Abstract
Francisella tularensis is a highly pathogenic gram negative bacterium that infects multiple sites in a host, including the skin and the respiratory tract, which can lead to the onset of a deadly disease with a 50% mortality rate. The live vaccine strain (LVS) of F. tularensis, while attenuated in humans but still virulent in mice, is not an option for vaccine use in the United States due to safety concerns, and currently no FDA approved vaccine exists. The purpose of the present work was to assess the ability of recombinant Francisella outer membrane protein A (FopA) to induce a protective response in mice. The gene encoding FopA from F. tularensis LVS was cloned and expressed in E. coli. The resulting recombinant protein was affinity-purified from the E. coli outer membrane, incorporated into liposomes and administered to mice via multiple routes. FopA-immunized mice produced FopA-specific antibodies and were protected against both lethal intradermal and intranasal challenges with F. tularensis LVS. The vaccinated mice had reduced bacterial numbers in their lungs, livers and spleens during infection, and complete bacterial clearance was observed by day 28 post infection. Passive transfer of FopA-immune serum protected naïve mice against lethal F. tularensis LVS challenge, showing that humoral immunity played an important role in vaccine efficacy. FopA-immunization was unable to protect against challenge with the fully virulent Schu S4 strain of F. tularensis, however, the findings demonstrate proof of principle that an immune response generated against a component of a subunit vaccine is protective against lethal respiratory and intradermal tularemia.
1. Introduction
Tularemia is caused by the highly infectious gram-negative bacterium Francisella tularensis. The disease can take many forms depending upon the route of infection, the most common being the ulceroglandular form that is usually acquired through a skin bite from an infected arthropod. While the pneumonic form of tularemia is less common, inhalation of ≤10 colony forming units (CFU) of a fully virulent strain of F. tularensis can cause disease with a 50% mortality rate. Due to its extreme infectivity, ease of dissemination, and substantial capacity to cause illness and death, F. tularensis is classified as a Category A biothreat agent [1-4].
Currently, there is no vaccine against F. tularensis that is approved for use in humans [2-7]. The attenuated live vaccine strain (LVS) of F. tularensis can provide protection against subsequent challenge with a fully virulent Type A strain; however, the protection is incomplete, and since LVS is a live vaccine it has some level of reactogenicity [3, 4, 6, 7]. Attenuated mutants of F. tularensis LVS have also been generated and found to provide some protection in mice against challenge with the fully virulent SchuS4 strain of F. tularensis. However, subunit vaccines prepared from specific proteins isolated from the pathogen may be a safer and more efficacious approach [8-10]. Nevertheless, there has been limited progress in developing such a vaccine for tularemia [3, 4, 6, 7].
Francisella outer membrane protein A (FopA) is a logical candidate vaccine target since it is expressed on the bacterial surface in abundance and would presumably be accessible to antibodies [11-14]. Indeed, FopA-specific antibodies are commonly found in the sera of convalescent patients following tularemia, indicating a high level of immunogenicity [15]. Savitt et al. [16] demonstrated that FopA-specific mAbs can provide partial protection against disease, lending credence to the concept that FopA may serve as a protective vaccine candidate.
In order to test the efficacy of FopA as a subunit vaccine against F. tularensis, recombinant F. tularensis FopA was expressed in E. coli, affinity purified, and incorporated into liposomes to ensure retention of the native structure. The liposomal recombinant protein was then administered to mice in conjunction with interleukin 12 (IL-12) and Alum as adjuvants. The results suggest that FopA is a viable vaccine candidate for antibody-mediated protection against lethal tularemia.
2. Materials and Methods
2.1 Mice
5-8 week old C57BL/6 and BALB/c mice were purchased from Charles River Laboratories (Wilmington, MA) through a contract with the National Cancer Institute or from Taconic (Rensselaer, NY). The mice were housed at Albany Medical College and all experimental procedures were approved by the institutional animal care and use committee.
2.2 Bacteria
F. tularensis LVS and LVS FopA∷Tn5 that lacks FopA expression were grown in Mueller Hinton Broth as previously described [17, 18]. E. coli BL21(DE3)omp8 cells containing the pET1739:FopA plasmid were grown in Luria broth (LB) containing 100μg/ml ampicillin [19].
