Abstract
Secretins form large multimeric complexes in the outer membranes of many Gram-negative bacteria, where they function as dedicated gateways that allow proteins to access the extracellular environment. Despite their overall relatedness, different secretins use different specific and general mechanisms for their targeting, assembly, and membrane insertion. We report that all tested secretins from several type II secretion systems and from the filamentous bacteriophage f1 can spontaneously multimerize and insert into liposomes in an in vitro transcription-translation system. Phylogenetic analyses indicate that these secretins form a group distinct from the secretins of the type IV piliation and type III secretion systems, which do not autoassemble in vitro. A mutation causing a proline-to-leucine substitution allowed PilQ secretins from two different type IV piliation systems to assemble in vitro, albeit with very low efficiency, suggesting that autoassembly is an inherent property of all secretins.
INTRODUCTION
Secretins are large multimeric protein complexes in the outer membranes (OM) of Gram-negative bacteria (diderms) that form the portal to the extracellular milieu in type II secretion systems (T2SS), in type IV piliation and DNA uptake systems (T4PS), in needle formation by type III secretion systems (T3SS), and in filamentous phage secretion systems (reviewed in reference 29). Secretins share a domain organization comprising a relatively poorly conserved periplasmic N-terminal region, which confers substrate specificity (39) and interacts with inner membrane components of the secretion machinery (46), a more highly conserved secretin C domain (PFAM secretin, PF00263) involved in multimerization and membrane insertion (4, 19, 20), and, in at least some cases, a C-terminal region of variable length that is important for OM localization (33, 42).
Despite their similar primary structures and domain organization, secretins use different mechanisms for targeting and insertion into the OM. The archetypical secretin, the dodecameric PulD from the Klebsiella oxytoca (also called K. pneumoniae) T2SS (14), is targeted to the OM by a dedicated lipoprotein chaperone, the pilotin PulS, which binds to the intrinsically disordered C-terminal segment in PulD, the S domain (33, 42), and is itself targeted to the OM by the Lol machinery (8). Once docked at the OM, PulD assembly and insertion depend only on the core domain (the last repeat of the N-terminal domain and the C domain) (19, 21). Specific OM insertion factors, such as the Bam (beta-barrel assembly) machinery, are not required (7), as indicated by the ability of in vitro-synthesized PulD to insert into liposomes (19), the phenomenon referred to here as autoassembly. The closely related T2SS secretin OutD of Dickeya dadantii (Erwinia chrysanthemi) is also targeted and assembled by this mechanism (18, 19). The corresponding secretin XcpQ from the T2SS of Pseudomonas aeruginosa can also insert spontaneously into membranes in vitro (23), but its correct localization to the OM depends on a periplasmic protein, PA0943 (38). HxcQ, the secretin of a different P. aeruginosa T2SS, is a lipoprotein in which the lipid anchor is required for correct targeting to the OM but not for multimerization (44).
The BfpB secretin of an Escherichia coli T4PS is also a lipoprotein, but lipid modification is not required for targeting to the OM (31). The secretin PilQ from the T4PS of Neisseria meningitidis requires a lipoprotein chaperone, PilW, for multimer stability (3) and the Bam complex for correct assembly in the OM (45) and cannot assemble in vitro (20). Likewise, PilQ from P. aeruginosa is also unable to assemble in vitro (23) and requires a PilW orthologue, PilF, for multimerization and OM localization (27), but it appears to be only minimally dependent on BamA and is negatively impacted by inactivation of the Lol pathway (23). In the Myxococcus xanthus T4PS, assembly of stable PilQ multimers is dependent on another PilW orthologue, Tgl, but targeting and membrane insertion of the complex have not been thoroughly investigated (35).
Stability and multimerization of the Shigella flexneri T3SS secretin MxiD are dependent on another lipoprotein, MxiM (37). Although PulS, PilF/PilW, and MxiM are all lipoproteins and have related functions, they have completely different structures (27, 36, 42, 43). Efficient assembly of the P. aeruginosa T3SS secretin PscC depends on both BamA and the Lol machinery, and this secretin does not autoassemble in vitro (23).
In contrast to all of these secretins, the pIV secretin from the E. coli filamentous bacteriophage f1 does not appear to need any accessory factor for targeting and assembly, although targeting is inefficient compared to that of other secretins (11, 24).
In this report, we extend the analysis of the capacity of T2SS and pIV secretins to autoassemble and correlate this capacity with their phylogenetic relatedness and their relation to T4PS and T3SS secretins. We also describe attempts to identify mutations that allow autoassembly of T4PS secretins.
MATERIALS AND METHODS
Materials.
Unless otherwise indicated, chemicals were of molecular biology grade and were purchased from Sigma. DNA polymerases, restriction endonucleases, and DNA modification enzymes were from New England BioLabs, Invitrogen, or Roche. Plasmid purification, PCR purification, and gel extraction kits were purchased from Qiagen and used according to manufacturers' recommendations.
Bacterial strains and growth conditions.
Information on bacterial strains and plasmids is outlined in Table S1 in the supplemental material. E. coli K-12 strain PAP105 [Δ(lac-proAB) F′ (lacIq1 ΔlacZM15 proAB+ Tn10)] was used for plasmid manipulations, and E. coli B strain BL21 (F− ompT hsdS gal endA) (λDE3) carrying pDIA17 (pACYC184 derivative carrying lacIq under tet promoter control) was used for incytotoxicity experiments (19). Cultures were grown under aeration at 30°C or 37°C in LB broth or LB with 10% M63 minimal medium (32) containing, when appropriate, 0.4% maltose to induce expression of pul genes encoding the T2SS machinery, and appropriate antibiotics (chloramphenicol, 25 μg·ml−1; ampicillin, 100 μg·ml−1; kanamycin, 50 μg·ml−1). For secretion assays, PAP105 carrying pCHAP8243 (a derivative of pCHAP231 [14] carrying the complete pul operon with a pulD deletion and pulA encoding a soluble nonacylated variant of PulA) and pCHAP3635 (pulD+) or its derivatives encoding PulD variants were grown to an optical density at 600 nm (OD600) of approximately 1.5 and centrifuged to separate cells and supernatant.
