Abstract
In this study, we demonstrate that the lack of retinoic acid-related orphan receptor (ROR) γ or α expression in mice significantly reduced the peak expression level of Cry1, Bmal1, E4bp4, Rev-Erbα and Per2 in an ROR isotype- and tissue-selective manner without affecting the phase of their rhythmic expression. Analysis of RORγ/RORα double knockout mice indicated that in certain tissues RORγ and RORα exhibited a certain degree of redundancy in regulating clock gene expression. Reporter gene analysis showed that RORγ was able to induce reporter gene activity through the RORE-containing regulatory regions of Cry1, Bmal1, Rev-Erbα and E4bp4. Co-expression of Rev-Erbα or addition of a novel ROR antagonist repressed this activation. ChIP-Seq and ChIP–Quantitative real-time polymerase chain reaction (QPCR) analysis demonstrated that in vivo RORγ regulate these genes directly and in a Zeitgeber time (ZT)-dependent manner through these ROREs. This transcriptional activation by RORs was associated with changes in histone acetylation and chromatin accessibility. The rhythmic expression of RORγ1 by clock proteins may lead to the rhythmic expression of RORγ1 target genes. The presence of RORγ binding sites and its down-regulation in RORγ−/− liver suggest that the rhythmic expression of Avpr1a depends on RORγ consistent with the concept that RORγ1 provides a link between the clock machinery and its regulation of metabolic genes.
INTRODUCTION
The retinoic acid-related orphan receptors α-γ (RORα-γ or NR1F1-3) constitute a subfamily of the nuclear receptor superfamily (1). Through alternative splicing and promoter usage, each ROR gene produces several isoforms that are expressed in a highly tissue-specific manner. RORs have been implicated in the regulation of embryonic development and in several metabolic and immunological processes (1–12).
RORs are among a number of nuclear receptors involved in the regulation of circadian behavior and clock gene expression (1,8,13–18). In mammals, the suprachiasmatic nucleus (SCN) functions as the central circadian pacemaker that integrates light–dark cycle input and synchronizes the autonomous oscillators in peripheral tissues (19–21). At the molecular level the clockwork consists of an integral network of several interlocking positive and negative transcriptional and translational feedback loops. The heterodimeric complex consisting of brain and muscle ARNT-like (Bmal1) and circadian locomotor output cycles kaput (Clock) or its paralog neuronal PAS domain protein 2 (Npas2), are involved in the positive control of the oscillator, while two cryptochrome (Cry) and three period proteins (Per) are part of the negative control mechanism. Several accessory pathways, in one of which the Rev-Erb nuclear receptors play a role (19–21), further regulate the core loop. Although several studies have provided evidence for a regulatory function of RORs, particularly roles for RORα and RORβ in the SCN, in the regulation of circadian rhythm and clock gene expression, their precise function and regulation are not yet fully understood (13,15,16,18,22–27). Less is known about the role of RORγ in the regulation of circadian rhythm. Recently, we reported that Npas2 is directly regulated by RORγ suggesting that it may be an important modulator of the circadian clock in peripheral tissues (25).
To obtain greater insights into the roles of RORs in the regulation of circadian rhythm, we examined the effects of the loss of RORα and/or RORγ on clock gene expression in several peripheral tissues of ROR knockout mice. Our data showed that particularly the loss of RORγ reduced the peak expression level of several clock genes in a tissue-selective manner without significantly affecting the phase of their rhythmic expression pattern. The effect of RORα deficiency on the expression of these clock genes was limited largely to the kidney. Loss of both RORα and RORγ in double knockout (DKO) mice had a greater impact on the peak expression levels of these clock genes than single knockouts suggesting a certain degree of redundancy between RORα and RORγ. We demonstrated that RORs regulate the transcription of Cry1, Bmal1, Rev-Erbα and E4bp4 directly as indicated by reporter gene and mutation analysis. This transcriptional activation was inhibited by ROR antagonists. Chromatin immunoprecipitation sequencing (ChIP-Seq) and ChIP-QPCR analyses indicated that RORs were associated with these ROR response elements (ROREs) in vivo supporting the conclusion that these clock genes are directly regulated by RORs. We further showed that this transcriptional regulation was ZT-dependent and associated with changes in histone acetylation and chromatin structure. Recent studies demonstrated that clock proteins and Rev-Erbs are important regulators of energy homeostasis and lipid/glucose metabolism indicating a connection between the controls of circadian clock and various metabolic pathways (14,20,28,29). Although RORs regulate the expression of several metabolic genes, whether and what role they play in this interrelationship has yet to be determined (4,14,30,31). The rhythmic expression of RORγ1 by Bmal1/Clock and Rev-Erb might lead to the rhythmic expression of RORγ1 target genes. The in-phase expression pattern of RORγ1 and the arginine vasopressin receptor 1a gene (Avpr1a), the dramatic decrease in Avpr1a expression in RORγ−/− liver, and the ChIP-Seq data are consistent with the hypothesis that RORs function as intermediate regulators, providing a link between clock proteins and their regulation of metabolic genes.
MATERIALS AND METHODS
Experimental animals
C57BL/6 staggerer (RORαsg/sg) mice, a natural mutant strain containing a 6.5 kb deletion in RORα that prevents translation of the LBD, were purchased from Jackson Laboratories (Bar Harbor, ME, USA) (32). These mice exhibit a very similar phenotype as mice with a targeted disruption of RORα (1,33). C57BL/6 RORγ−/− and RORαsg/sgRORγ−/− DKO mice were described previously (4,10). Mice were supplied ad libitum with NIH-A31 formula and water and maintained at 25°C on a constant 12 h light:12 h dark cycle. Littermate wild-type (WT) mice were used as controls. All animal protocols followed the guidelines outlined by the NIH Guide for the Care and Use of Laboratory Animals were approved by the Institutional Animal Care and Use Committee at the NIEHS.
RNA isolation
To study gene expression during circadian time, tissues were excised from WT, RORγ−/− and RORαsg/sg mice every 4 h over a period of 24 h, processed overnight in RNAlater® solution (Ambion, Austin, TX, USA) at 4°C, and then stored at –80°C until use. Tissues from DKO and littermate WT mice were collected every 6 h over a period of 24 h. Tissues were homogenized in RNeasy lysis buffer (RLT) (Qiagen, Valencia, CA, USA) with a Polytron PT-3000 (Brinkmann Instruments, Westbury, NY, USA). RNA was then extracted using a QIAshredder column and RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. Hepa1-6 stable cell lines and brown adipocytes were directly lysed in RLT buffer and extracted as described for tissues. Cells were treated for 24 h with and without 10 µM of the inverse ROR agonist T0901317 (Sigma-Aldrich, St Louis, MO, USA) (34) or with 1 µM the RORγ-selective antagonist ‘A’ (R)-N-(1-((4-methoxy-phenyl)sulfonyl)-4-methyl-1,2,3,4-tetrahydroquinolin-7-yl)-2,4,6-trimethylbenzene-sulfonamide provided by GlaxoSmithKline (V. Birault, in preparation) before RNA extraction.
QRT–PCR analysis was performed by SYBR Green I or the TaqMan system (Applied Biosystems, Foster City, CA, USA) to quantify gene expression. The RNA was reverse-transcribed using High-Capacity cDNA Archive Kit (Applied Biosystems). The reactions were carried out in triplicate in a 7300 Real Time PCR system (Applied Biosystems) using 20 ng of cDNA and the following conditions: 2 min at 45°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. All the results were normalized by the amount of 18 S rRNA or Gapdh mRNA. Product specificity was confirmed by melting curve analysis. All QRT–PCR primer and probe sequences are listed in Supplementary Table S1.
Hepa1-6, HepG2 and brown preadipocyte stable cell lines
Brown adipocytes were isolated from postnatal day 2 WT and RORγ−/− mice by collagenase digestion as described previously (25). Preadipocytes were immortalized by infection with pBabe retrovirus encoding SV40-Large T antigen as described previously (25). Brown preadipocyte cell lines BAT(RORγ1), BAT(RORα4), BAT(RORγ1E502Q) and BAT(RORα4ΔAF2) stably expressing Flag-RORγ1, Flag-RORα4, mutant Flag-RORγ1(E502Q) or Flag-RORα4ΔAF2 (35) and Hepa1-6(RORα) and Hepa1-6(RORγ) stably expressing Flag-RORα, Flag-RORγ, were generated by infection with the respective pLXIN retrovirus (Clontech, Palo Alto, CA, USA) and subsequent selection in G418 (25). The stable cell lines BAT(E) and Hep1-6(E) containing the pLXIN empty vector were used as controls. The expression and the nuclear localization of ROR proteins were confirmed by western blot analysis and confocal microscopy, respectively. Cell lines established from three to five individual clones were examined in QPCR and ChIP analysis.
ChIP assay
The ChIP assays were performed using a ChIP assay kit from Millipore (Billerica, MA, USA) according to the manufacturer’s protocol with minor modifications. In short, livers isolated from WT, RORαsg/sg and RORγ−/− mice at the circadian time indicated, were homogenized with a polytron PT 3000 (Brinkmann Instruments) and cross-linked by 1% formaldehyde for 20 min at room temperature. After a wash in phosphate buffered saline (PBS), an aliquot of the cross-linked chromatin was sonicated and incubated overnight with anti-RORα, anti-RORγ or anti-Clock antibody (sc-6062, sc-28559 or sc-6927, respectively; Santa Cruz Biotechnology, Santa Cruz, CA, USA) or anti-acetylated histone H3K9Ac antibody (07-352; Millipore). After incubation with protein G agarose beads for 2 h, and several washes, DNA–protein complexes were eluted. The cross-links were reversed by overnight incubation at 65°C in the presence of 25 mM NaCl and subsequent digestion with RNase A and proteinase K, followed by purification of the ChIPed-DNA. The ChIPed-DNA was then amplified by QPCR; all reactions were carried out in triplicate. The procedure for ChIP analysis using Hepa1-6 cells was similar as described for tissues except that 2 × 106 cells were cross-linked with 4% formaldehyde for 10 min and the immunoprecipitation was performed with anti-Flag M2 affinity gel (Sigma-Aldrich). The sequences of the primers used for ChIP-QPCR are listed in Supplementary Table S2.
ChIP-Seq analysis
ChIPed-DNA was prepared as described (36) using RORγ- and RORα-specific antibodies generated against amino acids 129–231 and 121–213 in mouse RORγ1 and RORα4, respectively. ChIP-Seq analysis was performed by the NIH Intramural Sequencing Center and data were analyzed as reported previously (36).