2.3. Cloning, expression, and purification of recombinant FopA
The FopA-encoding sequence from F. tularensis LVS was amplified by PCR using the following primers: GGTACCGCAGGTTCAGATAATATCGATACGTTAGC (FopA sense primer tailed with a Kpn1 restriction site) and CTCGAGCTATTAGTTAGCTTCTTTAAGTGGAGCTGATACG (FopA antisense primer tailed with an additional stop codon and an Xho restriction site). The amplified PCR fragment was cloned using the TOPO© TA 2.1 vector and One Shot© chemically competent E. coli (Invitrogen; Carlsbad CA). The cloned TOPO:FopA plasmid was isolated from transformed E. coli using the Purelink HiPure Plasmid DNA MidiPrep kit (Invitrogen) from which the 1.1 kb FopA coding sequence was liberated using Kpn1 and Xho1 restriction enzymes. The digested DNA was then cloned into the pET1739 expression vector, a modified pET17b vector which contains the multiple cloning sites of pET39b (including the dsbA gene) (Novagen/EMD4Biosciences, San Diego, CA) and the ampicillin resistance gene of pET17b (Novagen), using the Kpn1 and Xho1 restriction sites. The FopA encoding sequence was inserted 3’ and in frame with the E. coli dsbA coding sequence which is under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter. pET1739:FopA was then used to transform E. coli BL21(DE3)omp8 (omp8) cells which were first cultured in LB supplemented with ampicillin (100μg/ml) and then incubated in the presence of 0.2-0.5 mM IPTG for 2 hrs to induce expression of the recombinant His-tagged DsbA/FopA-fusion protein. E. coli BL21(DE3)omp8 was used because it contains several mutations in genes responsible for the expression of endogenous outer membrane proteins and thus, it is ideal for the expression of exogenous recombinant outer membrane proteins [19]. Once the fusion protein was sufficiently expressed, the bacteria were pelleted, resuspended in 50mM Tris/10mM NaCl, and treated with 0.3mg/ml lysozyme and 125 units of Benzonase Nuclease (EMD Chemicals, Gibbstown, NJ) for 12 hrs at 4°C to ensure total lysis of the bacteria. The lysate was then mixed with an equal volume of 10% Triton X-114 and was incubated at 30°C for 30 min. After centrifugation at 12,000g for 10 min at room temp, the detergent phase was collected. The remaining pellet was further fractionated by resuspension in 5% Sarkosyl in 50mM Tris/10mM NaCl and centrifuged at 12,000g for 10 min at room temp. The supernatant was collected and mixed with the detergent phase. The Sarkosyl/Triton-X 114 mixture was then diluted five-fold with 0.5% Elugent (EMD4Biosciences) in 50mM Tris/100 mM NaCl. One ml of 5 PRIME* PerfectPro* Ni-NTA Agarose (Fisher Scientific, Pittsburgh, PA) was added to the pooled factions and incubated at 4°C for 48 hrs. The Ni-NTA agarose beads were collected by passage through a cell strainer, washed twice with 0.5 % Elugent in 50mM Tris/100mM NaCl, and then washed two times with 1.0% n-Octyl-β-D-glucopyranoside (Fisher Scientific) in 50mM Tris/100mM NaCl, before resuspension in 1ml of 1% n-Octyl-β-D-glucopyranoside in 50mM Tris/100mM NaCl. The Ni-NTA agarose beads were incubated in the presence of 20U thrombin protease for 24 hrs at room temp to liberate the FopA-portion of the His-tagged fusion protein, and the Ni-NTA beads were then removed by passage through a cell strainer. The concentration of rFopA was determined using the BCA protein assay (Thermo Scientific, Waltham, MA).
2.4. Incorporation of FopA into liposomes
A synthetic blend of phospholipids, DOPE:DOPS:DOPC (5:3:2 w/w in chloroform), was purchased from Avanti-Polar Lipids (Alabaster, AL), allowed to dry for 12 hrs, and resuspended in PBS to a final concentration of 40mg/ml. A 10-20μl aliquot was then added to 100μl of 1% n-octyl-β-D-glucopyranoside (34mM) (Fisher Scientific) in 50mM Tris/100mM NaCl containing the cleaved rFopA to a final phospholipid:protein molar ratio of 250:1. The mixture was sonicated until it became translucent, and was then dried in a speed vacuum. The resulting pellet was resuspended in 100 μl sterile water and allowed to equilibrate at 4°C for at least 12 hrs. One ml of PBS was added to the mixture to dilute the n-octyl-β-D-glucopyranoside below a critical micelle concentration (~25mM), and the liposomes resulting from the aggregation of the phospholipid/protein complexes were pelleted by centrifugation at 24,000g for 30 min. The liposome pellet was washed with PBS and resuspended in 100μl of PBS. SDS-PAGE was used to confirm that rFopA was successfully incorporated into the liposomes.
2.5. ELISA
96-well Maxisorb ELISA plates (Nalge Nunc International, Rochester, NY) were coated with ~5 × 106 CFU/well of F. tularensis LVS or fopA∷Tn5 [17], or with 0.3μg of recombinant FopA protein in 100μl of 0.05 M carbonate buffer, pH 9.4, and incubated for overnight at 4°C. The plates were washed with PBS containing 0.05% Tween 20, and then blocked with 10% FCS in PBS at 37°C for at least 2 hrs. After washing, two-fold dilutions of FopA-immune sera, or sera collected from mice receiving a sham-immunization of empty liposomes were incubated in the wells for 2 hrs at 37°C. 100μl of HRP-conjugated goat-anti mouse Ig (Invitrogen) was then used together with TMB peroxidase substrate (BD Biosciences, San Jose, CA) to detect bound antibody at 450nm. The titers were calculated as the serum dilution yielding an OD value that was 50% of the maximum OD obtained in the assay.
