Abstract
Natural antisense transcripts (NATs) exist ubiquitously in mammalian genomes and play roles in the regulation of gene expression. However, both the existence of bidirectional antisense RNA regulation and the possibility of protein-coding genes that function as antisense RNAs remain speculative. Here, we found that the protein-coding gene, deoxyhypusine synthase (DHPS), as the NAT of WDR83, concordantly regulated the expression of WDR83 mRNA and protein. Conversely, WDR83 also regulated DHPS by antisense pairing in a concordant manner. WDR83 and DHPS were capable of forming an RNA duplex at overlapping 3′ untranslated regions and this duplex increased their mutual stability, which was required for the bidirectional regulation. As a pair of protein-coding cis-sense/antisense transcripts, WDR83 and DHPS were upregulated simultaneously and correlated positively in gastric cancer (GC), driving GC pathophysiology by promoting cell proliferation. Furthermore, the positive relationship between WDR83 and DHPS was also observed in other cancers. The bidirectional regulatory relationship between WDR83 and DHPS not only enriches our understanding of antisense regulation, but also provides a more complete understanding of their functions in tumor development.
Keywords: bidirectional regulation, natural antisense transcript, gastric cancer
Introduction
Natural antisense transcripts (NATs) are defined as RNA molecules that are complementary to their endogenous sense genes 1, 2. They are generally classified into two types: cis-NATs that are transcribed from the opposite strands of their sense genes 3 and trans-NATs that are transcribed at different genomic loci from their sense partners 4. With advances in delineating the transcriptome, a number of NATs have been systematically identified in mammalian genomes; for example, up to 72% of transcripts have been demonstrated to have antisense partners in mammalian transcriptomes 1, 5, 6. NATs play important roles in many physiological and pathological processes, including genomic imprinting, alternative splicing, X inactivation, mRNA stability, and translational regulation 6, 7. Many reports have demonstrated that antisense transcripts can regulate the expression of sense genes. However, few reports have investigated whether bidirectional regulation can exist between antisense RNAs and sense protein-coding genes. In addition, recent efforts have paid close attention to noncoding RNAs (ncRNAs) as antisense transcripts of sense genes 8, 9. To date, no systematic studies have been published, which discuss antisense protein-coding genes that can affect sense protein-coding genes. Considering the widespread amount of antisense transcription and the pivotal role of antisense RNA regulation, this field is of great interest.
WDR83 (also known as MORG1) functions as a module in the assembly of a multi-component scaffold for components of the ERK-MAPK pathway, including ERK1/2, MEK1/2, Raf-1, and B-Raf; it also selectively facilitates ERK1/2 activation in response to a subset of agonists 10, 11. The aberrant activation of the ERK signaling, which contributes to tumorigenesis by promoting cell proliferation 12, 13, has been frequently observed in cancers. WDR83 also interacts with eglnine homolog 3 (EGLN3, also known as PHD3 (prolyl-hydroxylase 3)) and regulates the expression of hypoxia-inducible factor 1 (HIF-1) 14. As the major regulator of the hypoxic adaptive response, HIF-1 plays key roles in tumorigenesis 15. Considering the essential role of WDR83 in ERK signaling and tumorigenesis, the regulation of WDR83 at either the protein or RNA level may influence its associated pathways and the overall process of tumorigenesis.
In this study, we described a cis-sense/antisense (cis-SA) pair that consisted of two protein-coding genes, WDR83 and deoxyhypusine synthase (DHPS). This cis-SA pair was originally identified by the Natural Antisense Transcript Database (NATsDB) 16. We showed that DHPS, as the cis-NAT for WDR83, was increased in parallel with WDR83 in gastric cancer (GC). DHPS could increase the mRNA and protein expression of WDR83 through their overlapping 3′ untranslated regions (3′UTRs). WDR83 also regulated DHPS expression in a concordant manner. The underlying mechanisms and consequences of this regulatory process were further investigated.
Results
WDR83 and DHPS were upregulated simultaneously and positively correlated in GC
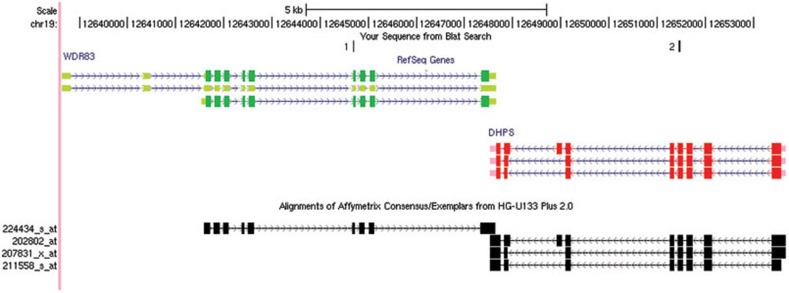
From NATsDB, a database that is based on millions of human ESTs and mRNAs, we identified that DHPS and WDR83 are transcribed from opposite strands of the same region of chromosome 19. Both WDR83 and DHPS had three splice variants, and they formed a 'tail-to-tail' pairing pattern, including 113 nucleotides with full complementarity to their constitutive 3′UTRs (Figure 1).
Figure 1.
Genomic organization of WDR83 and DHPS. DHPS and WDR83 are transcribed from opposite strands of the same region on chromosome 19. The coding regions and non-coding regions of WDR83 are marked with dark green and light green, respectively. The coding regions and noncoding regions of DHPS are marked with red and pink, respectively. The siRNA target sites of WDR83 (site 1) and DHPS (site 2) are located outside the overlapping regions to avoid off-target effects. The black boxes represent Affymetrix HG-U133 Plus2.0 array probe sets that correspond to WDR83 and DHPS. Arrows show the direction of transcription and blocks indicate exons.
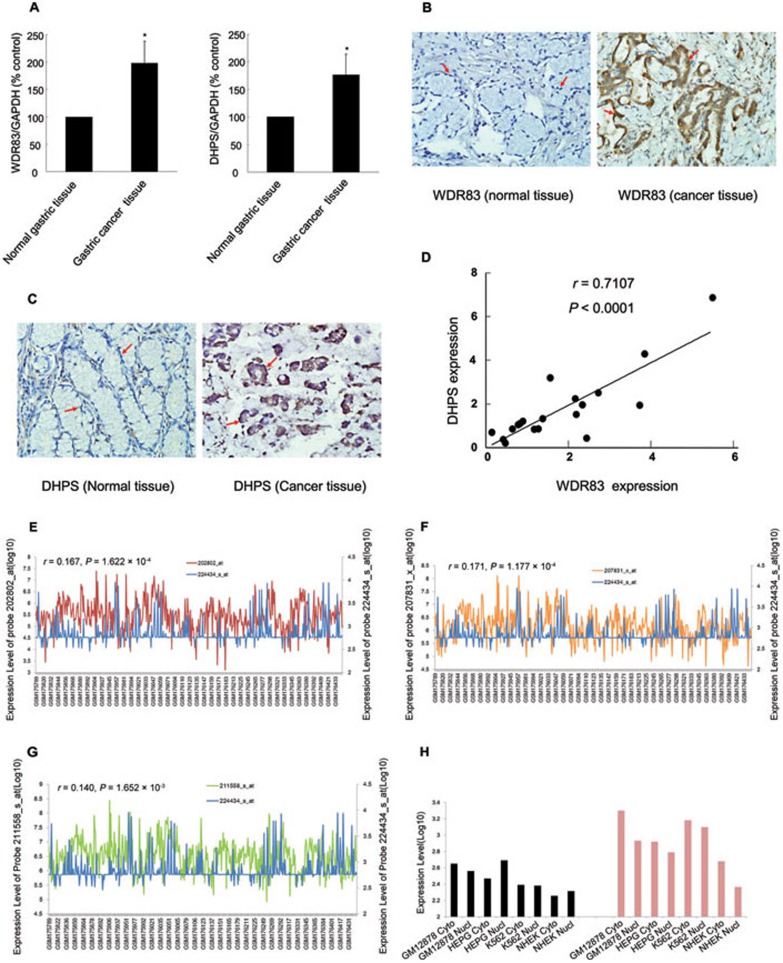
WDR83 and DHPS may be subjected to complicated antisense regulation. To investigate this hypothesis, their expression was examined by quantitative real-time PCR (qPCR) analyses between 19 pairs of human GC tissues and matched normal tissues. Both WDR83 and DHPS mRNA expressions were significantly increased in GC samples compared with their nontumorous counterparts (P = 0.0198 and P = 0.0286, respectively; Figure 2A). We found that the protein expression levels of WDR83 and DHPS were also elevated in primary GC tissues using immunohistochemistry (IHC; Figure 2B and 2C). These observations suggested that the expression of WDR83 and DHPS at both the mRNA and protein levels may be regulated concordantly. Interestingly, linear regression analysis revealed a strong positive correlation between DHPS and WDR83 mRNA expression in GC (r =0.7107, P < 0.0001, Figure 2D). Furthermore, transcription expression profiling in another 504 normal tissues using the Affymetrix U133 Plus2.0 array also confirmed the significantly positive correlation between WDR83 and DHPS (P < 0.001, Figure 2E-2G). The positive correlation coefficient was small but strongly significant, indicating that the concordant regulation between WDR83 and DHPS expression was of importance. Finally, subcellular location profiling of four different cell lines using the Affymetrix Tiling Array revealed that WDR83 and DHPS transcripts were widely expressed in both the cytoplasm and nucleus (Figure 2H), suggesting that these two genes were expressed in the same subcellular locations, which may be convenient for antisense regulation to occur. However, the exact underlying molecular mechanisms remain unclear.