Formaldehyde-assisted isolation of regulatory elements
Formaldehyde-assisted isolation of regulatory elements (FAIRE) was performed as previously described (37). Briefly, mouse livers were cross-linked and sonicated as described for the ChIP assay. Samples were centrifuged and the DNA in the supernatants was isolated by three consecutive extractions with phenol–chloroform–isoamyl alcohol (25:24:1). After the final extraction, the FAIRE samples were reverse cross-linked as described for the ChIP assay. The enrichment of fragmented genomic DNA in the FAIRE samples relative to each input DNA was measured in triplicate by QPCR.
Reporter gene assay
The RORγ1(–1338/–1) and RORγ1(–1338/–968) promoter regions, and the RORE-containing regulatory regions of Cry1 (+22976/+23214), E4bp4 (+5828/+6150), Bmal1 (–650/+105), Rev-Erbα (–657/+38) and Avpr1a (–68969/–67651), and Avpr1a (–63512/–61359) were inserted into the pGL4.10 or pGL4.27 reporter plasmids. Corresponding mutant pGL4.10/27 plasmids, containing mutations within the E-boxes (CACGTG into CAATTG) or ROREs were generated using a Quickchange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). The sequences were verified by DNA sequencing. Human hepatoma Huh-7 cells were co-transfected with pCMVβ-Gal, pCMV-Sport6-Bmal1, pCMV-Sport6-Clock, pCMV-Sport6-Cry1 and the pGL4.10 RORγ1(–1338/–1) or pGL4.27-RORγ1(–1338/–968) reporter plasmid as indicated using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After 24 h incubation, the luciferase and β-galactosidase were analyzed with a Luciferase Assay Substrate (Promega) and Luminescent β-galactosidase Detection Kit II (Clontech). All transfections were performed in triplicate, and each experiment was repeated at least twice. Except for Rev-Erbα, all ROREs were mutated from A/GGGTCA to A/GAATCA. The RORE1 and RORE2 within the Rev-Erbα promoter were mutated from GTGTCACTGGGGCA to ATATCACTGGGGCA and from GTGTCACTGGGGCA to GTGTCACTGAAGCA, respectively.
RESULTS
RORγ1, not RORα, exhibited a strong oscillatory pattern of expression in peripheral tissues
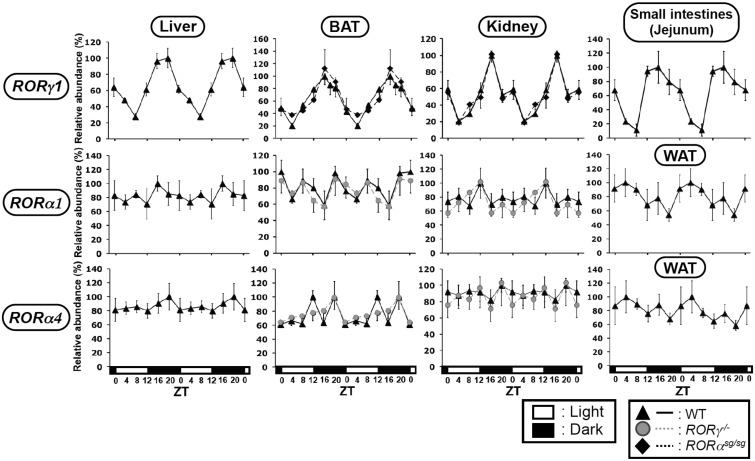
As shown in Figure 1, RORγ1 exhibited a strong oscillatory pattern of expression in mouse liver consistent with previous results with samples from a different set of mice (25). RORγ1 also exhibited a robust oscillatory pattern of expression in brown adipose tissue (BAT), kidney and small intestines (jejunum) with peak expression around ZT16 (Figure 1), a few hours earlier than in liver. RORα1 and RORα4 expression did not show a strong circadian regulation in liver, BAT, WAT or kidney (Figure 1). Because of its negligible level of expression, the circadian regulation of RORα was not analyzed in the small intestines. The oscillatory pattern of RORγ expression was not changed in BAT or kidney of RORαsg/sg mice. The loss of RORγ did not affect RORα4 expression in kidney and only slightly reduced its expression in BAT, while it had little effect on the expression of RORα1 (Figure 1). These data indicated that RORα and RORγ are regulated largely independently from each other and that RORγ1 rather than RORα1 or RORα4 exhibits a robust oscillatory pattern of expression in these peripheral tissues.
Figure 1.
Oscillatory pattern of expression of RORγ1, RORα1 and RORα4 in mouse BAT, kidney, WAT and small intestines. Liver, BAT, kidney, WAT and small intestines (jejunum) from WT, RORαsg/sg and RORγ−/− mice (n = 4) were isolated every 4 h over a period of 24 h. Subsequently, the expression of RORγ1, RORα1 and RORα4 was analyzed by QRT-PCR. The oscillatory expression patterns in liver were similar to those previously reported (25) using samples from different WT mice. The 24 h expression pattern was double-plotted. The open and solid boxes indicate the 12 h light and dark periods, respectively. Data represent mean ±SD; *P < 0.05, **P < 0.01, ***P < 0.001 by ANOVA.
Circadian regulation of clock gene expression in RORα and RORγ null mice
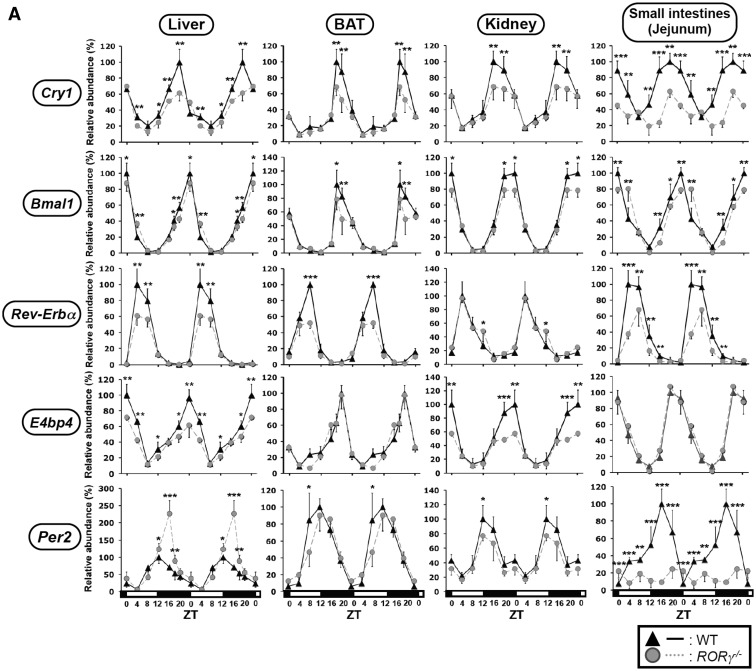
A number of studies have provided evidence for a role of RORs in the transcriptional regulation of several clock genes (13–18,21,23,24). Two recent studies reported on the transcriptional regulation of Npas2 in RORγ−/− and RORαsg/sg mice (24,25). However, information on the regulation of clock genes by RORs in vivo has remained rather limited. We, therefore, compared the circadian expression of several clock genes in more detail in a few peripheral tissues of WT, RORαsg/sg and RORγ−/– mice. In liver, BAT, WAT, kidney and small intestines of WT mice, Bmal1, E4bp4, Cry1 and Rev-Erbα mRNA reached an optimal level of expression at ZT0 (Bmal1 and E4bp4), ZT16-20 (Cry1) and ZT4-ZT8 (Rev-Erbα) in agreement with previous studies (Figure 2A and B) (13,18,38–40). In RORγ−/− mice, Cry1 mRNA expression was consistently suppressed up to 30–40% in all four tissues with the largest decrease seen in the small intestines (Figure 2A). These observations suggested that RORγ plays an important role in the regulation of Cry1 expression across several peripheral tissues. The oscillatory pattern of expression of Bmal1 in liver, BAT, kidney and small intestines from RORγ−/− mice was very similar to those in WT mice or slightly reduced at peak levels in some tissues (Figure 2A). A small shift in the circadian phase of Bmal1 expression was observed in the RORγ−/− small intestines. The peak expression of Rev-Erbα was reduced in the liver, BAT and small intestines of RORγ−/− mice, but was not significantly altered in RORγ−/− kidney. E4bp4 expression was selectively suppressed by about 40–50% at peak levels in RORγ−/− liver and kidney, but changed little in BAT and the small intestines of RORγ−/− mice. Peak expression of Per2 was somewhat elevated in RORγ−/− liver, slightly diminished in BAT and kidney, and very significantly reduced in the small intestines of RORγ−/− mice. These observations support the hypothesis that RORγ regulates the circadian expression of Cry1, Bmal1, Rev-Erbα, E4bp4 and Per2 in a tissue-selective manner.
Figure 2.
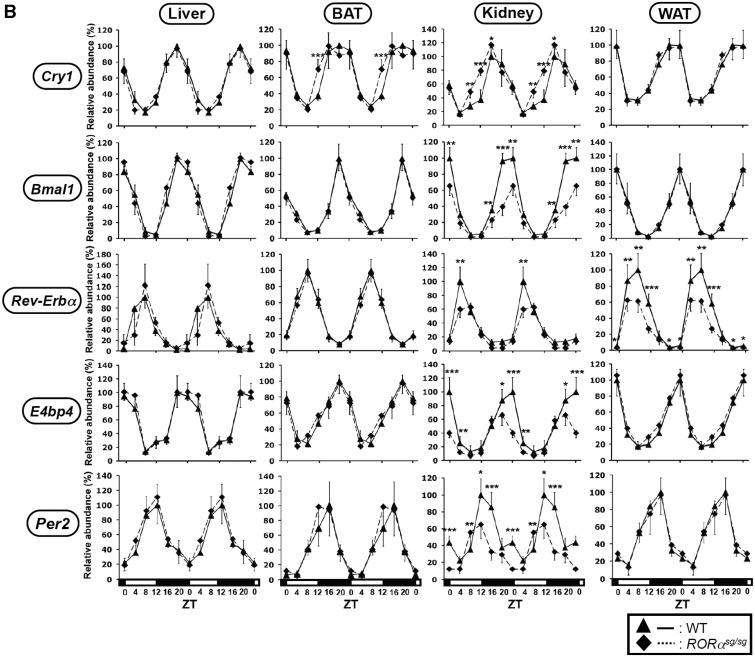
Comparison of the circadian expression pattern of Cry1, Bmal1, Rev-Erbα, E4bp4 and Per2 in several peripheral tissues from WT, RORγ−/− and RORαsg/sg mice. Liver, BAT, kidney and small intestines from WT, RORγ−/− (A) and RORαsg/sg (B) mice (n = 4) were isolated every 4 h over a period of 24 h and expression of was analyzed by QRT-PCR as indicated. The 24 h expression pattern was double-plotted. Data represent mean ±SD; *P < 0.05, **P < 0.01, ***P < 0.001 by ANOVA.
