Abstract
The genetic links between p53 and metabolic processes such as oxidative phosphorylation are being studied with increasing interest as cellular metabolism appears to play an important role in tumorigenesis. This review focuses on how p53 regulation of various metabolic genes may influence redox homeostasis as the genome is constantly susceptible to oxidative damage, a consequence of living in an aerobic environment. As p53-like genetic sequences are also found in life forms that may not necessarily benefit from tumor suppression, an evolutionary introduction is given in an attempt to understand why p53 might regulate a basic cellular activity such as metabolism. The presented epidemiologic and experimental data suggest that one reason may be for the homeostatic regulation of oxygen, the essential substrate for reactive oxygen species (ROS) generation.
Keywords: p53, oxygen, oxidative stress, redox, antioxidant, metabolism, cancer, mitochondria, altitude
Introduction
The challenges of studying the complex history of atmospheric molecular oxygen faced by geologists appear to extend to cancer biologists [1]. Oxygen has been shown both to promote and inhibit tumorigenesis, depending on various factors including its level and the experimental model [2–6]. Even the basic question of whether oxidative stress, fueled by molecular oxygen, is causally linked to aging and tumorigenesis is debated [7]. However, studying a fundamental principle such as that governing the evolution of aerobic life may provide useful lessons for advancing our understanding of cancer biology.
p53 is commonly referred to as one of the most important tumor suppressor genes and the “guardian of the genome” [8]. However, p53 gene-like sequences are also found in unicellular forms of life that would not be expected to benefit from its sophisticated tumor surveillance function [9]. Thus, it has been proposed that p53 may have provided basic adaptive functions for cell survival prior to its adoption for tumor suppression. One such primordial connection may be that between p53 and numerous metabolic functions including mitochondrial respiration [10–13]. Although the growing number of disparate cellular processes regulated by p53 may appear to be of unclear significance for its tumor suppressor function [14], examining them from an evolutionary perspective may provide important lessons about tumorigenesis.
With the rapid increase in atmospheric oxygen over the past half billion years, life is thought to have evolved from a relatively anoxic to an oxygen-rich environment [15]. Protection from oxygen toxicity, or oxidative stress, has thus been proposed as a driving evolutionary force underlying the symbiotic incorporation of the oxygen consuming purple bacteria that were the progenitors of mitochondria [16]. Notably, the mitochondrion is generally regarded as the major source of reactive oxygen species (ROS), but a critical review of the supporting data is not entirely convincing of this concept [17]. In fact, cells with defective respiration due to the disruption of a critical mitochondrial gene regulated by p53 actually display increased oxidative stress and genomic DNA damage [18]. If p53 regulation of metabolism represents an adaptive function, it could also be speculated that the other p53 family members, p63 and p73, may regulate some aspect of metabolism. Despite the well characterized role of p63 and p73 in apoptosis and development, their direct effect on metabolic pathways remains to be determined [19].
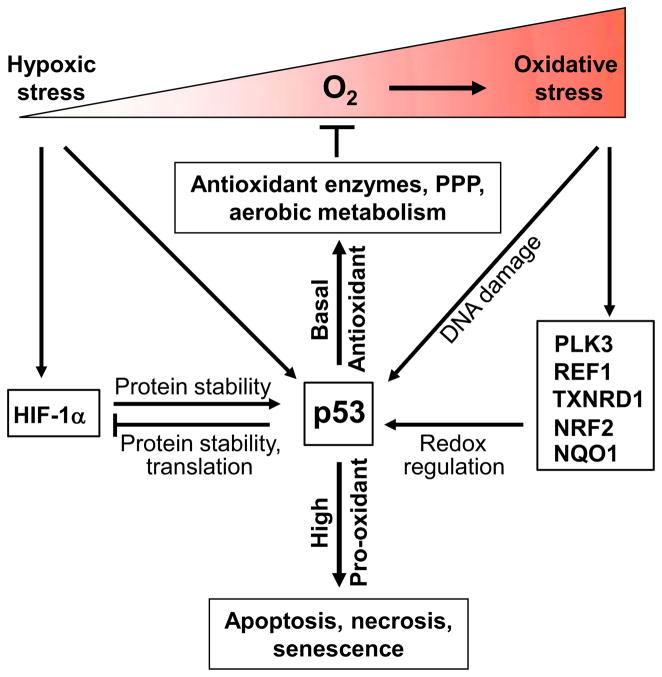
Based on the following review of epidemiologic and basic experimental data, the possibility arises that regulation of aerobic metabolism by p53 may contribute to oxygen and redox homeostasis for preventing oxidative DNA damage and maintaining genomic stability. p53 has been shown to have both pro-oxidant and antioxidant functions depending on its level, activity and context of induction; however, both activities are directed at tumor suppression [10, 13, 20, 21]. The antioxidant function of p53 at basal levels may protect against genomic DNA damage while the pro-oxidant effect at high p53 levels may induce apoptosis or senescence to eliminate cells with irreparable genomic DNA damage (Fig. 2). The pro-oxidant effects can be through both direct and indirect mechanisms as briefly summarized in this review. Because there is already extensive literature supporting the pro-oxidant function of p53, this review will mainly focus on the role of basal level p53-regulated metabolic pathways in redox homeostasis. A major objective of this review is to propose that p53 promotion of aerobic metabolism reduces oxidative stress which in turn may contribute to preventing DNA damage and genomic instability.
Fig. 2.
Redox regulation of p53 and its dual role in cellular redox homeostasis. Oxygen serves as the essential substrate for reactive oxygen species generation and oxidative stress. Varying intracellular oxygen availability can regulate p53 activity and protein level through a number of different mechanisms including oxidative DNA damage, various redox regulatory genes, severe hypoxic stress, and HIF-1α. Basal levels of p53 have antioxidant function that prevents oxidative damage while high levels of p53 are pro-oxidant and cause apoptosis, necrosis or senescence to eliminate cells with irreversible DNA damage.
