Abstract
We present a testable model for the origin of the nucleus, the membrane-bounded organelle that defines eukaryotes. A chimeric cell evolved via symbiogenesis by syntrophic merger between an archaebacterium and a eubacterium. The archaebacterium, a thermoacidophil resembling extant Thermoplasma, generated hydrogen sulfide to protect the eubacterium, a heterotrophic swimmer comparable to Spirochaeta or Hollandina that oxidized sulfide to sulfur. Selection pressure for speed swimming and oxygen avoidance led to an ancient analogue of the extant cosmopolitan bacterial consortium “Thiodendron latens.” By eubacterial-archaebacterial genetic integration, the chimera, an amitochondriate heterotroph, evolved. This “earliest branching protist” that formed by permanent DNA recombination generated the nucleus as a component of the karyomastigont, an intracellular complex that assured genetic continuity of the former symbionts. The karyomastigont organellar system, common in extant amitochondriate protists as well as in presumed mitochondriate ancestors, minimally consists of a single nucleus, a single kinetosome and their protein connector. As predecessor of standard mitosis, the karyomastigont preceded free (unattached) nuclei. The nucleus evolved in karyomastigont ancestors by detachment at least five times (archamoebae, calonymphids, chlorophyte green algae, ciliates, foraminifera). This specific model of syntrophic chimeric fusion can be proved by sequence comparison of functional domains of motility proteins isolated from candidate taxa.
Keywords: Archaeprotists, spirochetes, sulfur syntrophy, Thiodendron, trichomonad
Two Domains, Not Three
All living beings are composed of cells and are unambiguously classifiable into one of two categories: prokaryote (bacteria) or eukaryote (nucleated organisms). Here we outline the origin of the nucleus, the membrane-bounded organelle that defines eukaryotes. The common ancestor of all eukaryotes by genome fusion of two or more different prokaryotes became “chimeras” via symbiogenesis (1). Long term physical association between metabolically dependent consortia bacteria led, by genetic fusion, to this chimera. The chimera originated when an archaebacterium (a thermoacidophil) and a motile eubacterium emerged under selective pressure: oxygen threat and scarcity both of carbon compounds and electron acceptors. The nucleus evolved in the chimera. The earliest descendant of this momentous merger, if alive today, would be recognized as an amitochondriate protist. An advantage of our model includes its simultaneous consistency in the evolutionary scenario across fields of science: cell biology, developmental biology, ecology, genetics, microbiology, molecular evolution, paleontology, protistology. Environmentally plausible habitats and modern taxa are easily comprehensible as legacies of the fusion event. The scheme that generates predictions demonstrable by molecular biology, especially motile protein sequence comparisons (2), provides insight into the structure, physiology, and classification of microorganisms.
Our analysis requires the two- (Bacteria/Eukarya) not the three- (Archaea/Eubacteria/Eukarya) domain system (3). The prokaryote vs. eukaryote that replaced the animal vs. plant dichotomy so far has resisted every challenge. Microbiologist's molecular biology-based threat to the prokaryote vs. eukaryote evolutionary distinction seems idle (4). In a history of contradictory classifications of microorganisms since 1820, Scamardella (5) noted that Woese's entirely nonmorphological system ignores symbioses. But bacterial consortia and protist endosymbioses irreducibly underlie evolutionary transitions from prokaryotes to eukaryotes. Although some prokaryotes [certain Gram-positive bacteria (6)] are intermediate between eubacteria and archaebacteria, no organisms intermediate between prokaryotes and eukaryotes exist. These facts render the 16S rRNA and other nonmorphological taxonomies of Woese and others inadequate. Only all-inclusive taxonomy, based on the work of thousands of investigators over more than 200 years on live organisms (7), suffices for detailed evolutionary reconstruction (4).