Figure 2.
Expression analysis and the subcellular location of WDR83 and DHPS in GC tissues and different cell lines. (A) WDR83 and DHPS mRNA levels were quantified by qPCR in 19 pairs of GC specimens and their matched normal tissues. WDR83 and DHPS expression were significantly increased in GC tissues compared with their normal tissues (P = 0.0198 and 0.0286, respectively, n = 19; *P< 0.05, **P< 0.01, ANOVA). (B, C) Representative expression of WDR83 and DHPS in GC specimens and their matched normal tissues as determined by IHC (magnification 200×); in GC tissues, high-level expression of WDR83 or DHPS was shown by strong staining. (D) Regression analysis of WDR83 and DHPS expression revealed that WDR83 and DHPS were positively correlated (r = 0.7107, P < 0.0001). The correlation coefficient (r) measured the reliability and the P-value measured the statistical significance of the correlation between the x and y variables. (E-G) Expression correlation analysis of WDR83 and DHPS in 504 normal tissues: (E) the expression of WDR83 (probe 224434_s_at, blue) and DHPS (probe 202802_at, red) exhibited a significantly positive correlation (r = 0.167, P< 0.00016); (F) the expression of WDR83 (probe 224434_s_at, blue) and DHPS (probe 207831_at, orange) also exhibited a significantly positive correlation (r = 0.171, P< 0.00012); (G) the expression of WDR83 (probe 224434_s_at, blue) and DHPS (probe 211558_s_at, green) exhibited concordant expression patterns (r = 0.140, P< 0.00017). (H) Subcellular location profiling of WDR83 and DHPS using the Affymetrix Tiling Array revealed that both genes were widely expressed in the cytoplasm and nucleus in different cell lines.
DHPS mRNA regulated WDR83 through antisense overlapping
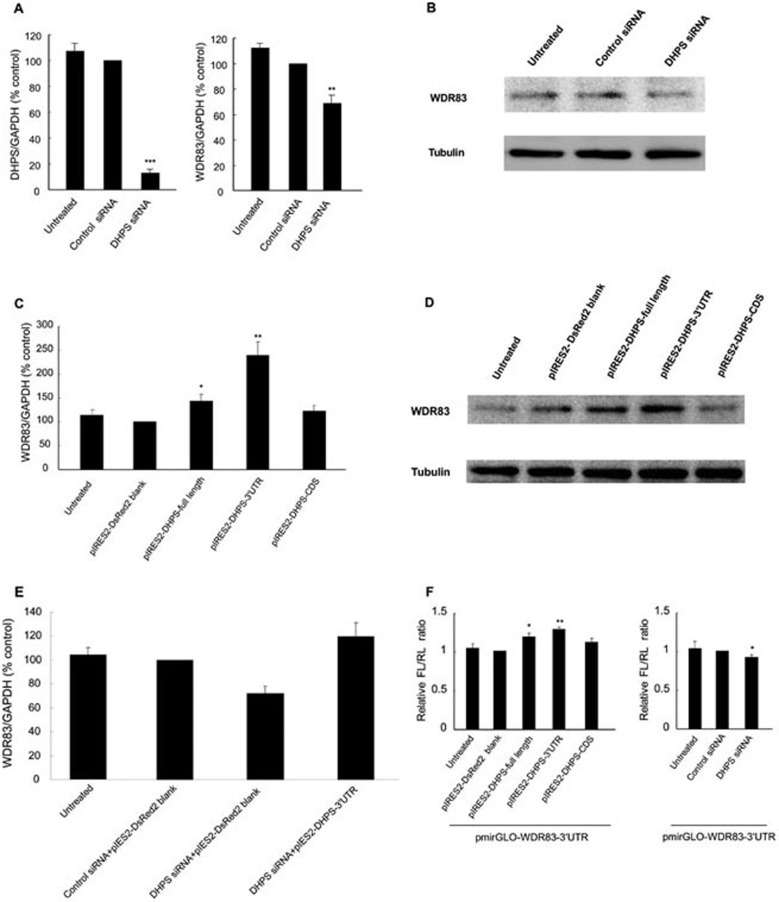
The common function of antisense transcripts is to regulate the expression of sense transcripts 1. To investigate whether DHPS could modulate the expression of WDR83, specific siRNAs that target the non-overlapping regions of DHPS were transfected into the MGC803 GC cell line. The results showed that knockdown of DHPS by siRNA led to a significant decrease not only in the level of the targeted DHPS mRNA but also in the level of WDR83 mRNA (Figure 3A). Western blotting analysis displayed that siRNAs targeting against DHPS also reduced the expression of WDR83 protein (Figure 3B). Thus, the data suggested that DHPS could regulate WDR83 at both mRNA and protein expression levels.
Figure 3.
DHPS upregulated WDR83 mRNA and protein expression through the 3′UTRs. (A) Knockdown of DHPS by siRNAs in MGC803 cells reduced WDR83 mRNA levels by 31.4% (n ≥ 3, P = 0.0039). (B) Western blot analysis demonstrated that the siRNAs knockdown of DHPS decreased WDR83 protein expression. (C) Quantification of WDR83 mRNA levels after overexpression of three different plasmids in MGC803 cells. qPCR analysis indicated that overexpression of DHPS-full length or DHPS-3′UTR resulted in a significant increase in WDR83 mRNA levels (n ≥ 3, P = 0.0110 and 0.0080, respectively). (D) Western blot showing that WDR83 protein levels were significantly increased in MGC803 cells transfected with DHPS-full length or DHPS-3′UTR plasmids. (E) WDR83 mRNA levels were measured by qPCR in MGC803 cells where endogenous DHPS were first knocked down by siRNA for 24 h and then restored by exogenous expression of DHPS-3′UTR (pIRES2-DHPS-3′UTR) for further 24 h. (F) The luciferase reporter assay showed that the WDR83 3′UTR-containing construct induced a 24.4% increase in luciferase activity in cells that were cotransfected with the DHPS 3′UTR plasmid (P = 0.0020), and the WDR83 full-length construct also showed an increase in luciferase activity (P = 0.0310). The DHPS CDS-containing construct did not have an obvious effect on pmirGLO-WDR83-3′UTR (P = 0.7845). Meanwhile, silencing DHPS decreased luciferase activity (P = 0.0158). Relative firefly luciferase activity over renilla luciferase activity: FL/RL, n ≥ 3, ANOVA; *P< 0.05, **P< 0.01, ***P< 0.001.
We further examined whether DHPS mRNA or protein perform the regulation of WDR83. We constructed three different plasmids: (1) pIRES2-DHPS-full length, containing the full-length DHPS transcript; (2) pIRES2-DHPS-CDS, containing the coding region of the DHPS transcript; and (3) pIRES2-DHPS-3′UTR containing only the 3′UTR of the DHPS transcript (described in Materials and Methods). These plasmids were transfected into MGC803 cells separately. Compared with the control vector, the overexpression of plasmids that contained the DHPS 3′UTR (pIRES2-DHPS-full length or pIRES2-DHPS-3′UTR) resulted in a significant increase in WDR83 mRNA and protein levels, whereas plasmids that lacked overlapping 3′UTR regions (pIRES2-DHPS-CDS) did not have an obvious effect on WDR83 expression (Figure 3C and 3D). These data indicated that DHPS mRNA, rather than DHPS protein, regulated WDR83, and that the antisense 3′UTR of DHPS might play an important role in this regulation. Furthermore, the transfection of DHPS 3′UTR plasmids (pIRES2-DHPS-3′UTR) into MGC803 cells rescued the downregulation of WDR83 after DHPS knockdown (P = 0.0145, Figure 3E). Thus, the 3′UTR of DHPS was essential for the regulation of WDR83 mRNA expression.