Oxygen toxicity and tumorigenesis
The antioxidant mechanisms used by cells for protection from ROS have been extensively delineated, but less is known about how cells may actively decrease exposure to oxygen. Even in bioenergetically demanding tissues such as the heart, molecular oxygen has previously been thought to be non-limiting for mitochondrial respiration under normal physiologic conditions [22]. However, direct measurements show that oxygen concentrations vary significantly among tissue types suggesting that the levels of oxygen are dependent on its local delivery and utilization [23]. A recent paper measuring the oxidation state of mitochondria has revealed a significant oxygen concentration gradient from the site of delivery (capillary) to the site of utilization (core of skeletal muscle) [24]. As might be predicted from such an observation, it has also been shown that increased mitochondrial biogenesis decreases oxygen concentration in skeletal muscle, presumably by increasing its consumption, and supports the concept that modulation of mitochondrial activity has the potential to affect intracellular oxygen homeostasis [25].
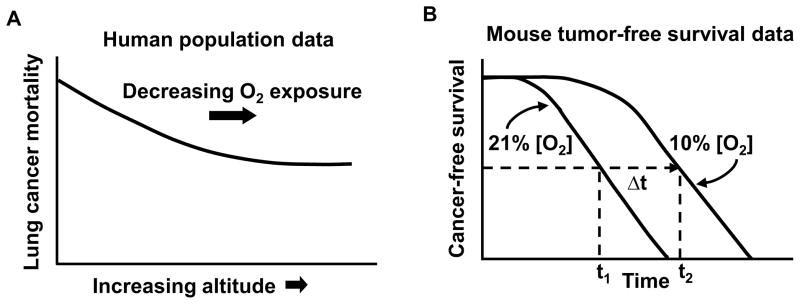
While on one hand oxygen is necessary for cellular energy production by oxidative phosphorylation (OXPHOS), it can also serve as the substrate for the generation of ROS that causes oxidative DNA damage. In fact, oxygen has been shown to be a mutagen in bacteria and proposed to be toxic to humans [15, 26]. A large epidemiologic study supports the concept that oxidative stress, as followed by exposure to ambient oxygen, may indeed be associated with tumorigenesis. Using data from 80 cities in the United States, the age-adjusted cancer mortality rates for a number of common cancers, including lung cancer, showed a significant negative relationship with increasing altitude even after adjusting for background radiation (Fig. 1A) [27]. Therefore, it has been proposed that changes in exposure to oxygen may be an important factor contributing to the striking inverse relationship between altitude and age-adjusted cancer mortality.
Fig. 1.
Relationship between cancer and oxygen exposure. A) A schematic representation of the inverse relationship between age-adjusted male lung cancer mortality and altitude (oxygen exposure). Figure adapted from [27] with publisher’s permission. B) A schematic representation of the increase in cancer-free survival of p53−/− mice by chronic exposure to 10% oxygen versus 21% (room air) (difference in median cancer-free survival time, ~40–50%) [6].
The epidemiologic observation that oxygen may be pro-tumorigenic has recently been supported by experimental data. Chronically lowering the ambient oxygen exposure of p53−/− mice, a well established model of accelerated de novo tumorigenesis, dramatically increased cancer-free survival (Fig. 1B) [18]. Lowering the exposure to oxygen was associated with an increase in the antioxidant capacity of blood and a concomitant decrease in oxidative DNA damage [6]. Furthermore, in the APCMin/+ mouse model of intestinal polyposis, reduced oxygen exposure decreased the appearance of polyps which serve as a marker of genomic instability at the wild-type APC gene locus [6, 28]. It is notable that most of the polyps in the APCMin/+ mouse model appear in the small intestine which is known to have a higher intraluminal oxygen tension compared to the large intestine [29, 30]. A recent publication has provided genetic evidence that increased mitochondrial ROS caused by mitochondrial transcription factor A (TFAM) haploinsufficiency is associated with increased polyp number and growth in the small intestine of APCMin/+ mice [31]. Using human colon cancer cells, the genetic disruption of mitochondrial respiration altered intracellular oxygen homeostasis and markedly increased oxidative DNA damage which could be prevented by decreasing ambient oxygen [18]. Taken together, these data suggest the critical role that the mitochondria and oxygen homeostasis may play in tumorigenesis. We speculate that although most of the DNA damage caused by oxygen can be repaired by response proteins such as p53, they are unlikely to prevent all of the cumulative mutations over a life-time that manifest as the inverse relationship between cancer incidence and altitude (oxygen exposure).
p53 and mitochondria are oxygen-responsive
It is well established that mammalian cells utilize hypoxia inducible factors (HIF genes) to sense and respond to alterations in cellular oxygen availability. Mitochondrial respiration has the potential to increase HIF-1α protein levels by at least two different mechanisms: 1) relative hypoxia caused by the consumption and redistribution of intracellular oxygen [32, 33]; and/or 2) ROS production associated with respiratory electron transfer under hypoxic conditions [34, 35]. Potentially as feedback under conditions of limiting oxygen, HIF-1α can inhibit respiration by transactivating the PDK gene and can decrease mitochondrial biogenesis by repressing the expression of C-MYC, an activator of PGC-1β transcription [36–38]. In contrast, increased oxygen exposure may stabilize p53 through multiple mechanisms and promote mitochondrial biogenesis. Among the possible scenarios, molecular oxygen could regulate p53 via redox sensitive proteins as discussed in the next section or it could serve as a substrate to increase ROS levels which damage DNA or activate signaling enzymes such as polo-like kinase 3 (PLK3) with resultant N-terminal phosphorylation and stabilization of p53 (Fig. 2) [18, 39]. As evidence of interaction between these two oxygen sensitive transcription factors, p53 has been shown to decrease HIF-1α level by inhibiting its translation through p53-induced microRNA-107 or by promoting its degradation by MDM2-mediated ubiquitination [40, 41]. In parallel, HIF-1α may prevent p53 degradation by inhibiting MDM2-mediated p53 ubiquitination under severe stress such as anoxia [42]. Such regulatory relationships between p53 and HIF-1α as well as other HIF members have been extensively studied and are reviewed in greater detail elsewhere [43–45].
Redox regulation of p53
Although p53 stability and activity are regulated by a host of different posttranslational modifications [46], the primary structure of p53 confers redox sensitivity to its DNA binding and transactivating properties [47, 48]. The hydrophobic DNA binding domain of p53 has 10 cysteine amino acid residues, the majority of which are also present in the two well-studied p53 isoforms delta133p53 and delta40p53, both associated with oxidative stress and aging [48]. Cysteine residues 176, 238 and 242 of p53 are known to coordinate zinc and play a critical role in its DNA binding and transactivating activities [49]. Cysteine 124, 141 and 182 are sites of glutathionylation that may modulate p53 DNA binding and tetramerization [50, 51]. The formation of intramolecular disulfide bonds between cysteine pairs 275/277 and 135/141 both modulate DNA binding of p53 [52, 53]. In addition, ROS has been shown to induce p53 phosphorylation and activation [54], while reactive nitrogen species (RNS) can cause p53 nitration at critical tyrosine residues to disrupt its interaction with DNA [55, 56]. The specific redox modifications of p53 and its isoforms are extensive and have been expertly reviewed elsewhere [48].
