Abstract
Cellular signaling processes depend on specific spatiotemporal distributions of their molecular components. Multi-color high-resolution microscopy now permits detailed assessment of such distributions, providing the input for fine-grained computational models that explore the mechanisms governing dynamic assembly of multi-molecular complexes and their role in shaping cellular behavior. However, incorporating into such models both complex molecular reaction cascades and the spatial localization of signaling components within dynamic cellular morphologies presents substantial challenges. Here we introduce an approach that addresses these challenges by automatically generating computational representations of complex reaction networks based on simple bi-molecular interaction rules embedded into detailed, adaptive models of cellular morphology. Using examples of receptor-mediated cellular adhesion and signal-induced localized MAPK activation in yeast, we illustrate the capacity of this simulation technique to provide insights into cell biological processes. The modeling algorithms, implemented in a version of the Simmune tool set, are accessible through intuitive graphical interfaces as well as programming libraries.
Introduction
The increasing spatial resolution of optical microscopy, in concert with novel technologies for labeling cell constituents, is continuously revealing new details about how cells actively tailor the chemical composition of membrane domains during signaling or generate intracellular concentration gradients in response to localized external signals 1–3. Reflecting these insights, modern simulation approaches no longer assume well-mixed biochemistry but instead simulate signaling processes within computational representations of cellular geometries (see Supplementary Note 1 for a detailed discussion of available approaches). In parallel, experimentalists and theorists have begun to explore the physiological role of multi-component molecular complexes. Systematic analyses of the spectrum of binding partners of receptor tyrosine kinases such as the EGF receptor 4 or of the assembly of ‘signalosomes’ around scaffold proteins 5 have revealed that signaling pathways are intricate factories, dynamically assembling and disassembling molecular complexes with specific functions.
Defining computational models for exploring the reaction dynamics of networks of these large multi-molecular complexes requires approaches that differ from the traditional strategy of explicitly formulating all mass-action equations for the complete set of reactants in a system. The reason is that reaction dynamics in systems with many molecular components, or with components that have many binding sites, may involve very large numbers of alternative interactions and complexes, a phenomenon sometimes referred to as ‘combinatorial explosion’ 6. However, because all the reactions within such networks are based on interactions between pairs of molecular binding sites, much of the complexity can be reduced to defining the rules that describe these binary interactions. Taking advantage of this underlying reality, ‘rule-based’ approaches leave the task of actually assembling the complexes and reaction networks to the computer, once the investigator has defined the fundamental components and their pair-wise interactions 7–12. Rule-based simulation approaches have, however, so far been limited with regard to their ability to take into account the cellular morphologies in which the molecular networks operate and had to assume static geometries or well-stirred conditions.
Here, we introduce a computational approach that can be used to simulate complex signaling processes in the context of realistic and even dynamic cellular morphologies. This novel capacity is based on the ability to generate reaction networks automatically, taking into account the location-dependent molecular interaction possibilities determined by the dynamic membrane geometries and 3-dimensional shapes of the simulated cells (Fig. 1, Supplementary Note 2). The approach is a major extension and improvement of the modeling tool Simmune7. Most of its functionalities are available to users who prefer a graphical user interface (GUI) to writing model definition scripts. It also permits modelers to simulate aspects of biology that would be difficult to address with conventional methods, such as the local dynamics of multi-molecular signaling complexes in large networks. We use the approach to simulate two well-studied biological systems, the reaction-diffusion processes of adhesion receptors at dynamic cell-cell contacts, and the formation of MAPK activity gradients by intracellular multi-molecular signaling complexes in the yeast mating pheromone response.
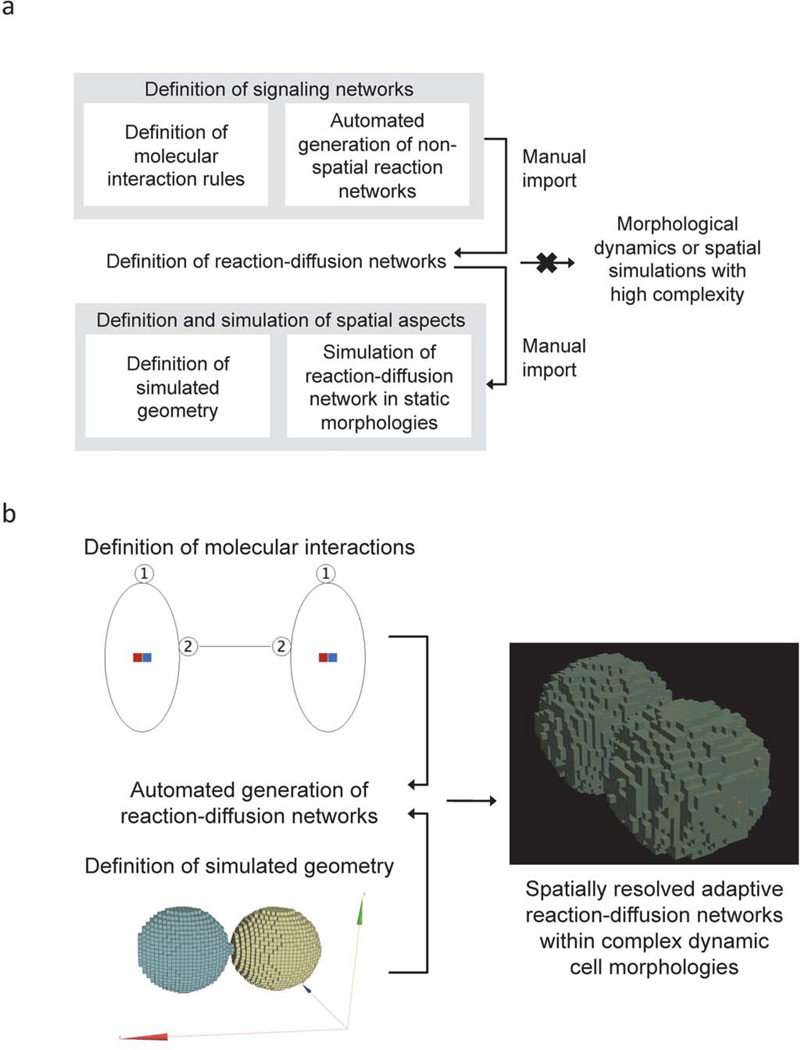
Figure 1. Automated spatially-resolved generation of reaction networks to simulate cellular signaling in realistic morphologies.
a) The schematic illustrates that non-spatial ‘rule-based’ modeling methods can generate signaling networks based on pairwise molecular interactions but cannot take into account the specific localizations of the molecules. Automatically-generated non-spatial networks would have to be translated manually (‘imported’) into reaction-diffusion networks and then inserted into the simulated space by assigning molecular species to specific locations. Non-rule-based spatial simulation tools, on the other hand, allow users to define simulated geometries and simulate pre-defined reaction-diffusion networks within them. Extending the latter simulations to dynamic morphologies or to spatial simulations of highly complex reaction networks is, however, not possible because the reaction-diffusion networks must be fully specified prior to performing the spatially-resolved simulation. b) Simmune’s spatially-resolved automated network generator permits the definition of reaction networks based on molecular interaction rules and then generates and simulates reaction-diffusion networks while adapting them to potentially dynamic cellular morphologies such as shown here for the simulation of cell-cell contact formation.
