Abstract
Genetic screens of the collection of ∼4500 deletion mutants in Saccharomyces cerevisiae have identified the cohort of nonessential genes that promote maintenance of genome integrity. Here we probe the role of essential genes needed for genome stability. To this end, we screened 217 tetracycline-regulated promoter alleles of essential genes and identified 47 genes whose depletion results in spontaneous DNA damage. We further showed that 92 of these 217 essential genes have a role in suppressing chromosome rearrangements. We identified a core set of 15 genes involved in DNA replication that are critical in preventing both spontaneous DNA damage and genome rearrangements. Mapping, classification, and analysis of rearrangement breakpoints indicated that yeast fragile sites, Ty retrotransposons, tRNA genes, early origins of replication, and replication termination sites are common features at breakpoints when essential replication genes that suppress chromosome rearrangements are downregulated. We propose mechanisms by which depletion of essential replication proteins can lead to double-stranded DNA breaks near these features, which are subsequently repaired by homologous recombination at repeated elements.
Keywords: DNA replication, essential gene, chromosome rearrangement, Ty retrotransposon, genome stability
ACCURATE transmission of the genome is essential for normal cell growth and survival. As such, cells have developed elaborate mechanisms to prevent errors in replication and to respond to spontaneous DNA damage that can lead to genomic instability (Kolodner et al. 2002; Branzei and Foiani 2007, 2009, 2010; Harper and Elledge 2007; Cimprich and Cortez 2008). The failure to repair the genome in an error-free manner can result in chromosome abnormalities that underlie many human diseases, including cancers (Kolodner et al. 2002; McKinnon and Caldecott 2007; Aguilera and Gomez-Gonzalez 2008). Therefore, defining the genes that contribute to genome maintenance will be useful in understanding disease development and in designing new strategies for therapeutics. However, to date, a comprehensive curation of genes that function to suppress genome instability is incomplete.
Yeast is an ideal model for genomic studies due to the conservation of gene functions and biological pathways between yeast and humans. Phenotypic screens conducted with the Saccharomyces cerevisiae nonessential gene deletion collection (Giaever et al. 2002) have aided in the annotation and functional characterization of nonessential genes involved in the suppression of spontaneous DNA damage (Huang et al. 2003; Huang and Kolodner 2005; Shor et al. 2005; Alvaro et al. 2007) and in the suppression of spontaneous chromosome rearrangements (Smith et al. 2004; Yuen et al. 2007; Andersen et al. 2008). However, since the deletion of essential genes causes lethality, similar genome-wide screening approaches to identify the complete set of genes that suppress spontaneous DNA damage and chromosome rearrangements require collections of conditional alleles of essential genes.
Systematic collections of conditional alleles have been generated in several ways, including the replacement of native promoters with a tetracycline-regulated promoter (Mnaimneh et al. 2004; Yu et al. 2006), destabilization of target gene mRNAs through the insertion of a selectable marker in the 3′-UTR of essential genes (Schuldiner et al. 2005), systematic addition of a heat-inducible degron to the amino terminus of the protein product (Labib et al. 2000), systematic generation of novel temperature-sensitive alleles (Ben-Aroya et al. 2008), and systematic integration of existing temperature-sensitive alleles (Li et al. 2011). Despite the availability of several essential gene collections, no one collection is complete, suggesting that complementary approaches using a number of screening strategies and multiple types of conditional alleles will be necessary to identify all of the essential genes that function to suppress genomic instability.
Here we describe a series of screens to identify essential genes that function to suppress genome instability, using the collection of tetracycline-regulated promoter replacement alleles (Tet alleles) of essential genes (Mnaimneh et al. 2004). We screened 217 Tet alleles of essential genes whose depletion caused accumulation in S or G2 phases of the cell cycle (Yu et al. 2006) and identified 47 with elevated levels of spontaneous DNA damage. A second screen performed with the same Tet alleles identified 92 essential genes that suppress the formation of chromosome rearrangements, whole chromosome deletions, and gene conversions. We quantified the levels of each type of mutation in 15 strains that exhibited both elevated levels of spontaneous DNA damage and chromosome rearrangements following the depletion of an essential gene. Mapping of rearrangement breakpoints in seven representative mutants from this set revealed several unique rearrangement structures. Sequence features, including Ty retrotransposons and DNA replication origins and termination zones, correlated with the rearrangements identified. We propose a central role for DNA replication proteins in suppressing the formation of chromosome breaks that promote chromosome rearrangements.
Materials and Methods
Yeast strains and media
Tet allele strains were constructed as described previously (Mnaimneh et al. 2004). The genotype of the wild-type Tet allele strain, R1158, is MATa URA3::CMV-tTA his3Δ1 leu2Δ0 met15Δ0. Using standard genetic methods, 217 MATα Tet allele strains were engineered to contain YFP-Ddc2 marked with a nourseothricin (Nat) resistance gene. Genotypes for strains used in this study are listed in Table S6. The essential genes that were studied are listed in Table S1 and Table S2. Standard yeast media and growth conditions were used unless otherwise specified (Sherman 1991).
Fluorescence microscopy
Tet allele strains were grown in YPD liquid media at 30°. Samples were divided into two cultures and grown in parallel in the presence and absence of 10 μg/ml doxycycline for 4 additional hours at 23°. Intracellular localization of Ddc2-YFP was determined by fluorescence microscopy as previously described for Rad52-YFP (Lisby et al. 2004; Lisby and Rothstein 2004; Chang et al. 2005). Ddc2 foci were quantified in at least 100 cells for each strain. Ddc2 foci in wild-type cells were analyzed four times and used to calculate a standard deviation. Tet allele strains that had Ddc2 foci levels that were at least three standard deviations greater than wild type were scored as positive.
Illegitimate mating assays
Tet allele strains and the R1158 wild-type strain were grown in parallel for 24 hr on YPD solid media either containing or lacking 10 μg/ml of doxycycline. A standard mating assay was performed with tester strains MCY13 (MATα , legitimate mating) and MCY14 (MATa, illegitimate mating) on the same media conditions that the strains were grown. Diploids were isolated by replica plating on minimal media.
In the quantitative form of this mating assay, Tet allele strains and R1158 wild-type strain were grown in parallel for 24 hr in YPD liquid media containing or lacking 10 μg/ml doxycycline. Strains were mixed with fivefold excess of MCY13, MCY14, or 1225α (MATα his4thr4) tester strains and plated on YPD solid media. After 5 hr, cells were collected, resuspended in water, and plated on diploid selection media. Independent illegitimate diploids were isolated after the mating of the Tet allele strains with the 1225α tester strain. For each mating experiment, ∼100 diploids were isolated and tested for their ability to grow in the presence or absence of histidine or threonine. This assay was repeated two times. Viability of each strain following growth in doxycycline was confirmed by plating on YPD. Only MCM7 (10%), NUF2 (30%), and UBC9 (50%) had <100% viability following growth in doxycycline.
Array comparative genome hybridization
Genomic DNA was extracted (Qiagen) from independent illegitimate diploids and wild-type diploids isolated from the mating assay. CGH on a microarray was performed as previously described (Dion and Brown 2009) using S. cerevisiae whole genome tiling microarrays (Affymetrix). Signal intensities of the experimental and wild-type control samples were normalized and compared using tiling analysis software (Affymetrix). Genomic patterns were mapped and analyzed using the integrated genome browser software (Affymetrix).
CHEF gel electrophoresis and Southern blot analysis
Contour-clamped homogenous electric field (CHEF) gels were used to examine intact chromosomes of illegitimate diploids isolated from the mating assay. CHEF gel analysis was performed as described previously (Desany et al. 1998). A 1.2% agarose gel was run at 8 V/cm using pulse times of 120 sec for 30 hr at 14° in 0.5× TBE buffer. PCR-purified fragments were radio labeled by random priming (Stratagene) and used as hybridization probes for Southern blot analysis. PCR primers designed for probe construction are listed in Table S7.
Restriction digestion and sequencing analysis of FS1 and FS2
Genomic DNA was isolated (Qiagen) from wild-type strains R1158 and BY4741 and digested with EcoRI and XbaI (New England Biolabs) using the suggested conditions. Digested fragments were separated on a 1% agarose gel and hybridized with FEN2 and FS2-2 probes for Southern blot analysis (Table S7). 5′ and 3′ ends of fragile site 1 (FS1) and FS2 were PCR amplified and sequenced. PCR primers used for both amplification and sequencing are listed in Table S8.
Enrichment analyses
S. cerevisiae chromosomes were broken into 5-kb bins. For each bin, the presence or absence of breakpoints and genomic features was tabulated. Various genomic features (Di Rienzi et al. 2009) and replication termination sites (Fachinetti et al. 2010) from previous datasets were used for analysis. For each feature, the total number of bins with both the feature and a breakpoint was determined. To test for enrichment of breakpoints and each feature, a hypergeometric distribution was assumed. P-values <0.05 were considered as evidence of a correlation and P-values <0.05 after a false discovery rate (FDR) correction were considered strongly significant.
