Background: Integrin αv promotes melanoma cell survival in in vivo and in vitro three-dimensional environments.
Results: PKCα is up-regulated in an integrin αv- and three-dimensional collagen-dependent manner and promotes p53 relocalization and melanoma survival.
Conclusion: PKCα functions downstream of integrin αv to promote melanoma cell survival.
Significance: PKCα has been identified as a key component of the integrin αv-mediated melanoma survival pathway.
Keywords: Apoptosis, Cancer Biology, Integrin, Melanoma, p53, Protein Kinase C (PKC), 3D Tissue Culture
Abstract
Protein kinase C α (PKCα) is overexpressed in numerous types of cancer. Importantly, PKCα has been linked to metastasis of malignant melanoma in patients. However, it has been unclear how PKCα may be regulated and how it exerts its role in melanoma. Here, we identified a role for PKCα in melanoma cell survival in a three-dimensional collagen model mimicking the in vivo pathophysiology of the dermis. A pathway was identified that involved integrin αv-mediated up-regulation of PKCα and PKCα-dependent regulation of p53 localization, which was connected to melanoma cell survival. Melanoma survival and growth in three-dimensional microenvironments requires the expression of integrin αv, which acts to suppress p53 activity. Interestingly, microarray analysis revealed that PKCα was up-regulated by integrin αv in a three-dimensional microenvironment-dependent manner. Integrin αv was observed to promote a relocalization of endogenous p53 from the nucleus to the cytoplasm upon growth in three-dimensional collagen as well as in vivo, whereas stable knockdown of PKCα inhibited the integrin αv-mediated relocalization of p53. Importantly, knockdown of PKCα also promoted apoptosis in three-dimensional collagen and in vivo, resulting in reduced tumor growth. This indicates that PKCα constitutes a crucial component of the integrin αv-mediated pathway(s) that promote p53 relocalization and melanoma survival.
Introduction
Protein kinase C α (PKCα) is a widely expressed, conventional member of the PKC family of serine/threonine kinases (1), which participates in numerous cellular functions such as proliferation, migration, survival, and apoptosis (2). Roles for PKCα have been identified in numerous cancer types, including breast (3), glioblastoma (4), and prostate (5). Strong links to melanoma have also been established (6), particularly to melanoma metastasis in patients (7–10). Interestingly, PKCα was required for integrin αvβ3-mediated melanoma invasion (9) and in integrin αvβ3-mediated murine primary osteoclast bone resorption and migration (11). Additional possible roles in cancer come from the ability of PKCα to influence apoptosis in a number of cell types where it is regarded as an anti-apoptotic member of the PKC family (12).
PKCα may exercise its anti-apoptotic function through phosphorylation of Bcl-2 (13). However, the ability of PKCα to influence the tumor suppressor p53 may also be of importance. Upon activation, p53 can promote apoptosis via the transcriptional up-regulation of proapoptotic factors, including PUMA, Apaf 1, and Bax (14) or via direct effects at the mitochondria (15). PKCα can bind and phosphorylate the C terminus of p53 (16, 17), although its effects upon p53 activity appear cell type specific (18, 19). Meanwhile, a recent yeast model identified PKCα as an inhibitor of p53-induced growth arrest (20). Nevertheless, it has remained unclear how PKCα may be regulated and exert its role in malignant melanoma.
Melanoma progression toward a malignant phenotype is characterized by a shift from a radial to a vertical and invasive growth pattern, which includes the up-regulation of integrin αvβ3 expression (21). Integrin αvβ3 promotes melanoma cell invasion, growth, and survival in both three-dimensional dermal collagen and in vivo in mice and in human dermis (22–26). Integrin αv-mediated regulation of melanoma cell survival is dependent upon cell growth within a three-dimensional environment, emphasizing the importance of three-dimensional models that mimic physiological conditions (22, 27). Indeed, integrin-mediated signal transduction and cell behavior in three-dimensional matrices are often different to that observed in two-dimensional tissue culture, and a closer approximation to physiological behavior (28–31). Previous data using a three-dimensional collagen model indicated that integrin αv expression promoted melanoma survival by inhibiting p53 activity and thereby apoptosis (22). Overcoming p53-mediated apoptosis is critical for cancer progression and although p53 is mutated in a significant number of cancers, the p53 gene is rarely mutated in melanoma (32). Despite this, malignant melanomas are typically extremely radio- and chemo-resistant and elevated p53 levels did not induce apoptosis (32, 33). However, it has remained unclear how integrin αv may mediate p53 inhibition.
To clarify how integrin αv may inhibit p53-mediated apoptosis, a three-dimensional collagen model was utilized. As the dermal extracellular matrix is comprised of ∼90% collagen type I, this model provides an environment comparable to physiological conditions (25). Through microarray-based expression profiling, we identified PKCα to be up-regulated downstream of integrin αv in a three-dimensional collagen-dependent manner. We also found that p53 translocation to the cytoplasm in melanoma cells was three-dimensional, integrin αv-, and PKCα-dependent. Furthermore, PKCα knockdown inhibited integrin αv-mediated melanoma survival in three-dimensional collagen and in vivo tumor growth. Our results indicate that in melanoma, PKCα up-regulation is critical for integrin αvβ3-mediated p53 regulation and suppression of apoptosis.
EXPERIMENTAL PROCEDURES
Culture Conditions
Human M21 melanoma cells and a FACS sorted integrin αv-negative subpopulation, M21L cells (23), were maintained in RPMI 1640 + HEPES containing 2 mm l-glutamine, 1.5% NaHCO3, and 5% FBS (all from Invitrogen). M21 cells stably expressing p53 siRNA (M21sip53) were maintained in M21 growth medium containing 600 μg/ml G418 (Invitrogen). M21shGFP, M21LshGFP, and M21shPKCα cells were maintained in M21 growth medium containing 3 μg/ml puromycin (Sigma Aldrich), whereas M21LPKCα cells were maintained in M21 growth medium containing 5 μg/ml blasticidin (Sigma Aldrich). Human melanoma FM88 cells stably expressing control siRNA (FM88sicon) and integrin αv siRNA (FM88siαv) were also maintained in M21 growth medium containing 600 μg/ml G418. FM88sicon, shPKCα, and FM88sicon, shGFP cells were maintained in M21 growth medium containing 600 μg/ml G418 and 3 μg/ml puromycin. FM88 cells express wild-type (WT) p53, whereas M21 cells express p53 with a G266E substitution that behaves as WT p53 (22). Prior to growth in three-dimensional collagen, cells were adapted with RPMI 1640 medium containing l-glutamine and 1% Nutridoma-SP (Roche Diagnostics) for 10 days (M21-derived cells) or 2 days (FM88-derived cells). Adapted cells were cultured in 2.5 mg/ml three-dimensional collagen type I gels (Advanced BioMatrix, San Diego, CA) prepared as described previously (25) and maintained with three-dimensional medium (RPMI 1640 + l-glutamine). To extract cells from the collagen, 0.25% clostridial collagenase (Worthington, Lakewood, NJ) in PBS was used for collagen digestion at 37 °C for 15 min. Cells were washed in PBS and used in downstream applications.
Analysis of p53 Localization by Immunofluorescence
For imaging of p53, 20 μl of collagen mixed with cells was pipetted into each well of an eight-well chamber slide (Nunc, Rochester, NY). Slides were inverted to produce a hanging drop of collagen and incubated at 37 °C, 5% CO2 for 30 min. Once set, the slide was placed the correct way up, and three-dimensional medium was added. After the period of time stated, cells were fixed in 4% paraformaldehyde for 10 min at room temperature and then permeabilized with 0.1% Triton X-100/PBS for 1 min at room temperature. Cells were blocked in 5% BSA/PBS for 20 min at room temperature and then probed with anti-p53 mAb (DO-1, Santa Cruz Biotechnology, Santa Cruz, CA; 1/200) in 5% BSA/PBS for 1 h at room temperature. After washing, Alexa Fluor 568 goat anti-mouse (Invitrogen, 1/200) and FITC-phalloidin (Invitrogen, 1/1000) were added for 1 h at room temperature in the dark. After further washing, cells were incubated with DRAQ5 DNA stain (Biostatus Ltd., Shepshed, UK, 1/1000) at 37 °C for 3 min in the dark. DRAQ5 was aspirated off, and the collagen was mounted in fluorescent mounting medium (DAKO, Glostrup, Denmark) and covered with a glass coverslip. For two-dimensional analysis of p53 localization, nutridoma-treated cells were cytospun onto slides, fixed, and prepared for imaging as in three dimensions.
