significant correlation between DNA methylation levels of the tandem repeat Sat2 and the markers measured here. The Spearman correlation coefficients for Sat2 and [3H]-methyl acceptance DNA methylation levels, and LUMA were -0.03 and 0.00, respectively. These results suggest that the decrease in WBC DNA methylation observed here is the result of overall genomic levels and not of specific genomic sequences such as repeated elements. Our findings indicate that in breast carcinogenesis overall genomic demethylation might increase disease susceptibility by alternative mechanisms that do not only include Sat2 tandem repeat activation. We did not assess genomic instability directly; therefore, future studies should characterize the separate contributions of Sat2 and global DNA methylation to breast cancer.
Our study specifically focuses on differences within families using sister sets from a registry of high-risk breast cancer families, a small percentage of which carry known genetic mutations in BRCA1 or BRCA2. Previous studies on global DNA methylation have focused on unrelated individuals. An additional strength is the sibling design, which allows us to examine these biomarkers within families and, through the design, control for fixed family-level effects that may confound the association between the markers and breast cancer. We were also able to use the same source of DNA within sisters so the source of DNA cannot explain our within-family findings.
The main limitation of our study is that the bloods were collected after diagnosis. Most epidemiologic studies of cancer and DNA methylation have also used this retrospective approach. Although our findings need to be replicated in prospective studies, we did observe similar elevated associations for the [3H]-methyl acceptance assay in WBC regardless of years since diagnosis. The results were stronger, and statistically significant, for those individuals at least two years post diagnosis, which suggests that the effect observed in our study is present after treatment has ended. Nevertheless, our study cannot address the effect current disease might have on DNA methylation levels in peripheral tissue.
Although the detailed mechanisms through which decreases in peripheral tissue global DNA methylation leads to increased cancer risk are not completely understood, possible mechanisms might include an increase in genomic instability resulting from lower global DNA methylation levels. Our study adds to the growing evidence that DNA methylation measured in peripheral blood may be an important biomarker even in high-risk families. Prospective studies will have to be performed to assess the temporality of this association and to better understand the role of epigenetic markers in blood and breast cancer risk.
Materials and Methods
Study participants and specimens
The New York site of the BCFR recruited high-risk breast and/or ovarian cancer families from clinical and community settings within the metropolitan New York area (for details see refs. 36-41). All family members who participated in the Registry completed an epidemiologic questionnaire to provide information on demographics, ethnicity, smoking, alcohol consumption, reproductive history, hormone use, weight, height and physical activity and a self-administered food frequency questionnaire. We collected blood from participants at the time of recruitment to permit the isolation of plasma and WBC fractions. The study was approved by Columbia University’s Institutional Review Board.
For this study, we selected female-only sibling sets including at least one sister affected with breast cancer (n = 273) and her unaffected sister(s) (n = 335) from whom blood specimens were available. For a subset of sibling sets, we isolated granulocytes using a Ficoll-density gradient, while for the remaining participants we collected the total white blood cells by simple centrifugation. DNA was extracted from either source depending on which blood cell type was available for each subject using the salting-out method; concentration and quality was determined by absorbance at 260 nm and 280 nm using Nanodrop technology. We conducted all laboratory assays blinded to case status and epidemiological data.
