Abstract
The connections between various nuclear processes and specific histone posttranslational modifications are dependent to a large extent on the acquisition of those modifications after histone synthesis. The reestablishment of histone posttranslational modifications after S phase is especially critical for H3K9 and H3K27 trimethylation, both of which are linked with epigenetic memory and must be stably transmitted from one cellular generation to the next. This report uses a proteomic strategy to interrogate how and when the cell coordinates the formation of histone posttranslational modifications during division. Paramount among the findings is that H3K9 and H3K27 trimethylation begins during S phase but is completed only during the subsequent G1 phase via two distinct pathways from the unmodified and preexisting dimethylated states. In short, we have systematically characterized the temporal origins and methylation pathways for histone posttranslational modifications during the cell cycle.
INTRODUCTION
The basic unit of chromatin is the core nucleosome, composed of a histone H3/H4 tetramer and a pair of histone H2A/H2B dimers coiling approximately 147 bp of DNA (31). The intimate relationship between DNA and histones enables a strong functional correlation between numerous nuclear processes and specific histone posttranslational modifications (PTMs), such as lysine methylation. Most histone PTMs do not directly affect first-order chromatin structure. Rather, histone PTMs recruit other proteins which themselves are direct regulators of various nuclear processes. Such a paradigm for histone PTM-directed regulation is encapsulated in the histone code hypothesis (26).
Many investigations have identified the proteins that modify specific histone residues, for instance, the methyltransferases and demethylases, which add and remove lysine methylation, respectively. In addition, several investigations have identified the proteins that recognize histone PTMs, for instance, the chromodomain-containing proteins that bind to lysine methylation. Based on the nuclear processes mediated by histone-modifying and -binding proteins, the functions of specific PTMs are inferred. One such process is transcriptional elongation. Dimethylation at lysine 36 on histone H3 (H3K36me2) by SET2 methyltransferase occurs during the elongation of transcriptionally engaged RNA polymerase II and marks transcribed genes for subsequent histone deacetylation by Rpd3S deacetylase (11, 32). Similarly, during cell cycle progression, monomethylation at lysine 20 on histone H4 (H4K20me1) by PR-Set7 methyltransferase is recognized by both L3MBTl1 and condensin II and is linked with S-phase progression (33, 48). Subsequent methylation that gives rise to H4K20me2/me3 by SUV4-20H1/2 methyltransferases is linked with M-phase progression (27, 43, 46).
The state of a cell is defined in part by global transcriptional and proliferation programs. Specific cellular states are also defined by the expression of specific genes. The transcriptional outcomes of a subset of these genes are stably inherited across division without alterations in their nucleotide sequences. These genes are epigenetically regulated. While transcriptional and mitotic regulation is linked with the histone PTMs mentioned above, epigenetic regulation is linked with other histone PTMs. Trimethylation at lysine 9 on histone H3 (H3K9me3) is catalyzed by SUV39H1 methyltransferase (42) and is implicated in centromeric and constitutive heterochromatin formation (35). Additionally, trimethylation at lysine 27 on histone H3 (H3K27me3) is catalyzed by EZH2 methyltransferase and is implicated in Polycomb group silencing (7, 10).
For genes that govern cellular identity and are epigenetically regulated, such as the homeotic genes governing segmentation, the inheritance of epigenetic information is poorly understood (22). Even less well understood is the transmission not only of the histone PTMs linked to epigenetic regulation but also of other PTMs linked to nonepigenetic events. While propagation of the genetic code is clearly defined, propagation of the histone code remains largely unaddressed. What has been established is the process of histone synthesis. The canonical histones are transcribed and translated during S phase (17, 19, 47). Preexisting canonical nucleosomes are randomly allocated to each strand, and new nucleosomes are assembled between the gaps (25, 52). The mechanisms of transmitting both epigenetic and nonepigenetic information contained in histone PTMs can thus be interpreted as how and when histones acquire PTMs following synthesis at S phase. The temporal issue of formation and inheritance pertains to all PTMs. Consequently, knowledge of histone PTM formation after histone synthesis provides a framework for unifying the many disparate functions linked to histone PTMs.
Predictions of the establishment of histone PTMs can be made based on their biological functions. New H4K20 methylation would likely be restricted to specific stages, while new H3K36 methylation would likely occur throughout the cell cycle. New H3K9 and H3K27 methylation is more complex to predict. Epigenetic silencing is a plastic yet ultimately stable state (45). After histone synthesis, the cell must reestablish the histone PTMs linked with epigenetic silencing for stable remembrance. Neither RNA polymerase II elongation nor cell cycle progression is a stable state that persists across a cell's lifetime. Instead, both processes act more as transitions between different stages of transcription or the cell cycle. Because of their association with epigenetic silencing, H3K9me3 and H3K27me3 are thus the critical PTMs to understand with respect to the mechanisms of their establishment.
One hypothesis would be replication-coupled H3K9me3 formation, similar to DNA CpG methylation (3). H3K9me3 is a hallmark of constitutive heterochromatic regions that are likely maintained across division. Additional evidence includes the dynamic association of heterochromatin protein 1 (HP1) with the origin replication complex (12, 40) and chromatin. HP1 binds to both H3K9me3 and SUV39H1 (36). Thus, the association between HP1 and the replication machinery may recruit SUV39H1 to form H3K9me3 on the replicated chromatin.
In contrast to H3K9me3, it is unlikely that replication-coupled methylation applies to H3K27me3, given examples of poised bivalent domains in embryonic stem cells and asymmetric division of neuroblasts into a self-renewing cell and a more differentiated lineage-specific cell. Both developmental processes are regulated by Polycomb group silencing (6). When the daughter cells do not possess the same developmental fate and histone PTM profile as each other or as the parent cell, equal templating of H3K27me3 during S phase must be altered. Further evidence includes the G2/M-phase phosphorylation of EZH2. Phosphorylation promotes enzymatic binding with noncoding RNA and is essential for EZH2-mediated silencing (29). Thus, an alternative hypothesis would be postreplication formation of H3K27me3.
One reason why the mechanisms governing histone PTM formation have remained unclear is the inability to distinguish between PTMs existing before S phase and PTMs newly formed after S phase on a residue-specific basis. Mass spectrometry provides an ideal assay for unambiguously monitoring new methylation at specific residues that are pulse-labeled with isotopically heavy methyl groups. This report builds on past experiments (53) in order to determine when and how histone PTMs are established during division. The crucial result is that the formation of both H3K9me3 and H3K27me3 begins during S phase but is fully completed only in the G1 phase following mitosis. The formation of H3K9me3 and H3K27me3 proceeds via two distinct pathways during distinct cell cycle phases. In summary, this report advances our understanding of histone methylation and makes novel suggestions about how cells propagate different chromatin domains and epigenetic information across the cell cycle.
MATERIALS AND METHODS
Cell cycle synchronization with double thymidine or thymidine-nocodazole.
Asynchronously growing HeLa S3 cells were cultured as described previously (53). In brief, HeLa S3 cells were maintained in Joklik modified Dulbecco's modified Eagle medium supplemented with 1% GlutaMAX (Invitrogen), 10% newborn calf serum (HyClone), and 1% penicillin-streptomycin at 2 × 105 to 8 × 105 cells/ml. Double-thymidine synchronizations were performed with incubation in 2 mM thymidine for 19 h, in a thymidine-free medium for 10 h, in 2 mM thymidine again for 17 h, and finally in a thymidine-free medium. For stable isotope labeling of amino acids in cell culture (SILAC), cells were released after the second thymidine block into thymidine-free Joklik medium lacking 12CH3-methionine and containing an equimolar quantity of 13CD3-methionine (Cambridge Isotope Laboratories, Inc.) and 5% dialyzed fetal bovine serum (Gemini Bioproducts). Thymidine-nocodazole synchronizations were performed with incubation in 2 mM thymidine for 18 to 24 h, in a thymidine-free medium for 3 to 4 h, in 100 ng/ml nocodazole for 12 h, and finally in standard or SILAC nocodazole-free Joklik medium. Following release from double-thymidine and thymidine-nocodazole blocks, aliquots were collected by ethanol fixation for flow cytometric analysis in order to assay synchronization efficiency, or by freezing in liquid nitrogen for gene expression and mass spectrometric analysis.
Propidium iodide staining and flow cytometry.
