Abstract
Exposure to DNA-damaging agents triggers signal transduction pathways that are thought to play a role in maintenance of genomic stability. A key protein in the cellular processes of nucleotide excision repair, DNA recombination, and DNA double-strand break repair is the single-stranded DNA binding protein, RPA. We showed previously that the p34 subunit of RPA becomes hyperphosphorylated as a delayed response (4–8 h) to UV radiation (10–30 J/m2). Here we show that UV-induced RPA-p34 hyperphosphorylation depends on expression of ATM, the product of the gene mutated in the human genetic disorder ataxia telangiectasia (A-T). UV-induced RPA-p34 hyperphosphorylation was not observed in A-T cells, but this response was restored by ATM expression. Furthermore, purified ATM kinase phosphorylates the p34 subunit of RPA complex in vitro at many of the same sites that are phosphorylated in vivo after UV radiation. Induction of this DNA damage response was also dependent on DNA replication; inhibition of DNA replication by aphidicolin prevented induction of RPA-p34 hyperphosphorylation by UV radiation. We postulate that this pathway is triggered by the accumulation of aberrant DNA replication intermediates, resulting from DNA replication fork blockage by UV photoproducts. Further, we suggest that RPA-p34 is hyperphosphorylated as a participant in the recombinational postreplication repair of these replication products. Successful resolution of these replication intermediates reduces the accumulation of chromosomal aberrations that would otherwise occur as a consequence of UV radiation.
INTRODUCTION
Unrepaired DNA damage can lead to mutagenesis and carcinogenesis. Exposure to DNA-damaging agents triggers signal transduction pathways that are thought to play a role in activating repair and recovery processes (Fornace, 1992; Lane, 1992; Fuks et al., 1993; Herrlich and Rahmsdorf, 1994). In mammalian cells, the activation of some of these pathways appears to be initiated at the cell membrane, whereas others appear to be triggered by the DNA damage itself (Karin and Herrlich, 1989; Karin and Hunter, 1995; Smith et al., 1999b). Responses to ionizing radiation (IR) appear to be initiated by the DNA damage itself, leading to activation of serine/threonine kinases (e.g., DNA-PK and ATM) and resulting in the phosphorylation of key regulatory proteins such as p53 and CHK1 (Sanchez et al., 1997; Smith et al., 1999b). ATM-dependent phosphorylation events have been shown to be required for normal DNA damage-induced checkpoint control (Lavin and Khanna, 1999). The DNA-binding protein replication protein A (RPA), which plays an essential role in DNA replication, recombination, and repair (Wold, 1997; Iftode et al., 1999) is also a substrate for these kinases (Zernik-Kobak et al., 1997; Gately et al., 1998; Chan et al., 2000). RPA becomes phosphorylated as a delayed response to UV radiation (Carty et al., 1994). However, the signal transduction pathway involved is thought to differ from that induced by IR because IR-induced phosphorylation of RPA was shown to be ATM dependent, whereas UV-induced phosphorylation of RPA appeared to occur normally in ataxia telangiectasia (A-T) cells (defective in ATM; Liu and Weaver, 1993). Because of the importance of RPA in processes required for maintaining genomic stability, such as DNA repair and recombination, we were interested in understanding more about the pathway that results in UV-induced RPA phosphorylation.
RPA is a heterotrimeric, single-stranded DNA-binding protein (subunits p70, p34, and p14) that functions in DNA replication, nucleotide excision repair, DNA recombination, double-strand break (DSB) repair, and transcriptional regulation (Wold, 1997; Iftode et al., 1999). Presumably, RPA participates in these diverse functions through its strong affinity for ssDNA and its ability to interact with numerous DNA replication proteins, including DNA polymerase α and proliferating cell nuclear antigen (PCNA); nucleotide excision repair proteins XPA, XPF, and XPG; DNA recombination and DSB repair proteins Rad51, Rad52, and DNA-PK; and transcription factors such as p53 (Dutta et al., 1993; He et al., 1995; Matsuda et al., 1995; Braun et al., 1997; Loor et al., 1997; Miller et al., 1997; Golub et al., 1998; New et al., 1998; Stigger et al., 1998; Shao et al., 1999). The p34 subunit of RPA becomes phosphorylated during the normal cell cycle (Din et al., 1990). Cell cycle-dependent phosphorylation of RPA-p34 begins at the onset of S phase and continues into mitosis, and dephosphorylation occurs in the latter part of mitosis, suggesting a physiological role for RPA-34 phosphorylation in cell cycle regulation (Din et al., 1990; Fotedar and Roberts, 1992). These cell cycle-dependent phosphorylation events occur primarily at Ser-23 and Ser-29 (Dutta and Stillman, 1992), which are consensus sites for Cdc2 cyclin-dependent kinase (Pan et al., 1994; Niu et al., 1997). These phosphorylated forms of RPA-p34 can be observed on immunoblots of polyacrylamide gels as more slowly migrating forms (Din et al., 1990; Carty et al., 1994).
RPA-p34 is hyperphosphorylated in response to DNA damage, converting a significant amount of the protein to its slowest migrating form (Carty et al., 1994; Zernik-Kobak et al., 1997; Shao et al., 1999). The kinetics of RPA-p34 hyperphosphorylation differ depending on the DNA-damaging agent. With low dose IR (2–10 Gy), hyperphosphorylation first appears at ∼1–2 h and peaks at 3–4 h after treatment (Liu and Weaver, 1993; Cheng et al., 1996); in contrast, with low dose UVC radiation (10–30 J/m2 UV) the hyperphosphorylated form does not begin to appear until 2–4 h after treatment and reaches a maximum at around 8–10 h (Carty et al., 1994). These results suggest that either the inducing signal or the signal transduction pathway leading to RPA-p34 hyperphosphorylation differs, depending on the genotoxic agent. We have shown that UV-induced hyperphosphorylation results in the phosphorylation of at least six serines and one threonine within the N-terminus of RPA-p34 (Zernik-Kobak et al., 1997). All of these sites within RPA-p34 can be phosphorylated in vitro when the RPA complex (p70, p34, and p14) is incubated with purified DNA-PK and double-stranded DNA (Zernik-Kobak et al., 1997). However, it is not clear what cellular enzymes are responsible for UV-induced hyperphosphorylation of RPA in vivo.
To investigate the role of the ATM kinase, we examined the capacity of A-T cells to carry out UV-induced RPA-p34 hyperphosphorylation. Here we demonstrate that A-T cells are defective in UV-induced hyperphosphorylation of RPA. When normal cells were irradiated with 10 J/m2 UVC, maximal RPA hyperphosphorylation was observed at 8–12 h after exposure. In A-T cells, we did not observe the most highly phosphorylated form of RPA for 24 h after UV exposure. In addition, we used recently developed inducible cell lines (Zhang et al., 1997, 1998) to show that expression of recombinant ATM protein in A-T cells corrected defective UV-induced hyperphosphorylation of RPA-p34. Furthermore, normal cells expressing antisense ATM cDNA became defective in UV-induced hyperphosphorylation of RPA-p34. A direct role for ATM in RPA-p34 hyperphosphorylation in vivo was suggested by the demonstration that purified ATM phosphorylated RPA-p34 in the purified three-subunit RPA complex in vitro on many of the same sites as are phosphorylated in vivo after UV radiation.
The time delay in UV radiation-induced RPA-p34 hyperphosphorylation suggests that this pathway is not induced directly by UV-induced photoproducts. Furthermore, because hyperphosphorylation occurs normally in nucleotide excision repair (NER)- deficient xeroderma pigmentosum group A (XPA) cells, the inducing signal does not appear to be an NER intermediate. Studies with synchronized cells suggest instead that replication of UV-damaged templates is required for generation of the inducing signal. On the basis of these observations, we postulate that the regions of single-stranded DNA and/or DNA strand breaks that are known to occur at replication forks blocked by UV-induced DNA template damage serve as the inducing signal(s) for ATM activation and RPA hyperphosphorylation.
MATERIALS AND METHODS
Cell Culture
The human skin-derived SV40-transformed fibroblast GM00637G (normal), AT5BIVA (A-T homozygous), and GM04429 (XP12BE; XPA) cell lines were obtained from the National Institute of General Medical Sciences Human Genetic Mutant Cell Repository (Coriell Institute for Medical Research, Camden, NJ). The SV40-transformed LM217 (normal) and AT3BISV (A-T homozygous) cell lines were generous gifts of Dr. Leon Kapp at the University of California at San Francisco. MO59K and MO59J (DNA-PKcs deficient) glioblastoma cells were generous gifts of Dr. Joan Allalunis-Turner (Cross Cancer Institute, Edmonton, Alberta, Canada). Recently, the two “normal” cell lines (LM217 and GM00637) were shown to be heterozygous for mutations in ATM (Wright et al., 1996). We have detected ATM protein by Western immunoblotting in these two normal lines but not in the two A-T lines used here (our unpublished results). AT1ABR is an Epstein Barr virus (EBV)-transformed A-T lymphoblastoid human cell line that contains a homozygous in-frame 9-bp deletion (codons 2546–2548) located upstream of the PI-3 kinase domain (codons 2855–2875), which causes a lack of kinase activity (Savitsky et al., 1995). These cells have been transfected with the ATM cDNA expression vector, pMAT1 with a metallothionein II promoter (Zhang et al., 1997), allowing CdCl2-inducible expression of ATM. C3ABR is an EBV-transformed normal lymphoblastoid cell line. These cells were transfected with a CdCl2-inducible antisense ATM cDNA expression vector, pMAT2 (Zhang et al., 1998).
Cell cultures were maintained at 37°C in a humidified atmosphere containing 5% CO2. Normal and A-T fibroblast cells were grown in minimal essential medium (MEM) supplemented with antibiotics, 15–20% fetal bovine serum (FBS), 2% essential amino acids, 1% nonessential amino acids, and 1% vitamins. MEM and all supplements were obtained from Life Technologies (Gaithersburg, MD). Human HeLa and XP12BE cells were grown in Dulbecco's minimal essential medium (DMEM), containing 10% FBS and antibiotics, and MO59J and MO59K cells were grown in DMEM/F12 supplemented with 10% FBS and antibiotics at 37°C and 5% CO2. Lymphoblasts were maintained in RPMI 1640 medium with 10% FBS and hygromycin B (0.2 mg/ml). Cells were maintained by subculturing at a ratio of 1:3, every 3 to 4 days, for at least 2 passages (∼24 h doubling time) before plating for experimentation. Cells were not maintained in culture longer than 15–20 passages. Induction of the ATM cDNA expression vector in pMAT1-transfected AT1ABR cells and the antisense ATM expression vector in pMAT2-transfected C3ABR cells was achieved by treatment with 5 μM CdCl2 for 8 h. After either CdCl2 induction or mock induction, the cells were washed twice with Hanks' buffered saline solution (HBBS), and the growth medium was replaced with fresh medium. After 8 h of CdCl2 induction the cells were treated with UVC radiation as described below.
Radiation Exposure
UVC radiation was delivered using a low-pressure mercury lamp (Mineralight lamp, model UVG-11; UVP, Inc., San Gabriel, CA) with maximal output at 254 nm. Before UV radiation exposure, medium was removed from the cells, and cells were washed twice in HBSS or phosphate-buffered saline (PBS; 58 mM Na2HPO4, 17 mM NaHPO4, 68 mM NaCl, 3 mM KCl). Cells were irradiated in buffer, which was then removed, and the growth medium previously removed from the cells (held at 37o during irradiation) was returned. The cell lines AT5BIVA, GM00637, LM217, MO59J, and MO59K all had similar clonigenic survival after UVC radiation; the XP12BE line had the expected increased sensitivity to UVC-induced cell killing.