[3H]-methyl acceptance assay
We used the [3H]-methyl acceptance assay as described by Balaghi and Wagner21 and Pilsner et al.28 In this assay, the DNA is incubated with [3H] S-adenosyl methionine in the presence of the SssI prokaryotic methylase enzyme, which indiscriminately methylates all unmethylated CpG sequences. Therefore, in this assay the ability to incorporate [3H] methyl groups in vitro is inversely related to endogenous DNA methylation. Briefly, 200 ng of DNA were incubated with 3U of SssI methylase (New England Biolabs); 3.8 µmol/L (1.1 µCi) [3H]-labeled S-adenosyl methionine (Perklin-Elmer); and EDTA, DTT, and Tris-HCL (pH 8.2) in a 30 µL mixture for 1 h at 37°C. The reaction was terminated on ice and 15 µL of the reaction mixture were applied onto a Whatman DE81 filter paper. The filter was washed on a vacuum filtration apparatus three times with 5 mL of 0.5 mol/L sodium phosphate buffer (pH 8.0), followed by 2 mL each of 70% and 100% ethanol. Dried filters were each placed in a vial with 5 mL of scintillation fluid (Scintisafe, Fisher), analyzed by a Packard scintillation counter and a measurement of disintegrations per minute (DPM) was obtained for each sample. Samples were processed in duplicate. Universal methylated and unmethylated DNA was used as negative and positive controls. DNA was quantified using PicoGreen double-strand DNA quantification reagent (Molecular Probes). Results of this assay are expressed as DPM per μg of DNA. A pooled sample of DNA from 5 controls was used as a quality control and analyzed with each plate of test samples to control for batch effects. Intra- and inter-assay CVs were 2.5% and 5.9%, respectively.
Luminometric methylation assay (LUMA)
The luminometric methylation assay was initially developed by Karimi et al.22,23 The assay is based on the ability of two isoschizomers to digest sequences differentially depending on the methylation status of the CpG site contained within the sequence. Isoschizomers MspI and HpaII target the same sequence, 5′-CCGG-3′; however, MspI will digest all CCGG sites and HpaII digestions will be blocked by the presence of a 5-mC in the second position. Samples are digested with one isoschizomer and EcoRI. EcoRI provides the reference for the DNA amount. Pyrosequencing is used to sequence the overhangs left by both enzymes. We ran a modified version of the original assay as reported.29 In brief, 200 ng of genomic DNA were cleaved at 37°C overnight with HpaII/EcoRI or MspI/EcoRI in two separate 10 μl reactions containing 33 mM TRIS-acetate, 10 mM Mg-acetate, 66 mM K-acetate pH 7.9, 0.1 mg/ml BSA and 5 units of each restriction enzyme. Ten μl of annealing buffer (20 mM TRIS-acetate, 2 mM Mg-acetate pH 7.6) were mixed with the digested samples and placed in a PyroMark Q24 system. The dispensation order used for sequencing was: GTGTCACAGTGT. For calculations, the peak heights of dispensations 9 through 12 were used. Samples with peaks lower than 3, the cut-off value for DNA quality, were discarded. Samples with values higher than 3 in the first two dispensations were indicative of low DNA quality, possibly DNA degradation, and were also discarded. Percentage of DNA methylation was expressed as [1-(HpaII/EcoRIΣG/ΣT)/ (MspI/ EcoRIΣG/ΣT)]*100. DNA extracted from a lymphoblastoid cell line established in our laboratory was run in every plate to assess batch and plate effects. The inter- and intra-assay CVs were 1.5% and 4.6%, respectively.
Statistical analysis
We used two measures of global DNA methylation, the [3H]-methyl acceptance assay, for which results are given in DPM/μg, and the LUMA assay, for which results are reported as percent DNA methylation; higher values from the [3H]-methyl acceptance assay in DPM/μg indicate lower global DNA methylation levels. We categorized each DNA methylation marker by quartiles based on the distribution of the marker in the unaffected sisters. We also modeled DNA methylation as a continuous variable. We have previously reported differences in DNA methylation levels by source of WBC DNA,19 so we performed all statistical analyses stratified by source of DNA (granulocytes vs. total WBC) as well as pooled.
We examined the correlation of each marker between sisters discordant on breast cancer status using Spearman’s correlation coefficient. We used conditional logistic regression to estimate associations between DNA methylation and breast cancer within sister sets; by design, all fixed-level family effects are eliminated by conditioning on family set number. In addition, we used generalized estimating equations (GEE) to estimate the overall average association between the markers and breast cancer.42 Both conditional logistic and GEE approaches led to similar inferences, so we present only the former.