Results
Molecular reaction networks in their spatial contexts
Modeling approaches that focus on phenomena of individual complexes (for instance, receptor clustering 13) and that simulate the motion and reactions performed by discrete particles incorporate spatial constraints naturally (Methods and Supplementary Fig. 1): complexes can only interact when they share a common location and the rates of their interactions depend on their local abundances. However, the computational cost of particle-based simulations increases at least linearly (and typically much faster than linearly) with the abundance of the complexes. This means that simulating a system with 1,000,000 molecules (roughly corresponding to a micromolar concentration in a cell of 10 microns diameter) will take at least 1000 times longer than simulating a system with 1000 molecules. Since many cellular signaling phenomena involve large numbers of molecules and do not require approaches that track individual particles, they can be investigated far more efficiently by simulating the dynamics of local molecular concentrations instead of using particle-based simulators (Methods and Supplementary Fig. 1).
It should be noted that one of the most computationally expensive aspects of particle-based simulations, namely capturing the diffusional motion of particles in between molecular encounters and reactions, has recently been addressed with techniques that either avoid taking single Brownian diffusion steps (GFRD14) or that take large steps and correct for the introduced error (Smoldyn 15). Nevertheless, these simulations remain very time consuming 15 where particle numbers are large; their immediate value lies in applications with relatively low numbers of particles to be simulated.
Without taking into account spatial aspects, rule-based approaches to model cellular signaling networks can translate interaction rules directly into sets of complexes and the reactions between them and simulate them as sets of ordinary differential equations (ODEs). In contrast, for spatially-resolved simulations, the rates of these reactions and even the types of reactions that will actually occur depend on the morphology within which the biochemical components of the network operate. The new simulation algorithms in Simmune therefore apply the rules that specify how molecular interactions are translated into complexes and reaction networks for each volume or membrane element individually, based on its local biochemical composition. For membrane elements that are in contact with other membranes (for instance, as part of an intercellular contact) the network generation algorithm additionally determines whether the opposing membrane contains receptor complexes that can interact with the local molecular complexes. Each complex is then assigned a list of reaction processes (associations, dissociations or transformations). Finally, the diffusional exchange among neighboring volume or membrane elements is determined and added to the list of processes that now can be used to calculate how local concentrations of molecular complexes will change over time.
Rule-based network creation can be computationally expensive (Supplementary Note 3). For simulations with high spatial resolution, Simmune has to generate local networks for thousands of volume and membrane elements and therefore uses a multi-step strategy that first builds a non-spatial reaction network based on all possible interactions among the molecular species in the simulated biological system. This template network is then adapted to the specific local molecular environment of each volume or membrane element, which can be done much faster than creating networks de novo. Additionally, each newly generated local network is stored in a network repository that allows reuse when networks have to be built for locations with identical molecular composition. This strategy is efficient enough to permit the automatic creation of reaction networks even within the context of changing cellular shapes, for instance when simulating receptor interactions at dynamic cell-cell contact interfaces. The results of the simulations we report here emphasize the potentially important differences between morphologically dynamic versus static modeling of reaction-diffusion processes.
Static vs. dynamic simulations of intercellular E-cadherin
E-cadherin-mediated cell-cell adhesion is essential for the organization of multicellular organisms 16. Here we simulate cell-cell contact regulation involving just this one molecular species, under conditions in which both membranes express the same adhesion molecules and in which these molecules form homodimers or larger complexes based on these dimers 17, 18. Focusing just on this isolated set of interactions allowed us to explore a simple but important system with well-established phenomenology.
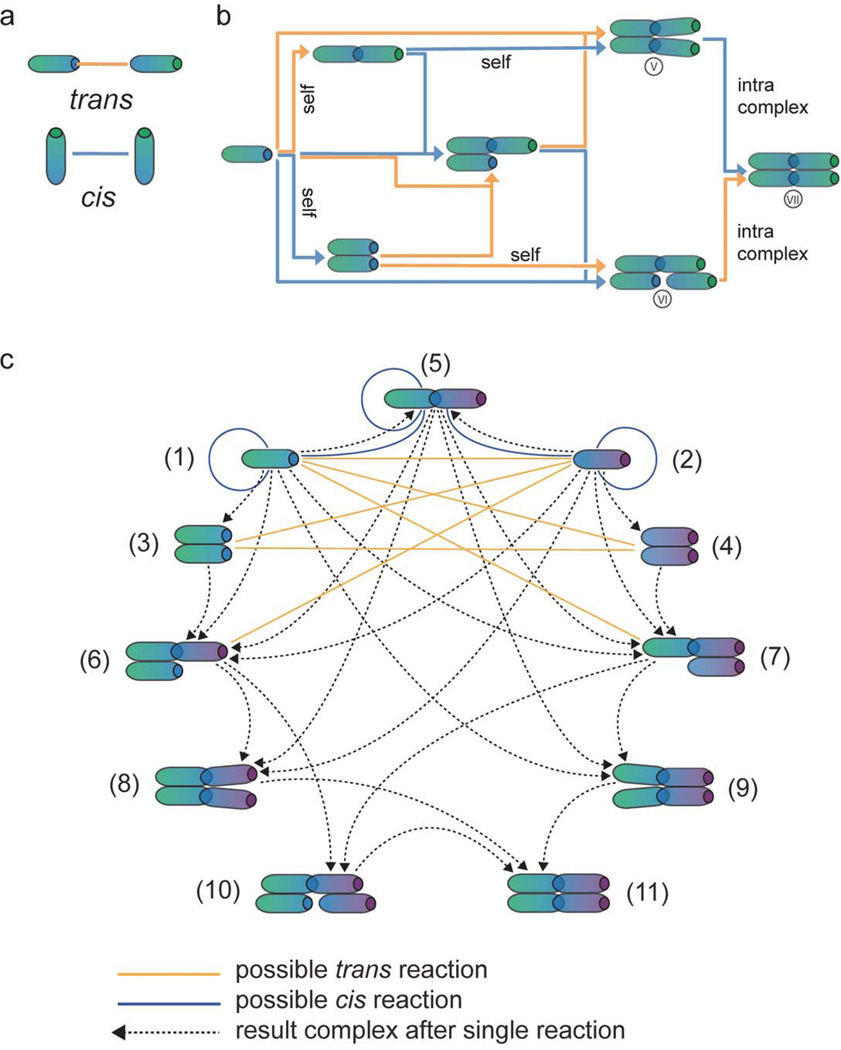
The purely biochemical network resulting from the fundamental interactions between E-cadherin monomers (Fig. 2a) contains only seven biochemically distinct complexes when spatial issues are ignored (Fig. 2b). In contrast, adding into the analysis the localization of the molecular components in two adjacent cell membranes yields a considerably more complicated picture (Fig. 2c). This is due to the fact that structurally equivalent complexes now can differ with regard to which side of the membrane-membrane interface their E-cadherin molecules belong. For example, the tetramer with an open cis-interface (labeled with a roman V in Fig. 2b) exists in two versions at the cell-cell interface (labeled (8) and (9) in Fig. 2c), facing either one or the other cell with its ‘open’ end. Even without including lateral diffusion within the membranes, a mathematical description of the possible reactions between the different types of complexes would require more than 100 reaction terms (Supplementary Note 2b). Incorporating spatial aspects thus transforms generation of the biochemically simple network of E-cadherin trans- and cis-dimerization into a nontrivial task, as is illustrated in Fig. 3 for one step of the local network generation process for two adjacent membrane elements.