Cell Synchronization
To obtain cells primarily in S phase, asynchronous cells were treated with 6 μM aphidicolin (Sigma-Aldrich, St. Louis, MO) for 16–20 h. The medium containing aphidicolin was removed, and cells were washed twice in serum-free medium and then incubated in serum-containing medium for an additional 2–4 h. The efficacy of the aphidicolin block and release protocol in LM217 and AT5BIVA cells was measured by labeling cells with bromodeoxyuridine (BrdU). Cells were fixed onto microscope slides, and BrdU uptake was detected with FITC-labeled anti-BrdU antibody and compared with propidium iodide (PI) staining by fluorescence microscopy. PI-stained nuclei were counted, and those that also stained positive for FITC were counted as positive for DNA synthesis. After 20 h of incubation with aphidicolin, <5% of both LM217 and AT5BIVA cells were labeled with anti-BrdU; 4 h after release from the aphidicolin block, the percentage of cells labeled with anti-BrdU in both cell lines had increased to 55–60%. To obtain cells synchronized in G1/S phase, HeLa cells were treated with 0.3 μM nocodazole for 16 h. Mitotic cells were collected by shaking the cells off the dish and pelleting them. Mitotic cells were released from nocodazole treatment for 7.5 h in fresh medium. The efficacy of the nocodazole block and release protocol was measured by double labeling cells with 3H- and 14C-thymidine as described below.
DNA Synthesis Assay
Thymidine incorporation was assayed essentially as described previously (Shao et al., 1997). Briefly, cells were prelabeled with 0.01 μCi/ml 14C-thymidine (NEN Life Science Products, Inc., Boston, MA) for 24 h at 37°C. After incubation in fresh medium for 1 h, cells were treated with aphidicolin as above. The rate of DNA synthesis after release from aphidicolin treatment was measured by 3H-thymidine incorporation during a 30 min pulse with 10 μCi/ml 3H-thymidine (NEN Life Science Products, Inc., Boston, MA). After labeling with 14C- and 3H-thymidine, cells were washed with PBS twice and lysed with 500 μl of 0.2 M NaOH. Triplicate samples (150 μl each) were collected onto squares of Whatman No. 3 filters (Whatman International, Maidstone, England) and rinsed with 5% trichloroacetic acid and 100% ethanol. The radioactivity of each sample was counted by dual-label liquid scintillation, and the ratio of 3H/14C reflected the DNA synthesis activity.
Immunoblotting
For Western immunoblots, cells were lysed in 2 ml PBSTDS (58 mM Na2HPO4, 17 mM NaHPO4, 68 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) that included 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml aprotinin, 0.1 mM PMSF, 5 mM NaF, and 1 mM Na vanadate. The insoluble fraction was pelleted by centrifugation at 3600 × g for 15 min at 4°C, and the supernatant was removed and stored at −70°C until use. Total protein concentration of cell lysates was determined by using the Coomassie Plus Protein assay (Pierce, Rockford, IL). Samples (50 μg total protein for RPA detection and 100 μg total protein for ATM and DNA-PK detection) were solubilized in Laemmli sample loading buffer, boiled at 100°C for 5 min, and then separated on 12% (for RPA) or 6% (for ATM and DNA-PK) denaturing SDS polyacrylamide gels (monomer to cross-linker ratio, 37:5:1) using the Laemmli buffer system (Laemmli, 1970). Proteins were transferred to Immobilon-P polyvinyl-divinyl fluoride (PVDF) transfer membranes (Millipore Corp., Bedford, MA) using a semidry apparatus (Bio-Rad Laboratories, Hercules, CA) at a maximum of 150 mA and 20 V for 1.5–2 h. The nontransferred portion of the gel was stained with 0.15% Coomassie blue to ensure equal loading of the protein samples. The membranes were blocked for 0.5–1 h with TTBS (100 mM Tris-HCl [pH 7.5], 0.9% NaCl, 0.3% Tween-20) containing 5% powdered milk and then probed with anti-ATM (1:2500; Novus Biologicals, Littleton, CO or Ab-3, Oncogene Science), anti-DNA-PK (1 μg/ml Ab-1; Oncogene Science, Cambridge, MA), anti-ATR (2 μg/ml Ab-1; Oncogene Science), or anti-RPA-p34 (2 μg/ml Ab-3, Oncogene Science; or a 1:1500 dilution of monoclonal antibody 34A [Kenny et al., 1990]) for 1–2 h. After washing four times with TTBS, the membranes were incubated with horseradish peroxidase–linked secondary antibody; sheep anti-mouse secondary antibody was used for RPA (1:2000), and donkey anti-rabbit secondary antibody was used for ATM and DNA-PK (1:1500 and 1:3000, respectively; Amersham Life Science Inc., Arlington Heights, IL). Membranes were washed three times in TTBS, and the proteins were visualized using the ECL chemoluminescent method (Amersham Life Science Inc.).
Nuclear Extract Preparation
Nuclear extracts were prepared as previously described (Andrews and Faller, 1991) with slight modifications, using hypotonic lysis followed by high salt extraction of nuclei. Briefly, frozen cell pellets were resuspended in 400-1000 μl of buffer A (10 mM HEPES-KOH [pH 7.9] at 4°C, 1.5 mM MgCl2, 10 mM KCl, 1 mM dithiothreitol [DTT], 0.5 mM PMSF, 20 mM β-glycerophosphate, 10 mM p-nitrophenylphosphate, 0.1 mg/ml pepstatin, and aprotinin). Cells were allowed to swell for 10 min, vortexed for 10 s, and centrifuged for 10 s at 23,000 × g. The pellet was resuspended in twice its volume of buffer C (10 mM HEPES-KOH [pH 7.9] at 4°C, 5% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.5 mM PMSF, 20 mM β-glycerophosphate, 10 mM p-nitrophenylphosphate) and incubated on ice for 20 min. The nuclear protein fraction was isolated by centrifugation for 10 min at 23,000 × g and collected as the supernatant fraction. Total protein concentration of nuclear extracts was determined by using the Coomassie Plus Protein assay (Pierce)
Purification of RPA
The RPA protein was expressed and purified from Escherichia coli BL21 (DE3) cells transformed with p11d-tRPA vector (a gift from Dr. Marc Wold, University of Iowa, Iowa City, IA) as described previously (Henricksen et al., 1994). The p11d-tRPA vector coexpresses all three RPA subunits (RPA-p70, RPA-p34, and RPA-p14).
Purification of ATM
ATM was purified according to recently published methods (Smith et al., 1999a) with minor modifications. Briefly, HeLa nuclear extracts were prepared according to Dignam et al. (1983) from 20 liter of HeLa S3 cells (Cell Culture Center, Minneapolis, MN) and equilibrated in buffer D (22 mM HEPES/KOH [pH 7.6], 20% glycerol, 2 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 0.5 mM PMSF, and 1 mM Na metabisulfite) containing 50 mM KCl. Nuclear extract (16 ml) was loaded onto a Q-Sepharose column (20 ml, 1.6 × 10 cm; Amersham Pharmacia Biotech, Piscataway, NJ) that was equilibrated in 50 mM KCl buffer D and washed with 2.5 column volumes (50 ml) of buffer D containing 100 mM KCl. The column was eluted with a 100-ml continuous salt gradient of 50–500 mM KCl in buffer D, and 2-ml fractions were collected. The Q-Sepharose column fractions containing ATM as detected by immunoblotting were dialyzed against 2 liters of 50 mM KCl buffer D overnight. The dialysate was fractionated by size exclusion chromatography on a gel filtration column (1.6 × 100 cm with Sephacryl SR-400 HR, 10,000–2000,000; Sigma-Aldrich) equilibrated with buffer A (50 mM Tris-HCl [pH 7.5], and 100 mM KCl), collecting 1-ml fractions. Fractions containing ATM eluting in the 350- to 450-kDa range were identified by immunoblotting, pooled (16 ml total), and dialyzed overnight against 50 mM KCl buffer D. The pooled fractions were loaded onto a heparin agarose column (5 ml, 1.6 × 2.5 cm; Amersham Pharmacia Biotech) equilibrated with 50 mM KCl buffer D and washed after sample application with eight column volumes of buffer D containing 100 mM KCl. The column was eluted with a 50 ml KCl continuous gradient from 50 to 500 KCl, and 1-ml fractions eluting at 230–240 mM KCl were identified, pooled, and dialyzed against 2 liters of 50 mM KCl buffer D. The pooled ATM fractions were incubated with 150 μg of biotinylated 50-bp dsDNA conjugated to streptavidin-coated iron oxide beads (Dynal, Lake Success, NY) on a rotator at 4°C for 1 h. The ATM-DNA complexes were collected using a magnetic stand (Dynal) and washed five times with 0.5 ml of 50 mM KCl buffer D. ATM protein was eluted with 250 mM KCl buffer D (200 μl).
Protein Kinase Assays
Kinase reactions were performed by incubating purified ATM or 10 U of DNA-PKcs/Ku (Promega, Madison, WI) or 20 U of Cdc2p34/Cyclin B (New England Biolabs, Beverly, MA) at 37°C for 30 min. in 30 μl of kinase buffer (20 mM HEPES [pH 7.4], 10 mM MgCl2, 100 μM ATP, 2 mM DTT, 0.2 μg of single-stranded φX174 virion DNA (ssDNA), 10 μCi [γ-32P]ATP, and 0.5 μg of purified RPA). The kinase reaction was stopped by the addition of 1× Laemmli sample loading buffer. Proteins were separated on 12% denaturing SDS-PAGE gels and exposed to film.
Two-dimensional Phosphopeptide Mapping
Purified recombinant RPA complex was phosphorylated by either purified ATM or 10 U of DNA-PK for 30 min. Hyperphosphorylated RPA-p34 was separated by SDS-PAGE and transferred to a PVDF membrane. The hyperphosphorylated form was detected by phosphorimager analysis and excised from the membrane. The PVDF membrane pieces containing phosphorylated RPA-p34 were blocked with polyvinylpyrrolidone 360 (0.5% in 100 mM acetic acid) and then subjected to chymotrypsin/trypsin digestion (2 × 10 μg of chymotrypsin for 2 h at 37°C; Boehringer Mannheim, Indianapolis, IN; Luo et al., 1990). After digestion, the peptides were oxidized with performic acid for 1 h at 25°C. Phosphorylated RPA-p34 chymotryptic/tryptic peptides were separated on TLC plates by electrophoresis in pH 1.9 buffer in the first dimension (anode on the left, cathode on the right) by using a model HRH gel electrophoresis apparatus (International Biotechnologies, Inc., New Haven, CT) and ascending chromatography in isobutyric acid buffer (62.5% isobutyric acid, 4.8% pyridine, 2.9% glacial acetic acid, 1.9% n-butanol) in the second dimension (Luo et al., 1990). The phosphorylated peptides were visualized by phosphorimager analysis (Storm model 860 phosphorimager; Molecular Dynamics, Sunnyvale, CA).