Plasmid construction and mutagenesis.
Constructed plasmids are outlined in Table S1 in the supplemental material, and primers used for cloning are listed in Table S2. pilQ amplified from Neisseria meningitidis strain 8013 genomic DNA using primers PNN30 and PNN31 and pIV amplified from f1 filamentous phage DNA using primers PNN59 and PNN60 were cloned into NdeI-BamHI sites of pIVEX2.3MCS (Roche Applied Science). DNA encoding T2SS secretins from Legionella pneumophila strain Paris (lspD), Shewanella oneidensis MR-1 (gspD), and Xanthomonas campestris pv. campestris (xcsD) were amplified with the appropriate primers (see Table S2) and likewise cloned into pIVEX2.3MCS. For reasons explained in Results, linear templates containing the essential T7 promoter elements were created for T2SS secretin genes from Vibrio cholerae El Tor O1 (epsD) and Aeromonas hydrophila subsp. hydrophila (exeD) using the LinTempGenSet kit (5 Prime) with the appropriate primers (see Table S2), thereby allowing their expression in the rapid translation system (RTS) without cloning.
Random-site mutagenesis of DNA encoding PilQMC58 in pIVEX2.3MCS (Roche Applied Science) (pCHAP3783 [19]) was preformed by error-prone PCR using the GeneMorph II EZClone domain mutagenesis kit (Stratagene) with pilQ-specific primers PNN5 and PNN6. Mutated DNA was electroporated into E. coli strain BL21(pDIA17), and clones of interest were selected based on their reduced growth at 37°C versus 30°C. Site-directed mutagenesis was performed with complementary pairs of mutagenic primers using an adapted two-step PCR based on the QuikChange site-directed mutagenesis kit protocol (Stratagene). All constructs were verified by sequencing.
In vitro synthesis reactions.
To test their ability to self-assemble in vitro, secretins were synthesized in a 50-μl E. coli HY 100 RTS reaction mixture (5 Prime) in the presence of 100 μg of asolectin liposomes (Sigma) and 0.5 μg of vector DNA (pIVEX2.3MCS) at 30°C for 6 h (19). To check for multimer assembly and insertion into liposomes, the reaction mixture was diluted in 5 volumes of phosphate-buffered saline (PBS) before centrifugation in a Beckman TLA-55 rotor at 23,000 × g for 10 min at 20°C. The pellet was resuspended in 50 μl of the same buffer, and an aliquot was treated with phenol to dissociate multimers and then analyzed together with an untreated aliquot by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with or without prior heating to 100°C. Another aliquot of the resuspended pellet was centrifuged and resuspended in 4 M urea in the same buffer, incubated for 15 min at room temperature, and centrifuged again. The urea-washed pellet was resuspended, and one aliquot of each fraction was prepared for analysis as described above.
SDS-PAGE and immunoblotting.
Total cell extracts and aliquots from in vitro reaction mixtures were diluted in SDS sample buffer and, where appropriate, were heated at 100°C for 5 min. Secretin multimers were dissociated by phenol treatment (22), precipitated by acetone at −20°C, and resuspended in sample buffer. Proteins solubilized in sample buffer were separated by electrophoresis in Tris-glycine-buffered 10% polyacrylamide gels or 4-to-20% gradient gels (Bio-Rad) with SDS (SDS-PAGE) and either stained with Coomassie brilliant blue or electroblotted onto nitrocellulose membrane (GE Healthcare). Immunoblotting for PilQ from N. meningitidis (PilQNm) was performed with either N-terminal or C-terminal specific antibodies (17), antibodies specific for PilQ from P. aeruginosa (PilQPa) (Stephen Lory), or PulD- and PulA-specific antibodies from the laboratory collection. Secondary antibodies conjugated to horseradish peroxidase (GE Healthcare) were detected by enhanced chemiluminescence (Pierce).
Electron microscopy.
Liposomes were centrifuged at 23,000 × g for 10 min, resuspended in 4 M urea for 15 min to dissociate liposome aggregates and loosely bound material, diluted to 1 M urea by adding 20 mM Tris containing 250 mM NaCl (TN buffer), centrifuged at 250,000 × g for 30 min, and resuspended in TN buffer. For imaging of negatively stained samples, aliquots of 5 μl were adsorbed on carbon film-coated copper grids, washed with 10 droplets of pure water, and subsequently stained with 2% uranyl acetate. Images were recorded on a charge-coupled device (CCD) camera (2K by 2K pixel resolution; Veleta; Olympus) at a ×92,000 magnification using a Philips CM10 transmission electron microscope operating at 80 kV.
Phylogenetic analysis of the secretin family.
A selection of secretin sequences from a published data set (6) supplemented with additional sequences corresponding to the proteins examined experimentally were aligned by the Muscle program with default parameters (15). Informative positions were selected using the BMGE program using the following parameters (10): BLOSUM30 similarity matrix, a gap rate cutoff of 0.20, a sliding window size of 3, and an entropy score cutoff of 0.5. Phylogenetic trees were built with the maximum-likelihood search implemented in RAxML (40) (model LG+G+F). The tree with the highest likelihood was selected among 200 starting tree inferences. One thousand bootstrap replicates were constructed, and the relationships in these bootstrap trees were used to display supports on the branches of the selected maximum-likelihood tree.
RESULTS
The f1 filamentous phage pIV secretin can autoassemble.