Confocal images were taken using a Zeiss LSM510 microscope with a C-Apochromat 63×/1.2 numerical aperture water objective and LSM510 software. p53 fluorescent intensities in the cell cytoplasm and nucleus were analyzed using NIH ImageJ software and converted into a cytoplasmic/nuclear ratio for comparison between cells and experiments.
In Vivo p53 Imaging
p53 localization was established in vivo using the chick chorioallantoic membrane (CAM)3 model as described previously (34). Briefly, a small window was made in the shell above the CAM, and 5 × 106 M21 (αv+) or M21L (αv−) cells in 30 μl of sterile PBS were pipetted onto the membrane. The window was covered using tape and incubated for 7 days to allow tumor growth. Tumors were removed from the CAM, fixed with 4% paraformaldehyde overnight, rinsed in 70% ethanol, and then stored in fresh 70% ethanol overnight at 4 °C. Tumors were dehydrated, embedded in paraffin, and sectioned. Samples were mounted on glass slides at 60 °C for 30 min and then rehydrated in xylene followed by 100, 95, and 70% ethanol steps. Antigen retrieval was performed in citrate buffer, pH 6.0, heated for 4 min. After cooling and washing, the sample was permeabilized in 0.1% Triton X-100/PBS and blocked in 20% FBS/PBS for 1 h at room temperature. Samples were incubated with anti-p53 mAb (DO-1, Santa Cruz Biotechnology, 1/100) and anti-β-catenin pAb (Invitrogen, 1/250) in 10% FBS/PBS for 1 h at room temperature. After washing, Cy-5 goat anti-mouse (Jackson ImmunoResearch Laboratories, West Grove, PA, 1/200) and Rhodamine Red-X goat anti-rabbit antibodies (Jackson ImmunoResearch Laboratories, 1/250) in 10% FBS/PBS were added for 1 h at room temperature. Hoechst DNA stain (Invitrogen, 1/4000) was added in PBS for 3 min at room temperature. Samples were mounted in fluorescent mounting medium (DAKO) and imaged using an Andor spinning disc confocal with an Olympus UPlanSApo 60×/1.35 numerical aperture oil objective. NIH ImageJ software was used to determine the p53 localization ratio by identifying cytoplasmic (β-catenin) and nuclear (Hoechst) areas of cells before the mean p53 intensity of each area was measured. Mean p53 intensities were converted into a cytoplasmic/nuclear ratio to allow cellular and population comparisons.
In vivo localization of p53 was also established in mouse xenograft tumors. FM88sicon, shGFP; and FM88sicon, shPKCα tumors were stored at −80 °C upon excision and later embedded in Cryomount medium (Histolab, Gothenburg, Sweden), frozen, and sliced into 10-μm sections. Sections were placed onto coverslips and fixed (4% paraformaldehyde, 10 min at room temperature), washed, and blocked in 10% donkey serum/PBS containing 0.3% Triton X-100 for 1 h at room temperature. Sections were probed with FITC-conjugated anti-p53 mAb (DO-1, 1/100) and anti-β-catenin pAb (Invitrogen, 1/250) in 10% donkey serum/PBS overnight at 4 °C. After washing, Rhodamine Red-X goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, 1/500) in 10% donkey serum/PBS was added for 1 h at room temperature. DRAQ5 (Biostatus Ltd., 1/1000) was added for 3 min at 37 °C before the sections were mounted in fluorescent mounting medium (DAKO). Images were acquired using a Nikon A1R microscope with a Plan-Apochromat VC 60×/1.4 numerical aperture oil objective and Nikon A1R software. Quantitative analysis was performed as for the chick CAM images.
Immunoblotting
Cell lysates were prepared using lysis buffer (20 mm Tris, pH 7.6, 1 mm EDTA, 1 mm EGTA, 150 mm NaCl, and 1% Triton X-100 supplemented with 1 mm Na3VO4, 4% Complete protease inhibitor mixture (Roche Applied Science), 0.1% phosphatase inhibitor mixture 1 (Sigma Aldrich)). Soluble protein concentrations were ascertained using the BCA protein assay (Pierce), whereas non-soluble pellet fractions were sonicated in 3× sample buffer (187.5 mm Tris, pH 6.8, 6% SDS, 30% glycerol, 150 mm DTT, 0.03 g of bromphenol blue). Equivalent amounts of protein were separated by SDS-PAGE and then transferred onto polyvinylidene fluoride membrane (Millipore, Billerica, MA). Membranes were blocked in 5% nonfat milk for 1 h at room temperature and then incubated at 4 °C for 16 h in 5% nonfat milk containing anti-p53 mAb (DO-1), anti-β-tubulin pAb (H-235), anti-GAPDH mAb (Millipore), anti-PKCα pAb (Cell Signaling Technology, Beverly, MA), antiphospho-histone H3 (Millipore) and anti-AKT (Cell Signaling Technology). After washing, membranes were incubated with horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibodies (Jackson ImmunoResearch Laboratories) for 1 h at room temperature and then developed using Western Lightning Plus enhanced chemiluminescence (PerkinElmer Life Sciences, Waltham, MA).
Determination of Apoptosis by Flow Cytometry
Cells were cultured in three-dimensional collagen for 3 or 5 days before being extracted as described above. The cells were resuspended in 0.05% BSA/PBS and fixed with ice cold 70% ethanol on ice for 30 min. Cells were spun down, resuspended in PBS with 1 μg/ml RNase A and 10 μg/ml propidium iodide (both from Sigma Aldrich), and incubated at 37 °C for 1 h. Cells were analyzed using the FACSCalibur flow cytometer and CellQuest software (both from Becton Dickinson, Franklin Lakes, NJ). The sub-G1 population of cells was regarded as apoptotic.
Determination of Apoptosis by Cleaved Caspase-3 Immunofluorescence
Cells were cultured in three-dimensional collagen on chamber slides and prepared as for p53 localization imaging. Apoptotic cells were visualized using anti-cleaved caspase-3 Asp-175 pAb (Cell Signaling Technology, 1/2000) and subsequent incubation with Alexa Fluor 568 goat anti-rabbit (Invitrogen, 1/200), FITC-phalloidin (Invitrogen, 1/1000), and DRAQ5 (Biostatus, Ltd., 1/1000). Once mounted, slides were imaged using a Nikon A1R confocal microscope with a Plan Apochromat 20×/0.75 numerical aperture air objective. 3 × 3 montage images were acquired for each planar view with two planes imaged per collagen drop. Analysis was performed using Nikon NIS-Elements software, nuclear outlines were identified using DRAQ5 staining, and the mean cleaved caspase-3 intensity within the nucleus was ascertained.
Cleaved caspase-3 determination of apoptosis in vivo utilized mouse xenograft tumors mounted as for p53 localization studies. Permeabilized sections were probed with anti-cleaved caspase-3 Asp-175 (Cell Signaling Technology, 1/500) and subsequent incubation with Rhodamine Red-X goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, 1/500) and DRAQ5 (Biostatus Ltd., 1/1000). Images were acquired using a Nikon A1R confocal microscope with a Plan Apochromat 20×/0.75 numerical aperture air objective and analyzed using ImageJ software. Nuclear regions were identified from the DRAQ5 staining, and the mean intensity of cleaved caspase-3 in each region was measured.
Determination of Apoptosis by Scoring for Blebbing Phenotype
Dimethyl sulfoxide (DMSO) or 100 nm of the PKCα inhibitor Gö6983 (Merck KGaA, Darmstadt, Germany) was mixed with collagen prior to addition of cells. Collagen containing cells was added to a 24-well plate. Cells were cultured for 5 days after which apoptotic cells, using membrane blebbing as a read out, were counted on an inverted light microscope.