RESULTS
RPA-p34 Hyperphosphorylation in Asynchronous Cell Cultures
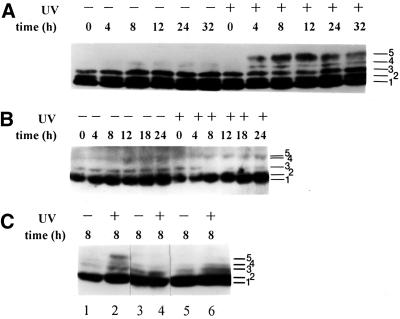
We showed previously that hyperphosphorylation of the p34 subunit of RPA occurs 4–8 h after exposure of HeLa cells to UVC radiation (10–30 J/m2; Carty et al., 1994). Because phosphorylated forms of RPA-p34 migrate slower on SDS-PAGE than do unphosphorylated forms (Carty et al., 1994; Brush et al., 1996), changes in phosphorylation can be measured by Western immunoblotting of cell lysates. To determine whether UV-induced hyperphosphorylation of RPA-p34 occurs normally in A-T cells, we compared the gel migration pattern of RPA-p34 from SV40-transformed skin fibroblast cell lines from normal individuals (LM217 and GM00637) and A-T patients (AT5BIVA and AT3BISV). Rapidly growing subconfluent monolayer cultures were either irradiated with 10 J/m2 UVC or mock-irradiated. At various times after irradiation, whole cell lysates or nuclear extracts were prepared, and proteins were separated on SDS-PAGE, blotted to membranes, and probed with antibody against RPA-p34. In Figure 1A, mock-irradiated LM217 cells show three major cell cycle–dependent forms of RPA-p34 (bands 1–3), and the pattern of forms did not change during a 32-h time course. Lysates prepared from LM217 cells 4 h after UV exposure (10 J/m2) contained at least two additional slower migrating forms of RPA-p34 (bands 4 and 5). The intensity of the slowest migrating form (band 5, the hyperphosphorylated form) increased to a maximum at 12 h postexposure. Hyperphosphorylation of RPA-p34 was also seen in a time- (our unpublished results) and dose-dependent manner in another normal cell line, GM00637, that was exposed to 10 J/m2 UVC (Figure 1C, lanes 1 and 2). The general pattern of UV-induced phosphorylation of RPA-p34 that we observed was comparable among HeLa (Carty et al., 1994), LM217, and GM00637 cells.
Figure 1.
RPA-p34 phosphorylation in asynchronous normal and A-T cells exposed to 10 J/m2 UVC. Cell lysates (A and B) or nuclear extracts (C) were prepared from asynchronously growing cells at indicated times after 10 J/m2 UV radiation or mock exposure. The p34 subunit of RPA was detected by Western immunoblotting with monoclonal antibody 34A. Phosphorylation causes a slower migration on the gel; at least five different forms of the p34 subunit of RPA can be visualized. (A) RPA-p34 in LM217 cells; (B) RPA-p34 in AT5BIVA cells; (C) RPA-p34 in nuclear lysates of GM00637 (lanes 1 and 2), AT5BIVA (lanes 3 and 4), and AT3BISV (lanes 5 and 6) cells. Band 1 is unphosphorylated RPA-p34; bands 2 and 3 are cell cycle–dependent phosphorylated forms; and band 5 is the DNA damage–induced hyperphosphorylated form.
Figure 1B shows that the three faster-migrating forms of RPA-p34 seen in LM217 cells are also observed in mock-irradiated A-T cells, although form 3 appears to be less intense than in the normal cells. However, after exposure of A-T cells to 10 J/m2 UVC, there was very little change in the banding pattern of RPA-p34. Only a very faint band was observed at the position of the hyperphosphorylated form (band 5). A similar result (Figure 1C) was obtained from the analysis of UVC-induced RPA-p34 hyperphosphorylation in nuclear extracts from another normal cell line (GM00637; lanes 1 and 2) and another A-T cell line (AT3BISV; lanes 5 and 6). These data suggest that UV-induced hyperphosphorylation of RPA depends on expression of ATM.
UV-induced RPA Phosphorylation in Cells Expressing Inducible Recombinant ATM Protein or ATM cDNA Antisense Transcript
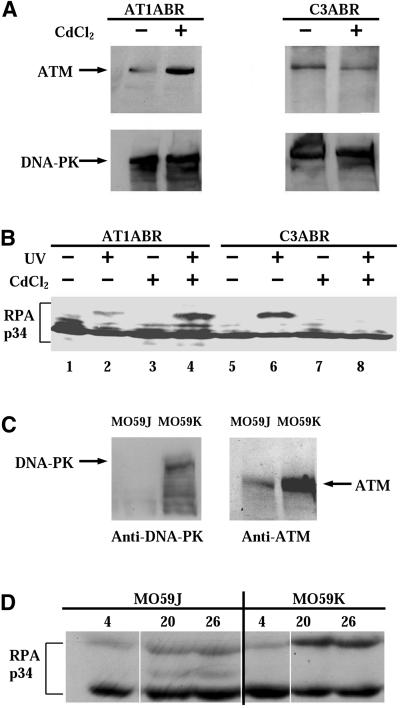
In order to confirm that the difference in UV-induced hyperphosphorylation of RPA-p34 observed in A-T cells versus normal cells is due to the presence or absence of ATM, we used A-T cells expressing recombinant ATM protein or normal cells expressing ATM antisense cDNA both under control of the metallothionein promoter and inducible with CdCl2. We used the A-T lymphoblastoid cell line AT1ABR, and the normal cell line C3ABR transfected with either an inducible full-length ATM cDNA or an antisense ATM cDNA, respectively (Zhang et al., 1997, 1998). We confirmed by Western immunoblotting that expression of ATM was induced in the A-T cells and repressed in the normal cells by incubation in the presence of CdCl2. The AT1ABR cell line contains a homozygous 9-bp in-frame deletion starting at codon 2546 of the open reading frame (Zhang et al., 1997). This enables AT1ABR cells to produce a near full-length but nonfunctional protein and is consistent with our detection by immunoblotting of endogenous ATM protein in uninduced AT1ABR cells (Figure 2A, top left lane 1). As shown in Figure 2A, top left lane 2, the amount of ATM protein increased in AT1ABR cells after CdCl2 treatment. The induction of antisense ATM cDNA by CdCl2 in transfected C3ABR cells reduced the level of ATM protein detected at 8 h postinduction (Figure 2A, top right lane 4). The expression of DNA-PK remained constant after CdCl2 treatment in both A-T cells and normal cells (Figure 2A, bottom panels).
Figure 2.
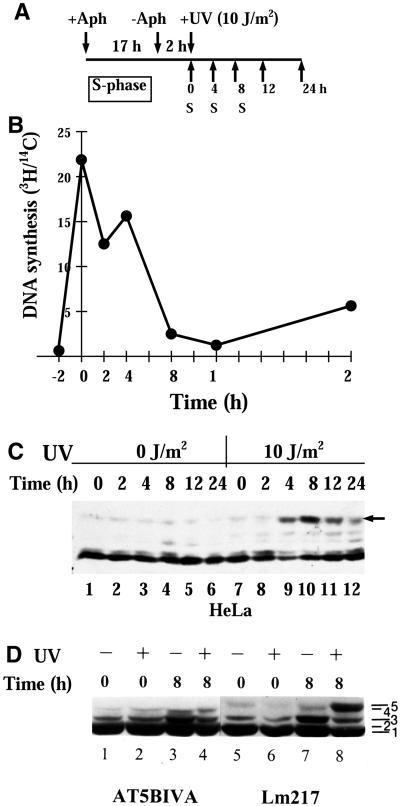
(A) Expression of ATM and DNA-PK proteins in pMAT1 stably transfected A-T cells (AT1ABR) and pMAT2 stably transfected normal C3ABR cells. Cells were treated with 5 μM CdCl2 for 8 h. Whole-cell lysates (100 μg) were analyzed by SDS-PAGE and immunoblotted with anti-ATM (Ab-3; Oncogene Science) antibodies (top panels). AT1ABR is the parental A-T cell line containing a homozygous 9-bp in-frame deletion and is capable of producing near full-length but nonfunctional protein. Upon induction of the transfected line, a functional full-length recombinant ATM protein is produced. C3ABR is a control lymphoblastoid cell line expressing normal ATM. Induction of the transfected line reduces ATM expression. Membranes were reblotted with anti-DNA-PKcs antibody (Ab-1; Oncogene Science; bottom panels). (B) Phosphorylation of RPA-p34 after exposure of cells to UVC radiation. AT1ABR cells, transfected with a CdCl2-inducible ATM cDNA expression vector pMAT1, and C3ABR cells, transfected with an inducible antisense ATM cDNA expression vector pMAT2, were treated with 5 μM CdCl2 for 8 h (+) or mock-treated (−). After 8 h of CdCl2 treatment, cells were UVC irradiated (30 J/m2) or mock-irradiated. Eight hours after UV treatment, whole-cell lysates were prepared and RPA-p34 was examined by gel electrophoresis, followed by immunoblotting with anti-RPA-p34 (Ab-3; Oncogene Science) antibody. (C) DNA-PKcs (left) and ATM (right) expression in MO59J and MO59K cells detected in whole-cell lysates by immunoblotting. (D) MO59J and MO59K cells were irradiated with 30 J/m2 UV, and whole-cell lysates were prepared at the indicated times after UV irradiation. RPA-p34 hyperphosphorylation was detected by immunoblotting using anti-RPA-p34 (Ab-3; Oncogene Science) antibody. To ensure equal loading and transfer of the protein samples, the gels and the PVDF membranes were stained with Coomassie blue and colloidal gold solution, respectively.
First, we examined whether expression of ATM in A-T cells could affect UV-induced hyperphosphorylation of RPA-p34. Cells were grown in the presence or absence of CdCl2, whole cell lysates were prepared 8 h after UVC (30 J/m2) treatment, and lysate proteins were subjected to Western blot analysis using anti-RPA-p34 antibody. After UV irradiation, a weak band was detectable at the position of the hyperphosphorylated form of RPA-p34 in AT1ABR cells in the absence of expression of recombinant ATM protein (Figure 2B, lane 2). However, after induction of expression of full-length recombinant ATM protein in AT1ABR cells, the UV-induced hyperphosphorylation of RPA increased substantially (Figure 2B, lane 4), indicating that the expression of full-length ATM protein increased RPA-p34 phosphorylation. As expected, the mock-irradiated AT1ABR cells did not exhibit a hyperphosphorylated form regardless of CdCl2 induction, although cell cycle–dependent forms of phosphorylated RPA-p34, having intermediate gel mobility, could be detected in these cells (Figure 2B, lanes 1 and 3).
We next investigated whether decreased expression of ATM protein in normal cells could affect RPA-p34 hyperphosphorylation. Cell lysates prepared from UV-irradiated (30 J/m2), uninduced C3ABR cells showed hyperphosphorylation of RPA, as demonstrated by the presence of the slow-migrating form of RPA-p34 (Figure 2B, lane 6). This band was not observed in mock-irradiated C3ABR cells (Figure 2B, lane 5). After induction of expression of ATM cDNA antisense transcripts in the transfected C3ABR cells, hyperphosphorylation of RPA-p34 was not detected at 8 h after UV irradiation (Figure 2B, lane 8) or after mock irradiation (Figure 2B, lane 7). These data demonstrate that UV-induced hyperphosphorylation of RPA-p34 is dependent on ATM expression.