To further test whether the DHPS 3′UTR regulated WDR83 expression through antisense pairing, we generated a luciferase reporter construct (pmirGLO-WDR83-3′UTR) that carried the WDR83 3′UTR. A 113-bp fragment of the WDR83 3′UTR, which covered the entire overlapping region, was inserted downstream of the luciferase reporter gene in the pmirGLO vector. We examined the effects of DHPS overexpression or knockdown on pmirGLO-WDR83-3′UTR activity. Strikingly, the luciferase activity of pmirGLO-WDR83-3′UTR significantly increased in cells that were cotransfected with the DHPS-3′UTR plasmid (P = 0.0020, Figure 3F), and the overexpression of the DHPS full-length construct also induced an increase in luciferase activity (P = 0.0310, Figure 3F). The DHPS CDS-containing construct did not have an obvious effect on pmirGLO-WDR83-3′UTR (P = 0.7845, Figure 3F). Meanwhile, silencing DHPS decreased the luciferase activity of pmirGLO-WDR83-3′UTR (P = 0.0158, Figure 3F). These results were consistent with our observation that the overexpression of the DHPS 3′UTR resulted in a significant increase in WDR83 mRNA and protein levels. Taken together, our data demonstrated that the DHPS overlapping 3′UTR mediated the regulation of WDR83 expression through antisense pairing, which supported the notion that although the 3′UTR had no coding potential, it exerted a biological effect 17.
WDR83 mRNA also regulated DHPS through antisense pairing
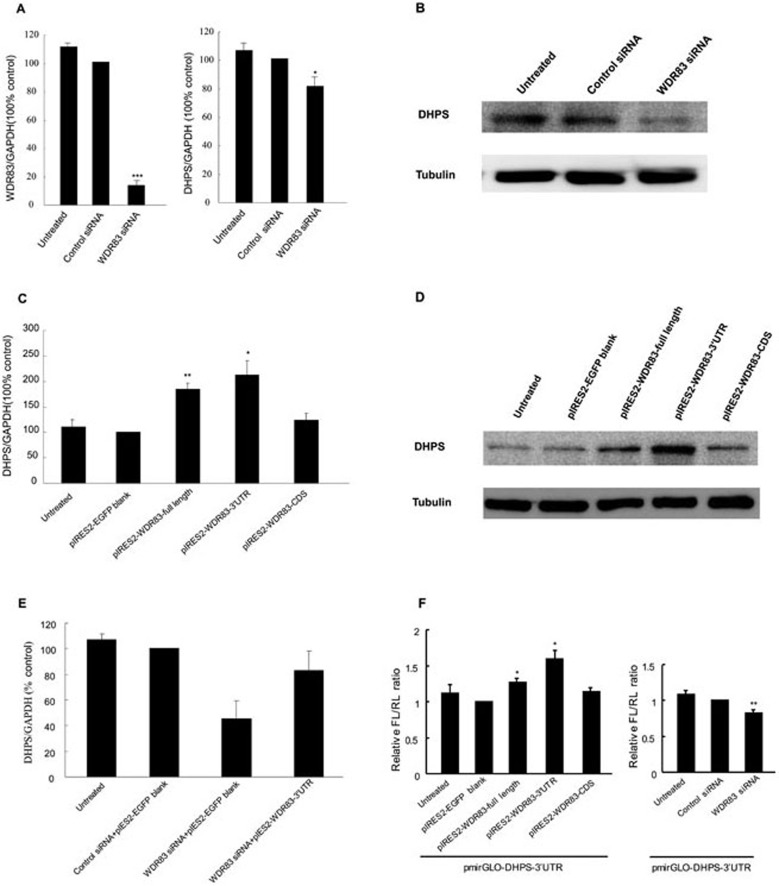
To study whether there was a possible regulation of DHPS by WDR83, DHPS mRNA and protein expression were examined by silencing WDR83. Unexpectedly, we observed that the knockdown of WDR83 resulted in significant reductions of DHPS mRNA and protein expression levels (Figure 4A and 4B). To further examine this regulatory process, we constructed three different plasmids (described in Materials and Methods) and transfected them separately into MGC803 cells. Interestingly, the data showed that the pIRES2-WDR83-3′UTR and pIRES2-WDR83-full length plasmids dramatically increased levels of DHPS mRNA and protein, whereas the pIRES2-WDR83-CDS plasmid did not affect the expression of DHPS significantly (Figure 4C and 4D). These observations indicated that WDR83 mRNA, rather than the WDR83 protein, performed the regulation. Furthermore, the transfection of WDR83 3′UTR plasmids (pIRES2-WDR83-3′UTR) rescued the downregulation of DHPS after the WDR83 knockdown (P = 0.0233, Figure 4E). Simultaneously, we observed that the luciferase activity of pmirGLO-DHPS-3′UTR was significantly increased in cells that were cotransfected with either the WDR83 3′UTR plasmid or the WDR83 full-length plasmid (P = 0.0481 and 0.0490, respectively, Figure 4F). The luciferase activity of pmirGLO-DHPS-3′UTR was markedly decreased in cells with the WDR83 knockdown (P = 0.0059, Figure 4F). Our observations further confirmed that the regulation of DHPS mRNA and protein expression by WDR83 was dependent on 3′UTR antisense pairing. Collectively, there was a bidirectional regulation between WDR83 and DHPS, which was consistent with our observations that the expression levels of WDR83 and DHPS were positively correlated in GC.
Figure 4.
WDR83 also upregulated DHPS mRNA and protein expression. (A) Knockdown of WDR83 by siRNAs in MGC803 cells resulted in an 18.7% decrease in DHPS mRNA expression (n ≥ 3, P = 0.0257). (B) Western blot analysis indicated that WDR83 downregulation also caused a decrease in DHPS protein expression. (C) Quantification of DHPS mRNA levels after overexpression of three different plasmids in MGC803 cells. qPCR analysis revealed that overexpression of only the transcripts containing the overlapping regions (WDR83-full length or WDR83-3′UTR) resulted in a significant increase in DHPS mRNA levels (n ≥ 3, P = 0.0014 and 0.0165, respectively). (D) Western blot analysis demonstrated that WDR83-full length or WDR83-3′UTR significantly increased DHPS protein levels in MGC803 cells. (E) Endogenous WDR83 was first knocked down by siRNA for 24 h and then restored by the exogenous expression of WDR83-3′UTR (pIRES2-WDR83-3′UTR) for further 24 h. Cells were harvested and the levels of DHPS mRNA were measured by qPCR. (F) The luciferase activity of pmirGLO-DHPS-3′UTR was increased significantly in cells that were cotransfected with the WDR83 3′UTR plasmid or the WDR83 full-length plasmid (P = 0.0481 and 0.0490, respectively), and the luciferase activity of pmirGLO-DHPS-3′UTR was markedly decreased in cells with knocked down WDR83 (P = 0.006; n ≥ 3, ANOVA; *P< 0.05, **P< 0.01, ***P< 0.001).