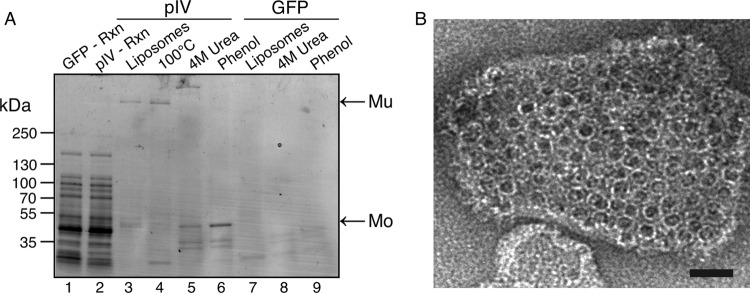
Previous studies revealed that the T2SS secretins PulD, OutD, and XcpQ assemble into membrane-embedded secretin rings when produced in an in vitro transcription-translation system supplemented with liposomes, whereas two PilQ secretins (T4PS) and PscC (T3SS) do not (19, 23). Here, we report that pIV secretin from f1 filamentous phage produced without its signal peptide behaves like PulD, OutD, and XcpQ in this system, forming SDS-, heat- and urea-resistant, phenol-sensitive multimers detectable by SDS-PAGE (Fig. 1A). In this experiment, the multimer (Mu) was identified by its dissociation following phenol treatment and the monomer (Mo) by its corresponding increased abundance. Neither of these two bands was detected when green fluorescent protein (GFP) DNA was used. Electron microscopy of in vitro synthesized pIV revealed membrane-embedded secretin-like rings (Fig. 1B).
Fig 1.
Phage pIV secretin can autoassemble and insert into liposomes in vitro. (A) Products (Rxn; lane 1 and lane 2) and centrifuged liposomes (pIV, lanes 3 to 5; GFP control, lanes 7 to 9) from in vitro synthesis reactions in which either pIV secretin or GFP control was synthesized were dissolved in SDS sample buffer and then separated by SDS-PAGE on a gradient gel and stained with Coomassie blue. Separate samples were heated in SDS to 100°C (lane 3) or were treated with 4 M urea (lanes 5 and 8) or phenol (lanes 6 and 9) prior to dissolution in SDS. Samples enriched for liposomes (lanes 3 to 9) correspond to five times more material than was in the total reaction mixture. Mu, pIV multimer; Mo, pIV monomer. We cannot explain the reduced mobility of the pIV multimer after urea treatment (lane 5). (B) Negatively stained liposomes with inserted pIV secretin multimers examined by electron microscopy. The average diameter of the secretin particles is 14 nm. Bar, 25 nm.
Sequences of T2SS and filamentous phage secretins are closely related.
The C domain of ∼200 to 250 residues and encompassing the membrane-spanning and gate regions is the most conserved region of secretins (29). A secretin sequence data set was constructed with secretins from T2SS, T3SS, T4PS, and filamentous phage secretion systems, and after poorly aligned regions had been filtered out, a maximum-likelihood phylogeny was constructed (Fig. 2). This tree revealed well-supported groups for the different secretin subfamilies: the T3SS secretins (100% bootstrap support), the T4PS secretins (100% bootstrap), and the T2SS secretins (99% bootstrap). Within the T2SS group, X. campestris XpsD-related secretins formed a clade (90% bootstrap) that was the first to emerge, followed by the filamentous phage secretins (98% bootstrap). This tree topology is consistent with a previously published tree (6) and groups filamentous phage secretins together with T2SS secretins. These observations suggest that autoassembly could be a specific characteristic of the T2SS-phage pIV secretin subfamily.
Fig 2.
Phylogeny of the secretin family. The maximum-likelihood tree shows the relationships between secretins of the different machineries in which they are found. Species names and protein names are indicated for protein sequences tested experimentally in the in vitro synthesis system. + and −, secretins that are able and unable to multimerize and insert into liposomes in vitro, respectively. Bootstrap supports for the tree are displayed only when below 80%, except for branches corresponding to key relationships mentioned in the text (in bold). Proteins mentioned in the main text but not included in the analysis are given in parentheses below the name of the system to which they belong.
Spontaneous assembly and membrane insertion are general characteristics of T2SS secretins.
We next tested whether other secretins of the T2SS-phage pIV subfamily (LspD from Legionella pneumophila, GspD from Shewanella oneidensis, ExeD from Aeromonas hydrophila, EpsD from Vibrio cholerae, and XcsD from Xanthomonas campestris) share with the previously tested members of this subfamily their capacity for autoassembly and membrane insertion. PCR-amplified DNA for each secretin without its signal peptide was cloned into pIVEX2.3MCS under the control of the T7 promoter. DNA encoding the secretins from Aeromonas and Vibrio could not be cloned into pIVEX2.3MCS. Previous studies have shown that pulD thus cloned is toxic except in the presence of multiple copies of the lac repressor gene lacI and, in cells grown at 30°C, a phenomenon referred to as incytotoxicity (21). DNA encoding the Aeromonas and Vibrio secretins could not be cloned even under these conditions, suggesting that they are even more toxic than PulD. Thus, DNA encoding these two secretins was produced as linear templates with the necessary T7 promoter elements.