RPA-p34 Hyperphosphorylation in a DNA-PKcs–deficient Cell Line
We showed previously that purified DNA-PK can hyperphosphorylate RPA-p34 in vitro (Zernik-Kobak et al., 1997). To determine whether DNA-PK also participates in UV-induced RPA-p34 hyperphosphorylation in vivo, we compared RPA-p34 phosphorylation in DNA-PKcs–deficient MO59J glioblastoma cells and DNA-PKcs–expressing MO59K cells derived from the same tumor specimen (Lees-Miller et al., 1995). Western blot analysis of cell lysates from DNA-PKcs–deficient MO59J cells confirmed that these cells lack detectable DNA-PKcs (Figure 2C). However, these cells showed only low levels of ATM protein expression as well (Figure 2C), consistent with previous reports (Chan et al., 1998; Gately et al., 1998). In MO59K cells, we observed normal levels of both DNA-PKcs and ATM (Figure 2C). As shown in Figure 2D, RPA-p34 hyperphosphorylation after UV irradiation was observed in both MO59J and MO59K cells. The hyperphosphorylated band appears to be slightly less intense in the MO59J cells, suggesting that either DNA-PK participates in UV-induced hyperphosphorylation of RPA-p34 or that the reduced levels of ATM in these cells causes a reduction in RPA-p34 hyperphosphorylation. The fact that RPA-p34 hyperphosphorylation occurs at all in DNA-PKcs–deficient cells (Fried et al., 1996) suggests that in vivo, a kinase in addition to DNA-PK is involved in UV-induced RPA-p34 hyperphosphorylation.
Hyperphosphorylation of RPA-p34 In Vitro by Purified ATM
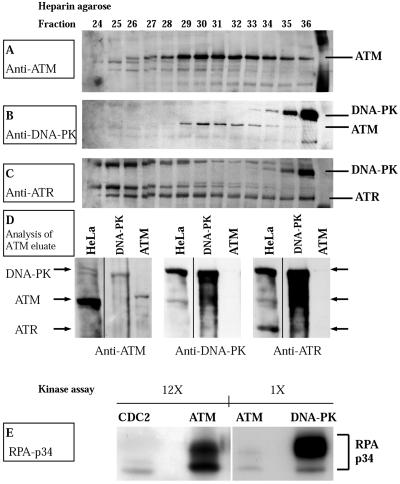
We next asked whether ATM kinase itself might be responsible for phosphorylation of RPA-p34. To address this question, we tested the capacity of purified ATM to phosphorylate the p34 subunit of purified RPA complex in vitro. We purified ATM from HeLa nuclei according to recently published procedures (Smith et al., 1999a). We confirmed separation of ATM from DNA-PK at the heparin agarose chromatography step of the purification by Western immunoblotting of fractions from the column (Figure 3,A and B). Fractions eluting at 230–250 mM KCl (e.g., fractions 29–33, Figure 3, A–C) were pooled for further purification. ATM was further separated from ATR in the final DNA-binding step of the purification. Immunoblotting of the ATM eluate for the presence of DNA-PK and ATR (ATM- and RAD3-related protein) demonstrated that these proteins were absent from the conjugated ATM-DNA purified fractions (Figure 3D).
Figure 3.
Purification and kinase activity of ATM. (A) Western immunoblot of fractions from heparin agarose affinity column. A 100-μl aliquot of the indicated fractions was analyzed on a 6% SDS-polyacrylamide gel, transferred to PVDF membrane, and probed with antibody to ATM (Novus Biologicals). (B) The PVDF membrane was reprobed without stripping with antibody to DNA-PKcs (Ab-1; Oncogene Science). (C) ATR was detected with anti-ATR (Ab-1; Oncogene Science) without stripping the membrane. (D) Western immunoblot of HeLa cell nuclear extract (70 μg), DNA-PKcs/Ku (100 U; Promega), and purified ATM (35 μl). The samples were analyzed on a 6% SDS-polyacrylamide gel, transferred to PVDF membrane, and probed with antibody to ATM (left; Novus Biologicals). The PVDF membrane was reprobed without stripping with antibody to DNA-PKcs (center, Ab-1; Oncogene Science), and ATR was detected with anti-ATR (right, Ab-1; Oncogene Science). (E) Phosphorylation of RPA by Cdc2p34/cyclin B (New England Biolabs) purified ATM and DNA-PKcs/Ku (Promega). Purified recombinant RPA heterotrimer (0.5 μg) was incubated with either Cdc2p34/cyclin B (20 U), purified ATM or DNA-PKcs/Ku (10 U) in a 30-μl reaction containing 20 mM HEPES (pH 7.4), 10 mM MgCl2, 100 μM ATP, 2 mM DTT, 0.2 μg of single-stranded φX174 virion circular DNA, and 10 μCi [γ-32P]ATP. Samples were incubated for 30 min at 37°C. The kinase reaction was stopped by the addition of 1× Laemmli sample loading buffer, and samples were separated on 12% SDS-polyacrylamide gels followed by autoradiography. The designations 1× and 12× refer to the relative exposure time of the films shown.
Purified RPA complex was incubated with purified ATM and [γ-32P]dATP in the presence of single-stranded DNA. DNA-PK and cyclin B/cdc2 kinase, which are known to phosphorylate RPA in vitro (Dutta and Stillman, 1992; Brush et al., 1994; Henricksen et al., 1996) were included as controls. The results in Figure 3E demonstrate that RPA-p34 becomes hyperphosphorylated in the presence of ATM. Furthermore, this kinase activity is sensitive to 50 nM wortmanin (our unpublished results), ruling out the ATR kinase, which is only sensitive to much higher concentrations (Sarkaria et al., 1998). Although ATM is less active than DNA-PK, the shift in mobility of RPA-p34 is comparable with ATM and DNA-PK, suggesting that a similar number of sites on RPA-p34 are phosphorylated by the two kinases. In contrast, cyclin B/cdc2 kinase-mediated phosphorylation resulted primarily in a single labeled band, consistent with the known specificity of this kinase for only two sites (serines 23 and 29) on RPA-p34 (Pan et al., 1994; Niu et al., 1997). These results demonstrate that ATM has the capacity to phosphorylate the p34 subunit of RPA complex in vitro, suggesting that ATM could play a direct role in UV-induced hyperphosphorylation in vivo. These results are consistent with those of Chan et al. (2000), who demonstrated that ATM, purified from human placenta, had the capacity to phosphorylate RPA-p34.
Mapping of the Sites of Phosphorylation of RPA-p34 by ATM Kinase
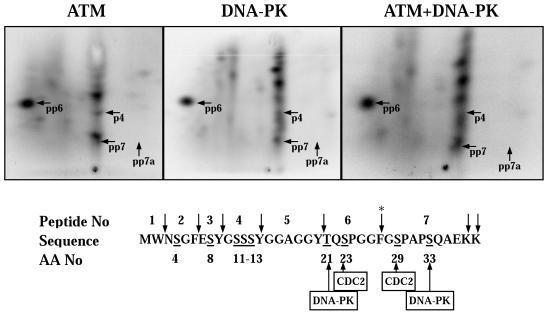
We showed previously that DNA-PK phosphorylates the p34 subunit of the RPA complex in vitro at many of the same sites that are phosphorylated in vivo after UV-irradiation (Zernik-Kobak et al., 1997). To determine whether ATM has similar substrate specificity to DNA-PK, we carried out two-dimensional peptide mapping of the phosphorylated RPA-p34 protein bands. The hyperphosphorylated form was separated by SDS-PAGE, transferred to PVDF membrane, excised, and digested with trypsin/chymotrypsin. The resulting peptides were separated in two dimensions and visualized by phosphorimager analysis (Figure 4). Overall, a similar pattern of phosphorylation was observed with ATM as with DNA-PK. Cochromatography of ATM- and DNA-PK-phosphorylated RPA-p34 confirmed those similarities (Figure 4, right panel). Both kinases exhibited strong phosphorylation of peptide 6 on two sites (pp6). As reported previously (Zernik-Kobak et al., 1997), phosphorylation of peptide 7 on two sites (pp7) by DNA-PK could also be detected; however, with ATM it appears to be phosphorylated to a lesser extent. Although both DNA-PK and ATM appeared to phosphorylate peptide 4 (p4), DNA-PK phosphorylated other peptides (probably peptides 2 and 3) more strongly. These results demonstrate that ATM phosphorylates RPA-p34 at multiple sites, and many of these sites appear to be the same as those phosphorylated by DNA-PK. However, there appear to be additional phosphopeptides near peptide 6 (pp6) and in the top right quadrant unique to the ATM map, suggesting some differences in the site specificities of the two kinases in vitro. The functional significance of these phosphorylation events remains to be determined. However, published reports suggest a role for RPA phosphorylation in DNA replication and interaction with other regulatory proteins (Iftode et al., 1999).
Figure 4.
Chymotryptic/tryptic phosphopeptide maps of in vitro–phosphorylated recombinant RPA-p34. (Top) Five hundred nanograms of purified recombinant RPA complex was phosphorylated by either purified ATM or 10 U of DNA-PKcs/Ku for 30 min. Hyperphosphorylated RPA-p34 was separated by SDS-PAGE and transferred to a PVDF membrane. The hyperphosphorylated form of RPA-p34 was detected by phosphorimager analysis and excised from the membrane. The hyperphosphorylated RPA-p34 was digested twice with 10 μg of chymotrypsin/trypsin and oxidized with performic acid. The digested peptides were loaded onto TLC plates and separated by electrophoresis at pH 1.9 in the first dimension, followed by ascending chromatography in the second dimension. The labeled peptides were detected by phosphorimager analysis. To verify identical and unique peptides, equal ra-dioactivity from digests of DNA-PKcs/Ku and ATM-phosphorylated RPA-p34 were loaded onto the same chromatography plate and subjected to two-dimensional separation. Numbered tryptic/chymotryptic peptides indicated on the maps with arrows refer to the peptide sequence number (lower panel) with a letter “p” designating a phosphorylated serine or threonine on the peptide. (bottom) The amino acid sequence of the N-terminus of RPA-p34 is shown along with the sites for cleavage by trypsin/chymotrypsin (↓), sites phosphorylated by Cdc2p34/cyclin B and consensus sites for DNA-PKcs/Ku. The asterisk denotes the cleavage site that is blocked by adjacent phosphorylated amino acids.
The next series of experiments were carried out to understand more about the inducing signal for ATM-dependent, UV-induced hyperphosphorylation of RPA-p34. The fact that the time of appearance of the hyperphosphorylated form of RPA-p34 is delayed (appears at 4–8 h) after UV radiation compared with IR (appears at 1–3 h) suggests that UV-induced DNA damage (i.e., primarily pyrimidine cyclobutane dimers and 6-4 photoproducts) does not induce this pathway directly. With IR, the inducing signal is thought to be DNA strand breaks (Nelson and Kastan, 1994), although there is no direct evidence supporting this mechanism.
RPA p34 Hyperphosphorylation Does Not Require Nucleotide Excision Repair
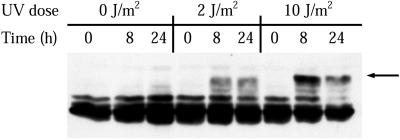
We first tested the possibility that the signal for UV-induced hyperphosphorylation of RPA-p34 was generated as a result of nucleotide excision repair (NER) of UV photoproducts. To test whether NER was necessary for induction of RPA-p34 hyperphosphorylation after UV radiation, we used a XPA cell line, which is deficient in the recognition and excision steps of NER. UV irradiation of XPA cells led to a strong induction of hyperphosphorylated RPA-p34 (Figure 5). These results indicate that the induction of RPA-p34 hyperphosphorylation by UV irradiation does not require active NER.