Results
Depletion of essential gene products causes spontaneous DNA damage
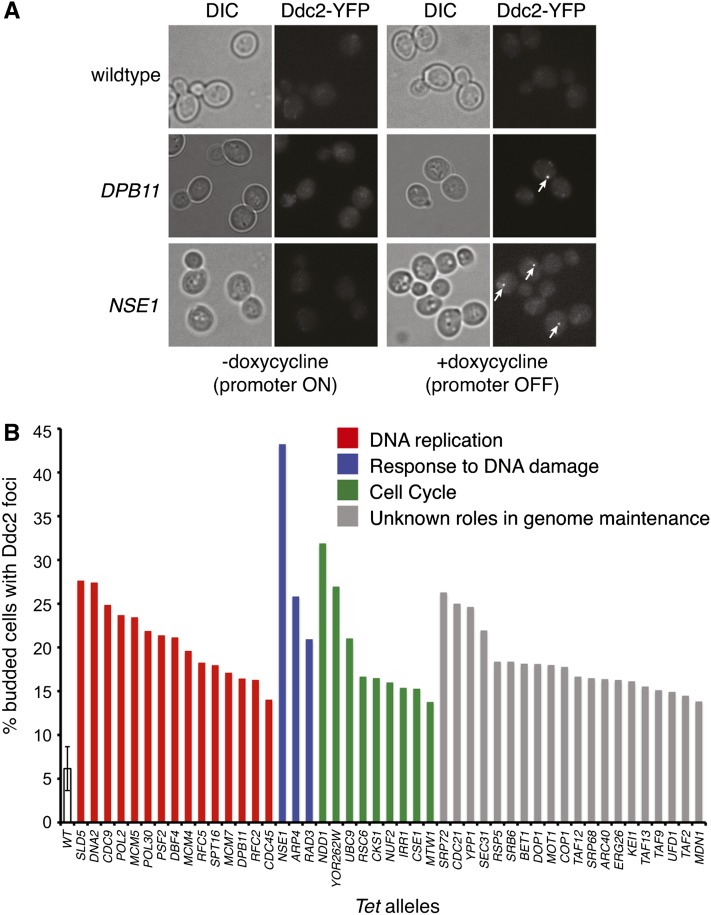
We used a collection of tetracycline-regulated promoter alleles (Tet alleles) (Mnaimneh et al. 2004; Yu et al. 2006) of essential genes to systematically identify genes that suppress spontaneous DNA damage. Since elevated levels of spontaneous DNA damage should elicit a checkpoint response and cause cell cycle delay, we screened the 217 strains that accumulated in S phase or G2 phase of the cell cycle following gene-product depletion by promoter shut off (Yu et al. 2006). Spontaneous DNA damage was measured by the relocalization of the DNA damage checkpoint protein Ddc2 from a diffuse nuclear pattern to discrete subnuclear foci (Figure 1A) (Melo et al. 2001; Lisby et al. 2004). Following growth of these strains in doxycycline to repress essential gene expression, the fraction of cells with Ddc2 foci was quantified (Supporting Information, Table S1). We determined that the individual depletion of 47 essential gene products caused an increase of Ddc2 foci relative to wild-type levels, using a cutoff of three standard deviations from the wild-type mean (Figure 1B). The gene ontology (GO) processes of the essential genes that were identified are varied (Table S2), but on average the highest levels of Ddc2 foci were observed following the depletion of gene products involved in DNA replication, response to DNA damage stimuli, and cell cycle progression, indicating the importance of these essential processes in the maintenance of genome integrity (Figure 1B). In addition to the identification of essential genes with defined roles in genome maintenance, 20 essential genes with previously unrecognized contributions to the suppression of spontaneous DNA damage were also identified (Figure 1B, gray bars).
Figure 1 .
Depletion of yeast essential genes results in elevated levels of spontaneous Ddc2 foci formation. (A) A total of 217 Tet alleles that express Ddc2-YFP and display a G2/M or S phase cell cycle arrest phenotype were grown in the presence of doxycycline (10 μg/ml) for 4 hr to inhibit the transcription of each essential gene. Representative DIC and YFP images are shown for the wild-type, DPB11 and NSE1 strains. Ddc2-YFP foci are indicated with white arrows. (B) The percentage of cells with Ddc2-YFP foci is plotted for 47 Tet alleles that showed an increase in Ddc2 foci of at least three standard deviations above the average observed in wild type. Bars are shaded according to the GO process annotation of each gene of interest.
Depletion of essential gene products causes chromosome loss and rearrangement
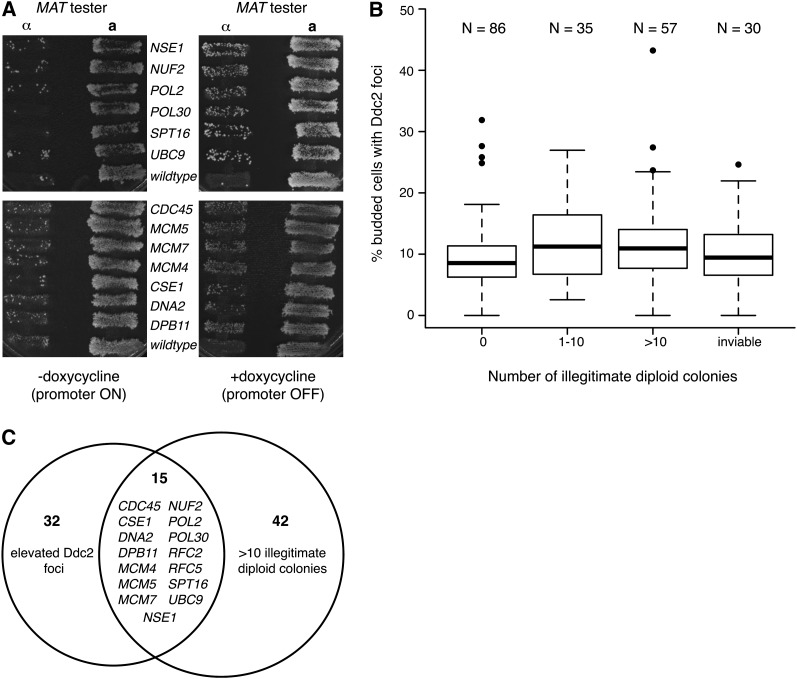
Increased levels of Ddc2 foci could reflect increased spontaneous DNA damage, defective repair of spontaneous DNA damage, or a combination of both. An increase in spontaneous DNA damage may not impact genome integrity if the damage is repaired accurately. To directly identify essential genes that suppress chromosome rearrangements and chromosome loss, we used an illegitimate mating assay (Strathern et al. 1981; Lemoine et al. 2005, 2008) that measures loss of genetic information from chromosome III to screen the same 217 Tet allele strains. Mutation or deletion of the MATα locus on chromosome III in haploid cells results in a reversion to the default MATa mating type, termed a-like fakers, allowing these MATα cells to mate illegitimately with strains of the MATα mating type (Strathern et al. 1981). We determined the levels of a-like faker formation using a patch mating assay (Figure 2A). We found that the depletion of 92 essential genes caused elevated illegitimate mating frequencies both relative to the minus doxycycline control and relative to the wild-type control, indicating loss of genetic information at the MAT locus in these strains (Table S3). Thirty strains did not form colonies in the presence of doxycycline and 9 strains could not be constructed with the MATα mating type and therefore could not be evaluated. Strains were subcategorized into groups with high (>10 colonies; 57 strains), moderate (1–10 colonies; 35 strains), or wild-type (0 colonies; 86 strains) levels of illegitimate mating and the distributions of Ddc2 foci formation for each category were compared (Figure 2B). Both the high and medium categories had greater Ddc2 foci formation when compared to the wild-type category (P-value of 0.022 for high vs. wild type and P-value of 0.028 for medium vs. wild type; one-sided Mann–Whitney test), indicating a relationship between the extents of Ddc2 focus formation and the illegitimate mating phenotype. Additionally, strains with spontaneous Ddc2 foci formation above our cutoff were more likely to have increased illegitimate mating (P-value of 0.00073; hypergeometric test), although the overlap between the two screens, at 29 genes, was not absolute.
Figure 2 .
Depletion of yeast essential genes results in elevated levels of illegitimate mating. (A) MATα Tet alleles were grown on YPD or YPD containing doxycycline (10 μg/ml) for 24 hr and a standard mating test was performed using MATα and MATa tester strains. Representative images of strains with elevated levels of illegitimate diploid formation following growth in doxycycline are shown. (B) The resulting number of illegitimate diploid colonies that grew without doxycycline treatment was subtracted from the number that grew with doxycycline treatment and was used to subcategorize the strains into four groups. For each group, the distribution of percentage of budded cells with Ddc2-YFP foci was plotted. Bold lines represent the median values, boxes represent the upper and lower quartiles, whiskers represent 1.5 times the interquartile range, and outliers are indicated by circles. (C) Comparison of Tet alleles with elevated levels of Ddc2 foci and >10 illegitimate mating diploid colonies.
We focused on the strains with the most robust chromosome instability phenotype, the 15 strains with both elevated Ddc2 foci and high levels of illegitimate mating (Figure 2C). These strains were subjected to a quantitative illegitimate mating assay (Table 1). In the presence of doxycycline, all of these strains exhibited significantly elevated levels of illegitimate mating relative to the wild-type strain. Increases in illegitimate mating ranged from <2-fold wild type (CSE1) to 62-fold wild type (MCM7). Previous studies of GAL promoter-regulated conditional alleles of DNA polymerases α and δ found increases of ∼200-fold and 50-fold, respectively (Lemoine et al. 2005, 2008). Although DNA polymerases α and δ were not assayed in our screens, we identified a role for DNA polymerase ε (POL2 and DPB11) in suppressing illegitimate mating. Additionally, we found that disrupting a wide range of replication functions (CDC45, MCM4, MCM5, MCM7, DPB11, POL2, POL30, RFC2, and RFC5) caused increased illegitimate mating. DNA2, which functions in Okazaki fragment processing (Budd et al. 2000; Lee et al. 2000) and in DNA repair (Zhu et al. 2008) resulted in increased illegitimate mating, as did repression of the DNA repair genes NSE1 (Santa Maria et al. 2007; Pebernard et al. 2008) and UBC9 (Branzei et al. 2006). Genes with functions outside of DNA replication and repair were also identified. CSE1 is responsible for nuclear shuttling of the nuclear transporter importin α (Hood and Silver 1998; Kunzler and Hurt 1998; Solsbacher et al. 1998), and roles for CSE1 in DNA replication (Yu et al. 2006) and in chromosome segregation (Xiao et al. 1993), likely reflecting effects on importin α cargos, have been described. NUF2 is a kinetochore component and functions in chromosome segregation (Osborne et al. 1994). Depletion of DNA replication (CDC45, MCM4, MCM7, and POL2) and segregation (NUF2) gene products had the most striking effect (>20-fold difference).