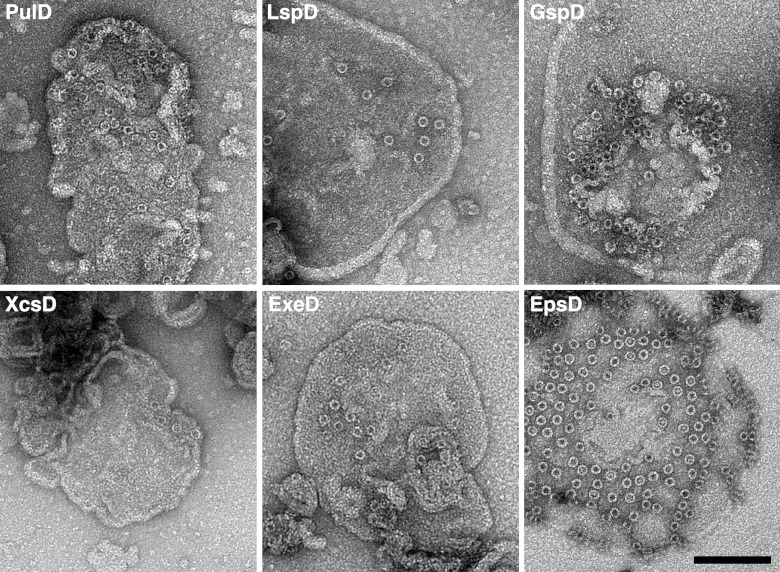
SDS-PAGE (see Fig. S1 in the supplemental material) and electron microscopy (Fig. 3) demonstrated that all five newly tested secretins formed typical secretin rings that were associated with liposomes when produced in the in vitro transcription-translation system with lecithin liposomes. These and previous results are summarized in Table 1 and outlined on the phylogenetic tree (Fig. 2). Thus, all tested T2SS and phage secretins can autoassemble, whereas the two previously tested T4PS secretins and the single T3SS secretin tested cannot (Fig. 2). The separation between these two groups of secretins is highly supported by the phylogenetic tree (99% bootstrap value). The autoassembly-competent secretins are thus closely related and have a common origin (see Discussion). These observations provide strong support for the idea that all secretins in the T2SS-phage secretion group are able to multimerize and insert spontaneously into liposomes.
Fig 3.
In vitro autoassembly is a general feature of T2SS secretins. Negatively stained liposomes from the respective samples were visualized by electron microscopy. Samples tested were from Klebsiella oxytoca (PulD), Aeromonas hydrophila (ExeD), Legionella pneumophila (LspD), Shewanella oneidensis (GspD), Vibrio cholerae (EpsD), and Xanthomonas campestris (XcsD). Bar, 100 nm.
Table 1.
Origins and in vitro assembly of analyzed secretins
| Secretin (species) | Secretion system | Accession no. | In vitro multimerizationa |
|---|---|---|---|
| PulD (Klebsiella oxytoca) | T2SS | P15644 | + |
| OutD (Dickeya dadantii) | T2SS | P31700 | + |
| XcpQ (Pseudomonas aeruginosa) | T2SS | NP_251795 | + |
| pIV (f1 filamentous phage) | f1 phage | P03666 | + |
| ExeD (Aeromonas hydrophila) | T2SS | P31780 | + |
| LspD (Legionella pneumophila) | T2SS | YP_123599 | + |
| EpsD (Vibrio cholerae) | T2SS | YP_001218221 | + |
| XcsD (Xanthomonas campestris) | T2SS | NP_638771 | + |
| GspD (Shewanella oneidensis) | T2SS | NP_715808 | + |
| PscC (Pseudomonas aeruginosa) | T3SS | NP_250407 | − |
| PilQ (Neisseria meningitidis) | T4PS | NP_274809 | − |
| PilQ (Pseudomonas aeruginosa) | T4PS | NP_253727 | − |
In vitro multimerization and insertion into liposomes was tested in the cell-free coupled transcription and translation system and analyzed by SDS-PAGE and electron microscopy.
Substitution of a single proline allows in vitro autoassembly of T4PS PilQ.
Since secretins share ancestry, we reasoned that secretins that cannot autoassemble might have recently lost the ability to do so and might, through mutation, easily reacquire it. To test this hypothesis, we mutagenized DNA encoding a signal peptide-less form of PilQ from the T4PS of N. meningitidis strain MC58 (19) and expressed it in E. coli. E. coli cells producing PulD in this way grow well at 30°C but, because of the incytotoxicity phenomenon likely caused by the insertion of PulD into the inner membrane, grow poorly at 37°C (21). After screening a bank of mutants, we identified one that consistently grew less well than the parent strain at 37°C. The PilQNm protein encoded by DNA in this clone was able to form multimers in the in vitro transcription-translation system (Fig. 4A). The efficiency of multimerization of this PilQ variant was difficult to establish because unassembled monomers were apparently degraded, giving rise to the multiple bands below that identified as the PilQ monomer. However, visual inspection of the immunoblot in Fig. 4A indicates that multimerization efficiency is considerably lower than that of PulD, which is estimated to multimerize at >50% efficiency under identical conditions.
Fig 4.
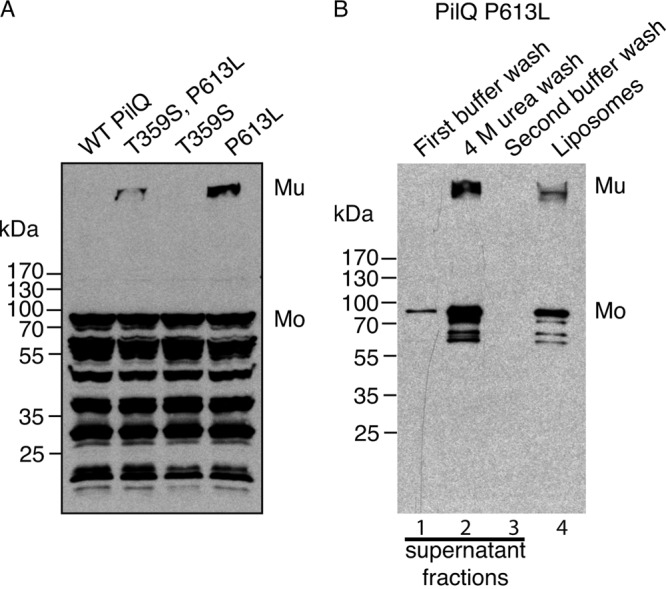
PilQNm* can multimerize and insert into liposomes spontaneously. (A) Total in vitro synthesis reactions of wild-type PilQNm (WT) and its derivatives with the indicated amino acid substitutions were analyzed by SDS-PAGE and immunoblotting with PilQNm antiserum. (B) Liposomes from the in vitro synthesis reaction of the PilQNm* mutant (P613L) were washed with PBS (supernatant fraction in lane 1) and 4 M urea treatment (supernatant in lane 2) and then again with PBS (supernatant in lane 3). The supernatant fractions and the washed liposomes (lane 4) were analyzed by SDS-PAGE and immunoblotting with PilQNm antisera. Mu, PilQ multimer; Mo, PilQ monomer.