We modeled the association adjusted for age, and then assessed confounding of the association for both markers. Potential confounders included age at blood draw, BMI (< 25 kg/m2 vs ≥ 25 kg/m2) and smoking status. We also evaluated the impact of further adjustment by other breast cancer risk factors including age at menarche, age at first pregnancy, oral contraceptive use, hormone replacement therapy use, alcohol intake, BRCA1/2 mutation status and menopausal status. We assessed confounding based on whether inclusion of the variable was associated with at least a 10% change in the association between DNA methylation and breast cancer compared with the age-adjusted model, or whether inclusion of the variable changed the statistical significance of the association between DNA methylation and breast cancer risk. Only age and smoking status empirically confounded our estimates so these were the only variables we kept in the final models. Because blood samples were taken after breast cancer diagnosis, we stratified our final models by whether the blood sample was drawn within 2 y of diagnosis or more than 2 y after diagnosis. We performed all analyses using SAS software 9.2 (SAS Institute). All statistical tests were based on 95% confidence limits.
Acknowledgments
This work was supported by an award from the Breast Cancer Research Foundation and National Institute of Health grants U01 CA69398, P30 CA13696, P30 ES009089 and K07 CA131094. This work was also supported by the National Cancer Institute, National Institutes of Health under RFA # CA-06–503 and through cooperative agreements with members of the Breast Cancer Family Registry (BCFR) and Principle Investigators, including Columbia University (U01 CA69398). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the BCFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the BCFR.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/20830
Reference List
- 1.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–9. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr. 2009;21:243–51. doi: 10.1097/MOP.0b013e32832925cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alves G, Tatro A, Fanning T. Differential methylation of human LINE-1 retrotransposons in malignant cells. Gene. 1996;176:39–44. doi: 10.1016/0378-1119(96)00205-3. [DOI] [PubMed] [Google Scholar]
- 4.Florl AR, Löwer R, Schmitz-Dräger BJ, Schulz WA. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br J Cancer. 1999;80:1312–21. doi: 10.1038/sj.bjc.6690524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lapeyre JN, Becker FF. 5-Methylcytosine content of nuclear DNA during chemical hepatocarcinogenesis and in carcinomas which result. Biochem Biophys Res Commun. 1979;87:698–705. doi: 10.1016/0006-291X(79)92015-1. [DOI] [PubMed] [Google Scholar]
- 6.Lu LJ, Randerath E, Randerath K. DNA hypomethylation in Morris hepatomas. Cancer Lett. 1983;19:231–9. doi: 10.1016/0304-3835(83)90159-3. [DOI] [PubMed] [Google Scholar]
- 7.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 8.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 9.Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics. 2011;6:828–37. doi: 10.4161/epi.6.7.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cash HL, Tao L, Yuan JM, Marsit CJ, Houseman EA, Xiang YB, et al. LINE-1 hypomethylation is associated with bladder cancer risk among nonsmoking Chinese. Int J Cancer. 2012;130:1151–9. doi: 10.1002/ijc.26098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi JY, James SR, Link PA, McCann SE, Hong CC, Davis W, et al. Association between global DNA hypomethylation in leukocytes and risk of breast cancer. Carcinogenesis. 