DHPS and WDR83 formed an RNA duplex and mutually increased their stability
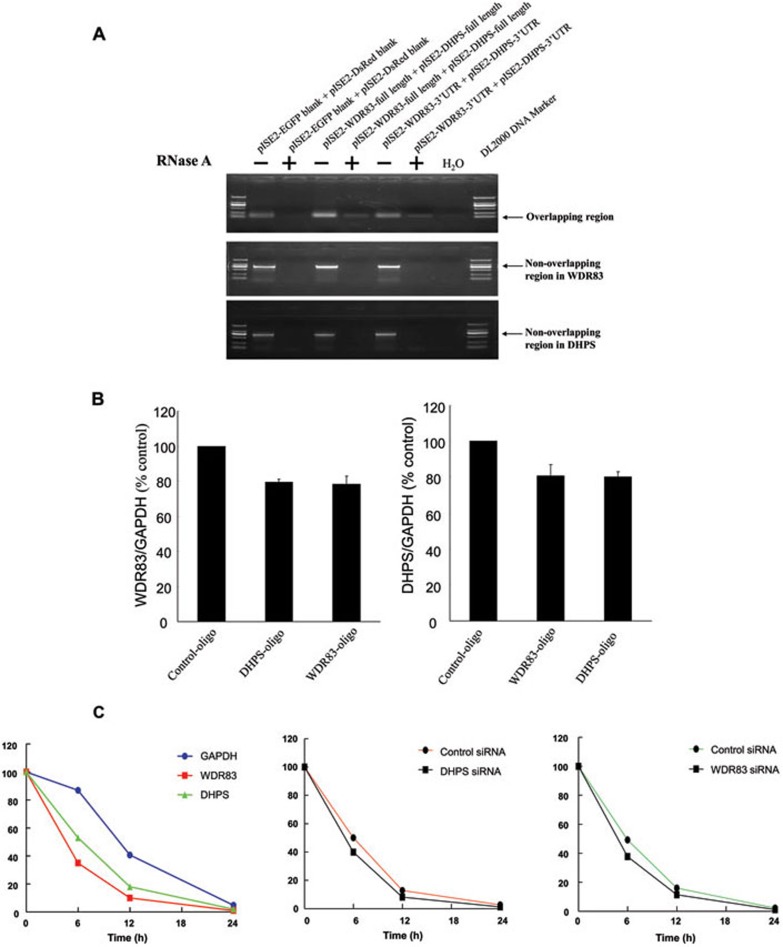
The above data revealed that the overlapping 3′UTR was critical in a bidirectional regulation between WDR83 and DHPS. Sense and antisense transcripts may potentially form double-stranded RNA structures that regulate the stability, transport or translation of the sense transcript 7. To examine the possibility of RNA duplex formation between WDR83 and DHPS, we cotransfected the WDR83 3′UTR plasmids and DHPS 3′UTR plasmids into MGC803 cells, and performed an RNase protection assay on RNA from MGC803 cells. Single-stranded RNA was digested with RNase A and the remaining double-stranded RNA was analyzed by RT-PCR using primers that corresponded to different regions of both transcripts. Only primers within the overlapping region of WDR83 and DHPS generated a PCR product (Figure 5A). Therefore, the overlapping part of both transcripts was protected from degradation. It was therefore expected that blocking the RNA/RNA interaction could inhibit the bidirectional regulation between these two genes. We designed 2′-O-methyl oligoribonucleotides to inhibit the formation of the WDR83/DHPS RNA duplex. These oligoribonucleotides were small single-stranded RNA fragments that were utilized previously to block SA pairing 18 and miRNA binding to miRNA targets 19. As shown in Figure 5B, the transfection of DHPS-specific oligonucleotides resulted in a decrease of WDR83 transcripts (P = 0.0192); meanwhile, the transfection of WDR83-specific oligonucleotides reduced DHPS transcripts (P < 0.001). These data further confirmed that the RNA-RNA duplex was required for the bidirectional regulation between WDR83 and DHPS.
Figure 5.
DHPS and WDR83 formed RNA duplex and increased stability of each other. (A) RPA was performed on RNA samples from MGC803 cells. The plasmids of pIRES2-WDR83-full length and pIRES2-DHPS-full length were cotransfected in the same reaction, and the plasmids of pIRES2-WDR83-3′UTR and pIRES2-DHPS-3′UTR were also cotransfected in the same reaction. Total RNA was extracted and purified, single-stranded RNA was digested with RNase A, and the remaining double-stranded RNA was subjected to RT-PCR to amplify the overlapping or non-overlapping regions of WDR83 and DHPS. (B) Blocking the WDR83/DHPS RNA-RNA interaction with 2′-O-methyl oligoribonucleotides inhibited the bidirectional regulation between them. (C) Stability of WDR83 and DHPS over time was measured by qPCR relative to time 0 after blocking new RNA synthesis with α-amanitin (25 mM). MGC803 cells were transfected with siRNA against WDR83 or DHPS or control siRNA for 24 h, and were then further exposed to 25 mM α-amanitin for 6, 12 and 24 h. Cells were harvested and the stability of the WDR83 and DHPS mRNA was analyzed by qPCR. 18S RNA, which was a product of RNA polymerase I and was unchanged after α-amanitin treatment, was used as the control.
It was reported that the RNA-RNA duplex could protect mRNA from RNase degradation and enhance mRNA stability 9, 20. To investigate whether the overlapping 3′UTR of WDR83 and DHPS RNA duplex affected the stability of the mRNA, we blocked any new mRNA synthesis with α-amanitin (an RNA polymerase II inhibitor) and analyzed the expression of WDR83, DHPS, GAPDH and 18S RNA over a 24-h period. 18S RNA, which was a product of RNA polymerase I and was unchanged after α-amanitin treatment, served as the internal control 9. As shown in Figure 5C, compared with control siRNAs, treatment with the specific siRNAs against DHPS resulted in a decreased stability of WDR83 mRNA; WDR83 knockdown also caused a reduced stability of DHPS mRNA. Taken together, these results suggested that the RNA duplex formed between WDR83 and DHPS increased the stability of both transcripts, which was required for their bidirectional regulation.
The above data proved that bidirectional antisense regulation occurred, at least at the post-transcriptional level. To examine whether this regulation might occur at the transcriptional level, we designed specific primers (listed in Supplementary information, Table S1) to detect the expression of DHPS or WDR83 pre-mRNA. Neither the expression of WDR83 pre-mRNA nor the expression of DHPS pre-mRNA was significantly changed when the partner gene was overexpressed or silenced (Supplementary information, Figure S1), which indicated that transcriptional regulation might not participate in the DHPS-WDR83 antisense control.
DHPS and WDR83 participated in the regulation of ERK signaling and E2F1 in GC cells
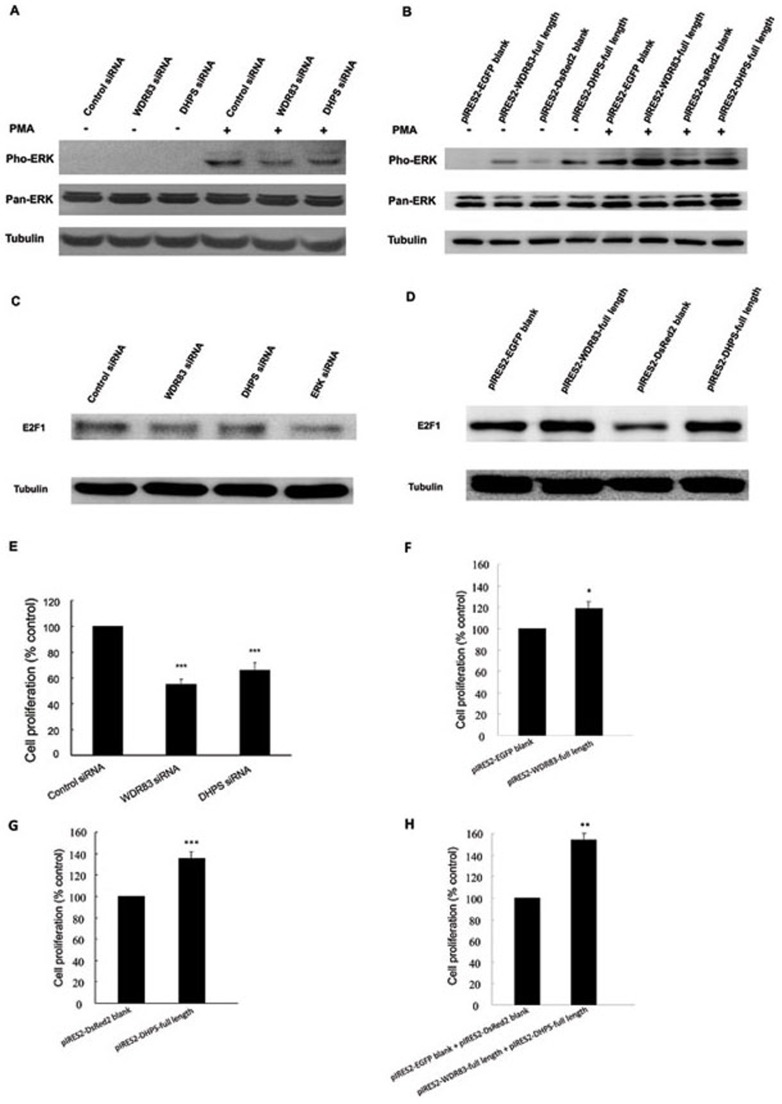
It has been reported that WDR83 selectively facilitates the activation of ERK signaling in an agonist-specific manner 10. The aberrant activation of the ERK pathway is frequently observed in human cancers 21, 22. To test whether WDR83 participated in agonist-specific ERK signaling in GC, MGC803 cells were transfected with WDR83 siRNAs. After serum starvation, cells were stimulated with 5 nM phorbol 12-myristate 13-acetate (PMA) and ERK activation was assessed by western blotting using an anti-phospho-ERK antibody. As shown in Figure 6A, the WDR83 knockdown dramatically decreased the PMA-stimulated phosphorylation of ERK1/2 in MGC803 cells; however, no detectable changes in total ERK1/2 protein were seen in WDR83 siRNAs-treated lysates. Thus, these findings suggested that WDR83 could facilitate agonist-specific ERK activation in GC and were consistent with results previously reported in HeLa cells 10. We also observed that the DHPS knockdown decreased ERK1/2 phosphorylation in response to PMA (Figure 6A). In contrast, the overexpression of either WDR83 or DHPS in combination with PMA enhanced ERK1/2 phosphorylation (Figure 6B). Taken together, these data suggested that both WDR83 and DHPS enhanced the activation of ERK signaling.