After ethanol fixation, cells were washed with phosphate-buffered saline. Cells were incubated with 0.08 mg/ml propidium iodide (Roche) and 0.02 mg/ml RNase in phosphate-buffered saline for >1 h with minimal light exposure at room temperature before flow cytometric analysis, performed by the Princeton University Flow Cytometry Core Facility (see File S1 in the supplemental material).
Histone isolation and derivatization.
Histones were isolated and derivatized as described elsewhere (54). Frozen cell pellets were lysed with 0.4% Nonidet P-40, 1 mM dithiothreitol, 10 mM sodium butyrate, 5 μM microcystin, and 300 μM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride. The insoluble chromatin pellet was extracted with 0.4 N H2SO4. Histones were precipitated with 25% (final volume) trichloroacetic acid. Histones were washed with acetone prior to solubilization in water for the Bradford protein assay and for propionic anhydride derivatization, performed as described previously (54). Peptides were desalted using homemade C18 Stage Tips (3M) prior to mass spectrometric analysis.
MS data analysis.
Mass spectrometric (MS) analysis was performed as described elsewhere (54). In brief, desalted histone peptides were autosampler loaded (Eksigent) onto a fused silica capillary column (inside diameter [i.d.], 75 μm) packed with Magic C18 reversed-phase 5-μm particles with a 200-Å pore size (Michrom BioResources Inc.) and were electrosprayed into a hybrid linear-quadrupole Orbitrap mass spectrometer (Thermo Scientific). Peptides were resolved by high-performance liquid chromatography (HPLC) with a reversed-phase gradient as described previously (54), except that buffer B was 95% acetonitrile in 0.1 M acetic acid. The mass spectrum (MS1) was collected in the Orbitrap at 30,000 resolution, followed by the collection of 5 data-dependent tandem mass spectra (MS2) in the linear quadrupole ion trap. Tandem mass spectra for each modified and isotopically labeled histone peptide were manually verified. Chromatographic peak integration was performed manually to quantify the relative abundances of the various labeled and modified histone peptides (see File S2 in the supplemental material). We added the suffix meX:Y to the designation of each histone peptide, where X refers to the total number of methyl groups and Y refers to the total number of 13CD3 methyl groups (53). Furthermore, we normalized the abundances of each labeled peptide with respect to all labeled states of the same modified peptide (53). For example, we normalized H3K27me1:0 with respect to H3K27me1:0 and H3K27me1:1. Unpaired and paired t tests or the Wilcoxon signed-rank test was used for statistical analysis depending on whether the data were normally distributed or not, respectively (MATLAB, version 7.8). Mass spectrometric data can be publicly accessed at https://proteomecommons.org with the hash code wysm6o6xY2QgaoaNPcGNJpXy+hrPfdqUcCefIfA2ishWXPwYNsmxEdj0AJrQxzsHuB1vOW/d8beymTchrL4D0Peq+UUAAAAAAAABZA==.
Microarray and quantitative reverse transcription-PCR (qRT-PCR) analysis.
RNA was purified by TRIzol (Invitrogen) extraction, ethanol precipitation, and RNeasy column (Invitrogen) from synchronized cell pellets using the manufacturer's supplied protocols. For RNA labeling, 175 ng of RNA was incubated with the T7 promoter primer. First-strand cDNA synthesis was performed with Moloney murine leukemia virus (MMLV) reverse transcriptase. cRNA synthesis from the cDNA templates was performed with T7 RNA polymerase and inorganic pyrophosphatase. cRNA from human universal reference RNA (Stratagene) was labeled using Cy3 dye, and cRNA from synchronized cells was labeled using Cy5 dye. Equal concentrations of cRNA from reference and synchronized samples were fragmented and hybridized onto an in-house-designed Agilent DNA oligonucleotide array containing probes for cyclin CCNA2 and the SUV39H1 and EZH2 methyltransferases (data available on request). Slides were scanned with an Agilent DNA microarray scanner, and image analysis was performed using Agilent Feature Extraction software (Agilent Technologies). Microarray analyses for each cell cycle phase were performed with two biological replicates, in which one biological replicate had 3 technical replicates for G1 and G2/M phase and 2 technical replicates for S phase. Data from all probes corresponding to a specific gene in each chip and between technical replicates were merged. We considered the expression of a gene to be significantly changing across the cell cycle if, first, it exhibited a statistically significant change in levels from G1 to S phase or from S to G2/M phase as determined by a nonparametric Kruskal-Wallis test; second, it exhibited a ≥1.5-fold change across each cell cycle stage; and third, it met both of these criteria in both biological replicates.
Quantitative RT-PCR was performed using SYBR green PCR master mix (Applied Biosystems) with the Mastercycler realplex thermocycler (Eppendorf). Primers used for PCR analysis were as follows: for human β-2-microglobulin, 5′-TTCTGGCCTGGAGGCTATC-3′ (forward) and 5′-TCAGGAAATTTGACTTTCCATTC-3′ (reverse); for human RPL11, 5′-GCC AAA CAC AGA ATCAGCAA-3′ (forward) and 5′-CTTTTGGATAGAAACGGGAATTT-3′ (reverse); for human glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-AGCCACATCGCTCAGACAC-3′ (forward) and 5′-GCCCAATACGACCAAATCC-3′ (reverse); for human EZH2, 5′-CGCTTTTCTGTAGGCGATGT-3′ (forward) and 5′-TGGGTGTTGCATGAAAAGAA-3 (reverse); and for human SUV39H1, 5′-TTGGAATCAGCTGCAGGAC-3′ (forward) and 5′-CGACTTCAAAGTCATAGAGGTTCC-3′ (reverse). Human EZH2 and SUV39H1 expression levels were normalized to those of three housekeeping genes: β-2-microglobulin, RPL11, and GAPDH.
SRM.
S-Adenosylmethionine (SAM) was extracted with 40% acetonitrile, 40% methanol, and 0.1% formic acid on ice and was centrifuged at 5,000 RCF (relative centrifugal force) to collect the supernatant. The supernatant was clarified by centrifugation at 16,000 RCF prior to MS analyses. MS analyses were performed on a TSQ Quantum Ultra triple quadrupole mass spectrometer (Thermo Fisher), coupled with an LC-10A HPLC system (Shimadzu). 12CH3- and 13CD3-SAM were detected in selected reaction monitoring (SRM) mode, with SRM being m/z 399 → 250 at 13 eV and 403 → 250 at 13 eV, respectively. The MS parameters were as follows: positive ionization mode; spray voltage, 3.2 kV; nitrogen as the sheath gas at 30 lb/in2 and as the auxiliary gas at 10 lb/in2; argon as the collision gas at 1.5 mtorr; and a capillary temperature of 325°C. The scan time was 0.1 s, with a scan width of 1 m/z.
Chromatographic separation was achieved on a Luna aminopropyl column (250 by 2 mm; particle size, 5 μm; Phenomenex) in hydrophilic interaction chromatography (HILIC) mode (4). Solvent A was 20 mM ammonium acetate plus 20 mM ammonium hydroxide in 95% acetonitrile, pH 9.40. Solvent B was acetonitrile. The gradient was 85% B at 0 min, 0% B at 15 min, 0% B at 28 min, 85% B at 30 min, and 85% B at 40 min. Other LC parameters were as follows: autosampler temperature, 4°C; column temperature, 15°C; injection volume, 20 μl; and solvent flow rate, 150 μl/min. The retention time for S-adenosyl-methionine is 13.6 min.
RESULTS
The majority of histone methylation sites, except H4K20, do not appear to change significantly across the cell cycle.