Table 1. Frequencies of illegitimate mating in tetracycline-regulatable promoter conditional alleles grown in the presence and absence of doxycycline.
| Frequencies of illegitimate mating (10−4) |
||||||
|---|---|---|---|---|---|---|
| Tet allele | −doxycycline |
+doxycycline |
||||
| Wild type | 0.83 | (0.20) | [1] | 0.8 | (0.26) | [1] |
| CDC45 | 11 | (6.0) | [14] | 17 | (1.6) | [22] |
| CSE1 | 0.5 | (0.04) | [0.6] | 1.4 | (0.40) | [1.8] |
| DNA2 | 4.3 | (2.7) | [5.2] | 4.7 | (2.5) | [5.9] |
| DPB11 | 0.94 | (0.1) | [1.1] | 9.2 | (3.4) | [12] |
| MCM4 | 11 | (9.8) | [13] | 36 | (27) | [45] |
| MCM5 | 3.4 | (0.8) | [4.1] | 12 | (3.0) | [15] |
| MCM7 | 4.1 | (2.9) | [4.9] | 34 | (24) | [62] |
| NSE1 | 17 | (11) | [21] | 39 | (7.3) | [48] |
| NUF2 | 2.5 | (1.1) | [3.0] | 18 | (8.9) | [22] |
| POL2 | 2.2 | (0.9) | [2.6] | 26 | (21) | [33] |
| POL30 | 5.4 | (4.9) | [6.5] | 8.3 | (2.7) | [10] |
| RFC2 | 2.2 | (0.03) | [2.6] | 5.9 | (0.14) | [7.4] |
| RFC5 | 1.5 | (0.76) | [1.8] | 3.0 | (0.64) | [3.8] |
| SPT16 | 2.4 | (1.7) | [2.9] | 11 | (11) | [14] |
| UBC9 | 2.4 | (0.4) | [2.9] | 3.9 | (2.2) | [4.8] |
Tet allele strains and wild-type strain were grown in parallel for 24 hr on YPD liquid media containing or lacking 10 μg/ml doxycycline. Strains were mixed with fivefold excess of a MATα tester strain and plated on YPD solid media. After 5 hr, cells were resuspended in water and plated on illegitimate diploid selection media. This assay was repeated two times. Numbers in parentheses represent standard deviations. Numbers in brackets represent the frequency normalized to wild type.
Chromosome III rearrangements in essential genome stability mutants
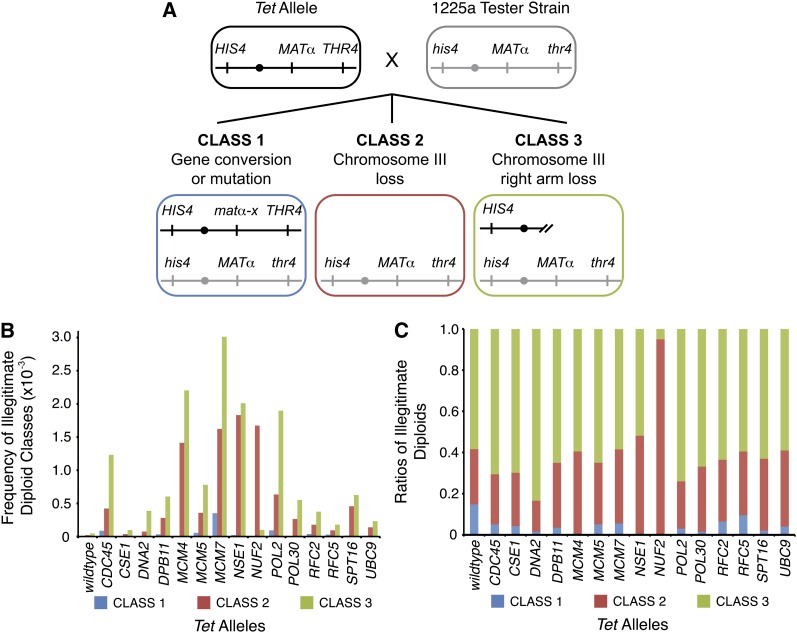
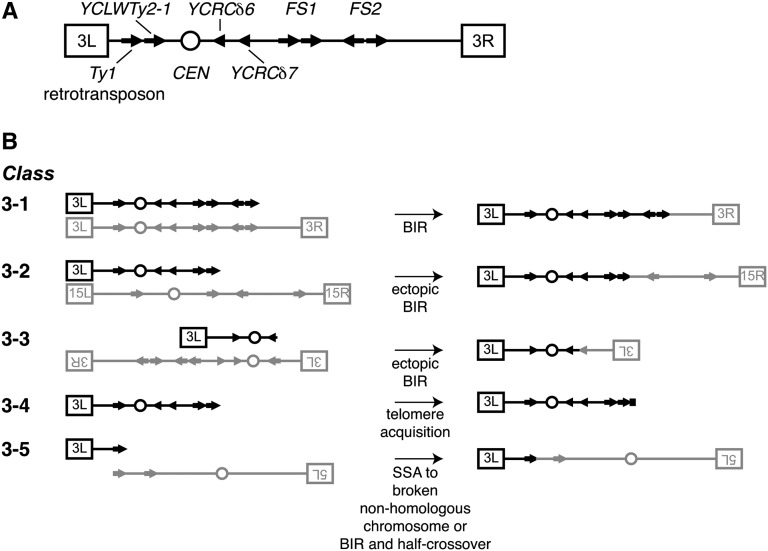
Three common classes of information loss on chromosome III in the illegitimate mating assay can be distinguished by performing the assay with a strain with nutritional markers flanking the MAT locus on chromosome III (Lemoine et al. 2005) (Figure 3A). We used this modified assay to classify chromosome instability events in the 15 strains with both increased spontaneous DNA damage and high levels of illegitimate mating. Class 1 mating events result from a gene conversion or mutation at the MATα locus. Class 2 events result from the loss of chromosome III. Class 3 events result from chromosome rearrangements that lead to the loss of the MATα locus and distal regions of the right arm of chromosome III (Figure 3A). For each strain we classified the chromosome rearrangements in ∼200 illegitimate diploids and measured the frequencies and ratios of the three classes (Figure 3, B and C).
Figure 3 .
Classification of rearrangement events that lead to illegitimate mating. (A) Schematic diagram of the three expected classes of genetic events resulting in illegitimate mating. Using diagnostic selection media, mutations in the MAT locus, whole chromosome III loss, and loss of the right arm of chromosome III can be distinguished as class 1, 2, and 3 genetic events, respectively (Lemoine et al. 2005, 2008). (B) We classified ∼200 illegitimate diploids for each strain. The frequencies of the three classes of rearrangements are plotted for the 15 strains with the most elevated levels of illegitimate mating. (C) Ratios of the three classes of rearrangements are plotted for the indicated strains.
Increases in class 2 (whole chromosome loss) and 3 (chromosome arm loss) rearrangements were evident for all 15 genes tested (Figure 3B). Depletion of CSE1, DPB11, MCM4, MCM5, POL30, SPT16, and UBC9 resulted in ratios of the three classes of chromosome instability that were not significantly different than that observed in the wild-type strain (P-value >0.01 by the Fisher exact test) (Figure 3C). This could indicate that depletion of these gene products exacerbates a condition already present in wild-type cells. By contrast, repression of CDC45, DNA2, MCM7, RFC2, RFC5, and POL2 resulted in a significant difference in ratios of the three classes relative to wild type (P-value <0.01 by the Fisher exact test) with a preferential increase in class 3 (chromosome arm loss) events. Depletion of NUF2, a kinetochore-associated protein, resulted in a dramatic increase in class 2 (whole chromosome loss) events, consistent with the function of this gene in chromosome segregation (Osborne et al. 1994; Wigge and Kilmartin 2001). Finally, we observed that the depletion of NSE1, a subunit of the structural maintenance of chromosome (Smc5/6) complex (Fujioka et al. 2002), resulted in the loss of class 1 (gene conversion or point mutation) events and in similar levels of class 2 (whole chromosome loss) and class 3 (chromosome arm loss) events. Our data suggest that NSE1 contributes to both the DNA repair (class 3) and chromosome segregation (class 2) functions of the SMC5/6 complex (Santa Maria et al. 2007; Behlke-Steinert et al. 2009; Irmisch et al. 2009; Outwin et al. 2009). We conclude that depletion of different essential gene functions can cause different patterns of genomic instability.