Figure 2. From non-spatial to spatial representations of E-cadherin interactions.
a) E-cadherin receptors can interact in cis (within one membrane) and in trans (linking two adjacent membranes). b) The schematic shows the E-cadherin reaction network assuming one cis- and one trans- binding site per receptor, without taking into account the membrane localization of the receptors. The roman numerals V, VI, VII label complexes for the discussion in Supplementary Note 2. Reactions in cis are represented by blue arrows, interactions in trans by orange arrows. Associations between identical components are labeled ‘self’.c) The schematic shows the consequence of embedding the network of (b) into the spatial context of two adjacent membranes. The solid lines indicate reaction possibilities (blue: cis, yellow: trans), the dotted arrows connect initial and resulting complexes for association reactions. For example, complex (1) can associate with (4) (as indicated by a solid yellow line between (1) and (4)) to form (6) (dotted arrow from (1) to (6)). The color gradients of the E-cadherin molecules (ranging from green to blue or from purple to blue) indicate that the complexes are treated as embedded in two adjacent membranes; blue indicates the intercellular domains. The number labels correspond to the numbering of the differential equations in Supplementary Note 2b.
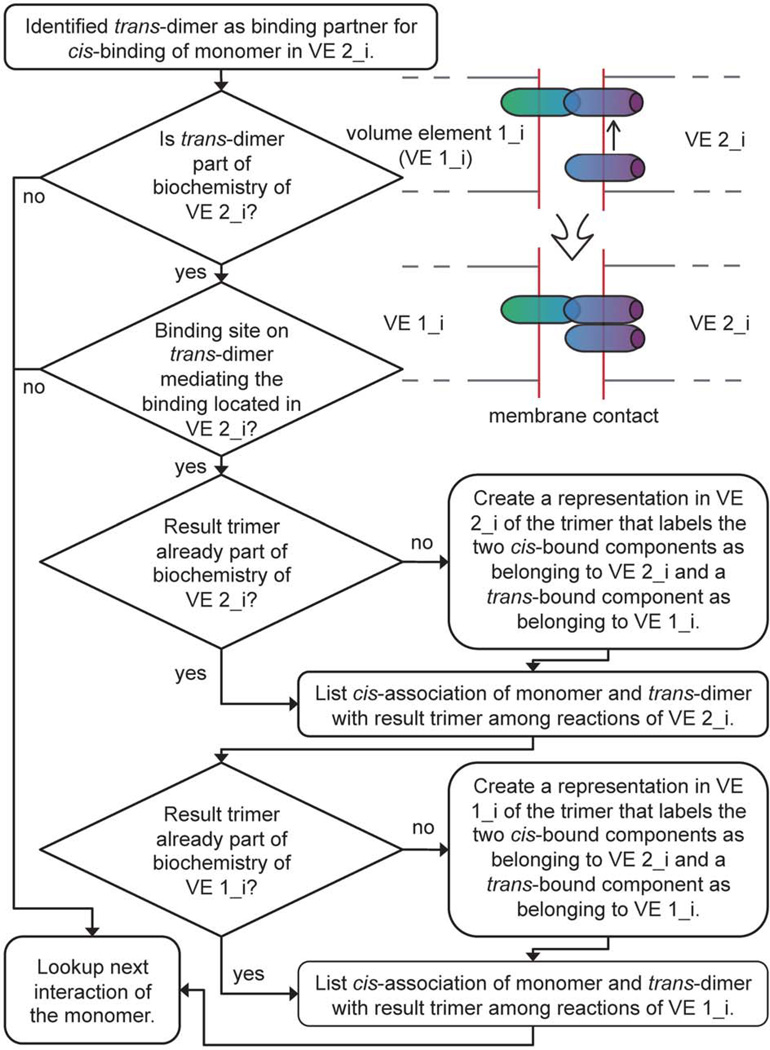
Figure 3. Automatic creation of an E-cadherin trimer for a membrane contact reaction network.
The depicted flow diagram illustrates how the algorithms that are part of our approach automatically create spatially resolved reactions and complexes based on simple user-provided rules specifying molecular interactions. The membrane of a volume element with index i in cell 2 (VE 2_i) contains a monomer that can bind through a cis-interaction to a trans-dimer. If no trans-dimer is present in VE 2_i the algorithm will look for the next potential interaction partner. If it is present in VE 2_i the algorithm will check whether the site capable of mediating the cis-interaction with the monomer is located in VE 2_i (it could be that the trans-dimer has no available cis binding site). If the binding site is available the algorithm will determine whether such a trimer (with two molecules located in the membrane of VE 2_i and one in the adjacent membrane of VE 1_i) is already represented as a complex within the biochemistry of VE 2_i in which case this complex representation will be used as the result complex of the association. Otherwise, a result trimer complex will be built within the biochemistry of VE 2_i. Since the result trimer also is ‘visible’ in the membrane element of VE 1_i it has to be inserted or identified in that reaction network as well. Finally, the reaction information is completed by assembling a code for the association of the monomer with the trans-dimer resulting in the formation of the trimer as result complex and with the reaction rate as specified by the user-supplied association rate between the involved binding sites.
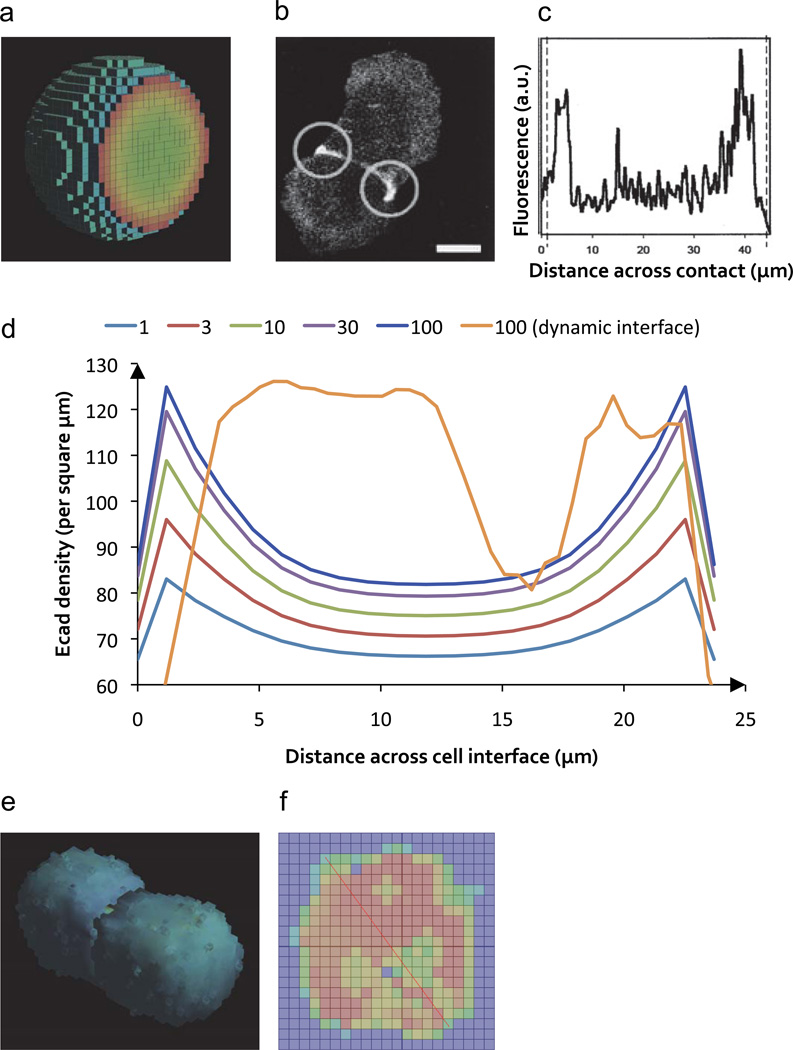
We first investigated the accumulation of E-cadherin at the interfaces between models of morphologically static cells that were simulated as coming into close proximity prior to the formation of receptor contacts, mimicking cellular crowding that arises when cells enter into a pre-existing epithelial layer involved in rearrangements or repair. It is currently assumed that cis interactions are much weaker than trans interactions 19 and that their main function may be to stabilize trans interactions 20. Varying the lifespan of cis-bonds within trans complexes between 1 and 100 seconds, we observed an accumulation of E-cadherin at the periphery of the contact zones (Fig. 4a), reminiscent of the experimentally observed accumulation of E-cadherin at the edges of adhesive cell contacts (Fig. 4b,c) 21. This accumulation was markedly modulated by the cis bond lifespan and reached up to 1.5 fold the concentration of E-cadherin outside of the contact zones (Fig. 4d). These results are consistent with the findings of a previous computational model of a cell in contact with a small sphere acting as a completely absorbing surface for E-cadherin 22.