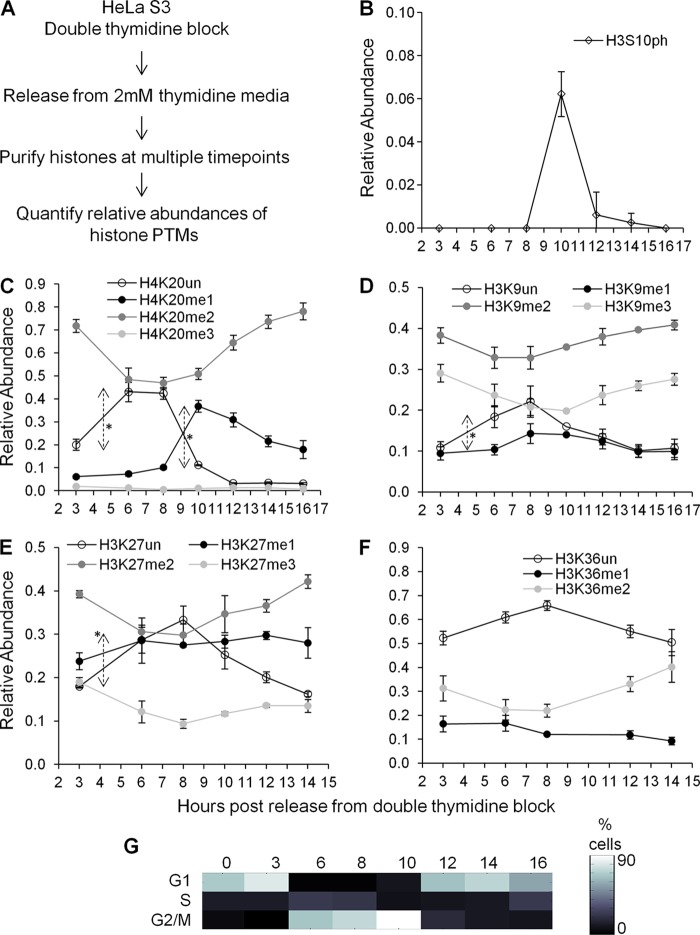
In the following analyses, we will discuss and synthesize the results for when and how histone PTMs are formed. We first investigated the overall trend in total histone PTM abundance during cell cycle progression. HeLa cells were released as a synchronized population from the G1/S boundary (Fig. 1A). As expected (23), histone H3S10 phosphorylation (H3S10ph) increased exclusively during M phase (Fig. 1B). H3S10ph is necessary for proper chromosomal segregation during mitosis (50). Methylation of H4K20 also changed dramatically, as expected (1, 39). In particular, expression of unmodified H4K20 (H4K20un) increased significantly during S phase (P = 0.0013; time [t] = −8.0552; df = 4), a finding consistent with replication-dependent synthesis of histone H4 unmodified at K20. This was followed by a significant increase in H4K20me1, initiating in late S phase and lasting into G2 phase (P = 0.0003; t = −18.7827; df = 3), and then an increase in H4K20me2 during M phase (Fig. 1C). Thus, our synchronization and mass spectrometric approaches captured known cell cycle-dependent PTM patterns.
Fig 1.
(A) Experimental strategy to measure changes in histone methylation levels across the cell cycle. (B) Relative abundance of histone H3S10 phosphorylation coupled with H3K9 dimethylation. Note that 1.0 stands for 100%. (C) Relative abundances of unmodified and mono-, di-, and trimethylated H4K20 peptides across the cell cycle. Double-headed arrows indicate significant changes in levels between time points. (D) Relative abundances of unmodified and mono-, di-, and trimethylated H3K9 peptides across the cell cycle. The double-headed arrow indicates a significant increase in H3K9un levels between 3 and 6 h. Note that H3K14 is unmodified. (E) Relative abundances of unmodified and mono-, di-, and trimethylated H3K27 peptides across the cell cycle. The double-headed arrow indicates a statistically significant change in H3K27un levels between 3 and 6 h. Note that H3K36 is unmodified. (F) Relative abundances of unmodified, monomethylated, and dimethylated H3K36 peptides across the cell cycle. Note that H3K27 is unmodified. Symbols in panels B to F represent averages of three technical replicates, and error bars represent standard deviations. (G) Heat map of synchronization efficiency of cells at G1, S, and G2/M phases, as determined by Watson Pragmatic extrapolation of flow cytometry data (see File S1 in the supplemental material).
When we examined cell cycle-dependent changes in H3K9 and H3K27 methylation levels, we found significant increases in unmodified H3K9 (P = 0.0129; t = −4.2722; df = 4) and unmodified H3K27 (P = 0.0034; t = −6.2207; df = 4) from G1 to S phase (Fig. 1D and E), consistent with another report (51). After S phase, we observed a small increase in H3K9me1 during G2, along with decreases in both H3K9me2 and H3K9me3 (Fig. 1D). Similar trends were observed for H3K27 mono-, di-, and trimethylation (Fig. 1E). Both H3K9 and H3K27 methylation change within approximately 10% in abundance across the cell cycle (Fig. 1D and E), in contrast with H4K20 methylation, which changes within a 30% range in a synchronized population (Fig. 1C). Additionally, H3K36 monomethylation and dimethylation do not fluctuate as much as H4K20 methylation (Fig. 1F). In our experiments, we were unable to reliably quantify H3K36 trimethylation due to its low abundance. In summary, global levels of H3K9, H3K27, and H3K36 methylation do not fluctuate dramatically across the cell cycle.
Heavy-methionine labeling reveals histone protein synthesis and methylation dynamics.
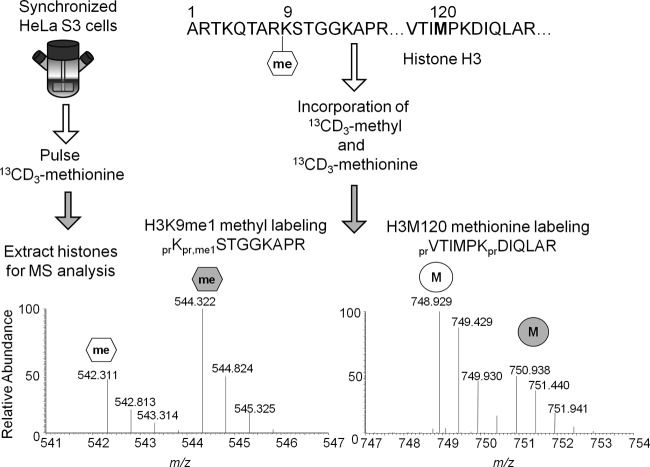
One limitation to a purely unlabeled quantification of histone PTMs (Fig. 1) is the inability to distinguish between “old” and “new” methylation events with respect to histone synthesis. A more precise analysis can be attained by pulse-labeling all methyl groups with a heavy isotope that can be differentiated from the naturally occurring light isotope by a 4.021-Da shift (Fig. 2). 13CD3-methionine is metabolized into 13CD3-S-adenosylmethionine (13CD3-SAM) as the sole methyl donor for all methylation events. 13CD3-methionine also labels any newly translated proteins containing methionine (Fig. 2). Such an experiment is termed heavy-methyl stable isotope labeling by amino acids in cell culture (SILAC) (38). Selected reaction monitoring of 12CH3- and 13CD3-SAM revealed that our heavy-methyl SILAC experiments saturated the intracellular SAM pools by approximately 94% within 3 h, which sets an upper limit for the maximum heavy-methyl labeling that could be observed (data available on request). Incomplete saturation could be due to 12CH3-methionine originating from autophagosomal degradation of proteins translated prior to the heavy-methionine pulse and being metabolized into 12CH3-SAM. Furthermore, peptides containing 13CD3-methionine or 13CD3-methyl groups elute and ionize similarly to peptides containing 12CH3-methionine or 12CH3-methyl groups. Finally, 13CD3-methionine is nontoxic and captures the in vivo methylation dynamics at a specific residue resulting from all endogenous methyltransferases.
Fig 2.
Illustration of experimental strategy. The partial sequence for histone H3 monomethylated on lysine 9 is provided. Both the methyl group at lysine 9 and methionine at position 120 on histone H3 will be replaced with the heavy isotope during new methylation and protein synthesis, respectively. At a given time point, one can assay both events from the mass spectrum and quantify the isotopic distributions corresponding to the light and heavy peptides. Abbreviations: pr, propone group from derivatization; me1, monomethyl.
We performed heavy-methyl SILAC in HeLa cells synchronized at the G1/S boundary to monitor new histone synthesis and methylation (Fig. 3A). Neither the H3 peptide comprising amino acids 117 to 128, VTIMPKDIQLAR, nor the H4 peptide comprising amino acids 79 to 92, KTVTAMDVVYALKR, is observed to be posttranslationally modified in these experiments. These peptides serve as references for histone synthesis, since new histones H3 and H4 will incorporate 13CD3-methionine at M120 and M84, respectively. Both H3 and H4 heavy peptides exhibited identical labeling efficiencies across the cell cycle (Fig. 3B) (P = 0.9514; t = 0.0640; df = 5). In contrast to the logarithmic labeling efficiency observed in asynchronous HeLa populations by use of heavy 13CD3-methionine (53) or 13C615N2-lysine (54), a logistic labeling efficiency is observed in synchronized cells, suggesting maximum synthesis rates at S phase. A logistic curve would have a single point of inflection, whereas a logarithmic curve would have no points of inflection.