We next examined the expression of these clock genes in liver, BAT, WAT and kidney of RORαsg/sg mice. As shown for RORγ−/− mice, the phase of the oscillatory pattern of expression of Bmal1, Cry1, Rev-Erbα, E4bp4 and Per2 was not significantly altered in any of the RORαsg/sg tissues analyzed (Figure 2B). In contrast, a significant reduction in the peak level of expression of Bmal1, E4bp4 (ZT18-20) and Per2 (ZT12) was observed in the kidneys of RORαsg/sg mice. Rev-Erbα peak expression at ZT8 was reduced in the kidney and WAT of RORαsg/sg mice, but was unaltered in the liver and BAT. No significant changes in the rhythmic expression pattern of Cry1 were observed in any of the four tissues of RORαsg/sg mice. These results demonstrated that the loss of RORα affected the expression of certain clock genes in a limited and tissue-selective manner. RORα regulated clock gene expression particularly in the kidney, a tissue in which it is relatively highly expressed (Y. Takeda, data not shown).
RORα and RORγ redundancy
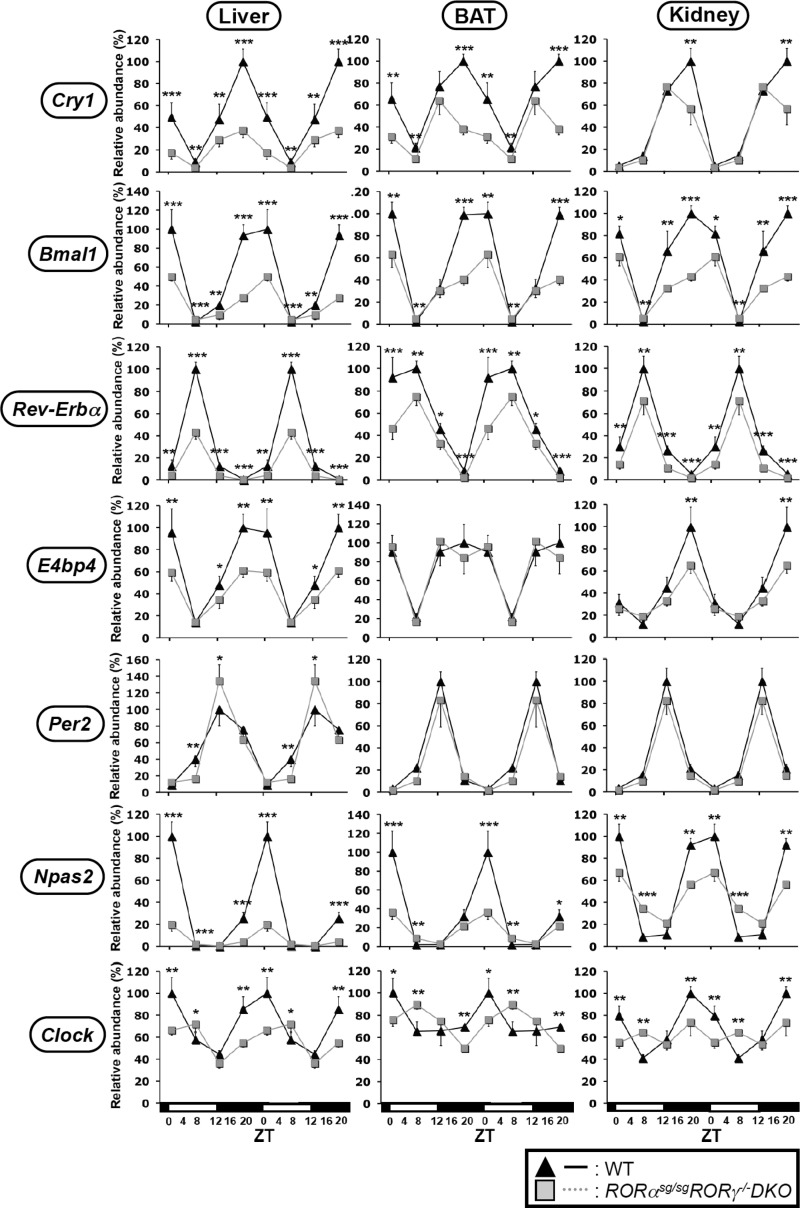
Previous studies have indicated that RORα and RORγ exhibited a certain degree of redundancy with respect to their effects on gene expression (4,7). To examine whether RORα and RORγ exhibited any redundancy in regulating clock gene expression, we examined the circadian expression of clock genes in liver, BAT and kidney collected from RORαsg/sgRORγ−/− DKO mice. As shown in Figure 3 and Supplementary Figure S1, the expression of several clock genes was reduced to a greater extent in certain tissues of DKO mice than those of RORγ−/− or RORαsg/sg single knockout mice. Particularly, Cry1 and Bmal1 mRNA expression were down-regulated to a significantly greater extent in the liver and BAT of DKO mice than in single knockout mice. The peak expression of Npas2 and Clock was also notably more reduced in DKO liver compared with their reported expression in single knockout mice (25). Little redundancy between RORα and RORγ was observed in the kidney. These results indicated that in some peripheral tissues RORα and RORγ exhibit a certain degree of redundancy regulating clock gene expression.
Figure 3.
Analysis of the circadian expression pattern of Cry1, Bmal1, Rev-Erbα, E4bp4, Per2, Npas2 and Clock in DKO mice. Liver, BAT and kidney were isolated at ZT2, ZT8, ZT14 and ZT20 from WT and RORαsg/sg RORγ−/− DKO mice (n = 4) and the expression of clock genes examined by QRT-PCR. The level of expression was normalized to the expression peak of littermate WT mice controls. Data represent mean ±SD; *P < 0.05, **P < 0.01, ***P < 0.001 by ANOVA.
RORs regulate clock gene expression through ROREs
RORs bind as monomers to ROREs consisting of the consensus sequence AGGTCA preceded by a 6-bp A/T rich region, in the promoter regulatory region of target genes (1). ROREs have been identified in several clock genes; Cry1, Bmal1 and Rev-Erbα contain two putative ROREs, while the E4bp4 and Clock genes contain one site (Supplementary Figure S2A) (13,15,16,18,22,23,38,41,42). However, the significance of these ROREs and the role of RORγ1 in clock gene regulation in vivo are still far from understood. Both RORα and RORγ were able to induce the Luc reporter driven by the RORE-containing regulatory region of Cry1, E4bp4, Bmal1, Rev-Erbα and Clock in Huh-7 cells. RORα and RORγ induced promoter activity of Cry1, respectively, 24- and 8-fold, Bmal1 16- and 8-fold, Rev-Erbα 7- and 5-fold, 3- and 2-fold (Supplementary Figure S2B) and Clock 4- and 2-fold (Supplementary Figure S3A) compared with cells transfected with the empty vector. Except for the RORE2 in Rev-Erbα, point mutations in each of these ROREs greatly reduced the activation of the Luc reporter by either RORα or RORγ. Double mutations in RORE1 and RORE2 in Cry1 and Bmal1 almost totally abolished the activation by RORs. These observations suggest that both ROREs are important in the regulation of Cry1 and Bmal1 and that RORE1 is most critical for the ROR-mediated regulation of Rev-Erbα, E4bp4 and Clock consistent with previous observations (13,15,22). Similar results as for Huh-7 cells were obtained in HEK293 cells (Y. Takeda, data not shown).
Inhibition of ROR-mediated activation of clock gene promoters by Rev-Erbα and ROR antagonists
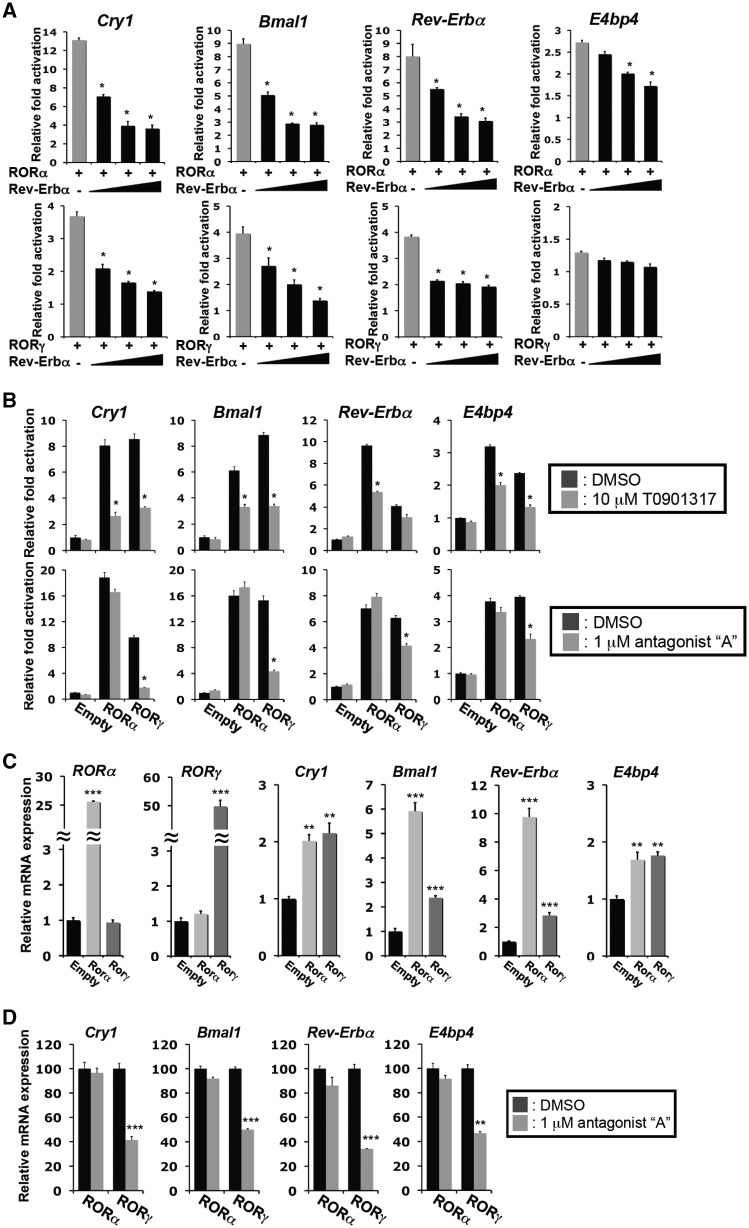
Previous studies reported that the transcriptional repressor Rev-Erbα can compete with RORs for the binding to ROREs (14,15,38,43). Figure 4A shows that exogenous Rev-Erbα expression effectively inhibited the Cry1(RORE1/2), Bmal1(RORE1/2), Rev-Erbα(RORE1)-dependent activation by either RORγ or RORα in a dose-dependent manner, likely by competing with RORα and RORγ for the same ROREs. Rev-Erbα had only a small effect on the E4bp4(RORE)-dependent activation by RORs (Figure 4A).