Besides its structural modifications by cellular redox state, p53 also interacts with regulatory redox proteins (Fig. 2). As an enzyme that can both repair DNA and regulate the redox state of proteins, apurinic endonuclease/redox factor 1 (APE/REF1, APEX1) directly interacts with p53 to stimulate its DNA binding and transcriptional activity [57, 58]. The redox-dependent mechanism by which REF1 increases basal p53 activity involves the thioredoxin system without the N-terminal phosphorylation of p53 associated with DNA damage [59]. It has been shown that depleting thioredoxin reductase 1 (TXNRD1) increases the DNA binding capacity of p53 by sequestering oxidized thioredoxin in the cytoplasm and preventing its oxidation of p53 in the nucleus. NAD(P)H:quinoneoxidoreductase-1 (NQO1), another redox enzyme that is responsive to oxidative stress through nuclear factor erythroid 2-like 2 (NRF2, NFE2L2), physically interacts with p53 and inhibits its degradation [60]. The growing number of redox factors interacting with and regulating p53 points to an important of p53 in redox homeostasis.
p53 can regulate redox homeostasis through the mitochondrion
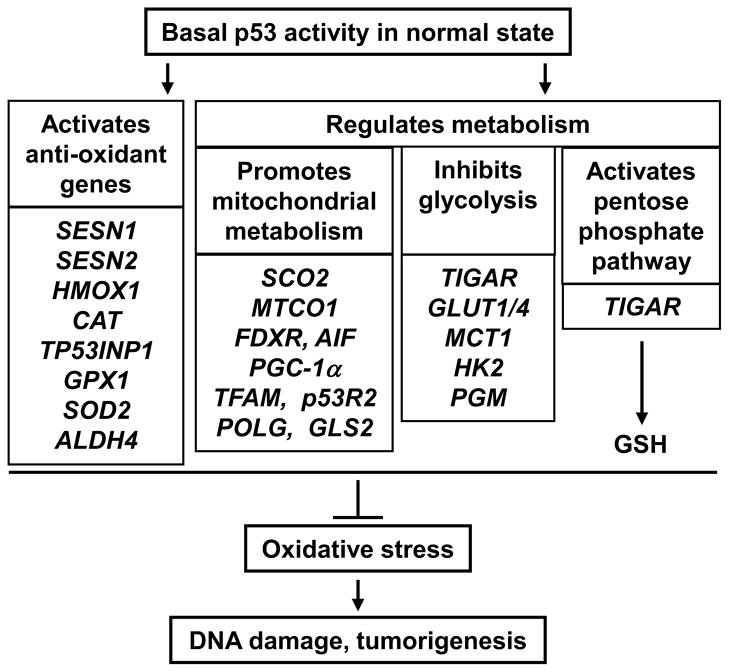
Increasing evidence indicates a dual role for p53 in redox homeostasis that depends on its activity and expression level as well as the context of the cell (Figs. 3 and 4) [10, 13, 20, 21]. A genetic link between p53 and mitochondrial respiration was established by using isogenic human colon cancer cells with p53 disruption and by the identification of synthesis of cytochrome c oxidase 2 (SCO2) as a transcriptional target of p53 required for complex IV assembly [61, 62]. It now appears that p53 promotes mitochondrial function through multiple pathways involving genes required for the assembly of other respiratory chain complexes, maintenance of the mitochondrial genome, regulation of mitochondrial dynamics, and metabolism (SCO2, cytochrome c oxidase subunit 1 (MTCO1), apoptosis-inducing factor (AIF), ferredoxin reductase (FDXR), PGC-1α, TFAM, p53-inducible ribonucleotide reductase (p53R2), DNA polymerase γ (POLG), mitochondrial glutaminase 2 (GLS2)) (Fig. 3) [12, 13, 63–65]. Although many of these metabolic pathways are transcriptionally regulated by p53, it is also evident that p53 can regulate metabolism by protein-protein interactions. For example, p53 can directly stabilize TFAM, interact with POLG, facilitate the translocation of p53R2 to the nucleus, and inactivate glucose-6-phosphate dehydrogenase (G6PD) at the protein level [66–69].
Fig. 3.
Basal levels of p53 under physiologic or normal states regulate multiple pathways to prevent oxidative DNA damage and tumorigenesis. p53 can counteract oxidative stress by inducing the expression of anti-oxidant enzymes and by increasing cellular antioxidant glutathione (GSH) biosynthesis through the pentose phosphate pathway. The promotion of aerobic metabolism by the concurrent stimulation of mitochondrial respiration and inhibition of glycolysis decreases the levels of ROS generating factors oxygen and high energy reducing equivalent (NADH), respectively.
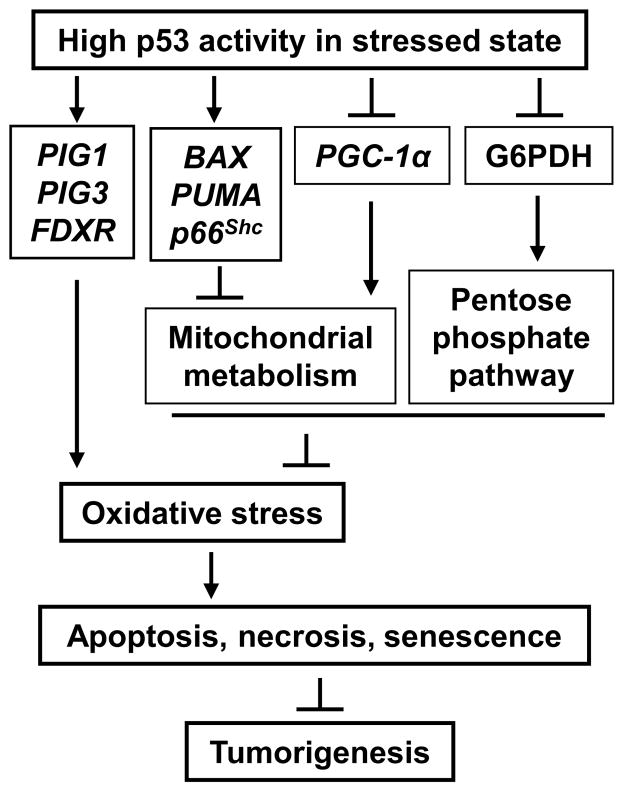
Fig. 4.