Essential gene product depletion causes genome rearrangements with boundaries at Ty retrotransposons
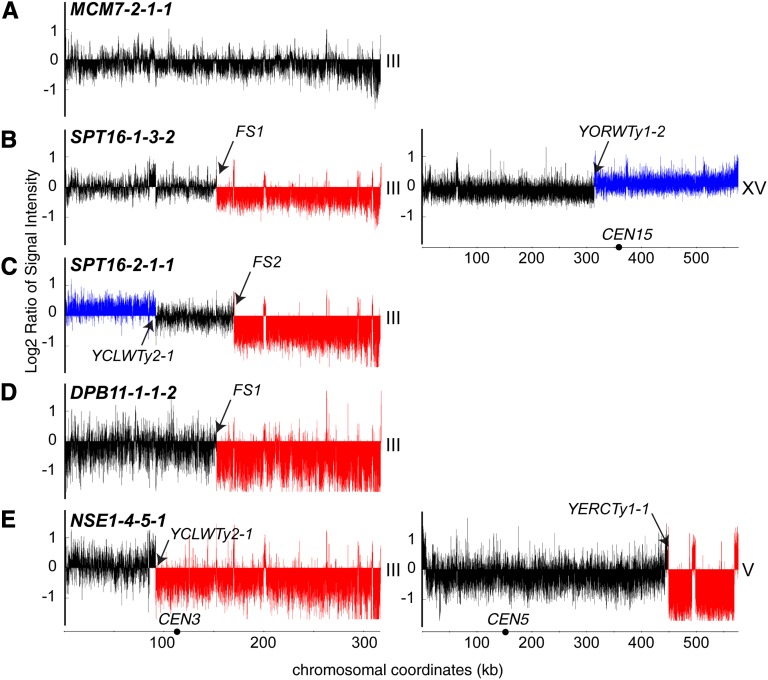
To obtain a comprehensive assessment of the chromosome rearrangement breakpoint locations in the illegitimate diploids that were isolated following essential gene depletion in our classification experiment, we used comparative genome hybridization on tiling microarrays to map rearrangement breakpoints (Dion and Brown 2009). Genomic DNA was isolated from six illegitimate diploid colonies that exhibited a chromosome III arm loss (class 3) phenotype following the depletion of each of seven essential genes (CDC45, DPB11, MCM7, NSE1, RFC2, SPT16, and UBC9), chosen to represent the functional diversity present in the quantitative illegitimate mating assay. Genomic DNA was hybridized to a S. cerevisiae whole genome tiling microarray and copy number variation was determined by comparison to genomic DNA isolated from a wild-type a/α diploid. Representative copy number profiles are depicted in Figure 4 and the boundaries of each rearrangement are shown in Table 2. Figure 5, A and B summarizes the breakpoints observed on chromosome III and the resulting subclasses of rearrangements that were observed.
Figure4 .
Comparative genome hybridization microarray analysis of class 3 illegitimate diploids. Genomic DNA was isolated from class 3 (chromosome III arm loss) illegitimate diploids and hybridized to a Saccharomyces cerevisiae whole genome tiling microarray to identify copy number variations. In each histogram, the y-axis represents log2 ratios of probe signal intensities, comparing the indicated strain to a legitimate MATa/α diploid, and the x-axis represents chromosome coordinates. Black arrows indicate breakpoint locations on each chromosome, black circles represent the locations of centromeres, and the chromosome number is indicated to the right of each histogram. A representative histogram for each of the major types of rearrangements observed is shown. (A) Class 3-1 diploid, in which no copy number variation of chromosome III was evident. (B) Class 3-2 diploids have a loss of sequence (red) from the right arm of chromosome III and duplication of sequences (blue) from chromosome XV. (C) Class 3-3 diploids have an amplification of the left arm sequence and a deletion of the right arm sequence of chromosome III. (D) Class 3-4 diploids have a loss of sequence from the right arm of chromosome III without copy number variation on nonhomologous chromosomes. (E) Class 3-5 diploids exhibit a loss of sequence from the right arm of chromosome III and loss of sequence from the right arm of chromosome V.
Table 2. Classification of class 3 illegitimate diploid chromosome rearrangements.
| Classa | Strain | Essential gene depleted | Type of chromosome rearrangement | Chromosomes involved | Size of altered chromosome (kb) | Boundaries of chromosome rearrangements |
|---|---|---|---|---|---|---|
| 3A(i)b | MCM7-2-57-1 | MCM7 | Translocation | III/VII | 690 | FS2; |
| YGRCTy1-2, YGRCTy2-1 | ||||||
| 3A(i) | RFC2-1-1-1 | RFC2 | Translocation | III/VII | 710 | FS1; YGRWTy1-1 |
| 3A(i) | RFC2-1-1-2 | RFC2 | Translocation | III/VII | 710 | FS1; YGRWTy1-1 |
| 3A(i)b | SPT16-1-3-2 | SPT16 | Translocation | III/XV | 640 | FS1; YORWTy1-2 |
| 3A(ii)b | CDC45-4-4-1 | CDC45 | Ectopic BIR | III/III | 120 | YCLWTy2-1; |
| YCRCδ6 | ||||||
| 3A(ii) | CDC45-4-52-1 | CDC45 | Ectopic BIR | III/III | 140 | YCLWTy2-1; |
| YCRCδ7 | ||||||
| 3A(ii)b | MCM7-6-1-1 | MCM7 | Ectopic BIR | III/III | 140 | YCLWTy2-1; |
| YCRCδ7 | ||||||
| 3A(ii) | RFC2-2-56-1 | RFC2 | Ectopic BIR | III/III | 140 | YCLWTy2-1; |
| YCRCδ7 | ||||||
| 3A(ii) | RFC2-2-6-1 | RFC2 | Ectopic BIR | III/III | 120 | YCLWTy2-1; |
| YCRCδ6 | ||||||
| 3A(ii) | RFC2-2-6-2 | RFC2 | Ectopic BIR | III/III | 120 | YCLWTy2-1; |
| YCRCδ6 | ||||||
| 3A(ii) | SPT16-1-55-1 | SPT16 | Ectopic BIR | III/III | 120 | YCLWTy2-1; |
| YCRCδ6 | ||||||
| 3A(ii)b | SPT16-2-1-1 | SPT16 | Ectopic BIR and interstitial deletion | III/III and V | 170 (chrIII) 520 (chrV) | YCLWTy2-1; FS2 (chrIII) and YERCTy1-1; YERCTy1-2 (chrV) |
| 3Bb | DPB11-1-1-2 | DPB11 | Arm deletion | III | 150 | FS1 |
| 3B | MCM7-4-1-2 | MCM7 | Arm deletion | III | 170 | FS2 |
| 3B | SPT16-2-51-1 | SPT16 | Arm deletion | III | 120 | YCRCδ6 |
| 3B | SPT16-2-51-2 | SPT16 | Arm deletion | III | 120 | YCRCδ6 |
| 3Db | RFC2-1-52-1 | RFC2 | Hawthorne deletion | III | 220 | MATα, HMR |
| 3Fb | DPB11-2-56-1 | DPB11 | Chromosome fusion | III/XVI | 970 | YCLWTy2-1; |
| YPLWTy1-1 | ||||||
| 3Fb | NSE1-4-5-1 | NSE1 | Chromosome fusion | III/V | 530 | YCLWTy2-1; YERCTy1-1 |
| 3F | NSE1-6-51-1 | NSE1 | Chromosome fusion | III/V | 580 | YCLWTy2-1; YERCTy1-2 |
| 3Fb | UBC9-1-51-1 | UBC9 | Chromosome fusion | III/XVI | 930 | YCLWTy2-1; YPRWTy1-3, YPRCTy1-4 |
| 3F | UBC9-1-51-2 | UBC9 | Chromosome fusion | III/XVI | 930 | YCLWTy2-1; YPRWTy1-3, YPRCTy1-4 |
| 3F | UBC9-1-51-3 | UBC9 | Chromosome fusion | III/XVI | 930 | YCLWTy2-1; YPRWTy1-3, YPRCTy1-4 |
All strains were examined by comparative genome hybridization on a microarray.
Genome rearrangements predicted from microarray analysis were confirmed by Southern blot analysis.
Figure 5 .
Mechanisms of repair in replication deficient mutants. (A) Schematic of boundaries of rearrangement that occur on chromosome III of replication mutants are shown. (B) Following a double stranded break and resection to Ty retrotransposons (arrows) or δ long terminal repeats (triangles), chromosome fragments can be repaired in illegitimate diploids through several mechanisms. Class 3-1: Chromosome III is repaired by BIR using the homologous chromosome of the tester strain. Class 3-2: Ectopic break-induced replication (BIR) mediated by strand invasion at a Ty retrotransposon on a nonhomologous chromosome XV of the tester strain (gray) yields nonreciprocal translocations. Class 3-3: Ectopic BIR involving strand invasion at a different locus of chromosome III results in shortened fragments of chromosome III with two left arms. Class 3-4: Chromosome III fragments are directly repaired by telomere acquisition. Class 3-5: Chromosome fusions can be created through single stranded annealing (SSA) of chromosome III and chromosome V fragments with boundaries at Ty retrotransposons or through a BIR and half-crossover event.
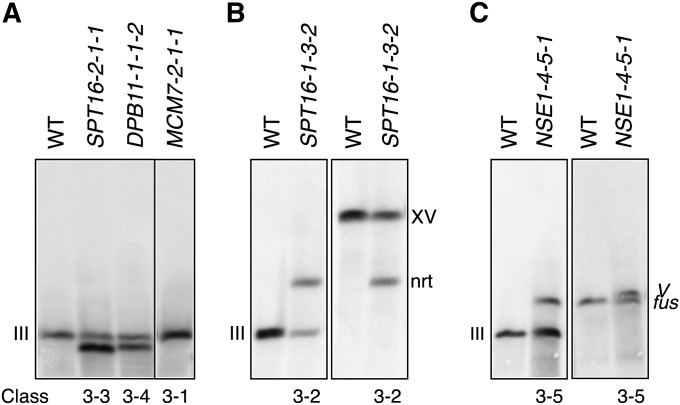
Four of the 42 class 3 illegitimate diploids that we tested exhibited poor hybridization profiles and were not analyzed further. The remaining samples were divided into five different subclasses on the basis of the rearrangement profiles of chromosome III. The most frequent subclass, class 3-1, comprised microarray profiles that lacked deletions or duplications (Figure 4A). This type of profile was observed in 15 of 38 (39%) illegitimate diploids and was present following the depletion of all essential genes tested, with the exception of RFC2 and SPT16. As previously suggested, this genome profile likely represents successful repair of chromosome III by break-induced replication (BIR) using the 1225α strain chromosome III as a template (Lemoine et al. 2005, 2008), resulting in full restoration of chromosome III sequences (Figure 5B). Southern blot analysis following separation of chromosomes on a contour-clamped homogeneous electric field (CHEF) gel, using a probe specific for the HIS4 locus on the left arm of chromosome III, revealed a single DNA band corresponding to the size of chromosome III, confirming that class 3C diploids have two intact copies of chromosome III (Figure 6A, lane 4).