Figure 6.
WDR83 and DHPS were involved in the regulation of cell proliferation, and the downregulation of WDR83 or DHPS inhibited ERK1/2 activation and E2F1 expression in MGC803 cells. (A) The knockdown of either WDR83 or DHPS decreased PMA-stimulated ERK1/2 phosphorylation compared with negative controls in MGC803 cells. (B) Overexpression of WDR83 or DHPS in combination with PMA enhanced ERK1/2 phosphorylation. (C) The knockdown of WDR83, DHPS, or ERK1/2 resulted in a significant decrease in E2F1 protein levels compared with the control cells. (D) Overexpression of WDR83 or DHPS induced an increase in E2F1 protein levels. (E) Downregulation of WDR83 or DHPS significantly inhibited cell proliferation in MGC803 cells (P ≤ 0.0001 and P = 0.0006, respectively). (F, G) In contrast, the overexpression of WDR83 or DHPS enhanced cell proliferation (P = 0.0148 and P = 0.0003, respectively). (H) Interestingly, the joint overexpression of WDR83 and DHPS enhanced cell proliferation even further (P = 0.0037). Cell proliferation was determined by the CCK-8 assay (n ≥ 3, ANOVA; *P< 0.05, **P< 0.01, ***P< 0.001).
Activated ERK1/2 might lead to the release and activation of the transcriptional factor, E2F1 23, 24. Indeed, silencing ERK1/2 in MGC803 cells led to reduced E2F1 protein levels compared to control siRNAs (Figure 6C), suggesting that ERK1/2 could mediate E2F1 activation in GC, which was consistent with previous reports in HECs cells 25. Considering that WDR83 and DHPS could facilitate ERK signaling activation, we further examined whether E2F1 could be modulated by WDR83 or DHPS in GC cells. As shown in Figure 6C, the knockdown of either WDR83 or DHPS resulted in a decrease in E2F1 protein expression compared with control siRNAs, and the overexpression of either WDR83 or DHPS induced an increase in E2F1 protein levels (Figure 6D).
Both WDR83 and DHPS enhanced cell proliferation
Our finding that WDR83 and DHPS expression was significantly increased in GC samples prompted us to explore their functional significance in GC. As illustrated in Figure 6E, the downregulation of WDR83 or DHPS inhibited the cell proliferation in GC cells. In contrast, the overexpression of either WDR83 or DHPS enhanced cell proliferation (Figure 6F and 6G). The joint overexpression of WDR83 and DHPS enhanced cell proliferation even further (Figure 6H). Therefore, WDR83 and DHPS might be involved in the control of cell proliferation in GC cells.
The interaction between WDR83 and DHPS existed in other cancers and species
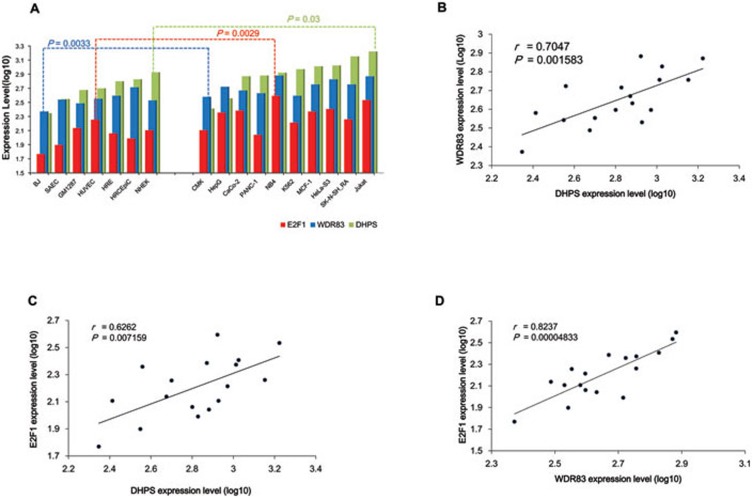
To extend our studies beyond GC, we examined the expression of WDR83 and DHPS in other cancer types. Transcriptional profiling of normal and cancer cell lines from the University of Washington Encyclopedia of DNA Elements (ENCODE) group (downloaded from the University of California, Santa Cruz (UCSC) database 26) was performed. Compared with normal cell lines, the expressions of WDR83 and DHPS in cancer cell lines (described in Supplementary information, Data S1) were markedly increased (P = 0.0033 and 0.03, respectively, Figure 7A) and positively correlated (r =0.7047, P = 0.0016, Figure 7B). These results provided evidence that WDR83 and DHPS play important roles in tumor progression and the positive correlation between them was common to other cancers. Moreover, a significant increase in E2F1 expression was observed in cancer cells compared with normal cells (P = 0.0029, Figure 7A), which indicated the essential role of E2F1 in cancer biogenesis. Meanwhile, the expression of E2F1 and DHPS, as well as E2F1 and WDR83, exhibited significantly positive correlations in multiple cell lines (r =0.6262, P = 0.007159, Figure 7C and r =0.8237, P = 0.0004833, Figure 7D). This finding supported our observations that WDR83 and DHPS might contribute to the enhancement of cell proliferation in GC cells.
Figure 7.
Positive correlations between WDR83, DHPS, and E2F1 in other cancers. The expressions of WDR83, DHPS, and E2F1 were analyzed in normal (n = 7) and cancer (n = 10) cell lines. Transcriptional profiles were derived from the ENCODE Transcriptome Project with the Affymetrix Human Exon 1.0 ST array. (A) The expression of WDR83, DHPS, and E2F1 was increased significantly in cancer cells compared with normal cells (P = 0.0033, 0.030, and 0.0029, respectively). (B-D) The expression of WDR83, E2F1, and DHPS exhibited positive correlation in multiple cell lines (P = 0.0016, 0.0071, and 0.00005, respectively).
Next, we studied the evolution of the interaction between WDR83 and DHPS in other species. Based on the HomoloGene annotation 27, WDR83 and DHPS were found to be well conserved at the protein level. They were widely expressed in eukaryotes (HomoloGene 12194 and 1453, respectively). The ensemble 64 gene tree homology annotation confirmed that both genes had one-to-one orthologs in most eukaryotes, with the exception of at least two DHPS paralogs in stickleback and puffer fish. As we showed that DHPS and WDR83 exhibited bidirectional RNA-RNA interaction through their overlapping 3′UTRs, we were interested in investigating whether their overlapping regions were as well conserved as their protein products. By tracking their genomic regions in well-annotated genomes, we found that DHPS and WDR83 were not located at the same locus until the origin of tetrapoda. Furthermore, in order to track when these two genes formed a cis-sense/antisense pair, we selected representative species from distinct phylogenetic branches of tetrapoda. The DHPS and WDR83 genes in these species contained identical 3′ end structures based on the transcriptome data (Supplementary information, Table S2 and Figure S2). These results showed that they overlapped with each other in all species, suggesting that they had formed cis-sense/antisense pairs since the origin of tetrapoda. Both the genomic alignment and transcriptional signals indicated that although WDR83 and DHPS were well conserved in the coding region across eukaryotes, the cis-RNA-RNA interactions between WDR83 and DHPS only existed in tetrapoda and were relatively young in terms of evolution compared to the coding region.
Discussion
In this study, we characterized a cis-SA pair that consisted of two protein-coding genes, WDR83 and DHPS, which were upregulated simultaneously and positively correlated in GC and other cancers. The regulation between DHPS and WDR83 was bidirectional. They mutually affected both of their mRNA and protein expression levels through their overlapping 3′UTRs. Different from previous unidirectional cis-SA regulation 18, the bidirectional regulation between WDR83 and DHPS enriches the understanding of post-transcriptional antisense regulation, as it indicates that SA interactions are much more complicated than previously thought.
There were two types of regulation between sense and NATs: (1) discordant regulation, where the NATs inhibit the sense RNAs or proteins; and (2) concordant regulation, where NATs increase the level of the sense RNAs or the corresponding proteins 9, 28. Here, the regulation between WDR83 and DHPS occurred in a concordant manner. This could at least be explained that the cis-SA pairing was capable of forming a sense-antisense RNA duplex at their overlapping regions, which might reduce mRNA degradation at the post-transcriptional level. Although endogenous RNA duplexes have been difficult to identify in human cells 18, 29, blocking the WDR83/DHPS RNA-RNA interaction with 2′-O-methyl oligoribonucleotides and in vitro RNA-duplex detection provided solid evidence that these two genes could form an RNA duplex. Other possible mechanisms for their concordant expression require further investigation.