High levels of p53 caused by severe stress such as irreversible DNA damage increase oxidative stress to promote the elimination of damaged cells and prevent tumorigenesis. p53 can transactivate genes capable of generating ROS during apoptosis. In contrast to the antioxidant effects of p53 shown in Fig. 3, the inhibition of both mitochondrial metabolism and the pentose phosphate pathway may also contribute to increase cellular oxidative stress.
The substrates used for mitochondrial metabolism can play an important role in cellular redox homeostasis. p53-stimulated glutaminolysis via GLS2 lowers oxidative stress levels in association with increased mitochondrial respiration and glutathione production [70, 71]. Furthermore, compared to succinate, the oxidation of fatty acids promoted by p53 through its target gene guanidinoacetate methyltransferase (GAMT) produces less ROS due to low reverse electron transfer [72, 73]. ROS-activated ataxia telangiectasia mutated (ATM) can phosphorylate p53 at Ser-18 which increases fatty acid oxidation via the transactivation of the gene encoding phosphatidic acid phosphohydrolase (LPIN1) [74]. As fatty acid oxidation consumes more oxygen per ATP generated compared to glucose utilization and also provides NADPH needed for the biosynthesis of glutathione [75], the choice of substrate for mitochondrial respiration may be another level at which p53 may moderate oxidative stress.
As might be expected in a homeostatic relationship, mitochondrial function in turn influences p53 levels. Mitochondrial dysfunction has been shown to increase DNA damage [76], and the specific disruption of respiratory complex IV increases p53 protein levels as well as oxidative DNA damage in an oxygen dependent manner [18]. In contrast, the disruption of complex I has been shown to impair p53 stabilization suggesting that the interaction between the electron transport chain and p53 is likely to be rather complex [77]. Interestingly, it also appears that p53 can inhibit mitochondrial function and increase oxidative stress for eliminating cells with irreparable DNA damage (Fig. 4). p53 target genes, such as BCL-2 binding component 3 (BBC3 or PUMA) and BCL2-associated X protein (BAX), are upregulated in association with high ROS levels and cytochrome c release during p53-mediated apoptosis [13, 78]. Telomere shortening can promote premature aging by activating p53 [79]. A recent study has shown that p53 activation by telomerase deficiency can suppress PGC-13 expression thereby inhibiting mitochondrial biogenesis and increasing cellular stress and senescence [76, 80]. Consistent with this observation, p53 can also promote mitochondrial damage and oxidative stress via the regulation of p66shc [81, 82]. Furthermore, p53 transcriptionally regulates FDXR, a mitochondrial cytochrome P-450 NADPH reductase that mediates ROS generation during p53-dependent apoptosis [83] but is also essential for heme biogenesis and normal mitochondrial function [84, 85].
p53 regulates oxidative stress via nonmitochondrial pathways
Adenoviral vector mediated overexpression studies first associated p53 with oxidative stress and resulted in the identification of a number of non-mitochondrial genes thought to have a pro-oxidant function (Fig. 4) [86, 87]. These genes include the pro-apoptotic galectin family member LGALS7 (p53-induced gene 1, PIG1) and the NADPH-quinone oxidoreductase homolog TP53I3 (PIG3), both of which have ROS generating capacity [88, 89]). In contrast, basal p53 expression levels were subsequently shown to have a striking antioxidant effect [78]. Depletion of p53 caused increased oxidative DNA damage and chromosomal instability while dietary supplementation with the antioxidant N-acetylcysteine (NAC) markedly delayed de novo tumorigenesis in p53−/− mice, suggesting that the antioxidant function of p53 plays an important role in tumor suppression [78]. p53-regulated sestrins (SESN), essential for regenerating peroxide-reducing peroxiredoxins (PRX), were implicated in this specific study [90, 91].
Consistent with the above findings, mouse and human cells lacking p53 show increased sensitivity to oxidative and nitrosative stress in association with decreased heme oxygenase-1 (HO-1), a p53-dependent antioxidant gene [92–94]. However, there are additional ROS detoxifying pathway genes directly regulated by p53 both in the cytosol, such as peroxisomal catalase (CAT) and tumor protein 53-induced nuclear protein 1 (TP53INPI), as well as in the mitochondria, such as glutathione peroxidase 1 (GPX1), manganese superoxide dismutase (SOD2) and aldehyde dehydrogenase 4 (ALDH4) [95–98]. The relative contributions and responses of these various enzymatic systems likely depend on various factors including the type of oxidative stress, cell type and even the species of interest.
Glycolytic metabolism results in the generation of high energy reducing equivalents that can contribute to superoxide production. Consistent with an antioxidant role for p53, it is notable that p53 inhibits glycolysis through more than one regulatory pathway (Fig. 3). It can down-regulate glucose transporters (GLUT1 and GLUT4) thereby decreasing the availability of glucose for glycolysis [99]. The lactate produced during glycolysis to regenerate oxidized NAD by lactate dehydrogenase, an enzyme implicated in tumorigenicity [100], requires elimination by transport across the plasma membrane. p53 represses the expression of the monocarboxylate transporter gene (MCT1) that would decrease lactate efflux out of the cell and inhibit glycolysis [101]. p53 mutational status can also affect the activity of glycolytic enzymes such as hexokinase 2 (HK2) and phosphoglycerate mutase (PGM) [102, 103]. PGM is down-regulated by wild-type p53, and mutated p53 appears to facilitate cellular immortalization by increasing glycolysis [103].
p53 can modulate the activity of the key gylcolysis entry enzyme phosphofructokinase 1 (PFK1) by transactivating TP53-induced glycolysis and apoptosis regulator (TIGAR) which hydrolyzes the allosteric activator fructose-2,6-bisphosphate [104]. Notably, the decrease in glycolysis results in increased flux through the pentose phosphate pathway (PPP) which lowers apoptosis and ROS levels through increased glutathione synthesis. However, more recent data show that p53 can directly interact with and inactivate G6PD, the rate limiting enzyme of the PPP (Fig. 4) [69]. The biologic significance of these two seemingly opposing effects appears unclear but may also be reflective of the complexities surrounding the pro-oxidant and antioxidant effects of p53 depending on cellular context.