2.6. Western blot analysis
Lysates of F. tularensis LVS and F. tularensis LVS fopA∷Tn5 were prepared by sonication of 2 × 109 bacteria/ml for 20 sec. Aliquots containing 3μg of total protein from each lysate were mixed with SDS sample buffer and were either boiled for 10 min or left at room temp, loaded into wells of a NUPAGE ® NOVEX 4-12% polyacrylamide gel (Invitrogen), and separated by SDS-PAGE. After transfer to PVDF membranes, the blots were incubated in 10% FCS/0.05% Tween 20 in PBS overnight at room temp. The whole cell lysates were probed with antisera prepared against recombinant FopA at a dilution of 1:10,000 in 10% FCS/0.05% Tween 20 in PBS. Following incubation for 1 hr at room temp, bound antibody was detected using HRP-conjugated goat-anti-mouse Ig at a dilution of 1:5000, and LUMI-Light PLUS substrate (Roche, Branchburg, NJ).
2.7. Animal immunization and challenge
Mice received priming doses of 10μg liposomal rFopA mixed with 2mg aluminum hydroxide and 0.25μg IL-12, each given subcutaneously and intraperitoneally (i.p.) (20μg total), followed by two intranasal (i.n.) booster inoculations of 20μg liposomal rFopA mixed with 0.5μg IL-12. Following a rest of at least 3 weeks, the mice were challenged intradermally (i.d.) or i.n. with a lethal doses of either F. tularensis LVS or F. tularensis Schu S4. As controls, mice were immunized with 108 UV-inactivated F. tularensis LVS, a sublethal dose of live F. tularensis LVS, or with empty liposomes (sham). All immunizations were performed under xylazine-ketamine anesthesia.
2.8. Bacterial burden analysis
FopA- and sham-immunized mice were challenged i.n. with a lethal dose of F. tularensis LVS and sacrificed on days 1, 5, and 7 and 28 post infection. The lungs, livers and spleens were collected and were individually homogenized in 700μl PBS. The LVS CFU were enumerated by incubating 10-fold dilutions on chocolate agar plates for 48 hrs at 37°C.
2.9. Passive serum transfer
250 μL of FopA-immune sera were injected i.p. into naïve mice 24 hrs before and 24 hrs after i.d. challenge with F. tularensis LVS. Control mice received either normal mouse serum or immune sera derived from mice sublethally infected with LVS.
2.10. Statistics
Statistical analysis was performed using Graph Pad 4 software (San Diego, CA). All survival data were analyzed by Kaplan-Meier Log Rank test. Bacterial burden data were transformed into log10 values, compared using the Students t-test, and expressed as the mean ± standard deviation. A P value less than 0.05 was considered to be statistically significant.
3. Results
3.1. Expression and purification of recombinant FopA
In order to ensure that rFopA was properly folded and retained its native conformation when administered as a vaccine, we expressed the recombinant protein in the E. coli outer membrane, which mimics it natural environment, and incorporated the final purified protein into liposomes in order to maintain rFopA in an outer-membrane-like environment.
Recombinant F. tularensis FopA fused to the His-tagged E. coli periplasmic protein Disulfide bond formation protein A (DsbA) was successfully expressed in E. coli omp8 cells and affinity purified from the Triton X-114 and Sarkosyl soluble fractions of bacterial lysates using Ni-NTA beads (Figs. 1A and 1B). The FopA portion of the fusion protein was isolated by thrombin cleavage and elution into n-Octyl-β-D-glucopyranoside.
Figure 1. Expression and purification of rFopA.
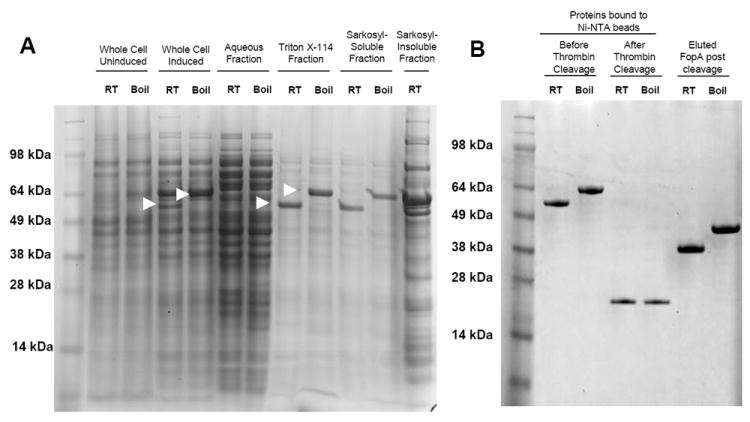
A. Coomassie-stained polyacrylamide gel of rFopA-expressing E. coli BL21(DE3)omp8 preparations. The fraction tested is indicated above the gel. Arrows indicate the FopA/DsbA fusion protein. B. Coomassie-stained polyacrylamide gel of the Triton X-114 and Sarkosyl-soluble fractions bound to Ni-NTA beads, before and after thrombin treatment, and the purified FopA protein obtained after thrombin cleavage. “Boiled” samples were heated in SDS loading buffer for 10 min before being loaded onto the polyacrylamide gel, while “RT” samples were left at room temp. “Induced” samples were derived from bacteria incubated in the presence of 0.3 mM IPTG, while “Uninduced” samples were derived from bacteria that were incubated in the absence of IPTG.