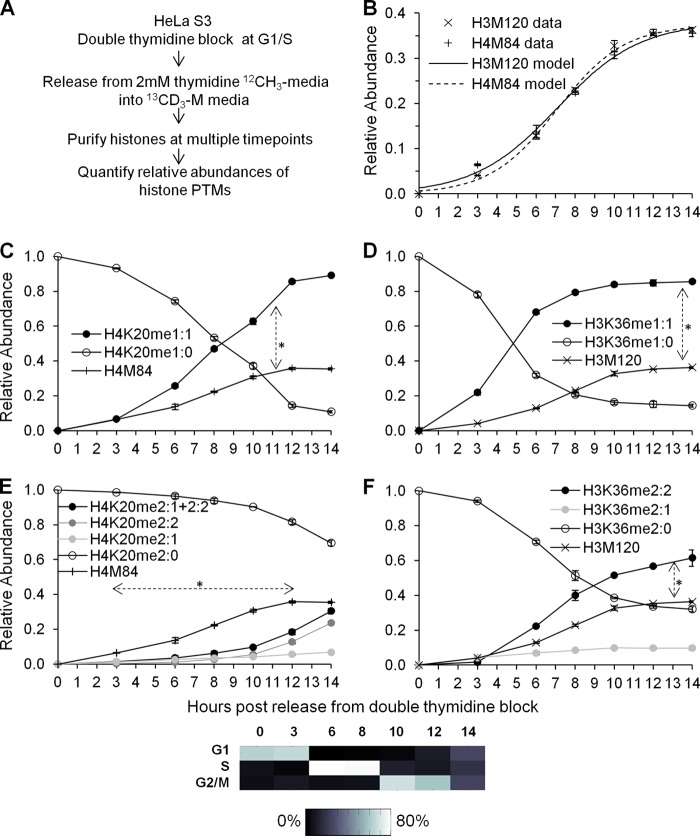
Fig 3.
(A) Experimental strategy to measure new methylation and histone synthesis in a G1/S synchronized HeLa population. (B) Histone H3 and H4 protein synthesis exhibits identical logistic-like methionine-labeling efficiency across the cell cycle. The same histone H3 and H4 protein synthesis data are portrayed in Fig. 3, 4, and 6 for each respective replicate. Curves represent logistic modeling of histone H3 and H4 protein synthesis. (C) Relative abundances of preexisting H4K20 monomethylation (H4K20me1:0) and newly formed H4K20 monomethylation (H4K20me1:1) across the cell cycle. Histone H4 synthesis is provided as a reference. The double-headed arrow indicates significantly greater levels of H4K20me1:1 than of the H4M84 peptide from 3 to 14 h. (D) Relative abundances of preexisting H3K36 monomethylation (H3K36me1:0) and newly formed H3K36 monomethylation (H3K36me1:1) across the cell cycle. Histone H3 synthesis is provided as a reference. The double-headed arrow indicates significantly greater levels of H3K36me1:1 than of the H3M120 peptide from 3 to 14 h. (E) Relative abundances of preexisting H4K20 dimethylation (H4K20me2:0), new H4K20 dimethylation from a preexisting monomethylated peptide (H4K20me2:1), new H4K20 dimethylation from an unmodified state (H4K20me2:2), and the sum of all new dimethylation events (H4K20me2:1 + 2:2) across the cell cycle. Histone H4 synthesis is provided as a reference. The dashed double-headed arrow indicates significantly lower levels of H4K20me2:1 + 2:2 than of the H4M84 peptide from 3 to 12 h. (F) Relative abundances of preexisting H3K36 dimethylation (H3K36me2:0), new H3K36 dimethylation from a preexisting monomethylated peptide (H3K36me2:1), and new H3K36 dimethylation from an unmodified state (H3K36me2:2) across the cell cycle. Histone H3 synthesis is provided as a reference. The double-headed arrow indicates significantly greater levels of H3K36me2:2 than of the H3M120 peptide from 3 to 14 h. Symbols in panels B to F represent averages of three independent experiments; error bars represent standard deviations. A heat map of synchronization efficiency is shown below the graphs (see also File S1 in the supplemental material).
One would expect 50% 13CD3-methionine labeling of histones after S phase, corresponding to new synthesis of half of the histones after DNA replication. Instead, approximately 40% labeling efficiency is observed for histones H3 and H4 (Fig. 3B). Assuming that the extent of S-adenosyl-methionine saturation determined by the SRM experiments reflects the extent of methionine saturation, we believe that the saturation index represents an upper limit for labeling efficiency. Not all the cells in our experiments are cycling as a synchronized population through S phase. Thus, not all cells would be synthesizing histone proteins, so the apparent labeling efficiency would be reduced. Similar labeling efficiencies have been observed in previous experiments (53) and are attributed to these reasons.
Histone methylation dynamics at H4K20 is cell cycle stage specific.
The ability to quantify histone protein synthesis provides an informative context for understanding new histone methylation. If 13CD3-methyl labeling matches or exceeds 13CD3-methionine labeling, new methylation events sufficiently compensate for the increase in histone abundance during S phase. As a result, methylation at that residue has reached its steady-state level. Another interpretation of excess 13CD3-methyl labeling is the addition of newly methylated histones to the pool of preexisting methylated histones without turnover, which would result in a net increase in total methylated histones. These two alternatives can be distinguished by examining whether total methylation changes across the cell cycle (Fig. 1). When 13CD3-methyl labeling falls short of 13CD3-methionine labeling, methylation has not reached its steady-state abundance when normalized to total histone levels.
We first examined 13CD3-methyl labeling of H4K20 mono- and dimethylation, two posttranslational modifications that fluctuated significantly across the cell cycle. The suffix meX:Y stands for X number of methyl groups and Y number of 13CD3-methyl groups. New H4K20 monomethylation (i.e., H4K20me1:1) significantly exceeded new histone H4 synthesis (P = 0.0200; t = −3.3643; df = 5) (Fig. 3C; also data available on request). This could be due to upregulated PR-Set7 methyltransferase activity (1). In contrast, the level of new H4K20 dimethylation (i.e., H4K20me2:1 and H4K20me2:2) was significantly lower than that of new histone H4 synthesis during the S and G2 phases (P = 0.0079; t = 4.9241; df = 4) (Fig. 3E).
During the late-G2 and M phases, there was a significant increase in H4K20 dimethylation, which reached approximately the levels of newly synthesized histone H4 (Fig. 3E; also data available on request). Most newly dimethylated H4K20 peptides contained two heavy 13CD3-methyl groups (i.e., H4K20me2:2), while a smaller fraction contained one light 12CH3-methyl group and one heavy 13CD3-methyl group (i.e., H4K20me2:1). Given that the unmodified H4K20 peptide is less abundant than both the H4K20me1 and H4K20me2 peptides during the late-G2 and M phases (Fig. 1C), newly dimethylated H4K20 (i.e., H4K20me2:2) more likely arises from a conversion of a newly monomethylated histone (i.e., H4K20me1:1) than from an unmodified histone.
Histone methylation dynamics at H3K36 is generally cell cycle stage independent.
The formation of new H4K20 mono- and dimethylation is restricted to specific cell cycle stages. We hypothesized that H3K36 methylation, as a global mark for transcriptional elongation, would not be restricted to specific stages. Indeed, we observed that new H3K36 monomethylation (H3K36me1:1) (P = 0.0313) and dimethylation (H3K36me2:2) (P = 0.0143; t = −3.6795; df = 5) significantly exceeded the formation of new histone H3 throughout the cell cycle (Fig. 3D and F; also data available on request). Since total H3K36 methylation levels do not fluctuate across the cell cycle (Fig. 1F), excess 13CD3-methyl labeling at H3K36 is likely attributable to methylation turnover. Turnover could occur, for instance, when the PTM state of a recently transcribed gene is reset to a poised inactive state (11). The turnover of histone proteins correlates with transcriptional activity (14, 15). Our experiments also show that H3K36 methylation, presumably enriched on active genes, can turn over in the absence of histone protein turnover.