Summary
The association between ambient oxygen exposure and tumorigenesis as revealed by both human epidemiologic studies and experimental mouse models suggests that life in an aerobic environment requires dynamic regulation of the cellular redox state. Delineating the specific effects of p53 on the redox state of the cell has been challenging because of its multi-faceted activities, however, a common emerging theme is that p53 plays an essential role in redox homeostasis. In the context of a normal cell under basal conditions, p53 regulates antioxidant enzymes and metabolism, such as promoting mitochondrial respiration while inhibiting glycolysis to reduce cellular ROS levels and prevent DNA damage. In senescent or DNA damaged cells, p53 induces pro-oxidant genes while suppressing mitochondrial respiration to increase oxidative stress and promote cell death. Together, the dual effect of p53 on redox homeostasis may significantly contribute to its overall function of maintaining genomic stability and tumor suppression.
Highlights.
Human and animal data suggest that oxygen promotes tumorigenesis
Oxygen consumption by mitochondrial respiration affects redox homeostasis
p53 has both pro-oxidant and antioxidant activities via multiple pathways
p53 regulates mitochondrial respiration as part of its antioxidant function
Acknowledgments
We wish to thank Ping-yuan Wang for invaluable discussions and assistance. This work was supported by the intramural program of National Heart, Lung and Blood Institute (NHLBI), NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hayes JM. Geochemistry. Earth’s redox history. Science. 2011;334:1654–1655. doi: 10.1126/science.1216481. [DOI] [PubMed] [Google Scholar]
- 2.Lindenschmidt RC, Tryka AF, Witschi HP. Inhibition of mouse lung tumor development by hyperoxia. Cancer Res. 1986;46:1994–2000. [PubMed] [Google Scholar]
- 3.Raa A, Stansberg C, Steen VM, Bjerkvig R, Reed RK, Stuhr LE. Hyperoxia retards growth and induces apoptosis and loss of glands and blood vessels in DMBA-induced rat mammary tumors. BMC Cancer. 2007;7:23. doi: 10.1186/1471-2407-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galluzzo M, Pennacchietti S, Rosano S, Comoglio PM, Michieli P. Prevention of hypoxia by myoglobin expression in human tumor cells promotes differentiation and inhibits metastasis. J Clin Invest. 2009;119:865–875. doi: 10.1172/JCI36579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sung HJ, Ma W, Starost MF, Lago CU, Lim PK, Sack MN, Kang JG, Wang PY, Hwang PM. Ambient oxygen promotes tumorigenesis. PLoS One. 2011;6:e19785. doi: 10.1371/journal.pone.0019785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benz CC, Yau C. Ageing, oxidative stress and cancer: paradigms in parallax. Nat Rev Cancer. 2008;8:875–879. doi: 10.1038/nrc2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 9.Lu WJ, Amatruda JF, Abrams JM. p53 ancestry: gazing through an evolutionary lens. Nat Rev Cancer. 2009;9:758–762. doi: 10.1038/nrc2732. [DOI] [PubMed] [Google Scholar]
- 10.Olovnikov IA, Kravchenko JE, Chumakov PM. Homeostatic functions of the p53 tumor suppressor: regulation of energy metabolism and antioxidant defense. Semin Cancer Biol. 2009;19:32–41. doi: 10.1016/j.semcancer.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottlieb E, Vousden KH. p53 regulation of metabolic pathways. Cold Spring Harb Perspect Biol. 2010;2:a001040. doi: 10.1101/cshperspect.a001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lago CU, Sung HJ, Ma W, Wang PY, Hwang PM. p53, Aerobic Metabolism and Cancer. Antioxid Redox Signal. doi: 10.1089/ars.2010.3650. Epub ahead of print; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galluzzi L, Morselli E, Kepp O, Vitale I, Pinti M, Kroemer G. Mitochondrial liaisons of p53. Antioxid Redox Signal. 2011;15:1691–1714. doi: 10.1089/ars.2010.3504. [DOI] [PubMed] [Google Scholar]
- 14.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 15.Sagan C. On the origin and planetary distribution of life. Radiat Res. 1961;15:174–192. [PubMed] [Google Scholar]
- 16.Margulis L. Symbiosis in Cell Evolution. New York: W. H. Freeman and Company; 1993. [Google Scholar]
- 17.Brown GC, Borutaite V. There is no evidence that mitochondria are the main source of reactive oxygen species in mammalian cells. Mitochondrion. 2012;12:1–4. doi: 10.1016/j.mito.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Sung HJ, Ma W, Wang P-y, Hynes J, O’Riordan TC, Combs CA, McCoy JP, Bunz F, Kang J-G, Hwang PM. Mitochondrial respiration protects against oxygen-associated DNA damage. Nat Commun. 2010;1:1–8. doi: 10.1038/ncomms1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dotsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol. 2010;2:a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bensaad K, Vousden KH. p53: new roles in metabolism. Trends Cell Biol. 2007;17:286–291. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Liu B, Chen Y, St Clair DK. ROS and p53: a versatile partnership. Free Radic Biol Med. 2008;44:1529–1535. doi: 10.1016/j.freeradbiomed.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arai AE, Kasserra CE, Territo PR, Gandjbakhche AH, Balaban RS. Myocardial oxygenation in vivo: optical spectroscopy of cytoplasmic myoglobin and mitochondrial cytochromes. Am J Physiol. 1999;277:H683–697. doi: 10.1152/ajpheart.1999.277.2.H683. [DOI] [PubMed] [Google Scholar]
- 23.Braun RD, Lanzen JL, Snyder SA, Dewhirst MW. Comparison of tumor and normal tissue oxygen tension measurements using OxyLite or microelectrodes in rodents. Am J Physiol Heart Circ Physiol. 2001;280:H2533–2544. doi: 10.1152/ajpheart.2001.280.6.H2533. [DOI] [PubMed] [Google Scholar]