Figure 5.
Time course of RPA phosphorylation after irradiation of XP12BE cells (XP complementation group A). XPA cells were either mock-irradiated or treated with the indicated doses of UVC. At the indicated times after treatment, cell lysates were prepared and the RPA-p34 phosphorylation pattern was analyzed by gel electrophoresis, followed by immunoblotting. The arrow to the right indicates the major hyperphosphorylated form of RPA-p34.
RPA p34 Hyperphosphorylation Is Associated with Replication of UV-damaged DNA
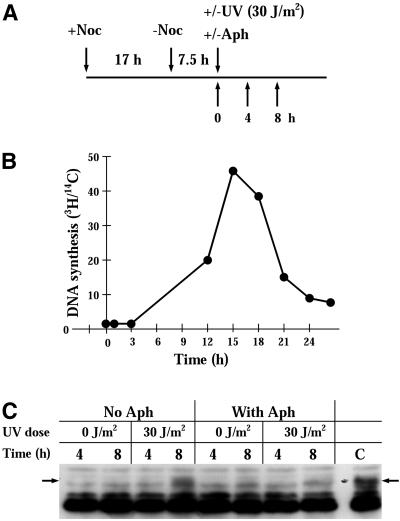
Replication of UV-damaged DNA templates leads to replication fork blockage at sites of damage (Park and Cleaver, 1979; Bierne and Michel, 1994; Seigneur et al., 1997). If blockage occurs on the leading strand, continued replication on the lagging strand leads to the accumulation of long stretches of single-stranded parental DNA (Wang and Smith, 1986; Kaufmann, 1989; Michel et al., 1997). In addition, DNA double-strand breaks occur at blocked replication forks (Wang and Smith, 1986; Kaufmann, 1989; Michel et al., 1997). If these abnormal DNA structures serve as signals for induction of RPA-p34 hyperphosphorylation, we would expect that induction would occur more intensely and more rapidly if synchronized cells were irradiated at the beginning of S phase. To test this hypothesis, cells were synchronized by nocodazole and then released for 7.5 h to place them at the G1/S border (Figure 6B). Cells were then irradiated with 30 J/m2 UV and incubated further in the presence or absence of aphidicolin (Figure 6C). In the absence of aphidicolin, hyperphosphorylation of RPA-p34 was observed by 8 h after UV irradiation. In the presence of aphidicolin, the UV-induced hyperphosphorylation RPA-p34 was absent. These data support the conclusion that UV-induced RPA-p34 hyperphosphorylation is associated with replication of UV-damaged DNA.
Figure 6.
UVC-induced RPA-p34 hyperphosphorylation in the presence of aphidicolin. (A) Protocol for treatment. HeLa cells were treated with 0.3 μM nocodazole for 16 h. Mitotic cells were collected by shaking the cells off the dish and pelleting them. To obtain cells synchronized in G1/S phase, mitotic cells were released from nocodazole treatment for 7.5 h in fresh medium. Cells were either mock-irradiated or irradiated with 30 J/m2 UVC in the presence or absence of aphidicolin (6 μM). (B) The rate of DNA synthesis after release from nocodazole treatment was measured as 3H-thymidine incorporation during a 30-min pulse with 10 μCi/ml 3H-thymidine (NEN Life Science Products, Inc.). Cells were prelabeled with 0.01 μCi/ml 14C-thymidine (NEN Life Science Products, Inc.) for 24 h at 37°C, followed by 3H-thymidine pulse labeling. After labeling, cells were washed with PBS twice and lysed with 500 μl of 0.2 M NaOH per dish. The radioactivity of each sample was counted by dual-label liquid scintillation, and the ratio of 3H/14C reflected the DNA synthesis activity. (C) RPA-p34 phosphorylation in nocodazole-synchronized HeLa cells exposed to UVC. Cells in G1/S phase were either mock-irradiated or irradiated with 30 J/m2 UVC in the presence or absence of aphidicolin (6 μM). After preparation of whole-cell lysates, RPA-p34 was visualized by immunoblotting with anti-RPA-p34 antibody and ECL chemiluminescent detection.
ATM Dependence of RPA-p34 Hyperphosphorylation in Aphidicolin-synchronized Cell Cultures
The next experiment was carried out to determine whether the hyperphosphorylation of RPA-p34 observed in aphidicolin-treated cells after UV-irradiation was ATM dependent. To test this hypothesis, we treated cells for 17 h with aphidicolin, which inhibits the elongation step of DNA replication and blocks cell cycle progression at the G1/S border (Pedrali-Noy et al., 1982). Cells were then released from the block by transferring them to fresh medium without aphidicolin. After an additional 2-h incubation, cells were irradiated with 10 J/m2 UVC or mock-irradiated (Figure 7A). Cell lysates were prepared at the indicated times thereafter, and the phosphorylation state of RPA-p34 was analyzed by Western immunoblotting. We confirmed that the cells released from the aphidicolin block had a rapid increase in DNA synthetic activity, by measuring the incorporation of [3H]thymidine (Figure 7B). We also confirmed that the DNA content of cells increased by 5 h after the release of aphidicolin block (our unpublished results). Strong hyperphosphorylation of RPA p34 was observed by 4 h after UV irradiation of aphidicolin-synchronized HeLa cells (Figure 7C). Immediately after irradiation or mock irradiation, the majority of RPA-p34 in lysates prepared from LM217 or AT5BIVA cells was in faster migrating forms of the subunit (particularly in the doublet of bands 1 and 2; Figure 7D, lanes 1, 2, 5, and 6). Eight hours after mock irradiation, this pattern did not change appreciably (Figure 7D, lanes 3 and 7). At 8 h after 10 J/m2 UVC exposure, however, a slow-mobility form (band 5) was apparent in lysates from LM217 cells (Figure 7D, lane 8) but not from A-T cells (Figure 7D, lane 4). These data support the observation that UV-induced hyperphosphorylation of RPA-p34 does not occur normally in A-T cells.
Figure 7.
UV-induced RPA-p34 phosphorylation in aphidicolin-synchronized cells. (A) Protocol for treatment. Cells were treated with 6 μM of aphidicolin for 16–20 h. After the medium containing aphidicolin was removed; the cells were washed twice in serum-free medium and further incubated in serum-containing medium without aphidicolin for 2–4 h. After 2–4 h in fresh medium, the cells were mock-irradiated or irradiated with 10 J/m2 and then incubated in the same medium for the indicated times. (B) The rate of DNA synthesis after release from aphidicolin treatment was measured as 3H-thymidine incorporation during a 30-min pulse with 10 μCi/ml 3H-thymidine (NEN Life Science Products, Inc.). Cells were prelabeled with 0.01 μCi/ml 14C-thymidine (NEN Life Science Products, Inc.) for 24 h at 37°C followed by 3H-thymidine pulse labeling. After labeling, cells were washed with PBS twice and lysed with 500 μl of 0.2 M NaOH per dish. The radioactivity of each sample was counted by dual-label liquid scintillation and the ratio of 3H/14C reflected the DNA synthesis activity. (C) Time course of RPA-p34 phosphorylation in HeLa cells. Whole cell lysates were prepared at various times after mock irradiation or irradiation with 10 J/m2 UVC. Proteins were separated on a 12% denaturing polyacrylamide gel. After transfer of the proteins to a PVDF membrane RPA-p34 phosphorylation pattern was analyzed by immunoblotting using an antibody specific for RPA-p34 (monoclonal antibody 34A). (D) RPA-p34 phosphorylation in aphidicolin synchronized normal Lm217 and A-T cells exposed to UVC. Cells were irradiated with 10 J/m2 UVC or mock-irradiated 2 h after release from the aphidicolin block. Cell lysates were prepared 0 or 8 h after irradiation. The p34 subunit of RPA was analyzed by immunoblotting.
DISCUSSION
We have investigated the requirements for UV radiation–induced RPA-p34 hyperphosphorylation. Western immunoblotting revealed that the p34 subunit of the RPA protein complex was hyperphosphorylated in a time- and dose-dependent manner in normal, asynchronously growing cells after exposure to UVC (Carty et al., 1994). This response was both delayed and reduced in intensity in A-T cells. We have also used recombinant ATM and antisense ATM expression vectors to demonstrate that UV-induced RPA-p34 hyperphosphorylation is dependent on the expression of ATM. Reduction of ATM levels in normal C3ABR cells, by the expression of an antisense cDNA, decreased UV-induced RPA-p34 hyperphosphorylation. Furthermore, expression of a full-length recombinant ATM protein in AT1ABR cells fully restored UV-induced RPA-p34 hyperphosphorylation in these cells. We conclude that ATM expression is required for UV-induced RPA-p34 hyperphosphorylation.
Our conclusions appear to differ from those of Liu and Weaver (1993), who observed that A-T cells were deficient in hyperphosphorylation of RPA-p34 in response to IR but not UV radiation. With IR, hyperphosphorylation of RPA-p34 was reduced and delayed in A-T cells compared with normal cells. UV-induced hyperphosphorylation of RPA-p34 was examined only up to 3 h after irradiation (Liu and Weaver, 1993). At these early times, little difference was observed between A-T cells and normal cells; after 500 J/m2 UV, strong hyperphosphorylation of RPA-p34 was observed in both A-T and normal cells; after 50 J/m2 UV, hyperphosphorylation of RPA-p34 was observed in some A-T and normal cells but not others; and after 10 J/m2 UV, little hyperphosphorylation was observed in either cell type. Because the ATM-dependent phosphorylation pathway we are investigating was induced at later times (4–8 h after UV irradiation) and at lower UV fluences (10–30 J/m2), our results are not, in fact, in direct conflict with those of Liu and Weaver (1993).
Our observation that purified ATM has the capacity to phosphorylate RPA-p34 in vitro suggests that the ATM kinase may participate directly in UV-induced RPA-p34 hyperphosphorylation in vivo. However, it is possible that the requirement of ATM for UV-induced RPA-p34 hyperphosphorylation is indirect and other kinases may be involved. We showed previously that DNA-PK phosphorylates in vitro many of the same sites within the N-terminus of RPA-p34 that are phosphorylated in vivo after UV-irradiation (Ser-11, Ser-12, or Ser-13 on peptide 4; Thr-21and Ser-23 on peptide 6; Ser-29 and Ser-33 on peptide 7; also probably Ser 4 on peptide 2, and Ser 8 on peptide 3; Zernik-Kobak et al., 1997). However, several lines of evidence suggest that DNA-PK is not the only kinase required for RPA-p34 phosphorylation after DNA damage. For example, human DNA-PKcs–deficient glioblastoma cells M059J hyperphosphorylated RPA-p34 after IR (Fried et al., 1996). This is in agreement with our observation that RPA-p34 hyperphosphorylation occurred in UV-irradiated M059J cells, albeit to a lesser extent. Our observations agree with other reports that M059J cells have extremely low levels of ATM protein. The lower levels of ATM, rather than the lack of DNA-PK, may account for the decreased levels of hyperphosphorylated RPA-p34 observed in these cells after genotoxic insult (Chan et al., 1998; Gately et al., 1998). A role for ATM in RPA-p34 phosphorylation is also supported by the demonstration that ATM immunoprecipitates from MO59J cells retain in vitro kinase activity toward RPA-p34 (Gately et al., 1998). Here we show that the sites of phosphorylation of RPA-p34 by ATM in vitro are similar to those of DNA-PK and therefore similar to those that occur in vivo. It is possible that both DNA-PK and ATM may participate in some way in UV-induced RPA-p34 hyperphosphorylation.
