Abstract
Pyrazinamide (PZA) is a first-line antitubercular drug known for its activity against persistent Mycobacterium tuberculosis bacilli. We set out to systematically determine the PZA susceptibility profiles and mutations in the pyrazinamidase (pncA) gene of a collection of multidrug-resistant tuberculosis (MDR-TB) clinical isolates and PZA-resistant (PZAr) spontaneous mutants. The frequency of acquired resistance to PZA was determined to be 10−5 bacilli in vitro. Selection at a lower concentration of PZA yielded a significantly larger number of spontaneous mutants. The methodical approach employed allowed for determination of the frequency of the PZAr phenotype correlated with mutations in the pncA gene, which was 87.5% for the laboratory-selected spontaneous mutants examined in this study. As elucidated by structural analysis, most of the identified mutations were foreseen to affect protein activity through either alteration of an active site residue or destabilization of protein structure, indicating some preferential mutation site rather than random scattering. Twelve percent of the PZAr mutants did not have a pncA mutation, strongly indicating the presence of at least one other mechanism(s) of PZAr.
INTRODUCTION
Pyrazinamide (PZA) was identified based on its structural activity relationship to nicotinamide, known for its antitubercular properties (28). PZA has the distinction of being identified directly in vivo, first in Mycobacterium tuberculosis-infected mice and guinea pigs and later in clinical cases (15, 33, 62). The in vitro activity of PZA was demonstrated subsequently, using media adjusted to a pH of 5.0 or 5.5 (36, 51, 66) because PZA has no detectable inhibitory activity against replicating bacilli at neutral pH.
PZA differs from most antitubercular drugs in having sterilizing activity on semidormant (persistent) M. tuberculosis bacilli. Its combination with rifampin and isoniazid in the standard tuberculosis (TB) treatment has reduced the duration of therapy to 6 months instead of the previous 9 to 12 months for otherwise healthy patients (65). PZA, like isoniazid and ethionamide, is a prodrug which must be converted into its active form for activity (12, 64). This enzymatic activation of PZA is catalyzed by the pyrazinamidase (PZase) encoded by the pncA gene in M. tuberculosis (54), and the active metabolite is pyrazinoic acid (POA). Interestingly, POA is active against PZA-resistant (PZAr) isolates in vitro but displays no in vivo activity (19). The overall mode of action of PZA is rather unusual and remains poorly understood. No specific target has yet been identified for either PZA or POA, although a recent report indicating a possible interference in the trans-translation pathway shows a potential promising mechanism of action (57). The fatty acid synthase (FAS I) was proposed and challenged as a possible target of PZA and analogs (6, 68). This mechanism of action proved to be valid in vitro but not in vivo. Although the final target of activated PZA has yet to be found, a model for the mechanism of action has been proposed: PZA crosses the mycobacterial cell wall by passive diffusion and is converted by the PZase in the cytoplasm into POA, which is then released through either passive diffusion or a weak efflux pump (65). If the extracellular medium presents an acidic pH, the acidic POA form is protonated in part into an uncharged HPOA form, which is easily reabsorbed by the cell and redissociated intracellularly, releasing H+ protons into the cytoplasm. Because the efflux mechanism of POA is inefficient or defective in M. tuberculosis, HPOA accumulates in the cytoplasm of the bacterium and causes cellular damage, resulting in cell death due to intracellular acidification. More recently, Shi and colleagues proposed that HPOA inhibits ribosomal protein S1 (RpsA). Inhibition of RpsA required for the trans-translation pathway leads to a decrease in stalled ribosome rescue and possibly an increase in the accumulation of toxic peptide waste (57). This model could explain the atypical characteristics of pyrazinamide, particularly its activity only at an acidic pH.
The PZase is a small protein of 186 amino acids that is encoded by the pncA gene. Mutations of the pncA gene or its putative promoter region are associated with most reported cases of PZA resistance in M. tuberculosis (54). Multiple mutations (substitutions, deletions, and insertions) have been described for this gene-promoter region (49, 52, 54, 59). A PZase-deficient strain can no longer metabolize the prodrug, resulting in PZAr (54), an observation first reported by Konno and coworkers in the early 1960s (25); the relationship was confirmed through quantification of PZase activity (8). As such, Mycobacterium bovis strains are intrinsically resistant to PZA due to a distinctive phylogenetic single nucleotide polymorphism (SNP) (57His → Asp [C169G]) of pncA resulting in an inactive PncA protein and hence in PZAr (54). The number of reported pncA mutants associated with PZAr varies from 70 to 100% (9, 17, 21, 22, 29, 58, 59), taking into account that PZA susceptibility testing has proven challenging (16).
Here we investigated the frequency of mutations in the pncA gene associated with PZAr in a collection of well-characterized M. tuberculosis clinical isolates comprising a 14-year complete capture of multidrug-resistant (MDR) isolates and PZAr spontaneous mutants. The correlations between PncA mutations, drug susceptibility, and structural analysis of the PncA protein were determined for selected PZAr mutants. Most notably, the frequency of spontaneous acquired resistance to PZA was determined and found to be concentration dependent.
MATERIALS AND METHODS
M. tuberculosis clinical isolates.
One hundred thirty-eight of 174 strains were selected from the MDR-TB collection maintained at the Tuberculosis & Mycobacteria Centre of the Scientific Institute of Public Health, Belgium. This collection of 174 isolates comprises the first isolate from each MDR-TB patient identified in Belgium between 1994 and 2008. Twenty-three isolates were eliminated from the study because of poor growth, contamination, or nonviability (unable to confirm drug susceptibility testing [DST] results). A further 13 isolates with the PZAr phenotype but carrying wild-type pncA were also eliminated from the study, as the DST results could not be reconfirmed due to accidental elimination of the isolates. The remaining 138 samples were included in the study regardless of PZA susceptibility profile. All were genotyped by spoligotyping and mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) typing (24 loci) in order to establish their genetic diversity (unrelated clinical isolates) or relatedness (clustered clinical isolates).
The resistance profiles of the clinical isolates for first- and second-line antibiotics were determined on solid medium by the proportion method of Canetti et al. (10) or in liquid medium in a radiometric Bactec 460 TB system (Becton, Dickinson Microbiology Systems, Cockeysville, MD) (since 2000) according to the manufacturer's instructions and the methods of Pfyffer et al. (47). Since 2005, a Bactec MGIT960 system has been used for DST of isoniazid, rifampin, and ethambutol, and it was expanded to PZA susceptibility testing in 2007 with the commercial availability of a PZA drug kit (Becton, Dickinson Microbiology Systems, Cockeysville, MD). Prior to 2007, all DST for PZA was performed by the method of Canetti et al. (10). DST confirmation of phenotypes was carried out by use of a Bactec MGIT960 PZA drug kit. MIC determination was performed by using this PZA drug kit, with the PZA provided in the kit diluted in a 1/4 volume (400 μg/ml) and subjected to 2-fold dilutions. The PZA susceptibilities of 15 samples (1 clinical isolate and 14 spontaneous mutants) displaying the PZAr phenotype while carrying wild-type pncA and rpsA were reconfirmed by the method described above.