The overexpression of DHPS 3′UTR or DHPS full-length construct significantly increased WDR83 expression and the luciferase activity of pmirGLO-DHPS-3′UTR. The same result was obtained in the regulation of DHPS by WDR83. However, the change of gene expression level or the luciferase activity in cells that were transfected with the partner gene full-length plasmids was lower than those achieved with 3′UTR plasmids. This difference was possibly due to the secondary structure formed by the full-length sequence. The intra-molecular secondary structure might constrain the accessibility of nucleotides, which participated in inter-molecular interactions 30. This might interfere with inter-molecular antisense regulation. Using Mfold 31, we predicted the secondary structures of DHPS and WDR83 full-length sequences and indeed found that the antisense overlapping regions participated in secondary structure formation with other regions (Supplementary information, Figure S3). Nevertheless, the results still supported the conclusion that the antisense 3′UTR played essential roles in bidirectional RNA regulation.
The overlapping region of WDR83 and DHPS was shown to exist since tetrapoda. The conservation of genomic elements suggested functional importance. We investigated the expression pattern of murine Wdr83 and Dhps in four mouse tissue types following acute caloric restriction 32, and found a strongly positive correlation between Dhps and Wdr83 mRNA expression (Supplementary information, Figure S4), which was similar to the pattern observed in human cancers. The expression pattern indicated that Dhps and Wdr83 might also perform a concordant antisense regulatory function in the mouse. Although the overlapping regions and coding regions of WDR83 and DHPS were highly conserved across species, the neighboring regions of these two genes differed between the human and mouse genomes. In humans, WDR83 shares a putative bidirectional promoter with another gene, MAN2B1. In the mouse, Wdr83 shares a putative bidirectional promoter with the BC056474 gene, which is not homologous to the human MAN2B1, indicating a different gene chain between species 33, 34. As a bidirectional promoter might lead to the coexpression of neighboring genes 35, the interspecies gene structure differences within this genomic locus might result in the coexpression of distinct gene triplets.
The bidirectional regulation between WDR83 and DHPS might at least be due to the WDR83-DHPS RNA duplex that forms at their overlapping 3′UTRs, where the deadenylation-dependent mRNA decay pathway is initiated and the poly(A)-tail is shortened 36. The RNA duplex might protect their 3′ ends from being attacked by exosomes. Another mechanism for the bidirectional regulation might be associated with RNA duplex masking miRNA-binding sites in both genes. It has been reported that antisense transcripts might compete for binding sites against miRNAs and obstruct miRNA-mediated regulation of target genes 37. Indeed, based on the microcosm database 38, we found 27 miRNA target sites in WDR83 and 11 miRNA-binding sites in DHPS at their overlapping regions (Supplementary information, Tables S3 and S4). Therefore, antisense pairing between WDR83 and DHPS might also interrupt the miRNA-mediated inhibition of target genes, which would increase the mRNA stability or the translation efficiency of both transcripts. However, further study on the exact mechanism is necessary.
In addition, we demonstrated that DHPS and WDR83 mRNA displayed two functions: they delivered protein-coding information by protein synthesis and exerted endogenous post-transcriptional regulatory roles in their mutual expression through their overlapping 3′UTRs. The canonical role of mRNA is to deliver protein-coding information to sites of protein synthesis, but our findings have shown that mRNAs also exert regulatory roles in their targeting genes, independently of their protein-coding functions. Thus, our observations emphasize the need to move beyond the confines of protein-coding mRNA and to highlight the regulatory roles of mRNA.
We also evaluated the biological function of WDR83 and DHPS in GC. WDR83 and DHPS played important roles in driving GC pathophysiology. The downregulation of WDR83 or DHPS inhibited cell proliferation in GC cells; conversely, the overexpression of WDR83 or DHPS enhanced cell proliferation. Therefore, WDR83 and DHPS drove GC pathophysiology by promoting cell proliferation. It has been reported that aberrant activation of ERK signaling was frequently seen in human cancers and clearly contributed to enhancing cell proliferation and survival 21, 22. E2F1 was also an important component of cell proliferation control 39, 40. Thus, the enhanced cell proliferation in GC cells by WDR83 and DHPS was perhaps possible as both genes were involved in the regulation of ERK signaling activation and E2F1 expression. Moreover, we extended our studies to other cancers. An analysis of transcriptional profiles from ENCODE group indicated that the expression of WDR83 and DHPS was also markedly increased in other cancers, which supported our hypothesis that both DHPS and WDR83 have oncogenic roles.
In this study, we studied the influence of antisense regulation on ERK signaling. WDR83 also attenuated expression of the HIF-1α by activating or stabilizing PHD3 14, 41. This antisense interaction might also indirectly regulate HIF-1α. More interestingly, HIF-1α was reported to be regulated by its two NATs 42, 43. Therefore, the regulation of HIF-1α may be more complex than previously thought, as it might comprise a gene network that includes multiple cis-sense/antisense pairs.
In conclusion, we have provided the first evidence of bidirectional regulation between the WDR83 and DHPS genes in human cells. This pair of protein-coding cis-SA transcripts could play an important role in the tumorigenesis of GC, as it appears to be involved in the regulation of ERK signaling and E2F1 expression. Further studies of the cis-SA pair may provide a more complete understanding of their functions in tumor development and eventually open avenues for novel therapeutic interventions in cancer.
Materials and Methods
Clinical and histological evaluation of human samples in this study
The human samples in this study were approved by the local ethics committee at the Shanghai Jiao-Tong University School of Medicine Renji Hospital. This research was performed according to the provisions of the Helsinki Declaration of 1975. None of the patients received preoperative treatment, including radiotherapy or chemotherapy. All tumor samples were examined histologically and all nontumorous tissues were taken at a distance of at least 5 cm from the tumor, which were confirmed to be normal and free of tumor cells using hematoxylin-eosin staining.
qPCR analysis
Total RNA was extracted from 19 human GC specimens (cancer and adjacent nontumorous tissues). Primary GC tissues and their matched normal tissues were obtained with informed consent from 19 patients in the Department of Surgery, Shanghai Jiao-Tong University School of Medicine Renji Hospital, the methods for estimating the sample number was described in Supplementary information, Data S1.
RNA samples were treated with RNase-free DNase (Ambion, Austin, TX, USA) according to the manufacturer's instructions. Reverse transcription was performed with random primers using the First Strand cDNA Synthesis Kit (Takara, Shiga, Japan). qPCR was performed using an ABI Prism 7300HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) with SYBR Premix Ex Taq II (Takara) and the 2−ΔΔCT method as described previously 44. The expression of each target gene was normalized to that of GAPDH. The primer sequences used are listed in Supplementary information, Table S1.
Subcellular transcriptional analysis of WDR83 and DHPS
The subcellular transcription of WDR83 and DHPS was analyzed using Affymetrix ENCODE Tiling Array, a tiling 91-array set containing probes against the non-repetitive portions of the human genome (details are described in Supplementary information, Data S1).
Immunohistochemistry
Immunohistochemistry (IHC) was performed using a standard streptavidin-biotin-peroxidase complex method 45. In brief, slides with paraffin sections were deparaffinized and rehydrated. Endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide for 15 min. For antigen retrieval, slides were microwave treated and boiled in 10 mM citrate buffer (pH 6.0) for 10 min. Nonspecific binding was blocked with 10% normal rabbit serum for 30 min. The slides were incubated with anti-WDR83 (1:500, Abcam, Cambridge, UK) or anti-DHPS (1:500, Sigma-Aldrich, St Louis, MO, USA) antibody overnight at 4 °C. A negative control was obtained by replacing the primary antibody with PBS.
Cells and culture conditions
The human GC cell line MGC803 was cultured in RPMI-1640 medium (Gibco, Gaithersburg, MD, USA) supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin and maintained at 37 °C in a humidified atmosphere with 5% CO2. For the stimulation experiments, PMA (Sigma) was dissolved in DMSO and diluted with tissue culture media to a final concentration of 5 nM. α-Amanitin (Sigma) was dissolved in distilled water and diluted to a final concentration of 25 mM.