This pilQ DNA carried two mutations, causing T359S and P613L substitutions. When the two mutations were created separately, incytotoxicity (poor growth at 37°C) and autoassembly in vitro were associated exclusively with the P613L substitution (Fig. 4A). Indeed, the T359S substitution appeared to diminish multimerization efficiency, since fewer multimers were observed with the T359S-P613L substitutions than with P613L alone (Fig. 4A). The P613L variant is referred to here as PilQNm*. Like other autoassembly-competent secretins, PilQNm* remained partially associated with the liposomes after a washing with 4 M urea (Fig. 4B), was dissociated by phenol treatment (data not shown), and appeared as typical secretin rings when examined by electron microscopy (Fig. 5), confirming that PilQNm* multimerizes and inserts into liposomes in vitro.
Fig 5.
PilQNm* inserted into liposomes. Negatively stained liposomes with inserted PilQNmP613L were visualized by electron microscopy. Arrows indicate the presence of PilQNm* rings inserted into liposomes. Bar, 100 nm.
To test the specificity of the P613L substitution, proline 613 in PilQNm was replaced by glycine, alanine, valine, phenylalanine, and cysteine. These substitutions all resulted in lower levels of autoassembly than P613L, except for the glycine substitution, which did not result in any detectable multimer formation but caused smearing that might indicate incomplete multimerization or multimer instability (Fig. 6A). Replacing the amino acid residues on either side of the proline by those found in the identical position in PulD (N612T and R614S) did not increase the efficiency of PilQNm* autoassembly (data not shown).
Fig 6.
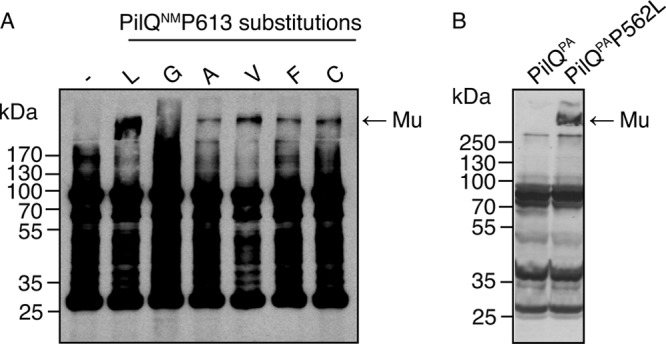
Effects of proline 613 substitutions in PilQNm and of the corresponding P562L substitution in PilQPa on autoassembly in vitro. (A) Total in vitro synthesis reactions producing wild-type PilQNm or its variants were analyzed by SDS-PAGE and immunoblotting with N. meningitidis PilQ antisera. (B) Total in vitro synthesis reactions producing wild-type PilQPa and PilQPa(P562L) from P. aeruginosa were analyzed by SDS-PAGE and immunoblotting with PilQPa antisera. Mu, PilQ multimer.
The corresponding substitution in P. aeruginosa PilQ also promotes autoassembly.
P613 is found in all analyzed secretins, irrespective of their ability to autoassemble. To test whether the autoassembly caused by the P613L substitution in PilQNm* is specific to this particular secretin, the same substitution was constructed in PilQNm from N. meningitidis strain 8013 (P605L). PilQ8013(P605L) behaved in the same manner as PilQNm* (data not shown). The same proline-to-leucine change in P. aeruginosa PilQ (P562L), creating PilQPa*, also resulted in the ability to autoassemble in vitro (Fig. 6B).
The corresponding proline-leucine substitution in PulD reduces multimerization but does not drastically affect function.
As mentioned above, proline at position 613 in PilQNm is completely conserved in all secretins, including, paradoxically, those that are able to assemble spontaneously. To determine the role of this residue in these secretins, the proline-to-leucine substitution was made in full-length signal peptide-less PulD (PulD28–660) and in a truncated version corresponding to the core and S domains (PulD28–42/259-660). Both PulD(P443L) variants exhibited reduced in vitro assembly (see Fig. S2A in the supplemental material), but electron microscopy of purified PulD(P443L) secretin produced in vitro did not reveal any gross physical changes compared to wild-type protein (see Fig. S2B).
PulD(P443L) produced in vivo also exhibited less efficient multimerization (Fig. 7A, top) and caused cells to produce somewhat higher levels of the phage shock protein PspA, probably indicative of increased misrouting to the inner membrane (2, 20), than wild-type PulD (Fig. 7A, bottom). Despite its reduced capacity to multimerize, however, PulD(P443L) could substitute for wild-type PulD in complementation assays for pullulanase (PulA) secretion, albeit with reduced efficiency compared to the wild-type protein (Fig. 7B).
Fig 7.