Selection of spontaneous PZAr M. tuberculosis mutants.
Spontaneous mutants resistant to PZA were selected on 7H11 plates containing either 100 or 500 μg/ml of PZA (Sigma-Aldrich) at pH 6.0. The protocol for selection of spontaneous mutants was adapted from the work of Luria and Delbruck, Mathys et al., and Morlock et al. (32, 35, 40). Briefly, a single colony of strain CDC1551 was cultured under standard conditions in 7H9 medium. At 3 weeks, the culture density was determined, adjusted to an optical density at 600 nm (OD600) of ∼0.02 (approximately 1 × 105 to 1 × 106 CFU/ml), and divided into 40 individual culture flasks (25 ml in each) in the absence of PZA or any other selective pressure. At day 32 postinoculation, the 40 cultures were adjusted to an OD600 of 1, and the cell density for 4 random samples was confirmed by serial dilution and plating for enumeration. All cultures were plated on 7H11 plates containing either 100 or 500 μg/ml PZA (pH 6.0), at concentrations of 107, 105, and 103 CFU/ml. Resistant colonies were picked and subcultured on 7H11 plates, pH 6.0, containing the same concentration used for the selection process, followed by stocking and DNA extraction. A maximum of 3 colonies per plate were picked and labeled according to the plate number for this study. A total of 112 colonies were sequenced. Statistical analysis was carried out using GraphPad Prism software. P values of <0.05 were considered significant.
Sequencing of drug target regions.
The sense and antisense strands of the pncA gene and the corresponding 100 nucleotides (nt) upstream were sequenced for all isolates, using the primers P6 (5′-−101CGCTTGCGGCGAGCGCTCCA−81-3′) and P1 (5′-+36GGTCATGTTCGCGATCGT+56-3′) (adapted from reference54). Nucleotide sequences were analyzed by using Sequencher software (Gene Codes Corporation).
Structural analysis and evaluation of mutation effects.
The X-ray structure of the M. tuberculosis PncA enzyme was recently determined at 2.2-Å resolution (46) (Protein Data Bank entry 3PL1). This three-dimensional (3D) structure was used to estimate the impact of selected point mutations on structure and/or function of the enzyme. The prediction of changes in thermal protein stability for each observed PncA mutant was obtained from the CUPSAT (44) (http://cupsat.tu-bs.de) and PoPMuSiC (13) (http://babylone.ulb.ac.be/popmusic/) websites. Both programs evaluate the change in free energy of the protein folding-unfolding process upon mutations, i.e., the ΔΔG. A positive or negative ΔΔG value indicates that the mutation is thermodynamically stabilizing or destabilizing, respectively, while the magnitude of ΔΔG indicates the extent of the alteration. Since mutations of a buried residue generally have more drastic consequences on protein structure, the solvent accessibility of mutated residues was calculated using the CUPSAT program.
RESULTS
Phenotypic resistance to PZA.
Among the 138 clinical MDR isolates evaluated for PZA susceptibility, 60 proved to be PZA resistant and 78 were found to be susceptible. PZA susceptibility testing by use of a Bactec MGIT960 PZA drug kit was used to repeat DST for all isolates with borderline resistance or unexpected phenotypic-genotypic or clustering correlations. Since the MDR collection comprised 80.6% of all positive cultures identified in Belgium in the period 1994 to 2008 and reached 95.5% in 2003 to 2008, this 43% PZAr observation for the 138 isolates is clinically significant.
Frequency of spontaneous mutations conferring pyrazinamide resistance on M. tuberculosis.
Samples originating from 40 individual drug-free CDC1551 cultures were plated in parallel on 7H11 plates (adjusted to a pH of 6.0) containing 100 or 500 μg/ml of PZA in order to select for PZAr spontaneous mutants. The frequencies of spontaneous mutants with resistance to PZA determined for all 40 flasks were found to be highly consistent, at ∼1 and 1.5 mutants per 105 bacilli for selection on 100 and 500 μg/ml PZA, respectively. This difference in mutation frequency is statistically significant (P < 0.001) (see Table S1 in the supplemental material).
Polymorphism in the pncA gene and its correlation with PncA structure and activity.
The pncA gene and corresponding ∼100 nucleotides upstream were sequenced for the 151 MDR-TB clinical isolates and 112 PZAr spontaneous mutants selected in vitro (43 selected on 100 μg/ml PZA and 69 selected on 500 μg/ml PZA). In total, the pncA gene and putative promoter region were sequenced for 263 samples. Thirteen of the 151 clinical samples had to be removed from the study postsequencing, as the original cultures were unavailable for DST reconfirmation.
The genetic analysis showed that 98.3% (59/60 isolates) of the Belgian MDR clinical isolates with the PZAr phenotype presented a mutation in the pncA gene. We found that 1.7% (1/60 isolates) of the PZAr MDR isolates carried wild-type pncA and its flanking region. A total (PZAr and PZAs) of 41 different amino acid changes, 3 protein truncations, and 5 frameshifts were observed, including 8 mutations previously not reported in the literature: Asp8Ala, Phe13Leu, Tyr64Ser, Glu107stop, Ala143Pro, Leu172Arg, and frameshifts starting in codons 55 and 82. For clarity and analytical purposes, samples were grouped according to mutation type and fingerprint cluster (Table 1), shared mutation type and strain diversity (Table 2), or susceptible isolates carrying mutations within pncA (Table 3). In this study, all cases with a shared mutation type also grouped genotypically (Table 1). Demographic data showed that the isolates in clusters were obtained from patients from the same country of origin (or residing there) (clusters 1-Georgia, 2-Chechnya, 4-Rwanda, 5-Belgium, and 6-Rwanda) or from patients living together or in proximity of each other (clusters 1, 2, and 5). It is noteworthy that one cluster of two isolates shared the same SNPs within pncA yet one was resistant and the other was susceptible to PZA. The implicated mutation, Met175Ile, has been described previously in the literature on PZAr strains. The phenotypes were confirmed in two additional independent experiments with a Bactec MGIT960 PZA drug kit. Finally, 5 different pncA SNPs were identified in 7 susceptible isolates, with 4 of these SNPs previously correlated with drug resistance in the literature (Table 3). Given the incongruence in results for these isolates, the sequence and susceptibility profile were retested by use of the Bactec MGIT960 system and 100 μg/ml PZA and confirmed the original susceptibility result. In addition, the MIC was determined by use of 2-fold dilutions of PZA (12.5 μg to 200 μg/ml) and the Bactec MGIT960 system and was found to be >200 μg/ml or between 100 and 200 μg/ml. Three of these mutants carry a Cys14Gly substitution which was also observed in one PZAr isolate.