Figure 6 .
Southern blot confirmation of chromosome rearrangements predicted in microarray analysis. (A) Intact chromosomal DNA was isolated from class 3 (chromosome arm loss) illegitimate diploids. Genomic DNA was separated on a contour-clamped homogenous electric field (CHEF) gel and chromosome III was detected through hybridization with a radio-labeled probe specific to the left arm of chromosome III. Smaller chromosome III fragments were also detected in samples with chromosome rearrangements. Representative Southern blots for a wild-type diploid, classes 3-3, 3-4, and 3-1 diploids of the indicated strains are shown, respectively. (B) Representative Southern blots of a wild-type and a class 3-2 illegitimate diploid. Chromosomes on the left and right were detected with probes for chromosomes III and XV, respectively. In addition to chromosome III and XV, a nonreciprocal translocation (nrt) was visualized. (C) Representative Southern blot of a wild-type and a class 3-5 illegitimate diploid. Chromosomes on the left and right were detected with probes for chromosome III and V, respectively. In addition to chromosomes III and V, a chromosome fusion (fus) event was detected.
Class 3-2 diploids had both a deletion of sequences from the right arm of chromosome III and a duplication of sequences from a non-homologous chromosome, suggesting the presence of a nonreciprocal chromosome translocation (Figures 4B and 5B). Illegitimate diploids isolated following the depletion of MCM7, RFC2, and SPT16 displayed this type of rearrangement. As previously described following depletion of DNA polymerase α or δ (Lemoine et al. 2005, 2008), this class had breakpoints either at FS1 (Fragile Site 1) or FS2 of chromosome III and at Ty1 retrotransposons of a nonhomologous chromosome (chromosome VII or XV, in our case). Previous sequencing analyses and restriction mapping of FS1 and FS2 regions in several strain backgrounds, including the S288C strain background that we used in our study, indicated the presence of a direct pair of tandem Ty1 retrotransposons and an inverted pair of Ty1 retrotransposons, respectively, that are unannotated in the Saccharomyces Genome Database (SGD) (Umezu et al. 2002; Lemoine et al. 2005, 2008; Hoang et al. 2010). We confirmed that this same arrangement of Ty1 retrotransposons at FS1 and FS2 is present in the wild-type Tet allele strain using PCR analyses (Figure S1A). We further verified the arrangement of Ty1 retrotransposons at FS2 using Southern blot analyses (Figure S1B). In keeping with the mechanism proposed by Lemoine et al. (2005, 2008), we predict that class 3-2 represents ectopic BIR with strand invasion at a Ty1 retrotransposon element of a nonhomologous chromosome (Figure 5B). Southern blot analysis of a representative SPT16 illegitimate diploid revealed one band consistent with the size of chromosome III as well as a second band consistent with the predicted size of a nonreciprocal translocation between chromosome III and chromosome XV (Figure 6B, lane 2). Hybridization of the same blot with a chromosome XV probe confirmed the presence of a chromosome containing sequences from both chromosome III and chromosome XV (Figure 6B, lane 4).
Class 3-3 illegitimate diploids had both an amplification of sequences proximal to the Ty2 retrotransposon, YCLWTy2-1, and a deletion of right arm sequences distal to FS2 or δ elements YCRCδ6 or YCRCδ7 of chromosome III (Figures 4C and 5B). Although class 3-2 was the most common class of rearrangement following depletion of DNA polymerase α or δ (Lemoine et al. 2005, 2008), class 3-3 was more common in our study. Sequencing of chromosome III in the S288C strain background used in our study indicates a Ty1 retrotransposon insertion directly upstream of YCLWTy2-1 in the forward orientation that is unannotated in the SGD, resulting in a configuration similar to FS1 (Hoang et al. 2010). This was the second most abundant profile and was observed in eight samples, following the depletion of CDC45, MCM7, RFC2, or SPT16. One model for this rearrangement (Figure 5B) is that it results from a BIR event with inaccurate strand invasion at the Ty retrotransposons on the left arm of chromosome III, which have homology to the Ty retrotransposons at the breakpoints on the right arm of chromosome III. The high frequency of this type of rearrangement could reflect the abundance and orientation of long terminal repeats (LTRs) and Ty retrotransposons on chromosome III or the spatial proximity of the relevant Ty retrotransposons and LTRs within the nucleus (Duan et al. 2010). Southern blot analysis using a probe within the amplified region of chromosome III resulted in the detection of two distinct chromosome sizes (Figure 6A, lane 2). One corresponds to the expected size of an intact chromosome III contributed by the tester strain and the other (of lower molecular weight) corresponds to the size of the inaccurately repaired chromosome predicted by comparative genome hybridization (CGH) data. Additionally, the intensity of the rearranged chromosome band was approximately twofold higher than that of the intact chromosome III band. Band sizes and intensities are consistent with the position of the probe in the amplified region of the left arm and therefore support the indicated structure of this class of rearrangement (Figure 5B).
Class 3-4 illegitimate diploids have only a deletion of the right arm of chromosome III (Figures 4D and 5B). We observed four of these events after the depletion of genes involved in DNA replication (MCM7, DPB11, and SPT16). Two of these events had breakpoints at FS1 and FS2 (MCM7 and DPB11), the same breakpoints observed following depletion of DNA polymerases α and δ (Lemoine et al. 2005, 2008). The other two, from SPT16 illegitimate diploids, had breakpoints at YCRCδ6. As suggested previously, this class might represent chromosome fragments that persist through direct telomere capping (Figure 5B) or by acquisition of telomeric sequences by BIR utilizing a telomere-proximal Ty element (Lemoine et al. 2005, 2008). These rearrangement profiles were confirmed by Southern blot analysis, where two bands of equal intensity were visualized, one corresponding to intact chromosome III and the other to the predicted chromosome III fragment (Figure 6A, lane 3).
The class 3-5 rearrangement pattern includes a deletion of all but the left arm of chromosome III in addition to an arm deletion on a nonhomologous chromosome (chromosome XVI or V) (Figures 4E and 5B). By contrast to the four subclasses of rearrangements described above, class 3-5 profiles were not observed following depletion of DNA polymerases α and δ (Lemoine et al. 2005, 2008). However, this profile has been documented upon the deletion of a nonessential gene required for strand invasion in homologous recombination, RAD52 (Casper et al. 2009). In our studies, class 3-5 profiles were observed following the depletion of NSE1 and UBC9, which also function in DNA repair (Branzei et al. 2006; Santa Maria et al. 2007; Irmisch et al. 2009). We found one additional class 3-5 rearrangement following the depletion of DPB11 (Table 2). In each case, the breakpoint on chromosome III corresponds with the only Ty retrotransposon on the left arm and the rearrangement results in the loss of one copy of the chromosome III centromere. We predict that this acentric chromosome III fragment would be fused to the nonhomologous chromosome to allow this chromosome fragment to persist (Figure 5B). The chromosome fusion could result from an interruption of a BIR event and subsequent resolution of the strand invasion intermediate with a nonhomologous chromosome, resulting in a half crossover chromosome. Alternatively, since the breakpoints on the nonhomologous chromosomes coincide with Ty retrotransposon sites, it is possible that a homology-mediated repair mechanism, such as single-strand annealing (SSA) (Mieczkowski et al. 2006), is involved in the formation of this chromosome fusion (Figure 5B). This mutant chromosome was confirmed by Southern blot analysis of a representative illegitimate diploid isolated following the depletion of NSE1 (Figure 6C, lanes 2 and 4). A probe specific for chromosome III detected two chromosome sizes, one corresponding to intact chromosome III and another to the predicted size of the chromosome III–chromosome V fusion. We also detected two chromosome bands following rehybridization with a probe specific for chromosome V, one corresponding to the size of intact chromosome V and the other corresponding to the expected size of the chromosome fusion.
Finally, depletion of RFC2 resulted in one example of a “Hawthorne deletion” (class 3-6), an interstitial deletion between repeated regions of the MATα and HMRa loci (Hawthorne 1963). We did not observe any examples of class 3-7, the hallmark of which is amplification of sequences between FS1 and FS2 (Casper et al. 2009). The total ratio of the seven rearrangement classes was 15:4:8:4:6:1:0 (3-1: 3-2: 3-3: 3-4: 3-5: 3-6: 3-7).
Boundaries of rearrangements correlate with Ty retrotransposons, LTRs, tRNA genes, early replication origins, and replication termination sites
To determine whether the boundaries of rearrangements are correlated with particular genomic features, we performed an enrichment test. We segmented the genome into 5-kb bins and scored each bin for the presence or absence of genomic features and breakpoints. For each feature, we determined the fold enrichment of bins that contain both the feature and a breakpoint in comparison to what would be expected if breakpoints were randomly placed into bins (Table 3). Consistent with other studies of rearrangement breakpoints in yeast (Dunham et al. 2002; Lemoine et al. 2005, 2008; Argueso et al. 2008; Li et al. 2009), our boundaries of rearrangement were significantly enriched at loci with Ty retrotransposons, LTRs, and tRNA genes (Table 3). As these sites have repetitive sequences, they may represent endpoints of resection that facilitate recombination repair between nonhomologous loci. We also found that boundaries of rearrangement were significantly enriched near early replication origins and replication termination sites (Table 3). One possibility is that misregulation of replication firing and replication fork convergence causes double-stranded DNA breaks that promote rearrangement events.