- 24.Schroeder JL, Luger-Hamer M, Pursley R, Pohida T, Chefd’hotel C, Kellman P, Balaban RS. Short communication: Subcellular motion compensation for minimally invasive microscopy, in vivo: evidence for oxygen gradients in resting muscle. Circ Res. 2010;106:1129–1133. doi: 10.1161/CIRCRESAHA.109.211946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Hagan KA, Cocchiglia S, Zhdanov AV, Tambuwala MM, Cummins EP, Monfared M, Agbor TA, Garvey JF, Papkovsky DB, Taylor CT, Allan BB. PGC-1alpha is coupled to HIF-1alpha-dependent gene expression by increasing mitochondrial oxygen consumption in skeletal muscle cells. Proc Natl Acad Sci U S A. 2009;106:2188–2193. doi: 10.1073/pnas.0808801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shanklin DR. A general theory of oxygen toxicity in man. Perspect Biol Med. 1969;13:80–100. doi: 10.1353/pbm.1969.0044. [DOI] [PubMed] [Google Scholar]
- 27.Weinberg CR, Brown KG, Hoel DG. Altitude, radiation, and mortality from cancer and heart disease. Radiat Res. 1987;112:381–390. [PubMed] [Google Scholar]
- 28.Luongo C, Moser AR, Gledhill S, Dove WF. Loss of Apc+ in intestinal adenomas from Min mice. Cancer Res. 1994;54:5947–5952. [PubMed] [Google Scholar]
- 29.He G, Shankar RA, Chzhan M, Samouilov A, Kuppusamy P, Zweier JL. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc Natl Acad Sci U S A. 1999;96:4586–4591. doi: 10.1073/pnas.96.8.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colgan SP, Taylor CT. Hypoxia: an alarm signal during intestinal inflammation. Nat Rev Gastroenterol Hepatol. 2010;7:281–287. doi: 10.1038/nrgastro.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woo DK, Green PD, Santos JH, D’Souza AD, Walther Z, Martin WD, Christian BE, Chandel NS, Shadel GS. Mitochondrial genome instability and ROS enhance intestinal tumorigenesis in APC(Min/+) mice. Am J Pathol. 2012;180:24–31. doi: 10.1016/j.ajpath.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagen T, Taylor CT, Lam F, Moncada S. Redistribution of intracellular oxygen in hypoxia by nitric oxide: effect on HIF1alpha. Science. 2003;302:1975–1978. doi: 10.1126/science.1088805. [DOI] [PubMed] [Google Scholar]
- 33.Taylor CT. Mitochondria and cellular oxygen sensing in the HIF pathway. Biochem J. 2008;409:19–26. doi: 10.1042/BJ20071249. [DOI] [PubMed] [Google Scholar]
- 34.Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci U S A. 1998;95:11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guzy RD, Hoyos B, Robin E, Chen H, Liu L, Mansfield KD, Simon MC, Hammerling U, Schumacker PT. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 36.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 37.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 39.Xie S, Wu H, Wang Q, Cogswell JP, Husain I, Conn C, Stambrook P, Jhanwar-Uniyal M, Dai W. Plk3 functionally links DNA damage to cell cycle arrest and apoptosis at least in part via the p53 pathway. J Biol Chem. 2001;276:43305–43312. doi: 10.1074/jbc.M106050200. [DOI] [PubMed] [Google Scholar]
- 40.Yamakuchi M, Lotterman CD, Bao C, Hruban RH, Karim B, Mendell JT, Huso D, Lowenstein CJ. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci U S A. 2010;107:6334–6339. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng Q, Dillehay LE, Madan A, Semenza GL, Bedi A. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev. 2000;14:34–44. [PMC free article] [PubMed] [Google Scholar]
- 42.Chen D, Li M, Luo J, Gu W. Direct interactions between HIF-1 alpha and Mdm2 modulate p53 function. J Biol Chem. 2003;278:13595–13598. doi: 10.1074/jbc.C200694200. [DOI] [PubMed] [Google Scholar]
- 43.Schmid T, Zhou J, Brune B. HIF-1 and p53: communication of transcription factors under hypoxia. J Cell Mol Med. 2004;8:423–431. doi: 10.1111/j.1582-4934.2004.tb00467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sermeus A, Michiels C. Reciprocal influence of the p53 and the hypoxic pathways. Cell Death Dis. 2011;2:e164. doi: 10.1038/cddis.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olsson A, Manzl C, Strasser A, Villunger A. How important are post-translational modifications in p53 for selectivity in target-gene transcription and tumour suppression? Cell Death Differ. 2007;14:1561–1575. doi: 10.1038/sj.cdd.4402196. [DOI] [PubMed] [Google Scholar]
- 47.Hainaut P, Milner J. Redox modulation of p53 conformation and sequence-specific DNA binding in vitro. Cancer Res. 1993;53:4469–4473. [PubMed] [Google Scholar]
- 48.Hafsi H, Hainaut P. Redox control and interplay between p53 isoforms: roles in the regulation of basal p53 levels, cell fate, and senescence. Antioxid Redox Signal. 2011;15:1655–1667. doi: 10.1089/ars.2010.3771. [DOI] [PubMed] [Google Scholar]
- 49.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 50.Sun XZ, Vinci C, Makmura L, Han S, Tran D, Nguyen J, Hamann M, Grazziani S, Sheppard S, Gutova M, Zhou F, Thomas J, Momand J. Formation of disulfide bond in p53 correlates with inhibition of DNA binding and tetramerization. Antioxid Redox Signal. 2003;5:655–665. doi: 10.1089/152308603770310338. [DOI] [PubMed] [Google Scholar]
- 51.Velu CS, Niture SK, Doneanu CE, Pattabiraman N, Srivenugopal KS. Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-binding domain during oxidative stress. Biochemistry. 2007;46:7765–7780. doi: 10.1021/bi700425y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buzek J, Latonen L, Kurki S, Peltonen K, Laiho M. Redox state of tumor suppressor p53 regulates its sequence-specific DNA binding in DNA-damaged cells by cysteine 277. Nucleic Acids Res. 2002;30:2340–2348. doi: 10.1093/nar/30.11.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Augustyn KE, Merino EJ, Barton JK. A role for DNA-mediated charge transport in regulating p53: Oxidation of the DNA-bound protein from a distance. Proc Natl Acad Sci U S A. 2007;104:18907–18912. doi: 10.1073/pnas.0709326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie S, Wang Q, Wu H, Cogswell J, Lu L, Jhanwar-Uniyal M, Dai W. Reactive oxygen species-induced phosphorylation of p53 on serine 20 is mediated in part by polo-like kinase-3. J Biol Chem. 2001;276:36194–36199. doi: 10.1074/jbc.M104157200. [DOI] [PubMed] [Google Scholar]