Figure 4. E-cadherin accumulation at static or dynamic cell contacts.
a) View of one of the two simulated cells interacting at a static interface with color-coded (from blue: low to red: high) local concentrations of E-cadherin. b, c) Experimental data showing that interacting cells accumulate fluorescently labeled E-cadherin at the periphery of their interfaces (© Rockefeller University Press, 1998. Originally published in J. Cell Biol. 142:1105–1119, reproduced with permission). Scale bar, 15 microns. Panel (c) shows the fluorescence line profile along the length of the cell-cell interface. d) The plot shows simulated E-cadherin density at the cell-cell interface (after 30 minutes of contact) with varying lifespan (1–100 seconds) of the cis interaction within E-cadherin complexes. For a lifespan of 100 seconds, both a static and dynamic interface are shown. e, f) Cell-cell contact after 1.5 simulated hours based on cellular Potts model dynamics. Panel (f) shows the color-coded E-cadherin concentration (red: high, blue: low) at the dynamic cellular interface. The red line was used to determine the concentration profile shown as the orange curve in part (d) of this figure. One voxel has a side length of 1.29 micrometers.
To study the evolution of local concentrations of intercellular E-cadherin complexes during the more physiological setting of gradual growth of receptor-mediated cellular adhesive contacts involving membranes of flexible topology, we performed simulations with receptor interaction networks embedded into the dynamic morphology of two interacting cells using a three-dimensional version of the ‘cellular Potts model’ (CPM) method 23 This method has been applied successfully to the study of cellular movement in models of biological 3D environments 24. It simulates the dynamic geometries of biological cells as coherent structures consisting of two- or three-dimensional basic spatial elements (for example, squares, hexagons or, in 3D, cubes) through stochastic updates that calculate the probabilities for adding or removing volume elements based on their particular states or geometric neighborhoods. The coupling between biochemistry and morphological dynamics was implemented through Potts energy terms that describe the binding status of membrane receptors (Methods and Supplementary Note 3). In contrast to simulations with static morphologies, combining receptor interactions and morphological dynamics requires rebuilding the local molecular reaction networks every time the morphology changes to take into account new receptor interaction possibilities or to discard those that are no longer supported by the membrane geometry.
The overall simulation flow after the initialization of cellular biochemistry and morphology proceeded according to the following pattern: 1) local network (reaction-diffusion) generation/adjustment, 2) integration of the corresponding system of coupled differential equations for the local concentrations of all molecular complexes, 3) Potts model update step, repeat until user event or end time point is reached (Supplementary Fig. 2). We note that the local network generation method introduced here can be embedded into any algorithm generating morphological dynamics, not just Potts model simulations. The network generation could in principle be combined with more sophisticated approaches for shape and surface dynamics (than just cubic extension and retraction) such as finite element methods or level sets but the coupling between shape dynamics and biochemistry would be highly nontrivial.
Cells were simulated as initially coming into contact at small membrane regions. These small contacts and the shape of the membranes in which the molecules were embedded were then allowed to evolve, driven by the formation of intercellular E-cadherin contacts. The simulations incorporated Potts model parameters chosen to reproduce previous work reporting experimentally determined growth rates of cell-cell contacts, which began at 30 microns/hour 21 and resulted over 1.5 hours in cells joined through a contact interface spanning almost the entire cellular diameter (Fig. 4e and Supplementary Movie 1). Even for receptor-interaction parameters that had led to pronounced peripheral cadherin accumulation along the edges of cellular contacts in the morphologically static case, these dynamic simulations instead showed an overall increase in cadherin density in the central contact zone modulated by the morphological features of the interface (such as regions of smooth or rugged, interrupted cell-cell contacts) (Fig. 4f). This qualitatively different behavior of the models is a consequence of the rapid (relative to the time scale governing the growth of cell-cell contacts) E-cadherin diffusion that allows fast recruitment of receptors that stabilize nascent contacts. The results of the simulations thus show that, due to the fast diffusion of E-cadherin, cells have to employ active transport mechanisms (as opposed to passive diffusion, as modeled here) to generate a high-concentration adhesive ring at the periphery of cellular contacts by removing receptors from the central contact zone in order to finally form mature adherens junctions 25. Indeed, performing morphologically dynamic simulations with a diffusion coefficient typical for larger trans-membrane receptors (tenfold lower than has been reported for E-cadherin), or with contacts that grew five times faster than experimentally reported (thereby shifting the balance between diffusion and morphological dynamics), we obtained a peripheral accumulation of these receptors (Supplementary Fig. 3a, b). In contrast, just the accumulation of E-cadherin in intercellular contact zones did not require active transport, in accord with previous experimental findings 26.
Computational exploration of MAPK activation in yeast
We next illustrate the application of our modeling method to exploring spatial aspects of MAPK activation in yeast, which requires the generation of a signaling network with a high degree of combinatorial complexity within a characteristic cellular morphology. MAPK activation downstream of the yeast pheromone receptor is organized around the scaffold protein Ste5. The scaffold interacts with a variety of partners, among them the G protein subunit Gβγ, the phospholipid PIP2 (phosphatidylinositol-(4,5)-bisphosphate), the kinase Ste20 initiating activation of the MAPK cascade, its interaction partner Ste11 (MAPKKK), the MAPKK Ste7 and, finally, the MAP Kinase Fus327. Many of these binding partners may exist in several different states of activation. Taking into account all possible combinations of membrane-bound and cytosolic Ste5 molecules in complex with one, two, or three of its interaction partners in phosphorylated or unphosphorylated states, as well as interactions with other membrane-bound components, yields local reaction networks equivalent to more than 150 coupled differential equations (ODEs) in the membrane regions and 70 coupled ODEs in the cytoplasmic regions. Diffusional coupling between the membrane and volume elements of the discretized cellular morphology has to be handled additionally. Capturing the large numbers of distinct molecular complexes involved in Fus3 activation is therefore a very difficult task that has been undertaken only a few times 28, 29 using conventional modeling techniques that assemble the components of a mathematical model in a non-automated manner.
Importantly, the shape of the yeast cells as they respond to pheromone stimulation has a significant effect on the signaling events: Saccharomyces cerevisiae responds to pheromone by assuming a polarized pear-like shape and accumulating signaling molecules in its narrow tip, the so-called ‘shmoo’ 27. Recently, a microscopy study elucidated the spatial distribution of the molecular components of the yeast pheromone response with enough quantitative detail to permit spatially-resolved simulations 30. Note that due to the necessity to account for both spatial biochemical inhomogeneity and the influence of intracellular diffusion, a non-spatial rule-based method would require a manual mapping of each one of the automatically generated sets of reactions – describing the biochemistry at one particular point in space – onto the volume elements of the simulated cellular geometry.
