Abstract
Otx2 plays essential roles in rostral brain development, and its counteraction with Gbx2 has been suggested to determine the midbrain-hindbrain boundary (MHB) in vertebrates. We previously identified the FM enhancer that is conserved among vertebrates and drives Otx2 transcription in forebrain/midbrain from the early somite stage. In this study, we found that the POU homeodomain of class III POU factors (Brn1, Brn2, Brn4, and Oct6) associates with noncanonical target sequence TAATTA in the FM enhancer. MicroRNA-mediated knockdown of Brn2 and Oct6 diminished the FM enhancer activity in anterior neural progenitor cells (NPCs) differentiated from P19 cells. The class III POU factors associate with the FM enhancer in forebrain and midbrain but not in hindbrain. We also demonstrated that the Gbx2 homeodomain recognizes the same target TAATTA in the FM enhancer, and Gbx2 associates with the FM enhancer in hindbrain. Gbx2 misexpression in the anterior NPCs repressed the FM enhancer activity and inhibited Brn2 association with the enhancer, whereas Gbx2 knockdown caused ectopic Brn2 association in the posterior NPCs. These results suggest that class III POU factors and Gbx2 share the same target site, TAATTA, in the FM enhancer and that their region-specific binding restricts Otx2 expression at the MHB.
INTRODUCTION
One of the most important events in the initial brain regionalization is the formation of the midbrain-hindbrain boundary (MHB), where isthmus, a local organizer for midbrain and hindbrain development, is formed. A homeobox transcription factor gene, Otx2, plays essential roles in the formation and patterning of the rostral brain, while a homeobox gene, Gbx2, does so in caudal brain development (2, 12, 56, 65). Their expression initially overlaps, but segregates around the 6-somite stage; the mutual suppression between them has been suggested to establish the MHB at embryonic day 8.5 (E8.5) and to maintain the isthmic structure at later stages (23, 53); however, the details of the molecular mechanisms of the suppressive interaction between the Otx2 and Gbx2 genes remain to be determined.
We have made efforts to elucidate the molecular mechanisms of the transcriptional regulation of the Otx2 gene and identified a series of cis-regulatory elements (26, 28–;31). Among these, the Otx2 expression in the anterior neural plate at the presomite stage is regulated by the AN enhancer, which is located 90 kb upstream (31, 60); its activity covers the entire anterior neural plate, the caudal limits being obscured and overlapping with the anterior part of the Gbx2 expression. The AN enhancer loses its activity at an early somite stage, and the subsequent Otx2 expression in rostral brain is regulated by the FM and FM2 enhancers, which are located 75 kb upstream and 115 kb downstream, respectively (28). The FM and FM2 enhancers lack activities in the most rostral part that correspond to future telencephalon and hypothalamus, and the caudal limit of their activities coincides with the MHB. Genetic analysis by enhancer mutants suggested that the FM enhancer, rather than the FM2 enhancer, plays major roles in the Otx2 expression in forebrain and midbrain. Moreover, the FM enhancer is conserved among vertebrate Otx2 orthologues, from skate to mammal and teleost orthologues, but the FM2 enhancer is unique to rodent Otx2 orthologues (28, 30). The antagonistic interaction between Otx2 and Gbx has been suggested not only in mice, but also in chicks (19, 24), Xenopus (61, 62), and zebrafish (11, 25). Therefore, it is probable that the FM enhancer is the target of the Gbx2 regulation of the Otx2 expression for MHB formation. The sequences essential to the FM enhancer and conserved among vertebrates include bicoid-type homeobox protein (BHP), two Tcf/Lef recognition sequences, and AGAATTTGCCTTCTAATTAAAAAGGATAA (X29) (28, 30). In this study, we have first tried to identify the factors that bind to the X29 sequences.
Brn1, Brn2, and Brn4 are class III POU factors that are expressed in the neural tube (16). The other member of the class III POU factors, Oct6, is also found in the anterior neuroectoderm (58, 68). The POU factors have been known to interact with canonical octamer sequence ATGCAAAT, through its POU-specific (POUS) domain and POU homeodomain (POUH). Their roles in initial brain development have not been demonstrated by each single mutant or by the Brn1 Brn2 double mutant (7, 37, 41–43, 57), suggesting their complementary functions in early brain development. In this study, we first demonstrate that the POU homeodomain of the class III POU factors interacts with noncanonical target sequence TAATTA in the X29 region. The target sequence TAATTA is highly conserved among vertebrates and is indispensable for the FM enhancer activity. We further demonstrate that Gbx2 also binds to this sequence, inhibiting the enhancer activity. The findings suggest that the Otx2 expression is activated by the class III POU factors in the forebrain and midbrain, whereas it is repressed by Gbx2 in the hindbrain, through their direct binding to the same target TAATTA sequence in the FM enhancer.
MATERIALS AND METHODS
Plasmid construction.
The 157-bp wild-type FM sequence (157FM-wt) was amplified by PCR with primers (5′-CCCCGCGGGGCTGAACAGTGTTCAAAGG-3′ and 5′-CGGGATCCCAAAAACTTCAGCCTGTGAA-3′ [underlines indicate SacII and BamHI sites, respectively]), using the 1.4-kb plasmid AH1.4kb (28) as a template, and inserted into SacII and BglII sites of the 1.8-kb plasmid 1.8kb-LacZ, which harbors Otx2 promoter sequences (31). In 157FM-X29mt, the X29 sequence was replaced with XbaI linker sequence (TCTAGA); to generate this, 157FM-wt was cloned into pBluescript SK(−), and an inverse PCR was conducted using primers oppositely oriented at the X29 site. The primers used were 5′-GATCTAGACAGACCGCCAGGCCTAA-3′ at the 5′ end of the X29 sequence and 5′-GATCTAGAAAGATAGAAGTAGCATTAGACT-3′ at its 3′ end. Underlines indicate XbaI sites. The PCR products were digested with XbaI and self-ligated. The 157FM-X29mt fragment in pBluescript SK(−) created by the SacII and BamHI digestion was inserted into the SacII and BglII sites of plasmid 1.8kb-LacZ. 157FM-M2, in which TCTAATTAAA was replaced with GAGCCGGCCC, was also obtained by an inverse PCR using 157FM-wt in pBluescript SK(−) as a template. The 5′-phosphorylated primers used were 5′-Phos-ggctcAGGCAAATTCTCAGACCGC-3′ and 5′-Phos-ggcccAAGGATAAAAGATAGAAGTAGC-3′. Lowercase letters indicate mutated sequences. The amplified products were self-ligated. 157FM-TCFmt was obtained by PCR with primers 5′-CCCCGCGGGGCTGAACAGTGggaccctG-3′ and 5′-CGGGATCCCAAAAACTTCAGCCTGTGAA-3′ (where underlines indicate SacII and BamHI sites, respectively, and lowercase letters indicate mutated sequences in the Tcf/Lef binding site), using plasmid AH1.4kb TCFmt (28), which has mutations in two Tcf/Lef binding sites, as a template. 157FM-BHPmt was obtained by PCR with 5′-CCCCGCGGGGCTGAACAGTGTTCAAAGG-3′ and 5′-CGGGATCCCAAAAACTTCAGCCTGTGAA-3′ (where underlines indicate SacII and BamHI sites, respectively), using plasmid AH1.4kb BHPmt (28) as a template. 157FM-TCFmt and 157FM-BHPmt fragments were inserted into the SacII and BamHI sites of pBluescript SK(−). To construct luciferase reporters, four tandem repeats of 157FM-wt, 157FM-X29mt, 157FM-TCFmt, and 157FM-BHPmt were cloned into pGL3-TK, which has a firefly luciferase gene conjugated with herpes simplex virus thymidine kinase (HSV TK) promoter (60) or pGL4.23, which has a firefly luciferase gene conjugated with a minimal promoter (Promega).
Brn1, Brn2, Brn4, and Oct6 cDNAs were isolated by reverse transcription-PCR (RT-PCR) with total RNAs extracted from E10.5 mouse embryos and subcloned into pTnT (Promega) for the electrophoretic mobility shift assay (EMSA). They were also subcloned into pCAGGS-Flag (20) and pCS2+MT (48) to be tagged with Flag and six copies of Myc, respectively, for the chromatin immunoprecipitation (ChIP) analysis. The primers used in the RT-PCR are shown in Table S2 in the supplemental material. The POUS and POUH fragments of Brn1, Brn2, Brn4, and Oct6 were amplified by PCR using pTnT-Brn1, pTnT-Brn2, pTnT-Brn4, and pTnT-Oct6, respectively, as a template. The reverse primer to amplify each POUH fragment was 5′-GCCTCTTCGCTATTACGCCA-3′, which is located downstream of the multicloning site in pTnT vector. Amplified fragments were digested with the appropriate restriction enzymes (EcoRI and NotI for Brn1POUH; EcoRI and XbaI for Brn1POUS; KpnI and NotI for Brn2POUH, Brn4POUH, and Oct6POUH; and KpnI and XhoI for Brn2POUS, Brn4POUS, and Oct6POUS, respectively) and subcloned into pTnT for EMSA. Each microRNA (miRNA) expression vector was constructed using a BLOCK-iT polymerase II (Pol II) miR RNA interference (RNAi) expression vector kit (Invitrogen) according to the manufacturer's instructions. Pre-miRNA oligonucleotides were synthesized (see Table S5 in the supplemental material), annealed, and inserted into the pcDNA6.2-GW/EmGFP-miR vector. Negative-control plasmid pcDNA6.2-GW/EmGFP-miR-neg (Invitrogen), which is thought not to target any known vertebrate genes, was provided by the manufacturer. To generate a chained miR-Oct6/Brn2 pre-miRNA vector (pcDNA6.2-GW/EmGFP-miR-Oct6/Brn2), Brn2 pre-miRNA was excised from its expression vector (pcDNA6.2-GW/EmGFP-miR-Brn2) by BamHI and XhoI digestion and inserted into BglII and XhoI sites immediately downstream of the Oct6 pre-miRNA in its expression vector (pcDNA6.2-GW/EmGFP-miR-Oct6). The chained negative-control vector (pcDNA6.2-GW/EmGFP-miR-neg/neg) was constructed similarly. The efficacy of these miRNA expression plasmids was examined by Western blot analyses with anti-lamin A/C (3267-100; Biovision), anti-Brn2 (sc-28594; Santa Cruz Biotechnology), or anti-Oct6 (ab31766; Abcam) antibody. Brn2 and Oct6 genes with silent mutations (Brn2sm and Oct6sm, respectively) were generated by the inverse PCR of Brn2 or Oct6 in pGEM-T as a template, followed by digestion with HindIII or MluI and self-ligation. The primers used were 5′-CGGATCAAgCTtGGcTTTACTCAAGCAGAC-3′ and 5′-AAATCCaAGcTTaATCCGCCTCTGCTTGAA-3′ for Brn2sm and 5′-GGTcGTaCGcGTaTGGTTCTGCAACCGG-3′ and 5′-CCAtACgCGtACgACCTCCTTCTCCAGTTG-3′ for Oct6sm. Underlines indicate HindIII and MluI sites, respectively, and lowercase letters indicate mutated sequences.