When Woese (8) insists “there are actually three, not two, primary phylogenetic groupings of organisms on this planet” and claims that they, the “Archaebacteria” (or, in his term that tries to deny their bacterial nature, the “Archaea”) and the “Eubacteria” are “each no more like the other than they are like eukaryotes,” he denies intracellular motility, including that of the mitotic nucleus. He minimizes these and other cell biological data, sexual life histories including cyclical cell fusion, fossil record correlation (9), and protein-based molecular comparisons (10, 11). The tacit, uninformed assumption of Woese and other molecular biologists that all heredity resides in nuclear genes is patently contradicted by embryological, cytological, and cytoplasmic heredity literature (12). The tubulin-actin motility systems of feeding and sexual cell fusion facilitate frequent viable incorporation of heterologous nucleic acid. Many eukaryotes, but no prokaryotes, regularly ingest entire cells, including, of course, their genomes, in a single phagocytotic event. This invalidates any single measure alone, including ribosomal RNA gene sequences, to represent the evolutionary history of a lineage.
As chimeras, eukaryotes that evolved by integration of more than a single prokaryotic genome (6) differ qualitatively from prokaryotes. Because prokaryotes are not directly comparable to symbiotically generated eukaryotes, we must reject Woese's three-domain interpretation. Yet our model greatly appreciates his archaebacterial-eubacterial distinction: the very first anaerobic eukaryotes derived from both of these prokaryotic lineages. The enzymes of protein synthesis in eukaryotes come primarily from archaebacteria whereas in the motility system (microtubules and their organizing centers), many soluble heat-shock and other proteins originated from eubacteria (9). Here we apply Gupta's idea (from protein sequences) (10) to comparative protist data (13) to show how two kinds of prokaryotes made the first chimeric eukaryote. We reconstruct the fusion event that produced the nucleus.
The Chimera: Archaebacterium/Eubacterium Merger
Study of conserved protein sequences [a far larger data set than that used by Woese et al. (3)] led Gupta (10) to conclude “all eukaryotic cells, including amitochondriate and aplastidic cells received major genetic contributions to the nuclear genome from both an archaebacterium (very probably of the eocyte, i.e., thermoacidophil group and a Gram-negative bacterium … [t]he ancestral eukaryotic cell never directly descended from archaebacteria but instead was a chimera formed by fusion and integration of the genomes of an archaebacerium and a Gram-negative bacterium” (p. 1487). The eubacterium ancestor has yet to be identified; Gupta rejects our spirochete hypothesis. In answer to which microbe provided the eubacterial contribution, he claims: “the sequence data … . suggest that the archaebacteria are polyphyletic and are close relatives of the Gram-positive bacteria” (p. 1485). The archaebacterial sequences, we posit, following Searcy (14), come from a Thermoplasma acidophilum-like thermoacidophilic (eocyte) prokaryote. This archaebacterial ancestor lived in warm, acidic, and sporadically sulfurous waters, where it used either elemental sulfur (generating H2S) or less than 5% oxygen (generating H2O) as terminal electron acceptor. As does its extant descendant, the ancient archaebacterium survived acid-hydrolysis environmental conditions by nucleosome-style histone-like protein coating of its DNA (14) and actin-like stress-protein synthesis (15). The wall-less archaebacterium was remarkably pleiomorphic; it tended into tight physical association with globules of elemental sulfur by use of its rudimentary cytoskeletal system (16). The second member of the consortium, an obligate anaerobe, required for growth the highly reduced conditions provided by sulfur and sulfate reduction to hydrogen sulfide. Degradation of carbohydrate (e.g., starch, sugars such as cellobiose) and oxidation of the sulfide to elemental sulfur by the eubacterium generated carbon-rich fermentation products and electron acceptors for the archaebacterium. When swimming eubacteria attached to the archaebacterium, the likelihood that the consortium efficiently reached its carbon sources was enhanced. This hypothetical consortium, before the integration to form a chimera (Fig. 1), differs little from the widespread and geochemically important “Thiodendron” (17, 18).
Figure 1.
Origin of the chimeric eukaryote with karyomastigonts from a motile sulfur-bacteria consortium.