Figure 4.
Rev-Erbα and ROR antagonists inhibited the activation of the RORE-containing regulatory regions by RORs. (A) Rev-Erbα co-expression inhibited the activation of the Cry1, Bmal1, Rev-Erbα and E4bp4 regulatory regions by RORs. Huh-7 cells were transfected with p3xFlag-CMV10-RORγ or p3xFlag-CMV10-RORα, pGL4.27 reporter plasmid driven by the RORE-containing regulatory region of Cry1, Bmal1, Rev-Erbα or E4bp4, and increasing concentrations (ROR:Rev-Erbα 1:0.3; 1:1; 1:2) of p3xFlag-CMV10-Rev-Erbα. Twenty-four hours later cells were assayed for reporter gene activity. (B) Inhibition of transcriptional activation by the ROR antagonists, T0901317 and compound A. Huh-7 cells transfected with p3xFlag-CMV10-RORγ or p3xFlag-CMV10-RORα, and the pGL4.27 reporter plasmid driven by the RORE-containing regulatory region of Cry1, Bmal1, Rev-Erbα or E4bp4, were treated with the indicated antagonist. Data represent mean ±SEM; *P < 0.01 by ANOVA. (C) Exogenous expression of RORγ1 or RORα4 in Hepa1-6 cells increased the expression of Cry1, Bmal1, Rev-Erbα and E4bp4. The expression of clock genes in Hepa1-6 cells (n = 5) stably expressing the empty vector, Flag-RORα4 or Flag-RORγ1 was examined by QRT-PCR analysis. The level of clock gene expression in Hepa1-6(Empty) was normalized to 1. (D) Treatment of Hepa1-6 cells with the RORγ-selective antagonist compound A (1 µM) repressed the induction of clock gene expression by RORγ, but not that by RORα. Data represent mean ±SEM; **P < 0.01, ***P < 0.001 by ANOVA.
Recently several ligands have been identified that function either as ROR agonists or antagonists (34,44–46). To examine the regulation of clock genes by RORs further, we examined the effect of the RORα and RORγ inverse agonist, T0901317, and the RORγ-selective antagonist ‘A’ on the activation of the RORE-containing regulatory region of several clock genes by RORs. T0901317 significantly inhibited Cry1(RORE1/2)-, Bmal1(RORE)-, Rev-Erbα(RORE)-, E4bp4(RORE)- and Clock(RORE)-dependent trans-activation of the reporter by both RORα and RORγ in Huh-7 cells (Figure 4B and Supplementary Figure S3B). In contrast, the RORγ-selective antagonist ‘A’ effectively inhibited the RORγ-induced trans-activation at concentrations 10× lower than T0901317, but had little effect on the activation mediated by RORα (Figure 4B).
To analyze the role of RORs and the effect of the RORγ-selective antagonist ‘A’, on the endogenous expression of Cry1, Bmal1, Rev-Erbα and E4bp4, we generated Hepa1-6 cells stably expressing the empty vector, Flag-RORα4 or Flag-RORγ1 (25). Expression of RORα and RORγ mRNA was greatly elevated in Hepa1-6(RORα4) and Hepa1-6(RORγ1), respectively (Figure 4C). Exogenous expression of RORα4 did not affect the levels of RORγ1 mRNA, neither did RORγ1 affect the expression of RORα, consistent with our conclusion that RORα and RORγ1 are regulated independently from each other. Most importantly, RORγ1 and RORα4 significantly increased the endogenous expression of Cry1, Bmal1, Rev-Erbα, E4bp4 and Clock compared with Hepa1-6(Empty) cells (Figure 4C and Supplementary Figure S3C). The addition of the RORγ-selective antagonist ‘A’ significantly reduced the expression of clock genes in Hepa1-6(RORγ1) cells, but had little effect in Hepa1-6(RORα4) cells (Figure 4D).
As shown for Hepa1-6 cells, Baml1, Cry1, Rev-Erbα and E4bp4 expression was also increased in brown preadipocyte cell lines, BAT(RORα4) and BAT(RORγ1), stably expressing Flag-RORα4 and Flag-RORγ1, respectively (Supplementary Figure S4). However, the expression of these clock genes was not changed in BAT(Flag-RORαΔAF2) and BAT(RORγE502Q), expressing Flag-RORαΔAF2 or Flag-RORγE502Q, mutants defective in their trans-activation function. Treatment of BAT(RORγ1) cells with the inverse agonist T0901317 inhibited the increased expression of Cry1, Bmal1, Rev-Erbα and E4bp4. All together, these observations are consistent with the conclusion that the expression of these clock genes is regulated by RORγ and RORα and is dependent on the activation domain of RORs.
Enhanced association of RORα and RORγ with the RORE-containing regulatory regions of other clock genes in vivo correlated with increased chromatin accessibility
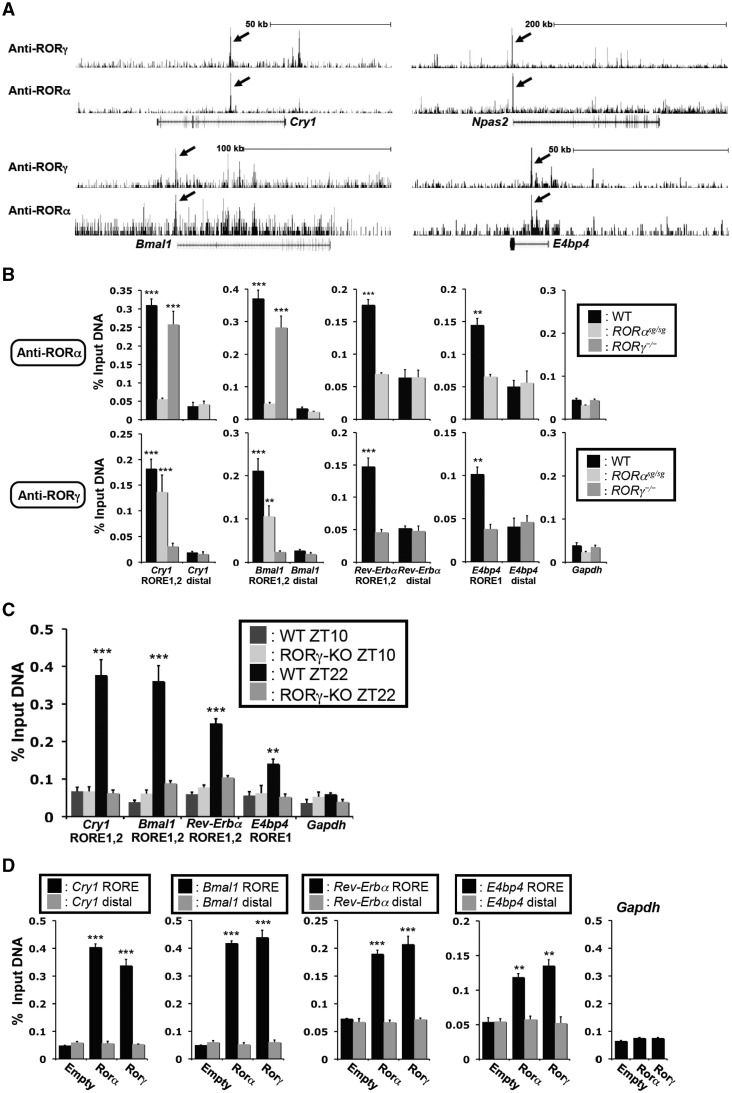
To determine whether these clock genes were directly regulated by RORα/γ in vivo, we analyzed our genome-wide maps of RORα/γ binding sites derived from ChIP-Seq analysis using ChIPed-chromatin from ZT22 mouse liver, for clock gene-associated ROR binding sites. This analysis identified RORα and RORγ binding peaks at RORE-containing regions near or within the Cry1, Npas2, Bmal1 and E4bp4 genes (Figure 5A). The strongest peaks were associated with Cry1 and Npas2, while a moderate association was found with Bmal1 and E4bp4. Low associations were found with ROREs within Rev-Erbα and Clock (data not shown). Most importantly, the strong ROR-binding peaks (indicated by arrows) for Cry1, Bmal1 and E4bp4 corresponded to the RORE-containing regions studied in Supplementary Figure S2. In the case of Npas2, the strong ROR-binding peak corresponded to the Npas2-RORE described recently (24,25).
Figure 5.
RORα and RORγ were associated with the RORE-containing regulatory regions of Cry1, Bmal1, Rev-Erbα and E4bp4 in vivo. (A) Representative view of ChIP-Seq results using either anti-RORγ or -RORα antibody in mouse liver tissue in Cry1, Npas2, Bmal1 and E4bp4 genes. Arrows indicate the peaks corresponding to ROREs studied in this article. Gene tracks were taken from the UCSC Genome Browser using mouse mm9 reference genome. (B) Recruitment of RORα and RORγ to the ROREs in Cry1, Bmal1, Rev-Erbα and E4bp4 in vivo. ChIP-QPCR analysis was performed with chromatin isolated from livers of WT, RORαsg/sg and RORγ−/− mice (n = 4) collected at ZT22 and anti-RORα or -RORγ antibodies. QPCR amplification of distal sites and Gapdh were used as negative controls. (C) The recruitment of RORγ to the ROREs in Cry1, Bmal1, Rev-Erbα and E4bp4 was ZT-dependent. ChIP-QPCR was performed using an anti-RORγ antibody and chromatin from WT livers collected at ZT10 (low expression of RORγ) or ZT22 (high expression of RORγ). (D) Recruitment of RORα and RORγ to the RORE-containing regulatory regions of Cry1, Bmal1, Rev-Erbα and E4bp4 in Hepa1-6 cells. ChIP analysis was performed with chromatin isolated from Hepa1-6(Empty), Hepa1-6(RORα4) and Hepa1-6(RORγ1) cells and anti-Flag M2 antibody. QRT-PCR amplification in samples from Hepa1-6(Empty) cells and of the Gapdh gene served as negative controls. Data represent mean ±SEM; **P < 0.01, ***P < 0.001 by ANOVA.