RNA interference
Commercial siRNAs that specifically targeted the coding regions in either WDR83 (GenBank accession number: NM_001099737) or DHPS (GenBank accession number: NM_001930) were purchased from Qiagen (Hilden, Germany). ERK1/2 siRNAs were used as described previously 46. The uniqueness of the siRNA target sequences for the human genome was confirmed by the UCSC BLAT program. The siRNA sequences are listed in Supplementary information, Table S1. Briefly, MGC803 cells were plated in regular growth medium at 40%-50% confluence in six-well tissue plates 24 h before transfection, and then transfected with 5 nM siRNA using HiPerFect Transfection Reagent (Qiagen) in accordance with the manufacturer's instructions. Nonspecific siRNAs (Qiagen) was used as the negative control. After 48 h, the cells were collected for various assays. The selective silencing was confirmed by qPCR and western blotting analysis.
2′-O-methyl oligoribonucleotides inhibition
Cells were plated in regular growth medium at 40%-50% confluence in 12-well tissue plates 24 h before transfection and then transfected with 2′-O-methyl oligoribonucleotides (Invitrogen) at a 40 pmol/ml concentration using Lipofectamine 2000 (Invitrogen). After 24 h, the cells were collected for qPCR analysis. The 2′-O-methyl oligoribonucleotide sequences are listed in Supplementary information, Table S1.
Stability and α-amanitin treatment
MGC803 cells were plated in regular growth medium at 50%-60% confluence in six-well tissue plates 24 h before transfection and then transfected with 5 nM siRNA using HiPerFect Transfection Reagent (Qiagen) in accordance with the manufacturer's instructions. Nonspecific siRNA (Qiagen) was used as the negative control. After further 24 h incubation, cells were treated with 25 mM of α-amanitin and harvested for RNA purification and RT-PCR at 6, 12 and 24 h post treatment. We used three independent samples for each data point; all samples had untreated and untransfected matching samples for RNA purification and data analysis, as previously described 9.
Plasmids construction and transfection
Of the three splice variants of WDR83 transcripts, NM_001099737 and NM_032332 have coding potentials at the same coding regions. Full-length WDR83 cDNA (GenBank accession number: NM_001099737) was synthesized, and the CDS and 3′UTR were amplified by PCR from full-length WDR83 (the primers are listed in Supplementary information, Table S1). The pIRES2-EGFP vector (Clontech, Santa Clara, CA, USA) was used to construct three plasmids: pIRES2-WDR83-full length (the plasmid containing full-length WDR83), pIRES2-WDR83-CDS (the plasmid containing only the CDS region of WDR83) and pIRES2-WDR83-3′UTR (the plasmid containing only the 3′UTR of WDR83).
Of the three DHPS transcript variants, only NM_001930 encodes a protein with high DHPS activity 47, 48. Full-length DHPS cDNA (GenBank accession number: NM_001930) was synthesized, and the CDS and 3′UTR were amplified by PCR from full-length DHPS (the primers are listed in Supplementary information, Table S1). Three plasmids were constructed: pIRES2-DHPS-full length (the plasmid containing full-length DHPS), pIRES2-DHPS-CDS (the plasmid containing the CDS region of DHPS) and pIRES2-DHPS-3′UTR (the plasmid containing the 3′UTR of DHPS).
All constructs were verified by DNA sequencing. For plasmid transfection, cells were seeded in six-well tissue culture dishes 24 h before transfection and then transfected with plasmids (3-4 μg) using LipofectamineTM 2000 Reagent (Invitrogen) following the manufacturer's instructions. After 48 h of standard incubation, cells were trypsinized and used for various assays.
Luciferase assays
To further investigate whether the regulation between WDR83 and DHPS was mediated directly through their overlapping 3′UTRs, we constructed two luciferase reporter vectors: pmirGLO-WDR83-3′UTR and pmirGLO-DHPS-3′UTR (Supplementary information, Figure S5). The 3′UTR of WDR83 was amplified by PCR from the pIRES2-WDR83-3′UTR plasmid and inserted into the pmirGLO vector between the XhoI and SalI sites (Promega, Madison, WI, USA). The 3′UTR of DHPS was amplified by PCR from the pIRES2-DHPS-3′UTR plasmid and inserted into the pmirGLO vector between the SacI and XhoI sites. All constructs were verified by DNA sequencing. The plasmids were transfected into MGC803 cells with Lipofectamine 2000 (Invitrogen) as described previously. After 48 h of standard incubation, cells were assayed by the Dual-Luciferase Reporter Assay System Kit (Promega). All experiments were performed in triplicate with data pooled from at least three independent experiments.
RNase protection assay
The total RNA was purified by TRIzol reagent (Invitrogen). The RNA was divided into three parts: the sample of interest and the positive and negative controls. All samples were treated with DNase I (Invitrogen) in DNase I buffer. The negative control was heat-denatured at 94 °C for 15 min. Then, the sample of interest and negative control were treated with RNase A (Invitrogen) and the positive control was treated with RNase Out (Invitrogen) as previously described 9, 18. Following the RPA assay, we used RT-PCR to detect duplex formation by primers within the overlapping region or non-overlapping regions (the primers are listed in Supplementary information, Table S1).
Western blotting
Western blotting was performed according to standard protocols as described previously 49. Antibody information is listed in Supplementary information, Data S1.
Cell viability assay
Cell viability was assessed by a tetrazolium salt (WST-8)-based colorimetric assay using the Cell Counting Kit 8 (CCK-8; Dojindo, Rockville, MD, USA). Briefly, control and treated GC cells were seeded into 96-well plates at an initial density of 5 × 103 cells/well. At the specified time points, 10 μl of CCK-8 solution was added to each well and incubated for 1 h. Cell viability was determined by scanning with a microplate reader at 450 nm. Data were calculated as the percentage of viable cells as follows: relative viability (%) = (A450 (treated) − A450 (blank))/(A450 (control) − A450 (blank)) × 100%.
WDR83, DHPS, and E2F1 expression analysis in other cancers
The expression of WDR83, DHPS, and E2F1 in other cancers was analyzed using the Affymetrix Human Exon 1.0 ST array platform. The detailed protocol is shown in Supplementary information, Data S1.
Statistical analysis
Data are expressed as mean ± SD. Statistical differences between the two groups were determined by the Student's t-test. Differences between multiple groups were tested using analysis of variance (ANOVA). Regression and correlation coefficient analyses were performed using GraphPad Prism (GraphPad Software, Inc., La Jolla, CA, USA). All experiments were repeated at least three times.
Acknowledgments
We are grateful to Dr Tim (Qing-Rong) Liu (National Institutes of Health, USA) for insightful suggestions. We thank the ENCODE Consortium, including the University of Washington ENCODE group, the transcriptome group at Affymetrix, and Cold Spring Harbor Laboratories, for providing the open-accessible transcriptome data and the labs of Michael Snyder, Mark Gerstein, and Sherman Weissman at Yale University and Peggy Farnham at University of California, Davis. This work was supported by grants from the National Basic Research Program of China (973 program; 2010CB5293 to JYF and 2007CB946904 to LPW), the National Natural Science Foundation of China (30971330 to JYF; National Outstanding Young Investigator Award (31025014) to LPW), the China Ministry of Science and Technology (863 Hi-Tech R&D Program; 2007AA02Z165 to LPW), and the Doctor Innovation Foundation of Shanghai Jiao-Tong University School of Medicine (BXJ201117 to WYS).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
Overlapping sequence between DHPS and WDR83 in representative species in tetraodon
miRNA binding sites in DHPS overlapping regions, which starts from the 5th base of DHPS 3′UTR
miRNA binding sites in WDR83 overlapping regions, which starts from the 44th base of WDR83 3′UTR.
(A, B) To analyze WDR83 or DHPS pre-mRNA, intron specific primers were designed (Supplementary information, Table S1).
Sequence alignment of antisense regions in representative species with Clustalw 1.
(A) The second structure of DHPS transcript predicted by Mfold 1.
The positive correlation between Dhps and Wdr83 in mouse.
Two luciferase reporter vectors pmirGLO-WDR83-3′UTR and pmirGLO-DHPS-3′UTR were constructed.