Table 1.
Clustered clinical isolates sharing the same genotypes and pncA mutationsd
| Strain ID | PZA susceptibilitya | nt polymorphism | aa changeb | MIRU type | Spoligotype | Familyc | SIT groupc | Cluster |
|---|---|---|---|---|---|---|---|---|
| 01MY0495 | R | G436A | A146T | 335247232234425113323832 | 034777777420771 | ND | ND | 1 |
| 06MY0899 | R | G436A | A146T | 335247232234425113323832 | 034777777420771 | ND | ND | 1 |
| 04MY1499 | R | A146C | D49A | 244233352644425173353723 | 000000000003771 | Beijing | SIT1 | 2 |
| 08MY1099 | R | A146C | D49A | 244233352644425173353723 | 000000000003771 | Beijing | SIT1 | 2 |
| 95MY0609 | R | Del C530 | FS | 234125132234425113333732 | 777777404760731 | LAM7_TUR | SIT1261 | 3 |
| 96MY0658 | R | Del C530 | FS | 234125132234425113333732 | 777777404760731 | LAM7_TUR | SIT1261 | 3 |
| 01MY1170 | R | A128C | H43P | 225233332244225153343822 | 777777777760731 | T2 | SIT52 | 4 |
| 06MY0371 | R | A128C | H43P | 225233332244225153343822 | 777777777760731 | T2 | SIT52 | 4 |
| 01MY0507 | R | T515G | L172R | 223263342334425143233613 | 777777777760771 | T1 | SIT53 | 5 |
| 03MY0423 | R | T515G | L172R | 223263342334425143233613 | 777777777760771 | T1 | SIT53 | 5 |
| 06MY0096 | R | T515G | L172R | 223263342334425143233613 | 777777777760771 | T1 | SIT53 | 5 |
| 03MY1173 | R | G525A | M175I | 225233332244225153343822 | 777777777760731 | T2 | SIT52 | 6 |
| 05MY1333 | S | G525A | M175I | 225233332244225153343822 | 777777777760731 | T2 | SIT52 | 6 |
R, resistant; S, susceptible.
FS, frameshift. Phenotypic determination was reconfirmed in 3 independent experiments for the M175I substitution.
SIT, spoligo-international type; ND, not determined.
There was an epidemiological link within every cluster identified.
Table 2.
Strains with distinct genotypes but common pncA mutations
| Strain ID | PZA susceptibility | nt polymorphism | aa change | MIRU type | Spoligotype | Family | SIT group | Mutation cluster |
|---|---|---|---|---|---|---|---|---|
| 02MY1010 | R | A188C | D63A | 223273342334425143233613 | 777777777760771 | T1 | SIT53 | 1 |
| 97MY0936 | R | A188G | D63G | 224233331334225153332422 | 777777743760771 | LAM10 | SIT61 | 1 |
| 08MY1582 | R | G22A | D8N | 2242333224322261433324?2 | 777777777760771 | T1 | SIT53 | 2 |
| 96MY0316 | R | G22A | D8N | 243243332234425123333832 | 700076777760771 | X3 | SIT92 | 2 |
| 97MY0555 | R | G22T | D8Y | 224253122334225153335522 | 777777770000771 | U | SIT602 | 2 |
| 95MY0036 | R | C211A | H71N | 224223422424225143333422 | 757777037760771 | ND | ND | 3 |
| 02MY0668 | R | C211T | H71Y | 223235332534425251334432 | 777777777720771 | Haarlem3 | SIT50 | 3 |
| 03MY0478 | R | C211T | H71Y | 244233362644425153353623 | 000000000003771 | Beijing | SIT1 | 3 |
| 03MY0092 | R | C244G | H82D | 224214132324116152532722 | 775777606060731 | LAM11_ZWE | SIT1549 | 4 |
| 08MY1150 | R | A245G | H82R | 244233352644425153353823 | 000000000003371 | Beijing | SIT265 | 4 |
| 01MY0507 | R | T515G | L172R | 223263342334425143233613 | 777777777760771 | T1 | SIT53 | 5a |
| 02MY0982 | R | T515G | L172R | 233245332434422153333732 | 777777777720771 | Haarlem3 | SIT50 | 5 |
| 96MY0800 | R | A29C | Q10P | 223245332634425153233532 | 777741777720671 | ND | ND | 6 |
| 99MY0820 | R | C28T | Q10Stop | 225233332244225153343822 | 777777777760731 | T2 | SIT52 | 6 |
| 01MY0222 | R | C260T | T87M | 224223322224225143324422 | 777777777760731 | T2 | SIT52 | 7 |
| 99MY1429 | R | C260T | T87M | 224223322224225143324412 | 777777777760771 | T1 | SIT53 | 7 |
| 02MY1134 | R | T464G | V155G | 244212232424126163332222 | 601775607760771 | LAM9 | SIT1545 | 8 |
| 08MY1426 | R | T464G | V155G | 243233352644425173353723 | 000000000003771 | Beijing | SIT1 | 8 |
| 05MY1333 | S | G525A | M175I | 225233332244225153343822 | 777777777760731 | T2 | SIT52 | 9a |
| 06MY0999 | R | A523G | M175V | 244233352644425153353823 | 000000000003371 | Beijing | SIT265 | 9 |
Cluster 5, family T1, and cluster 9, family T2, include 3 and 2 samples, respectively.
Table 3.
PZA-susceptible clinical isolates carrying mutations within the pncA gene
| Strain ID | nt polymorphism | aa change | MIRU type | Spoligotype | Family | SIT groupa |
|---|---|---|---|---|---|---|
| 02MY1182 | T40G | C14G | 225233332234225154343723 | 777777777760731 | T2 | SIT52 |
| 09MY0190 | T40G | C14G | 234243162623235152332522 | 777777777760731 | T2 | SIT52 |
| 09MY0191 | T40G | C14G | 234243162623235152332522 | 777777777760731 | T2 | SIT52 |
| 06MY0826 | A191C | Y64S | 244233352644425173353723 | 000000000003771 | Beijing | SIT1 |
| 05MY1333 | G525A | M175I | 225233332244225153343822 | 777777777760731 | T2 | SIT52 |
| 08MY1755 | A403C | T135P | 2242421626?4225153332522 | 777777777760731 | T2 | SIT52 |
| 01MY1015 | G427A | A143T | 223245332434415153233732 | 777737777720171 | ND | ND |
SIT, spoligo-international type.