Dimethylation at H3K9 proceeds in two distinct kinetic pathways.
Although methylation events at H4K20 and H3K36 are linked to disparate nuclear processes, the timing of methylation at both residues can nevertheless be rationalized. H4K20 methylation occurs at specific cell cycle stages, while H3K36 methylation occurs independently of the cell cycle. We next examined the methylation dynamics of H3K9. New monomethylation at H3K9 (i.e., H3K9me1:1) was significantly elevated (P = 0.0313) relative to new histone H3 synthesis (Fig. 4A). In this respect, monomethylation at H3K9 behaves similarly to monomethylation at H3K36 and H4K20. The elevated H3K9 monomethylation rate could reflect both turnover of H3K9me1 proteins and a net increase of relative monomethylated H3K9 levels during S phase (Fig. 1D).
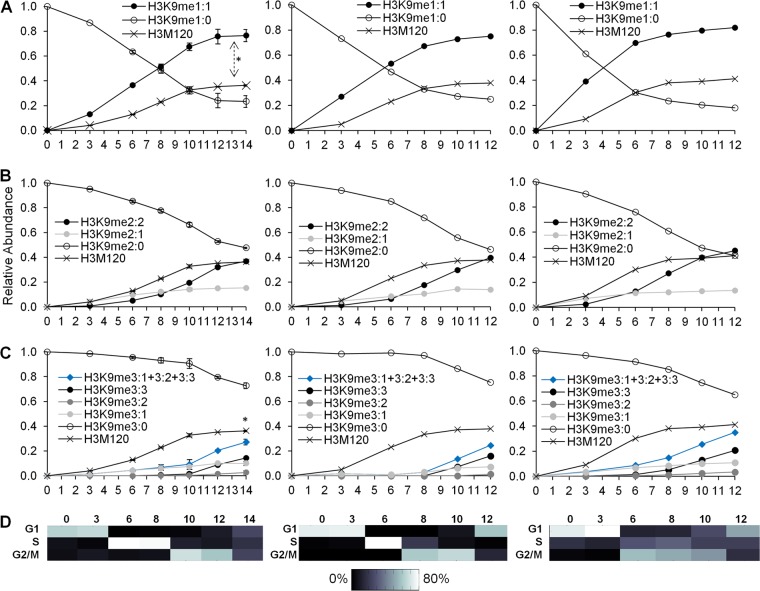
Fig 4.
(A) Three biological replicates of the relative abundances of preexisting H3K9 monomethylation (H3K9me1:0) and new H3K9 monomethylation (H3K9me1:1). The double-headed arrow indicates significantly greater levels of H3K9me1:1 than of the H3M120 peptide from 3 to 14 h. (B) Three replicates of the relative abundances of preexisting H3K9 dimethylation (H3K9me2:0), new H3K9 dimethylation from a preexisting monomethylated peptide (H3K9me2:1), and new H3K9 dimethylation from an unmodified state (H3K9me2:2). (C) Three replicates of the relative abundances of preexisting H3K9 trimethylation (H3K9me3:0), new H3K9 trimethylation from a preexisting monomethylated peptide (H3K9me3:2), new H3K9 trimethylation from a preexisting dimethylated peptide (H3K9me3:1), and new H3K9 trimethylation from an unmodified state (H3K9me3:3). The sum of all new H3K9 trimethylation events is also provided (H3K9me3:1 + 3:2 + 3:3). The asterisk indicates significantly lower levels of H3K9me3:1 + 3:2 + 3:3 than of the H3M120 peptide at 14 h. In panels A to C, symbols for the first biological replicate (left) represent averages of three technical replicates, and error bars represent standard deviations. Symbols for the second biological replicate (center) represent one technical measurement. Symbols for the third biological replicate (right) represent averages of two technical replicates. (D) Heat maps of synchronization efficiency for each respective biological replicate (see File S1 in the supplemental material).
New H3K9 dimethylation comprises two differently labeled peptides, H3K9me2:1 and H3K9me2:2, which contain one and two 13CD3-methyl groups, respectively. The most abundant newly dimethylated peptide at the late-G1 and S phases is H3K9me2:1, which does not increase further in abundance during G2/M phase (Fig. 4B). At the G2 and M phases, the most abundant newly dimethylated peptide is H3K9me2:2. During M phase, the level of formation of H3K9me2:2 reached that of new histone H3, suggesting that H3K9 dimethylation reaches its steady-state abundance by mitosis. Our data support a scenario where the preexisting monomethylated H3K9 acquires one heavy-methyl group to form H3K9me2:1 during the late-G1 and S phases. Additionally, our data suggest that a newly synthesized H3K9 peptide acquires two heavy-methyl groups to form H3K9me2:2 during the G2 and M phases. Thus, H3K9 dimethylation originates via two distinct pathways from two distinct substrates: from a preexisting monomethylated peptide during G1 and from an unmodified peptide during the G2 and M phases.
One alternative interpretation is that H3K9 dimethylation arises from a common unmodified substrate and acquires either a 12CH3-methyl or a 13CD3-methyl group in a probabilistic manner. The SRM experiments determined a 94% saturation of the S-adenosyl-methionine pool with the heavy isotope (data available on request). Thus, one can estimate the likelihood of formation of H3K9me2:2 to be 88% (i.e., 94% × 94%), that of H3K9me2:1 to be 11% (i.e., 2 × 6% × 94%), and that of H3K9me2:0 to be 1%. H3K9me2:2 would be preferentially formed over H3K9me2:1 in a ratio of approximately 8:1 if H3K9 dimethylation arises from a common substrate. The data do not support such a model, since H3K9me2:1 is preferentially formed over H3K9me2:2 during late-G1 phase (at 3 h) (P < 0.0001; t = −23.0520; df = 4) and S phase (at 6 h) (P < 0.0004; t = −10.6738; df = 4). Instead, the most likely model is two different origins of H3K9 dimethylation to account for the two different newly dimethylated peptides, the relevance of which remains unknown.
Trimethylation at H3K9 is not coupled to DNA replication and occurs via two pathways.
The heavy-methyl SILAC experiments revealed two kinetically distinct H3K9 dimethylation pathways. The experiments also revealed two distinct H3K9 trimethylation pathways with two different substrates. as represented by the H3K9me3:1 and H3K9me3:3 peptides (Fig. 4C). H3K9me3:1 originated from a preexisting dimethylated peptide (i.e., H3K9me2:0) that acquired one heavy-methyl group. This pathway occurs during the G1 and S phases. H3K9me3:3 originated from an unmodified H3K9 peptide that acquired three heavy-methyl groups. H3K9me3:3 appears mostly during the G2 and M phases. Interestingly, H3K9me3:2 was the least abundant newly trimethylated peptide across the cell cycle. This suggests that the addition of two new methyl groups to preexisting monomethylated H3K9 proteins does not significantly contribute to new H3K9 trimethylation, in contrast to other experiments suggesting that the monomethylated peptide acts as a substrate for trimethylation (34).
The crucial finding is that none of the heavy-methyl-labeled trimethylated H3K9 peptides individually (i.e., H3K9me3:1, H3K9me3:2, or H3K9me3:3) reached the levels of new histone H3 synthesized (Fig. 4C). This contrasts with H3K9 dimethylation, where new dimethylation from the unmodified state alone (i.e., H3K9me2:2) was sufficient to restore steady-state levels. When all newly trimethylated H3K9 peptides are considered together (i.e., H3K9me3:1 plus H3K9me3:2 plus H3K9me3:3), new trimethylation of H3K9 still remains significantly below new histone H3 levels (at 14 h, P < 0.0028; t = −6.5856; df = 4). Consequently, new H3K9 trimethylation begins at S phase but does not reach steady-state levels by mitosis.