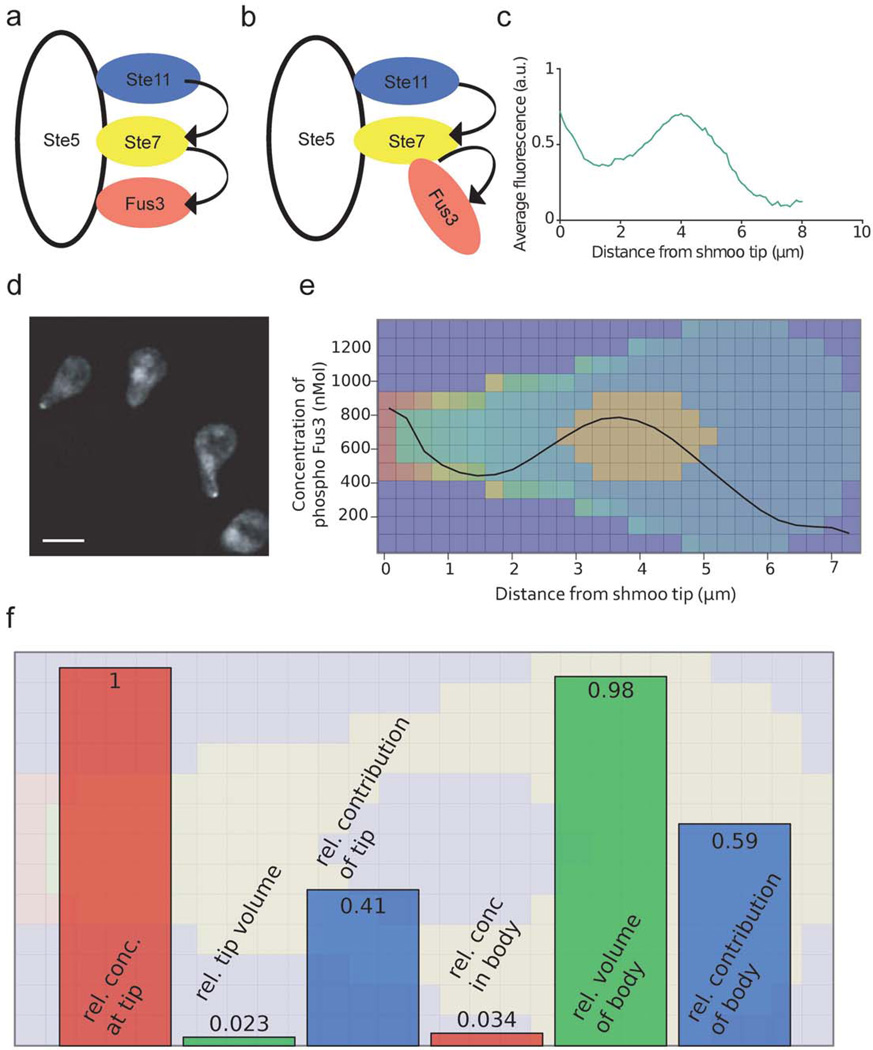
Phosphorylated Ste7 (pSte7) is thought to phosphorylate Fus3 in a Ste5-dependent scaffold-guided process (Fig. 5a). Given the low affinity of Fus3 for Ste5 (Kd ~ 1 micromole 30, 31), the concentration of complexes involving pSte7 and Fus3 bound to Ste5 must be low, posing the question how the cells achieve a phosphorylation of 40% of their Fus3 pool within less than an hour after pheromone stimulation30. After initial simulations, we therefore replaced the classical mechanism of intra-scaffold activation of Fus3 (Fig. 5a), by a mechanism according to which Ste5-bound, activated Ste7 directly binds to and activates Fus3 (Fig. 5b), taking advantage of the fact that the modeling GUI makes even such major structural changes quite painless. This mechanism was suggested in a recent structural study that revealed that binding to Ste5 significantly enhances Ste7 phosphorylation of Fus3 and that this does not require direct interaction of Fus3 with Ste5 31. This finding is in accord with the unexpected previous observation that suppressing the interaction of Fus3 with Ste5 actually promotes Fus3 activation 32.
Figure 5. Experimental and simulated patterns of Fus3 phosphorylation.
a, b) The schematic shows the conventional ‘in-scaffold’ activation scheme (a) for Fus3 and an alternative activation scheme (b) according to which scaffold-bound Ste7 activates Fus3 that is not bound to Ste5. Arrows indicate kinase activity. c) Experimentally reported concentration profile of Fus3 (© Nature Publishing Group, originally published in Nature Cell Biology 9(11):1319–26, 2007, reproduced with permission). d) Yeast cell with Fus3 accumulation in the shmoo after stimulation with pheromone. Scale bar, 5 microns. (© Nature Publishing Group, originally published in Nature Cell Biology 9(11):1319–26, 2007, reproduced with permission). e) Overlay of the simulated concentration profile of phosphorylated Fus3 as a function of the distance from the shmoo tip and color-coded concentration (from red: high to blue: low) within a central plane through the simulated yeast cell. Side length of one square is 0.28 micrometers. f) The schematic shows the relative concentrations of the pFus3 producing complexes at the shmoo tip and in the cell body (red squares), the relative volumes of the two spatial regions (green squares) and the resulting relative contributions to the total pFus3 production (blue squares).
In addition, we included a tip-localized binding partner for pFus3 representing one of the targets of its kinase activity as suggested by the authors of the experimental study 30. This allowed the model to account for an experimentally reported imbalance between Fus3 and Ste5 at the tip and to satisfy an additional constraint provided by the experimental data (the shmoo tip contained 1.7 fold less Ste5 than Fus3) that would have been impossible to satisfy without these Fus3 binding partners (Supplementary Fig. 4). With this second modification, simulations reproduced both the experimentally observed Fus3 concentration and phosphorylation profile (Fig. 5c,d,e) and the phosphorylation of 40% of the total Fus3 pool. As in the original experiments30 the Fus3 concentration profiles were calculated using a spatial averaging over cells with different localizations of the cell nucleus (Supplementary Fig. 5).
Among the advantages of the automated generation of reaction networks is the possibility to identify molecular complexes with common properties. Summing over the concentrations of all complexes containing the ‘Ste5-pSte7-Fus3’ pattern we could very efficiently determine the spatial concentration profile of the complexes that generate pFus3. Even though the concentration of these complexes was much lower in the body than at the shmoo tip, the total number of cytoplasmic sources for pFus3 was greater than those at the tip, since the shmoo tip encompasses only a few percent of the cell’s volume (Fig. 5f). Thus, our spatial rule-based approach was required for the simulation of a complex signaling cascade embedded into its specific geometry and was also very useful for analyzing the consequences of the spatial distribution. In this case, the simulations suggested that the fact that much of the pFus3 was produced in the cell body imposes a limit on the steepness of the gradient that can be produced by the highly concentrated source of this active kinase at the shmoo tip (Supplementary Note 4). Finally, the ability to include spatial aspects into quantitative models of cellular behavior considerably increases the stringency of the constraints provided by experimental data; this should facilitating instructive simulations even in cases where the data are less complete than for the yeast MAPK pathway.
Discussion
In addition to solving long-standing technical modeling issues through its ability to automatically generate spatially resolved reaction-diffusion networks, the approach presented here also offers an opportunity for experimentalists with limited computational experience to utilize modeling and simulation in their studies. As demonstrated in the step-by-step guide through the models, (re-) building, modifying and exploring the models and performing the simulations does not require a strong computational background or the creation of elaborate computer scripts, but instead can be done with the help of an intuitive graphical interface. Theorists will furthermore find it useful that the localized generation of reaction networks performed by our approach permits access to more details of local dynamics than is typically provided by ‘black box’ partial differential equation (PDE) solvers. This makes it straightforward to analyze the different reactions contributing to the kinetics with which local complex concentrations change over time. Finally, computational modelers can use the programming interface provided for all algorithms to create single- or multi-cellular simulations combining biochemical (reaction-diffusion) and morphological dynamics, using the different components of our tool set as building blocks for customized projects.
