Abstract
Treatment with demethylating drugs can induce demethylation and reactivation of abnormally silenced tumor suppressor genes in cancer cells, but it can also induce potentially deleterious loss of methylation of repetitive elements. To enable the observation of unwanted drug effects related to loss of methylation of repetitive DNA, we have developed a novel biosensor capable of reporting changes in DNA accessibility via luminescence, in living cells. The biosensor design comprises two independent modules, each with a polydactyl zinc finger domain fused to a half intein and to a split-luciferase domain that can be joined by conditional protein splicing after binding to adjacent DNA targets. We show that an artificial zinc finger design specifically targeting DNA sequences near the promoter region of the L1PA2 subfamily of Line-1 retroelements is able to generate luminescent signals, reporting loss of epigenetic silencing and increased DNA accessibility of retroelements in human cells treated with the demethylating drugs decitabine or 5-azacytidine.
Introduction
Demethylating drugs are being increasingly utilized in cancer therapeutics. These drugs are relatively nonspecific, and their actions are known to involve complex effects on transcriptional programs that go far beyond their action on gene targets of interest. It has been established that the demethylating activity is accompanied by a complex response involving downregulation and activation of a large number genes (Gius et al., 2004; Hagemann et al., 2011). Recently it was demonstrated that the demethylating drugs azacytidine and decitabine (both drugs are in clinical use and in several ongoing clinical trials) can dramatically alter the expression of the cMet ongogene in a number of different cancer cell lines. The abnormal expression of the cMet gene under the influence of either of the drugs is driven by an alternative promoter, and results in an alternatively spliced transcript (Weber et al., 2010). This abnormal cMet transcript had also been observed in tumor tissues of patients with bladder cancer (Wolff et al., 2010). In both cases the alternative promoter driving abnormal transcription is a Line-PA2 retroelement sequence of primate origin (Lizardi, 2010), which becomes active under the demethylating action of the drugs, or in the case of bladder cancers is demethylated during the course of tumor evolution. Therefore, an important problem in human epigenetics is to attain a better mechanistic understanding of the abnormal pharmacological activation of promoters derived from the ubiquitous retroelement sequences in the human genome. While it is well established that silencing of retroelements during gametogenesis is based on homology-driven mechanism involving piRNAs (Halic and Moazed, 2009; Moazed, 2009), the mechanisms responsible for maintenance of silencing in somatic cells remain to be elucidated in detail.
Powerful, sequencing-based methods are available to measure genome-wide changes in chromatin states, including transcription factor binding, histone modifications, or DNA methylation (Park, 2009; Hawkins et al., 2010). However, there are few approaches that enable observation of locus-specific epigenetic changes by imaging living cells or tissues. The success in monitoring the expression of genes involved in photoperiod processes via luminescence, in real time, over several days of observation, has set the stage for the use of similar approaches for the study of chromatin remodeling. Artificial fluorescent or luminescent fusion proteins introduced in the genomes of model organisms can be used to address this need, however, these powerful approaches are not applicable to human tissue experimentation due to ethical issues. Thus, there is an emerging need for a new class of nonintegrating biosensors specifically designed for imaging applications in human tissues, and for enabling time-resolved observation of specific epigenetic alterations in living cells.
To begin addressing this emerging need, we identified as an attractive biological model the abnormal loss of epigenetic silencing of retroelements under the influence of pharmacological agents. Long interspersed nuclear elements belonging to the Line-1 family of repeats represent a large fraction of the human genome (Lander et al., 2001), and a small subset of these elements remain transpositionally active. During normal gametogenesis, silencing of retrotransposon sequences by DNA methylation is established under the direction of piRNAs and the Piwi family of proteins (Aravin et al., 2008; Kuramochi-Miyagawa et al., 2008). The first observations of widespread Line-1 retrotransposon expression were reported for adult testicular tumors (Bratthauer and Fanning, 1992) and for pediatric germ cell tumors (Bratthauer and Fanning, 1993). More recently, the loss of epigenetic silencing of Line-1 elements has been reported in a multiplicity of cancers (e.g., Schulz et al., 2006; Belancio et al., 2009), and in the context of environmental exposures to chemicals and other toxicants. Notably, the loss of methylation of Line-1 elements, and the activation of the Line-1 antisense promoter can be induced by drugs (Weber et al., 2010) or by inflammatory cytokines, such as Interleukin-6 (Gasche et al., 2011), and these epigenetic changes can be observed after a few days in cell culture. It would be of considerable interest to be able to observe the abnormal loss of methylation of Line-1 elements, and the resulting increase in DNA accessibility, in living cells or tissues, as inflammatory processes evolve over time, or during disease progression or different drug treatments. Here we describe a proof-of-concept for the experimental use of a new class of nonintegrating, sequence-specific biosensors specifically designed for imaging applications in human cells and tissues, and for enabling direct observation of the time course of retroelement chromatin remodeling.