The heat-inducible shift in SDS-PAGE migration that is characteristic of bacterial outer membrane proteins, including endogenous FopA [12, 20, 21], was observed with the expressed fusion protein throughout all stages of the purification, with the exception of the Sarkosyl-insoluble fraction. The majority of the fusion protein present in this fraction ran at an apparent higher molecular weight, indicating that the protein was either improperly folded during expression or had become denatured during purification. The Sarkosyl-insoluble fraction was therefore not used for the final purification.
The final product that was isolated from the Triton X-114 and Sarkosyl-soluble fractions, and cleaved from the Ni-NTA beads, demonstrated a characteristic heat-modification in apparent molecular weight, both before and after liposome incorporation (Fig. 1B). This indicated that the rFopA maintained native structure following the purification process.
3.2. Induction of antibody responses following liposomal-rFopA immunization
Mice were immunized with liposomal FopA using IL-12 and alum as a vaccine adjuvants for induction of antibody responses, essentially as described in our previous studies [22-25]. Fourteen days after the final immunization, the sera were tested for their ability to recognize purified recombinant (rFopA) and endogenous FopA within F. tularensis LVS using ELISA and western blotting. It was found that antibodies in the FopA-immune sera were able to bind rFopA (average titer of 2000) as well as wildtype F. tularensis LVS as measured by ELISA (Fig. 2A and 2B). Sera collected from sham - immunized animals did not show appreciable binding to rFopA, and there was no significant binding of anti-FopA antisera to the F. tularensis fopA:Tn5 mutant, which lacks FopA expression, demonstrating the antigen specificity of the immune sera. These results were confirmed using western blotting of LVS and LVS fopA∷Tn5 cell lysates probed with anti-rFopA antisera. Anti-rFopA serum reacted with both the heat-modified and unmodified forms of endogenous FopA present within wildtype bacteria (Figure 2C). No other proteins were reactive on the blots, demonstrating the antigen specificity of the antiserum. No reactivity was detected with LVS fopA∷Tn5 lysates, further demonstrating the specificity of the FopA-specific sera.
Figure 2. Antibody responses in mice immunized with liposomal rFopA.
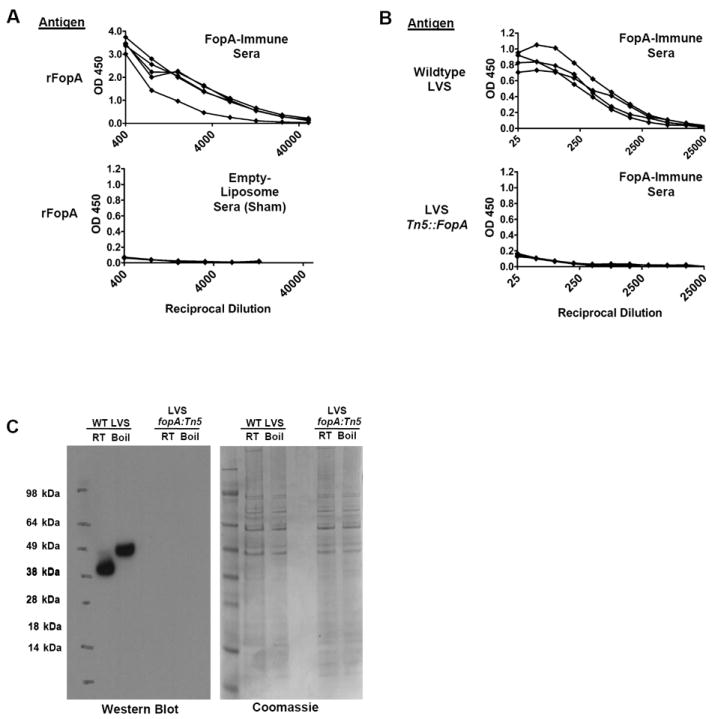
A. ELISA results showing the reactivity of either FopA immune sera, or sera collected from mice receiving sham immunizations with empty liposomes, with wells coated with rFopA. Each line represents the binding curve of an individual mouse. B. ELISA results showing the reactivity of sera from liposomal rFopA-immunized mice with wells coated with wildtype F. tularensis LVS (top) or F. tularensis LVS fopA:Tn5 mutant (bottom). Each line represents the binding curve of an individual mouse. C. Western blot (left) and coomassie-stained polyacrylamide gel (right) analysis of reactivity of pooled anti-rFopA serum against wildtype and fopA:Tn5 LVS cell lysates. 3μg of total protein from each lysate were loaded into each well. The PAGE analysis of the samples demonstrates equal loading of protein in each lane.