- 55.Chazotte-Aubert L, Hainaut P, Ohshima H. Nitric oxide nitrates tyrosine residues of tumor-suppressor p53 protein in MCF-7 cells. Biochem Biophys Res Commun. 2000;267:609–613. doi: 10.1006/bbrc.1999.2003. [DOI] [PubMed] [Google Scholar]
- 56.Bragado P, Armesilla A, Silva A, Porras A. Apoptosis by cisplatin requires p53 mediated p38alpha MAPK activation through ROS generation. Apoptosis. 2007;12:1733–1742. doi: 10.1007/s10495-007-0082-8. [DOI] [PubMed] [Google Scholar]
- 57.Jayaraman L, Murthy KG, Zhu C, Curran T, Xanthoudakis S, Prives C. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 1997;11:558–570. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- 58.Meira LB, Cheo DL, Hammer RE, Burns DK, Reis A, Friedberg EC. Genetic interaction between HAP1/REF-1 and p53. Nat Genet. 1997;17:145. doi: 10.1038/ng1097-145. [DOI] [PubMed] [Google Scholar]
- 59.Seemann S, Hainaut P. Roles of thioredoxin reductase 1 and APE/Ref-1 in the control of basal p53 stability and activity. Oncogene. 2005;24:3853–3863. doi: 10.1038/sj.onc.1208549. [DOI] [PubMed] [Google Scholar]
- 60.Asher G, Lotem J, Cohen B, Sachs L, Shaul Y. Regulation of p53 stability and p53-dependent apoptosis by NADH quinone oxidoreductase 1. Proc Natl Acad Sci U S A. 2001;98:1188–1193. doi: 10.1073/pnas.021558898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou S, Kachhap S, Singh KK. Mitochondrial impairment in p53-deficient human cancer cells. Mutagenesis. 2003;18:287–292. doi: 10.1093/mutage/18.3.287. [DOI] [PubMed] [Google Scholar]
- 62.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 63.Saleem A, Adhihetty PJ, Hood DA. Role of p53 in mitochondrial biogenesis and apoptosis in skeletal muscle. Physiol Genomics. 2009;37:58–66. doi: 10.1152/physiolgenomics.90346.2008. [DOI] [PubMed] [Google Scholar]
- 64.Li J, Donath S, Li Y, Qin D, Prabhakar BS, Li P. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 2010;6:e1000795. doi: 10.1371/journal.pgen.1000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maddocks OD, Vousden KH. Metabolic regulation by p53. J Mol Med. 2011;89:237–245. doi: 10.1007/s00109-011-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xue L, Zhou B, Liu X, Qiu W, Jin Z, Yen Y. Wild-type p53 regulates human ribonucleotide reductase by protein-protein interaction with p53R2 as well as hRRM2 subunits. Cancer Res. 2003;63:980–986. [PubMed] [Google Scholar]
- 67.Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, Keating MJ, Huang P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. Embo J. 2005;24:3482–3492. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong TS, Rajagopalan S, Freund SM, Rutherford TJ, Andreeva A, Townsley FM, Petrovich M, Fersht AR. Biophysical characterizations of human mitochondrial transcription factor A and its binding to tumor suppressor p53. Nucleic Acids Res. 2009;37:6765–6783. doi: 10.1093/nar/gkp750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, Yang X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011;13:310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A. 2010;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y, Sugano S, Sato E, Nagao T, Yokote K, Tatsuno I, Prives C. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A. 2010;107:7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ide T, Brown-Endres L, Chu K, Ongusaha PP, Ohtsuka T, El-Deiry WS, Aaronson SA, Lee SW. GAMT, a p53-inducible modulator of apoptosis, is critical for the adaptive response to nutrient stress. Mol Cell. 2009;36:379–392. doi: 10.1016/j.molcel.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73.Schonfeld P, Wieckowski MR, Lebiedzinska M, Wojtczak L. Mitochondrial fatty acid oxidation and oxidative stress: lack of reverse electron transfer-associated production of reactive oxygen species. Biochim Biophys Acta. 2010;1797:929–938. doi: 10.1016/j.bbabio.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 74.Assaily W, Rubinger DA, Wheaton K, Lin Y, Ma W, Xuan W, Brown-Endres L, Tsuchihara K, Mak TW, Benchimol S. ROS-mediated p53 induction of Lpin1 regulates fatty acid oxidation in response to nutritional stress. Mol Cell. 2011;44:491–501. doi: 10.1016/j.molcel.2011.08.038. [DOI] [PubMed] [Google Scholar]
- 75.Pike LS, Smift AL, Croteau NJ, Ferrick DA, Wu M. Inhibition of fatty acid oxidation by etomoxir impairs NADPH production and increases reactive oxygen species resulting in ATP depletion and cell death in human glioblastoma cells. Biochim Biophys Acta. 2011;1807:726–734. doi: 10.1016/j.bbabio.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 76.Liu J, Cao L, Chen J, Song S, Lee IH, Quijano C, Liu H, Keyvanfar K, Chen H, Cao LY, Ahn BH, Kumar NG, Rovira, Xu XL, van Lohuizen M, Motoyama N, Deng CX, Finkel T. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459:387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Compton S, Kim C, Griner NB, Potluri P, Scheffler IE, Sen S, Jerry DJ, Schneider S, Yadava N. Mitochondrial dysfunction impairs tumor suppressor p53 expression/function. J Biol Chem. 2011;286:20297–20312. doi: 10.1074/jbc.M110.163063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, Greider CW, DePinho RA. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 80.Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, Maser RS, Tonon G, Foerster F, Xiong R, Wang YA, Shukla SA, Jaskelioff M, Martin ES, Heffernan TP, Protopopov A, Ivanova E, Mahoney JE, Kost-Alimova M, Perry SR, Bronson R, Liao R, Mulligan R, Shirihai OS, Chin L, DePinho RA. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Trinei M, Giorgio M, Cicalese A, Barozzi S, Ventura A, Migliaccio E, Milia E, Padura IM, Raker VA, Maccarana M, Petronilli V, Minucci S, Bernardi P, Lanfrancone L, Pelicci PG. A p53-p66Shc signalling pathway controls intracellular redox status, levels of oxidation-damaged DNA and oxidative stress-induced apoptosis. Oncogene. 2002;21:3872–3878. doi: 10.1038/sj.onc.1205513. [DOI] [PubMed] [Google Scholar]