The “Thiodendron” Stage
The “Thiodendron” stage refers to an extant bacterial consortium that models our idea of an archaebacteria-eubacteria sulfur syntrophic motility symbiosis. The partners in our view merged to become the chimeric predecessor to archaeprotists. The membrane-bounded nucleus, by hypothesis, is the morphological manifestation of the chimera genetic system that evolved from a Thiodendron-type consortium. Each phenomenon we suggest, from free-living bacteria to integrated association, enjoys extant natural analogues.
Study of marine microbial mats revealed relevant bacterial consortia in more than six geographically separate locations. Isolations from Staraya Russa mineral spring 8, mineral spring Serebryani, Lake Nizhnee, mud-baths; littoral zone at the White Sea strait near Veliky Island, Gulf of Nilma; Pacific Ocean hydrothermal habitats at the Kurile Islands and Kraternaya Bay; Matupi Harbor Bay, Papua New Guinea, etc. (17) all yielded “Thiodendron latens” or very similar bacteria. Samples were taken from just below oxygen-sulfide interface in anoxic waters (17, 18). Laboratory work showed it necessary to abolish the genus Thiodendron because it is a sulfur syntrophy. A stable ectosymbiotic association of two bacterial types grows as an anaerobic consortium between 4 and 32°C at marine pH values and salinities. Starch, cellobiose, and other carbohydrates (not cellulose, amino acids, organic acids, or alcohol) supplemented by heterotrophic CO2 fixation provide it carbon. Thiodendron appears as bluish-white spherical gelatinous colonies, concentric in structure within a slimy matrix produced by the consortium bacteria. The dominant partner invariably is a distinctive strain of pleiomorphic spirochetes: they vary from the typical walled Spirochaeta 1:2:1 morphology to large membranous spheres, sulfur-studded threads, gliding or nonmotile cells of variable width (0.09–0.45 μm) and lengths to millimeters. The other partner, a small, morphologically stable vibrioid, Desulfobacter sp., requires organic carbon, primarily acetate, from spirochetal carbohydrate degradation. The spirochetal Escherichia coli-like formic acid fermentation generates energy and food. Desulfobacter sp. cells that reduce both sulfate and sulfur to sulfide are always present in the natural consortium but in far less abundance than the spirochetes. We envision the Thiodendron consortium of “free-living spirochetes in geochemical sulfur cycle” (ref. 18, p. 456) and spirochete motility symbioses (19) as preadaptations for chimera evolution. Thiodendron differs from the archaebacterium-eubacterium association we hypothesize; the marine Desulfobacter would have been replaced with a pleiomorphic wall-less, sulfuric-acid tolerant soil Thermoplasma-like archaebacterium. New thermoplasmas are under study. We predict strains that participate in spirochete consortia in less saline, more acidic, and higher temperature sulfurous habitats than Thiodendron will be found.
When “pure cultures” that survived low oxygen were first described [by B. V. Perfil'ev in 1969, in Russian (see refs. 17 and 18] a complex life history of vibrioids, spheroids, threads and helices was attributed to “Thiodendron latens”. We now know these morphologies are artifacts of environmental selection pressure: Dubinina et al. (ref. 17, p. 435), reported that “the pattern of bacterial growth changes drastically when the redox potential of the medium is brought down by addition of 500 mg/l of sodium sulfide.” The differential growth of the two tightly associated partners in the consortium imitates the purported Thiodendron bacterial developmental patterns. The syntrophy is maintained by lowering the level of oxygen enough for spirochete growth. The processes of sulfur oxidation-reduction and oxygen removal from oxygen-sensitive enzymes, we suggest, were internalized by the chimera and retained by their protist descendants as developmental cues.
Metabolic interaction, in particular syntrophy under anoxia, retained the integrated prokaryotes as emphasized by Martin and Müller (20). However, we reject their concept, for which no evidence exists, that the archaebacterial partner was a methanogen. Our sulfur syntrophy idea, by contrast, is bolstered by observations that hydrogen sulfide is still generated in amitochondriate, anucleate eukaryotic cells (mammalian erythrocytes) (21).