3.3. Protection against lethal F. tularensis challenge
To assess protection against lethal LVS infection, the rFopA-immunized mice were challenged i.d. with a lethal dose of F. tularensis LVS. I.d. infection is the most common way of acquiring tularemia in nature [2]. Mice were primed with liposomal rFopA, boosted twice and challenged with LVS 56 days following the final vacccine inoculation. It was found that 80% (4/5) of the rFopA-immunized animals were protected from a lethal i.d. LVS challenge, while 83% (5/6) of the unvaccinated mice succumbed to infection (Fig. 3A). As expected, all mice that had recovered from a sublethal LVS infection survived the challenge.
Figure 3. Protection of rFopA-immunized mice from lethal LVS infection.
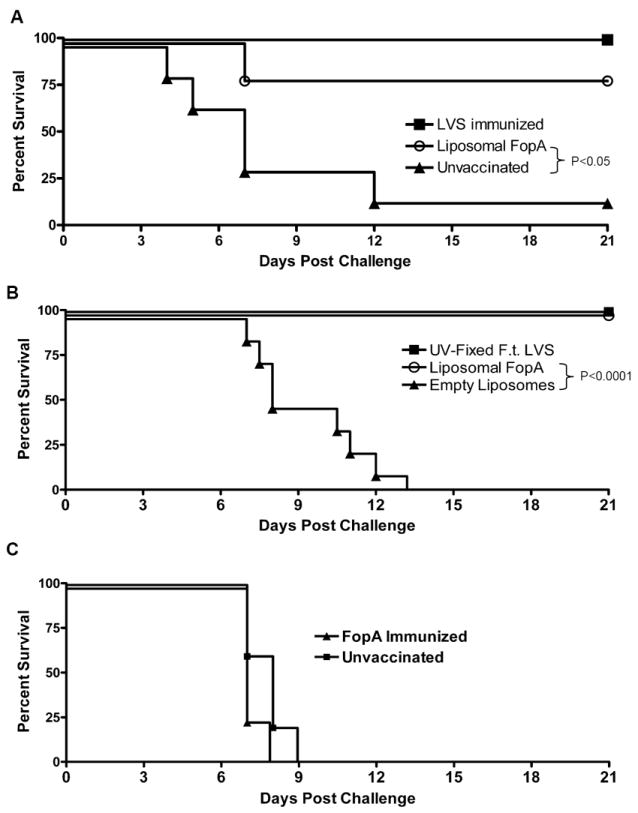
A. Survival of unvaccinated C57BL/6 mice or mice immunized with a sublethal dose of F.tularensis LVS, or with liposomal rFopA after i.d. challenge with a lethal dose of F. tularensis LVS. Challenge was performed 35 days after the final immunization. The FopA-vaccinated and unvaccinated groups consisted of 5 and 6 mice, respectively, while the LVS-immune group consisted of 3 mice. B. Survival of C57BL/6 mice immunized with either 108 CFU of UV-fixed F. tularensis LVS, liposomal FopA, or empty liposomes, after i.n challenge with F. tularensis. Challenge was performed 21 days after the final immunization. Each group consisted of 8 mice. C. Survival of unvaccinated C57BL/6 mice or mice immunized with liposomal rFopA after i.d. challenge with a lethal dose of F. tularensis SchuS4. The FopA-vaccinated and unvaccinated groups consisted of 4 and 5 mice, respectively.
The ability of rFopA-immunization to protect against lethal pulmonary LVS challenge was also assessed as a more stringent test of effective vaccination [2, 26]. Again, it was found that 100% of rFopA-immunized animals were protected from lethal i.n. LVS challenge, similar to animals immunized against inactivated LVS (Fig. 3B). All sham immunized mice inoculated with empty liposomes succumbed to infection by day 14.
Finally, protection against lethal SchuS4 challenge was examined. However, no FopA immunized animals survived a lethal i.d. challenge with Schu S4 (Fig. 3C).
3.4. Reduction of LVS bacterial burden in FopA-immunized mice
It was next determined whether the increased survival in rFopA-immunized mice was due to control of bacterial growth and dissemination during infection. FopA- and sham (empty liposome)-immunized mice were challenged i.n. with a lethal LVS dose and bacterial CFU were determined in lungs, livers, and spleens at various times after challenge. No significant differences in CFU were observed in lungs harvested from the two groups on day 1 post infection, and no bacteria were detected in the livers or spleens (Fig, 4). By day 5 post infection, however, the lungs and livers from FopA-immunized mice were found to contain significantly fewer bacteria than those from sham-immunized mice. By day 7, bacteria in the livers and spleens of FopA-immunized animals were beginning to be cleared and were significantly lower than those in the sham-immunized animals. No F. tularensis was detected in any organs harvested from rFopA-immunized and i.d. or i.n. challenged animals by day 28 post infection (data not shown).