- 82.Orsini F, Migliaccio E, Moroni M, Contursi C, Raker VA, Piccini D, Martin-Padura I, Pelliccia G, Trinei M, Bono M, Puri C, Tacchetti C, Ferrini M, Mannucci R, Nicoletti I, Lanfrancone L, Giorgio M, Pelicci PG. The life span determinant p66Shc localizes to mitochondria where it associates with mitochondrial heat shock protein 70 and regulates trans-membrane potential. J Biol Chem. 2004;279:25689–25695. doi: 10.1074/jbc.M401844200. [DOI] [PubMed] [Google Scholar]
- 83.Hwang PM, Bunz F, Yu J, Rago C, Chan TA, Murphy MP, Kelso GF, Smith RA, Kinzler KW, Vogelstein B. Ferredoxin reductase affects p53-dependent, 5-fluorouracil-induced apoptosis in colorectal cancer cells. Nat Med. 2001;7:1111–1117. doi: 10.1038/nm1001-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 85.Shi Y, Ghosh M, Kovtunovych G, Crooks DR, Rouault TA. Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis. Biochim Biophys Acta. 2012;1823:484–492. doi: 10.1016/j.bbamcr.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnson TM, Yu ZX, Ferrans VJ, Lowenstein RA, Finkel T. Reactive oxygen species are downstream mediators of p53-dependent apoptosis. Proc Natl Acad Sci U S A. 1996;93:11848–11852. doi: 10.1073/pnas.93.21.11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 88.Rao PV, Krishna CM, Zigler JS., Jr Identification and characterization of the enzymatic activity of zeta-crystallin from guinea pig lens. A novel NADPH:quinone oxidoreductase. J Biol Chem. 1992;267:96–102. [PubMed] [Google Scholar]
- 89.Kuwabara I, Kuwabara Y, Yang RY, Schuler M, Green DR, Zuraw BL, Hsu DK, Liu FT. Galectin-7 (PIG1) exhibits pro-apoptotic function through JNK activation and mitochondrial cytochrome c release. J Biol Chem. 2002;277:3487–3497. doi: 10.1074/jbc.M109360200. [DOI] [PubMed] [Google Scholar]
- 90.Velasco-Miguel S, Buckbinder L, Jean P, Gelbert L, Talbott R, Laidlaw J, Seizinger B, Kley N. PA26, a novel target of the p53 tumor suppressor and member of the GADD family of DNA damage and growth arrest inducible genes. Oncogene. 1999;18:127–137. doi: 10.1038/sj.onc.1202274. [DOI] [PubMed] [Google Scholar]
- 91.Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 92.Yu J, Zhang L, Hwang PM, Rago C, Kinzler KW, Vogelstein B. Identification and classification of p53-regulated genes. Proc Natl Acad Sci U S A. 1999;96:14517–14522. doi: 10.1073/pnas.96.25.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meiller A, Alvarez S, Drane P, Lallemand C, Blanchard B, Tovey M, May E. p53-dependent stimulation of redox-related genes in the lymphoid organs of gamma-irradiated--mice identification of Haeme-oxygenase 1 as a direct p53 target gene. Nucleic Acids Res. 2007;35:6924–6934. doi: 10.1093/nar/gkm824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nam SY, Sabapathy K. p53 promotes cellular survival in a context-dependent manner by directly inducing the expression of haeme-oxygenase-1. Oncogene. 30:4476–4486. doi: 10.1038/onc.2011.150. [DOI] [PubMed] [Google Scholar]
- 95.Tan M, Li S, Swaroop M, Guan K, Oberley LW, Sun Y. Transcriptional activation of the human glutathione peroxidase promoter by p53. J Biol Chem. 1999;274:12061–12066. doi: 10.1074/jbc.274.17.12061. [DOI] [PubMed] [Google Scholar]
- 96.Hussain SP, Amstad P, He P, Robles A, Lupold S, Kaneko I, Ichimiya M, Sengupta S, Mechanic L, Okamura S, Hofseth LJ, Moake M, Nagashima M, Forrester KS, Harris CC. p53-induced up-regulation of MnSOD and GPx but not catalase increases oxidative stress and apoptosis. Cancer Res. 2004;64:2350–2356. doi: 10.1158/0008-5472.can-2287-2. [DOI] [PubMed] [Google Scholar]
- 97.O’Connor JC, Wallace DM, O’Brien CJ, Cotter TG. A novel antioxidant function for the tumor-suppressor gene p53 in the retinal ganglion cell. Invest Ophthalmol Vis Sci. 2008;49:4237–4244. doi: 10.1167/iovs.08-1963. [DOI] [PubMed] [Google Scholar]
- 98.Cano CE, Gommeaux J, Pietri S, Culcasi M, Garcia S, Seux M, Barelier S, Vasseur S, Spoto RP, Pebusque MJ, Dusetti NJ, Iovanna JL, Carrier A. Tumor protein 53-induced nuclear protein 1 is a major mediator of p53 antioxidant function. Cancer Res. 2009;69:219–226. doi: 10.1158/0008-5472.CAN-08-2320. [DOI] [PubMed] [Google Scholar]
- 99.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 100.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 101.Boidot R, Vegran F, Meulle A, Le Breton A, Dessy C, Sonveaux P, Lizard-Nacol S, Feron O. Regulation of Monocarboxylate Transporter MCT1 Expression by p53 Mediates Inward and Outward Lactate Fluxes in Tumors. Cancer Res. 72:939–948. doi: 10.1158/0008-5472.CAN-11-2474. [DOI] [PubMed] [Google Scholar]
- 102.Mathupala SP, Heese C, Pedersen PL. Glucose catabolism in cancer cells. The type II hexokinase promoter contains functionally active response elements for the tumor suppressor p53. J Biol Chem. 1997;272:22776–22780. doi: 10.1074/jbc.272.36.22776. [DOI] [PubMed] [Google Scholar]
- 103.Kondoh H, Lleonart ME, Gil J, Wang J, Degan P, Peters G, Martinez D, Carnero A, Beach D. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–185. [PubMed] [Google Scholar]
- 104.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]