To examine the specificity of the recruitment of RORs to the RORE-containing regulatory region of the Bmal1, Cry1, Rev-Erbα and E4bp4 genes in vivo, ChIP-QPCR was carried out using ChIPed liver DNA from WT, RORαsg/sg and RORγ−/− mice (Figure 5B and Supplementary Figure S3D). Both RORα and RORγ were found to be associated with these RORE-containing regulatory regions in WT liver. As expected, ChIP analysis with the RORγ antibody did not show a significant increase in amplification of these regulatory regions in RORγ−/– liver; however, RORγ was still recruited to these RORE regions in RORαsg/sg liver in agreement with the specificity of the RORγ antibody. ChIP analysis with RORα antibody yielded the inverse results. Non-RORE containing regions within Gapdh and clock genes, which served as negative controls, showed low amplification.
Because of the strong oscillatory pattern of expression of RORγ1, we compared ChIP-QPCR analysis with liver tissues collected at ZT22, a time close to the peak expression of RORγ1 and Cry1 and Bmal1, with those isolated at ZT10, when these genes are expressed at lower levels. We hypothesized that RORγ1 would be associated with the Bmal1(RORE1/2) and Cry1(RORE1/2) regions particularly at the time when they are most highly expressed. Figure 5C confirmed that RORγ1 was more efficiently recruited to these regulatory regions in ZT22 liver than in ZT10 liver. No increased amplification was observed with samples from RORγ−/− liver at either ZT. RORγ1 showed a weaker association with the Rev-Erbα(RORE1) and E4bp4(RORE1) regions (Figure 5C). The recruitment of RORγ1 to the Rev-Erbα(RORE1) was higher at ZT22 than ZT10 and correlated inversely with the level of Rev-Erbα expression.
The association of RORα and RORγ with the RORE-containing regulatory regions of the Bmal1, Cry1, Rev-Erbα and E4bp4 was supported by ChIP analysis using an anti-Flag M2 antibody and chromatin isolated from Hepa1-6(Empty), Hepa1-6(RORα4) Hepa1-6(RORγ1) cells. As shown in Figure 5D, RORα and RORγ were strongly associated with ROREs in the Bmal1(RORE1/2) and Cry1(RORE1/2) regions and to a lesser extend to those of Rev-Erbα and E4bp4. No significant recruitment of RORs was observed in Hepa1-6(Empty) cells, to distal sites or the Gapdh promoter, which served as negative controls. Collectively, the ChIP-Seq and ChIP-QPCR data are in agreement with the concept that both RORα and RORγ regulate several clock genes directly through their interaction with specific ROREs.
Chromatin structure
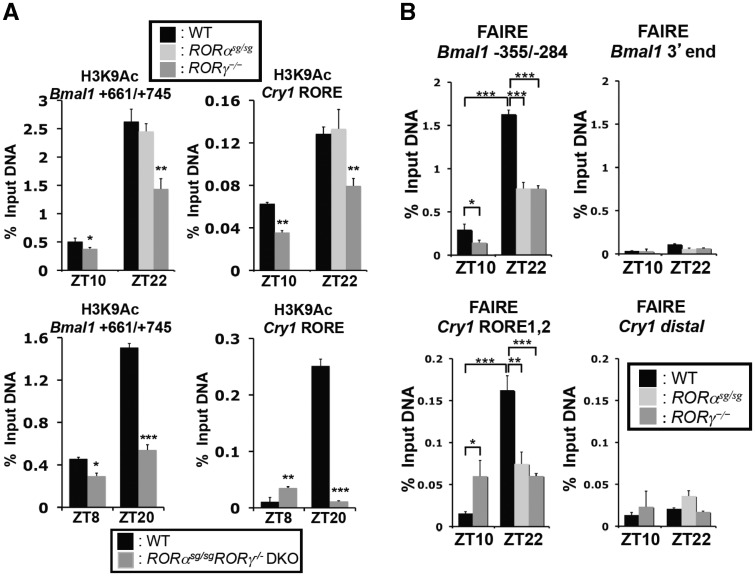
Changes in gene expression are often associated with alterations in chromatin structure, histone methylation and acetylation. We, therefore, examined whether the association of RORs with the Bmal1(RORE1/2) and Cry1(RORE1/2) regions correlated with changes in histone acetylation and chromatin accessibility. Levels of histone acetylation, including H3K9Ac have been reported to correlate positively with actively transcribed genes (47). ChIP analysis with DNA samples from livers isolated at ZT10 and ZT22 showed that the level of H3K9Ac associated with the Cry1(RORE1/2) and Bmal1(RORE1/2) regions was considerably higher at ZT22 than at ZT10 (Figure 6A). This correlated with the high level of association of RORα and RORγ with these regions (Figure 5A and B) and the elevated expression of these clock genes at ZT22. The H3K9Ac signal was significantly reduced in RORγ−/− liver, not affected in RORαsg/sg liver, and further decreased in DKO mice. These observations indicate that the recruitment of particularly RORγ is associated with actively transcribed chromatin and elevated expression of Bmal1 and Cry1.
Figure 6.
The recruitment of RORα and RORγ to the RORE-containing regulatory regions of Cry1 and Bmal1 was associated with chromatin modifications. (A) The association of H3K9Ac with the RORE-containing regulatory region of Cry1 and Bmal1 was analyzed by ChIP analysis using chromatin samples prepared from liver of WT, RORαsg/sg, RORγ−/− and RORαsg/sgRORγ−/− DKO mice (n = 4) and an anti-H3K9Ac antibody. (B) DNA accessibility at the RORE sites was assessed by FAIRE-QPCR analysis. FAIRE-QPCR at the RORE-containing region indicated and at a downstream distal site was performed using chromatin prepared from WT, RORαsg/sg and RORγ−/− livers collected at ZT10 or ZT22. Data represent mean ±SEM; *P < 0.05, ***P < 0.001 by ANOVA.
To assess chromatin accessibility, we performed FAIRE, which has been used as a tool to identify actively transcribed genes (37). FAIRE analysis on the Cry1(RORE1/2) and Bmal1(RORE1/2) regions with chromatin isolated from liver at ZT10 and ZT22 showed that the FAIRE signals were markedly higher at ZT22, a time at which Cry1 and Bmal1 are highly expressed, compared with that of ZT10 (Figure 6B). There was no significant change in FAIRE signal at the distal region of the Cry1 gene. Loss of RORγ or RORα expression led to a reduction in FAIRE signal at ZT22. This observation is consistent with our H3K9Ac data and suggests that the binding of RORs may promote a more open chromatin structure at these regulatory sites.
Transcriptional activation of the RORγ1 promoter by Clock/Bmal1 heterodimers was repressed by Cry1 and correlated with changes in chromatin accessibility
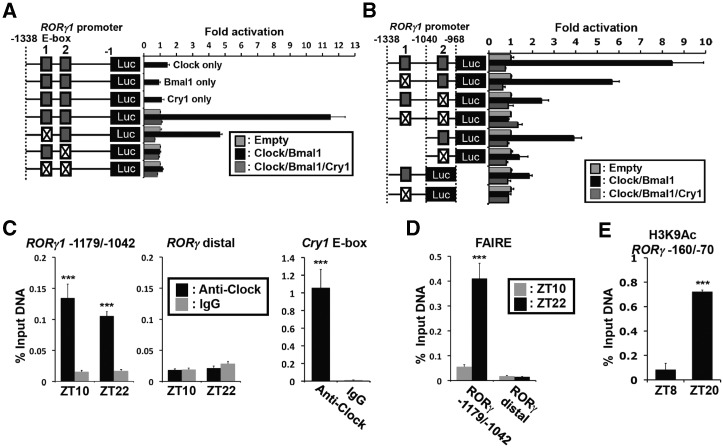
As shown in Figure 1, RORγ1 has a strong oscillatory pattern of expression. Previous studies reported that the RORγ1 promoter contains two successive E-boxes (E1 and E2) and is activated by Clock/Bmal1 heterodimers (18,48). Reporter gene analyses in human hepatoma Huh-7 cells confirmed that Clock/Bmal1 greatly enhanced RORγ1(–1338/–1) proximal promoter activity and that mutation of either E1 or E2 significantly reduced the activation, while the double mutation totally abolished the induction by Clock/Bmal1. Co-expression with the transcriptional repressor Cry1 almost completely blocked the activation of the RORγ1 promoter by Clock/Bmal1 (Figure 7A and B). To determine whether Clock protein was associated with the E-box-containing RORγ1(–1179/–1042), we performed ChIP analysis using chromatin isolated from mouse liver and an anti-Clock antibody. Our data showed that Clock protein was recruited to the E-box region of the proximal RORγ1(–1179/–1042) promoter region in vivo (Figure 7C). The recruitment of Clock protein to the Cry1 E-box (38) was used as a positive control. Relatively little association of Clock protein was observed with a distant region (at –6.2 kb) of the RORγ1 promoter, which served as a negative control. The level of Clock recruitment to the RORγ1(–1179/–1042) region was not very different between ZT10 and ZT22 similar to what has been reported previously for the recruitment of Clock/Bmal1 to the Cry1(E-box) promoter region (38). These data strengthen the previous conclusions (18,48) that Clock/Bmal1 and Cry1 complexes regulate RORγ1 expression through their interaction with E-boxes in the RORγ1 promoter.
Figure 7.