Figure 4. Bacterial burdens in lungs, livers, and spleens following infection.
Mice were immunized with liposomal rFopA or empty liposomes in the presence of IL-12 as adjuvant and mean CFU were measured in the indicated tissues on days 1, 5, and 7 post i.n challenge. The error bars represent standard deviation from groups of 5-6 mice.
3.5. Ability of FopA-specific antibodies to protect mice against lethal LVS challenge
To directly determine the role of FopA-specific antibodies in mediating protection, passive transfer experiments were conducted. Naïve mice inoculated i.p. with either LVS immune- or FopA-specific sera were able to survive lethal i.d. bacterial challenge, while nearly all mice receiving non-immune sera succumbed to infection (Fig. 5). These results demonstrate that passive transfer of rFopA-specific antibodies can protect against lethal F. tularensis LVS challenge.
Figure 5. Protection by passive transfer of anti-rFopA antiserum.
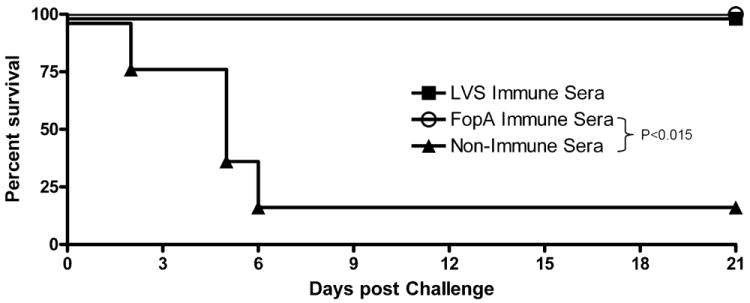
Survival curves of BALB/c mice that were injected i.p. with 250 μl of pooled F. tularensis LVS-immune serum, FopA-immune serum or normal serum. Serum injections were performed 24 hrs before and 24 hrs after i.d. challenge with 2LD50 of F. tularensis LVS. Each group consisted of 5 mice.
4. Discussion
The results described here demonstrate a proof of principle that a subunit vaccine prepared from specific F. tularensis proteins can induce protection against lethal Francisella challenge. FopA immunization not only protected against both lethal i.d. and i.n. infections, but also induced a sterilizing immune response that resulted in complete clearance of bacteria by day 28 post-infection. Moreover, FopA-immune sera could protect naïve mice against challenge, indicating that humoral immunity significantly contributed to the protective response.
FopA was used as a candidate protein not only because of its location on the bacterial surface but also because of its abundant expression [11, 12]. Fulop et. al. [13, 14] previously examined FopA as a potential vaccine candidate using FopA-expressing Salmonella. However, they failed to observe protection, concluding that FopA was not likely to be a useful target for subunit vaccination against tularemia. However, it is not clear from those studies how well Francisella FopA was expressed on the surface of the vector in the presence of other Salmonella outer membrane components, including the proinflammatory Salmonella LPS. The inability to test FopA in isolation may have been due in part to the potential difficulty of purifying the protein from Salmonella without denaturing its porin structure. It is important to note that when expressed in E. coli BL21(DE3)omp8, much of the FopA that is produced migrates on SDS-PAGE gels at an higher apparent molecular weight regardless of previous boiling, indicating that the major portion is not properly folded This same phenomenon may also have occurred when FopA was expressed in Salmonella and could have resulted in FopA molecules being in a non-native form during the immunization studies, ultimately accounting for the apparent lack of protection observed. This potential problem, however, was avoided in our studies, since the misfolded rFopA present in the Sarkosyl-insoluble fraction was removed and discarded. Only properly folded rFopA, collected from the Triton X-114- and Sarkosyl-soluble fractions, was used for our immunization studies. In contrast to the negative results of Fulop et al., FopA-specific mAbs that were generated from mice during infection with live LVS, in which FopA is properly folded and in its native environment, were able to provide partial protection in a mouse challenge model [16]. Other studies have also demonstrated that isolated LPS, as well as preparations of isolated outer membrane proteins from F. tularensis can induce protective responses in mice [11, 13, 14]. These findings, when considered together, provide credibility to the concept of developing a subunit vaccine against tularemia based upon outer membrane components.
Passive transfer experiments demonstrated an important role for FopA-specific antibodies in mediating the observed protection against infection. Previous work from our laboratory and others have demonstrated the importance of antibodies in combating F. tularensis infection [16, 27-32]. It is likely that T cells were required for induction of these protective antibodies but whether FopA-specific T cells also play a role in the effector phase of protection is unknown and the subject of our ongoing investigations.