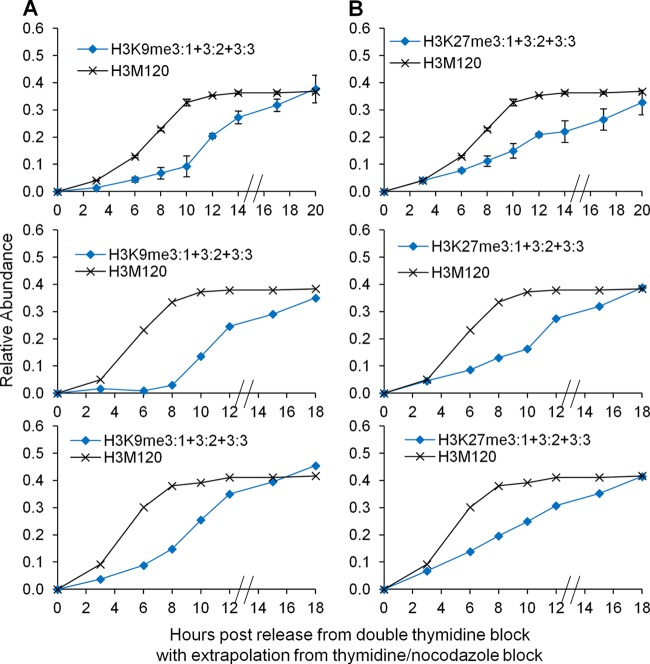
We hypothesized that a change in SUV39H1 methyltransferase levels could account for the latency in H3K9 trimethylation during the cell cycle. Microarray and RT-PCR analyses revealed nonsignificant fluctuations in SUV39H1 methyltransferase transcript levels between the G1, S, and G2/M phases (data available on request). We next considered thymidine-nocodazole treatment, which synchronizes cells at the metaphase boundary. Thus, the thymidine-nocodazole experiments tested the hypothesis that new H3K9 trimethylation in early- and mid-G1 phase, when combined with new trimethylation in the S and G2/M phases of the previous cell cycle, would reach steady-state levels. When we pooled all new trimethylated H3K9 peptides formed in the previous cell cycle with new trimethylated peptides formed in the G1 phase of the next cell cycle (i.e., H3K9me3:1 plus H3K9me3:2 plus H3K9me3:3), the levels of new trimethylation were indeed sufficient to reach steady-state levels (Fig. 5A). Our results revealed that H3K9 trimethylation is not replication coupled and is completed during the G1 phase of the next cell cycle, primarily from the unmodified and preexisting dimethylated histones.
Fig 5.
(A) All new H3K9 trimethylation events (H3K9me3:1 + 3:2 + 3:3) prior to S phase from the thymidine-nocodazole experiments were added to those from the double-thymidine replicate experiments (Fig. 4). (B) All new H3K27 trimethylation events (H3K27me3:1 + 3:2 + 3:3) from the thymidine-nocodazole experiments were added to those from the double-thymidine replicate experiments (Fig. 6). In both panels, hatched lines on x axes represent temporal extrapolation between the different synchronization methods. The respective histone H3 synthesis data for each biological replicate are shown as a reference.
Histone methylation dynamics at H3K27 proceeds in two distinct kinetic pathways.
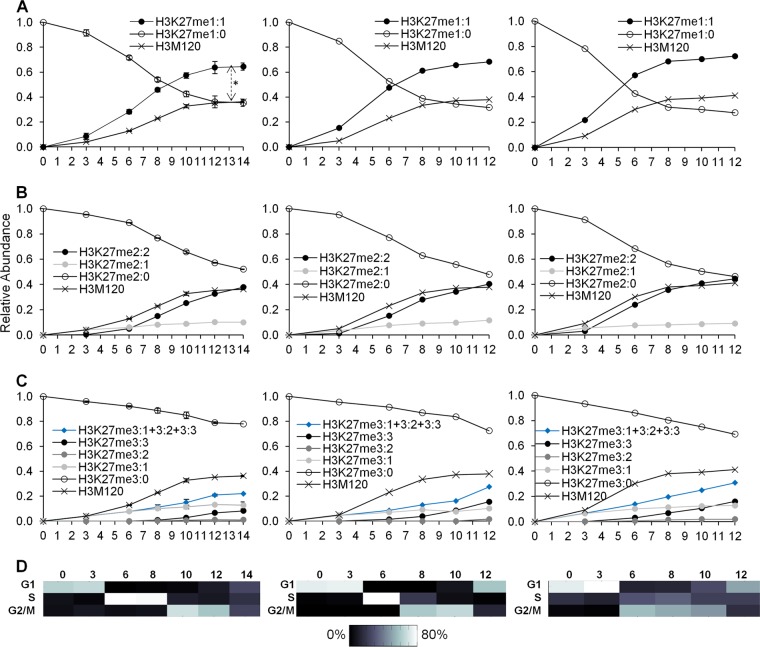
We finally examined the methylation of H3K27 across the cell cycle, which proceeds similarly to H3K9 methylation. The levels of new H3K27 monomethylation (i.e., H3K27me1:1) exceeded new histone H3 levels (P = 0.0028; t = −5.4714; df = 5) (Fig. 6A). Interestingly, H3K27 dimethylation proceeds in two kinetically distinct pathways in a processive manner (Fig. 6B). Processive dimethylation occurs when the methyltransferase binds to the unmodified substrate without releasing the histone at each methylation step. Nonprocessive dimethylation occurs when the methyltransferase releases the histone after transferring a methyl group and rebinds the newly monomethylated substrate to form a dimethylated peptide (13).
Fig 6.
(A) Three biological replicates of the relative abundances of preexisting (H3K27me1:0) and new (H3K27me1:1) H3K27 monomethylation. The double-headed arrow indicates significantly greater levels of H3K27me1:1 than of the H3M120 peptide at each time point from 3 to 14 h. (B) Three replicates of the relative abundances of preexisting H3K27 dimethylation (H3K27me2:0) and new H3K27 dimethylation from a preexisting monomethylated peptide (H3K27me2:1) or from an unmodified state (H3K27me2:2). (C) Three replicates of the relative abundances of preexisting H3K27 trimethylation (H3K27me3:0) and of new H3K27 trimethylation from a preexisting monomethylated (H3K27me3:2) or dimethylated (H3K27me3:1) peptide or from an unmodified state (H3K27me3:3). The sum of all new H3K27 trimethylation events is also provided (H3K27me3:1 + 3:2 + 3:3). Data in panels A to C are as described in the legend to Fig. 4. (D) Heat maps of synchronization efficiency for each respective biological replicate (see File S1 in the supplemental material).
One would predict that a nonprocessive methyltransferase would rebind H3K27me1:0 and H3K27me1:1 with a bias determined by their abundances relative to each other. If H3K27me1:0 is more abundant than H3K27me1:1, a nonprocessive methyltransferase will bind to H3K27me1:0 to form H3K27me2:1 more often than with H3K27me1:1 to form H3K27me2:2. When H3K27me1:1 becomes more abundant than H3K27me1:0, we expect a preferential formation of H3K27me2:2 over H3K27me2:1. A processive methyltransferase would show no dependency on the relative levels of light and heavy H3K27, since it would remain bound to the original substrate. The experiments revealed that the time when the abundance of H3K27me2:2 exceeds that of H3K27me2:1 precedes the time when the levels of H3K27me1:1 exceed those of H3K27me1:0 by a few hours (Fig. 6A and B). Thus, the data support processive dimethylation at H3K27.
Like new H3K9 trimethylation, new H3K27 trimethylation is mostly composed of methylation from the preexisting dimethylated substrate to form H3K27me3:1 during G1 phase and methylation from the newly synthesized substrate to form H3K27me3:3 during the S and G2/M phases (Fig. 6C). EZH2 methyltransferase transcripts did not fluctuate significantly across the cell cycle (data available on request). Also similarly to H3K9 trimethylation, all new trimethylation pathways (i.e., H3K27me3:1 plus H3K27me3:2 plus H3K27me3:3) were necessary to re-form steady-state H3K27 trimethylation levels during the G1 phase of the next cell cycle (Fig. 5C). Thus, these results support the hypothesis that H3K27 trimethylation is not replication coupled and requires methylation primarily from the unmodified and preexisting dimethylated histones to reach steady-state levels.