Microarray
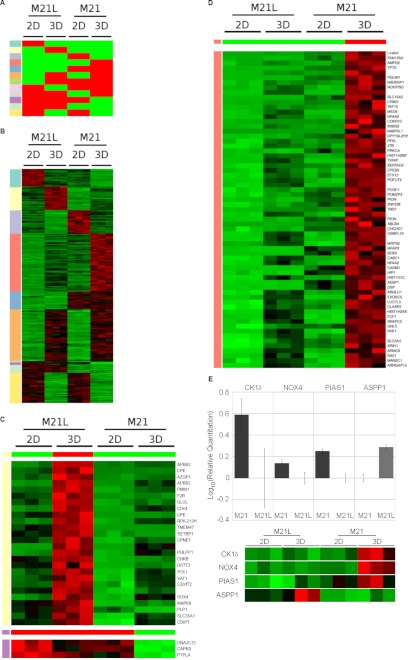
Total RNA from 2 × 106 M21 (αv+) and M21L (αv−) cells grown for 72 h in three-dimensional collagen was isolated using the RNeasy kit (Qiagen, Solna Sweden). Three independent biological replicates were used for each condition. Raw data from Affyetrix hgu133a chips (Affymetrix Inc., Santa Clara, CA, USA) were normalized using the robust multiarray average with GC correction (GCRMA) procedure, implemented in the R package gcrma (quantile-based normalization with background correction based on gene chip content of probes (35)). Differential expression between conditions was estimated using significance analysis of microarrays procedure (36) implemented in the R package siggenes. Each gene found to be differentially expressed was assigned to the most similar pattern of expression. First, the gene profile was normalized to the 0–1 range, and then the Euclidean distance to each of the possible expression patterns (also range normalized, a schematic is shown in Fig. 1A) was computed. The gene was assigned to the pattern with the smallest distance.
FIGURE 1.
Microarray analysis of M21 (αv+) and M21L (αv−) cells grown in two- (2D) and three-dimensional (3D) collagen tissue culture identified a number of patterns of gene up- and down-regulation. A, diagram illustrating the 12 different categories of gene regulation observed. Red indicates high relative expression, and green indicates low relative expression. The identity of each expression pattern (1–8) is also indicated by its row-side color code (leftmost column), which is used in B–D and supplemental Table S1 to identify the patterns. B, genes significantly altered between any conditions were detected using the significance analysis of microarrays procedure (38), using a False Discovery Rate cut-off of 0.001, resulting in 262 genes identified. Genes were grouped based upon which of the 12 categories their regulation falls within. Each row is color-coded according to its corresponding pattern in A. Individual categories and the genes that constitute each category can be seen in supplemental Table S1. The expression profiles for each gene and condition shows the levels determined in three separate experiments. C, to select genes that follow a specific pattern of expression, the significant genes were range-normalized to the 0–1 range and ranked by similarity to a pattern of regulation (see “Experimental Procedures”). Shown are the gene expression patterns of the genes that were most similar to the pattern shown in the top panel A with the corresponding color code: pale yellow (1), low expression in M21L (αv−) and M21 (αv+) cells in two dimensions, but high relative expression in M21L cells in three dimensions; purple (2), high expression in M21L (αv−) and M21 (αv+) cells in two dimensions, but low relative expression in M21 cells in three dimensions. Relative expression is indicated by color, where green represents low expression and red indicates high expression. D, gene expression patterns (as detailed in C) showing genes with low expression in M21L (αv−) and M21 (αv+) in two dimensions but high relative expression in M21 cells in three dimensions (corresponding to the light red pattern in A). E, RT-PCR verification of microarray data showing sample gene transcriptional modification upon growth for M21 (αv+) versus M21L (αv−) in three-dimensional collagen with or without integrin αv. Graphs shown are mean log10 relative change of three separate experiments. Microarray data are shown alongside the graphs for comparison and are prepared in the manner detailed in C.
qPCR Determination of mRNA Levels
RNA was isolated from cells cultured in three-dimensional collagen for 3 days using the RNeasy mini kit (Qiagen). cDNA was produced by random priming, and RNase was treated using the cDNA ProtoScript First Strand cDNA Synthesis kit (New England Biolabs). Primers targeting ASPP1 were as follows: (5′-AGCAAGCCACACCACCTAAGAATTA-3′ (forward) and 5′-TGAACCCGAAGGTAAAACGGG-3′ (antisense)), CK1δ (5′-GCTCCACATTGAGAGCAAAA-3′ (forward) and 5′-ATCTGATGGTGGGGATGC-3′ (antisense)), NOX4 (5′-ACCGGCA GAGTTTACCCAGCACA-3 (forward) and 5′-TCTTTCGGCACAGTACAGGCACAA-3′ (antisense)), PIAS1 (5′-AGCCGACCAATTAATATCACCTCA-3′ (forward) and 5′-TATTCCCTTTGCTCGTAACCTCTG-3′) (antisense), PKCα (5′-CCTATGGCGTCCTGTTGTATGAA-3′ (forward) and 5′-CCGCTTGGCTGGGTGTTT-3′ (antisense)), and HPRT (5′-TGACACTGGCAAAACAATGCT-3′ (forward) and 5′-GTCCTTTTCACCAGCAAGCT-3′ (antisense)) as an internal control were used in a SYBR green qPCR assay (Applied Biosystems Invitrogen). DNA amplification was measured and analyzed using the Applied Biosystems 7500 Fast real-time PCR system and 7000 SDS software.
Stable Cell Lines
Full-length PKCα cDNA was cloned into the pLenti6/V5-d-TOPO vector (Invitrogen), whereas the expression vectors pLKO.1-puro shPKCα and pLKO.1-puro shGFP were purchased from Sigma Aldrich. MISSION® PKCα shRNA sequences TRCN0000001692 and TRCN0000001692 were utilized for stable PKCα knockdown (Sigma Aldrich, NM_002737). The target gene plasmids along with the pMDLg/RRE, pCMV-VSVG, and pRSV-Rev plasmids required for viral production were transfected into HEK293FT cells using Lipofectamine 2000 (Invitrogen). The cells outlined above were infected with viruses containing the appropriate plasmids, and positive cells were selected 48 h post infection using 5 μg/ml blasticidin (Invitrogen) or 3 μg/ml puromycin (Sigma Aldrich).
In Vivo Tumor Growth
Animal experiments were performed according to a permit from the Stockholm South ethical committee in Sweden. Balb/c nude female mice (6–8 weeks) were obtained from Taconic. 5 × 105 FM88sicon, shGFP, FM88sicon; and shPKCα melanoma cells were mixed with Matrigel (Becton, Dickinson) in a 1:1 ratio and injected subcutaneously at the back of the mice. The wet weight of dissected tumors was determined at the experiment's conclusion, and tumors were utilized in downstream applications as described.
RESULTS
Identification of Integrin αv- and Three-dimensional Collagen-dependent Transcriptional Changes
Integrin-mediated outside-in signaling can activate a number of downstream pathways that regulate gene transcription (37). To establish integrin αv-mediated changes to gene transcription, the metastatic melanoma cell line M21 (αv+) and the integrin αv-negative M21-derived M21L cells (23) were utilized. M21 (αv+) and M21L (αv−) cells were cultured in two- and three-dimensional collagen for 72 h to assess the integrin αv- and three-dimensional collagen dependence of gene transcriptional changes (Fig. 1, A and B, and supplemental Table S1). A number of genes showed significant change between two- and three-dimensional culture conditions, which were independent of the integrin αv status (orange group, Fig. 1, A and B), whereas the transcription of other genes was identified to be integrin αv-dependent but two- and three-dimensional collagen independent (blue group, Fig. 1, A and B). The transcription of other genes was specifically up-regulated in three-dimensional collagen upon lack of integrin αv expression (pale yellow group, Fig. 1, A–C), whereas some genes were down-regulated in an integrin αv and three-dimensional collagen-dependent manner (purple group, Fig. 1, A–C). Genes undergoing significant integrin αv and three-dimensional collagen-dependent transcriptional up-regulation (pink group, Fig. 1, A, B, and D) were also identified. A number of targets linked to p53 function that displayed differential regulation on the microarray were validated by quantitative real-time PCR (qPCR). ASPP1, CK1δ, NOX4, and PIAS1 qPCR showed the same regulation as observed with the microarray (Fig. 1E). The microarray data also identified a number of integrin αv-dependent transcriptional changes that specifically occur upon growth on a two-dimensional substrate or within three-dimensional-collagen (supplemental Table S1, full microarray data available at the GEO database (www.ncbi.nlm.nih.gov/geo/, accession no. GSE34219). This information aids our understanding of how integrin αv promotes melanoma cell survival and highlights specific hits that can clarify the molecular pathways implicated in this process.