The activation of the RORγ1 promoter by Bmal1/Clock was repressed by Cry1, and associated with recruitment of Clock and increased accessibility to the RORγ1(E-box1, 2) site. (A, B) Activation of the RORγ1 promoter by Bmal1/Clock. Huh-7 cells were co-transfected with pGL4 reporter plasmids containing the wild type or mutant RORγ1(–1338/–1) (A) or RORγ1(–1338/–968) (B) promoter region, pCMV-β-Gal, empty vector, and pCMV-Sport6 expression plasmids for Clock, Bmal1 and Cry1 as indicated. The E-box 1 and 2 (solid boxes) were mutated from CACGTG to CAATTG (crossed box) as indicated. (C) Recruitment of Clock protein to the RORγ1(E-box1,2) promoter site in vivo. ChIP-QPCR at the RORγ1(–1179-/–1042) site was performed using an anti-Clock antibody and chromatin isolated from mouse livers collected at ZT10 or ZT22. ChIP with an anti-IgG and QPCR amplification of a distal site (–6.2 kb) of RORγ1 were used as negative controls. ChIP analysis targeting the E-box in Cry1 was used as a positive control. The experiment was repeated twice independently. (D) Chromatin accessibility at the RORγ1(E-box1, 2) region was assessed by FAIRE-QPCR analysis using chromatin samples prepared from WT livers isolated at ZT10 or ZT22. FAIRE-QPCR at a distal RORγ1 site was used as a negative control. (E) Activation of the RORγ promoter correlated with an increase in the association of H3K9Ac. ChIP-QPCR analysis was performed using an anti-H3K9Ac antibody and chromatin isolated from mouse livers collected at ZT8 or ZT20. Data represent mean ±SEM; ***P < 0.001 by ANOVA.
To assess chromatin accessibility in relation to the level of RORγ1 expression, we performed FAIRE on the RORγ1(–1179/–1042) promoter region with chromatin isolated from liver at ZT10 and ZT22. As shown in Figure 7D, the FAIRE-QPCR signal was markedly higher in samples isolated from liver at ZT22, a time at which RORγ1 is highly expressed, compared with ZT10 when RORγ1 expression is low. A low FAIRE-QPCR signal was obtained at a distal site. Our results are consistent with the concept that at ZT22 this region of the RORγ1 promoter is more accessible and associated with functionally active enhancers. This was supported by ChIP analysis using an H3K9Ac antibody. The level of H3K9Ac associated with the RORγ1(–160/–70) region was considerably higher at ZT20 than at ZT8 (Figure 7E). This is consistent with the high level of RORγ1 mRNA expression at ZT20 and a more open chromatin structure at this site.
Regulation of the rhythmic expression of RORγ1 target genes
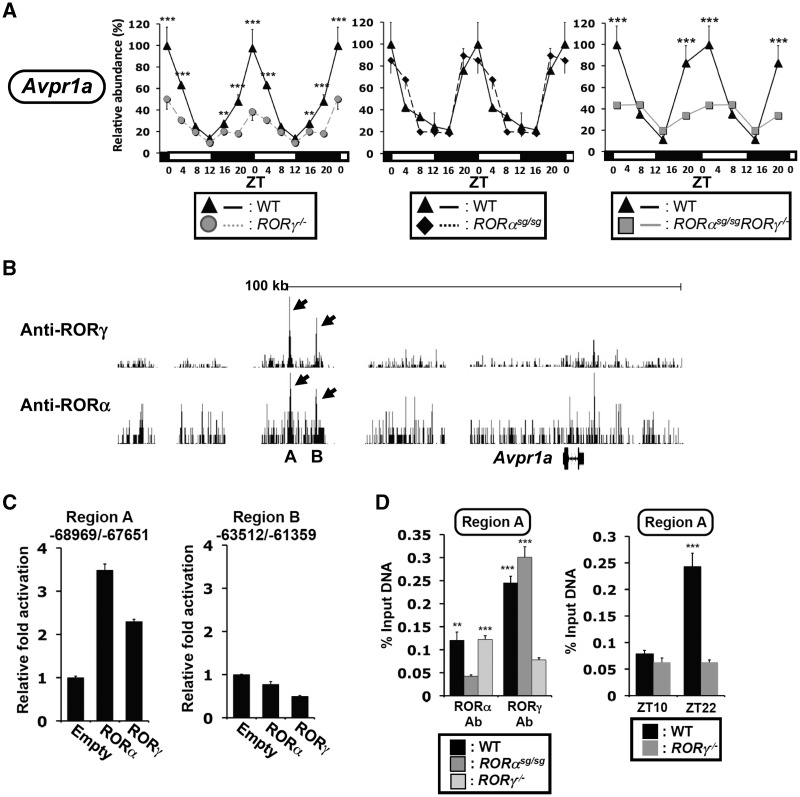
We previously proposed that in addition to their modulation of clock gene expression, RORs might function as a link between the clock machinery and the circadian regulation of down-stream genes (1). The loss of RORs has been shown to affect the rhythmic expression of several genes involved in glucose and lipid metabolism, including perilipin 2 (Plin2 or Adfp) and arginine vasopressin receptor 1a (Avpr1a) (17,31). Figure 8A shows that as for Npas2 and Cry1, the peak expression of Avpr1a at ZT0, a time at which RORγ1 is most highly expressed, was greatly diminished in RORγ−/− liver. The loss of RORα had little effect on Avpr1a expression, while the pattern of Avpr1a expression in DKO liver was very similar to that in RORγ−/− liver. These observations indicate that Avpr1a expression is selectively regulated by RORγ1. Analysis of our genome-wide map of RORγ binding sites showed strong association of RORγ with two regions, A and B, upstream from the Avpr1a transcription start site (Figure 8B). RORγ was able to induce region A-dependent trans-activation, but not region B-mediated trans-activation (Figure 8C). ChIP analysis showed that RORγ was recruited to region A at ZT22 (Figure 8D), but not at ZT10, as observed for clock genes (Figure 5C). Although overexpression of RORα induced region A-dependent trans-activation (Figure 8C), RORα was only weakly associated with region A and B (Figure 8B and D). Together, these observations suggest that RORγ1, rather than RORα, is involved in the direct regulation of the rhythmic Avpr1a expression.
Figure 8.
Regulation of the circadian expression of Avpr1a by RORγ. Liver from WT, RORγ−/−, RORαsg/sg and RORαsg/sgRORγ−/− DKO mice (n = 4) were isolated every 4 h over a period of 24 h and Avpr1a mRNA was quantified by QRT-PCR. The 24 h expression pattern was double-plotted. Data represent mean ±SD; **P < 0.01 and ***P < 0.001 by ANOVA. (B) Analysis of our genome-wide map of ROR binding sites showed strong association of RORγ with two regions, A and B, upstream from the Avpr1a transcription start and weak association of RORα. Data were derived from ChIP-Seq analysis using ChIPed chromatin from liver and an anti-RORγ or -RORα antibody. (C) Exogenous expression of RORα/γ was able to induce region A-dependent trans-activation, but not region B-mediated trans-activation in Huh-7 cells. (D) ChIP-QPCR analysis was performed with chromatin isolated from livers of WT, RORαsg/sg and RORγ−/− mice (n = 4) collected at ZT22 and an anti-RORα or -RORγ antibody (left panel). The recruitment of RORγ to region A was ZT-dependent. ChIP-QPCR was performed using an anti-RORγ antibody and chromatin from WT livers collected at ZT10 (low expression of RORγ) or ZT22 (high expression of RORγ) (right panel). Data represent mean ±SEM; **P < 0.01, ***P < 0.001 by ANOVA.
DISCUSSION
In this study, we demonstrate that RORα4 and RORα1 have a weak and that RORγ1 exhibits a robust oscillatory pattern of expression in BAT, WAT, kidney and jejunum as we reported previously for liver (25). We further show that the loss of RORγ did not significantly alter the expression pattern of RORα1 or RORα4, while the loss of RORα had little effect on the oscillatory expression pattern of RORγ1 in either BAT or kidney suggesting that RORα and RORγ are regulated independently from each other under the conditions tested. To obtain further insights into the roles of RORα and RORγ in circadian regulation in vivo, we compared the oscillatory expression pattern of Cry1, Baml1, E4bp4, Rev-Erbα and Per2 in several tissues from RORα and RORγ single knockout and RORα/RORγ DKO mice. The loss of either RORα or RORγ, or both had either a small or no effect on the phase of the rhythmic expression of these clock genes (Figures 2 and 3). However, the loss of RORα or RORγ affected the peak expression of several clock genes in a tissue- and ROR isotype-selective manner. Loss of RORγ significantly reduced the peak expression of Cry1, Rev-Erbα, E4bp4 and Per2 in most tissues tested and caused a small reduction in Bmal1 peak expression. The increased levels of Per2 in RORγ−/− liver might be related to the observed, reduced expression of E4bp4, which has been reported to function as a negative regulator of Per2 (49). The effect of the loss of RORα on the expression of Bmal1, Rev-Erbα, E4bp4 and Per2 was limited mainly to the kidney, a tissue in which RORα is relatively highly expressed, while Cry1 expression was little changed in RORαsg/sg kidney. We reported previously that the loss of RORγ, but not the loss of RORα, reduced peak expression of Npas2 and Clock (25). These observations indicated that both RORs, but particularly RORγ, are modulators of clock gene expression in several peripheral tissues. The tissue selectivity of clock gene regulation by RORα and RORγ might be due to differences in their level of expression and/or transcriptional activity.
We previously reported that RORα and RORγ exhibit a certain degree of functional redundancy in liver and Th17 cells. The hepatic expression of several Phase I and Phase II metabolic genes was affected to considerably greater extent in DKO than in single knockout mice (4) and both RORα and RORγ have been shown to promote Th17 differentiation and Il-17 expression (50). We, therefore, were interested in determining whether RORα and RORγ exhibited any redundancy in regulating clock gene expression in liver, kidney and BAT. Comparison of Cry1, Bmal1, Rev-Erbα, E4bp4, Per2, Npas2 and Clock expression between single knockout and DKO mice showed a complex picture. The expression of Cry1, Bmal1, Rev-Erbα, Npas2 and Clock in particular was reduced to a significantly greater degree in liver and BAT of DKO mice than in single knockout mice (Figure 3 and Supplementary Figure S1). A greater reduction in Cry1, Bmal1 and Rev-Erbα expression was also observed in DKO kidney; however, little redundancy was observed in the small intestines (jejunum), where RORα is not significantly expressed. Our data suggest that RORα and RORγ exhibit a degree of functional redundancy in their regulation of clock gene expression that explains some of the synergistic effects observed in some tissues of DKO mice. It is interesting to note that Rev-Erbα and Rev-Erbβ, which can compete with RORs for the same bindings sites, also work together in regulating clock gene expression (51,52).