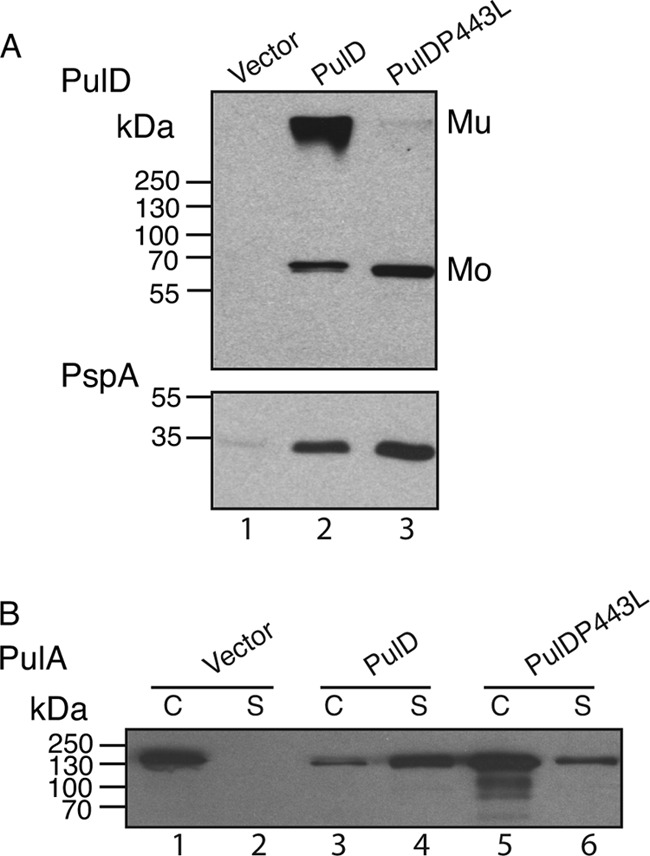
Effect of the P443L substitution on PulD assembly and function. (A) Total cell lysates of E. coli PAP105 in which either wild-type PulD or PulD(P443L) was produced. An equivalent of 0.05 absorbance unit of culture was separated by SDS-PAGE and immunoblotted with PulD- or PspA-specific antisera. Mu, PulD multimer; Mo, PulD monomer. (B) Total cell lysate (C) and culture supernatant (S) fractions of E. coli PAP105(pCHAP8243) producing all the Pul factors except PulD and a soluble variant of PulA (lanes 1 and 2), complemented with either wild-type PulD (lanes 3 and 4) or PulD(P443L) (lanes 5 and 6). An equivalent of 0.015 absorbance unit of culture was separated by SDS-PAGE and immunoblotted with PulA-specific antiserum.
DISCUSSION
Secretins are key components of many trans-envelope OM machineries in diderms that allow proteins to reach the cell surface and beyond. The large complexes that they form incorporate a gated channel to allow proteins or filamentous bacteriophages to pass (secretion) or structures (needles, pili) to be assembled without disrupting the OM barrier. Although significant advances have been made, understanding secretin assembly and function has been hindered by the lack of high-resolution structural information on the core domain and relies mainly on biochemical and genetic approaches.
Secretins can be classified into one of two groups: those that can autoassemble and insert into membranes in vitro (T2SS and filamentous phage secretins) and those that cannot (T4PS and T3SS secretins) (19, 23; also this study). The phylogenetic tree of secretins cannot be rooted because of the lack of an outgroup sequence; hence, we cannot determine the branch of the tree in which the last common ancestor of all secretins is placed. Consequently, we cannot determine which system-specific secretin emerged first or if the original secretin performed multiple functions. Of all of the machineries in which a secretin participates, only the T4PS has close homologs in monoderm (Gram-positive) bacteria and in archaea (5). It is therefore tempting to speculate that a secretin was added to this ancestral structure when it was acquired by diderms. However, the T4PS and T2SS are closely related and partially interchangeable (25), but self-assembling secretins are found only in the latter. Thus, in this scenario, the T4PS is the ancestor of the T2SS, T4PS secretins are the ancestors of T2SS secretins, and autoassembly evolved more recently than assisted assembly.
The filamentous phage secretins might have been acquired from a T2SS, an idea supported by the fact that at least one filamentous phage does not have its own secretin but instead uses the T2SS secretin of its host bacterium (12). This idea would be compatible with a potential selective advantage conferred by a minimal bacteriophage genome in which a secretin would be essential (for example, in E. coli strains that do not constitutively produce an endogenous secretin) but ancillary factors would not. However, this scenario does not explain all of the phenomena observed. For example, although genes for T2SS secretins are very frequently found sandwiched between pulC and pulE homologues, as in the K. oxytoca T2SS (13), suggesting highly conserved organization and function, they use different sets of specific cofactors for targeting to the OM (1, 26). This observation suggests that filamentous phage secretins, which do not have targeting and assembly factors, are ancestral and that different targeting cofactors evolved to facilitate efficient T2SS secretin targeting to the OM.
All secretins show substantial similarity in their C domains; in PulD, the C domain contains all the information needed for autoassembly and insertion (19–21). Presumably, sequence differences in this region explain the differing abilities of secretins to autoassemble. The fact that a single proline-to-leucine substitution in two different T4PS secretins enables them to assemble, albeit inefficiently, in vitro indicates that the structural or sequence differences underlying their ability or otherwise to autoassemble are likely quite minor. It might be significant that the P613G substitution in PilQNm apparently caused an intermediate situation leading to the appearance of incomplete multimers seen as multiple bands when examined by SDS-PAGE (Fig. 6A), suggesting that flexibility (induced by proline or glycine) rather than side chains (present only in proline) at this position impedes autoassembly. However, this proline residue is completely conserved in all secretins, and thus, its presence or absence cannot be a primary determinant of autoassembly. We hypothesize that residues elsewhere in PilQ might prevent autoassembly and that the P613L substitution partially compensates for their effect, or that autoassembling secretins have overcome the deleterious effect of this proline residue on autoassembly by compensatory adjustments elsewhere in the C domain.
One intriguing question that we were unable to answer is whether the autoassembly variants of PilQNm* and PilQPa* are functional and have lost their dependence on their respective cofactors (PilW and PilF, respectively) for multimerization. PilQNm* was produced in insufficient amounts in a nonpolar pilQ mutant of N. meningitidis to be able to test its ability to multimerize and function in vivo (V. Pelicic, personal communication). Similarly, PilQPa* was unstable when produced in a pilQ mutant of P. aeruginosa, again precluding complementation tests (S. Lory, personal communication). These results led us to construct and test the corresponding substitution in PulD. Surprisingly, the P-to-L substitution actually caused decreased multimerization of PulD in vitro (see Fig. S1 in the supplemental material) and in vivo (Fig. 7), presumably because this proline residue plays a positive role in some aspect of secretin assembly.