In order to better understand the phenotypic-genotypic correlation of the isolates, CDC1551-derived PZAr spontaneous mutants were selected at 100 and 500 μg/ml PZA in 7H11 medium adjusted to pH 6.0. Globally, 87.5% (98/112 mutants) of the PZAr mutants presented a pncA mutation, but when the mutants were segregated into those selected at 100 and 500 μg/ml PZA, we found that only 67% (29/43 mutants) of the former were mutated, whereas all of the strains (69/69 mutants) selected on 500 μg/ml PZA displayed a pncA or putative promoter mutation (see Table S2 in the supplemental material). The MICs of the 14 in vitro mutants without any mutation in the pncA region were determined by use of 2-fold dilutions of PZA (50 to 400 μg/ml) and the Bactec MGIT960 method, and all were found to be >400 μg/ml. All 14 spontaneous mutants were subjected to whole-genome sequencing and found to carry a single SNP each. Most isolates were found to carry different mutations, but they were all within the same operon. These data will be communicated at a later date.
Twelve and 24 amino acid alterations were found uniquely in the 100- and 500-μg/ml PZA selections, respectively, while 5 were shared between the 100- and 500-μg/ml selections. Comparison of amino acid mutations identified in clinical isolates with those determined for the spontaneous mutants showed 9 polymorphisms and 17 codons in common. No polymorphisms were identified in putative promoter regions of the clinical isolates; however, a Del−4A mutation and −6G → C and −7T → C substitutions were found in the spontaneous mutants selected at 500 μg/ml of PZA. The MICs for 2 of the isolates (Del−4A and Del−7C) carrying alterations within the putative promoter region were determined, and both were found to be resistant to >400 μg/ml of PZA with the MGIT960 system. Reverse transcription-PCR (RT-PCR) to evaluate the expression of the gene was not performed.
In spontaneous mutants, a mutation in codon 1 was the most frequently encountered mutation. The Met1Ile mutation was present in 10% (3/29 mutants) of the mutants selected on 100 μg/ml PZA and presenting a pncA mutation. Among the 69 mutants selected on 500 μg/ml PZA, 10 Met1Ile and 2 Met1Thr changes were encountered (17% of isolates). The second most frequently encountered mutated codon in the PZAr spontaneous mutants was codon 8, represented by 7 isolates (4 Asp8Glu, 1 Asp8Asn, 1 Asp8Tyr, and 1 frameshift mutant) of the 69 mutants selected on 500 μg/ml PZA (10% of isolates).
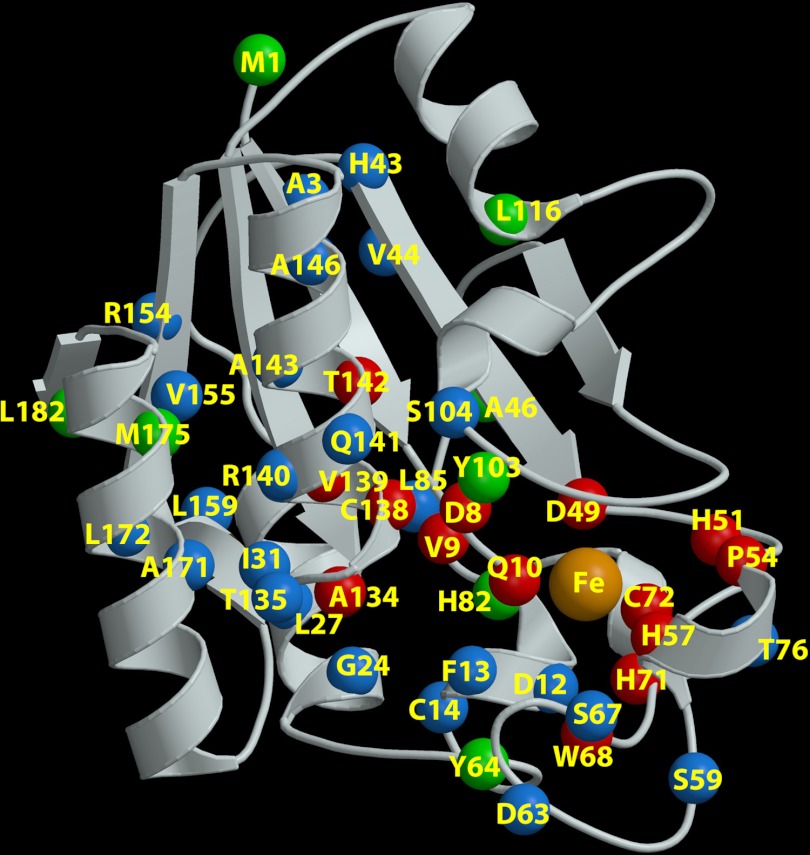
We analyzed the recently published high-resolution structure of the Mycobacterium tuberculosis PncA protein (46) coupled with predictions of protein stability using both the PoPMuSiC (13) and CUPSAT (44) programs. Table S2 in the supplemental material lists the observed mutation types in this study and the corresponding stabilizing/destabilizing effects on the PncA structure, while Fig. 1 illustrates the dispersion and positions of the mutations in a three-dimensional model of the PZase. The data indicate that mutations of amino acid residues associated with either iron or substrate binding or the catalytic active site are most directly implicated in resistance. Most of the mutations in Table S2 affect buried amino acids and were determined to be energetically destabilizing, suggesting a reduction of protein stability and thereby a diminished or depleted PZase activity. Substitutions at codons 46 and 64 (46Ala → Val and 64Tyr → Ser) were predicted to be stabilizing, while substitutions at positions 82, 87, 103, and 175 (82His → Asp, 82His → Arg, 87Thr → Met, 103Tyr → His, and 175Met → Ile) were found to be neutral. All except for the 46Ala → Val and 103Tyr → His substitutions were found in clinical isolates. The 175Met → Ile mutation, which is predicted to be neutral in the 3D model, was also found in both PZA-resistant and -susceptible strains in this study (Tables 1 and 3).
Fig 1.
3D structure of pyrazinamidase showing that mutations cover the entire protein structure. The ribbon is shown in cream. The α-carbon atom of each mutated residue is shown as a colored ball. Green balls indicate neutral or stabilizing mutations (M1, A46, Y64, H82, T87, Y103, L116, T135, M175, and L182); the green residues A46, H82, and Y103 are in the vicinity of the active site, which could correlate with the loss of enzyme activity. Blue balls indicate destabilizing residues (A3, D12, F13, C14, G24, L27, I31, L35, H43, V44, S59, D63, S67, W68, T76, L85, S104, T135, D136, R140, Q141, A143, A146, R154, V155, L159, A171, and L172). Red balls indicate residues located in the active site pocket or involved in chelating the Fe atom (orange) (D8, V9, Q10, D49, H51, P54, H57, W68, H71, C72, G132, A134, C138, V139, and T142). Note that 4 α-carbon atoms (residues L35, T87, G132, and D136) are not visible, as they are behind the ribbon (on the bottom), and therefore they are not labeled in the figure. The figure was produced by successively using the MolScript (26) and Raster3D (37) programs.