Gbx2 cDNA was amplified by PCR with primers 5′-GGCTCGAGATGAGCGCAGCGTTCCCG-3′ and 5′-GGCTCGAGTCAGGGTCGGGCCTGCTCC-3′ (where underlines indicate XhoI sites) using pGbx2 as a template (a kind gift from Michael A. Frohman [10]) and subcloned into pCAGGS-Flag. Gbx2HD was amplified by PCR with primers 5′-AGACTGGTACCAGTTAAGAAGACCCCGGCCAC-3′ and 5′-GGCTCGAGTCAGGGTCGGGCCTGCTCC-3′ (where underlines indicate KpnI and XhoI sites, respectively) using pGbx2 as a template and subcloned into pTnT.
Transient reporter analysis in transgenic mouse embryos.
Preparation of transgenes, DNA injection into zygotes, β-galactosidase (β-Gal) staining, and genotyping were performed as described previously (60).
Preparation of nuclear extracts.
Nuclear extracts were prepared from the forebrain/midbrain of E10.5 mouse embryos or neuralized P19 cells with NE-PER nuclear and cytoplasmic extraction reagents (Pierce) as described previously (60).
EMSA.
The biotin-labeled 157FM-wt or 157FM-X29mt probe was prepared by PCR with a biotinylated sense primer (5′-biotin-GGCTGAACAGTGTTCAAAGG-3′) and an antisense primer (5′-CAAAAACTTCAGCCTGTGAA-3′), using 157FM-wt or 157FM-X29mt in the 1.8kb-LacZ vector as a template. Biotinylated oligonucleotides of wild-type X29 (X29-wt), mutant X29 (X29-M1∼X29-M4), and reference PN-1 octamer were synthesized (see Fig. 3A and 4B) and annealed. Brn1, Brn2, Brn4, Oct6, Brn1POUH, Brn1POUS, Brn2POUH, Brn2POUS, Brn4POUH, Brn4POUS, Oct6POUH, Oct6POUS, and Gbx2 homeodomain proteins were synthesized in vitro using the TnT coupled-reticulocyte-lysate system (Promega). EMSAs were performed using a LightShift chemiluminescent EMSA kit (Pierce) according to the manufacturer's protocol, with minor modification. EMSAs with nuclear extracts were conducted using stabilized streptavidin-horseradish peroxidase conjugate, substrate equilibration buffer, and chemiluminescent substrate, whereas those with in vitro-synthesized proteins were conducted with streptavidin-alkaline phosphatase (AP) conjugate (Roche) and detection buffer (100 mM Tris-HCl [pH 9.5], 100 mM NaCl) and CDP-Star (Roche), respectively. In the supershift assay, 1 μg of antibody (anti-Brn2 [sc-28594; Santa Cruz Biotechnology] or rabbit control IgG (ab46540; Abcam) was added to the reaction mixtures.
Fig 3.
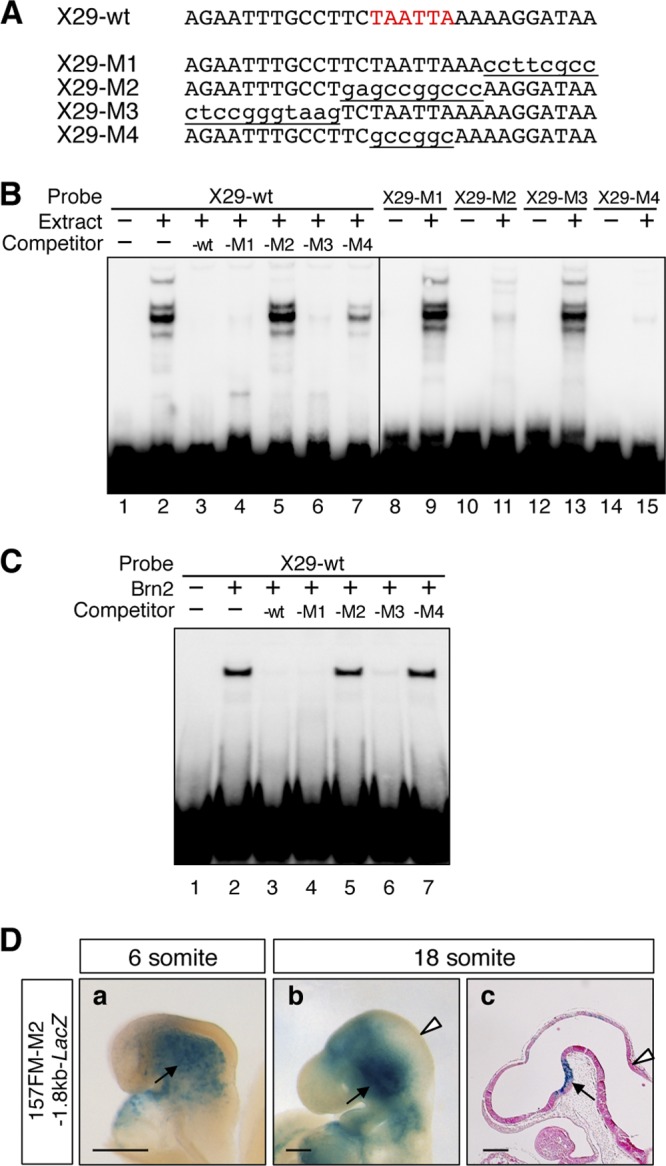
Class III POU factors associate with noncanonical target TAATTA in X29. (A) Sequences of oligonucleotides used in EMSA. Underlined lowercase letters indicate mutations. (B) EMSA indicated that X29 forms complexes with FM nuclear extracts, and the complexes are challenged by 50-fold molar excess of X29-wt, X29-M1, and X29-M3 competitors, but not by X29-M2 or X29-M4 competitors, in which the TAATTA sequence is mutated (left panel). Labeled X29-M1 and X29-M3 also form complexes with FM nuclear extracts, while labeled X29-M2 and X29-M4 do not (right panel). (C) EMSA also indicated that X29-wt forms a complex with Brn2 synthesized in vitro, and the complex is challenged by 50-fold molar excess of X29-wt, X29-M1, and X29-M3 but not by X29-M2 or X29-M4. (D) The X29-M2 mutation abolishes the 157FM enhancer activity in the forebrain and midbrain. Ten and 14 157FM-M2-1.8kb-LacZ transgenic embryos β-Gal positive in cephalic mesenchyme were examined at the 6-somite (a) and 18-somite (b and c) stages, respectively; typical examples of β-Gal expression are shown. Whole-mount lateral views (a and b) and a parasaggital section (c) are shown. Arrowheads indicate MHB, and arrows indicate the β-Gal expression in cephalic mesenchyme and ventral diencephalon by the 1.8-kb promoter. Scale bars, 200 μm.
Fig 4.
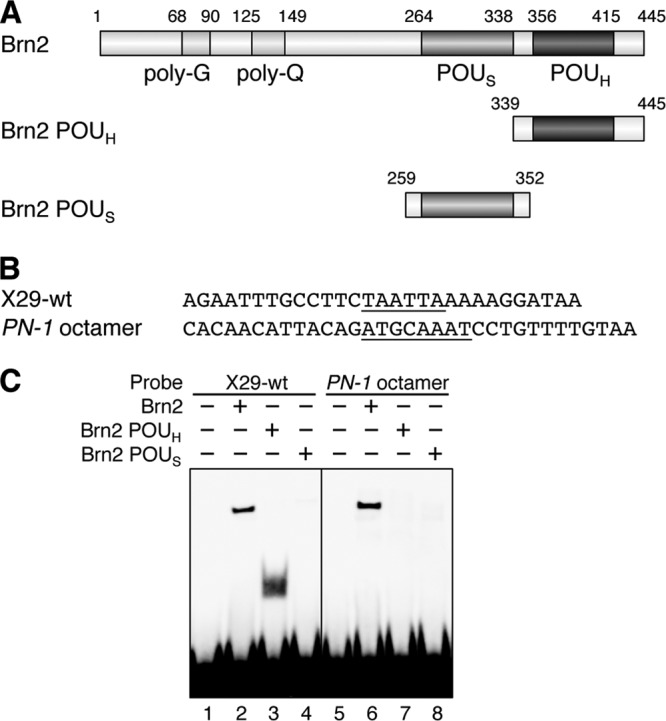
Brn2 associates with X29 by POUH. (A) Brn2 deletion constructs used in the EMSA. (B) X29 and PN-1 octamer sequences. (C) The EMSA indicated that Brn2 POUH, but not POUS, associates with X29 by itself (left panel). This contrasts with the Brn2 association with PN-1 octamer to which neither POUH or POUS binds alone (right panel).
DNA pulldown assay.
Fifty micrograms of nuclear extracts was incubated with 5 μg poly(dI-dC) (Roche) and 20 μg salmon sperm DNA (BioDynamics Laboratory) in DNA pulldown buffer (20 mM HEPES-KOH [pH 7.9], 80 mM KCl, 1 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol [DTT], 10% glycerol, 0.1% Triton X-100, complete protease inhibitor mixture [Roche]) for 10 min at 16°C. After the addition of 1 μg biotinylated DNA (157FM-wt or 157FM-X29mt), the mixture was incubated for another 10 min at 16°C. Fifty microliters of Dynabeads MyOne streptavidin T1 (Invitrogen), washed twice with DNA pulldown buffer, was added to the protein-DNA mixture and incubated for 15 min at 4°C with gentle agitation. Immobilized protein-DNA complexes were washed with DNA pulldown buffer five times and lysed in SDS sample buffer. The eluates were separated on an SDS-PAGE gradient gel (5 to 20%). The gel was stained with a negative gel stain mass spectrometry (MS) kit (Wako). The protein bands were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Cell culture and luciferase assay.