DISCUSSION
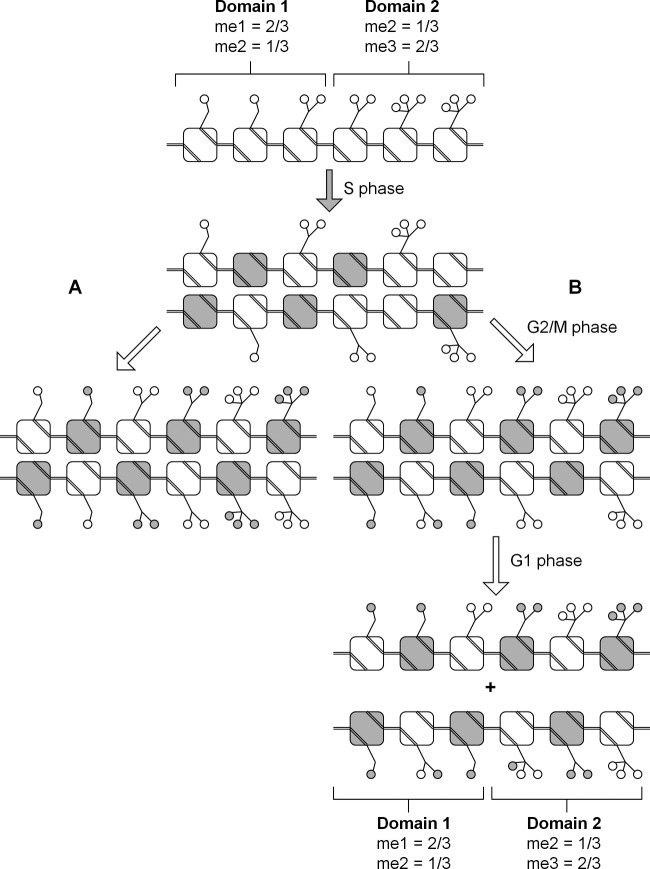
These experiments constitute some of the most highly time resolved analyses of histone methylation formation and turnover across the cell cycle and test the hypotheses that histone methylation is a replication-coupled or replication-independent process. Methylation at H4K20 is largely restricted to specific cell cycle stages, while methylation at H3K36 is generally independent of the cell cycle. H3K9 methylation and H3K27 methylation are formed similarly to one another, despite the initial hypotheses that H3K9 trimethylation would be replication coupled (41) while H3K27 trimethylation would be postreplication coupled. In particular, we provide evidence that trimethylation of H3K9 and H3K27 begins as early as S phase but is restored to steady-state levels only by the G1 phase of the next cell cycle (Fig. 7).
Fig 7.
Model summarizing the origin and formation of histone methylation across the cell cycle for H3K9 and H3K27. Preexisting histone modifications (open circles) are diluted following new histone synthesis (shaded squares) during S phase. For simplicity, only one histone H3 tail is shown per nucleosome (squares). Two hypothetical chromatin domains are shown, where the first is characterized by mostly monomethylated histones and the second is characterized by mostly trimethylated histones. (A) In the simplest hypothetical model, all the histone modifications are replaced following replication from the unmodified state. Our data do not support such a model. (B) In the model proposed on the basis of these experiments, a fraction of histones is modified from the unmodified state during G2/M phase. Some modifications are converted from preexisting methylated states, while K9 trimethylation and K27 trimethylation are left unformed. During the G1 phase of the next cell cycle, monomethylation is nearly fully turned over, while preexisting dimethylated histones have been converted to trimethylated histones. All the histone modifications are fully established at the correct relative stoichiometry in each chromatin domain.
An important discovery is that the preexisting dimethylated states for both residues contribute to the new trimethylated states following S phase (Fig. 7). De novo formation of H3K9 and H3K27 trimethylation from the unmodified state during the S and G2 phases (i.e., H3K9me3:3 and H3K27me3:3) reaches approximately 50% of the newly synthesized histone H3 levels. The remaining 50% is accounted for by the conversion of preexisting dimethylated residues, which acquire an additional methyl group during G1 phase to become new trimethylated residues (i.e., me3:1). Thus, new dimethylation must occur in excess of newly synthesized histone H3 proteins in order to replace the fraction of preexisting dimethylated histones that is converted to the trimethylated state.
The sum of all new H3K9 and H3K27 dimethylation events exceeds the levels of newly synthesized histone H3 proteins by >25% (i.e., 1.25 × 40% = 50%) (Fig. 5A and B) before M phase and by approximately 50% during the subsequent G1 phase (i.e., 1.5 × 40% = 60%) (data not shown). The excess production of newly dimethylated H3K9 and H3K27 proteins may replace the fraction of preexisting dimethylated histones (i.e., H3K9me2:0 and H3K27me2:0) that is being converted to the trimethylated pool (i.e., H3K9me3:1 and H3K27me3:1). Total H3K9me2 and H3K27me2 peptides occur at relative abundances approximately 1.3 and 2 times greater than those of total H3K9me3 and H3K27me3 peptides, respectively (Fig. 1D and E). Therefore, allocation of 50% of the preexisting dimethylated histones would be sufficient (i.e., 50% × 1.3 = 0.65 [>0.5]) to reach the steady-state trimethylated levels that otherwise could not be achieved with trimethylation from the unmodified state alone.
The two distinct trimethylation pathways suggested by our data are consistent with ChIP-Seq (high-throughput sequencing in combination with chromatin immunoprecipitation) experiments that have mapped the genomic localization of H3K9 and H3K27 methylation (5, 49). In particular, regions enriched in H3K9 and H3K27 monomethylation generally do not overlap regions enriched in H3K9 and H3K27 trimethylation. This mutual exclusion is logical given that H3K9 monomethylation and H3K27 monomethylation are generally considered euchromatic marks, while H3K9 trimethylation and H3K27 trimethylation are classically heterochromatic marks. ChIP-Seq reveals a much closer overall distribution between dimethylation and trimethylation at both residues, supporting our results suggesting that the preexisting dimethylated H3K9 and H3K27 pools do contribute to trimethylated H3K9 and H3K27 formation.
A second compelling finding is that H3K9 and H3K27 trimethylation is fully established much later than replication-dependent histone synthesis, during the G1 phase of the next cell cycle. Such a model has been predicted before on the basis of numerous other studies (45). Elegant immunofluorescence experiments have shown that Polycomb repressive complexes 1 and 2, which coordinate Polycomb silencing, are disrupted during mitosis. The complexes are re-formed on chromatin predominantly during mid- to late-G1 phase in Drosophila melanogaster embryos (9) and U2OS cells (2). These experiments support and complement our experiments on the timing and formation of H3K27me3 itself. In contrast, another series of experiments was unable to detect the complete synthesis of H3K9me2, H3K9me3, or H3K27me3 to match the newly synthesized histone H3 levels (51). As stated previously, this would imply a serial dilution of those histone modifications over successive cell cycles and would be difficult to reconcile with stable maintenance of epigenetic information across cell generations.
One possible mechanism for the completion of H3K9 trimethylation in the next cell cycle is that SUV39H1 methyltransferase activity does not fully restore H3K9me3 levels, due to antagonism from the mitotic occurrence of H3S10 phosphorylation. In particular, the surge in H3S10ph levels may prevent H3K9me3 from being fully synthesized by M phase. HP1 association with chromatin is diffuse in D. melanogaster embryos during metaphase and anaphase, and HP1 relocalizes on chromatin during the next interphase (30). This disruption in HP1 localization may lead to a disruption in SUV39H1 methyltransferase recruitment and activity, and thus in H3K9 trimethylation, during mitosis. A similar event may occur for H3K27me3, where another cell cycle-specific PTM on a nearby residue may occlude EZH2 binding. Neither SUV39H1 nor EZH2 mRNA levels were observed to be upregulated during G1. Thus, a change in methyltransferase protein levels or modulation of methyltransferase activity by other histone modifications or by modifications on the enzymes themselves could account for the latency of H3K9 and H3K27 trimethylation. The absence of appreciable turnover at these residues is similar to the lack of H4K20me3 turnover across the cell cycle (39), where H4K20me3 was initially implicated in position effect variegation silencing, such as that for H3K9me3, but has recently been questioned as an epigenetic silencing mark (44). Furthermore, ectopic overexpression of EZH2, and presumably increased H3K27 trimethylation, accelerates G1-phase progression and leads to increased accumulation in S phase (8). Thus, it is possible that the complete re-formation of H3K9me3 and H3K27me3 may provide a rate-limiting step restraining cells from beginning another round of histone synthesis and DNA replication.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paul Schedl for incisive and invaluable discussions; Eric Chan and Gary LeRoy for the design of the custom gene expression microarray; Mario Andres Blanco, Peter DiMaggio, and Donna Storton for microarray analysis assistance; Wenyun Lu and Joshua Rabinowitz for SRM analysis; Christina DeCoste and Christi O'Donnell for flow cytometry; Shu Lin, Shelby Blythe, and Olivier Devergne for qRT-PCR assistance; and Girish Deshpande, Michelle Gonzales-Cope, Shu Lin, Wei Li, Elizabeth Gavis, Yali Dou, Robert Schneider, and Or Gozani for reading the manuscript.