T. acidophilum in pure culture attach to suspended elemental sulfur. When sulfur is available, they generate hydrogen sulfide (16). Although severely hindered by ambient oxygen, they are microaerophilic in the presence of small quantities (<5%) of oxygen. The Thermoplasma partner thus would be expected to produce sulfide and scrub small quantities of oxygen to maintain low redox potential in the spirochete association. The syntrophic predecessors to the chimera is metabolically analogous to Thiodendron where Desulfobacter reduces sulfur and sulfate producing sulfide at levels that permit the spirochetes to grow. We simply suggest the replacement of the marine sulfidogen with Thermoplasma. In both the theoretical and actual case, the spirochetes would supply oxidized sulfur as terminal electron acceptor to the sulfidogen.
The DNA of the Thermoplasma-like archaebacterium permanently recombined with that of the eubacterial swimmer. A precedent exists for our suggestion that membrane hypertrophies around DNA to form a stable vesicle in some prokaryotes: the membrane-bounded nucleoid in the eubacterium Gemmata obscuriglobus (22). The joint Thermoplasma-like archaebacterial DNA package that began as the consortium nucleoid became the chimera's nucleus.
The two unlike prokaryotes together produced a persistent protein exudate package. This step in the origin of the nucleus—the genetic integration of the two-membered consortium to form the chimera—is traceable by its morphological legacy: the karyomastigont. The attached swimmer partner, precursor to mitotic microtubule system, belonged to genera like the nearly ubiquitous consortium-former Spirochaeta or the cytoplasmic tubule-maker Hollandina (19). The swimmer's attachment structures hypertrophied as typically they do in extant motility symbioses (19). The archaebacterium-eubacterium swimmer attachment system became the karyomastigont. The proteinaceous karyomastigont that united partner DNA in a membrane-bounded, jointly produced package, assured stability to the chimera. All of the DNA of the former prokaryotes recombined inside the membrane to become nuclear DNA while the protein-based motility system of the eubacterium, from the moment of fusion until the present, segregated the chimeric DNA. During the lower Proterozoic eon (2,500–1,800 million years ago), many interactions inside the chimera generated protists in which mitosis and eventually meiotic sexuality evolved. The key concept here is that the karyomastigont, retained by amitochondriate protists and later by their mitochondriate descendants, is the morphological manifestation of the original archaebacterial-eubacterial fused genetic system. Free (unattached) nuclei evolved many times by disassociation from the rest of the karyomastigont. The karyomastigont, therefore, was the first microtubule-organizing center.
Karyomastigonts Preceded Nuclei
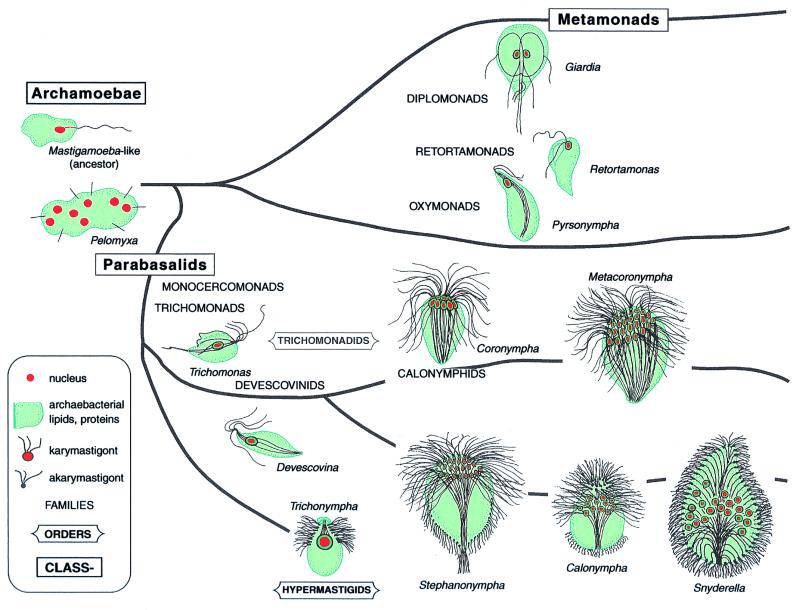
The term “karyomastigont” was coined by Janicki (23) to refer to a conspicuous organellar system he observed in certain protists: the mastigont (“cell whip,” eukaryotic flagellum, or undulipodium, the [9 (2) + (2)] microtubular axoneme underlain by its [9 (3) + 0)] kinetosome) attached by a “nuclear connector” or “rhizoplast” to a nucleus. The need for a term came from Janicki's work on highly motile trichomonad symbionts in the intestines of termites where karyomastigonts dominate the cells. When kinetosomes, nuclear connector, and other components were present but the nucleus was absent from its predictable position, Janicki called the organelle system an “akaryomastigont.” In the Calonymphidae, one family of entirely multinucleate trichomonads, numerous karyomastigonts, and akaryomastigonts are simultaneously present in the same cell (e.g., Calonympha grassii) (24).