P19 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 1% penicillin/streptomycin. The cells were neurally differentiated as described by Xia et al. (67). For luciferase assay, P19 cells plated at 80 to 90% confluence in a 6-well plate were transfected with 250 ng of reporter plasmids and 250 ng of pGL4.75, containing Renilla luciferase gene directed by cytomegalovirus (CMV) promoter, as an internal control using Lipofectamine LTX (Invitrogen). To knock down Oct6 and Brn2, 3 μg knockdown plasmids (pcDNA6.2-GW/EmGFP-miR-neg/neg or pcDNA6.2-GW/EmGFP-miR-Oct6/Brn2) was added to the reaction mixtures. To rescue Oct6 and Brn2 knockdown, in addition to the knockdown plasmid, 40 ng effector plasmids (pCAGGS-Flag, pCAGGS-Flag-Brn2sm, pCAGGS-Flag-Oct6sm, pCAGGS-Flag-Brn1, or pCAGGS-Flag-Brn4) was added to the reaction mixtures. To misexpress Gbx2, 50 ng or 200 ng pCS2-Gbx2 was added. After 3 days of culture in induction medium, the firefly and Renilla luciferase activities were measured with the Dual-Glo luciferase assay system (Promega) according to the manufacturer's instructions, and the firefly luciferase expression was corrected by the Renilla luciferase expression. Data are presented as means ± standard deviations (SD) of three independent experiments.
RT-PCR and RT-qPCR.
Total RNAs were extracted from cultured cells by using Isogen (Nippon Gene). Reverse transcription (RT) was performed using SuperScript III reverse transcriptase (Invitrogen). Quantitative PCR (qPCR) was performed using Power SYBR green PCR master mix (Applied Biosystems). The quantification was normalized by the amount of Tbp amplification (59). The primers used for PCR are shown in Tables S3 and S4 in the supplemental material. For all primer sets tested, correlation (r2) was >0.98, and the slope was −3.1 to −3.6 in each standard curve. Data are presented as means ± SD.
ChIP assay.
P19 cells plated at 80 to 90% confluence in a 10-cm dish were transfected with 15 μg of plasmids (pCS2-MT, pCS2-MT-Brn2, or pCS2-MT-Oct6) using Lipofectamine LTX (Invitrogen). For cotransfection, 15 μg effector plasmids (pCAGGS-Flag or pCAGGS-Flag-Gbx2) or knockdown plasmids (pcDNA6.2-GW/EmGFP-miR-neg, pcDNA6.2-GW/EmGFP-miR-Gbx2#1, or pcDNA6.2-GW/EmGFP-miR-Gbx2#2) was added. For ChIP analysis using embryonic brain, forebrain/midbrain and hindbrain tissues were dissected from 130 mouse embryos at E10.5 and minced as small as possible. The ChIP assay was performed as described previously, with minor modifications (55). In brief, 50 to 60 mg of neural progenitor cells (NPCs) or minced brain tissues was cross-linked with 1% formaldehyde for 15 min. ChIP DNA was sheared by sonication and immunoprecipitated with antibody (anti-Myc [ab9132; Abcam], anti-Flag [F3165; Sigma], anti-Brn2 [sc-6029; Santa Cruz Biotechnology], anti-Oct6 [sc-11661; Santa Cruz Biotechnology], anti-Tcf1 [2203; Cell Signaling], anti-Lef1 [17-604; Millipore], anti-Otx2 [ab21990; Abcam], anti-Gbx2 [SAB2104635; Sigma], or rabbit control IgG [ab46540; Abcam]) coupled to Dynabeads conjugated with protein G (Invitrogen). Precipitated DNAs were analyzed by qPCR with specific primers 5′-CTGGCGGTCTGAGAATTTGC-3′ and 5′-ACTTCAGCCTGTGAAGTATCACTG-3′ for the FM enhancer region (FMR) and 5′-CAGAGGTATCCCTATTCAGGATTCC-3′ and 5′-GTAACCACATGGGTTCCCTTCTG-3′ for the intergenic region (IGR). The quantities of precipitated DNA were calculated by comparison to a standard curve generated by serial dilutions of input DNA. ChIP assays were repeated at least twice independently; each PCR amplification was conducted three times.
RESULTS
X29 sequence is essential to FM enhancer activity.
Our previous study demonstrated that the 157-bp FM sequence (157FM) (Fig. 1A) that is found 75 kb upstream of the mouse Otx2 translation start site bears the enhancer activity for the Otx2 expression in the forebrain and midbrain at the somite stage (Fig. 1C) (28). It has no activity until the 3-somite stage, becomes active at the 6-somite stage, and has the caudal limit sharply delineated at the MHB (Fig. 1C, panels a to f); it lacks the activity in telencephalon and hypothalamus. The 157FM enhancer has two potential Tcf/Lef binding sequences, one potential bicoid type homeobox protein (BHP) binding sequence, and a 29-bp sequence (X29) that are highly conserved among vertebrate Otx2 orthologues, including those of mice, chicks, Xenopus, coelacanths, zebrafish, medaka, and skates (Fig. 1A). We previously demonstrated that mutations in the Tcf/Lef sites and the BHP site disrupt the FM enhancer activity (28). Indeed, a chromatin immunoprecipitation (ChIP) assay using forebrain/midbrain tissue from E10.5 mouse embryos demonstrated that Tcf1, Lef1, and Otx2 associate with the FM enhancer region (Fig. 1B). An electrophoretic mobility shift assay (EMSA) showed that in vitro-synthesized Lef1 and Otx2 bind to the 157FM probe but not to either the 157FM-TCFmt or 157FM-BHPmt probe (see Fig. S1 in the supplemental material). Furthermore, the replacement of the X29 sequence in the 157FM with the XbaI linker sequence (157FM-X29mt) also abolished the enhancer activity in the anterior neuroectoderm at the 6-somite stage and in forebrain and midbrain at the 18-somite stage (Fig. 1C, panels g to i). No sequences other than those of the BHP, Tcf/Lef, and X29 sites are scarcely conserved between mouse and zebrafish FM enhancers that have similar activities in forebrain and midbrain at somite stages (28, 30).
Fig 1.
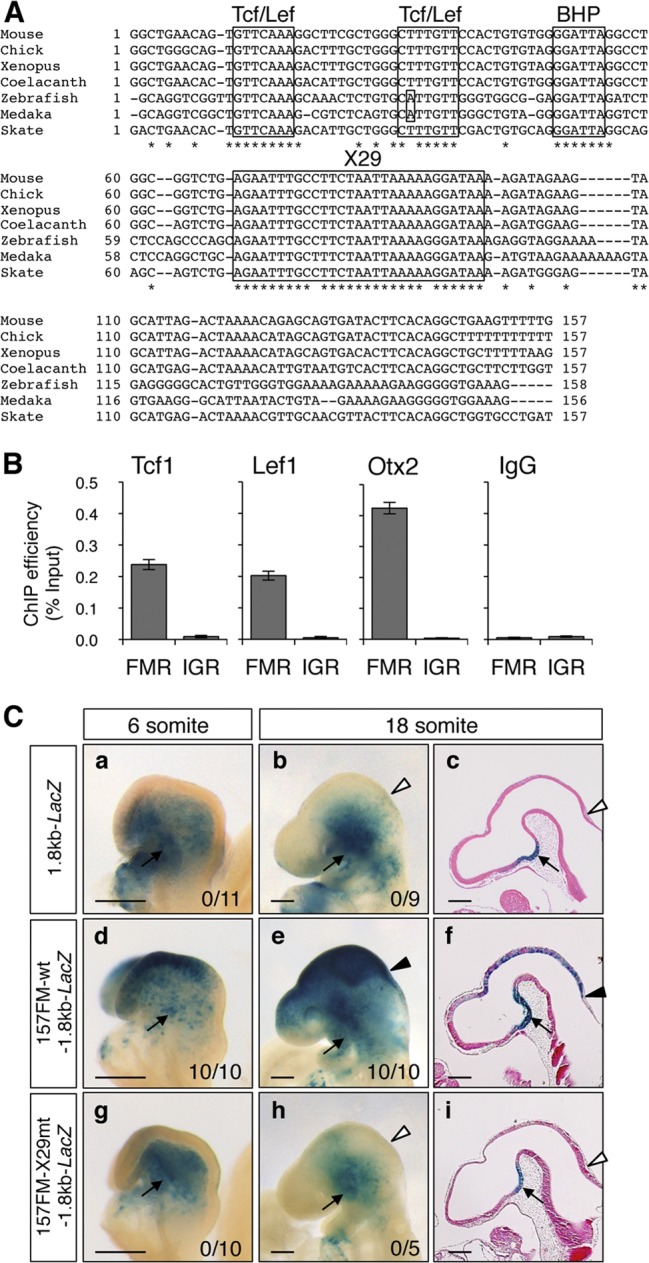
X29 is conserved among vertebrates and is essential to FM enhancer activity. (A) The 157FM sequence contains two Tcf/Lef sites, the BHP site, and X29, which are highly conserved in mouse, chick, Xenopus, coelacanth, zebrafish, medaka, and skate Otx2 gene loci. Asterisks indicate nucleotides identical among all of these animals. (B) The ChIP assay with E10.5 forebrain/midbrain indicated that Tcf1, Lef1, and Otx2 associate with the FM enhancer region (FMR) in forebrain/midbrain. Primer pairs against the FMR and intergenic region (IGR) were used for quantitative PCR (Fig. 6A). Relative amounts of PCR products are expressed as percentages of input chromatin. (C) 157FM-wt or 157FM-X29mt was conjugated with a 1.8-kb Otx2 promoter and lacZ gene (157FM-wt-1.8kb-LacZ or 157FM-X29mt-1.8kb-LacZ), and transient β-Gal expression was analyzed in mouse embryos at the 6- and 18-somite stages. The 1.8-kb promoter has activities in cephalic mesenchyme and ventral diencephalon (a to c) (26). 157FM-wt was sufficient to activate β-Gal expression in forebrain and midbrain (d to f), whereas 157FM-X29mt loses the enhancer activity (g to i). The number of embryos β-Gal positive in forebrain and midbrain among those that expressed β-Gal in the cephalic mesenchyme is given at the bottom right. Shown are whole-mount lateral views (a, b, d, e, g, and h) and parasaggital sections (c, f, and i) at the stages indicated. Arrowheads indicate MHB, and arrows represent the β-Gal expression in cephalic mesenchyme and ventral diencephalon by the 1.8-kb promoter. Scale bars, 200 μm.