DISCUSSION
Pyrazinamide is an essential first-line antitubercular drug used in TB treatment, yet much remains to be understood in regard to its biology and mechanisms of action. It has the distinct and unique characteristic that it was first discovered in an in vivo screen of nicotinamide derivatives in a structure-activity relationship study in 1948 (27). Today we know that PZA is a prodrug activated by the PZase encoded by pncA, a nonessential gene in M. tuberculosis (54). Like the case for some other nonessential target genes, mutations of pncA can often be deleterious for enzymatic activity, either due to a complete loss of function through a frameshift or truncation or due to SNPs in essential amino acid residues involved in catalysis, located at substrate or metal binding sites, or important for protein structure and stability. Surprisingly, PZAr isolates carrying pncA mutations do not seem to be associated with a loss of bacterial fitness, either in vivo or in vitro, and while no selective pressure for this gene has been identified, little evidence of genetic drift has been reported. The lack of detectable effect on the overall fitness of the organism might be due to the fact that PZase is important in the recycling pathway of NAD, not in its synthesis.
In this study, MDR isolates captured over a 14-year period were sequenced for the pncA gene and flanking sequence, regardless of PZA susceptibility profile. Interestingly, 43% of the MDR strains analyzed proved to be additionally resistant to PZA. This is in agreement with the results of similar studies from Japan (53%) (2), Thailand (49%) (22), and South Africa (52.1%) (41). A study from India showed ∼30.4% PZAr in MDR strains, and one from Lisbon, Portugal, showed 82.7% (48/58 isolates) PZAr in MDR isolates; however, the Portuguese isolates were also highly genotypically clustered and likely associated with outbreak situations (45, 60). Interestingly, one study showed ∼6% PZAr in otherwise pan-susceptible isolates (22).
The type and distribution of observed mutations were in agreement with previously published data and the data in the TBDReaMDB database (http://www.tbdreamdb.com) (52). Comparison of mutations identified in clinical isolates with those determined from the spontaneous mutants showed 16 substitutions or sites of polymorphism in common (see Table S2 in the supplemental material). All 16 have also been reported previously in the literature, further underlining the probable functional importance of these amino acids, as previously described by Scorpio and coworkers (53). Mutations in the putative promoter region have also been reported widely. The most common promoter mutation is −11A → G, which has been reported to occur in phylogenetically diverse strains (4, 11, 17, 18, 23, 29, 34, 38, 43, 48, 59). Other putative promoter mutations include −12T → C (41, 59), −10T → C (41), and −7T → C (43). In this study, no polymorphisms were identified in putative promoter regions of the clinical isolates; however, a Del−4A mutation and −6G → C and −7T → C substitutions were found in the spontaneous mutants selected at 500 μg/ml of PZA.
The mutations in the PncA protein associated with PZAr are so diverse that the identification of the same mutation in two different isolates may suggest possible transmission and merits further genotyping and epidemiological investigation. Correlations between genotypic clustering and pncA mutations were reported in studies from Brazil and Portugal (4, 45, 50). Strain clusters sharing the same pncA mutation in South Africa grouped together by spoligotype but differed by MIRU-VNTR type (41). In this study, isolates sharing the same genotype and pncA mutation also shared epidemiological or demographic characteristics (Table 1). In contrast, genotypically distinct isolates carrying mutations in the same codon, whether resulting in the same or different substitutions, facilitated the identification of essential residues involved in the activation of PZA into pyrazinoic acid.
A puzzling observation in this study and previous reports (the TBDReaMDB database) is the appearance of scattered mutations throughout the pncA gene and putative promoter region, suggesting multiple possibilities for inactivation or decreased activity and downregulation of the PZase. However, a closer look clearly identifies preferential sites. The mutations observed in the first codon have been reported in other studies (29, 39, 45, 48) and could very well interfere with the initiation of translation. The second most frequently encountered mutated codon in the PZAr spontaneous mutants was codon 8 (10% of the mutants obtained with 500 μg/ml PZA). Asp8 is part of the catalytic triad Cys138-Asp8-Lys96 (46, 63), and mutations of this residue have frequently been reported in the literature on PZAr strains. In order to better understand this “scattered” phenomenon, we analyzed the recently published high-resolution structure of the M. tuberculosis PncA protein (46) for the observed mutations in this study, coupled with predictions of protein stability. Most of the observed mutations affect buried amino acids and were determined to be energetically destabilizing, suggesting a reduction of protein stability and thereby a diminished or depleted PZase activity. Amino acids associated with either iron or substrate binding or catalytic active sites were most directly implicated in resistance. This is consistent with the literature and exemplified in the spontaneous mutants selected on 500 μg/ml PZA in this study. For instance, the four Fe-chelating residues (Asp49, His51, His57, and His71) were all found to be mutated among the PZAr isolates. His57 also happens to be the intrinsic phylogenetic mutation found in all M. bovis PZAr isolates. Likewise, modifications of the residues of the active site, such as Asp8, Val9, Gln10, Thr47, Gly132, Ala134, Cys138, Val139, and Thr142, were also all identified in the PZAr isolates and/or spontaneous mutants. Almost all of these residues have already been identified by mutagenesis as essential for the enzymatic activity of the protein (46, 63).
The effect of neutral or stabilizing mutations on protein function is less evident. Substitutions at codons 46 and 64 were predicted to be stabilizing, while the substitutions at positions 82, 87, 103, and 175 were found to be neutral. One plausible explanation could be that substitutions at positions 46 (46Ala → Val/Ser/Pro [22, 31, 42]), 82 (82His → Asp/Arg/Leu [42, 50, 58]), and 103 (103Tyr → His/Stop/Ser/Asp/Cys [29–31, 42, 50, 58, 61]), which have already been reported extensively in the literature, are located within close proximity of the substrate binding site and may impact the enzymatic activity while not affecting the overall protein structure. A puzzling challenge in this study was the observation that mutations at positions 14 and 175 (14Cys → Gly and 175Met → Ile) were found to be associated with both PZA-susceptible and -resistant isolates. Four clinical isolates were found to carry the 14Cys → Gly substitution, but only one of these was found to be resistant. Likewise, 2 clinical isolates were found to carry the 175Met → Ile substitution, with one being resistant and one being susceptible. Repeated reconfirmation by use of the standard Bactec MGIT960 PZA kit confirmed the phenotype, showing that the MICs for the 2 resistant isolates were borderline (>100 and <200 μg/ml), representing in part a possible source of the previously reported discrepancies. In the literature, these substitutions (14Cys → Gly [56] and 175Met → Ile [53]) have been associated with PZA resistance. These mutations were not identified in the spontaneous mutants (see Table S2 in the supplemental material). Furthermore, it is possible that these clinical isolates also carry other mutations interfering with PZA susceptibility. Interestingly, the latter mutation is predicted to confer a neutral effect, which might still allow for some enzymatic activity, although PZase activity was not explored in this study. Finally, it should be noted that the predicted values of stabilizing energy may not account for all influencing factors.