Integrin αv-mediated Up-regulation of PKCα in Three-dimensional Collagen Matrices
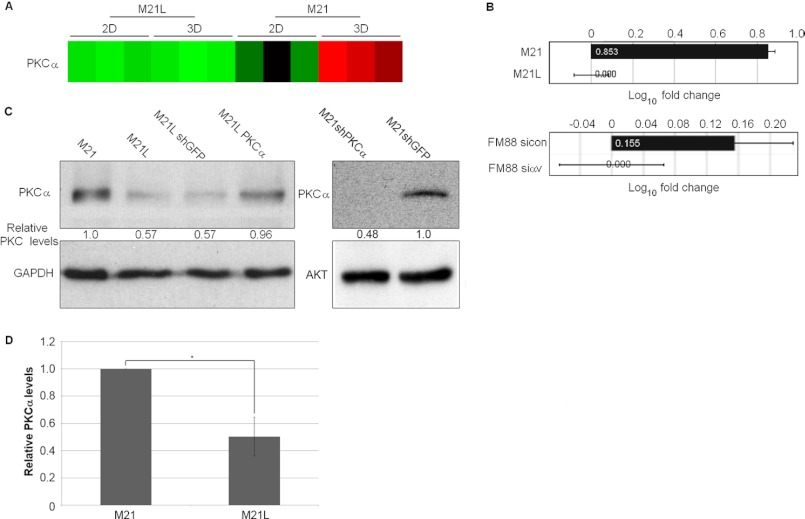
Interestingly, the PKCα gene was among the genes found to undergo transcriptional regulation in an integrin αv- and three-dimensional collagen-dependent manner (Figs. 1D and 2A). Importantly, PKCα has previously been identified to promote melanoma progression (6, 9). The transcriptional regulation of PKCα was verified by qPCR; M21 cells (αv+) showed a substantial increase in PKCα mRNA, whereas FM88sicon cells (αv+) also showed an increase in PKCα mRNA levels compared with FM88siαv cells (αv−), although the fold-change observed was smaller than that in M21 cells (Fig. 2B). PKCα protein levels were also significantly higher in M21 cells (αv+), compared with M21L cells (αv−) (Fig. 2, C and D). These results show that in three-dimensional collagen, PKCα is elevated in an integrin αv-dependent manner, suggesting a potential role for PKCα downstream of integrin αv in melanoma.
FIGURE 2.
Integrin αv up-regulates melanoma cell PKCα mRNA and protein levels. A, microarray analysis of PKCα transcription levels visualized as displayed in Fig. 1. B, RT-PCR quantification of PKCα mRNA levels in M21 versus M21L cells and FM88sicon versus FM88siαv cells cultured in three-dimensional (3D) collagen for 3 days. Log10 fold change graph shows the mean fold change of three separate experiments. C, levels of PKCα were examined by immunoblotting of lysates from M21, M21L, M21LshGFP, M21LPKC, M21shGFP, and M21shPKCα cells cultured in three-dimensional collagen for 3 days. GAPDH and Akt levels were determined as loading controls. D, quantification of PKCα expression levels as determined by immunoblot of lysates from M21 (αv+) and M21L (αv−) cells grown in three-dimensional collagen for 3 days. Graph shows mean PKCα expression levels corrected to loading controls ± S.E. among three independent experiments with M21 PKCα levels normalized to 1 (*, p < 0.05 using t test). 2D, two-dimensional.
PKCα Is Critical for Integrin αv-mediated Relocalization of p53
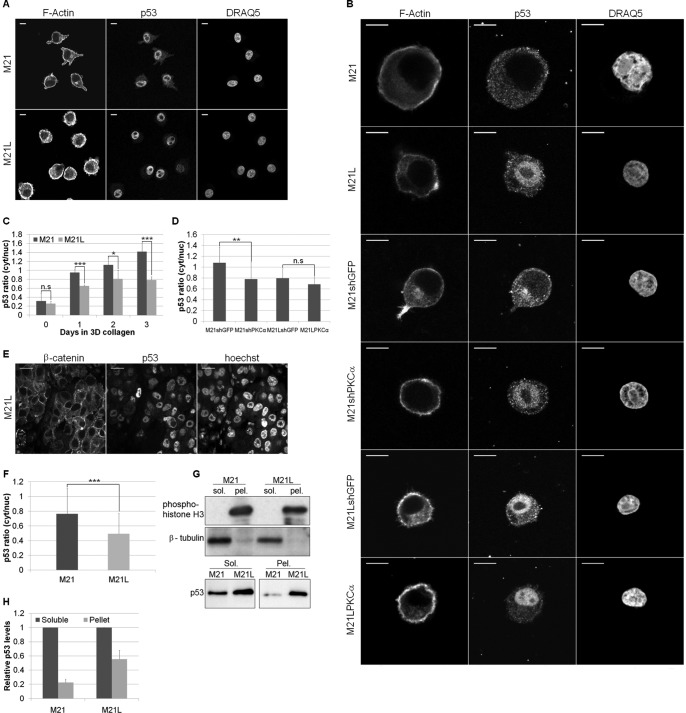
Previous data showed that integrin αv promoted melanoma cell survival in three-dimensional collagen through inhibition of p53-mediated apoptosis (22). Nuclear localization of p53 has been linked to its oligomerization state (38) and hence transcriptional activity. However, although cytoplasmic p53 is linked to mitochondrial-mediated apoptosis (15), promotion of a cytoplasmic p53 localization has also been observed to inhibit p53-mediated apoptosis in a number of survival mechanisms (39–41). Here, we identified a three-dimensional-dependent relocalization of p53. Under two-dimensional conditions, p53 was predominantly nuclear in both M21 (αv+) and M21L (αv−) cells (Fig. 3A) but in three-dimensional collagen, M21 (αv+) cells had a higher ratio of cytoplasmic to nuclear p53 as compared with two-dimensional collagen (Fig. 3, A and B). Importantly, M21 (αv+) cells displayed a higher cytoplasmic to nuclear p53 ratio as compared with M21L (αv−) cells (Fig. 3, B and C). This integrin αv-regulated relocalization of p53 was also observed in M21 (αv+) and M21L (αv−) tumors grown in vivo in the chick chorioallantoic membrane (CAM) (Fig. 3, E and F). Studies using in vivo FM88sicon and FM88siαv tumors, also grown in the chick CAM, showed similar results with integrin αv knockdown resulting in elevated p53 nuclear levels (data not shown). In addition, lysates from cells cultured in three-dimensional collagen showed elevated p53 levels in the nuclear fraction of M21L (αv−) cells as compared with M21 (αv+) cells (Fig. 3, G and H). However, analysis of p53 localization using lysate fractions of FM88 cells grown in three-dimensional collagen proved difficult, possibly due to the comparatively low p53 expression levels observed. To examine whether up-regulation of PKCα was important for p53 and integrin αv function, PKCα levels were modified by stable knockdown and overexpression in M21 (αv+) and M21L (αv−) cells, respectively (Fig. 2C). PKCα can phosphorylate p53 in vitro (16), although its effect upon p53 activity remains unclear (17, 20). Importantly, under three-dimensional collagen conditions, M21 (αv+) cells with stably knocked down PKCα exhibited a reduced p53 relocalisation with a greater proportion of p53 within the nucleus compared with control M21 cells stably expressing GFP knockdown sequences (Fig. 3, B and D). However, PKCα overexpression in M21L (αv−) cells, was not capable of rescuing the integrin αv-mediated p53 relocalization (Fig. 3, B and D), suggesting that PKCα is necessary but not sufficient to promote p53 relocalization and that one or more other cellular responses to integrin αv are also required.
FIGURE 3.