Class III POU factors interact with X29 in 157FM.
An EMSA was conducted to assess the presence of factors in nuclear extracts of E10.5 forebrain and midbrain tissues (FM nuclear extracts) that bind to the X29 sequence. The EMSA indicated the presence of nuclear proteins that form complexes with the wild-type 157FM probe (157FM-wt) (Fig. 2A, lanes 1 to 3) but not with the 157FM-X29mt probe (Fig. 2A, lanes 4 to 6). The complexes were lost with a 20- or 50-fold molar excess amount of 157FM-wt competitor, as well as mutant 157FM competitors in which Tcf/Lef sites or the BHP site was transversely mutated (157FM-TCFmt or 157FM-BHPmt) (Fig. 2B, lanes 1 to 8). However, the 157FM-X29mt did not compete with 157FM-wt for the complex formation efficiently (Fig. 2B, lanes 9 and 10).
Fig 2.
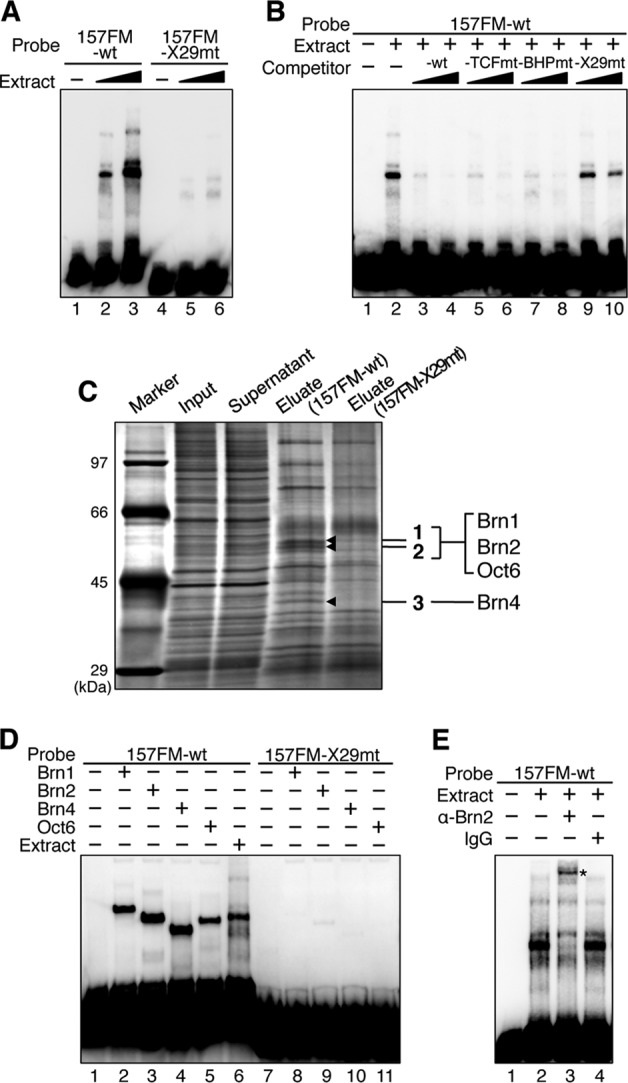
Class III POU factors bind to X29 in the 157FM enhancer. (A) EMSA indicated the presence of factors that associate with 157FM-wt, but not with 157FM-X29mt, in FM nuclear extracts. (B) The complexes between 157FM-wt and FM nuclear extracts are overcome by 20- and 50-fold molar excesses of 157FM-wt, 157FM-TCFmt, and 157FM-BHPmt but not 157FM-X29mt. (C) DNA pulldown assay and LC-MS/MS analysis identified Brn1, Brn2, Oct6, and Brn4 as factors that bind to X29. Brn1, Brn2, and Oct6 were detected from a slice containing bands 1 and 2, and Brn4 was identified from a slice containing band 3 (see Table S1 in the supplemental material). There are additional bands at around 97 kDa, 80 kDa, and 42 kDa (just above band 3) that exhibited a decrease of intensities in the 157FM-X29mt eluates; however, they were not reproducibly reduced in the mutant eluates. Sfpq (splicing factor, proline and glutamine rich) (46), Arid3a (AT-rich interactive domain-containing protein 3A) (18), and bovine serum albumin (BSA) were identified from these bands, respectively, though they were not reproducibly identified from the bands. BSA is included in magnetic bead suspension. It remains for future studies to determine whether Sfpq and Arid3a regulate the FM enhancer. (D) EMSA indicated that 157FM-wt complexes with Brn1, Brn2, Brn4, and Oct6 synthesized in vitro, while 157FM-X29mt does not. (E) The main band of the complexes between 157FM-wt and FM nuclear extracts was supershifted by anti-Brn2 antibody (asterisk) but not by normal IgG.
We then performed the DNA pulldown assay to identify the proteins interacting with the X29 sequence. FM nuclear extracts were treated with biotin-labeled 157FM-wt or 157FM-X29mt and with streptavidin-coated magnetic beads. When precipitated proteins were compared by SDS-PAGE, three bands (1 to 3) were reproducibly found that were precipitated with 157FM-wt but not with 157FM-X29mt (Fig. 2C). Mass spectrometric analysis of these bands detected Brn1, Brn2, Brn4, and Oct6 at high scores (see Table S1 in the supplemental material); all of these are members of the class III POU transcription factor. Indeed, a labeled 157FM-wt probe formed complexes with each of the class III POU proteins synthesized in vitro (Fig. 2D, lanes 1 to 5), while the 157FM-X29mt probe did not (lanes 7 to 11). The sizes of the shifted bands with each of the class III POU factors corresponded to those with the nuclear extracts (Fig. 2D, compare lanes 2 to 5 to lane 6); class III POU factors thus well explain the EMSA with E10.5 FM nuclear extracts. Furthermore, the main shift band of the 157FM-wt probe with the FM nuclear extracts was supershifted with anti-Brn2 antibody (Fig. 2E).
Class III POU factors associate with noncanonical target sequence TAATTA through its POU homeodomain.
In the 157FM enhancer, the X29 sequence was sufficient to form the complexes with the FM nuclear extracts (Fig. 3A and B, lanes 1 and 2). The sequence responsible for the class III POU factor binding in X29 was determined by a competition analysis using a series of mutant oligonucleotides, X29-M1∼X29-M4 (Fig. 3A). The complexes were lost by an excess amount of X29-M1 and X29-M3 as well as by wild-type X29 (X29-wt) (Fig. 3B, lanes 3, 4, and 6) but not by X29-M2, in which TCTAATTAAA was transversely mutated (Fig. 3B, lane 5). Furthermore, X29-M4, in which TAATTA sequence was mutated, failed to eliminate the complex formation, although it was less effective than X29-M2, suggesting a contribution of the two bases flanking TAATTA to the complex formation (Fig. 3B, lane 7). Furthermore, X29-M2 and X29-M4 failed to disrupt the complex formation between X29-wt and Brn2 synthesized in vitro (Fig. 3C). Consistent with this competition analysis, X29-M1 and X29-M3 formed DNA-protein complexes with the FM nuclear extracts (Fig. 3B, lanes 8, 9, 12, and 13), while X29-M2 and X29-M4 did not (Fig. 3B, lanes 10, 11, 14, and 15). The role of the TAATTA sequence in the enhancer activity was confirmed by the transient transgenic assay with 157FM-M2 which has the M2-type mutation in 157FM. 157FM-M2 indeed failed to express β-Gal in the forebrain and midbrain at the 6- and 18-somite stages (Fig. 3D) as 157FM-X29mt did (Fig. 1C). Thus, we conclude that the TAATTA sequence in the X29 is responsible for the class III POU factor binding to the FM enhancer and is critical for its activity.
The class III POU factors have been known to interact with canonical octamer sequence ATGCAAAT by two conserved DNA-binding domains, the POU-specific domain (POUS) and POU homeodomain (POUH) (Fig. 4A; see Fig. S2A in the supplemental material) (5, 33, 47). Since TAATTA does not match the octamer sequence, the details of Brn2 binding to the TAATTA were examined by EMSA; the octamer sequence in a brain-specific enhancer of the PN-1 gene was used as a typical target of Brn2 binding (Fig. 4B) (39). Full-length Brn2 bound to the octamer sequence (Fig. 4C, lanes 5 and 6), while POUS or POUH alone did not (Fig. 4C, lanes 7 and 8). In contrast, although POUS alone did not, POUH bound to X29 by itself (Fig. 4C, lanes 1 to 4), suggesting that the interaction of Brn2 with X29 is mainly mediated by its POUH. We also confirmed that the POUH domains of Brn1, Brn4, and Oct6 bound to X29, but their POUS domains do not; these domains did not bind to the PN-1 octamer sequence by themselves (see Fig. S2B). The study, however, does not exclude the possibility that the POUS domain contributes to the binding.
Brn2 and Oct6 knockdown inhibits FM enhancer activity in anterior NPCs.