Despite many efforts, it has not proven possible to isolate secretins in complex with other interacting components of the T2SS except pilotin (34) and GspC (28), suggesting that other interactions are transient. Thus, these secretins are not incorporated into a stable machinery and can be assembled independently of it, which can be easily achieved by autoassembly. This is also the case for filamentous phage secretions, for which the only known partner is an inner membrane protein (16). The requirement for the Bam complex to assist the assembly of N. meningitidis PilQ (45) might represent a further example of this targeting mechanism, since the core element of this system, BamA, is found only in the outer membrane, and thus, PilQ assembly can occur only at this membrane. Preliminary studies indicated that a soluble derivative of the N. meningitidis secretin chaperone PilW did not promote PilQNm assembly in the in vitro assay, probably because the appropriate Bam complex was not present in the in vitro reaction. However, P. aeruginosa PilQ exhibits only a minimal Bam requirement (23). The situation for T3SS secretins is probably similar, for although they are an integral part of a stable multicomponent structure, they can assemble independently (9). Thus, all secretins share two major requirements: to efficiently target the OM to prevent insertion into the inner membrane, which could have deleterious consequences for the bacterium (as exemplified by secretin pIV) (2, 20, 24), and to avoid assembling into mixed multimers. Both of these challenges are met by using a variety of different targeting and assembly routes.
Clones encoding signal peptide-less variants of the Vibrio and Aeromonas T2SS secretins epsD and exeD could not be constructed, even under conditions of very tight repression, suggesting that these secretins are even more cytotoxic than PulD (21). EpsD and ExeD are closely related and belong to a subclass of T2SSs possessing GspA and GspB proteins that are thought to form a complex in the inner membrane that creates space in the peptidoglycan to allow the transport and assembly of the secretin (30, 41). One explanation for the elevated cytotoxicity might be that the EpsD and ExeD have a more open channel in their resting state than does PulD. Since GspA and GspB are required for efficient multimerization, they might also play a role in channel gating or in maintaining the OM barrier.
The ability of secretins to autoassemble in the OM might have been responsible for their impressive evolutionary success that is reflected by their presence in protein secretion, phage extrusion, bacterial appendage, and DNA uptake systems. Further studies will be required to extend the present analyses to other secretins and to study factors that permit the multimerization of secretins that are unable to autoassemble.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Ingrid Guilvout and Séverine Collin for their helpful discussions and to members of the Molecular Genetics Unit for their support. We thank Andréa Dessen for the plasmid encoding soluble PilW, Vladimir Pelicic for N. meningitidis 8013 DNA and for performing PilQNm complementation experiments, Stephen Lory for vector pHH41, for PilQPa antibodies, and for performing PilQPa complementation experiments, Marjorie Russel for PspA antibodies, Carmen Buchrieser for Legionella pneumophila strain Paris genomic DNA, Didier Mazel for Vibrio cholerae El Tor O1 genomic DNA, Tracy Palmer for Shewanella oneidensis MR-1 genomic DNA, and the Biological Resource Centre of Institut Pasteur (CRBIP) for strains of Aeromonas hydrophila subsp. hydrophila (CIP 76.14) and Xanthomonas campestris pv. campestris (CIP 74.23).
This work was supported in part by grants ANR-05-0307-01 and ANR-09-BLAN-0291. N.N.N. was supported by a postdoctoral fellowship from the Canadian Louis Pasteur Foundation during part of this work.
Footnotes
Published ahead of print 6 July 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Bayan N, Guilvout I, Pugsley AP. 2006. Secretins take shape. Mol. Microbiol. 60:1–4 [DOI] [PubMed] [Google Scholar]
- 2. Brissette JL, Russel M, Weiner L, Model P. 1990. Phage shock protein, a stress protein of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 87:862–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carbonnelle E, Helaine S, Prouvensier L, Nassif X, Pelicic V. 2005. Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occurring after pilus assembly, essential for fibre stability and function. Mol. Microbiol. 55:54–64 [DOI] [PubMed] [Google Scholar]
- 4. Chami M, et al. 2005. Structural insights into the secretin PulD and its trypsin resistant core. J. Biol. Chem. 280:37732–37741 [DOI] [PubMed] [Google Scholar]
- 5. Chen I, Provvedi R, Dubnau D. 2006. A macromolecular complex formed by a pilin-like protein in competent Bacillus subtilis. J. Biol. Chem. 281:21720–21727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clock SA, Planet PJ, Perez BA, Figurski DH. 2008. Outer membrane components of the Tad (tight adherence) secreton of Aggregatibacter actinomycetemcomitans. J. Bacteriol. 190:980–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Collin S, Guilvout I, Chami M, Pugsley AP. 2007. YaeT-independent multimerization and outer membrane association of secretin PulD. Mol. Microbiol. 64:1350–1357 [DOI] [PubMed] [Google Scholar]
- 8. Collin S, Guilvout I, Nickerson NN, Pugsley AP. 2011. Sorting of an integral outer membrane protein via the lipoprotein-specific Lol pathway and a dedicated lipoprotein pilotin. Mol. Microbiol. 80:655–665 [DOI] [PubMed] [Google Scholar]
- 9. Crago AM, Koronakis V. 1998. Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol. Microbiol. 30:47–56 [DOI] [PubMed] [Google Scholar]
- 10. Criscuolo A, Gribaldo S. 2010. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Daefler S, Guilvout I, Hardie KR, Pugsley AP, Russel M. 1997. The C-terminal domain of the secretin PulD contains the binding site for its cognate chaperone, PulS, and confers PulS dependence on pIVf1 function. Mol. Microbiol. 24:465–475 [DOI] [PubMed] [Google Scholar]
- 12. Davis BM, et al. 2000. Convergence of the secretory pathways for cholera toxin and the filamentous phage, CTXphi. Science 288:333–335 [DOI] [PubMed] [Google Scholar]