2009;30:1889–97. doi: 10.1093/carcin/bgp143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hou L, Wang H, Sartori S, Gawron A, Lissowska J, Bollati V, et al. Blood leukocyte DNA hypomethylation and gastric cancer risk in a high-risk Polish population. Int J Cancer. 2010;127:1866–74. doi: 10.1002/ijc.25190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–14. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 14.Lim U, Flood A, Choi SW, Albanes D, Cross AJ, Schatzkin A, et al. Genomic methylation of leukocyte DNA in relation to colorectal adenoma among asymptomatic women. Gastroenterology. 2008;134:47–55. doi: 10.1053/j.gastro.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore LE, Pfeiffer RM, Poscablo C, Real FX, Kogevinas M, Silverman D, et al. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case-control study. Lancet Oncol. 2008;9:359–66. doi: 10.1016/S1470-2045(08)70038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pufulete M, Al-Ghnaniem R, Leather AJ, Appleby P, Gout S, Terry C, et al. Folate status, genomic DNA hypomethylation, and risk of colorectal adenoma and cancer: a case control study. Gastroenterology. 2003;124:1240–8. doi: 10.1016/S0016-5085(03)00279-8. [DOI] [PubMed] [Google Scholar]
- 17.Wilhelm CS, Kelsey KT, Butler R, Plaza S, Gagne L, Zens MS, et al. Implications of LINE1 methylation for bladder cancer risk in women. Clin Cancer Res. 2010;16:1682–9. doi: 10.1158/1078-0432.CCR-09-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–40. doi: 10.1016/S0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 19.Wu HC, Delgado-Cruzata L, Flom JD, Kappil M, Ferris JS, Liao Y, et al. Global methylation profiles in DNA from different blood cell types. Epigenetics. 2011;6:76–85. doi: 10.4161/epi.6.1.13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson HH, Marsit CJ, Kelsey KT. Global methylation in exposure biology and translational medical science. Environ Health Perspect. 2011;119:1528–33. doi: 10.1289/ehp.1103423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balaghi M, Wagner C. DNA methylation in folate deficiency: use of CpG methylase. Biochem Biophys Res Commun. 1993;193:1184–90. doi: 10.1006/bbrc.1993.1750. [DOI] [PubMed] [Google Scholar]
- 22.Karimi M, Johansson S, Ekström TJ. Using LUMA: a Luminometric-based assay for global DNA-methylation. Epigenetics. 2006;1:45–8. doi: 10.4161/epi.1.1.2587. [DOI] [PubMed] [Google Scholar]
- 23.Karimi M, Johansson S, Stach D, Corcoran M, Grandér D, Schalling M, et al. LUMA (LUminometric Methylation Assay)--a high throughput method to the analysis of genomic DNA methylation. Exp Cell Res. 2006;312:1989–95. doi: 10.1016/j.yexcr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Basten GP, Duthie SJ, Pirie L, Vaughan N, Hill MH, Powers HJ. Sensitivity of markers of DNA stability and DNA repair activity to folate supplementation in healthy volunteers. Br J Cancer. 2006;94:1942–7. doi: 10.1038/sj.bjc.6603197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fenech M, Aitken C, Rinaldi J. Folate, vitamin B12, homocysteine status and DNA damage in young Australian adults. Carcinogenesis. 1998;19:1163–71. doi: 10.1093/carcin/19.7.1163. [DOI] [PubMed] [Google Scholar]
- 26.Shelnutt KP, Kauwell GP, Gregory JF, 3rd, Maneval DR, Quinlivan EP, Theriaque DW, et al. Methylenetetrahydrofolate reductase 677C-->T polymorphism affects DNA methylation in response to controlled folate intake in young women. J Nutr Biochem. 2004;15:554–60. doi: 10.1016/j.jnutbio.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Pilsner JR, Hu H, Ettinger A, Sánchez BN, Wright RO, Cantonwine D, et al. Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect. 2009;117:1466–71. doi: 10.1289/ehp.0800497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pilsner JR, Liu X, Ahsan H, Ilievski V, Slavkovich V, Levy D, et al. Genomic methylation of peripheral blood leukocyte DNA: influences of arsenic and folate in Bangladeshi adults. Am J Clin Nutr. 2007;86:1179–86. doi: 10.1093/ajcn/86.4.1179. [DOI] [PubMed] [Google Scholar]