The karyomastigont, an ancestral feature of eukaryotes, is present in “early branching protists” (25–27). Archaeprotists, a large inclusive taxon (phylum of Kingdom Protoctista) (7) are heterotrophic unicells that inhabit anoxic environments. All lack mitochondria. At least 28 families are placed in the phylum Archaeprotista. Examples include archaemoebae (Pelomyxa and Mastigamoeba), metamonads (Retortamonas), diplomonads (Giardia), oxymonads (Pyrsonympha), and the two orders of Parabasalia: Trichomonadida [Devescovina, Mixotricha, Monocercomonas, Trichomonas, and calonymphids (Coronympha, Snyderella)] and Hypermastigida (Lophomonas, Staurojoenina, and Trichonympha). These cells either bear karyomastigonts or derive by differential organelle reproduction (simple morphological steps) from those that do (Table 1). When, during evolution of these protists, nuclei were severed from their karyomastigonts, akaryomastigonts were generated (31). Nuclei, unattached, at least temporarily, to undulipodia were freed to proliferate and occupy central positions in cells. Undulipodia, also freed to proliferate, generated larger, faster-swimming cells in the same evolutionary step.
Table 1.
Karyomastigont distribution in unicellular protoctists
|
Archaeprotista*
| |||
|---|---|---|---|
| Class | Karyomastigont | Kinetosome | Nucleus |
| Pelobiontids | +† | − | + |
| Metamonads | + | + | +/− |
| Parabasalids | + | + | + |
| Trichomonads | + | + | +/− |
| Hypermastigids | − | + | − |
| Chlorophyta | |||
| Genus | Karyomastigont | Kinetosome | Nucleus |
| Chlamydomonas | + | + | − |
| Chlorella | − | − | − |
| Acetabularia | + | + | + |
| Ciliophora | |||
| Subphyla | Karyomastigont | Kinetosome | Nucleus |
| Postciliodesmatophora | − | + | + |
| Rabdophora | − | + | + |
| Cyrtophora | − | + | + |
| Discomitochondria | |||
| Class | Karyomastigont | Kinetosome | Nucleus |
| Amoebomastigotes | + | − | +/− |
| Kinetoplastids | − | − | − |
| Euglenids | −/? | − | − |
| Pseudociliates | − | + | − |
| Granuloreticulosa | |||
| Class | Karyomastigont | Kinetosome | Nucleus |
| Reticulomixids | −/? | − | + |
| Foraminiferans | + | − | + |
| Hemimastigophora | |||
| Genus | Karyomastigont | Kinetosome | Nucleus |
| Stereonema | − | + | +/− |
| Spironema | − | + | − |
| Hemimastix | − | + | − |
| Zoomastigota | |||
| Class | Karyomastigont | Kinetosome | Nucleus |
| Jakobids | ? | − | − |
| Bicosoecids | +/? | + | − |
| Proteromonads | + | − | − |
| Opalinids | − | + | + |
| Choanomastigotes | + | − | − |
Bold entries are protoctist phyla. All species of Archaeprotists lack mitochondria. “Karyomastigont,” “kinetosome,” and “nucleus,” refer to relative proliferation of these organelles. Members of the phylum Archaeprotista group into one of three classes: Pelobiontid giant amoebae; Metamonads, which include three subclasses: Diplomonads (Giardia), Retortamonads (Retortamonas), and Oxymonads (such as Pyrsonympha and Saccinobaculus); and Parabasalia. The Class Parabasalia unites trichomonads, devescovinids, calonymphids, and hypermastigotes such as Trichonympha. The phylum Discomitochondria includes amoebomastigotes, kinetoplastids (Trypanosoma), euglenids, and pseudociliates (Stephanopogon). The Hemimastigophora comprise a new southern-hemisphere phylum of free-living mitochondriate protists (28). Hemimastigophorans probably evolved from members of the kinetoplastid-euglenid taxon (29). If so, they represent a seventh example of release of the nucleus from the karyomastigont and subsequent kinetosome proliferation. The phylum Granuloreticulosa includes the shelled (Class Foraminifera) and unshelled (Class Reticulomyxa) foraminiferans. The phylum Zoomastigota includes five classes of single-celled, free-living and symbiotrophic mitochondriate protists: Jakobids, Bicosoecids, Proteromonads, Opalinids, and Choanomastigotes. Details of the biology are in the work by Margulis et al. (30). A current phylogeny is depicted in Fig. 2.