In this study, 9% (7/78 strains) of PZA-susceptible clinical strains also carried mutations within pncA, suggesting that some mutations either are phylogenetic (not associated with the resistance phenotype) or indicate genetic drift, notions that need to be evaluated further. Phylogenetic linkage for some neutral or synonymous mutations can be elucidated in some instances, while for most other SNPs it is not possible to assess given the restricted genotypic information required to draw firm conclusions. As such, the synonymous mutation 65Ser → Ser has been identified in at least 7 different studies, but only 2 provide genotypic data which suggest that this mutation is found predominantly in some, but not all, members of the CAS spoligotype strain family (17, 60). The two CAS isolates investigated in our study did not have this silent mutation. In contrast, the PZAr phylogenetic SNP 57His → Asp (54, 59) demarcates the branching of M. bovis from M. tuberculosis. SNPs on codon 57 are not uniquely restricted to M. bovis, as they may also occur in M. tuberculosis isolates (17, 29). Alternative substitutions, such as 57His → Pro and 57His → Arg, are also possible (4, 7, 21). Another well-defined phylogenetic SNP in pncA is the synonymous substitution 46Ala → Ala in Mycobacterium canettii.
Other studies also report synonymous or neutral mutations in PZAs isolates (2, 9, 14, 17, 23, 55, 58). We identified 5 possible pncA alterations not associated with a drug resistance phenotype among 138 sequenced clinical isolates, even though the considered mutation or codon has been described in the literature as being associated with PZAr: 14Cys → Gly (56), 64Tyr → Ser (29, 42), 135Thr → Pro (17, 40, 56), 143Ala → Thr (17), and 175Met → Ile (29, 31, 53). The 175Met → Ile mutation (previously described for PZAr strains) (29, 31, 53) found in a Beijing SIT1 isolate was not present in another 29 strains belonging to the same strain family, suggesting a random event or subbranch rather than a phylogenetic demarcation.
Previously, the 47Thr → Ala SNP, first reported for the W-MDR strain (5, 59), was later proposed erroneously to be a neutral phylogenetic mutation (16). In the present study, as in other works, we found no correlation between the 47Thr → Ala mutation and the phylogeny of the wide W-Beijing strain family, whether strains were susceptible or resistant. Although the 47Thr → Ala substitution was associated with the W-MDR outbreak isolate from New York City (5, 59), other W-Beijing strains most often carry a wild-type 47Thr residue, and most unrelated PZAr Beijing isolates display a variety of different mutations within the pncA gene (17, 23, 60). Most recently, the 47Thr → Ala SNP was found to be the most common SNP associated with PZAr in a large systematic study carried out by the Centers for Disease Control and Prevention (CDC) (9).
In this study, 1/60 PZAr isolates was found to carry wild-type pncA and its flank. The resistant phenotype of this isolate was reconfirmed, and the MIC was found to be >400 μg/ml. Unfortunately, 13 other isolates sharing the same profile could not be included in the study, as the cultures were no longer available for reconfirmation. Consequently, an accurate determination of the frequency of PZAr isolates displaying wild-type pncA could not be performed. Only 1 PZAr clinical isolate was found to carry wild-type pncA and rpsA, involved in the recently identified additional mechanism of resistance (57).
The frequency of PZAr mutants carrying wild-type pncA has been a point of much contention and speculation, due primarily to the inherent complications in PZA susceptibility determinations, possible phylogenetic predisposition, and the limited number of unbiased population-based studies. Indeed, the frequency of PZAr associated with pncA mutations has been reported to range from 70 to 100%, with reports of 70.58% (17), 72% (59), 75% (22), 84.6% (9), 91.4% (21), 97% (29), and 99.94% (58). In this study, PZAr and PZAs isolates of the MDR collection were evaluated genotypically and phenotypically. Within the spontaneous mutants, wild-type pncA-carrying PZAr mutants were identified only among those selected at the lower concentration of 100 μg/ml of PZA, with none isolated at 500 μg/ml (solid medium). This difference may also be attributed to bias introduced in the selection of spontaneous mutants at different concentrations (100 versus 500 μg/ml PZA at pH 6.0) or to other unknown factors associated with the in vitro selection of PZAr spontaneous mutants. Subsequent tests and reconfirmation of the MIC for all 14 wild-type mutants showed MICs of >400 μg/ml in liquid medium, although the mutants were originally selected at 100 μg/ml PZA. Mutations selected at low concentrations of a given drug can often result in higher levels of resistance in subsequent MIC determinations, as commonly observed with other antitubercular drugs, such as rifampin, isoniazid, and fluoroquinolones. This study clearly indicates that other targets or mechanisms are associated with PZA resistance. The 14 PZAr mutants have been subjected to whole-genome sequencing, and mutations were found in a single operon, with none found within rpsA or pncA (57).
Finally, the simultaneous selection of PZAr spontaneous mutants originating from 40 independent pan-susceptible cultures allowed for determination of frequencies of 1.5 and 1 PZAr mutant per 105 bacilli for selection on 100 μg/ml and 500 μg/ml PZA, respectively, which are rather elevated values compared to those for rifampin or the fluoroquinolones. Different mutation frequencies between different concentrations of the same drug have been described previously for other antitubercular drugs, notably the fluoroquinolones (1, 24, 67). Our estimate differs radically from the value of 10−7 to 10−8 CFU/ml proposed by Bamaga et al. (3). This discrepancy is hard to explain, though various factors may have contributed, including pH, medium, and drug concentration. Scorpio et al. (53) and Hirano and coworkers (20) also generated spontaneous mutants but did not estimate the possible frequency of mutagenesis.
In conclusion, we obtained numerous interesting observations in this study: (i) the frequency of mutagenesis to PZAr at pH 6.0 was found to be relatively high, at 10−5 CFU/ml; (ii) approximately 12% of the PZA-resistant spontaneous mutants did not carry mutations within pncA or its flank; (iii) approximately 43% of all clinical MDR-TB isolates investigated were additionally resistant to PZA; (iv) 8 novel substitutions in pncA were discovered; (v) protein destabilization may explain resistance patterns; and (vi) observed substitutions which did not confer a resistant phenotype could not be linked with genetic drift. Further studies are necessary to clearly correlate all pncA mutations to a PZA phenotype and to improve the resolution of PZA DST determination by molecular biology and overcome or complement the limitations of phenotypic susceptibility determinations.
Supplementary Material
ACKNOWLEDGMENTS
K.S. is the recipient of a scholarship from the Belgian Federal Science Policy Office, granted to the Scientific Institute of Public Health, Belgium. R.W. is a research associate at the Belgian Fund for Scientific Research (FNRS). Part of this study was supported by grants from the Belgian National Research Fund Fond pour la Recherche Scientifique Médicale (FRSM) (grants 3.4.544.F and 3.4.511.07.F).