Integrin αv and PKCα regulate p53 localization in three-dimensional collagen. A, immunofluorescence with an anti-p53 antibody was utilized to analyze p53 localization in M21 (αv+) and M21L (αv−) cells grown in two dimensions. F-actin and nuclear staining (DRAQ5) were used to highlight the cell periphery and the nucleus. Scale bar, 10 μm. B, p53 localization in three-dimensional collagen was analyzed by immunofluorescence in M21, M21L, M21shGFP, M21shPKCα, M21LshGFP, and M21LPKCα cells. F-actin and nuclear (DRAQ5) staining were determined to highlight the cell periphery and the nucleus. Scale bar, 10 μm. C, image quantification of M21 and M21L cells in two dimensions (2D; 0 days) and after 1, 2, and 3 days in three-dimensional (3D) collagen. Representative areas of a uniform size were selected within the cell cytoplasm and nucleus, and the mean p53 intensity was determined. A cytoplasm/nucleus intensity ratio was determined to allow comparison across samples and plotted. Graph shows mean p53 ratio ± S.E. among three separate experiments (n = 24–45 cells/condition; n.s., not significant; *, p < 0.05; ***, p < 0.001 using t test). D, quantification of p53 localization in cells with PKCα levels manipulated and control cells after 3 days in three-dimensional collagen. Images for M21shGFP, M21shPKCα, M21LshGFP, and M21LPKCα were analyzed as in C. Graph shows mean p53 ratio ± S.E. among three separate experiments (**, p < 0.01 using t test). E, representative images of an in vivo M21L tumor section as analyzed in F. M21L cells were stained by immunofluorescence with an anti-p53 antibody to establish the p53 localization. β-Catenin and Hoechst were utilized to determine the cell periphery and nucleus, respectively. Scale bar, 10 μm. F, quantification of p53 localization from images of in vivo M21 and M21L tumors grown in a chick CAM assay. Images were analyzed as in C, and the graph shows the mean p53 ratio ± S.E. among three separate experiments (***, p < 0.001 using t test). G, upper: lysate fractionation was assessed by immunoblotting the soluble (sol.) and pellet (pel.) fractions of M21 and M21L lysates for phosphohistone H3 and β-tubulin. Immunoblots are representative of two separate experiments. Lower: soluble and pellet levels of p53 were determined by immunoblotting lysates from M21 and M21L cells. The immunoblot is representative of three separate experiments. H, quantification of soluble and pellet p53 levels as determined by immunoblot. Graph shows mean p53 levels ± S.E. among three independent experiments with soluble p53 levels normalized to 1.
PKCα Is Required for Integrin αv-dependent Melanoma Survival
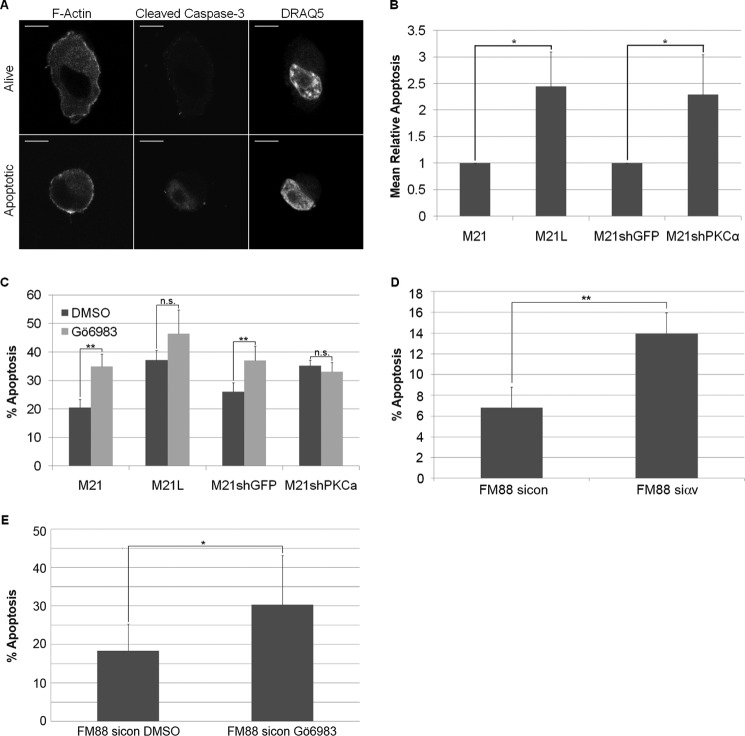
To identify apoptotic cells within three-dimensional collagen, cleaved caspase-3 levels were analyzed (Fig. 4A). As shown previously (22), a larger fraction of M21L (αv−) cells undergo apoptosis compared with M21 (αv+) cells in three-dimensional collagen (Fig. 4B). Furthermore, M21 cells with stably knocked down PKCα showed increased levels of apoptosis compared with control M21 cells (Fig. 4B). Addition of the PKCα inhibitor, Gö6983 also induced apoptosis in M21 cells in three-dimensional collagen (Fig. 4C), suggesting PKCα knockdown-mediated induction of apoptosis is not an artifact or a consequence of off-target effects. In addition, a second M21shPKCα stable cell line, expressing an alternative PKCα shRNA sequence, also exhibited increased apoptosis levels compared with M21shGFP cells as determined through visualization of the blebbing phenotype (data not shown). FM88 cells with stably knocked down integrin αv also showed higher levels of apoptosis compared with control FM88 (αv+) cells (Fig. 4D), indicating that survival in the FM88 melanoma cell line is also controlled by integrin αv in three-dimensional-collagen. In addition, inhibition of PKCα with Gö6983 also increased the observed incidence of apoptosis in FM88 cells (Fig. 4E). However, M21L (αv−) cells stably overexpressing PKCα were unable to promote melanoma survival (data not shown). This correlates with the inability of PKCα overexpression to rescue integrin αv-mediated relocalization of p53 in three-dimensional collagen and indicates it is insufficient to promote melanoma survival alone in the absence of integrin αv.
FIGURE 4.
PKCα regulates melanoma cell survival in three-dimensional collagen. A, apoptosis was measured by immunofluorescence with an anti-cleaved caspase-3 antibody. F-actin and nuclear (DRAQ5) staining was performed to visualize the cell periphery and nucleus, respectively. Representative images of a viable and an apoptotic M21shGFP cell in three-dimensional collagen are shown. Scale bar, 10 μm. B, quantification of apoptotic cells cultured in three-dimensional collagen for 5 days. Automated identification of nuclear regions was determined by DRAQ5 staining, and the mean nuclear cleaved caspase-3 intensities were measured. M21sip53 cells after 1 h in three-dimensional collagen were used as a control for cleaved caspase-3 intensity in non-apoptotic cells and used to define viable and apoptotic intensities in subsequent experiments. Graphs show mean percentage of apoptotic cells from three independent experiments ± S.E. (*, p < 0.001 using t test). C, light microscopy was used to manually count healthy versus apoptotic cells cultured for 5 days in three-dimensional collagen. M21, M21L, M21shGFP, and M21shPKCα cells were cultured with dimethyl sulfoxide or 100 nm Gö6983 and scored as healthy or blebbing. Graph shows mean levels of apoptosis ± S.E. among three independent experiments (n.s., not significant; **, p < 0.01 using chi squared test on arcsin normalized data). D, quantification of apoptotic cells cultured in three-dimensional collagen for 3 days. FM88sicon and FM88siαv were stained with PI, and the apoptotic population (sub-G1) was established using FACS. Graphs show mean percentage of apoptotic cells from three independent experiments ± S.E. (**, p < 0.01 using t test). E, quantification of apoptosis was determined using light microscopy to manually quantify the apoptotic cells cultured in three-dimensional collagen for 3 days. FM88sicon cells were cultured with dimethyl sulfoxide or 100 nm Gö6983 and scored as healthy or blebbing. Graph shows mean levels of apoptosis ± S.D. for 170 (dimethyl sulfoxide, DMSO) and 136 (Gö6983) cells (*, p < 0.05 using a Chi-squared test on arcsin normalized data).