Another candidate for in vivo phosphorylation of RPA-p34 is ATR. Expression of kinase-inactive ATR protein (ATRkd) in human fibroblasts increased sensitivity to UV and IR and led to loss of cell cycle checkpoint control (Wright et al., 1998). Although ATR kinase does not phosphorylate RPA-p34 in vitro (Hall-Jackson et al., 1999), the functional overlap between ATM and ATR suggests that RPA-p34 may be an in vivo substrate for ATR. Although ATM has been implicated in IR-induced RPA-p34 hyperphosphorylation and ATM-immune precipitates of IR-treated cells phosphorylated RPA-p34 (Gately et al., 1998), a direct role for ATM kinase in IR-induced hyperphosphorylation has also not yet been demonstrated.
The signal for IR-induced, ATM-dependent RPA-p34 hyperphosphorylation is thought to be IR-induced DNA strand breaks (Liu and Weaver, 1993), although no direct evidence for this hypothesis has been provided. UV radiation induces primarily cyclobutane pyrimidine dimers and 6-4 photoproducts. Because UV-induced RPA-p34 hyperphosphorylation only occurs several hours after UV irradiation, UV photoproducts are not likely to be the primary inducing signal. There are several different pathways by which cells process UV-induced DNA damage (Figure 8) that might generate the signal for ATM-dependent RPA-p34 hyperphosphorylation. In normal cells, most UV-induced DNA damage is repaired by NER, which generates DNA single-strand breaks as transient intermediates (Kaufmann and Wilson, 1990). However, nucleotide excision repair does not appear to be required for induction of RPA-p34 hyperphosphorylation by UV radiation, because we showed that hyperphosphorylation was induced normally in XPA cells.
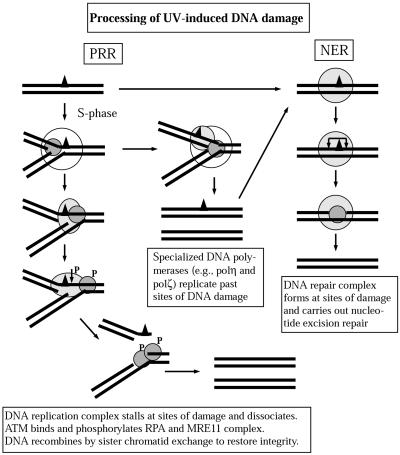
Figure 8.
Model for cellular processing of UV-induced DNA damage. UV radiation induces mainly pyrimidine cyclobutane dimers and 6-4 photoproducts. The majority of these are repaired by nucleotide excision repair (NER), which involves recognition of the site of damage by XPA with the assistance of RPA and the recruitment of additional repair proteins that incise the DNA on either side of the damage, release a single-stranded oligonucleotide containing the damage and fill in the resulting gap. If cells are in S phase and DNA replication occurs before all the damage is repaired, the DNA replication fork may encounter sites of damage. One of several specialized DNA polymerases may facilitate replication past the damage. If replication becomes stalled at sites of DNA damage, mechanisms of postreplication repair (PRR) are activated. This activation appears to involve the ATM kinase, which phosphorylates RPA and NBS1. The MRE11/RAD50/NBS1 complex may participate in processing the incompletely replicated DNA, facilitating recombinational repair.
Our observations and others (Rodrigo et al., 2000) that UV-induced RPA-p34 hyperphosphorylation was blocked when DNA replication was inhibited by aphidicolin argue that the inducing signal for ATM-dependent RPA-p34 hyperphosphorylation is generated during replication of UV-damaged DNA templates. Models for replication of UV damaged DNA templates and postreplication repair were first proposed in the early 1970s to explain the induction of chromosomal aberrations by UV and chemical carcinogens (Bender et al., 1973, 1974); these models have been refined more recently to include new information on mechanisms of postreplication repair (Seigneur et al., 1998). When DNA replication occurs while UV-photoproducts remain in the DNA, some of these lesions block the progression of the DNA replication fork (Park and Cleaver, 1979). This blockage is sometimes overcome by the action of DNA polymerases (e.g., polymerases η and ζ and the product of hREV1) that have the capacity to bypass UV photoproducts (Gibbs et al., 1998; Lin et al., 1999; Masutani et al., 1999). However, if the blockage persists, aberrant DNA replication intermediates are produced that contain long regions of single-stranded DNA; double-strand DNA breaks also accumulate (Wang and Smith, 1986; Kaufmann, 1989; Michel et al., 1997). Although the exact nature of the inducing signal remains to be elucidated, it is interesting to note that ATM-dependent RPA-p34 phosphorylation in vitro is activated by closed circular single-stranded DNA. If the double-strand breaks that accumulate after UV radiation are the inducing signal, this would suggest similarities between the signals for IR- and UV-induced ATM activation. Recently, it was shown that DNA replication was also required for induction of RPA-p34 hyperphosphorylation by adozelesin, a DNA-alkylating agent, but not by IR (Liu et al., 2000). Like UV, adozelesin would be expected to cause DNA replication–blocking lesions.
The aberrant replication intermediates that accumulate after UV radiation are thought to be resolved through a recombinational mechanism between sister chromatids (Bender et al., 1973, 1974; Kaufmann, 1989; Haber, 2000; Lowndes and Murguia, 2000). However, the enzymology of this process is not yet understood. Recently, Limoli et al. (2000) demonstrated colocalization of PCNA (involved in DNA replication) and the MRE11 complex (Mre11/Rad50/Nbs1, involved in recombinational repair) in foci 4–8 h after UV radiation of XPV human fibroblasts (deficient in DNA polymerase η). This result implicates the MRE11 complex in recognition and/or resolution of DNA replication intermediates after UV radiation. Indeed, it has been suggested that this complex performs a similar role during replication of undamaged templates (Petrini, 2000). Because NBS1 appears to be a substrate for the ATM kinase (Gatei et al., 2000; Lim et al., 2000), it is possible that MRE11 focus formation in response to UV damage also involves ATM (Petrini, 2000). ATM-dependent RPA-p34 hyperphosphorylation may occur in concert with phosphorylation of other members of this complex.
ATM-dependent phosphorylation events are involved in IR-induced checkpoint control. The radioresistant DNA synthesis observed in A-T cells is attributed to the loss of the S-phase checkpoint. Interestingly, mutation of the ATM kinase phosphorylation site on NBS1 also leads to loss of the IR-induced, S-phase checkpoint (Lim et al., 2000). This observation suggests that the MRE11 complex may play a role in IR-induced ATM-dependent cell signaling. Whether phosphorylation of NBS1 or RPA plays a role in UV-induced checkpoint control has not yet been determined.
Presently, there is no definitive evidence for a functional role of RPA hyperphosphorylation. However, a number of lines of evidence suggest that phosphorylation may alter the activity of RPA for DNA replication, NER, and double-strand break repair (reviewed in Iftode et al., 1999). The strong binding affinity of the three-subunit RPA complex for ssDNA is predominantly attributed to the RPA-p70 subunit; the 34-kDa subunit is thought to play a regulatory role (Wold, 1997; Bochkareva et al., 1998; Iftode et al., 1999). RPA also binds with high affinity to certain double-stranded DNA regulatory sequences (Singh and Samson, 1995; Tang et al., 1996) and to double-stranded DNA carrying UV photoproducts or cis-platin DNA cross links (Clugston et al., 1992; Burns et al., 1996; Patrick and Turchi, 1998). RPA has a double-stranded DNA unwinding activity, which is altered by phosphorylation (Georgaki et al., 1992; Georgaki and Hubscher, 1993). We have shown that the DNA replication capacity was reduced in extracts from UV-irradiated human cells (containing hyperphosphorylated RPA), but replication capacity could be restored by the addition of purified RPA (Carty et al., 1994). A similar observation was recently made with extracts from adozelesin-treated cells (Liu et al., 2000). Park et al. (1999) reported DNA synthesis inhibition in RPA-enriched replication extracts from UV-treated MO59K (DNA-PKcs+) cells but not in cell extracts from MO59J (DNA-PKcs−) cells, implying modulation of RPA by DNA-PK. However, interpretation of these data is complicated by the fact that the DNA-PKcs–deficient glioblastoma cells (MO59J) also have reduced amounts of ATM compared with the control line (MO59K). Rodrigo et al. (2000) reported a temporal parallel between RPA-p34 hyperphosphorylation and DNA synthesis inhibition after UVC irradiation, and Shao et al. (1999) reported a similar relationship after camptothecin treatment. In addition, Henricksen et al. (1996) demonstrated that phosphorylation of RPA-p34 modulates DNA replication. Also, RPA phosphorylation reduces the ability of RPA to interact with DNA polymerase α-primase (Iftode et al., 1999) and p53 (Abramova et al., 1997). It is not clear how these in vitro observations relate to RPA function in vivo and in particular to DNA repair functions or the S-phase checkpoint.
What is the biological significance of a role for ATM in cellular responses to UV-induced DNA damage? A-T patients are acutely sensitive to ionizing radiation, but no enhanced sensitivity to UV radiation has been documented (Paterson and Smith, 1979; Sedgwick and Boder, 1991; Woods and Taylor, 1992; Taylor et al., 1994). Likewise A-T cells exhibit enhanced sensitivity to killing by IR but not UV. However, there are abnormalities in the response of A-T cells to UV radiation. Both the accumulation of DNA strand breaks and the subsequent generation of chromosome aberrations are elevated in UV-irradiated A-T cells (Sasaki, 1980; Ejima and Sasaki, 1986; Kaufmann, 1989; Kaufmann and Wilson, 1994). Kaufmann and Wilson demonstrated that the greatest numbers of chromosomal aberrations occurred when normal cells were UV irradiated at the G1/S border and that A-T fibroblasts were hypersensitive to the induction of chromosomal aberrations by UV (Kaufmann and Wilson, 1994). The fact that UV induces mainly chromatid-type chromosomal aberrations suggests that they arise after DNA replication has occurred (Kaufmann, 1989). This abnormality in response to UV in A-T cells likely reflects an abnormality in response to DNA replication-blocking lesions in general, which would include those caused by carcinogens that form bulky adducts (e.g., benzo[a]pyrene) and alkylated bases (e.g., methyl methane sulfonate). Indeed a very similar response was observed in adozelesin-treated cells (Liu et al., 2000). Perhaps this defect in response to a broader range of DNA-damaging agents accounts for the appearance of a broad spectrum of cancers in adult A-T patients (Meyn, 1999). Recently ATM has been linked to other chromosome instability syndromes through its apparent interaction with the MRE11/RAD50/NBS1 complex. The MRE11 gene has been found to be mutated in two patients with an A-T-like syndrome (Stewart et al., 1999), and the NBS1 gene is mutated in Nijmegen breakage syndrome (Gatei et al., 2000; Lim et al., 2000; Petrini, 2000; Wu et al., 2000; Zhao et al., 2000). Perhaps all of these diseases share a postreplication repair defect.