Footnotes
Published ahead of print 23 July 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Alangaden GJ, Manavathu EK, Vakulenko SB, Zvonok NM, Lerner SA. 1995. Characterization of fluoroquinolone-resistant mutant strains of Mycobacterium tuberculosis selected in the laboratory and isolated from patients. Antimicrob. Agents Chemother. 39:1700–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ando H, et al. 2010. Pyrazinamide resistance in multidrug-resistant Mycobacterium tuberculosis isolates in Japan. Clin. Microbiol. Infect. 16:1164–1168 [DOI] [PubMed] [Google Scholar]
- 3. Bamaga M, Zhang H, Wright DJ. 2001. New mutations in pncA of in vitro selected pyrazinamide-resistant strains of Mycobacterium tuberculosis. Microb. Drug Resist. 7:223–228 [DOI] [PubMed] [Google Scholar]
- 4. Barco P, et al. 2006. pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis clinical isolates from the southeast region of Brazil. J. Antimicrob. Chemother. 58:930–935 [DOI] [PubMed] [Google Scholar]
- 5. Bifani PJ, et al. 1996. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA 275:452–457 [PubMed] [Google Scholar]
- 6. Boshoff HI, Mizrahi V, Barry CE., 3rd 2002. Effects of pyrazinamide on fatty acid synthesis by whole mycobacterial cells and purified fatty acid synthase I. J. Bacteriol. 184:2167–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown TJ, Tansel O, French GL. 2000. Simultaneous identification and typing of multi-drug-resistant Mycobacterium tuberculosis isolates by analysis of pncA and rpoB. J. Med. Microbiol. 49:651–656 [DOI] [PubMed] [Google Scholar]
- 8. Butler WR, Kilburn JO. 1983. Susceptibility of Mycobacterium tuberculosis to pyrazinamide and its relationship to pyrazinamidase activity. Antimicrob. Agents Chemother. 24:600–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Campbell PJ, et al. 2011. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 55:2032–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Canetti G, et al. 1963. Mycobacteria: laboratory methods for testing drug sensitivity and resistance. Bull. World Health Organ. 29:565–578 [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng SJ, Thibert L, Sanchez T, Heifets L, Zhang Y. 2000. pncA mutations as a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis: spread of a monoresistant strain in Quebec, Canada. Antimicrob. Agents Chemother. 44:528–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DeBarber AE, Mdluli K, Bosman M, Bekker LG, Barry CE., 3rd 2000. Ethionamide activation and sensitivity in multidrug-resistant Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 97:9677–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dehouck Y, et al. 2009. Fast and accurate predictions of protein stability changes upon mutations using statistical potentials and neural networks: PoPMuSiC-2.0. Bioinformatics 25:2537–2543 [DOI] [PubMed] [Google Scholar]
- 14. Denkin S, Volokhov D, Chizhikov V, Zhang Y. 2005. Microarray-based pncA genotyping of pyrazinamide-resistant strains of Mycobacterium tuberculosis. J. Med. Microbiol. 54:1127–1131 [DOI] [PubMed] [Google Scholar]
- 15. Dessau FI, Yeager RL, Burger FJ, Williams JH. 1952. Pyrazinamide (aldinamide) in experimental tuberculosis of the guinea pig. Am. Rev. Tuberc. 65:519–522 [PubMed] [Google Scholar]
- 16. Dormandy J, et al. 2007. Discrepant results between pyrazinamide susceptibility testing by the reference BACTEC 460TB method and pncA DNA sequencing in patients infected with multidrug-resistant W-Beijing Mycobacterium tuberculosis strains. Chest 131:497–501 [DOI] [PubMed] [Google Scholar]
- 17. Doustdar F, Khosravi AD, Farnia P. 2009. Mycobacterium tuberculosis genotypic diversity in pyrazinamide-resistant isolates of Iran. Microb. Drug Resist. 15:251–256 [DOI] [PubMed] [Google Scholar]
- 18. Escalante P, et al. 1998. Genotypic characterization of drug-resistant Mycobacterium tuberculosis isolates from Peru. Tuber. Lung Dis. 79:111–118 [DOI] [PubMed] [Google Scholar]
- 19. Gangadharam PR. 1986. Murine models for mycobacterioses. Semin. Respir. Infect. 1:250–261 [PubMed] [Google Scholar]
- 20. Hirano K, Takahashi M, Kazumi Y, Fukasawa Y, Abe C. 1997. Mutation in pncA is a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis. Tuber. Lung Dis. 78:117–122 [DOI] [PubMed] [Google Scholar]
- 21. Hou L, et al. 2000. Molecular characterization of pncA gene mutations in Mycobacterium tuberculosis clinical isolates from China. Epidemiol. Infect. 124:227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jonmalung J, Prammananan T, Leechawengwongs M, Chaiprasert A. 2010. Surveillance of pyrazinamide susceptibility among multidrug-resistant Mycobacterium tuberculosis isolates from Siriraj Hospital, Thailand. BMC Microbiol. 10:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jureen P, Werngren J, Toro JC, Hoffner S. 2008. Pyrazinamide resistance and pncA gene mutations in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 52:1852–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kocagoz T, et al. 1996. Gyrase mutations in laboratory-selected, fluoroquinolone-resistant mutants of Mycobacterium tuberculosis H37Ra. Antimicrob. Agents Chemother. 40:1768–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Konno K, Feldmann FM, McDermott W. 1967. Pyrazinamide susceptibility and amidase activity of tubercle bacilli. Am. Rev. Respir. Dis. 95:461–469 [DOI] [PubMed] [Google Scholar]
- 26. Kraulis PJ. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946–950 [Google Scholar]
- 27. Kushner S, et al. 1948. Experimental chemotherapy of tuberculosis; substituted nicotinamides. J. Org. Chem. 13:834–836 [DOI] [PubMed] [Google Scholar]
- 28. Kushner S, et al. 1952. Experimental chemotherapy of tuberculosis. II. The synthesis of pyrazinamides and related compounds. J. Am. Chem. Soc. 74:3617–3621 [Google Scholar]
- 29. Lee KW, Lee JM, Jung KS. 2001. Characterization of pncA mutations of pyrazinamide-resistant Mycobacterium tuberculosis in Korea. J. Korean Med. Sci. 16:537–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lemaitre N, Sougakoff W, Truffot-Pernot C, Jarlier V. 1999. Characterization of new mutations in pyrazinamide-resistant strains of Mycobacterium tuberculosis and identification of conserved regions important for the catalytic activity of the pyrazinamidase PncA. Antimicrob. Agents Chemother. 43:1761–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Louw GE, et al. 2006. Frequency and implications of pyrazinamide resistance in managing previously treated tuberculosis patients. Int. J. Tuberc. Lung Dis. 10:802–807 [PubMed] [Google Scholar]
- 32. Luria SE, Delbruck M. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Malone L, et al. 1952. The effect of pyrazinamide (aldinamide) on experimental tuberculosis in mice. Am. Rev. Tuberc. 65:511–518 [PubMed] [Google Scholar]