PKCα Promotes p53 Relocalization and Melanoma Survival in Vivo
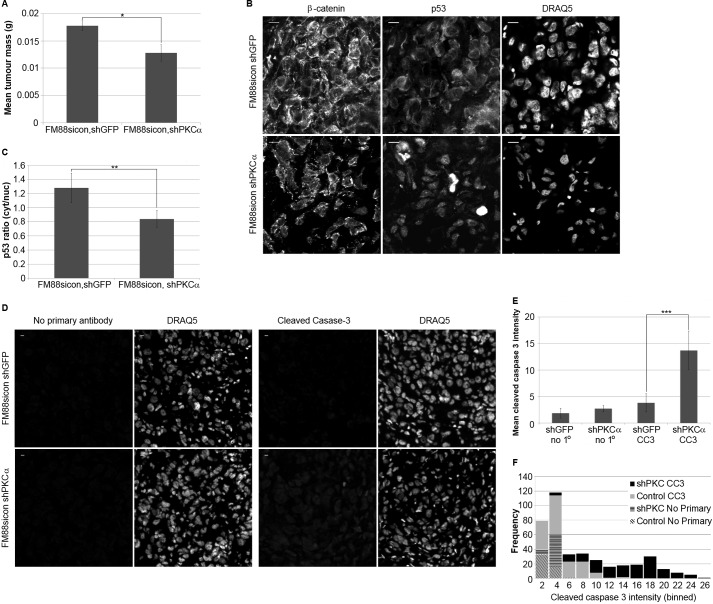
To determine whether PKCα may be required downstream of integrin αv for p53 relocalization and melanoma tumor growth in vivo, control FM88 cells (αv+), and control FM88 cells with stably knocked down PKCα, were injected subcutaneously into the back of balb/c nude mice. After 18 days of growth, FM88 cells with stably knocked down PKCα had a significantly reduced mass compared with control cells (Fig. 5A), identifying a role for PKCα in melanoma tumor growth. To establish whether the reduction in FM88sicon shPKCα tumor growth corresponded with the altered p53 localization observed in vitro, p53 immunofluorescence was performed on tumor sections. FM88sicon shPKCα tumor cells had a significantly higher nuclear p53 localization compared with FM88sicon shGFP tumor cells (Fig. 5, B and C), suggesting that PKCα is required for promoting a cytoplasmic relocalization of p53 both in vitro and in vivo. Furthermore, FM88sicon shPKCα tumors had significantly higher levels of cleaved caspase-3 (Fig. 5, D–F), suggesting increased rates of apoptosis within the FM88sicon shPKCα tumor. This supports a role for PKCα regulating melanoma tumor apoptosis and suggests that PKCα is an essential in vitro and in vivo downstream component of integrin αv-mediated cell survival.
FIGURE 5.
PKCα regulates p53 localization and apoptosis in vivo and promotes melanoma tumor growth. A, FM88sicon, shGFP (control) and FM88sicon, shPKCα (PKCα-) melanoma cells were subcutaneously injected into Balb/c nude mice. Tumors were removed from mice after 18 days of growth. Removed tumors were weighed on electronic scales. Graph shows mean tumor wet weight ± S.D. (n = 7, *, p < 0.05 using t test). B, localization of p53 was observed in FM88 tumor sections with an FITC-conjugated anti-p53 antibody. β-Catenin and nuclear (DRAQ5) staining was performed to visualize the cell periphery and nucleus, respectively. Representative images of FM88sicon, shGFP and FM88sicon, shPKCα tumor sections are shown. Scale bar, 10 μm. C, quantification of p53 localization in FM88 tumor sections. Representative areas of a uniform size were selected within the cell cytoplasm and nucleus, and the mean p53 intensity was determined. Graph shows mean p53 ratio ± S.D. (n = 60–120 cells/tumor, five tumors per cell type; **, p < 0.01 using t test). D, apoptosis in FM88 tumor sections was analyzed by immunofluorescence using an anti-cleaved caspase-3 antibody. Nuclear (DRAQ5) staining was performed to visualize the nucleus. Tumor sections stained with no primary antibody added were used as a background fluorescence control. E, representative areas of a uniform size were selected using the DRAQ5 channel, and the mean intensity of cleaved caspase-3 was determined. Graph shows mean cleaved caspase-3 intensity ± S.E. (n = 50 cells/tumor, three tumors/cell type; ***, p < 0.001 using t test). F, data from E plotted as a histogram showing frequency of cleaved caspase-3 intensity for each tumor type.
DISCUSSION
A role for PKCα in regulating p53 localization and melanoma survival is of particular interest given its strong association with melanoma (6). Although PKC may regulate integrin αvβ5 function in melanoma cells in two-dimensional culture (42), our results here link PKCα into signaling downstream of integrin αv in melanoma cells. The PKC protein family may regulate melanoma survival, as PMA treatment promoted anchorage-independent survival of melanoma cells (43). To this end, PKCα has been suggested to be anti-apoptotic (12), and differing mechanisms have been described, including PKCα-mediated phosphorylation of bcl2 (13, 44), although data in glioma cells suggested that the mechanism of PKCα was kinase-independent (4). However, our data indicate that PKCα kinase activity is critical for melanoma cell survival, as use of the PKC kinase inhibitor Gö6983 resulted in elevated levels of apoptosis. PKCα has also been associated with melanoma metastasis (8), possibly through the regulation of adhesion turnover and Rac activity (9). In addition, PKCα inhibition reduced ERK1/2 activity and was also implicated as a possible mechanism to inhibit melanoma invasion (10). This finding corresponds with our previous data implicating integrin αv in the regulation of MEK1 and ERK1/2 upon melanoma growth in three-dimensional collagen (22) and supports the finding that PKCα is a downstream effector of integrin αv.
Inhibition of apoptosis by integrin αv is achieved through the suppression of the DNA-binding activity of p53 (22). PKCα can phosphorylate the C-terminal region of p53 at Ser-376 and Ser-378 (16) and inhibit p53 in yeast (20), whereas PKCα overexpression promoted murine p53 transcriptional activity (17). The observed differences in the role of PKCα in p53 regulation could be a consequence of differences between the model systems used. The induction of apoptosis upon PKCα knockdown in this study, together with previous data (22), suggests that PKCα may function as an inhibitor of p53 in melanoma. However, we were unable to detect direct PKCα regulation of p53. The anti-p53 antibody epitope of pAb421 covers the region potentially phosphorylated by PKCα, and the binding of the antibody is inhibited by phosphorylation in this region (18). However, immunoblotting with pAb421 showed no clear difference between M21 and M21L cells after growth in three-dimensional collagen (data not shown). On the other hand, addition of Gö6983 promoted apoptosis, suggesting that PKCα kinase activity is required for melanoma cell survival. PKCα phosphorylation of p53 has been reported to promote p53 ubiquitination and degradation in unstressed fibroblasts (45). However, PKCα knockdown did not increase p53 levels in our melanoma cells, but we did observe an elevated level of nuclear p53. Interestingly, phosphorylation of p53 by GSK3β at Ser-315 and Ser-376, a PKCα phosphorylation site, promoted a p53 cytoplasmic localization and an inhibition of p53-mediated apoptosis during endoplasmic reticulum stress (41). This finding is in concert with our results in melanoma cells upon growth in three-dimensional collagen. Although it appears likely that PKCα does regulate p53 in a kinase-dependent manner, this may be an indirect regulation via other effectors or through phosphorylation at p53 sites other than Ser-376/378.
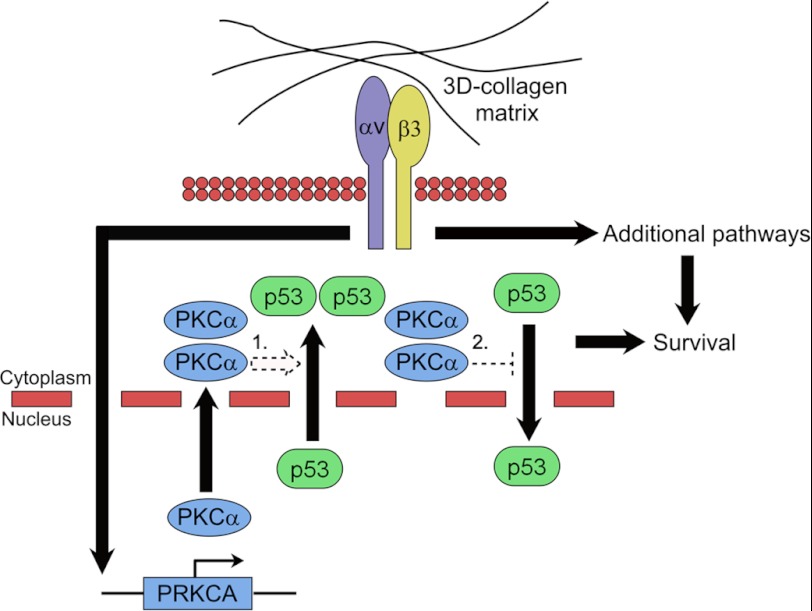
An alternative to directly regulating p53 transcriptional activity is to influence its cellular localization. Nuclear export signal sequences within p53 can drive export upon p53 monomerization (38) or after DNA damage-dependent phosphorylation of Ser-15 (46). In addition, p53 nuclear localization sequences and a cytoplasmic sequestration domain have been identified (47, 48). We found that in two-dimensional culture, p53 was almost entirely nuclear located in both M21 (αv+) and M21L (αv−) cells. However, within three-dimensional collagen and in vivo, p53 became progressively more cytoplasmic in a PKCα- and integrin αv-dependent manner. The development of three-dimensional tissue culture has furthered our understanding of tissue (30) and cancer development, including melanoma (25). Integrin αvβ3 expression is critical for the development of invasive melanoma (21, 24). However, it was the use of three-dimensional tissue culture that allowed unraveling of the role of integrin αv in promoting melanoma survival (25) through inhibition of p53 (22). The role of integrin αv in inhibiting apoptosis through blocking p53 function suggested an explanation to why p53 mutations are rarely observed in malignant melanoma (32). The translocation of p53 into the cytoplasm was identified as a three-dimensional microenvironment-, integrin αv-, and PKCα-dependent event. We propose that melanoma cells promote a cytoplasmic p53 localization to overcome apoptosis, avoiding the requirement for p53 mutation (Fig. 6). Our results also point out a pathway with multiple potential targets for development of melanoma therapy, including integrin αv (26), p53 (49), and PKCα.