In summary, we have demonstrated that the RPA-p34 hyperphosphorylation that occurs in response to UV radiation is dependent on DNA replication and expression of the ATM kinase. We postulate that this pathway is induced in response to abnormal DNA replication intermediates that accumulate as a result of DNA replication fork blockage by UV photoproducts. Further, we suggest ATM plays a role in signaling to components of the postreplication repair apparatus, including RPA, to trigger recombinational repair and prevent the formation of chromosomal aberrations resulting from unresolved replication intermediates.
ACKNOWLEDGMENTS
We thank the Center for Environmental Genetics Molecular Biology Core for expert technical assistance. This work was supported by grants R01-NS34782 and P30-ES06096 from the National Institutes of Health and by a research grant from the A-T Children's Project.
REFERENCES
- Abramova NA, Russell J, Botchan M, Li R. Interaction between replication protein A and p53 is disrupted after UV damage in a DNA repair-dependent manner. Proc Natl Acad Sci USA. 1997;94:7186–7191. doi: 10.1073/pnas.94.14.7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499–2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender MA, Griggs HG, Bedford JS. Mechanisms of chromosomal aberration production. 3. Chemicals and ionizing radiation. Mutat Res. 1974;23:197–212. doi: 10.1016/0027-5107(74)90140-7. [DOI] [PubMed] [Google Scholar]
- Bender MA, Griggs HG, Walker PL. Mechanisms of chromosomal aberration production. I. Aberration induction by ultraviolet light. Mutat Res. 1973;20:387–402. doi: 10.1016/0027-5107(73)90060-2. [DOI] [PubMed] [Google Scholar]
- Bierne H, Michel B. When replication forks stop. Mol Microbiol. 1994;13:17–23. doi: 10.1111/j.1365-2958.1994.tb00398.x. [DOI] [PubMed] [Google Scholar]
- Bochkareva E, Frappier L, Edwards AM, Bochkarev A. The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J Biol Chem. 1998;273:3932–3936. doi: 10.1074/jbc.273.7.3932. [DOI] [PubMed] [Google Scholar]
- Braun KA, Lao Y, He Z, Ingles CJ, Wold MS. Role of protein-protein interactions in the function of replication protein A (RPA): RPA modulates the activity of DNA polymerase alpha by multiple mechanisms. Biochemistry. 1997;36:8443–8454. doi: 10.1021/bi970473r. [DOI] [PubMed] [Google Scholar]
- Brush GS, Anderson CW, Kelly TJ. The DNA-activated protein kinase is required for the phosphorylation of replication protein A during simian virus 40 DNA replication. Proc Natl Acad Sci USA. 1994;91:12520–12524. doi: 10.1073/pnas.91.26.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush GS, Morrow DM, Hieter P, Kelly TJ. The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc Natl Acad Sci USA. 1996;93:15075–15080. doi: 10.1073/pnas.93.26.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JL, Guzder SN, Sung P, Prakash S, Prakash L. An affinity of human replication protein A for ultraviolet-damaged DNA. J Biol Chem. 1996;271:11607–11610. doi: 10.1074/jbc.271.20.11607. [DOI] [PubMed] [Google Scholar]
- Carty MP, Zernik-Kobak M, McGrath S, Dixon K. UV light-induced DNA synthesis arrest in HeLa cells is associated with changes in phosphorylation of human single-stranded DNA-binding protein. EMBO J. 1994;13:2114–2123. doi: 10.1002/j.1460-2075.1994.tb06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DW, Gately DP, Urban S, Galloway AM, Lees-Miller SP, Yen T, Allalunis-Turner J. Lack of correlation between ATM protein expression and tumor cell radiosensitivity. Int J Radiat Biol. 1998;74:217–224. doi: 10.1080/095530098141591. [DOI] [PubMed] [Google Scholar]
- Chan DW, Son SC, Block W, Ye R, Khanna KK, Wold MS, Douglas P, Goodarzi AA, Pelley J, Taya Y, Lavin MF, Lees-Miller SP. Purification and characterization of ATM from human placenta. A manganese-dependent, wortmannin-sensitive serine/threonine protein kinase. J Biol Chem. 2000;275:7803–7810. doi: 10.1074/jbc.275.11.7803. [DOI] [PubMed] [Google Scholar]
- Cheng X, Cheong N, Wang Y, Iliakis G. Ionizing radiation-induced phosphorylation of RPA p34 is deficient in ataxia telangiectasia and reduced in aged normal fibroblasts. Radiother Oncol. 1996;39:43–52. doi: 10.1016/0167-8140(96)01712-4. [DOI] [PubMed] [Google Scholar]
- Clugston CK, McLaughlin K, Kenny MK, Brown R. Binding of human single-stranded DNA binding protein to DNA damaged by the anticancer drug cis-diamminedichloroplatinum (II) Cancer Res. 1992;52:6375–6379. [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din S, Brill SJ, Fairman MP, Stillman B. Cell-cycle-regulated phosphorylation of DNA replication factor A from human and yeast cells. Genes Dev. 1990;4:968–977. doi: 10.1101/gad.4.6.968. [DOI] [PubMed] [Google Scholar]
- Dutta A, Ruppert JM, Aster JC, Winchester E. Inhibition of DNA replication factor RPA by p53. Nature. 1993;365:79–82. doi: 10.1038/365079a0. [DOI] [PubMed] [Google Scholar]
- Dutta A, Stillman B. cdc2 family kinases phosphorylate a human cell DNA replication factor, RPA, and activate DNA replication. EMBO J. 1992;11:2189–2199. doi: 10.1002/j.1460-2075.1992.tb05278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejima Y, Sasaki MS. Enhanced expression of X-ray- and UV-induced chromosome aberrations by cytosine arabinoside in ataxia telangiectasia cells. Mutat Res. 1986;159:117–123. doi: 10.1016/0027-5107(86)90120-x. [DOI] [PubMed] [Google Scholar]
- Fornace AJ. Mammalian genes induced by radiation: activation of genes associated with growth control. Annu Rev Genet. 1992;26:507–526. doi: 10.1146/annurev.ge.26.120192.002451. [DOI] [PubMed] [Google Scholar]
- Fotedar R, Roberts JM. Cell cycle regulated phosphorylation of RPA-32 occurs within the replication initiation complex. EMBO J. 1992;11:2177–2187. doi: 10.1002/j.1460-2075.1992.tb05277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried LM, Koumenis C, Peterson SR, Green SL, van Zijl P, Allalunis-Turner J, Chen DJ, Fishel R, Giaccia AJ, Brown JM, Kirchgessner CU. The DNA damage response in DNA-dependent protein kinase-deficient SCID mouse cells: replication protein A hyperphosphorylation and p53 induction. Proc Natl Acad Sci USA. 1996;93:13825–13830. doi: 10.1073/pnas.93.24.13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks Z, Haimovitz-Friedman A, Hallahan DE, Kufe DW, Weichselbaum RR. Stress response genes induced in mammalian cells by ionizing radiation. Radiat Oncol Invest. 1993;1:81–93. [Google Scholar]
- Gatei M, Young D, Cerosaletti KM, Desai-Mehta A, Spring K, Kozlov S, Lavin MF, Gatti RA, Concannon P, Khanna K. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat Genet. 2000;25:115–119. doi: 10.1038/75508. [DOI] [PubMed] [Google Scholar]
- Gately DP, Hittle JC, Chan GKT, Yen TJ. Characterization of ATM expression, localization, and associated DNA-dependent protein kinase activity. Mol Biol Cell. 1998;9:2361–2374. doi: 10.1091/mbc.9.9.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgaki A, Hubscher U. DNA unwinding by replication protein A is a property of the 70 kDa subunit and is facilitated by phosphorylation of the 32 kDa subunit. Nucleic Acids Res. 1993;21:3659–3665. doi: 10.1093/nar/21.16.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgaki A, Strack B, Podust V, Hubscher U. DNA unwinding activity of replication protein A. FEBS Lett. 1992;308:240–244. doi: 10.1016/0014-5793(92)81283-r. [DOI] [PubMed] [Google Scholar]
- Gibbs PE, McGregor WG, Maher VM, Nisson P, Lawrence CW. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase zeta. Proc Natl Acad Sci USA. 1998;95:6876–6880. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub EI, Gupta RC, Haaf T, Wold MS, Radding CM. Interaction of human rad51 recombination protein with single-stranded DNA binding protein, RPA. Nucleic Acids Res. 1998;26:5388–5393. doi: 10.1093/nar/26.23.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. Recombination: a frank view of exchanges and vice versa. Curr Opin Cell Biol. 2000;12:286–292. doi: 10.1016/s0955-0674(00)00090-9. [DOI] [PubMed] [Google Scholar]
- Hall-Jackson CA, Cross DA, Morrice N, Smythe C. ATR is a caffeine-sensitive, DNA-activated protein kinase with a substrate specificity distinct from DNA-PK. Oncogene. 1999;18:6707–6713. doi: 10.1038/sj.onc.1203077. [DOI] [PubMed] [Google Scholar]
- He Z, Henricksen LA, Wold MS, Ingles CJ. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- Henricksen LA, Carter T, Dutta A, Wold MS. Phosphorylation of human replication protein A by the DNA-dependent protein kinase is involved in the modulation of DNA replication. Nucleic Acids Res. 1996;24:3107–3112. doi: 10.1093/nar/24.15.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization [published erratum appears in J. Biol. Chem. 1994; 269, 16519] J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- Herrlich P, Rahmsdorf HJ. Transcriptional and post-transcriptional responses to DNA-damaging agents. Cell Biol. 1994;6:425–431. doi: 10.1016/0955-0674(94)90036-1. [DOI] [PubMed] [Google Scholar]
- Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. Crit Rev Biochem Mol Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- Karin M, Herrlich P. Cis and transacting genetic elements responsible for the induction of specific genes by tumor promoters, serum factors and stress. In: Colburn NH, editor. Genes and Signal Transduction in Multistage Carcinogenesis. New York: Marcel Dekker, Inc.; 1989. pp. 415–440. [Google Scholar]
- Karin M, Hunter T. Transcriptional control by protein phosphorylation: signal transmission from the cell surface to the nucleus. Curr Biol. 1995;5:747–757. doi: 10.1016/s0960-9822(95)00151-5. [DOI] [PubMed] [Google Scholar]
- Kaufmann WK. Pathways of human cell post-replication repair. Carcinogenesis. 1989;10:1–11. doi: 10.1093/carcin/10.1.1. [DOI] [PubMed] [Google Scholar]
- Kaufmann WK, Wilson SJ. DNA repair endonuclease activity during synchronous growth of diploid human fibroblasts. Mutat Res. 1990;236:107–117. doi: 10.1016/0921-8777(90)90038-7. [DOI] [PubMed] [Google Scholar]
- Kaufmann WK, Wilson SJ. G1 arrest and cell-cycle-dependent clastogenesis in UV-irradiated human fibroblasts. Mutat Res. 1994;314:67–76. doi: 10.1016/0921-8777(94)90062-0. [DOI] [PubMed] [Google Scholar]
- Kenny MK, Schlegel U, Furneaux H, Hurwitz J. The role of human single-stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J Biol Chem. 1990;265:7693–7700. [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lane DP. Cancer, p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Lavin MF, Khanna KK. ATM: the protein encoded by the gene mutated in the radiosensitive syndrome ataxia-telangiectasia. Int J Radiat Biol. 1999;75:1201–1214. doi: 10.1080/095530099139359. [DOI] [PubMed] [Google Scholar]