Table 3. Enrichment analysis of the correlation between boundaries of chromosome rearrangements (n = 14) and selected genomic features.
| Feature | Fold enrichment | P-value | False discovery rate corrected P-value |
|---|---|---|---|
| LTRs | 8.823 | 1.87 × 10−12 | 3.55 × 10−11 |
| tRNA genes | 7.495 | 3.76 × 10−9 | 3.57 × 10−8 |
| Ty retrotransposons | 7.988 | 0.005 | 0.0344 |
| Termination sites | 7.313 | 0.007 | 0.0333 |
| Early replication origins | 9.355 | 0.0184 | 0.0699 |
| High confidence ARSs | 1.739 | 0.186 | 0.505 |
Discussion
Comparison of conditional allele screens for genome instability mutants
The collection of tetracycline-regulated promoter conditional alleles (Tet alleles) encompasses 773 essential genes (63%), of a total of 1135 that are annotated in the SGD (Mnaimneh et al. 2004; Yu et al. 2006). We quantified the accumulation of Ddc2 damage foci in this set of strains after first filtering for the 217 strains that showed accumulation in S or G2 phase following promoter shutoff. We identified 47 genes that function to protect the genome from spontaneous DNA damage, 20 of which did not have previously annotated roles in the maintenance of genome stability. A similar screen for Ddc2 foci accumulation was recently reported, using a set of 592 temperature-sensitive conditional alleles representing 399 essential genes (Li et al. 2011). Of the 114 genes that were in common to both sets of conditional alleles (Figure S2A), mutants of 10 essential genes displayed elevated levels of Ddc2 foci in both screening efforts, a small but significant overlap (P-value of 0.0043; hypergeometric test) (Figure S2B, Table S4). Fifteen genes were identified in our screen that were negative in Li et al. (2011) and 15 genes were identified in Li et al. (2011) that were negative in our screen (Figure S2B). Altogether, we identified 37 genes with elevated Ddc2 foci that were not identified by Li et al. (2011).
We also screened for the a-like faker chromosome instability phenotype in 208 Tet alleles that we assayed for Ddc2 foci formation. Of the 68 genes that were in common with a recent a-like faker screening effort using ts alleles (Stirling et al. 2011) (Figure S2C), the overlap of 11 essential gene mutants with elevated levels of a-like fakers in both screening efforts was insignificant (P-value of 0.064 by the hypergeometric test) (Figure S2D, Table S5). We identified 59 essential genes that contribute to the suppression of genome instability that were not identified by Stirling et al. (2011). Focusing on the genes assayed in both screens, there were seven false negatives in our screen of Tet alleles and 22 false negatives in the Stirling et al. (2011) screen of 364 ts alleles. These, and similar false negatives in the Ddc2 foci screens, likely represent cases where the false negative allele was not sufficiently compromised to display a significant phenotype. Finally, a recent screen was performed to determine the extent of Rad52 foci in 305 essential chromosome instability genes. Comparison with our Ddc2 foci screen revealed that only the depletion of CDC9, CDC45, MCM5, and NSE1 and PSF2 resulted in elevated levels of both Rad52 and Ddc2 foci (Stirling et al. 2012). Given that each conditional allele collection is currently incomplete, that a positive score with one kind of allele is at best weakly predictive of a positive score with a distinct allele, that the overlap between screens of different allele collections is small, and that the functions of essential genes are likely perturbed to varying degrees within any one collection, screening complementary collections of different kinds of alleles will ultimately be necessary to define the complete cohort of essential genes that maintain genome stability.
A common theme in the overlap among these screens of essential gene collections is enrichment for genes that function in DNA replication, indicating that among essential processes, replication defects are strong contributors to genome instability. Both Ddc2 foci screens were enriched for genes with DNA replication as their GO process annotations, relative to the S. cerevisiae genome (14.5- and 25.3-fold enrichment for this study and Li et al. 2011, respectively; Bonferroni corrected P-values of 7.41 × 10−12 and 2.51 × 10−27). Similarly, both a-like faker screens displayed enrichment for DNA replication (10.8- and 17.5-fold enrichment for this study and Stirling et al. 2011, respectively; Bonferroni corrected P-values of 1.87 × 10−13 and 3.20 × 10−19).
We compared the functional differences between essential gene alleles that had elevated Ddc2 foci and those that had increased frequency of illegitimate mating, as this is the first time the same set of essential genes has been analyzed with both assays. Eighteen alleles displayed only elevated levels of Ddc2 foci and 63 alleles had only the a-like faker chromosome instability phenotype, while 29 alleles had elevated levels of both. This core of 29 genes was enriched for DNA replication function relative to the genome (20.4-fold enrichment; Bonferroni corrected P-value of 2.04 × 10−12), again indicating the primary role of replication errors in genome rearrangements. By contrast, the alleles that displayed only the a-like faker chromosome instability were enriched for genes involved in transcription initiation (15.9-fold enrichment; Bonferroni corrected P-value of 6.22 × 10−6). By analyzing the overlap between the screens, we found that strains with increased illegitimate mating tended to have a larger fraction of cells with spontaneous Ddc2 foci, and that strains with spontaneous Ddc2 foci formation above our cutoff were more likely to have increased illegitimate mating, which suggests some predictive value of one phenotype for the other. However, the overlap between the Ddc2 focus screen and the a-like faker screen was far from absolute. Therefore, consistent with a recent study (Stirling et al. 2011), we suggest that the complete set of genes with roles in genome maintenance will be obtained not only by screening different allele collections, but also by the application of multiple screening methods.
Essential genes involved in DNA replication are critical for genome stability
We observed a prominent role for DNA replication genes in the suppression of chromosome rearrangements. Defects in a range of distinct replication functions, including initiation (CDC45, DBF4, DPB11, MCM4, MCM5, MCM7, and PSF2), elongation (CDC45, DNA2, MCM4, MCM5, MCM7, POL2, POL30, PSF2, RFC2, and RFC5), and termination (UBC9) caused spontaneous DNA damage and chromosome rearrangements, suggesting that each stage of replication is crucia to the maintenance of genome stability. The rearrangements that we observed likely involve DNA double stranded breaks (DSBs) and there are several mechanisms by which defects in replication could contribute to DSB formation.
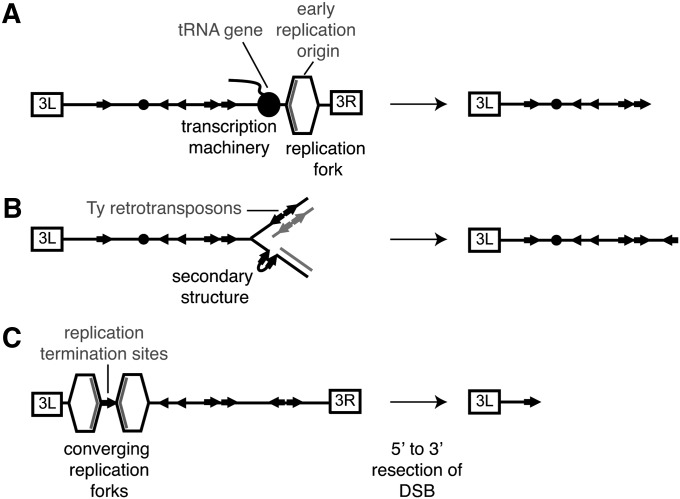
Reduced levels of proteins involved in prereplicative and preinitiation complex formation at replication origins likely result in fewer replication forks emanating from fewer origins, increasing the likelihood that regions of the genome might remain unreplicated and become susceptible to breakage. Consistent with this view, reduced levels of activated origins of replication and elevated frequencies of gross chromosomal rearrangements have been observed in strains with mutations in CDC6, CLN2, ORC2, or SIC1 genes involved in origin licensing (Bruschi et al. 1995; Lengronne and Schwob 2002; Shimada et al. 2002; Tanaka and Diffley 2002; Bielinsky 2003). Depletion of DNA replication elongation factors might disrupt the kinetics of replication in S phase, resulting in replication fork stalling and chromosome rearrangements as has been noted in RFA1 mutants (Chen et al. 1998). Defects in elongation could also disrupt the coordinated movements of replisomes and transcription machinery along the DNA, leading to increased levels of collision and DNA breakage (Deshpande and Newlon 1996; Ivessa et al. 2003) (Figure 7A). Another consequence of defective replication elongation could be the delay of Okazaki fragment synthesis resulting in the accumulation of single-stranded DNA (ssDNA) on the lagging strand, secondary structure formation, and blocks to replisome progression (Sogo et al. 2002) (Figure 7B). This type of mechanism has been proposed to allow the formation of hairpin structures at inverted Ty retrotransposon repeats, causing chromosome rearrangements when DNA polymerase α and δ are depleted (Lemoine et al. 2005, 2008; Casper et al. 2009). Depletion of polymerase ε (POL2) in our study could cause fragile site instability by a similar mechanism. Finally, when two replication forks converge in the last stages of replication, termination structures need to be accurately resolved prior to mitosis to prevent DSBs (Figure 7C). Both Ubc9 and the Nse1-containing Smc5/6 complex have connections to resolution of termination structures (Branzei et al. 2006) and we observe that rearrangement breakpoints are enriched at termination sites (Fachinetti et al. 2010) (Table 3), suggesting that defective termination could contribute to accumulation of chromosome rearrangements. Together, our results emphasize the importance of replication defects, in initiation, elongation, and termination, in causing DNA damage and chromosome rearrangements.