Barry M. Zee is supported by the National Science Foundation (NSF) Graduate Research Fellowship Program and Sigma Xi grants-in-aid research. Laura-Mae P. Britton is funded by the Department of Molecular Biology at Princeton University. Daniel Wolle is funded by an NIH training grant. Devorah M. Haberman is funded by the Department of Chemistry at Princeton University. Benjamin A. Garcia gratefully acknowledges support from an NSF Early Faculty Career award, NSF grant CBET-0941143, and an NIH Director's New Innovator award (DP2OD007447) from the Office of the Director, NIH.
Footnotes
Published ahead of print 30 April 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Abbas T, et al. 2010. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol. Cell 40: 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aoto T, Saitoh H, Sakamoto Y, Watanabe S, Nakao M. 2008. Polycomb group protein-associated chromatin is reproduced in post-mitotic G1 phase and is required for S phase progression. J. Biol. Chem. 283: 18905–18915 [DOI] [PubMed] [Google Scholar]
- 3. Araujo FD, Knox JD, Szyf M, Price GB, Zannis-Hadjopoulos M. 1998. Concurrent replication and methylation at mammalian origins of replication. Mol. Cell. Biol. 18: 3475–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bajad SU, et al. 2006. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J. Chromatogr. A 1125: 76–88 [DOI] [PubMed] [Google Scholar]
- 5. Barski A, et al. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–827 [DOI] [PubMed] [Google Scholar]
- 6. Bello B, Holbro N, Reichert H. 2007. Polycomb group genes are required for neural stem cell survival in postembryonic neurogenesis of Drosophila. Development 134: 1091–1099 [DOI] [PubMed] [Google Scholar]
- 7. Bernstein BE, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- 8. Bracken AP, et al. 2003. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 22: 5323–5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buchenau P, Hodgson J, Strutt H, Arndt-Jovin DJ. 1998. The distribution of polycomb-group proteins during cell division and development in Drosophila embryos: impact on models for silencing. J. Cell Biol. 141: 469–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cao R, et al. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- 11. Carrozza MJ, et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- 12. Cheutin T, et al. 2003. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299: 721–725 [DOI] [PubMed] [Google Scholar]
- 13. Chin HG, Patnaik D, Estève PO, Jacobsen SE, Pradhan S. 2006. Catalytic properties and kinetic mechanism of human recombinant Lys-9 histone H3 methyltransferase SUV39H1: participation of the chromodomain in enzymatic catalysis. Biochemistry 45: 3272–3284 [DOI] [PubMed] [Google Scholar]
- 14. Deal RB, Henikoff JG, Henikoff S. 2010. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328: 1161–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dion MF, et al. 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315: 1405–1408 [DOI] [PubMed] [Google Scholar]
- 16. Estève PO, et al. 2006. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 20: 3089–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Graves RA, Pandey NB, Chodchoy N, Marzluff WF. 1987. Translation is required for regulation of histone mRNA degradation. Cell 48: 615–626 [DOI] [PubMed] [Google Scholar]
- 18. Hagstrom K, Schedl P. 1997. Remembrance of things past: maintaining gene expression patterns with altered chromatin. Curr. Opin. Genet. Dev. 7: 814–821 [DOI] [PubMed] [Google Scholar]
- 19. Heintz N, Sive HL, Roeder RG. 1983. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol. Cell. Biol. 3: 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heit R, Rattner JB, Chan GK, Hendzel MJ. 2009. G2 histone methylation is required for the proper segregation of chromosomes. J. Cell Sci. 122: 2957–2968 [DOI] [PubMed] [Google Scholar]
- 21. Hirota T, Lipp JJ, Toh BH, Peters JM. 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438: 1176–1180 [DOI] [PubMed] [Google Scholar]
- 22. Holliday R. 1987. The inheritance of epigenetic defects. Science 238: 163–170 [DOI] [PubMed] [Google Scholar]
- 23. Hsu JY, et al. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102: 279–291 [DOI] [PubMed] [Google Scholar]
- 24. Huen MSY, Sy SMH, van Deursen JM, Chen J. 2008. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4–K20 methylation and DNA replication. J. Biol. Chem. 283: 11073–11077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jackson V. 1988. Deposition of newly synthesized histones: hybrid nucleosomes are not tandemly arranged on daughter DNA strands. Biochemistry 27: 2109–2120 [DOI] [PubMed] [Google Scholar]
- 26. Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- 27. Jørgensen S, et al. 2007. The histone methyltransferase SET8 is required for S-phase progression. J. Cell Biol. 179: 1337–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Julien E, Herr W. 2004. A switch in mitotic histone H4 lysine 20 methylation status is linked to M phase defects upon loss of HCF-1. Mol. Cell 14: 713–725 [DOI] [PubMed] [Google Scholar]
- 29. Kaneko S, et al. 2010. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 24: 2615–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kellum R, Raff JW, Alberts BM. 1995. Heterochromatin protein 1 distribution during development and during the cell cycle in Drosophila embryos. J. Cell Sci. 108: 1407–1418 [DOI] [PubMed] [Google Scholar]
- 31. Kornberg RD, Lorch Y. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98: 285–294 [DOI] [PubMed] [Google Scholar]
- 32. Krogan NJ, et al. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23: 4207–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu W, et al. 2010. PHF8 mediates histone H4 lysine 20 demethylation events in cell cycle progression. Nature 466: 508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loyola A, et al. 2009. The HP1α-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 10: 769–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martin C, Zhang Y. 2005. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6: 838–849 [DOI] [PubMed] [Google Scholar]
- 36. Melcher M, et al. 2000. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol. Cell. Biol. 20: 3728–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ng HH, Robert F, Young RA, Struhl K. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- 38. Ong SE, Mittler G, Mann M. 2004. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods 1: 119–126 [DOI] [PubMed] [Google Scholar]
- 39. Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. 2008. Certain and progressive methylation of histone H4 and lysine 20 during the cell cycle. Mol. Cell. Biol. 28: 468–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Prasanth SG, Shen Z, Prasanth KV, Stillman B. 2010. Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc. Natl. Acad. Sci. U. S. A. 107: 15093–15098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Probst AV, Dunleavy E, Almouzni G. 2009. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 10: 192–206 [DOI] [PubMed] [Google Scholar]
- 42. Rea S, et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferase. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- 43. Rice JC, et al. 2002. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev. 16: 2225–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sakaguchi A, Karachentsev D, Seth-Pasricha M, Druzhinina M, Steward R. 2008. Functional characterization of the Drosophila Hmt4-20/Suv4-20 histone methyltransferase. Genetics 179: 317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sarkies P, Sale JE. 2 October 2011. Propagation of histone marks and epigenetic memory during normal and interrupted DNA replication. Cell. Mol. Life Sci. [Epub ahead of print.] doi:10.1007/s00018-011-0824-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schotta G, et al. 2008. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 22: 2048–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sittman DB, Graves RA, Marzluff WF. 1983. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc. Natl. Acad. Sci. U. S. A. 80: 1849–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trojer P, et al. 2007. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129: 915–928 [DOI] [PubMed] [Google Scholar]
- 49. Wang Z, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. 1999. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 97: 99–109 [DOI] [PubMed] [Google Scholar]
- 51. Xu M, Wang W, Chen S, Zhu B. 4 November 2011. A model for mitotic inheritance of histone lysine methylation. EMBO Rep. [Epub ahead of print.] doi:10.1038/embor.2011.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu M, et al. 2010. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science 328: 94–98 [DOI] [PubMed] [Google Scholar]
- 53. Zee BM, et al. 2010. In vivo residue-specific histone methylation dynamics. J. Biol. Chem. 285: 3341–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zee BM, Levin RS, DiMaggio PA, Garcia BA. 2010. Global turnover of histone post-translational modifications and variants in human cells. Epigenetics Chromatin 3: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.