- 29.Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299:2877–83. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deneberg S, Grövdal M, Karimi M, Jansson M, Nahi H, Corbacioglu A, et al. Gene-specific and global methylation patterns predict outcome in patients with acute myeloid leukemia. Leukemia. 2010;24:932–41. doi: 10.1038/leu.2010.41. [DOI] [PubMed] [Google Scholar]
- 31.Lee JJ, Geli J, Larsson C, Wallin G, Karimi M, Zedenius J, et al. Gene-specific promoter hypermethylation without global hypomethylation in follicular thyroid cancer. Int J Oncol. 2008;33:861–9. [PubMed] [Google Scholar]
- 32.Römermann D, Hasemeier B, Metzig K, Schlegelberger B, Länger F, Kreipe H, et al. [Methylation status of LINE-1 sequences in patients with MDS or secondary AML] Verh Dtsch Ges Pathol. 2007;91:338–42. [PubMed] [Google Scholar]
- 33.Zhu ZZ, Sparrow D, Hou L, Tarantini L, Bollati V, Litonjua AA, et al. Repetitive element hypomethylation in blood leukocyte DNA and cancer incidence, prevalence, and mortality in elderly individuals: the Normative Aging Study. Cancer Causes Control. 2011;22:437–47. doi: 10.1007/s10552-010-9715-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oakeley EJ. DNA methylation analysis: a review of current methodologies. Pharmacol Ther. 1999;84:389–400. doi: 10.1016/S0163-7258(99)00043-1. [DOI] [PubMed] [Google Scholar]
- 36.John EM, Hopper JL, Beck JC, Knight JA, Neuhausen SL, Senie RT, et al. Breast Cancer Family Registry The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res. 2004;6:R375–89. doi: 10.1186/bcr801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zipprich J, Terry MB, Brandt-Rauf P, Freyer GA, Liao Y, Agrawal M, et al. XRCC1 polymorphisms and breast cancer risk from the New York Site of the Breast Cancer Family Registry: A family-based case-control study. J Carcinog. 2010;9:4. doi: 10.4103/1477-3163.62535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zipprich J, Terry MB, Liao Y, Agrawal M, Gurvich I, Senie R, et al. Plasma protein carbonyls and breast cancer risk in sisters discordant for breast cancer from the New York site of the Breast Cancer Family Registry. Cancer Res. 2009;69:2966–72. doi: 10.1158/0008-5472.CAN-08-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Machella N, Terry MB, Zipprich J, Gurvich I, Liao Y, Senie RT, et al. Double-strand breaks repair in lymphoblastoid cell lines from sisters discordant for breast cancer from the New York site of the BCFR. Carcinogenesis. 2008;29:1367–72. doi: 10.1093/carcin/bgn140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennedy DO, Agrawal M, Shen J, Terry MB, Zhang FF, Senie RT, et al. DNA repair capacity of lymphoblastoid cell lines from sisters discordant for breast cancer. J Natl Cancer Inst. 2005;97:127–32. doi: 10.1093/jnci/dji013. [DOI] [PubMed] [Google Scholar]
- 41.Shen J, Desai M, Agrawal M, Kennedy DO, Senie RT, Santella RM, et al. Polymorphisms in nucleotide excision repair genes and DNA repair capacity phenotype in sisters discordant for breast cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:1614–9. doi: 10.1158/1055-9965.EPI-06-0218. [DOI] [PubMed] [Google Scholar]
- 42.Zeger SL, Liang KY, Albert PS. Models for longitudinal data: a generalized estimating equation approach. Biometrics. 1988;44:1049–60. doi: 10.2307/2531734. [DOI] [PubMed] [Google Scholar]
- 43.Wu HC, Delgado-Cruzata L, Flom JD, Perrin M, Liao Y, Ferris J, et al. Repetitive element SDNA methylation levels in white blood cell DNA from sisters discordant for breast cancer from the New York site of the BCFR. Carcinogenesis. 2012 doi: 10.1093/carcin/bgs201. [DOI] [PMC free article] [PubMed] [Google Scholar]