All four class III POU factors are expressed in brain, and none of their single mutants or Brn1 Brn2 double mutants exhibited defects in early brain development, suggesting their complementary functions (7, 37, 41–43, 57). To examine the roles of the class III POU factors in the FM enhancer activity through the loss-of-function studies, we have chosen neural progenitor cells (NPCs) differentiated from P19 embryonic carcinoma cells. A retinoic acid (RA) treatment for 3 days generated posterior NPCs, including those with hindbrain-like characteristics, expressing Gbx2 and Hoxd4 (see Fig. S3 in the supplemental material). On the other hand, a suspension culture of P19 cells in serum-free medium supplemented with N2B27 for 3 days yielded anterior NPCs of forebrain-midbrain characteristics, expressing Six3, Pax6, Dmbx1, Otx1, and Otx2; they did not express Gbx2 or Hoxd4 significantly (see Fig. S3) (67). Lef1, Wnt1, and Meis2, encoding cofactors of Otx2 (4), and Tle4, encoding a corepressor of both Otx2 and Gbx2 (17), were expressed in both the anterior and posterior NPCs as they were expressed in vivo (see Fig. S3). Quantitative RT-PCR indicated that in the anterior NPCs Brn2 and Oct6 were expressed abundantly, while Brn1 was expressed less and Brn4 was expressed insignificantly (see Fig. S4A in the supplemental material). In the posterior NPCs, Brn1, Brn2, and Brn4 were expressed, while Oct6 was transiently expressed 24 h after induction, as reported previously (38). Wnt signal was active in both anterior and posterior NPCs, as evidenced by the Axin2 expression (22) and by the TOP-flash assay (see Fig. S4B). Embryonic stem cell marker Oct4 expression was lost after 48 h of culture in both anterior and posterior NPC populations. In the luciferase assay with the anterior NPCs, the quadruplet 157FM-wt (157FM-wtx4) exhibited about 90-fold activation, whereas there was little activation in the assay with the posterior NPCs or undifferentiated P19 cells (Fig. 5A). In addition, the 157FM enhancer activity in the anterior NPCs was lost by mutations at either X29 (157FM-X29mtx4), two Tcf/Lef sites (157FM-TCFmtx4), or the BHP site (157FM-BHPmtx4) (Fig. 5B). These are consistent with the 157FM activities in embryonic brain (Fig. 1C) (28, 30).
Fig 5.
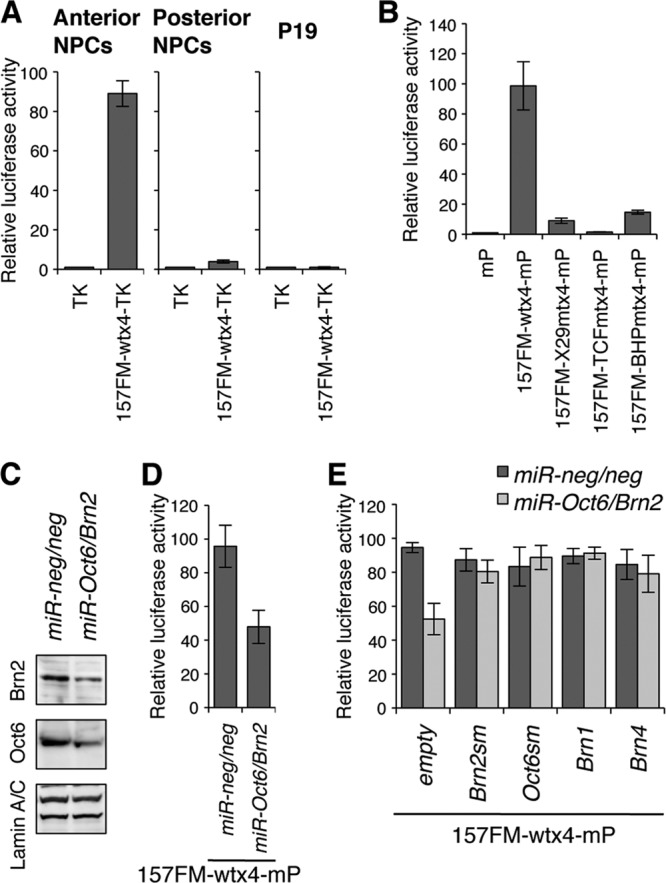
The 157FM enhancer activity is diminished by Brn2 and Oct6 knockdown in anterior NPCs. (A) The luciferase assay indicated that the 157FM enhancer is active in anterior NPCs but not in posterior NPCs or undifferentiated P19 cells. The luciferase gene was directed by the TK promoter alone (TK) or the promoter conjugated with the quadruplet 157FM-wt (157FM-wtx4-TK). P19 cells were transfected with TK or 157FM-wtx4-TK and differentiated into either anterior or posterior NPCs. (B) The 157FM enhancer activity in anterior NPCs was lost by mutations in either the X29 site (157FM-X29mtx4-mP), the two Tcf/Lef sites (157FM-TCFmtx4-mP), or the BHP site (157FM-BHPmtx4-mP); the luciferase gene was directed by a minimal promoter (mP) alone or the promoter conjugated with each 157FM. (C) The efficacy of Oct6 and Brn2 knockdown by miRNAs (miR-Oct6/Brn2 or miR-neg/neg) in anterior NPCs was examined by Western blot analysis. (D) The luciferase assay indicated that the Brn2 and Oct6 knockdown diminishes the 157FM enhancer activity. P19 cells were cotransfected with 157FM-wtx4-mP and either miR-Oct6/Brn2 or miR-neg/neg and differentiated into anterior NPCs. (E) The reduction was restored by the overexpression of Brn2, Oct6, Brn1, or Brn4. Silent mutations were introduced at the miRNA target sites of Brn2 and Oct6 (Brn2sm and Oct6sm, respectively) (see Fig. S5A and B in the supplemental material).
The functions of the class III POU factors on 157FM enhancer activities were next examined with the anterior NPCs by microRNA (miRNA)-mediated knockdown. A vector targeted to both Brn2 and Oct6 (miR-Oct6/Brn2) knocked down the levels of Brn2 and Oct6 protein expression to about 44 and 45%, respectively (Fig. 5C). Under this condition, in which expression of Brn1 and Brn4 is not knocked down, the miRNA reduced the luciferase expression by the 157FM enhancer to about half (Fig. 5D). Furthermore, the reduction was restored by the overexpression of Brn2sm and Oct6sm (Fig. 5E), in which silent mutations were introduced in the miRNA target 4 bases of Brn2 and Oct6, respectively (see Fig. S5A and B in the supplemental material). The reduction caused by miR-Oct6/Brn2 was also restored by Brn1 or Brn4 overexpression (Fig. 5E).
Class III POU factors associate with 157FM only in forebrain/midbrain and anterior NPCs.
Class III POU factors are expressed not only in forebrain and midbrain but also in hindbrain, raising a question as to whether the factors associate with the 157FM in hindbrain. A ChIP assay was first conducted with exogenous Brn2 and Oct6 tagged with Myc (MT-Brn2 and MT-Oct6) in anterior and posterior NPCs. Quantitative PCR using primers for the FM enhancer region or an intergenic region (IGR), as a negative control, on precipitates from cross-linked chromatin-DNA complexes with anti-Myc antibody indicated that MT-Brn2 and MT-Oct6 associate with the FM enhancer region in the anterior NPCs but not in the posterior NPCs (Fig. 6A and B). Moreover, the ChIP analysis with anti-Brn2 and anti-Oct6 antibodies also demonstrated that endogenous Brn2 and Oct6 associate with the FM enhancer region of the Otx2 locus in forebrain and midbrain but not in hindbrain (Fig. 6C). The question is then how the class III POU factors’ association with the FM enhancer is limited to the forebrain and midbrain, in spite of their expression in hindbrain.
Fig 6.
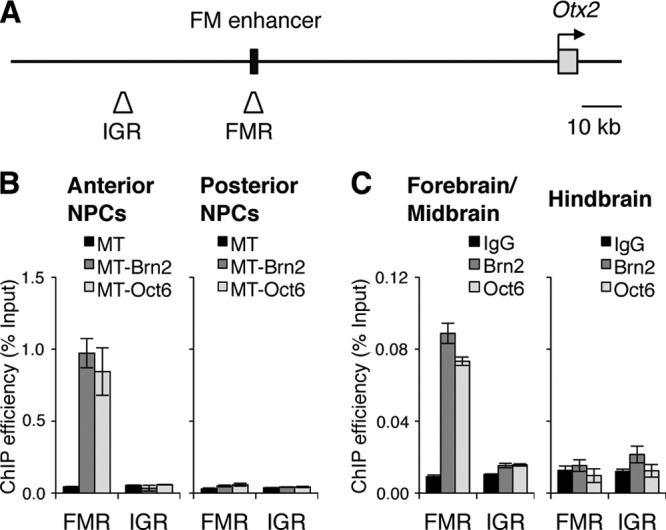
Brn2 and Oct6 associate with the FM enhancer region in anterior NPCs and embryonic forebrain/midbrain. (A) Locations of the FM enhancer region (FMR) and intergenic region (IGR) in the mouse Otx2 locus for the ChIP assay. (B) ChIP assay indicated that Myc-tagged Brn2 (MT-Brn2) and Oct6 (MT-Oct6) associate with the FMR in anterior NPCs but not in posterior NPCs. Primer pairs against the FMR and IGR were used for quantitative PCR. Relative amounts of PCR products are expressed as a percentage of input chromatin. (C) The ChIP assay also indicated that endogenous Brn2 and Oct6 specifically associate with FMR in embryonic forebrain/midbrain but not in hindbrain.
Gbx2 competes with class III POU factors for TAATTA to repress FM enhancer activity.
Gbx2 has been suggested to suppress the Otx2 expression for hindbrain formation (40, 65). Gbx2 misexpression repressed the FM enhancer activity in the anterior NPCs (Fig. 7A). ChIP analysis showed that exogenous Gbx2 tagged with Flag (FL-Gbx2) associated with the FM enhancer region not only in the posterior NPCs but also in the anterior NPCs (Fig. 7B), consistent with the suppressive effect of the Gbx2 misexpression on FM enhancer activity. Moreover, the ChIP assay with E10.5 hindbrain has demonstrated that endogenous Gbx2 associates with the FM enhancer (Fig. 7C). Thus, Gbx2 may suppress the Otx2 expression by directly binding to the FM enhancer.
Fig 7.