The regulation of Cry1, E4bp4, Baml1 and Rev-Erbα by RORα4 and RORγ1 was supported by data showing that exogenous expression of RORα4 or RORγ1 in Hepa1-6 and BAT cells increased the expression of all four clock genes (Figure 4C and Supplementary Figure S4). Mutant ROR receptors lacking trans-activating activity were unable to enhance expression of these clock genes indicating that the induction of clock genes by RORs is dependent on the activation function of RORs. This was supported by data showing that ROR antagonists significantly reduced the induction of Bmal1, Cry1, E4bp4 and Rev-Erbα mRNA by RORα and RORγ.
RORs regulate gene transcription directly by binding ROREs in the regulatory region of target genes (1). Previous studies have identified putative ROREs in a number of clock genes (13,15,18,22–25,41). Reporter gene analysis showed that both RORα and RORγ were able to activate the Luc reporter under the control of the RORE-containing regions of Cry1, Bmal1, Rev-Erbα, E4bp4 or Clock (Supplementary Figures S2 and S3). Mutation of either the RORE1 or RORE2 in Bmal1 or Cry1 partially reduced this activation, whereas mutation of both ROREs totally abolished the activation of the Luc reporter suggesting that both RORE1 and RORE2 are required for the optimal activation of these genes by RORα and RORγ. In the case of E4bp4, Rev-Erbα and Clock the activation by RORα and RORγ was mediated through RORE1. The activation of these regulatory regions was inhibited by co-expression with Rev-Erbα, a transcriptional repressor that can compete with RORs for RORE binding (14,15,41,43). Moreover, treatment with the RORα and RORγ inverse agonist T0901317 significantly inhibited the activation by either RORα or RORγ, while the RORγ-selective antagonist ‘A’ inhibited the induction by RORγ, but not that by RORα (Figure 4 and Supplementary Figures S3 and S4). These observations are consistent with the concept that RORα and RORγ enhance the expression of clock genes directly by binding ROREs in their promoter regulatory region and further confirm that this induction requires the activation function of RORs. The mouse and human Cry1(RORE1/2) are distal from the transcription start site (TSS) and located within introns at +23086/23141 and +66014/66068, respectively. ROR activator complexes bound to distal ROREs might exert coordinated control of Cry1 expression via a physical interaction with protein complexes at the proximal promoter or TSS (53).
Our genome-wide maps of RORα/γ binding sites in liver identified major RORα and RORγ binding peaks near or within the Cry1, Npas2, Bmal1 and E4bp4 genes (Figure 5A). These sites corresponded to the ROREs studied in this article or reported previously (18,24,25). The recruitment of RORs to these regions was further supported by ChIP-QPCR analysis using ChIPed DNA from liver tissue and Hepa1-6(RORγ1) or Hepa1-6(RORα4) cells. The specificity of ROR binding was indicated by data showing that no significant recruitment of RORα or RORγ was observed in, respectively, RORαsg/sg and RORγ−/– liver or to non-RORE containing distal regions of the respective clock gene or Gapdh. Because RORγ1 exhibits a robust circadian pattern of expression, we determined whether the recruitment of RORγ1 to the regulatory regions was ZT-dependent. This was supported by ChIP-QPCR analysis showing higher levels of association of RORγ1 with the RORE-containing regulatory regions of Cry1, Bmal1 and E4bp4 at ZT22, when RORγ as well as these clock genes are relatively highly expressed, than at ZT10, when these genes are expressed at low levels. Moreover, increased competition for RORE binding by Rev-Erbα/β, which are highly expressed at ZT10, is likely a major factor in the reduced interaction of RORs with ROREs at this ZT. Unexpectedly, the recruitment of RORγ1 to the Rev-Erbα(RORE) was also higher at ZT22 than ZT10 despite the low expression of Rev-Erbα at ZT22 indicating that binding of RORγ is not necessarily sufficient to enhance transcription of Rev-Erbα. ChIP-Seq analysis suggests that this association is relatively weak suggesting that RORs may play a lesser role in regulating Rev-Erb expression. Collectively, our promoter analyses, and ChIP-Seq data support the concept that the up-regulation of Cry1, Npas2, Bmal1 and E4bp4 transcription by RORs in vivo occurs by a direct mechanism and involves recruitment of RORγ1 and RORα to ROREs within the regulatory regions of these genes. It is interesting to note that the Rev-Erb binding sites associated with Bmal1 and E4bp4 identified by ChIP-Seq analysis are the same as the ROR binding sites identified by our ChIP-Seq analysis (52) consistent with competition and regulation through the same binding sites. However, for Cry1 the major ROR binding sites were different from those described for Rev-Erb suggesting that RORs and Rev-Erbs may not always regulate clock gene expression through competition for the same response elements.
In many instances activation of transcription is accompanied by changes in chromatin structure. Actively transcribed regions are associated with an open chromatin structure and accompanied by changes in histone acetylation and methylation. ChIP analysis with an antibody against H3K9Ac, a marker for actively transcribed chromatin, showed that the level of H3K9Ac association with the Bmal1 and Cry1 regulatory regions in WT liver was higher at ZT22 than at ZT10 (Figure 6A). This association was diminished in RORγ−/– liver, but not altered in RORαsg/sg liver. FAIRE, which has been used as a tool to map open chromatin (37), revealed that the FAIRE signal was significantly increased at the Cry1(RORE1/2) and Bmal1(RORE1/2) regulatory region in liver at ZT22 compared with ZT10 consistent with increased chromatin accessibility at these sites. This correlates with the increased association of H3K9Ac and the relatively high level of Cry1 and Bmal1 expression at ZT22 (Figure 6B). The loss of RORγ caused a significant reduction in FAIRE signal at the Cry1(RORE1/2) and Bmal1(RORE1/2) region consistent with a positive relationship between RORγ binding, chromatin remodeling and transcriptional activation of these genes.
In addition to the circadian regulation of clock genes by RORs, RORγ itself exhibits a robust rhythmic expression in several peripheral tissues that is controlled by clock proteins (Figure 1) (13,15,18,25). The positive regulation of RORγ1 is partially mediated through the binding of Bmal1-Clock heterodimers to two E-boxes in the proximal promoter of RORγ1 consistent with a previous study (48). Moreover, one of the E-boxes (E2) within RORγ has been reported to be able to drive the oscillation of a reporter in cultured cells, indicating the importance of this E-box in the circadian regulation of RORγ by Clock/Bmal1 dimers (18). In this study, we show that the repressor Cry1 effectively blocked this activation (Figure 7). Cry1 likely mediates this repression through inhibition of the transcriptional activity of Bmal1/Clock heterodimers (54). ChIP analysis using an anti-Clock antibody showed that Clock protein was recruited to the (E1/2-box) sites in the RORγ1 promoter in liver consistent with the hypothesis that RORγ1 transcription is regulated directly by Bmal1-Clock heterodimers in vivo. Unexpectedly little difference was observed in Clock recruitment between ZT10 and ZT22, times of low and high RORγ expression, respectively. Similarly, a previous study demonstrated that the degree of Bmal1-Clock binding did not correlate with the level of activation of Cry1 transcription (38). These observations suggested that binding of Bmal1-Clock is not necessarily sufficient to activate transcription of RORγ1. The latter may be due to a repressor function of the Bmal1-Clock complex and the type of co-repressors or co-activators recruited to the heterodimer, or crosstalk with other transcriptional factors (55). In contrast to Clock binding, the association of H3K9Ac and the FAIRE signal was significantly increased at the RORγ1(E-box) promoter region in liver at ZT22 compared with ZT10 indicating that the transcriptional activation of RORγ1 correlated with increased chromatin accessibility consistent with an association between open chromatin structure and functionally active enhancers. In addition to its regulation by Bmal1/Clock, Rev-Erbs are recruited to the regulatory region of RORγ (52), consistent with the concept that they are also involved in the regulation of the rhythmic expression of RORγ.
In summary, study of RORα, RORγ and DKO mice showed that ROR receptors modulate the expression of several clock genes in an ROR isoform- and tissue-dependent manner. Particularly RORγ1 appears to affect peak expression of clock genes in several peripheral tissues. Study of DKO mice, ChIP-Seq and ChIP-QPCR analysis indicated a certain degree of redundancy between RORα and RORγ and demonstrated that in vivo RORα/γ regulate these genes directly and in a ZT-dependent manner through their interaction with ROREs. We provided evidence that this transcriptional regulation is associated with changes in chromatin structure. Our results are consistent with the concept of a reciprocal relationship between the regulation of clock genes and RORγ1 (Supplementary Figure S5). Recent studies demonstrated that clock proteins are important regulators of energy homeostasis and lipid/glucose metabolism suggesting a connection between the regulation of the circadian clock and various metabolic pathways (4,14,20,30,31). We previously proposed that RORs might function as a link between clock proteins and their regulation of metabolic genes (1). One might predict that the Bmal1/Clock/Rev-Erb-mediated rhythmic expression of RORγ1 leads to the rhythmic expression of RORγ1 target genes, including metabolic genes, such as Avpr1a. This concept is supported by observations showing that the oscillatory expression of RORγ1 and Avpr1a are in phase, that the loss of RORγ greatly reduces Avpr1a peak expression, and the presence of RORγ binding sites (Figure 7) (17). In this scenario, the rhythmic expression of downstream target genes, such as Avpr1a, depends on the rhythmic expression of RORγ1 and to a certain degree on Rev-Erb through its regulation of RORγ1 and/or its competition with RORγ1 for Avpr1a(RORE) binding (Supplementary Figure S5). Analysis of additional RORγ1 target genes is required to obtain further support for this hypothesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–5.
FUNDING
Intramural Research Program of the National Institute of Environmental Health Sciences, National Institutes of Health [Z01-ES-101586]. Funding for open access charge: National Institute of Environmental Health Sciences, National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Kristin Lichti-Kaiser and Gary ZeRuth for their comments on the manuscript and Laura Miller (NIEHS) for her great assistance with the mice.