Methods
All models and the tools described here for generating the reported modeling results can be downloaded from the NIH website at: www.niaid.nih.gov/LabsAndResources/labs/aboutlabs/lsb/Pages/simmuneproject.aspx The tools run on Linux, MacIntosh®, and Windows® operating systems. The models are stored in SQL (Structured Query Language) databases to allow for flexible and efficient organization and for detailed queries about molecular properties and interactions. The database communicates with the simulation software through an API (Application Programming Interface) that can directly be accessed by computational modelers or programmers who want to implement their own simulation engines or supply programmatically generated models to the simulator. Model databases are stored as SQLite dbf files that can be easily exchanged between researchers.
Simulating cellular biochemistry in the context of intercellular receptor-mediated contacts
To illustrate the challenges associated with transitioning from a non-spatial to a spatial representation of a signaling network, we consider a simple model of receptor-mediated cellular communication with the following rules (Supplementary Fig. 1): R1 and R2 are two receptors, each with one extracellular and one intracellular binding site. They can bind to each other extracellularly and can bind to intracellular components, C1 and C2, respectively. C1 can interact with R1 regardless of the receptor’s extracellular binding state whereas C2 can bind to R2 only after this receptor has been bound by R1 (outside-in signaling). R1 can bind to R2 only after C1 has attached itself to intracellular domain of R1 (inside-out signaling). The network thus incorporates outside-in and inside-out signaling mechanisms representing typical binding event-induced changes in the behavior of transmembrane proteins responsible for transmitting and reacting to cellular contact signals 33.
Whereas the biochemical reaction possibilities resulting from the rules in the model can be described easily as a network graph when the geometry of the system is ignored (Supplementary Fig. 1a), taking into account the spatial localization of the network’s molecular components relative to two cytoplasmic membranes of adjacent cells expressing either R1 and C1 or R2 and C2 creates an substantial additional layer of complexity.
To illustrate this we depict components that need to be treated as cytosolic as green for cell 1 and as purple for cell 2. Extracellular membrane-bound components are depicted in blue (Supplementary Fig. 1b,c). C1 and C2 perform free 3D diffusion as unbound molecules. After their association with the transmembrane receptors R1 and R2, they are confined to 2D membrane-guided diffusion. Another important difference between a non-spatial and a spatially-resolved simulation of the model is that in the latter case molecular complexes need to be accounted for in multiple locations simultaneously. As one example, the receptor-receptor association (reaction (2)) creates a complex that is ‘visible’ in five distinct spatial regions: cytoplasm 1, membrane 1, intercellular space, membrane 2 and cytoplasm 2. In a simulation that tracks individual complexes, the interaction possibilities of these intercellular transmembrane complexes would follow naturally from their individual positions in space: complexes that are spatially very close to each other can react (Supplementary Fig. 1c). However, switching to what is usually a far more efficient approach by simulating the dynamics of molecular concentrations in two discretized, grid-like representations of the membranes and adjacent volumes of the two interacting cells means giving up the information about locations of individual molecule complexes (Supplementary Fig. 1d). The algorithms updating the concentrations over time of molecular complexes that are present in multiple adjacent membrane and volume elements thus need to be able to model these interdependencies on the ensemble level (local concentrations of complex species) even though, physically, they arise at the level of individual complexes. For example, the association of C1:R1 and R2 in two adjacent membrane grid elements depicted by the red squares in Fig. 1d increases the concentration of the product complex C1:R1:R2 in the two membrane grid elements and their associated volumes (sub-membrane and intercellular). It decreases the concentration of C1:R1 in the grid element on the left (cell 1) and of R2 in the grid element on the right (cell 2). Thus, while the rate of this reaction depends only on the concentrations of C1:R1 and R2 in the intercellular space hosting the interaction, the formation of the larger complex changes the concentrations of complexes in multiple additional locations.
To address these issues we developed algorithms that construct the computational representations of reaction networks for individual volume (or membrane) elements based on their local geometry and the locally available molecular components, as opposed to using interaction rules to generate a global reaction network and then mapping it onto discretized spatial structures. The latter approach would have difficulties simulating situations in which multimolecular complexes could exist in multiple versions that, while biochemically identical, nonetheless differ with regard to the roles they play within the modeled cellular geometry. The adhesion receptors ICAM-1 and LFA-1, both expressed on interacting T- and B-lymphocytes, are biological examples of this situation. Here, both cells express the equivalents of R1 and R2 (ICAM-1 and LFA-1). The complex ICAM-1:LFA-1 can thus mean two things: ICAM-1 is anchored on the T-cell and LFA-1 on the B-cell, or vice versa. The purely biochemical, theoretical interaction possibilities of the intercellular complex as part of a non-spatial signaling network are identical in both cases but the algorithm simulating the network’s reaction dynamics in space now needs two representations in each grid element with distinct local reaction rules. Using the biochemically simpler system of E-cadherin receptor homodimerization, we discuss in Fig. 1 and Supplementary Note 2a how our approach builds local reaction networks capable of correctly handling such situations by encoding the spatial localization of the components of multi-molecular complexes to be able to dynamically assemble and disassemble complexes connecting multiple distinct spatial regions. This approach solves the above-mentioned problem: components involved in the formation of trans-membrane complexes ‘know’ where they come from, thereby making it straightforward to assign target volume (or membrane) elements to the fragments of dissociating complexes. The information contained in the complexes (origins of their components, list of complex interactions that can generate them, list of decays that disintegrate them into fragments) is then translated into sets of equations encoding the concentration changes (and their kinetic rates) of the complexes in the system (Supplementary Note 3a).
All simulation methods described here are implemented in algorithms that are either accessible through graphical user interfaces or programmatically through application programming interface (API) files. Examples illustrating how to access the reported functionality in custom programs using the algorithms introduced here are provided in the documentation for the modeler/simulator API. The numerical parameters we used are given in Supplementary Note 5.
Generation of location-dependent reaction networks
Reactions between molecular complexes are based on binding possibilities between pairs of molecular binding sites. The rate constants of these reactions may be determined by the complexes to which the interacting molecules belong: Two molecules A and B may have different association (and dissociation) rates depending on whether they interact as mono-molecular ‘complexes’ A and B or are part of larger complexes, for instance, as A bound to C in a complex A:C. These properties of the molecules and the complexes they may form define the purely biochemical network mentioned at the beginning of the Results section. The modeler interface provides the functionality to specify these properties using a graphical interface. Once the computational representations of the cellular morphologies have been generated (see next section), local networks in the volume elements of the spatial discretization are generated based on the molecular complexes that are initially placed into these volume elements (as initial conditions of a particular simulation of the model) and their potential interactions and the complexes they can form. The local networks in each volume element will generate only those complexes that can actually be formed based on the locally available initial biochemical make-up in that element. For instance, if A can bind to B and C but a local volume element contains only A and C, the algorithm will generate a representation of complex A:C but not A:B instead of generating both and just setting the concentration of A:B to zero. This distinction ensures a maximally efficient (sparse) representation of the reaction kinetics. In all membrane locations, the molecular complexes are given additional tags that encode their volume element identity. This allows the algorithm to generate intercellular membrane-membrane complexes and identify which components of a complex (each belonging to one of two distinct membrane elements) they decay to when the binding that mediates the membrane-membrane link (in the E-cadherin example the trans-trans interaction) is lost. With the information about which interactions are possible locally and the location tagging, the algorithm can finally determine all reactions for each single local molecular complex: associations, where two complexes form a larger complex; dissociations, where a complex decays into two smaller ones; and transformations, where a complex changes the state of its molecular components, for instance switching from unphosphorylated to phosphorylated. In spite the overhead resulting from the additional information needed to generate localized spatial networks Simmune’s network generators compare favorably with other approaches for automated network generation (Supplementary Note 3b and Supplementary Figure 6).