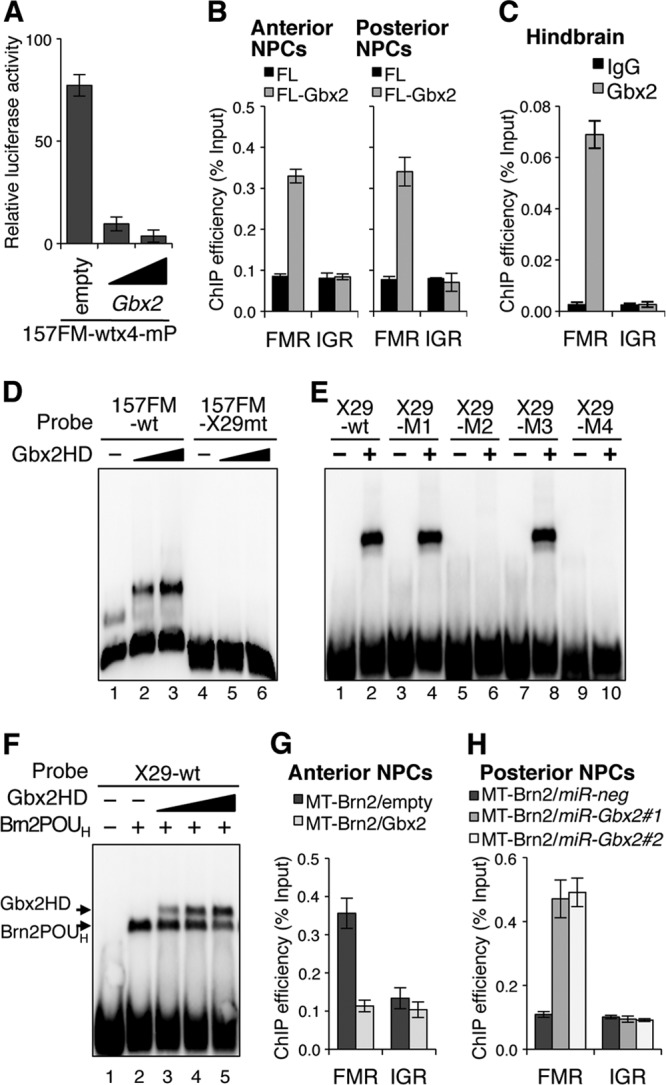
Gbx2 suppresses FM enhancer activity by competing with the class III POU factors for the binding to the TAATTA site. (A) A luciferase assay indicated that Gbx2 misexpression (50 ng or 200 ng) suppresses the 157FM enhancer activity in anterior NPCs. (B) ChIP analyses showed that Flag-tagged Gbx2 (FL-Gbx2) associates with FMR in both anterior and posterior NPCs. (C) The ChIP assay also indicated that endogenous Gbx2 associates with FMR in embryonic hindbrain. (D) EMSA indicated that the Gbx2 homeodomain (HD) associates with 157FM-wt but not with 157FM-X29mt. (E) Gbx2 HD associates with the X29-wt, X29-M1, and X29-M3 but not with X29-M2 or X29-M4, which has mutations in the TAATTA site. (F) The association between X29-wt and Brn2 POUH was overcome by increasing amounts of Gbx2 HD. (G) The ChIP assay indicated that the binding of MT-Brn2 to FMR is challenged by Gbx2 misexpression in anterior NPCs. (H) Gbx2 knockdown allowed MT-Brn2 binding to FMR in posterior NPCs. miR-Gbx2#1 and miR-Gbx2#2 represent two Gbx2 miRNAs designed for different target sites (see Fig. S5C in the supplemental material).
Gbx2 has a Q50-type homeodomain that is known to recognize TAATTG, TAATGG, or TAATTA (8, 51). Thus, we examined the possibility that Gbx2 competes with the class III POU factors for binding to the TAATTA site of the X29 in the 157FM. An EMSA showed that Gbx2 homeodomain bound to 157FM-wt but not to 157FM-X29mt (Fig. 7D). Furthermore, the Gbx2 homeodomain bound to X29-wt, X29-M1, and X29-M3 (Fig. 7E, lanes 1 to 4, 7, and 8) but not to X29-M2 or X29-M4 (Fig. 7E, lanes 5, 6, 9, and 10). This indicates that the Gbx2 homeodomain recognizes the TAATTA sequence with which the POUH domain of the class III POU factors associates. Indeed, the Gbx2 homeodomain competed with the Brn2 POUH for the binding to X29-wt (Fig. 7F). A ChIP assay using the anterior NPCs cotransfected with MT-Brn2 and Gbx2 also indicated that the association of MT-Brn2 with the FM enhancer region was inhibited by Gbx2 (Fig. 7G). On the other hand, Gbx2 knockdown in the posterior NPCs by miRNAs (miR-Gbx2#1 and miR-Gbx2#2) (see Fig. S5C in the supplemental material) allowed the MT-Brn2 association with the FM enhancer region (Fig. 7H). Thus, Gbx2 competes with the class III POU factors in its binding to the TAATTA of X29 in the FM enhancer.
DISCUSSION
The counteraction between Otx2 and Gbx2 has been suggested to establish the MHB. In Gbx2 mutants, the Otx2 expression expands posteriorly and anterior hindbrain is lost (65). Gbx2 misexpression in the midbrain under the Wnt1 promoter suppressed the Otx2 expression, thereby causing anterior expansion of the hindbrain (40). On the other hand, in Otx mutants, Gbx2-positive hindbrain expands anteriorly in a dosage-dependent manner (1, 28, 49, 56). Otx2 ectopic expression at the En1 locus caused the expansion of midbrain and the reduction of hindbrain (9). However, the details of molecular mechanisms of Otx2 and Gbx2 counteraction have remained uncertain. This study demonstrates that Gbx2 restricts the Otx2 expression to the forebrain/midbrain by directly binding to its FM enhancer, competing with the class III POU factors (Fig. 8).
Fig 8.
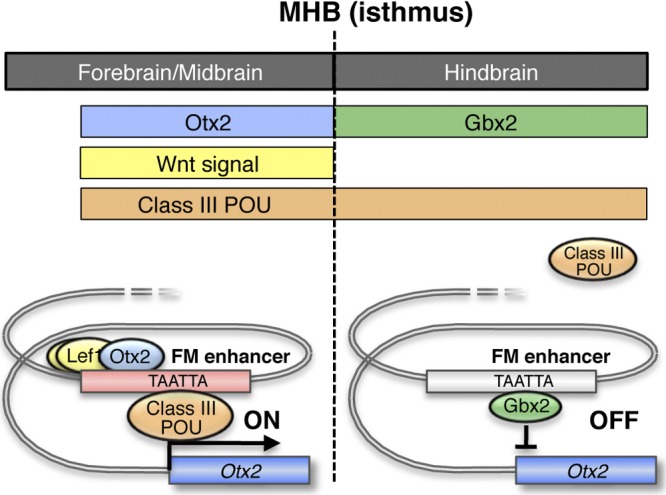
A model for transcriptional regulation of Otx2 gene by the FM enhancer in the early somite stage. At this stage, Tcf/Lef activity and Otx2 expression are found in forebrain/midbrain but not in rostral forebrain and hindbrain (upper panel). Gbx2 is expressed in hindbrain; the counteraction between Otx2 and Gbx2 has been suggested to establish the MHB. Class III POU factors are also not expressed in rostral forebrain at this stage, but are in hindbrain, as well as in forebrain/midbrain (upper panel). The FM enhancer is active in forebrain/midbrain by the association of Tcf/Lef, Otx2, and class III POU factors; these factors cooperatively activate Otx2 transcription (left lower panel). In rostral forebrain, the FM enhancer is inactive in the absence of these factors, and in hindbrain, Gbx2 competes with the class III POU factors to suppress the enhancer activity (right lower panel).
The expression of all four class III POU factors in brain precluded genetic confirmation of their regulation of the FM enhancer in vivo (7, 37, 41–43, 57), but the present analysis by miRNA knockdown in anterior NPCs strongly suggests this. In chicks, class III POU factors have not been examined, but in Xenopus, three Xenopus laevis genes, XlPOU1, XlPOU2, and XlPOU3, have been identified; they are also expressed in developing brain (3, 6). In zebrafish, four class III POU genes have been identified (zp-12, zp-23, zp-47, and zp-50), and all begin to be expressed at the early somite stage; although the expression patterns of each gene have diverged, their overall expression covers the broad region of the brain (13, 14, 54). The TAATTA site in the FM enhancer is highly conserved among vertebrate Otx2 orthologous loci, and the interaction of this site with the class III POU factors may be commonly crucial for vertebrate forebrain and midbrain development.
The class III POU factors are known to interact with the canonical octamer sequence ATGCAAAT or with ATGC(N)2–3AAAT (5, 33, 47). However, a few studies reported that Brn4 and Oct6 interact not only with the octamer sequence but also with AT-rich sequences that lack the ATGC site recognized by POUS (15, 45, 66). More recently, a ChIP-chip analysis of melanoma cells showed that Brn2 associates with many AT-rich sequences in the genome (27). The binding to the AT-rich sequences is reported to occur by the POUH alone, and this interaction was characterized in detail with the class II POU factor Oct1 by Verrijzer et al. (63). The EMSA in the present study demonstrated that the POUH of class III POU factors also associates with TAATTA of the FM enhancer. The ChIP analysis demonstrated that the Brn2 and Oct6 interaction with the FM region occurs not only in anterior NPCs in culture but also in forebrain and midbrain in vivo. However, the class III POU factors do not associate with the FM enhancer region in hindbrain. TAATTA is a sequence that is known to be recognized by the Q50-type homeodomain. Gbx2 was indeed found to associate with the FM enhancer region in hindbrain. Gbx2 is also expressed in chick and Xenopus hindbrain (44, 52, 64), and gbx1 and gbx2 are expressed in zebrafish hindbrain (25). Thus, the binding of the class III POU factors in forebrain and midbrain and of Gbx in hindbrain to the TAATTA site in the FM enhancer may be the conserved mechanism for vertebrate brain development.