Structure known but not demonstrated for all species at the electron microscopic level.
The karyomastigont is the conspicuous central cytoskeleton in basal members of virtually all archaeprotist lineages [three classes: Archamoeba, Metamonads, and Parabasalia (32)] (Fig. 2). In trichomonads, the karyomastigont, which includes a parabasal body (Golgi complex), coordinates the placement of hydrogenosomes (membrane-bounded bacterial-sized cell inclusions that generate hydrogen). The karyomastigont reproduces as a unit structure. Typically, four attached kinetosomes with rolled sheets of microtubules (the axostyle and its extension the pelta) reproduce as their morphological relationships are retained. Kinetosomes reproduce first, the nucleus divides, and the two groups of kinetosomes separate at the poles of a thin microtubule spindle called the paradesmose. Kinetosomes and associated structures are partitioned to one of the two new karyomastigonts. The other produces components it lacks such as the Golgi complex and axostyle.
Figure 2.
Biological phylogeny of chimeric eukaryotes taken to be primitively amitochondriate.
Nuclear α-proteobacterial genes were interpreted to have originated from lost or degenerate mitochondria in at least two archaeprotist species [Giardia lamblia (33); Trichomonas vaginalis (34, 35)] and in a microsporidian (36). Hydrogenosomes, at least some types, share common origin with mitochondria. In the hydrogen hypothesis (20), hydrogenosomes are claimed to be the source of eubacterial genes in amitochondriates. That mitochondria were never acquired in the ancestors we consider more likely than that they were lost in every species of these anaerobic protists. Eubacterial genes in the nucleus that are not from the original spirochete probably were acquired in amitochondriate protists from proteobacterial symbionts other than those of the mitochondrial lineage. Gram-negative bacteria, some of which may be related to ancestors of hydrogenosomes, are rampant as epibionts, endobionts, and even endonuclear symbionts—for example, in Caduceia versatilis (37).
Karyomastigonts freed (detached from) nuclei independently in many lineages both before and after the acquisition of mitochondria. Calonymphid ancestors of Snyderella released free nuclei before the mitochondrial symbiosis (13), and Chlamydomonas-like ancestors of other chlorophytes such as Acetabularia released the nuclei after the lineage was fully aerobic (38). In trophic forms of protists that lack mastigote stages, the karyomastigont is generally absent. An exception is Histomonas, an amoeboid trichomonad cell that lacks an axoneme but bears enough of the remnant karyomastigont structure to permit its classification with parabasalids rather than with rhizopod amoebae (39). This organellar system appears in the zoospores, motile trophic forms, or sperm of many organisms, suggesting the relative ease of karyomastigont development. The karyomastigont, apparently in some cells, is easily lost, suppressed, and regained. In many taxa of multinucleate or multicellular protists (foraminifera, green algae) and even in plants, the karyomastigont persists only in the zoospores or gametes.