FIGURE 6.
Model for integrin αv promotion of melanoma cell survival in three-dimensional collagen. Melanoma cells within the dermis of the skin are surrounded by a three-dimensional matrix predominantly consisting of type I collagen as shown at the top of the figure. Melanoma expression of integrin αv promotes transcriptional up-regulation of PKCα resulting in elevated PKCα protein levels. Increased PKCα levels promote a relocalization of endogenous p53 protein from a chiefly nuclear to a cytoplasmic localization. PKCα signaling could promote the relocalization of p53 by either 1) enhancing the nuclear export of p53 or 2) by inhibiting the translocation of p53 into the nucleus from the cytoplasm. PKCα-mediated relocalization of p53, in conjunction with other integrin αv-mediated signaling pathways, results in increased melanoma survival.
Sequestration of p53 in the cytoplasm has also been implicated in oxaliplatin-resistant cervical carcinoma cell survival (50), supporting the principle of cytoplasmic sequestration of p53 in the inhibition of apoptosis. However, it is unclear whether the shift in p53 localization in melanoma may be a consequence of increased nuclear export and/or decreased nuclear import/tethering within the cytoplasm.
In conclusion, we have identified PKCα as a downstream effector of integrin αv that promotes a p53 cytoplasmic relocalization and inhibits melanoma cell apoptosis both in vitro and in vivo, resultantly promoting melanoma tumor growth. This pathway is dependent upon cellular growth in a three-dimensional environment and is not apparent under standard two-dimensional conditions, stressing the importance of using as physiological conditions as possible. Although the role for PKCα in this process is apparent, further studies are needed to delineate in detail the signaling cascade that results in p53 cytoplasmic shuttling.
Supplementary Material
Acknowledgments
We thank Dr. Pablo Hernandez-Varas for expert technical assistance with confocal microscopy. Imaging was performed at the Unit for Live Cell Imaging at the Department of Biosciences and Nutrition, Karolinska Institutet, supported by the Knut and Alice Wallenberg foundation, The Swedish Research Council and the Center for Biosciences at Karolinska Institutet.
This work was supported by grants from the Swedish Cancer Society, EU-FP6-Active p53, and the Swedish Research Council (to S. S. and G. S.) and from the Center of Biosciences and the Foundation for Allergy and Cancer in Sweden (to S. S.).

This article contains supplemental Table S1.
- CAM
- chorioallantoic membrane
- pAb
- polyclonal antibody
- qPCR
- quantitative real-time PCR.
REFERENCES
- 1. Parker P. J., Murray-Rust J. (2004) PKC at a glance. J. Cell Sci. 117, 131–132 [DOI] [PubMed] [Google Scholar]
- 2. Nakashima S. (2002) Protein kinase C α (PKCα): Regulation and biological function. J. Biochem. 132, 669–675 [DOI] [PubMed] [Google Scholar]
- 3. Lønne G. K., Cornmark L., Zahirovic I. O., Landberg G., Jirström K., Larsson C. (2010) PKCα expression is a marker for breast cancer aggressiveness. Mol. Cancer 9, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cameron A. J., Procyk K. J., Leitges M., Parker P. J. (2008) PKCα protein but not kinase activity is critical for glioma cell proliferation and survival. Int. J. Cancer 123, 769–779 [DOI] [PubMed] [Google Scholar]
- 5. Koren R., Ben Meir D., Langzam L., Dekel Y., Konichezky M., Baniel J., Livne P. M., Gal R., Sampson S. R. (2004) Expression of protein kinase C isoenzymes in benign hyperplasia and carcinoma of prostate. Oncol. Rep. 11, 321–326 [PubMed] [Google Scholar]
- 6. Lahn M. M., Sundell K. L. (2004) The role of protein kinase C-α (PKC-α) in melanoma. Melanoma Res. 14, 85–89 [DOI] [PubMed] [Google Scholar]
- 7. Byers H. R., Boissel S. J., Tu C., Park H. Y. (2010) RNAi-mediated knockdown of protein kinase C-α inhibits cell migration in MM-RU human metastatic melanoma cell line. Melanoma Res. 20, 171–178 [DOI] [PubMed] [Google Scholar]
- 8. Dennis J. U., Dean N. M., Bennett C. F., Griffith J. W., Lang C. M., Welch D. R. (1998) Human melanoma metastasis is inhibited following ex vivo treatment with an antisense oligonucleotide to protein kinase C-α. Cancer Lett. 128, 65–70 [DOI] [PubMed] [Google Scholar]
- 9. Putnam A. J., Schulz V. V., Freiter E. M., Bill H. M., Miranti C. K. (2009) Src, PKCα, and PKCδ are required for αvβ3 integrin-mediated metastatic melanoma invasion. Cell Commun. Signal 7, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tsubaki M., Matsuoka H., Yamamoto C., Kato C., Ogaki M., Satou T., Itoh T., Kusunoki T., Tanimori Y., Nishida S. (2007) The protein kinase C inhibitor, H7, inhibits tumor cell invasion and metastasis in mouse melanoma via suppression of ERK1/2. Clin. Exp. Metastasis 24, 431–438 [DOI] [PubMed] [Google Scholar]
- 11. Rucci N., DiGiacinto C., Orrù L., Millimaggi D., Baron R., Teti A. (2005) A novel protein kinase Cα-dependent signal to ERK1/2 activated by αVβ3 integrin in osteoclasts and in Chinese hamster ovary (CHO) cells. J. Cell Sci. 118, 3263–3275 [DOI] [PubMed] [Google Scholar]
- 12. Gutcher I., Webb P. R., Anderson N. G. (2003) The isoform-specific regulation of apoptosis by protein kinase C. Cell Mol. Life Sci. 60, 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ruvolo P. P., Deng X., Carr B. K., May W. S. (1998) A functional role for mitochondrial protein kinase Cα in Bcl2 phosphorylation and suppression of apoptosis. J. Biol. Chem. 273, 25436–25442 [DOI] [PubMed] [Google Scholar]
- 14. Vousden K. H., Lu X. (2002) Live or let die: The cell's response to p53. Nat. Rev. Cancer 2, 594–604 [DOI] [PubMed] [Google Scholar]
- 15. Speidel D. (2010) Transcription-independent p53 apoptosis: An alternative route to death. Trends Cell Biol. 20, 14–24 [DOI] [PubMed] [Google Scholar]
- 16. Delphin C., Huang K. P., Scotto C., Chapel A., Vincon M., Chambaz E., Garin J., Baudier J. (1997) The in vitro phosphorylation of p53 by calcium-dependent protein kinase C–characterization of a protein kinase C-binding site on p53. Eur. J. Biochem. 245, 684–692 [DOI] [PubMed] [Google Scholar]
- 17. Youmell M., Park S. J., Basu S., Price B. D. (1998) Regulation of the p53 protein by protein kinase C α and protein kinase C ζ. Biochem. Biophys. Res. Commun. 245, 514–518 [DOI] [PubMed] [Google Scholar]
- 18. Delphin C., Baudier J. (1994) The protein kinase C activator, phorbol ester, cooperates with the wild-type p53 species of Ras-transformed embryo fibroblast growth arrest. J. Biol. Chem. 269, 29579–29587 [PubMed] [Google Scholar]
- 19. Price B. D., Calderwood S. K. (1993) Increased sequence-specific p53-DNA binding activity after DNA damage is attenuated by phorbol esters. Oncogene 8, 3055–3062 [PubMed] [Google Scholar]
- 20. Coutinho I., Pereira G., Leão M., Gonçalves J., Côrte-Real M., Saraiva L. (2009) Differential regulation of p53 function by protein kinase C isoforms revealed by a yeast cell system. FEBS Lett. 583, 3582–3588 [DOI] [PubMed] [Google Scholar]