Materials and Methods
References
- Katayama S, Tomaru Y, Kasukawa T, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Werner A, Sayer JA. Naturally occurring antisense RNA: function and mechanisms of action. Curr Opin Nephrol Hypertens. 2009;18:343–349. doi: 10.1097/MNH.0b013e32832cb982. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu XS, Liu QR, Wei L. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res. 2006;34:3465–3475. doi: 10.1093/nar/gkl473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JT, Zhang Y, Kong L, Liu QR, Wei L. Trans-natural antisense transcripts including noncoding RNAs in 10 species: implications for expression regulation. Nucleic Acids Res. 2008;36:4833–4844. doi: 10.1093/nar/gkn470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiter T, Reich E, Williams RW, Simon P. Antisense transcription: a critical look in both directions. Cell Mol Life Sci. 2009;66:94–112. doi: 10.1007/s00018-008-8381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A, Carlile M, Swan D. What do natural antisense transcripts regulate. RNA Biol. 2009;6:43–48. doi: 10.4161/rna.6.1.7568. [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Gius D, Onyango P, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vomastek T, Schaeffer HJ, Tarcsafalvi A, Smolkin ME, Bissonette EA, Weber MJ. Modular construction of a signaling scaffold: MORG1 interacts with components of the ERK cascade and links ERK signaling to specific agonists. Proc Natl Acad Sci USA. 2004;101:6981–6986. doi: 10.1073/pnas.0305894101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- Lee SH, Hu LL, Gonzalez-Navajas J, et al. ERK activation drives intestinal tumorigenesis in Apc(min/+) mice. Nat Med. 2010;16:665–670. doi: 10.1038/nm.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Zong CS, Xia W, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008;10:138–148. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfer U, Hopfer H, Jablonski K, Stahl RA, Wolf G. The novel WD-repeat protein Morg1 acts as a molecular scaffold for hypoxia-inducible factor prolyl hydroxylase 3 (PHD3) J Biol Chem. 2006;281:8645–8655. doi: 10.1074/jbc.M513751200. [DOI] [PubMed] [Google Scholar]
- Monti E, Gariboldi MB. HIF-1 as a target for cancer chemotherapy, chemosensitization and chemoprevention. Curr Mol Pharmacol. 2011;4:62–77. doi: 10.2174/1874467211104010062. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li J, Kong L, Gao G, Liu QR, Wei L.NATsDB: Natural Antisense Transcripts DataBase Nucleic Acids Res 200735Database issue:D156–D161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Nishizawa M, Ozaki T, et al. Natural antisense transcript stabilizes inducible nitric oxide synthase messenger RNA in rat hepatocytes. Hepatology. 2008;47:686–697. doi: 10.1002/hep.22036. [DOI] [PubMed] [Google Scholar]
- Mahmoudi S, Henriksson S, Corcoran M, Mendez-Vidal C, Wiman KG, Farnebo M. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol Cell. 2009;33:462–471. doi: 10.1016/j.molcel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stazic D, Lindell D, Steglich C. Antisense RNA protects mRNA from RNase E degradation by RNA-RNA duplex formation during phage infection. Nucleic Acids Res. 2011;39:4890–4899. doi: 10.1093/nar/gkr037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- Hong SK, Yoon S, Moelling C, Arthan D, Park JI. Noncatalytic function of ERK1/2 can promote Raf/MEK/ERK-mediated growth arrest signaling. J Biol Chem. 2009;284:33006–33018. doi: 10.1074/jbc.M109.012591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotayev K, Chaussepied M, Ginsberg D. ERK activation is regulated by E2F1 and is essential for E2F1-induced S phase entry. Cell Signal. 2008;20:1221–1226. doi: 10.1016/j.cellsig.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Lavoie JN, L'Allemain G, Brunet A, Muller R, Pouyssegur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- Pintus G, Tadolini B, Posadino AM, et al. PKC/Raf/MEK/ERK signaling pathway modulates native-LDL-induced E2F-1 gene expression and endothelial cell proliferation. Cardiovasc Res. 2003;59:934–944. doi: 10.1016/s0008-6363(03)00526-1. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information Nucleic Acids Res 201139Database issue:D38–D51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su WY, Xiong H, Fang JY. Natural antisense transcripts regulate gene expression in an epigenetic manner. Biochem Biophys Res Commun. 2010;396:177–181. doi: 10.1016/j.bbrc.2010.04.147. [DOI] [PubMed] [Google Scholar]
- Munroe SH, Zhu J. Overlapping transcripts, double-stranded RNA and antisense regulation: a genomic perspective. Cell Mol Life Sci. 2006;63:2102–2118. doi: 10.1007/s00018-006-6070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann SE, Menzel P, Backofen R, Gorodkin J. The PETfold and PETcofold web servers for intra- and intermolecular structures of multiple RNA sequences. Nucleic Acids Res. 2011;39 Web server issue:W107–111. doi: 10.1093/nar/gkr248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman C, Kerrison ND, Cooray A, et al. Coordinated multitissue transcriptional and plasma metabonomic profiles following acute caloric restriction in mice. Physiol Genomics. 2006;27:187–200. doi: 10.1152/physiolgenomics.00084.2006. [DOI] [PubMed] [Google Scholar]
- Engstrom PG, Suzuki H, Ninomiya N, et al. Complex loci in human and mouse genomes. PLoS Genet. 2006;2:e47. doi: 10.1371/journal.pgen.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N, Kasukawa T, Oyama R, et al. Transcript annotation in FANTOM3: mouse gene catalog based on physical cDNAs. PLoS Genet. 2006;2:e62. doi: 10.1371/journal.pgen.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins PJ, Kobayashi Y, Nguyen L, Trinklein ND, Myers RM. The ets-related transcription factor GABP directs bidirectional transcription. PLoS Genet. 2007;3:e208. doi: 10.1371/journal.pgen.0030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Zhang M, Huang J, et al. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ.miRBase: microRNA sequences, targets and gene nomenclature Nucleic Acids Res 200634Database issue:D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DG, Schwarz JK, Cress WD, Nevins JR. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- Real S, Meo-Evoli N, Espada L, Tauler A. E2F1 regulates cellular growth by mTORC1 signaling. PLoS One. 2011;6:e16163. doi: 10.1371/journal.pone.0016163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase D, Keiner S, Mawrin C, Wolf G. Reduced Morg1 expression in ischemic human brain. Neurosci Lett. 2009;455:46–50. doi: 10.1016/j.neulet.2009.03.048. [DOI] [PubMed] [Google Scholar]
- Rossignol F, Vache C, Clottes E. Natural antisense transcripts of hypoxia-inducible factor 1alpha are detected in different normal and tumour human tissues. Gene. 2002;299:135–140. doi: 10.1016/s0378-1119(02)01049-1. [DOI] [PubMed] [Google Scholar]
- Span PN, Rao JU, Oude Ophuis SB, et al. Overexpression of the natural antisense hypoxia-inducible factor-1alpha transcript is associated with malignant pheochromocytoma/paraganglioma. Endocr Relat Cancer. 2011;18:323–331. doi: 10.1530/ERC-10-0184. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Tang DJ, Dong SS, Ma NF, et al. Overexpression of eukaryotic initiation factor 5A2 enhances cell motility and promotes tumor metastasis in hepatocellular carcinoma. Hepatology. 2010;51:1255–1263. doi: 10.1002/hep.23451. [DOI] [PubMed] [Google Scholar]
- Dimitri CA, Dowdle W, MacKeigan JP, Blenis J, Murphy LO. Spatially separate docking sites on ERK2 regulate distinct signaling events in vivo. Curr Biol. 2005;15:1319–1324. doi: 10.1016/j.cub.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Yan YP, Tao Y, Chen KY. Molecular cloning and functional expression of human deoxyhypusine synthase cDNA based on expressed sequence tag information. Biochem J. 1996;315(Pt 2):429–434. doi: 10.1042/bj3150429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe YA, Wolff EC, Park MH. Cloning and expression of human deoxyhypusine synthase cDNA. Structure-function studies with the recombinant enzyme and mutant proteins. J Biol Chem. 1995;270:22386–22392. doi: 10.1074/jbc.270.38.22386. [DOI] [PubMed] [Google Scholar]
- Lu R, Wang X, Chen ZF, Sun DF, Tian XQ, Fang JY. Inhibition of the extracellular signal-regulated kinase/mitogen-activated protein kinase pathway decreases DNA methylation in colon cancer cells. J Biol Chem. 2007;282:12249–12259. doi: 10.1074/jbc.M608525200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overlapping sequence between DHPS and WDR83 in representative species in tetraodon
miRNA binding sites in DHPS overlapping regions, which starts from the 5th base of DHPS 3′UTR
miRNA binding sites in WDR83 overlapping regions, which starts from the 44th base of WDR83 3′UTR.
(A, B) To analyze WDR83 or DHPS pre-mRNA, intron specific primers were designed (Supplementary information, Table S1).
Sequence alignment of antisense regions in representative species with Clustalw 1.
(A) The second structure of DHPS transcript predicted by Mfold 1.
The positive correlation between Dhps and Wdr83 in mouse.
Two luciferase reporter vectors pmirGLO-WDR83-3′UTR and pmirGLO-DHPS-3′UTR were constructed.
Materials and Methods