- 13. d'Enfert C, Reyss I, Wandersman C, Pugsley AP. 1989. Protein secretion by Gram-negative bacteria: characterization of two membrane proteins required for pullulanase secretion by Escherichia coli K-12. J. Biol. Chem. 264:17462–17468 [PubMed] [Google Scholar]
- 14. d'Enfert C, Ryter A, Pugsley AP. 1987. Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J. 6:3531–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng JN, Model P, Russel M. 1999. A trans-envelope protein complex needed for filamentous phage assembly and export. Mol. Microbiol. 34:745–755 [DOI] [PubMed] [Google Scholar]
- 17. Frye SA, et al. 2006. Topology of the outer-membrane secretin PilQ from Neisseria meningitidis. Microbiology 152:3751–3764 [DOI] [PubMed] [Google Scholar]
- 18. Gu S, Rehman S, Wang X, Shevchik VE, Pickersgill RW. 2012. Structural and functional insights into the pilotin-secretin complex of the type II secretion system. PLoS Pathog. 8:e1002531 doi: 10.1371/journal.ppat.1002531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guilvout I, et al. 2008. In vitro multimerization and membrane insertion of bacterial outer membrane secretin PulD. J. Mol. Biol. 382:13–23 [DOI] [PubMed] [Google Scholar]
- 20. Guilvout I, Chami M, Engel A, Pugsley AP, Bayan N. 2006. Bacterial outer membrane secretin PulD assembles and inserts into the inner membrane in the absence of its pilotin. EMBO J. 25:5241–5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guilvout I, Nickerson NN, Chami M, Pugsley AP. 2011. Multimerization-defective variants of dodecameric secretin PulD. Res. Microbiol. 162:180–190 [DOI] [PubMed] [Google Scholar]
- 22. Hardie KR, Lory S, Pugsley AP. 1996. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15:978–988 [PMC free article] [PubMed] [Google Scholar]
- 23. Hoang HH, et al. 2011. Outer membrane targeting of Pseudomonas aeruginosa proteins shows variable dependence on the components of Bam and Lol machineries. mBio 2:e00246–11 doi: 10.1128/mBio.00246-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kazmierczak BI, Mielke DL, Russel M, Model P. 1994. pIV, a filamentous phage protein that mediates phage export across the bacterial cell envelope, forms a multimer. J. Mol. Biol. 238:187–198 [DOI] [PubMed] [Google Scholar]
- 25. Kohler R, et al. 2004. Structure and assembly of the pseudopilin PulG. Mol. Microbiol. 54:647–664 [DOI] [PubMed] [Google Scholar]
- 26. Koo J, Burrows LL, Howell PL. 2012. Decoding the roles of pilotins and accessory proteins in secretin escort services. FEMS Microbiol. Lett. 328:1–12 [DOI] [PubMed] [Google Scholar]
- 27. Koo J, et al. 2008. PilF is an outer membrane lipoprotein required for multimerization and localization of the Pseudomonas aeruginosa type IV pilus secretin. J. Bacteriol. 190:6961–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Korotkov K, et al. 2011. Structural and functional studies on the interaction of GspC and GspD in the type II secretion system. PLoS Pathog. 7:e1002228 doi:10.1371/journal.ppat.1002228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Korotkov KV, Gonen T, Hol WG. 2011. Secretins: dynamic channels for protein transport across membranes. Trends Biochem. Sci. 36:433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li G, Howard SP. 2010. ExeA binds to peptidoglycan and forms a multimer for assembly of the type II secretion apparatus in Aeromonas hydrophila. Mol. Microbiol. 76:772–781 [DOI] [PubMed] [Google Scholar]
- 31. Lieberman JA, et al. 2012. Outer membrane targeting, ultrastructure, and single molecule localization of the enteropathogenic Escherichia coli type IV pilus secretin BfpB. J. Bacteriol. 194:1646–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 33. Nickerson NN, et al. 2011. Outer membrane targeting of secretin PulD protein relies on disordered domain recognition by a dedicated chaperone. J. Biol. Chem. 286:38833–38843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nouwen N, et al. 1999. Secretin PulD: association with pilot protein PulS, structure and ion-conducting channel formation. Proc. Natl. Acad. Sci. U. S. A. 96:8173–8177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nudleman E, Wall D, Kaiser D. 2006. Polar assembly of the type IV pilus secretin in Myxococcus xanthus. Mol. Microbiol. 60:16–39 [DOI] [PubMed] [Google Scholar]
- 36. Okon M, et al. 2008. Structural characterization of the type-III pilot-secretin complex from Shigella flexneri. Structure 16:1544–1554 [DOI] [PubMed] [Google Scholar]
- 37. Schuch R, Maurelli AT. 2001. MxiM and MxiJ, base elements of the Mxi-Spa type III secretion system of Shigella, interact with and stabilize the MxiD secretin in the cell envelope. J. Bacteriol. 183:6991–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seo J, Brencic A, Darwin AJ. 2009. Analysis of secretin-induced stress in Pseudomonas aeruginosa suggests prevention rather than response and identifies a novel protein involved in secretin function. J. Bacteriol. 191:898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shevchik VE, Robert-Badouy J, Condemine G. 1997. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 16:3007–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 41. Strozen TG, et al. 2011. Involvement of the GspAB complex in assembly of the type II secretion system secretin of Aeromonas and Vibrio species. J. Bacteriol. 193:2322–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tosi T, et al. 2011. Pilotin-secretin recognition in the type II secretion system of Klebsiella oxytoca. Mol. Microbiol. 82:1422–1432 [DOI] [PubMed] [Google Scholar]
- 43. Trindade MB, Job V, Contreras-Martel C, Pelicic V, Dessen A. 2008. Structure of a widely conserved type IV pilus biogenesis factor that affects the stability of secretin multimers. J. Mol. Biol. 378:1031–1039 [DOI] [PubMed] [Google Scholar]
- 44. Viarre V, et al. 2009. HxcQ liposecretin is self-piloted to the outer membrane by its N-terminal lipid anchor. J. Biol. Chem. 284:33815–33823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299:262–265 [DOI] [PubMed] [Google Scholar]
- 46. Wang X, et al. 2012. Cysteine scanning mutagenesis and disulfide mapping analysis of the arrangement of GspC and GspD protomers within the T2SS. J. Biol. Chem. 287:19082–19093 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.