Generation of computational representations of cellular morphologies: Finite volume method
Partial differential equations (PDEs) are the continuous mathematical representation of the reaction-diffusion system given by interacting and diffusing molecular complexes. PDEs that describe the spatio-temporal behavior of conserved quantities such as diffusing molecules are amenable to discretization by the finite volume method. The method's central idea is to cover the spatial region on which the PDE is defined with a set of volume elements and express the rate of change of the conserved quantity in those elements in terms of the flux through their surface [Finite Volume Methods Robert Eymard, Thierry Gallouet and Raphaele Herbin October 2006. This manuscript is an update of the preprint n0 97-19 du LATP, UMR 6632, Marseille, September 1997 that appeared in Handbook of Numerical Analysis, P.G. Ciarlet, J.L. Lions eds, vol 7, pp 713–1020]. In the case of diffusion the flux through the surfaces is determined by Fick's law. Cytosolic and extracellular diffusion can be discretized in a natural way by choosing a piecewise constant approximation of concentrations, and an approximation of the gradients through the distances between the centers of the volume elements (these are cubic in the implementation used here). However, the surface elements of the cubic grid elements that cover a simulated cell do not provide an adequate approximation of the cell's surface. The distances and interface lengths entering Fick's law have to be inferred from the cell's shape. This will be explained in the following section.
Generation of modified surface geometry for membrane diffusion
The surface elements of the cubic grid elements that cover a simulated cell provide an inadequate approximation to the cell's surface (cf. Supplementary Note 3d). To illustrate this issue consider a sphere of radius 1. The circumference of that sphere is 2π and its area 4π. In contrast, the circumference of the discretized sphere at the equator is 8, independent of the resolution of the discretization. (Distinct parts of the circumference can be viewed from four directions, perpendicular to the front, back, left and right faces of the cubic grid elements defining the sphere. The visible circumference from each of these four viewpoints is 2 for a total of 8.) The surface area of the discretized sphere converges to 6π for a grid constant approaching 0. By an argument similar to the previous one the area can be viewed from 6 directions. In each direction a circle with area π is approximated by the surfaces of the grid elements, leading to a total area of 6π. Considering these discrepancies between the discretized and ideal sphere it is obvious that a naive approach using the surface of the cubic grid cells to solve the diffusion equation will not yield results converging to the true solution.
This can be remedied by locally adapting the area and shape as well as the distances between surface elements of the cubic grid elements (Supplementary Fig. 7). The approach taken here is based on a method 34 that was designed to provide accurate approximations to idealized smooth surfaces that do not change over the course of a simulation. Our simulation scheme is designed to allow for frequent changes of the surface geometry. This requires rebuilding the surface approximation and redistributing membrane molecules (Supplementary Note 3c and Supplementary Fig. 8) after each change of the surface geometry. We therefore modified the original method to reduce the computational cost for creating the approximation. In numerical tests the error of the much faster modified method was still in the sub-percent range (cf. Supplementary Note 6). The Supplementary Movies 1 and 2 use the surface normals generated by this method to display the curvature of the cell membranes.
Potts Model dynamics
We use a Cellular Potts model (CPM) 23 to simulate dynamic cell morphologies. During each morphology update, which occurs with a user-defined frequency, a subset of the volume elements that are adjacent to the cell's surface are randomly chosen. The energy cost for the extension of the element in a randomly chosen direction and the retraction of the element are evaluated. The change with the lower cost is chosen and will be accepted if the cost is smaller than zero or with a probability exp(-E) for a cost E. A volume element of one cell can extend into a volume element occupied by another cell; the total cost will be the sum of the costs for extending one volume element and retracting the other.
Note that this strategy as implemented here permits the development of models that couple membrane biochemical and morphological dynamics. However, this coupling is phenomenological in the sense that membrane dynamics are not simulated based on biomechanical calculations that describe surface deformations in terms of forces generated, for instance, by actin and myosin fibers that interact with the cell’s membrane. Accordingly, Potts model parameters have to be chosen heuristically to reproduce the experimentally observed dynamics of cellular morphology.
Our CPM utilizes four different energy terms. The relative strengths sx of the terms are tunable as part of a simulation definition. The total cost E is given by
where Ev and Ea are contribution controlling the cost of volume and surface changes, Eb determines the contribution of receptor binding between adjacent membranes and Ec represents a phenomenological term that penalizes small thin protrusions. These terms are defined in detail below. Their proportionality constants have been chosen to result in realistic behavior of the E-cadherin receptor mediated adhesion model for relative strengths close to one.
Additional user defined terms can be added through the programming interface (API), by deriving a new class from the PottsTerm-class and reimplementing its extendCost(Cube * c, DIRECTION d) and retractCost(Cube * c) methods as illustrated by the PottsTermExample class in the smun_PottsTerm.h header file. An implementation example is provided in the file “PottsExample.cpp”.
The volume term models the effects of an impermeable cell membrane. The potential energy of a cell with volume V and equilibrium Volume V0 is proportional to the squared volume difference. The energy cost for osmotic swelling of the cell, given the new Vnew and old Vold cell volumes, is thus
where Ve is the volume of a single volume element, n the number of volume elements, and n0 the number of volume elements at which the cell is in equilibrium.
The Volume V0 is specified as a fraction of the initial cell volume. This is done through the “CellDesigner” user interface. In the case of the E-cadherin model the fraction equals one, thus the cell is in equilibrium at the start of the simulation, with a relative strength sv=1.
The area term models energy cost of changes to the cells surface area. The area change ΔA is approximated by change of the area of the surface of the subset of the cubic grid that is covered by the cell
with Δs=s-s0 being the change of the number of surface elements and Ae the area of a single surface element. In analogy to the Volume term we model the energy cost for extending the membrane out of it's relaxed state by a harmonic potential. The energy cost for changing the morphology is thus
The number of surface elements of the relaxed membrane is specified as a fraction of the initial cell surface analogous to the relaxed volume. Setting the relaxed fraction to a value different from one allows the user to model phenomena such as the opening of membrane reservoirs. The relaxed surface of the cells used in the E-cadherin model equals their initial surface, the area term has a relative strength sa=1/2.
The binding term models the energy cost of adhesion due to the breaking of intercellular bonds due to changes of the morphology. The contribution of each type of bond is proportional to the number of bonds that are broken by the morphology change
where the first sum is over all surface elements that are removed and the second sums over all intercellular bonds. The area of a surface element is given by ASurface and the surface concentration of a bond type by CBond. Only bonds that are marked as trans in the model specification can be used as bonds that enter the adhesion term, the relative strength of the adhesion term is specified together with the other potts term during the setup of a simulation. Naming the binding sites in the components of the interacting molecules specifies bonds. The E-cadherin model has only one type of binding that can enter as a binding term; the trans interaction of the E-cadherin molecules, which is given a relative strength sb=2.
The bending resistance of the cell membrane is modeled by a phenomenological term that depends only on the number of direct neighbors of the changing volume element. The relative cost for extending a volume element is Erel={1, 0, −1, 10, −25} with one to five neighbors. This penalizes the formation of small, thin extensions while it favors closing of small invaginations. The cost terms for retracting a volume element have the opposite cost favoring the retraction of a small protrusion while penalizing the formation of invaginations. The asymmetric nature of the cost terms allow the cell to sample its surroundings while keeping the cell's surface from becoming very rough. The only parameter of this purely local term is its relative contribution to the total cost E. For the E-cadherin model we chose the relative contribution sc=1/2.