- 21. Albelda S. M., Mette S. A., Elder D. E., Stewart R., Damjanovich L., Herlyn M., Buck C. A. (1990) Integrin distribution in malignant melanoma: Association of the β3 subunit with tumor progression. Cancer Res. 50, 6757–6764 [PubMed] [Google Scholar]
- 22. Bao W., Strömblad S. (2004) Integrin αv-mediated inactivation of p53 controls a MEK1-dependent melanoma cell survival pathway in three-dimensional collagen. J. Cell Biol. 167, 745–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Felding-Habermann B., Mueller B. M., Romerdahl C. A., Cheresh D. A. (1992) Involvement of integrin αV gene expression in human melanoma tumorigenicity. J. Clin. Invest. 89, 2018–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hsu M. Y., Shih D. T., Meier F. E., Van Belle P., Hsu J. Y., Elder D. E., Buck C. A., Herlyn M. (1998) Adenoviral gene transfer of β3 integrin subunit induces conversion from radial to vertical growth phase in primary human melanoma. Am. J. Pathol. 153, 1435–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Montgomery A. M., Reisfeld R. A., Cheresh D. A. (1994) Integrin αvβ3 rescues melanoma cells from apoptosis in three-dimensional dermal collagen. Proc. Natl. Acad. Sci. U.S.A. 91, 8856–8860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Petitclerc E., Strömblad S., von Schalscha T. L., Mitjans F., Piulats J., Montgomery A. M., Cheresh D. A., Brooks P. C. (1999) Integrin α(v)β3 promotes M21 melanoma growth in human skin by regulating tumor cell survival. Cancer Res. 59, 2724–2730 [PubMed] [Google Scholar]
- 27. Pampaloni F., Reynaud E. G., Stelzer E. H. (2007) The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 8, 839–845 [DOI] [PubMed] [Google Scholar]
- 28. Cukierman E., Pankov R., Stevens D. R., Yamada K. M. (2001) Taking cell-matrix adhesions to the third dimension. Science 294, 1708–1712 [DOI] [PubMed] [Google Scholar]
- 29. Larsen M., Artym V. V., Green J. A., Yamada K. M. (2006) The matrix reorganized: Extracellular matrix remodeling and integrin signaling. Curr. Opin. Cell Biol. 18, 463–471 [DOI] [PubMed] [Google Scholar]
- 30. Wang F., Weaver V. M., Petersen O. W., Larabell C. A., Dedhar S., Briand P., Lupu R., Bissell M. J. (1998) Reciprocal interactions between β1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: A different perspective in epithelial biology. Proc. Natl. Acad. Sci. U.S.A. 95, 14821–14826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weigelt B., Lo A. T., Park C. C., Gray J. W., Bissell M. J. (2010) HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the three-dimensional microenvironment. Breast Cancer Res. Treat. 122, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Satyamoorthy K., Chehab N. H., Waterman M. J., Lien M. C., El-Deiry W. S., Herlyn M., Halazonetis T. D. (2000) Aberrant regulation and function of wild-type p53 in radioresistant melanoma cells. Cell Growth Differ. 11, 467–474 [PubMed] [Google Scholar]
- 33. Luu Y., Li G. (2003) The p53-stabilizing compound, CP-31398, does not enhance chemosensitivity in human melanoma cells. Anticancer Res. 23, 99–105 [PubMed] [Google Scholar]
- 34. Brooks P. C., Montgomery A. M., Rosenfeld M., Reisfeld R. A., Hu T., Klier G., Cheresh D. A. (1994) Integrin αvβ3 antagonists promote tumor regression by inducing apoptosis of angiogenic blood vessels. Cell 79, 1157–1164 [DOI] [PubMed] [Google Scholar]
- 35. Wu C. C., Huang H. C., Juan H. F., Chen S. T. (2004) GeneNetwork: An interactive tool for reconstruction of genetic networks using microarray data. Bioinformatics 20, 3691–3693 [DOI] [PubMed] [Google Scholar]
- 36. Tusher V. G., Tibshirani R., Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Legate K. R., Montañez E., Kudlacek O., Fässler R. (2006) ILK, PINCH, and parvin: The tIPP of integrin signaling. Nat. Rev. Mol. Cell Biol. 7, 20–31 [DOI] [PubMed] [Google Scholar]
- 38. Stommel J. M., Marchenko N. D., Jimenez G. S., Moll U. M., Hope T. J., Wahl G. M. (1999) A leucine-rich nuclear export signal in the p53 tetramerization domain: Regulation of subcellular localization and p53 activity by NES masking. EMBO J. 18, 1660–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Haller D., Mackiewicz M., Gerber S., Beyer D., Kullmann B., Schneider I., Ahmed J. S., Seitzer U. (2010) Cytoplasmic sequestration of p53 promotes survival in leukocytes transformed by Theileria. Oncogene 29, 3079–3086 [DOI] [PubMed] [Google Scholar]
- 40. Liu J., Yu G., Zhao Y., Zhao D., Wang Y., Wang L., Liu J., Li L., Zeng Y., Dang Y., Wang C., Gao G., Long W., Lonard D. M., Qiao S., Tsai M. J., Zhang B., Luo H., Li X. (2010) REGγ modulates p53 activity by regulating its cellular localization. J. Cell Sci. 123, 4076–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Qu L., Huang S., Baltzis D., Rivas-Estilla A. M., Pluquet O., Hatzoglou M., Koumenis C., Taya Y., Yoshimura A., Koromilas A. E. (2004) Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3β. Genes Dev. 18, 261–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lewis J. M., Cheresh D. A., Schwartz M. A. (1996) Protein kinase C regulates αvβ5-dependent cytoskeletal associations and focal adhesion kinase phosphorylation. J. Cell Biol. 134, 1323–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jørgensen K., Skrede M., Cruciani V., Mikalsen S. O., Slipicevic A., Flørenes V. A. (2005) Phorbol ester phorbol 12-myristate 13-acetate promotes anchorage-independent growth and survival of melanomas through MEK-independent activation of ERK1/2. Biochem. Biophys. Res. Commun. 329, 266–274 [DOI] [PubMed] [Google Scholar]
- 44. Jiffar T., Kurinna S., Suck G., Carlson-Bremer D., Ricciardi M. R., Konopleva M., Andreeff M., Ruvolo P. P. (2004) PKCα mediates chemoresistance in acute lymphoblastic leukemia through effects on Bcl2 phosphorylation. Leukemia 18, 505–512 [DOI] [PubMed] [Google Scholar]
- 45. Chernov M. V., Bean L. J., Lerner N., Stark G. R. (2001) Regulation of ubiquitination and degradation of p53 in unstressed cells through C-terminal phosphorylation. J. Biol. Chem. 276, 31819–31824 [DOI] [PubMed] [Google Scholar]
- 46. Zhang Y., Xiong Y. (2001) A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science 292, 1910–1915 [DOI] [PubMed] [Google Scholar]
- 47. Dang C. V., Lee W. M. (1989) Nuclear and nucleolar targeting sequences of c-erb-A, c-myb, N-myc, p53, HSP70, and HIV tat proteins. J. Biol. Chem. 264, 18019–18023 [PubMed] [Google Scholar]
- 48. Liang S. H., Clarke M. F. (1999) A bipartite nuclear localization signal is required for p53 nuclear import regulated by a carboxyl-terminal domain. J. Biol. Chem. 274, 32699–32703 [DOI] [PubMed] [Google Scholar]
- 49. Bao W., Chen M., Zhao X., Kumar R., Spinnler C., Thullberg M., Issaeva N., Selivanova G., Strömblad S. (2011) PRIMA-1Met/APR-246 induces wild-type p53-dependent suppression of malignant melanoma tumor growth in three-dimensional culture and in vivo. Cell Cycle 10, 301–307 [DOI] [PubMed] [Google Scholar]
- 50. Komlodi-Pasztor E., Trostel S., Sackett D., Poruchynsky M., Fojo T. (2009) Impaired p53 binding to importin: A novel mechanism of cytoplasmic sequestration identified in oxaliplatin-resistant cells. Oncogene 28, 3111–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.