During the initial brain regionalization, the caudal part of the Otx2 expression and the anterior part of the Gbx2 expression overlap until the 3-somite stage, and they segregate by the 6-somite stage (56). The Otx2-positive anterior neuroectoderm develops into forebrain and midbrain, and Gbx2-positive posterior neuroectoderm develops into hindbrain. Fgf8 expression is broad at the boundary region at the 3-somite stage, but it is confined to the MHB by the 6-somite stage; it subsequently establishes the expression in the dorso-ventral stripe at the hindbrain side of the MHB. Wnt1 expression occurs around the 1-somite stage and also establishes the expression in the dorso-ventral stripe at the midbrain side of the MHB; Wnt1 is also expressed in the dorsal midline of midbrain and caudal forebrain. En1, En2, Pax2, and Pax5 are also expressed in caudal midbrain and hindbrain or MHB region. En, Pax, Wnt1, and Fgf8 expression gradually becomes interdependent by the positive regulatory loop formation for the maintenance of isthmus. During these processes of the initial brain regionalization, the mechanisms of the Otx2 expression examined by the present and previous studies are summarized as follows (Fig. 8). Initially the Otx2 expression is directed by the AN enhancer, which is not regulated by Gbx2, and the caudal end of its activity overlaps with the Gbx2 expression (31). The enhancer that directs the Otx2 expression in anterior neural plate or rostral brain transits at the 3- to 6-somite stages from the AN enhancer to the FM enhancer (31). The upstream factors that regulate the AN enhancer largely remain to be identified (60); the mechanisms of how the AN enhancer activity is lost at the 3- to 6-somite stages are uncertain. A sequence for the bicoid-type homeobox protein binding is essential in the FM enhancer (28), and this study demonstrated that Otx2 indeed associates with the FM enhancer region in forebrain and midbrain. Otx2 expressed under the AN enhancer may activate the FM enhancer activity, and the activity may be maintained by Otx2 under the FM enhancer by itself. Tcf/Lef binding sites in the FM enhancer are also essential to its activity; this study also demonstrated that Tcf1 and Lef1 associate with the FM enhancer in forebrain and midbrain. The FM enhancer activity coincides with the area active in Wnt/β-catenin signaling in forebrain and midbrain, being absent in the rostral forebrain (36). Furthermore, Brn1, Brn2, and Brn4 expression occurs at the early somite stage, and Oct6 expression occurs at the egg cylinder stage (16, 58, 68) and upregulates the FM enhancer activity by binding to the TAATTA site in the FM enhancer. Of note is that Tcf/Lef, BHP, and the TAATTA site all are essential to activate the FM enhancer; a mutation in any of these binding sites abolishes the activity. Otx, Tcf/Lef, and the class III POU factor that binds to the FM enhancer must cooperate to activate the Otx2 expression. The class III POU factors are expressed not only in forebrain and midbrain, but also in hindbrain, but they are overcome by Gbx2 for the binding to the TAATTA site of the FM enhancer in hindbrain; the activity of the FM enhancer segregates from the Gbx2 expression and coincides with the MHB later than the 6-somite stage when the Otx2 expression is directed by the FM enhancer (Fig. 8). We consider that these findings largely explain the Otx2 expression at the early somite stage when initial brain regionalization takes place. The Otx, Tcf/Lef, and class III POU factor/Gbx2 binding sites are conserved among gnathostome Otx2 FM loci, and each component is similarly expressed in gnathostome brain. Their regulation of Otx2 expression would have been established in an ancestral gnathostome or vertebrate and conserved among all gnathostomes or vertebrates.
A question arises about the presence or absence of FM enhancer regulation by Fgf8 signaling. The signaling has been suggested to suppress Otx2 expression by directing Gbx2 expression (35, 50). However, several studies suggested that Fgf8 suppresses Otx2 expression even in the absence of Gbx2 (32, 34). No consensus sequence (GGAA) for the binding of ETS family transcriptional factors, such as Pea3 and Erm, in the Fgf signaling cascade, exists in the FM enhancer, and we have been unsuccessful in observing the Fgf suppression of the FM enhancer activity in anterior NPCs. The FM enhancer has a potential Pax binding sequence, but a mutation in this sequence did not affect the FM enhancer activity (28). The sequence is conserved among tetrapod Otx2 orthologues but not in teleost ones, and there has been no report that suggests the Pax regulation of Otx2 expression. En1 and En2 regulation of the FM enhancer remains to be examined, but there has also been no report that suggests such regulation. Another question comes from a report on a Gbx2 conditional mutant that the Gbx2 requirement to suppress the Otx2 expression changes with development (32). This study could not examine this possibility, but if true, such regulation should occur at the TAATTA site in the FM enhancer. A question also remains as to the presence of other factors that may bind to the X29 sequences other than TAATTA. Class III POU factors well explain the EMSA results from E10.5 FM nuclear extracts, and we have been unsuccessful in identifying other good candidates by the pulldown assay (Fig. 2). However, the site is tightly conserved among gnathostome Otx2 loci. The optimum conditions for the pulldown assay (salt concentration, the ratio between nuclear extract and bait DNA, reaction time, and temperature) depend on transcriptional factor binding, and it is probable that there are other factors that interact with this site but were not detected in the present assay. Finally, there is a question as to whether the Gbx expression is also directly suppressed by Otx2. Three enhancers (AMH1, -2, and -3) have been identified in zebrafish for gbx2 expression in hindbrain (21). The AMH1 enhancer harbors multiple BHP binding sites, but whether Otx2 indeed associates with these sites or whether Otx2 binding to these sites regulates gbx2 expression has not been determined; neither AMH2 nor AMH3 enhancer has been characterized. Moreover, no AMH1 to -3 orthologous enhancers exist in mouse or tetrapod Gbx2 loci. No analysis has been made of cis-regulatory sequences of the gbx1 gene that is expressed earlier at the stage of the MHB formation.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to K. Yamasu and N. Takasaki for critical discussions and thank M. A. Frohman for providing us with pGbx2 plasmid. We thank K. Shinmyozu (Mass Spectrometry Analysis Unit, RIKEN CDB) for LC-MS/MS analysis and are also grateful to the Laboratory for Animal Resource and Genetic Engineering for transgenic mouse production and the housing of mice.
This work was supported by a Grant-in-Aid for Creative Scientific Research from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print 7 May 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Acampora D, et al. 1999. Differential transcriptional control as the major molecular event in generating Otx1−/− and Otx2−/− divergent phenotypes. Development 126:1417–1426 [DOI] [PubMed] [Google Scholar]
- 2. Acampora D, Avantaggiato V, Tuorto F, Simeone A. 1997. Genetic control of brain morphogenesis through Otx gene dosage requirement. Development 124:3639–3650 [DOI] [PubMed] [Google Scholar]
- 3. Agarwal VR, Sato SM. 1991. XLPOU 1 and XLPOU 2, two novel POU domain genes expressed in the dorsoanterior region of Xenopus embryos. Dev. Biol. 147:363–373 [DOI] [PubMed] [Google Scholar]
- 4. Agoston Z, Schulte D. 2009. Meis2 competes with the Groucho co-repressor Tle4 for binding to Otx2 and specifies tectal fate without induction of a secondary midbrain-hindbrain boundary organizer. Development 136:3311–3322 [DOI] [PubMed] [Google Scholar]
- 5. Andersen B, Rosenfeld MG. 2001. POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endocr. Rev. 22:2–35 [DOI] [PubMed] [Google Scholar]
- 6. Baltzinger M, Relaix F, Remy P. 1996. Transcription of XLPOU3, a brain-specific gene, during Xenopus laevis early embryogenesis. Mech. Dev. 58:103–114 [DOI] [PubMed] [Google Scholar]
- 7. Bermingham JR, Jr, et al. 1996. Tst-1/Oct-6/SCIP regulates a unique step in peripheral myelination and is required for normal respiration. Genes Dev. 10:1751–1762 [DOI] [PubMed] [Google Scholar]
- 8. Billeter M. 1996. Homeodomain-type DNA recognition. Prog. Biophys. Mol. Biol. 66:211–225 [DOI] [PubMed] [Google Scholar]
- 9. Broccoli V, Boncinelli E, Wurst W. 1999. The caudal limit of Otx2 expression positions the isthmic organizer. Nature 401:164–168 [DOI] [PubMed] [Google Scholar]
- 10. Bulfone A, et al. 1993. Spatially restricted expression of Dlx-1, Dlx-2 (Tes-1), Gbx-2, and Wnt-3 in the embryonic day 12.5 mouse forebrain defines potential transverse and longitudinal segmental boundaries. J. Neurosci. 13:3155–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Foucher I, et al. 2006. Differentiation of cerebellar cell identities in absence of Fgf signalling in zebrafish Otx morphants. Development 133:1891–1900 [DOI] [PubMed] [Google Scholar]
- 12. Garda AL, Echevarria D, Martinez S. 2001. Neuroepithelial co-expression of Gbx2 and Otx2 precedes Fgf8 expression in the isthmic organizer. Mech. Dev. 101:111–118 [DOI] [PubMed] [Google Scholar]
- 13. Hauptmann G, Gerster T. 2000. Combinatorial expression of zebrafish Brn-1- and Brn-2-related POU genes in the embryonic brain, pronephric primordium, and pharyngeal arches. Dev. Dyn. 218:345–358 [DOI] [PubMed] [Google Scholar]
- 14. Hauptmann G, Gerster T. 1996. Complex expression of the zp-50 pou gene in the embryonic zebrafish brain is altered by overexpression of sonic hedgehog. Development 122:1769–1780 [DOI] [PubMed] [Google Scholar]
- 15. He X, et al. 1991. Tst-1, a member of the POU domain gene family, binds the promoter of the gene encoding the cell surface adhesion molecule P0. Mol. Cell. Biol. 11:1739–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. He X, et al. 1989. Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature 340:35–41 [DOI] [PubMed] [Google Scholar]