Figure 7 .
Mechanisms by which genome instability occurs in replication-deficient mutants. (A) tRNA genes and replication forks are clustered in proximity to Ty retrotransposons. The transcription machinery creates obstacles for replication fork progression and could lead to DSB formation and resection to the repeated elements. (B) Secondary structure formation involving repeated Ty retrotransposons (arrows) on the lagging strand causes replication fork stalling, subsequent replisome dissociation, and the formation of double stranded breaks (DSBs). (C) Failure to resolve termination structures at converging replication forks that flank Ty retrotransposons is a potential source of DSBs.
Ty retrotransposons and tRNA genes promote chromosome rearrangements
Each of the 38 rearrangement breakpoints that we mapped in this study, regardless of the specific function of the gene that was depleted, was proximal to a Ty retrotransposon element, highlighting the critical role of these repetitive elements in chromosome rearrangements in yeast. Chromosome rearrangements involving Ty retrotransposons are observed at a basal level in wild-type S. cerevisiae strains and a number of experimental connections between rearrangements and Ty elements have been made (reviewed in Garfinkel 2005; Lesage and Todeschini 2005; Mieczkowski et al. 2006).
Ty retrotransposons could function in at least two ways to promote chromosome rearrangements. They could represent sites of chromosome breakage (Lemoine et al. 2005) or they could provide homologous sequences for recombination-mediated repair of breaks that occur at distal sequences (Hoang et al. 2010). The yeast fragile site FS2 is thought to be a site of chromosome breakage, and a model in which ssDNA at inverted pairs of Ty retrotransposons, such as FS2, allows formation of secondary structures that inhibit the progression of the replisome, causing replication fork stalling and DNA breakage has been proposed (Lemoine et al. 2005, 2008; Casper et al. 2009) (Figure 7B). However, of the 38 chromosome rearrangement mutants that we mapped, we observed only 3 (7.9%) with boundaries within the FS2 region, suggesting that other modes of chromosome breakage predominate in our study.
Examination of the boundaries of chromosome rearrangements in replication mutants revealed significant enrichment of nearby early replication origins and tRNA genes (Table 3). Transcription complexes on tRNA genes can impede an oncoming replisome, thereby promoting replication fork pausing and DSB formation (Figure 7A) (Deshpande and Newlon 1996; Ivessa et al. 2003), suggesting that additional breakage could result from transcription–replication collisions. This combination of features surrounding breakpoints of rearrangement is consistent with those observed at natural evolutionary breakpoints when S. cerevisiae is compared to related yeasts, as well as breakpoints observed during artificial evolution of S. cerevisiae (Dunham et al. 2002; Kellis et al. 2003; Di Rienzi et al. 2009), suggesting that in addition to replication fidelity, these features are important determinants of instability.
Parallels with human common fragile sites
There are a number of parallels between common fragile sites in yeast and in humans. Inhibition or depletion of DNA polymerases (Glover et al. 1984; Lemoine et al. 2005, 2008) or DNA damage checkpoint proteins (Casper et al. 2002; Cha and Kleckner 2002; Arlt et al. 2004; Schwartz et al. 2005; Durkin et al. 2006; Raveendranathan et al. 2006; Vernon et al. 2008; Focarelli et al. 2009) can induce chromosome breaks at common fragile sites in both yeast and human. Although human common fragile sites lack distinctive sequence similarities, they have attributes that impair replication progression (Glover et al. 1984, 2005; Zlotorynski et al. 2003), a shared property of yeast fragile sites (Roeder and Fink 1980; Deshpande and Newlon 1996; Cha and Kleckner 2002; Ivessa et al. 2003; Lemoine et al. 2005; Admire et al. 2006; Raveendranathan et al. 2006). Additionally, recent studies of the human common fragile site FRA3B have suggested that instability at this site is not due to replication fork slowing or stalling, but rather is due to a paucity of replication initiation events (Letessier et al. 2011). In our studies, early firing origins of replication are enriched in regions with rearrangement breakpoints. Depletion of replication initiation factors could disrupt origin firing at these sites and thereby contribute to instability in a manner analogous to FRA3B. It will be of great interest to test the general role of replication proteins in suppressing chromosome rearrangements that we have observed in yeast in the maintenance of human common fragile sites.
Supplementary Material
Acknowledgments
We thank Philip Hieter for strains, Tao Qi for technical assistance, Lisa Yu for strain construction, and all members of the Brown laboratory for insightful discussions. This research was supported by the Canadian Institutes of Health Research (grant MOP-79368 to G.W.B.), the Canada Excellence Research Chair Program (F.P.R.), the Canadian Institute for Advance Research Fellowship (F.P.R.), and the Natural Sciences and Engineering Research Council of Canada (J.A.V.).
Footnotes
Communicating editor: J. Nickoloff
Literature Cited
- Admire A., Shanks L., Danzl N., Wang M., Weier U., et al. , 2006. Cycles of chromosome instability are associated with a fragile site and are increased by defects in DNA replication and checkpoint controls in yeast. Genes Dev. 20: 159–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A., Gomez-Gonzalez B., 2008. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 9: 204–217 [DOI] [PubMed] [Google Scholar]
- Alvaro D., Lisby M., Rothstein R., 2007. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 3: e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen M. P., Nelson Z. W., Hetrick E. D., Gottschling D. E., 2008. A genetic screen for increased loss of heterozygosity in Saccharomyces cerevisiae. Genetics 179: 1179–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso J. L., Westmoreland J., Mieczkowski P. A., Gawel M., Petes T. D., et al. , 2008. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc. Natl. Acad. Sci. USA 105: 11845–11850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt M. F., Xu B., Durkin S. G., Casper A. M., Kastan M. B., et al. , 2004. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol. Cell. Biol. 24: 6701–6709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlke-Steinert S., Touat-Todeschini L., Skoufias D. A., Margolis R. L., 2009. SMC5 and MMS21 are required for chromosome cohesion and mitotic progression. Cell Cycle 8: 2211–2218 [DOI] [PubMed] [Google Scholar]
- Ben-Aroya S., Coombes C., Kwok T., O’Donnell K. A., Boeke J. D., et al. , 2008. Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol. Cell 30: 248–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielinsky A. K., 2003. Replication origins: Why do we need so many? Cell Cycle 2: 307–309 [PubMed] [Google Scholar]
- Branzei D., Foiani M., 2007. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair (Amst.) 6: 994–1003 [DOI] [PubMed] [Google Scholar]
- Branzei D., Foiani M., 2009. The checkpoint response to replication stress. DNA Repair (Amst.) 8: 1038–1046 [DOI] [PubMed] [Google Scholar]
- Branzei D., Foiani M., 2010. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11: 208–219 [DOI] [PubMed] [Google Scholar]
- Branzei D., Sollier J., Liberi G., Zhao X., Maeda D., et al. , 2006. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509–522 [DOI] [PubMed] [Google Scholar]
- Bruschi C. V., McMillan J. N., Coglievina M., Esposito M. S., 1995. The genomic instability of yeast cdc6–1/cdc6–1 mutants involves chromosome structure and recombination. Mol. Gen. Genet. 249: 8–18 [DOI] [PubMed] [Google Scholar]
- Budd M. E., Choe W., Campbell J. L., 2000. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 275: 16518–16529 [DOI] [PubMed] [Google Scholar]
- Casper A. M., Nghiem P., Arlt M. F., Glover T. W., 2002. ATR regulates fragile site stability. Cell 111: 779–789 [DOI] [PubMed] [Google Scholar]
- Casper A. M., Greenwell P. W., Tang W., Petes T. D., 2009. Chromosome aberrations resulting from double-strand DNA breaks at a naturally occurring yeast fragile site composed of inverted ty elements are independent of Mre11p and Sae2p. Genetics 183: 423–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha R. S., Kleckner N., 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297: 602–606 [DOI] [PubMed] [Google Scholar]
- Chang M., Bellaoui M., Zhang C., Desai R., Morozov P., et al. , 2005. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 24: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Umezu K., Kolodner R. D., 1998. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol. Cell 2: 9–22 [DOI] [PubMed] [Google Scholar]
- Cimprich K. A., Cortez D., 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desany B. A., Alcasabas A. A., Bachant J. B., Elledge S. J., 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12: 2956–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A. M., Newlon C. S., 1996. DNA replication fork pause sites dependent on transcription. Science 272: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Dion B., Brown G. W., 2009. Comparative genome hybridization on tiling microarrays to detect aneuploidies in yeast. Methods Mol. Biol. 548: 1–18 [DOI] [PubMed] [Google Scholar]
- Di Rienzi S. C., Collingwood D., Raghuraman M. K., Brewer B. J., 2009. Fragile genomic sites are associated with origins of replication. Genome Biol. Evol. 1: 350–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z., Andronescu M., Schutz K., McIlwain S., Kim Y. J., et al. , 2010. A three-dimensional model of the yeast genome. Nature 465: 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham M. J., Badrane H., Ferea T., Adams J., Brown P. O., et al. , 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99: 16144–16149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin S. G., Arlt M. F., Howlett N. G., Glover T. W., 2006. Depletion of CHK1, but not CHK2, induces chromosomal instability and breaks at common fragile sites. Oncogene 25: 4381–4388 [DOI] [PubMed] [Google Scholar]
- Fachinetti D., Bermejo R., Cocito A., Minardi S., Katou Y., et al. , 2010. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol. Cell 39: 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focarelli M. L., Soza S., Mannini L., Paulis M., Montecucco A., et al. , 2009. Claspin inhibition leads to fragile site expression. Genes Chromosomes Cancer 48: 1083–1090 [DOI] [PubMed] [Google Scholar]