- 34. Marttila HJ, et al. 1999. pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis isolates from northwestern Russia. Antimicrob. Agents Chemother. 43:1764–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mathys V, et al. 2009. Molecular genetics of para-aminosalicylic acid resistance in clinical isolates and spontaneous mutants of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 53:2100–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McDermott DW, et al. 1954. Pyrazinamide-isoniazid in tuberculosis. Am. Rev. Tuberc. 69:319–333 [DOI] [PubMed] [Google Scholar]
- 37. Merritt EA, Murphy ME. 1994. Raster3D, version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D 50:869–873 [DOI] [PubMed] [Google Scholar]
- 38. Mestdagh M, et al. 1999. Relationship between pyrazinamide resistance, loss of pyrazinamidase activity, and mutations in the pncA locus in multidrug-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 43:2317–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miyagi C, Yamane N, Yogesh B, Ano H, Takashima T. 2004. Genetic and phenotypic characterization of pyrazinamide-resistant Mycobacterium tuberculosis complex isolates in Japan. Diagn. Microbiol. Infect. Dis. 48:111–116 [DOI] [PubMed] [Google Scholar]
- 40. Morlock GP, Plikaytis BB, Crawford JT. 2000. Characterization of spontaneous, in vitro-selected, rifampin-resistant mutants of Mycobacterium tuberculosis strain H37Rv. Antimicrob. Agents Chemother. 44:3298–3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mphahlele M, et al. 2008. Pyrazinamide resistance among South African multidrug-resistant Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 46:3459–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O'Sullivan DM, McHugh TD, Gillespie SH. 2005. Analysis of rpoB and pncA mutations in the published literature: an insight into the role of oxidative stress in Mycobacterium tuberculosis evolution? J. Antimicrob. Chemother. 55:674–679 [DOI] [PubMed] [Google Scholar]
- 43. Park SK, et al. 2001. pncA mutations in clinical Mycobacterium tuberculosis isolates from Korea. BMC Infect. Dis. 1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Parthiban V, Gromiha MM, Schomburg D. 2006. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 34:W239–W242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Perdigao J, et al. 2008. Multidrug-resistant tuberculosis in Lisbon, Portugal: a molecular epidemiological perspective. Microb. Drug Resist. 14:133–143 [DOI] [PubMed] [Google Scholar]
- 46. Petrella S, et al. 2011. Crystal structure of the pyrazinamidase of Mycobacterium tuberculosis: insights into natural and acquired resistance to pyrazinamide. PLoS One 6:e15785 doi:10.1371/journal.pone.0015785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pfyffer GE, et al. 1999. Multicenter laboratory validation of susceptibility testing of Mycobacterium tuberculosis against classical second-line and newer antimicrobial drugs by using the radiometric BACTEC 460 technique and the proportion method with solid media. J. Clin. Microbiol. 37:3179–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Portugal I, Barreiro L, Moniz-Pereira J, Brum L. 2004. pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis isolates in Portugal. Antimicrob. Agents Chemother. 48:2736–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ramaswamy S, Musser JM. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3–29 [DOI] [PubMed] [Google Scholar]
- 50. Rodrigues Vde F, et al. 2005. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis in Brazil. Antimicrob. Agents Chemother. 49:444–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Salfinger M, Heifets LB. 1988. Determination of pyrazinamide MICs for Mycobacterium tuberculosis at different pHs by the radiometric method. Antimicrob. Agents Chemother. 32:1002–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sandgren A, et al. 2009. Tuberculosis drug resistance mutation database. PLoS Med. 6:e2 doi:10.1371/journal.pmed.1000002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Scorpio A, et al. 1997. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 41:540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Scorpio A, Zhang Y. 1996. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 2:662–667 [DOI] [PubMed] [Google Scholar]
- 55. Sekiguchi J, et al. 2007. Detection of multidrug resistance in Mycobacterium tuberculosis. J. Clin. Microbiol. 45:179–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sheen P, et al. 2009. Sputum PCR-single-strand conformational polymorphism test for same-day detection of pyrazinamide resistance in tuberculosis patients. J. Clin. Microbiol. 47:2937–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shi W, et al. 2011. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 333:1630–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Somoskovi A, et al. 2007. Sequencing of the pncA gene in members of the Mycobacterium tuberculosis complex has important diagnostic applications: identification of a species-specific pncA mutation in “Mycobacterium canettii” and the reliable and rapid predictor of pyrazinamide resistance. J. Clin. Microbiol. 45:595–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sreevatsan S, Pan X, Zhang Y, Kreiswirth BN, Musser JM. 1997. Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob. Agents Chemother. 41:636–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stavrum R, Myneedu VP, Arora VK, Ahmed N, Grewal HM. 2009. In-depth molecular characterization of Mycobacterium tuberculosis from New Delhi—predominance of drug resistant isolates of the ‘modern’ (TbD1) type. PLoS One 4:e4540 doi:10.1371/journal.pone.0004540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tracevska T, et al. 2004. Spectrum of pncA mutations in multidrug-resistant Mycobacterium tuberculosis isolates obtained in Latvia. Antimicrob. Agents Chemother. 48:3209–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yeager RL, Munroe WG, Dessau FI. 1952. Pyrazinamide (aldinamide) in the treatment of pulmonary tuberculosis. Am. Rev. Tuberc. 65:523–546 [PubMed] [Google Scholar]
- 63. Zhang H, et al. 2008. Characterization of Mycobacterium tuberculosis nicotinamidase/pyrazinamidase. FEBS J. 275:753–762 [DOI] [PubMed] [Google Scholar]
- 64. Zhang Y, Heym B, Allen B, Young D, Cole S. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591–593 [DOI] [PubMed] [Google Scholar]
- 65. Zhang Y, Mitchison D. 2003. The curious characteristics of pyrazinamide: a review. Int. J. Tuberc. Lung Dis. 7:6–21 [PubMed] [Google Scholar]
- 66. Zhang Y, Scorpio A, Nikaido H, Sun Z. 1999. Role of acid pH and deficient efflux of pyrazinoic acid in unique susceptibility of Mycobacterium tuberculosis to pyrazinamide. J. Bacteriol. 181:2044–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhou J, et al. 2000. Selection of antibiotic-resistant bacterial mutants: allelic diversity among fluoroquinolone-resistant mutations. J. Infect. Dis. 182:517–525 [DOI] [PubMed] [Google Scholar]
- 68. Zimhony O, Cox JS, Welch JT, Vilcheze C, Jacobs WR., Jr 2000. Pyrazinamide inhibits the eukaryotic-like fatty acid synthetase I (FASI) of Mycobacterium tuberculosis. Nat. Med. 6:1043–1047 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.