- 17. Heimbucher T, et al. 2007. Gbx2 and Otx2 interact with the WD40 domain of Groucho/Tle corepressors. Mol. Cell. Biol. 27:340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Herrscher RF, et al. 1995. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev. 9:3067–3082 [DOI] [PubMed] [Google Scholar]
- 19. Hidalgo-Sanchez M, Simeone A, Alvarado-Mallart RM. 1999. Fgf8 and Gbx2 induction concomitant with Otx2 repression is correlated with midbrain-hindbrain fate of caudal prosencephalon. Development 126:3191–3203 [DOI] [PubMed] [Google Scholar]
- 20. Hirano M, Hashimoto S, Yonemura S, Sabe H, Aizawa S. 2008. EPB41L5 functions to post-transcriptionally regulate cadherin and integrin during epithelial-mesenchymal transition. J. Cell Biol. 182:1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Islam ME, et al. 2006. Three enhancer regions regulate gbx2 gene expression in the isthmic region during zebrafish development. Mech. Dev. 123:907–924 [DOI] [PubMed] [Google Scholar]
- 22. Jho EH, et al. 2002. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 22:1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Joyner AL, Liu A, Millet S. 2000. Otx2, Gbx2 and Fgf8 interact to position and maintain a mid-hindbrain organizer. Curr. Opin. Cell Biol. 12:736–741 [DOI] [PubMed] [Google Scholar]
- 24. Katahira T, et al. 2000. Interaction between Otx2 and Gbx2 defines the organizing center for the optic tectum. Mech. Dev. 91:43–52 [DOI] [PubMed] [Google Scholar]
- 25. Kikuta H, Kanai M, Ito Y, Yamasu K. 2003. gbx2 homeobox gene is required for the maintenance of the isthmic region in the zebrafish embryonic brain. Dev. Dyn. 228:433–450 [DOI] [PubMed] [Google Scholar]
- 26. Kimura C, et al. 1997. Cis-acting elements conserved between mouse and pufferfish Otx2 genes govern the expression in mesencephalic neural crest cells. Development 124:3929–3941 [DOI] [PubMed] [Google Scholar]
- 27. Kobi D, et al. 2010. Genome-wide analysis of POU3F2/BRN2 promoter occupancy in human melanoma cells reveals Kitl as a novel regulated target gene. Pigment Cell Melanoma Res. 23:404–418 [DOI] [PubMed] [Google Scholar]
- 28. Kurokawa D, et al. 2004. Regulation of Otx2 expression and its functions in mouse forebrain and midbrain. Development 131:3319–3331 [DOI] [PubMed] [Google Scholar]
- 29. Kurokawa D, et al. 2010. Evolutionary origin of the Otx2 enhancer for its expression in visceral endoderm. Dev. Biol. 342:110–120 [DOI] [PubMed] [Google Scholar]
- 30. Kurokawa D, et al. 2006. Evolutionary constraint on Otx2 neuroectoderm enhancers—deep conservation from skate to mouse and unique divergence in teleost. Proc. Natl. Acad. Sci. U. S. A. 103:19350–19355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kurokawa D, et al. 2004. Regulation of Otx2 expression and its functions in mouse epiblast and anterior neuroectoderm. Development 131:3307–3317 [DOI] [PubMed] [Google Scholar]
- 32. Li JY, Lao Z, Joyner AL. 2002. Changing requirements for Gbx2 in development of the cerebellum and maintenance of the mid/hindbrain organizer. Neuron 36:31–43 [DOI] [PubMed] [Google Scholar]
- 33. Li P, et al. 1993. Spacing and orientation of bipartite DNA-binding motifs as potential functional determinants for POU domain factors. Genes Dev. 7:2483–2496 [DOI] [PubMed] [Google Scholar]
- 34. Liu A, Joyner AL. 2001. EN and GBX2 play essential roles downstream of FGF8 in patterning the mouse mid/hindbrain region. Development 128:181–191 [DOI] [PubMed] [Google Scholar]
- 35. Liu A, Losos K, Joyner AL. 1999. FGF8 can activate Gbx2 and transform regions of the rostral mouse brain into a hindbrain fate. Development 126:4827–4838 [DOI] [PubMed] [Google Scholar]
- 36. Maretto S, et al. 2003. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc. Natl. Acad. Sci. U. S. A. 100:3299–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McEvilly RJ, de Diaz MO, Schonemann MD, Hooshmand F, Rosenfeld MG. 2002. Transcriptional regulation of cortical neuron migration by POU domain factors. Science 295:1528–1532 [DOI] [PubMed] [Google Scholar]
- 38. Meijer D, et al. 1990. The octamer binding factor Oct6: cDNA cloning and expression in early embryonic cells. Nucleic Acids Res. 18:7357–7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mihailescu D, Kury P, Monard D. 1999. An octamer-binding site is crucial for the activity of an enhancer active at the embryonic met-/mesencephalic junction. Mech. Dev. 84:55–67 [DOI] [PubMed] [Google Scholar]
- 40. Millet S, et al. 1999. A role for Gbx2 in repression of Otx2 and positioning the mid/hindbrain organizer. Nature 401:161–164 [DOI] [PubMed] [Google Scholar]
- 41. Minowa O, et al. 1999. Altered cochlear fibrocytes in a mouse model of DFN3 nonsyndromic deafness. Science 285:1408–1411 [DOI] [PubMed] [Google Scholar]
- 42. Nakai S, et al. 1995. The POU domain transcription factor Brn-2 is required for the determination of specific neuronal lineages in the hypothalamus of the mouse. Genes Dev. 9:3109–3121 [DOI] [PubMed] [Google Scholar]
- 43. Nakai S, et al. 2003. Crucial roles of Brn1 in distal tubule formation and function in mouse kidney. Development 130:4751–4759 [DOI] [PubMed] [Google Scholar]
- 44. Niss K, Leutz A. 1998. Expression of the homeobox gene GBX2 during chicken development. Mech. Dev. 76:151–155 [DOI] [PubMed] [Google Scholar]
- 45. Okazawa H, et al. 1996. Regulation of striatal D1A dopamine receptor gene transcription by Brn-4. Proc. Natl. Acad. Sci. U. S. A. 93:11933–11938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. 1993. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 7:393–406 [DOI] [PubMed] [Google Scholar]
- 47. Rhee JM, Gruber CA, Brodie TB, Trieu M, Turner EE. 1998. Highly cooperative homodimerization is a conserved property of neural POU proteins. J. Biol. Chem. 273:34196–34205 [DOI] [PubMed] [Google Scholar]
- 48. Rupp RA, Snider L, Weintraub H. 1994. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 8:1311–1323 [DOI] [PubMed] [Google Scholar]
- 49. Sakurai Y, et al. 2010. Otx2 and Otx1 protect diencephalon and mesencephalon from caudalization into metencephalon during early brain regionalization. Dev. Biol. 347:392–403 [DOI] [PubMed] [Google Scholar]
- 50. Sato T, Araki I, Nakamura H. 2001. Inductive signal and tissue responsiveness defining the tectum and the cerebellum. Development 128:2461–2469 [DOI] [PubMed] [Google Scholar]
- 51. Schier AF, Gehring WJ. 1993. Functional specificity of the homeodomain protein fushi tarazu: the role of DNA-binding specificity in vivo. Proc. Natl. Acad. Sci. U. S. A. 90:1450–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shamim H, Mason I. 1998. Expression of Gbx-2 during early development of the chick embryo. Mech. Dev. 76:157–159 [DOI] [PubMed] [Google Scholar]
- 53. Simeone A. 2000. Positioning the isthmic organizer where Otx2 and Gbx2 meet. Trends Genet. 16:237–240 [DOI] [PubMed] [Google Scholar]
- 54. Spaniol P, Bornmann C, Hauptmann G, Gerster T. 1996. Class III POU genes of zebrafish are predominantly expressed in the central nervous system. Nucleic Acids Res. 24:4874–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Suda Y, et al. 2010. The same enhancer regulates the earliest Emx2 expression in caudal forebrain primordium, subsequent expression in dorsal telencephalon and later expression in the cortical ventricular zone. Development 137:2939–2949 [DOI] [PubMed] [Google Scholar]
- 56. Suda Y, Matsuo I, Aizawa S. 1997. Cooperation between Otx1 and Otx2 genes in developmental patterning of rostral brain. Mech. Dev. 69:125–141 [DOI] [PubMed] [Google Scholar]
- 57. Sugitani Y, et al. 2002. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 16:1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Suzuki N, Rohdewohld H, Neuman T, Gruss P, Scholer HR. 1990. Oct-6: a POU transcription factor expressed in embryonal stem cells and in the developing brain. EMBO J. 9:3723–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Svingen T, Spiller CM, Kashimada K, Harley VR, Koopman P. 2009. Identification of suitable normalizing genes for quantitative real-time RT-PCR analysis of gene expression in fetal mouse gonads. Sex. Dev. 3:194–204 [DOI] [PubMed] [Google Scholar]
- 60. Takasaki N, Kurokawa D, Nakayama R, Nakayama J, Aizawa S. 2007. Acetylated YY1 regulates Otx2 expression in anterior neuroectoderm at two cis-sites 90 kb apart. EMBO J. 26:1649–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tour E, Pillemer G, Gruenbaum Y, Fainsod A. 2002. Gbx2 interacts with Otx2 and patterns the anterior-posterior axis during gastrulation in Xenopus. Mech. Dev. 112:141–151 [DOI] [PubMed] [Google Scholar]
- 62. Tour E, Pillemer G, Gruenbaum Y, Fainsod A. 2002. Otx2 can activate the isthmic organizer genetic network in the Xenopus embryo. Mech. Dev. 110:3–13 [DOI] [PubMed] [Google Scholar]
- 63. Verrijzer CP, et al. 1992. The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J. 11:4993–5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. von Bubnoff A, Schmidt JE, Kimelman D. 1996. The Xenopus laevis homeobox gene Xgbx-2 is an early marker of anteroposterior patterning in the ectoderm. Mech. Dev. 54:149–160 [DOI] [PubMed] [Google Scholar]
- 65. Wassarman KM, et al. 1997. Specification of the anterior hindbrain and establishment of a normal mid/hindbrain organizer is dependent on Gbx2 gene function. Development 124:2923–2934 [DOI] [PubMed] [Google Scholar]
- 66. Wierman ME, et al. 1997. Repression of gonadotropin-releasing hormone promoter activity by the POU homeodomain transcription factor SCIP/Oct-6/Tst-1: a regulatory mechanism of phenotype expression? Mol. Cell. Biol. 17:1652–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xia C, Wang C, Zhang K, Qian C, Jing N. 2007. Induction of a high population of neural stem cells with anterior neuroectoderm characters from epiblast-like P19 embryonic carcinoma cells. Differentiation 75:912–927 [DOI] [PubMed] [Google Scholar]
- 68. Zwart R, Broos L, Grosveld G, Meijer D. 1996. The restricted expression pattern of the POU factor Oct-6 during early development of the mouse nervous system. Mech. Dev. 54:185–194 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