- Fujioka Y., Kimata Y., Nomaguchi K., Watanabe K., Kohno K., 2002. Identification of a novel non-structural maintenance of chromosomes (SMC) component of the SMC5–SMC6 complex involved in DNA repair. J. Biol. Chem. 277: 21585–21591 [DOI] [PubMed] [Google Scholar]
- Garfinkel D. J., 2005. Genome evolution mediated by Ty elements in Saccharomyces. Cytogenet. Genome Res. 110: 63–69 [DOI] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., et al. , 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Glover T. W., Berger C., Coyle J., Echo B., 1984. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum. Genet. 67: 136–142 [DOI] [PubMed] [Google Scholar]
- Glover T. W., Arlt M. F., Casper A. M., Durkin S. G., 2005. Mechanisms of common fragile site instability. Hum. Mol. Genet. 14 Spec No. 2: R197–205. [DOI] [PubMed] [Google Scholar]
- Harper J. W., Elledge S. J., 2007. The DNA damage response: ten years after. Mol. Cell 28: 739–745 [DOI] [PubMed] [Google Scholar]
- Hawthorne D. C., 1963. A deletion in yeast and its bearing on the structure of the mating type locus. Genetics 48: 1727–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang M. L., Tan F. J., Lai D. C., Celniker S. E., Hoskins R. A., et al. , 2010. Competitive repair by naturally dispersed repetitive DNA during non-allelic homologous recombination. PLoS Genet. 6: e1001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood J. K., Silver P. A., 1998. Cse1p is required for export of Srp1p/importin-alpha from the nucleus in Saccharomyces cerevisiae. J. Biol. Chem. 273: 35142–35146 [DOI] [PubMed] [Google Scholar]
- Huang M. E., Kolodner R. D., 2005. A biological network in Saccharomyces cerevisiae prevents the deleterious effects of endogenous oxidative DNA damage. Mol. Cell 17: 709–720 [DOI] [PubMed] [Google Scholar]
- Huang M. E., Rio A. G., Nicolas A., Kolodner R. D., 2003. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl. Acad. Sci. USA 100: 11529–11534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmisch A., Ampatzidou E., Mizuno K., O’Connell M. J., Murray J. M., 2009. Smc5/6 maintains stalled replication forks in a recombination-competent conformation. EMBO J. 28: 144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa A. S., Lenzmeier B. A., Bessler J. B., Goudsouzian L. K., Schnakenberg S. L., et al. , 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell 12: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S., 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254 [DOI] [PubMed] [Google Scholar]
- Kolodner R. D., Putnam C. D., Myung K., 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297: 552–557 [DOI] [PubMed] [Google Scholar]
- Kunzler M., Hurt E. C., 1998. Cse1p functions as the nuclear export receptor for importin alpha in yeast. FEBS Lett. 433: 185–190 [DOI] [PubMed] [Google Scholar]
- Labib K., Tercero J. A., Diffley J. F., 2000. Uninterrupted MCM2–7 function required for DNA replication fork progression. Science 288: 1643–1647 [DOI] [PubMed] [Google Scholar]
- Lee K. H., Kim D. W., Bae S. H., Kim J. A., Ryu G. H., et al. , 2000. The endonuclease activity of the yeast Dna2 enzyme is essential in vivo. Nucleic Acids Res. 28: 2873–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine F. J., Degtyareva N. P., Lobachev K., Petes T. D., 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120: 587–598 [DOI] [PubMed] [Google Scholar]
- Lemoine F. J., Degtyareva N. P., Kokoska R. J., Petes T. D., 2008. Reduced levels of DNA polymerase delta induce chromosome fragile site instability in yeast. Mol. Cell. Biol. 28: 5359–5368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A., Schwob E., 2002. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1). Mol. Cell 9: 1067–1078 [DOI] [PubMed] [Google Scholar]
- Lesage P., Todeschini A. L., 2005. Happy together: the life and times of Ty retrotransposons and their hosts. Cytogenet. Genome Res. 110: 70–90 [DOI] [PubMed] [Google Scholar]
- Letessier A., Millot G. A., Koundrioukoff S., Lachages A. M., Vogt N., et al. , 2011. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470: 120–123 [DOI] [PubMed] [Google Scholar]
- Li X. C., Schimenti J. C., Tye B. K., 2009. Aneuploidy and improved growth are coincident but not causal in a yeast cancer model. PLoS Biol. 7: e1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Vizeacoumar F. J., Bahr S., Li J., Warringer J., et al. , 2011. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat. Biotechnol. 29: 361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Rothstein R., 2004. DNA damage checkpoint and repair centers. Curr. Opin. Cell Biol. 16: 328–334 [DOI] [PubMed] [Google Scholar]
- Lisby M., Barlow J. H., Burgess R. C., Rothstein R., 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- McKinnon P. J., Caldecott K. W., 2007. DNA strand break repair and human genetic disease. Annu. Rev. Genomics Hum. Genet. 8: 37–55 [DOI] [PubMed] [Google Scholar]
- Melo J. A., Cohen J., Toczyski D. P., 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15: 2809–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski P. A., Lemoine F. J., Petes T. D., 2006. Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amst.) 5: 1010–1020 [DOI] [PubMed] [Google Scholar]
- Mnaimneh S., Davierwala A. P., Haynes J., Moffat J., Peng W. T., et al. , 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 118: 31–44 [DOI] [PubMed] [Google Scholar]
- Osborne M. A., Schlenstedt G., Jinks T., Silver P. A., 1994. Nuf2, a spindle pole body-associated protein required for nuclear division in yeast. J. Cell Biol. 125: 853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outwin E. A., Irmisch A., Murray J. M., O’Connell M. J., 2009. Smc5-Smc6-dependent removal of cohesin from mitotic chromosomes. Mol. Cell. Biol. 29: 4363–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebernard S., Perry J. J., Tainer J. A., Boddy M. N., 2008. Nse1 RING-like domain supports functions of the Smc5-Smc6 holocomplex in genome stability. Mol. Biol. Cell 19: 4099–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveendranathan M., Chattopadhyay S., Bolon Y. T., Haworth J., Clarke D. J., et al. , 2006. Genome-wide replication profiles of S-phase checkpoint mutants reveal fragile sites in yeast. EMBO J. 25: 3627–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G. S., Fink G. R., 1980. DNA rearrangements associated with a transposable element in yeast. Cell 21: 239–249 [DOI] [PubMed] [Google Scholar]
- Santa Maria S. R., Gangavarapu V., Johnson R. E., Prakash L., Prakash S., 2007. Requirement of Nse1, a subunit of the Smc5-Smc6 complex, for Rad52-dependent postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 8409–8418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M., Collins S. R., Thompson N. J., Denic V., Bhamidipati A., et al. , 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123: 507–519 [DOI] [PubMed] [Google Scholar]
- Schwartz M., Zlotorynski E., Goldberg M., Ozeri E., Rahat A., et al. , 2005. Homologous recombination and nonhomologous end-joining repair pathways regulate fragile site stability. Genes Dev. 19: 2715–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., 1991. Getting started with yeast. Methods Enzymol. 194: 3–21 [DOI] [PubMed] [Google Scholar]
- Shimada K., Pasero P., Gasser S. M., 2002. ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev. 16: 3236–3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E., Weinstein J., Rothstein R., 2005. A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: four genes involved in error-free DNA repair. Genetics 169: 1275–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S., Hwang J. Y., Banerjee S., Majeed A., Gupta A., et al. , 2004. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101: 9039–9044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J. M., Lopes M., Foiani M., 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602 [DOI] [PubMed] [Google Scholar]
- Solsbacher J., Maurer P., Bischoff F. R., Schlenstedt G., 1998. Cse1p is involved in export of yeast importin alpha from the nucleus. Mol. Cell. Biol. 18: 6805–6815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling P. C., Bloom M. S., Solanki-Patil T., Smith S., Sipahimalani P., et al. , 2011. The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genet. 7: e1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling P. C., Chan Y. A., Minaker S. W., Aristizabal M. J., Barrett I., et al. , 2012. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 26: 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J., Hicks J., Herskowitz I., 1981. Control of cell type in yeast by the mating type locus. The alpha 1-alpha 2 hypothesis. J. Mol. Biol. 147: 357–372 [DOI] [PubMed] [Google Scholar]
- Tanaka S., Diffley J. F., 2002. Deregulated G1-cyclin expression induces genomic instability by preventing efficient pre-RC formation. Genes Dev. 16: 2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu K., Hiraoka M., Mori M., Maki H., 2002. Structural analysis of aberrant chromosomes that occur spontaneously in diploid Saccharomyces cerevisiae: retrotransposon Ty1 plays a crucial role in chromosomal rearrangements. Genetics 160: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon M., Lobachev K., Petes T. D., 2008. High rates of “unselected” aneuploidy and chromosome rearrangements in tel1 mec1 haploid yeast strains. Genetics 179: 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P. A., Kilmartin J. V., 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152: 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z., McGrew J. T., Schroeder A. J., Fitzgerald-Hayes M., 1993. CSE1 and CSE2, two new genes required for accurate mitotic chromosome segregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 4691–4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Pena Castillo L., Mnaimneh S., Hughes T. R., Brown G. W., 2006. A survey of essential gene function in the yeast cell division cycle. Mol. Biol. Cell 17: 4736–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen K. W., Warren C. D., Chen O., Kwok T., Hieter P., et al. , 2007. Systematic genome instability screens in yeast and their potential relevance to cancer. Proc. Natl. Acad. Sci. USA 104: 3925–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G., 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotorynski E., Rahat A., Skaug J., Ben-Porat N., Ozeri E., et al. , 2003. Molecular basis for expression of common and rare fragile sites. Mol. Cell. Biol. 23: 7143–7151 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.