The results presented here demonstrate that a subunit vaccine for human use may be possible, if presented in the correct context and with a suitable adjuvant. However, the rFopA immunization protocol used in this study was unable to protect mice from lethal i.d. challenge with the fully virulent SchuS4 strain of F. tularensis. Since the FopA molecules from LVS and SchuS4 share 99% nucleic acid homology, the lack of observed protection is unlikely to be due to an inability of LVS rFopA-specific antibodies to recognize SchuS4 FopA. However, it is possible that expression of FopA in the outer membranes of LVS and SchuS4 differ. Furthermore, the SchuS4 strain is significantly much more virulent than the LVS strain; therefore, it is reasonable to hypothesize that an immune response against a single outer membrane protein, although sufficient to protect against LVS, may not be fully effective against SchuS4. The fact that vaccination with rFopA, a single recombinant protein, is able to induce a protective response against lethal F. tularensis LVS infection, however, provides a useful model to further develop effective prophylactic and possibly even therapeutic treatments against this pathogenic biothreat.
Acknowledgments
We thank Dr. Jing Ren-Zhang for providing F. tularensis LVS FopA∷Tn5 and Shawn Roberts, Shannan Zilles, and Sharon Salmon for technical assistance. This work was supported by NIH PO1 A156320.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Oyston PC, Sjostedt A, Titball RW. Tularaemia: bioterrorism defense renews interest in Francisella tularensis. Nat Rev Microbiol. 2004;2:967–78. doi: 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- 2.Ellis J, Oyston PC, Green M, Titball RW. Tularemia. Clin Microbiol Rev. 2002;15:631–46. doi: 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pechous RD, McCarthy TR, Zahrt TC. Working toward the future: insights into Francisella tularensis pathogenesis and vaccine development. Microbiol Mol Biol Rev. 2009;73:684–711. doi: 10.1128/MMBR.00028-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salinas LJ, Greenfield RA, Little SE, Voskuhl GW. Tickborne infections in the southern United States. Am J Med Sci. 2010;340:194–201. doi: 10.1097/MAJ.0b013e3181e93817. [DOI] [PubMed] [Google Scholar]
- 5.Larsson P, Oyston PC, Chain P, Chu MC, Duffield M, Fuxelius HH, et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat Genet. 2005;37:153–9. doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- 6.Elkins KL, Cowley SC, Bosio CM. Innate and adaptive immunity to Francisella. Ann N Y Acad Sci. 2007;1105:284–324. doi: 10.1196/annals.1409.014. [DOI] [PubMed] [Google Scholar]
- 7.Isherwood KE, Titball RW, Davies DH, Felgner PL, Morrow WJ. Vaccination strategies for Francisella tularensis. Adv Drug Delivery Rev. 2005;57:1403–14. doi: 10.1016/j.addr.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 8.Bakshi CS, Malik M, Mahawar M, Kirimanjeswara GS, Hazlett KR, Palmer LE, et al. An improved vaccine for prevention of respiratory tularemia caused by Francisella tularensis SchuS4 strain. Vaccine. 2008;26:5276–88. doi: 10.1016/j.vaccine.2008.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakshi CS, Malik M, Regan K, Melendez JA, Metzger DW, Pavlov VM, et al. Superoxide dismutase B gene (sodB)-deficient mutants of Francisella tularensis demonstrate hypersensitivity to oxidative stress and attenuated virulence. J Bacteriol. 2006;188:6443–8. doi: 10.1128/JB.00266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia Q, Lee BY, Bowen R, Dillon BJ, Som SM, Horwitz MA. A Francisella tularensis live vaccine strain (LVS) mutant with a deletion in capB, encoding a putative capsular biosynthesis protein, is significantly more attenuated than LVS yet induces potent protective immunity in mice against F. tularensis challenge. Infect Immun. 2010;78:4341–55. doi: 10.1128/IAI.00192-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huntley JF, Conley PG, Hagman KE, Norgard MV. Characterization of Francisella tularensis outer membrane proteins. J Bacteriol. 2007;189:561–74. doi: 10.1128/JB.01505-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nano FE. Identification of a heat-modifiable protein of Francisella tularensis and molecular cloning of the encoding gene. Microb Pathog. 1988;5:109–19. doi: 10.1016/0882-4010(88)90013-7. [DOI] [PubMed] [Google Scholar]
- 13.Fulop M, Manchee R, Titball R. Role of lipopolysaccharide and a major outer membrane protein from Francisella tularensis in the induction of immunity against tularemia. Vaccine. 1995;13:1220–5. doi: 10.1016/0264-410x(95)00062-6. [DOI] [PubMed] [Google Scholar]
- 14.Fulop M, Manchee R, Titball R. Role of two outer membrane antigens in the induction of protective immunity against Francisella tularensis strains of different virulence. FEMS Immunol Med Microbiol. 1996;13:245–7. doi: 10.1111/j.1574-695X.1996.tb00245.x. [DOI] [PubMed] [Google Scholar]