In yeast, nematode, insect, and mammalian cells, nonkaryomastigont microtubule-organizing centers are “required to position nuclei at specific locations in the cytoplasm” (40). The link between the microtubule organizing center and the nuclei “is mysterious” (40). To us, the link is an evolutionary legacy, a remnant of the original archaebacterial-eubacterial connector. The modern organelles (i.e., centriole-kinetosomes, untethered nuclei, Golgi, and axostyles) derive from what first ensured genetic continuity of the chimera's components: the karyomastigont, a structure that would have been much more conspicuous to Proterozoic investigators than to us.
Acknowledgments
We thank our colleagues Ray Bradley, Michael Chapman, Floyd Craft, Kathryn Delisle (for figures), Ugo d'Ambrosio, Donna Reppard, Dennis Searcy, and Andrew Wier. We acknowledge research assistance from the University of Massachusetts Graduate School via Linda Slakey, Dean of Natural Science and Mathematics, from the Richard Lounsbery Foundation, and from the American Museum of Natural History Department of Invertebrates (New York). Our research is supported by National Aeronautics and Space Administration Space Sciences and Comision Interministerial de Ciencia y Tecnologia Project No. AMB98-0338 (to R.G.).
Footnotes
This paper was presented at the National Academy of Sciences colloquium “Variation and Evolution in Plants and Microorganisms: Toward a New Synthesis 50 Years After Stebbins,” held January 27–29, 2000, at the Arnold and Mabel Beckman Center in Irvine, CA.
References
- 1.Golding G B, Gupta R S. Mol Biol Evol. 1995;12:1–6. doi: 10.1093/oxfordjournals.molbev.a040178. [DOI] [PubMed] [Google Scholar]
- 2.Chapman, M., Dolan, M. F. & Margulis, L. (2000) Q. Rev. Biol., in press. [DOI] [PubMed]
- 3.Woese C R, Kandler O, Wheelis M L. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayr E. Proc Natl Acad Sci USA. 1998;95:9720–9723. doi: 10.1073/pnas.95.17.9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scamardella J M. Int Microbiol. 1999;2:207–216. [PubMed] [Google Scholar]
- 6.Gupta R S. Mol Microbiol. 1998;29:695–708. doi: 10.1046/j.1365-2958.1998.00978.x. [DOI] [PubMed] [Google Scholar]
- 7.Margulis L, Schwartz K V. Five Kingdoms: An Illustrated Guide to the Phyla of Life on Earth. New York: Freeman; 1998. [Google Scholar]
- 8.Woese C R. Proc Natl Acad Sci USA. 1998;95:11043–11046. doi: 10.1073/pnas.95.19.11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Margulis L. Proc Natl Acad Sci USA. 1996;93:1071–1076. doi: 10.1073/pnas.93.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta R S. Microbiol Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta R S. Theor Popul Biol. 1998;54:91–104. doi: 10.1006/tpbi.1998.1376. [DOI] [PubMed] [Google Scholar]
- 12.Sapp J. Hist Philos Life Sci. 1999;20:3–38. [PubMed] [Google Scholar]
- 13.Dolan M F, d'Ambrosio U, Wier A, Margulis L. Acta Protozool. 2000;39:135–141. [Google Scholar]
- 14.Searcy D G. In: The Origin and Evolution of the Cell. Hartman H, Matsuno K, editors. Singapore: World Scientific; 1992. pp. 47–78. [Google Scholar]
- 15.Searcy D G, Delange R J. Biochim Biophys Acta. 1980;609:197–200. doi: 10.1016/0005-2787(80)90212-9. [DOI] [PubMed] [Google Scholar]