REFERENCES
- 1.Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucleic Recept. Signal. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gold DA, Baek SH, Schork NJ, Rose DW, Larsen DD, Sachs BD, Rosenfeld MG, Hamilton BA. RORalpha coordinates reciprocal signaling in cerebellar development through sonic hedgehog and calcium-dependent pathways. Neuron. 2003;40:1119–1131. doi: 10.1016/s0896-6273(03)00769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinmayr M, Andre E, Conquet F, Rondi-Reig L, Delhaye-Bouchaud N, Auclair N, Daniel H, Crepel F, Mariani J, Sotelo C, et al. Staggerer phenotype in retinoid-related orphan receptor alpha-deficient mice. Proc. Natl Acad. Sci. USA. 1998;95:3960–3965. doi: 10.1073/pnas.95.7.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang HS, Angers M, Beak JY, Wu X, Gimble JM, Wada T, Xie W, Collins JB, Grissom SF, Jetten AM. Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol. Genomics. 2007;31:281–294. doi: 10.1152/physiolgenomics.00098.2007. [DOI] [PubMed] [Google Scholar]
- 5.Lau P, Fitzsimmons RL, Raichur S, Wang SC, Lechtken A, Muscat GE. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J. Biol. Chem. 2008;283:18411–18421. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- 6.Jaradat M, Stapleton C, Tilley SL, Dixon D, Erikson CJ, McCaskill JG, Kang HS, Angers M, Liao G, Collins J, et al. Modulatory role for retinoid-related orphan receptor alpha in allergen-induced lung inflammation. Am. J. Respir. Crit. Care Med. 2006;174:1299–1309. doi: 10.1164/rccm.200510-1672OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang XO, Nurieva R, Martinez GJ, Kang HS, Chung Y, Pappu BP, Shah B, Chang SH, Schluns KS, Watowich SS, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaeren-Wiemers N, Andre E, Kapfhammer JP, Becker-Andre M. The expression pattern of the orphan nuclear receptor RORbeta in the developing and adult rat nervous system suggests a role in the processing of sensory information and in circadian rhythm. Eur. J. Neurosci. 1997;9:2687–2701. doi: 10.1111/j.1460-9568.1997.tb01698.x. [DOI] [PubMed] [Google Scholar]
- 9.Jia L, Oh EC, Ng L, Srinivas M, Brooks M, Swaroop A, Forrest D. Retinoid-related orphan nuclear receptor RORbeta is an early-acting factor in rod photoreceptor development. Proc. Natl Acad. Sci. USA. 2009;106:17534–17539. doi: 10.1073/pnas.0902425106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurebayashi S, Ueda E, Sakaue M, Patel DD, Medvedev A, Zhang F, Jetten AM. Retinoid-related orphan receptor gamma (RORgamma) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc. Natl Acad. Sci. USA. 2000;97:10132–10137. doi: 10.1073/pnas.97.18.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, Mebius RE, Littman DR. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 12.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The Orphan Nuclear Receptor RORgammat Directs the Differentiation Program of Proinflammatory IL-17(+) T Helper Cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 13.Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- 14.Duez H, Staels B. Nuclear receptors linking circadian rhythms and cardiometabolic control. Arterioscler. Thromb. Vasc. Biol. 2010;30:1529–1534. doi: 10.1161/ATVBAHA.110.209098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillaumond F, Dardente H, Giguere V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 16.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 17.Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian clocks. Nat. Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 19.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat. Rev. Mol. Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
- 22.Raspe E, Mautino G, Duval C, Fontaine C, Duez H, Barbier O, Monte D, Fruchart J, Fruchart JC, Staels B. Transcriptional regulation of human Rev-erbalpha gene expression by the orphan nuclear receptor RORalpha. J. Biol. Chem. 2002;277:49275–49281. doi: 10.1074/jbc.M206215200. [DOI] [PubMed] [Google Scholar]
- 23.Delerive P, Chin WW, Suen CS. Identification of Reverb(alpha) as a novel ROR(alpha) target gene. J. Biol. Chem. 2002;277:35013–35018. doi: 10.1074/jbc.M202979200. [DOI] [PubMed] [Google Scholar]
- 24.Crumbley C, Wang Y, Kojetin DJ, Burris TP. Characterization of the core mammalian clock component, NPAS2, as a REV-ERBα/RORα target gene. J. Biol. Chem. 2010;285:35386–35392. doi: 10.1074/jbc.M110.129288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeda Y, Kang HS, Angers M, Jetten AM. Retinoic acid-related orphan receptor γ (RORγ) directly regulates neuronal PAS domain protein 2 (Npas2) transcription in vivo. Nucleic Acids Res. 2011;39:4769–4782. doi: 10.1093/nar/gkq1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.VanDunk C, Hunter LA, Gray PA. Development, maturation, and necessity of transcription factors in the mouse suprachiasmatic nucleus. J. Neurosci. 2011;31:6457–6467. doi: 10.1523/JNEUROSCI.5385-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masana MI, Sumaya IC, Becker-Andre M, Dubocovich ML. Behavioral characterization and modulation of circadian rhythms by light and melatonin in C3H/HeN mice homozygous for the RORbeta knockout. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007;292: R2357–2367. doi: 10.1152/ajpregu.00687.2006. [DOI] [PubMed] [Google Scholar]
- 28.Yin L, Wu N, Lazar MA. Nuclear receptor Rev-erbα: a heme receptor that coordinates circadian rhythm and metabolism. Nucl. Recept. Signal. 2010;8:e001. doi: 10.1621/nrs.08001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schibler U, Naef F. Cellular oscillators: rhythmic gene expression and metabolism. Curr. Opin. Cell Biol. 2005;17:223–229. doi: 10.1016/j.ceb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Lau P, Fitzsimmons RL, Pearen MA, Watt MJ, Muscat GE. Homozygous staggerer (sg/sg) mice display improved insulin sensitivity and enhanced glucose uptake in skeletal muscle. Diabetologia. 2011;54:1169–1180. doi: 10.1007/s00125-011-2046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang HS, Okamoto K, Takeda Y, Beak JY, Gerrish K, Bortner CD, DeGraff LM, Wada T, Xie W, Jetten AM. Transcriptional profiling reveals a role for RORalpha in regulating gene expression in obesity-associated inflammation and hepatic steatosis. Physiol. Genomics. 2011;43:818–828. doi: 10.1152/physiolgenomics.00206.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamilton BA, Frankel WN, Kerrebrock AW, Hawkins TL, FitzHugh W, Kusumi K, Russell LB, Mueller KL, van Berkel V, Birren BW, et al. Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature. 1996;379:736–739. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- 33.Dussault I, Fawcett D, Matthyssen A, Bader JA, Giguere V. Orphan nuclear receptor RORalpha-deficient mice display the cerebellar defects of staggerer. Mech. Dev. 1998;70:147–153. doi: 10.1016/s0925-4773(97)00187-1. [DOI] [PubMed] [Google Scholar]
- 34.Kumar N, Solt LA, Conkright JJ, Wang Y, Istrate MA, Busby SA, Garcia-Ordonez RD, Burris TP, Griffin PR. The benzenesulfoamide T0901317 [N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluorometh yl)ethyl]phenyl]-benzenesulfonamide] is a novel retinoic acid receptor-related orphan receptor-alpha/gamma inverse agonist. Mol. Pharmacol. 2010;77:228–236. doi: 10.1124/mol.109.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurebayashi S, Nakajima T, Kim SC, Chang CY, McDonnell DP, Renaud JP, Jetten AM. Selective LXXLL peptides antagonize transcriptional activation by the retinoid-related orphan receptor RORgamma. Biochem. Biophys. Res. Commun. 2004;315:919–927. doi: 10.1016/j.bbrc.2004.01.131. [DOI] [PubMed] [Google Scholar]
- 36.Narlikar L, Jothi R. ChIP-Seq data analysis: identification of protein-DNA binding sites with SISSRs peak-finder. Methods Mol. Biol. 2012;802:305–322. doi: 10.1007/978-1-61779-400-1_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 39.Ptitsyn AA, Zvonic S, Conrad SA, Scott LK, Mynatt RL, Gimble JM. Circadian clocks are resounding in peripheral tissues. PLoS Comput. Biol. 2006;2:e16. doi: 10.1371/journal.pcbi.0020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, Wu X, Goh BC, Mynatt RL, Gimble JM. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- 41.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 42.Ukai-Tadenuma M, Yamada RG, Xu H, Ripperger JA, Liu AC, Ueda HR. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144:268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 43.Austin S, Medvedev A, Yan ZH, Adachi H, Hirose T, Jetten AM. Induction of the nuclear orphan receptor RORgamma during adipocyte differentiation of D1 and 3T3-L1 cells. Cell Growth Differ. 1998;9:267–276. [PubMed] [Google Scholar]
- 44.Kallen JA, Schlaeppi J, Bitsch F, Geisse S, Geiser M, Delhon I, Fournier B. X-ray structure of the RORα LBD at 1.63A: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORα. Structure. 2002;10:1697–1707. doi: 10.1016/s0969-2126(02)00912-7. [DOI] [PubMed] [Google Scholar]
- 45.Jin L, Martynowski D, Zheng S, Wada T, Xie W, Li Y. Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma. Mol. Endocrinol. 2010;24:923–929. doi: 10.1210/me.2009-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huh JR, Leung MW, Huang P, Ryan DA, Krout MR, Malapaka RR, Chow J, Manel N, Ciofani M, Kim SV, et al. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORgammat activity. Nature. 2011;472:486–490. doi: 10.1038/nature09978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacDonald VE, Howe LJ. Histone acetylation: where to go and how to get there. Epigenetics. 2009;4:139–143. doi: 10.4161/epi.4.3.8484. [DOI] [PubMed] [Google Scholar]
- 48.Mongrain V, Ruan X, Dardente H, Fortier EE, Cermakian N. Clock-dependent and independent transcriptional control of the two isoforms from the mouse Rorgamma gene. Genes Cells. 2008;13:1197–1210. doi: 10.1111/j.1365-2443.2008.01237.x. [DOI] [PubMed] [Google Scholar]
- 49.Ohno T, Onishi Y, Ishida N. A novel E4BP4 element drives circadian expression of mPeriod2. Nucleic Acids Res. 2007;35:648–655. doi: 10.1093/nar/gkl868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miele A, Bystricky K, Dekker J. Yeast silent mating type loci form heterochromatic clusters through silencer protein-dependent long-range interactions. PLoS Genet. 2009;5:e1000478. doi: 10.1371/journal.pgen.1000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat. Rev. Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 55.Grimaldi B, Nakahata Y, Sahar S, Kaluzova M, Gauthier D, Pham K, Patel N, Hirayama J, Sassone-Corsi P. Chromatin remodeling and circadian control: master regulator CLOCK is an enzyme. Cold Spring Harb. Symp. Quant. Biol. 2007;72:105–112. doi: 10.1101/sqb.2007.72.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.