- 15.Bevanger L, Maeland JA, Naess AI. Competitive enzyme immunoassay for antibodies to a 43,000-molecular-weight Francisella tularensis outer membrane protein for the diagnosis of tularemia. J Clin Microbiol. 1989;27:922–6. doi: 10.1128/jcm.27.5.922-926.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savitt AG, Mena-Taboada P, Monsalve G, Benach JL. Francisella tularensis infection-derived monoclonal antibodies provide detection, protection, and therapy. Clin Vaccine Immunol. 2009;16:414–22. doi: 10.1128/CVI.00362-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su J, Yang J, Zhao D, Kawula TH, Banas JA, Zhang JR. Genome-wide identification of Francisella tularensis virulence determinants. Infect Immun. 2007;75:3089–101. doi: 10.1128/IAI.01865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazlett KR, Caldon SD, McArthur DG, Cirillo KA, Kirimanjeswara GS, Magguilli ML, et al. Adaptation of Francisella tularensis to the mammalian environment is governed by cues which can be mimicked in vitro. Infect Immun. 2008;76:4479–88. doi: 10.1128/IAI.00610-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prilipov A, Phale PS, Van Gelder P, Rosenbusch JP, Koebnik R. Coupling site-directed mutagenesis with high-level expression: large scale production of mutant porins from E. coli. FEMS Microbiol Lett. 1998;163:65–72. doi: 10.1111/j.1574-6968.1998.tb13027.x. [DOI] [PubMed] [Google Scholar]
- 20.Dekker N, Merck K, Tommassen J, Verheij HM. In vitro folding of Escherichia coli outer-membrane phospholipase A. Eur J Biochem. 1995;232:214–9. doi: 10.1111/j.1432-1033.1995.tb20801.x. [DOI] [PubMed] [Google Scholar]
- 21.Heller KB. Apparent molecular weights of a heat-modifiable protein from the outer membrane of Escherichia coli in gels with different acrylamide concentrations. J Bacteriol. 1978;134:1181–3. doi: 10.1128/jb.134.3.1181-1183.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arulanandam BP, O’Toole M, Metzger DW. Intranasal interleukin-12 is a powerful adjuvant for protective mucosal immunity. J Infect Dis. 1999;180:940–9. doi: 10.1086/314996. [DOI] [PubMed] [Google Scholar]
- 23.Baron SD, Singh R, Metzger DW. Inactivated Francisella tularensis live vaccine strain protects against respiratory tularemia by intranasal vaccination in an immunoglobulin A-dependent fashion. Infect Immun. 2007;75:2152–62. doi: 10.1128/IAI.01606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duckett NS, Olmos S, Durrant DM, Metzger DW. Intranasal interleukin-12 treatment for protection against respiratory infection with the Francisella tularensis live vaccine strain. Infect Immun. 2005;73:2306–11. doi: 10.1128/IAI.73.4.2306-2311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch JM, Briles DE, Metzger DW. Increased protection against pneumococcal disease by mucosal administration of conjugate vaccine plus interleukin-12. Infect Immun. 2003;71:4780–8. doi: 10.1128/IAI.71.8.4780-4788.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fortier AH, Slayter MV, Ziemba R, Meltzer MS, Nacy CA. Live vaccine strain of Francisella tularensis: infection and immunity in mice. Infect Immun. 1991;59:2922–8. doi: 10.1128/iai.59.9.2922-2928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirimanjeswara GS, Golden JM, Bakshi CS, Metzger DW. Prophylactic and therapeutic use of antibodies for protection against respiratory infection with Francisella tularensis. J Immunol. 2007;179:532–9. doi: 10.4049/jimmunol.179.1.532. [DOI] [PubMed] [Google Scholar]
- 28.Foshay L. A comparative study of the treatment of tularemia with immune serum, hyperimmune serum and streptomycin. Am J Med. 1946;1:180–8. doi: 10.1016/0002-9343(46)90036-8. [DOI] [PubMed] [Google Scholar]
- 29.Drabick JJ, Narayanan RB, Williams JC, Leduc JW, Nacy CA. Passive protection of mice against lethal Francisella tularensis (live tularemia vaccine strain) infection by the sera of human recipients of the live tularemia vaccine. Am J Med Sci. 1994;308:83–7. doi: 10.1097/00000441-199408000-00003. [DOI] [PubMed] [Google Scholar]
- 30.Fulop M, Mastroeni P, Green M, Titball RW. Role of antibody to lipopolysaccharide in protection against low- and high-virulence strains of Francisella tularensis. Vaccine. 2001;19:4465–72. doi: 10.1016/s0264-410x(01)00189-x. [DOI] [PubMed] [Google Scholar]
- 31.Rhinehart-Jones TR, Fortier AH, Elkins KL. Transfer of immunity against lethal murine Francisella infection by specific antibody depends on host gamma interferon and T cells. Infect Immun. 1994;62:3129–37. doi: 10.1128/iai.62.8.3129-3137.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klimpel GR, Eaves-Pyles T, Moen ST, Taormina J, Peterson JW, Chopra AK, et al. Levofloxacin rescues mice from lethal intra-nasal infections with virulent Francisella tularensis and induces immunity and production of protective antibody. Vaccine. 2008;26:6874–82. doi: 10.1016/j.vaccine.2008.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]