Supplementary Material
Acknowledgements
The authors are grateful for helpful comments from R. Schwartz, R. Varma, A. Nita-Lazar, I. Fraser, J. Tsang, and D. Cioffi as well as for insightful discussions with the members of the Laboratory for Systems Biology (LSB) at the National Institute of Allergy and Infectious Diseases. The authors also acknowledge comments from the (anonymous) reviewers that helped improving the clarity of the manuscript. The authors thank A. Meier-Schellersheim for creating Figure 2 and Supplementary Figure 1. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases of the National Institutes of Health.
Glossary
- MAPK
Mitogen-activated protein kinase
Footnotes
Author contributions
B.A. wrote the application for designing cellular morphologies. A.D.G wrote the simulator GUI. M.M.S. and F.K. developed the volume/membrane spatial discretization. B.A. developed the adaptive membrane diffusion discretization. B.A. and T.P. performed numerical tests. F.Z. wrote the application for defining molecular interactions. M.M.S. developed the spatially adaptive network generator. B.A. and M.M.S. developed the integrator drivers. All authors discussed the project on an ongoing basis and wrote the manuscript.
References
- 1.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 2.Kholodenko BN. Four-dimensional organization of protein kinase signaling cascades: the roles of diffusion, endocytosis and molecular motors. J Exp Biol. 2003;206:2073–2082. doi: 10.1242/jeb.00298. [DOI] [PubMed] [Google Scholar]
- 3.Delon J, Germain RN. Information transfer at the immunological synapse. Curr Biol. 2000;10:R923–R933. doi: 10.1016/s0960-9822(00)00870-8. [DOI] [PubMed] [Google Scholar]
- 4.Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 5.Brown MD, Sacks DB. Protein scaffolds in MAP kinase signalling. Cell Signal. 2009;21:462–469. doi: 10.1016/j.cellsig.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hlavacek WS, Faeder JR, Blinov ML, Perelson AS, Goldstein B. The complexity of complexes in signal transduction. Biotechnol Bioeng. 2003;84:783–794. doi: 10.1002/bit.10842. [DOI] [PubMed] [Google Scholar]
- 7.Meier-Schellersheim M, et al. Key role of local regulation in chemosensing revealed by a new molecular interaction-based modeling method. PLoS Comput Biol. 2006;2:e82. doi: 10.1371/journal.pcbi.0020082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hlavacek WS, et al. Rules for modeling signal-transduction systems. Sci STKE. 2006;2006:re6. doi: 10.1126/stke.3442006re6. [DOI] [PubMed] [Google Scholar]
- 9.Lok L, Brent R. Automatic generation of cellular reaction networks with Moleculizer 1.0. Nat Biotechnol. 2005;23:131–136. doi: 10.1038/nbt1054. [DOI] [PubMed] [Google Scholar]
- 10.Feret J, Danos V, Krivine J, Harmer R, Fontana W. Internal coarse-graining of molecular systems. Proc Natl Acad Sci U S A. 2009;106:6453–6458. doi: 10.1073/pnas.0809908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koschorreck M, Gilles ED. ALC: automated reduction of rule-based models. BMC Syst Biol. 2008;2:91. doi: 10.1186/1752-0509-2-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mallavarapu A, Thomson M, Ullian B, Gunawardena J. Programming with models: modularity and abstraction provide powerful capabilities for systems biology. J R Soc Interface. 2009;6:257–270. doi: 10.1098/rsif.2008.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varma R, Campi G, Yokosuka T, Saito T, Dustin ML. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity. 2006;25:117–127. doi: 10.1016/j.immuni.2006.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Zon JS, ten Wolde PR. Green's-function reaction dynamics: a particle-based approach for simulating biochemical networks in time and space. J Chem Phys. 2005;123:234910. doi: 10.1063/1.2137716. [DOI] [PubMed] [Google Scholar]
- 15.Andrews SS, Addy NJ, Brent R, Arkin AP. Detailed simulations of cell biology with Smoldyn 2.1. PLoS Comput Biol. 2010;6:e1000705. doi: 10.1371/journal.pcbi.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 17.Sivasankar S, Zhang Y, Nelson WJ, Chu S. Characterizing the initial encounter complex in cadherin adhesion. Structure. 2009;17:1075–1081. doi: 10.1016/j.str.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavey M, Lecuit T. Molecular bases of cell-cell junctions stability and dynamics. Cold Spring Harb Perspect Biol. 2009;1:a002998. doi: 10.1101/cshperspect.a002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Sivasankar S, Nelson WJ, Chu S. Resolving cadherin interactions and binding cooperativity at the single-molecule level. Proc Natl Acad Sci U S A. 2009;106:109–114. doi: 10.1073/pnas.0811350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Troyanovsky S. Cadherin dimers in cell-cell adhesion. Eur J Cell Biol. 2005;84:225–233. doi: 10.1016/j.ejcb.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Adams CL, Chen YT, Smith SJ, Nelson WJ. Mechanisms of epithelial cell-cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin-green fluorescent protein. J Cell Biol. 1998;142:1105–1119. doi: 10.1083/jcb.142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez TD, Tamada M, Sheetz MP, Nelson WJ. Immediate-early signaling induced by E-cadherin engagement and adhesion. J Biol Chem. 2008;283:5014–5022. doi: 10.1074/jbc.M705209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glazier JA, Graner F. Simulation of the differential adhesion driven rearrangement of biological cells. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1993;47:2128–2154. doi: 10.1103/physreve.47.2128. [DOI] [PubMed] [Google Scholar]
- 24.Beltman JB, Maree AF, Lynch JN, Miller MJ, de Boer RJ. Lymph node topology dictates T cell migration behavior. J Exp Med. 2007;204:771–780. doi: 10.1084/jem.20061278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Moreno M, Jamora C, Fuchs E. Sticky business: orchestrating cellular signals at adherens junctions. Cell. 2003;112:535–548. doi: 10.1016/s0092-8674(03)00108-9. [DOI] [PubMed] [Google Scholar]
- 26.Hong S, Troyanovsky RB, Troyanovsky SM. Spontaneous assembly and active disassembly balance adherens junction homeostasis. Proc Natl Acad Sci U S A. 2010;107:3528–3533. doi: 10.1073/pnas.0911027107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dohlman HG, Thorner JW. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu Rev Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- 28.Kofahl B, Klipp E. Modelling the dynamics of the yeast pheromone pathway. Yeast. 2004;21:831–850. doi: 10.1002/yea.1122. [DOI] [PubMed] [Google Scholar]
- 29.Shao D, Zheng W, Qiu W, Ouyang Q, Tang C. Dynamic studies of scaffold-dependent mating pathway in yeast. Biophys J. 2006;91:3986–4001. doi: 10.1529/biophysj.106.081661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maeder CI, et al. Spatial regulation of Fus3 MAP kinase activity through a reaction-diffusion mechanism in yeast pheromone signalling. Nat Cell Biol. 2007;9:1319–1326. doi: 10.1038/ncb1652. [DOI] [PubMed] [Google Scholar]
- 31.Good M, Tang G, Singleton J, Remenyi A, Lim WA. The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell. 2009;136:1085–1097. doi: 10.1016/j.cell.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhattacharyya RP, et al. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science. 2006;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- 33.Evans R, et al. Integrins in immunity. J Cell Sci. 2009;122:215–225. doi: 10.1242/jcs.019117. [DOI] [PubMed] [Google Scholar]
- 34.Novak IL, et al. Diffusion on a Curved Surface Coupled to Diffusion in the Volume: Application to Cell Biology. J Comput Phys. 2007;226:1271–1290. doi: 10.1016/j.jcp.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.