- Lees-Miller SP, Godbout R, Chan DW, Weinfeld M, Day R S, 3rd, Barron GM, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH, Kastan MB. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- Limoli CL, Giedzinski E, Morgan WF, Cleaver JE. Inaugural article: polymerase eta deficiency in the xeroderma pigmentosum variant uncovers an overlap between the S phase checkpoint and double-strand break repair. Proc Natl Acad Sci USA. 2000;97:7939–7946. doi: 10.1073/pnas.130182897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Xin H, Zhang Y, Wu X, Yuan F, Wang Z. The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res. 1999;27:4468–4475. doi: 10.1093/nar/27.22.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JS, Kuo SR, McHugh MM, Beerman TA, Melendy T. Adozelesin triggers DNA damage response pathways and arrests SV40 DNA replication through replication protein A inactivation. J Biol Chem. 2000;275:1391–1397. doi: 10.1074/jbc.275.2.1391. [DOI] [PubMed] [Google Scholar]
- Liu VF, Weaver DT. The ionizing radiation-induced replication protein A phosphorylation response differs between ataxia telangiectasia and normal human cells. Mol Cell Biol. 1993;13:7222–7231. doi: 10.1128/mcb.13.12.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loor G, Zhang SJ, Zhang P, Toomey NL, Lee MY. Identification of DNA replication and cell cycle proteins that interact with PCNA. Nucleic Acids Res. 1997;25:5041–5046. doi: 10.1093/nar/25.24.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowndes NF, Murguia JR. Sensing and responding to DNA damage. Curr Opin Genet Dev. 2000;10:17–25. doi: 10.1016/s0959-437x(99)00050-7. [DOI] [PubMed] [Google Scholar]
- Luo K, Hurley TR, Sefton BM. Transfer of proteins to membranes facilitates both cyanogen bromide cleavage and two-dimensional proteolytic mapping. Oncogene. 1990;5:921–923. [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Saijo M, Kuraoka I, Kobayashi T, Nakatsu Y, Nagai A, Enjoji T, Masutani C, Sugasawa K, Hanaoka F. DNA repair protein XPA binds replication protein A (RPA) J Biol Chem. 1995;270:4152–4157. doi: 10.1074/jbc.270.8.4152. [DOI] [PubMed] [Google Scholar]
- Meyn MS. Ataxia-telangiectasia, cancer and the pathobiology of the ATM gene. Clin Genet. 1999;55:289–304. doi: 10.1034/j.1399-0004.1999.550501.x. [DOI] [PubMed] [Google Scholar]
- Michel B, Ehrlich SD, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SD, Moses K, Jayaraman L, Prives C. Complex formation between p53 and replication protein A inhibits the sequence-specific DNA binding of p53 and is regulated by single- stranded DNA. Mol Cell Biol. 1997;17:2194–2201. doi: 10.1128/mcb.17.4.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WG, Kastan MB. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol. 1994;14:1815–1823. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A [see comments] Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- Niu H, Erdjument-Bromage H, Pan ZQ, Lee SH, Tempst P, Hurwitz J. Mapping of amino acid residues in the p34 subunit of human single-stranded DNA-binding protein phosphorylated by DNA-dependent protein kinase and Cdc2 kinase in vitro. J Biol Chem. 1997;272:12634–12641. doi: 10.1074/jbc.272.19.12634. [DOI] [PubMed] [Google Scholar]
- Pan ZQ, Amin AA, Gibbs E, Niu H, Hurwitz J. Phosphorylation of the p34 subunit of human single-stranded-DNA-binding protein in cyclin A-activated G1 extracts is catalyzed by cdk-cyclin A complex and DNA-dependent protein kinase. Proc Natl Acad Sci USA. 1994;91:8343–8347. doi: 10.1073/pnas.91.18.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Park SJ, Peng X, Wang M, Yu MA, Lee SH. Involvement of DNA-dependent protein kinase in UV-induced replication arrest. J Biol Chem. 1999;274:32520–32527. doi: 10.1074/jbc.274.45.32520. [DOI] [PubMed] [Google Scholar]
- Park SD, Cleaver JE. Recovery of DNA synthesis after ultraviolet irradiation of xeroderma pigmentosum cells depends on excision repair and is blocked by caffeine. Nucleic Acids Res. 1979;6:1151–1159. doi: 10.1093/nar/6.3.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson MC, Smith PJ. Ataxia telangiectasia: an inherited human disorder involving hypersensitivity to ionizing radiation and related DNA-damaging chemicals. Annu Rev Genet. 1979;13:291–318. doi: 10.1146/annurev.ge.13.120179.001451. [DOI] [PubMed] [Google Scholar]
- Patrick SM, Turchi JJ. Human replication protein A preferentially binds cisplatin-damaged duplex DNA in vitro. Biochemistry. 1998;37:8808–8815. doi: 10.1021/bi9730590. [DOI] [PubMed] [Google Scholar]
- Pedrali-Noy G, Belvedere M, Crepaldi T, Focher F, Spadari S. Inhibition of DNA replication and growth of several human and murine neoplastic cells by aphidicolin without detectable effect upon synthesis of immunoglobulins and HLA antigens. Cancer Res. 1982;42:3810–3813. [PubMed] [Google Scholar]
- Petrini JH. The Mre11 complex and ATM: collaborating to navigate S phase. Curr Opin Cell Biol. 2000;12:293–296. doi: 10.1016/s0955-0674(00)00091-0. [DOI] [PubMed] [Google Scholar]
- Rodrigo G, Roumagnac S, Wold MS, Salles B, Calsou P. DNA replication but not nucleotide excision repair is required for UVC-induced replication protein A phosphorylation in mammalian cells. Mol Cell Biol. 2000;20:2696–2705. doi: 10.1128/mcb.20.8.2696-2705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25 [see comments] Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- Sarkaria JN, Tibbetts RS, Busby EC, Kennedy AP, Hill DE, Abraham RT. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 1998;58:4375–4382. [PubMed] [Google Scholar]
- Sasaki MS. Chromosome aberration formation and sister chromatid exchange in relation to DNA repair in human cells. Basic Life Sci. 1980;15:285–313. doi: 10.1007/978-1-4684-3842-0_19. [DOI] [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- Sedgwick RP, Boder E. Ataxia-Telangiectasia (208900; 208910; 208920) In: deJong JMBV, editor. Handbook of Clinical Neurology. New York: Elsevier Science; 1991. pp. 347–423. [Google Scholar]
- Seigneur M, Bidnenko V, Ehrlich SD, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- Seigneur M, Ehrlich SD, Michel B. Blocking rolling circle replication with a UV lesion creates a deletion hotspot. Mol Microbiol. 1997;26:569–580. doi: 10.1046/j.1365-2958.1997.6021977.x. [DOI] [PubMed] [Google Scholar]
- Shao RG, Cao CX, Shimizu T, O'Connor PM, Kohn KW, Pommier Y. Abrogation of an S-phase checkpoint and potentiation of camptothecin cytotoxicity by 7-hydroxystaurosporine (UCN-01) in human cancer cell lines, possibly influenced by p53 function. Cancer Res. 1997;57:4029–4035. [PubMed] [Google Scholar]
- Shao RG, Cao CX, Zhang H, Kohn KW, Wold MS, Pommier Y. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 1999;18:1397–1406. doi: 10.1093/emboj/18.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Samson L. Replication protein A binds to regulatory elements in yeast DNA repair and DNA metabolism genes. Proc Natl Acad Sci USA. 1995;92:4907–4911. doi: 10.1073/pnas.92.11.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GC, Cary RB, Lakin ND, Hann BC, Teo SH, Chen DJ, Jackson SP. Purification and DNA binding properties of the ataxia-telangiectasia gene product ATM. Proc Natl Acad Sci USA. 1991a;96:11134–11139. doi: 10.1073/pnas.96.20.11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GC, Divecha N, Lakin ND, Jackson SP. DNA-dependent protein kinase and related proteins. Biochem Soc Symp. 1999b;64:91–104. [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- Stigger E, Drissi R, Lee SH. Functional analysis of human replication protein A in nucleotide excision repair. J Biol Chem. 1998;273:9337–9343. doi: 10.1074/jbc.273.15.9337. [DOI] [PubMed] [Google Scholar]
- Tang CM, Tomkinson AE, Lane WS, Wold MS, Seto E. Replication protein A is a component of a complex that binds the human metallothionein IIA gene transcription start site. J Biol Chem. 1996;271:21637–21644. doi: 10.1074/jbc.271.35.21637. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Byrd PJ, McConville CM, Thacker S. Genetic and cellular features of ataxia telangiectasia. Int J Radiat Biol. 1994;65:65–70. doi: 10.1080/09553009414550091. [DOI] [PubMed] [Google Scholar]
- Wang TC, Smith KC. Postreplication repair in ultraviolet-irradiated human fibroblasts: formation and repair of DNA double-strand breaks. Carcinogenesis. 1986;7:389–392. doi: 10.1093/carcin/7.3.389. [DOI] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA- binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Woods CG, Taylor AM. Ataxia telangiectasia in the British Isles: the clinical and laboratory features of 70 affected individuals. Q J Med. 1992;82:169–179. [PubMed] [Google Scholar]
- Wright J, Teraoka S, Onengut S, Tolun A, Gatti RA, Ochs HD, Concannon P. A high frequency of distinct ATM gene mutations in ataxia- telangiectasia. Am J Hum Genet. 1996;59:839–846. [PMC free article] [PubMed] [Google Scholar]
- Wright JA, Keegan KS, Herendeen DR, Bentley NJ, Carr AM, Hoekstra MF, Concannon P. Protein kinase mutants of human ATR increase sensitivity to UV and ionizing radiation and abrogate cell cycle checkpoint control. Proc Natl Acad Sci USA. 1998;95:7445–7450. doi: 10.1073/pnas.95.13.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Ranganathan V, Weisman DS, Heine WF, Ciccone DN, O'Neill TB, Crick KE, Pierce KA, Lane WS, Rathbun G, Livingston DM, Weaver DT. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response [see comments] Nature. 2000;405:477–482. doi: 10.1038/35013089. [DOI] [PubMed] [Google Scholar]
- Zernik-Kobak M, Vasunia K, Connelly M, Anderson CW, Dixon K. Sites of UV-induced phosphorylation of the p34 subunit of replication protein A from HeLa cells. J Biol Chem. 1997;272:23896–23904. doi: 10.1074/jbc.272.38.23896. [DOI] [PubMed] [Google Scholar]
- Zhang N, Chen P, Gatei M, Scott S, Khanna KK, Lavin MF. An anti-sense construct of full-length ATM cDNA imposes a radiosensitive phenotype on normal cells. Oncogene. 1998;17:811–818. doi: 10.1038/sj.onc.1202007. [DOI] [PubMed] [Google Scholar]
- Zhang N, Chen P, Khanna KK, Scott S, Gatei M, Kozlov S, Watters D, Spring K, Yen T, Lavin MF. Isolation of full-length ATM cDNA and correction of the ataxia-telangiectasia cellular phenotype. Proc Natl Acad Sci USA. 1997;94:8021–8026. doi: 10.1073/pnas.94.15.8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Weng YC, Yuan SS, Lin YT, Hsu HC, Lin SC, Gerbino E, Song MH, Zdzienicka MZ, Gatti RA, Shay JW, Ziv Y, Shiloh Y, Lee EY. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products [see comments] Nature. 2000;405:473–477. doi: 10.1038/35013083. [DOI] [PubMed] [Google Scholar]