- 16.Searcy D, Hixon W G. BioSystems. 1994;10:19–28. [Google Scholar]
- 17.Dubinina G A, Leshcheva N V, Grabovich M Y. Microbiology. 1993;62:432–444. [Google Scholar]
- 18.Dubinina G A, Grabovich M Y, Lesheva N V. Microbiology. 1993;62:450–456. [Google Scholar]
- 19.Margulis L. Symbiosis in Cell Evolution. New York: Freeman; 1993. [Google Scholar]
- 20.Martin W, Müller M. Nature (London) 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 21.Searcy D, Lee S H. J Exp Zool. 1998;282:310–322. doi: 10.1002/(sici)1097-010x(19981015)282:3<310::aid-jez4>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 22.Fuerst J A, Webb R I. Proc Natl Acad Sci USA. 1991;88:8184–8188. doi: 10.1073/pnas.88.18.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janicki C. Z Wiss Zool. 1915;112:573–691. [Google Scholar]
- 24.Kirby H, Margulis L. Symbiosis. 1994;16:7–63. [PubMed] [Google Scholar]
- 25.Dacks J B, Redfield R. J Eukaryotic Microbiol. 1998;45:445–447. doi: 10.1111/j.1550-7408.1998.tb05097.x. [DOI] [PubMed] [Google Scholar]
- 26.Delgado-Viscogliosi P, Viscogliosi E, Gerbod D, Juldo J, Sogin M L, Edgcomb V. J Eukaryotic Microbiol. 2000;47:70–75. doi: 10.1111/j.1550-7408.2000.tb00013.x. [DOI] [PubMed] [Google Scholar]
- 27.Edgcomb V, Viscogliosi E, Simpson A G B, Delgado-Viscogliosi P, Roger A J, Sogin M L. Protist. 1998;149:359–366. doi: 10.1016/S1434-4610(98)70042-2. [DOI] [PubMed] [Google Scholar]
- 28.Foissner W, Blatterer H, Foissner I. Eur J Protistol. 1988;23:361–383. doi: 10.1016/S0932-4739(88)80027-0. [DOI] [PubMed] [Google Scholar]
- 29.Foissner W, Foissner I. J Eukaryotic Microbiol. 1993;40:422–438. [Google Scholar]
- 30.Margulis L, McKhann H I, Olendzenski L, editors. Illustrated Glossary of the Protoctista. Sudbury, MA: Jones & Bartlett; 1993. [Google Scholar]
- 31.Kirby H. Rev Soc Mex Hist Nat. 1949;10:57–79. [Google Scholar]
- 32.Brugerolle G. Protoplasma. 1991;164:70–90. [Google Scholar]
- 33.Roger A J, Srard S G, Tovar J, Clark C G, Smith M W, Gillin F D, Sogin M L. Proc Natl Acad Sci USA. 1998;95:229–234. doi: 10.1073/pnas.95.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roger A J, Clark C G, Doolittle W M. Proc Natl Acad Sci USA. 1996;93:14618–14622. doi: 10.1073/pnas.93.25.14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Germot A, Philippe H, Le Guyader H. Proc Natl Acad Sci USA. 1996;93:14614–14617. doi: 10.1073/pnas.93.25.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sogin M L. Curr Opin Gen Dev. 1997;7:792–799. doi: 10.1016/s0959-437x(97)80042-1. [DOI] [PubMed] [Google Scholar]
- 37.d'Ambrosio U, Dolan M, Wier A, Margulis L. Eur J Protistol. 1999;35:327–337. doi: 10.1016/S0932-4739(99)80011-X. [DOI] [PubMed] [Google Scholar]
- 38.Hall J, Luck D J L. Proc Natl Acad Sci USA. 1995;92:5129–5133. doi: 10.1073/pnas.92.11.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dyer B. In: Handbook of Protoctista. Margulis L, Corliss J O, Melkonian M, Chapman D J, editors. Sudbury, MA: Jones & Bartlett; 1990. pp. 252–258. [Google Scholar]
- 40.Raff J W. Curr Biol. 1999;9:R708–R710. doi: 10.1016/s0960-9822(99)80446-1. [DOI] [PubMed] [Google Scholar]