Materials and Methods
Selection of the zinc finger target sites on L1PA2
The L1PA2 5′UTR promoter region was analyzed by selecting, from the total 4600 L1PA2 sequences (obtained from Human Genome Build 16 using RepeatMasker), a subset of 2230 sequences that contained an intact 5′-UTR. These 2230 sequences were aligned, using CLUSTALX, to generate a consensus sequence of the 912 base L1PA2 5′-UTR (Supplementary Fig. S1; Supplementary Data are available online at www.liebertonline.com/dna). The consensus was used for selection of a pair of pentadactyl zinc finger 15 bp target sites using the Zinc Finger Tools program (www.scripps.edu/mb/bar bas/zfdesign/zfdesignhome.php). The sequence was analyzed on either the same strand or on the opposite strand. The candidate sites containing ANN and TNN triplets were excluded as these have been empirically shown to be less specific relative to GNN or CNN triplets. The two target sites for zinc finger-1 (ZF1) and zinc finger-2 (ZF2) were selected based on their high scores as determined by the program, close distance in between and relative high allele frequencies of more than 1000 copies in the human genome. The target site sequences are: CCTGACCCCCGAGCA (1) and GACCCACTTGAGGAG (2), which are separated by 7 bp in the consensus sequence.
Cloning of the L1PA2-targeting zinc fingers ZF1 and ZF2
The two zinc finger constructs (Supplementary Figs. S2 and S3) were assembled using the Zinc Finger Tools program (www.scripps.edu/mb/barbas/zfdesign/zfdesignhome.php) and cloned into the bacterial pMAL-c4× expression vector using assembly polymerase chain reaction (PCR) as previously described (Beltran et al., 2007). Each construct contains a N-terminal Maltose binding protein (Mbp) tag and a fusion of five zinc finger domains of 549 amino acid residues in length.
Bacterial expression of the ZF1 and ZF2
The plasmids were transformed into XL1-blue cells, grown in 5 mL TB media at 37°C over night, and induced 37°C with 1500 μM isopropyl-beta-D-1-thiogalactopyranoside (IPTG) for 3 h. Cells were harvested and resuspended in 300 μL zinc finger binding assay (ZnBA) buffer containing 10 mM Tris, pH 7.5, 90 mM KCl, 1 mM MgCl2, 90 μM ZnCl2, and 5 mM DTT. Cells were lysed by freeze/thaw in dry ice/ethanol bath and the supernatants were collected and stored at −80°C until use. Zinc finger protein expression was analyzed by Western blot using the mouse anti-Mbp antibody (Sigma-Aldrich, St. Louis, MO).
Zinc finger-DNA binding ELISA
The biotinylated DNA target hairpin oligos (Supplementary Table S1) harboring ZF1 and ZF2 target sites were obtained from Integrated DNA Technologies (IDT, Coralville, IA). The DNA oligos were resuspended in ZnBA buffer to 1 μg/μL. Flat bottom 96-well plates were coated with 0.4 μg streptavidin (Pierce, Rockford, IL) per well in 100 μL carbonate buffer at 4°C overnight. After washing two times with water, 100 μL 4.0 ng/μL DNA oligo was added to each well in ZnBA buffer and incubated 37°C for 1 h. The wells were then washed with water and blocked with 200 μL 3% bovine serum albumin in ZnBA buffer for 1 h at 37°C. Unpurified bacterial expression lysate (100 μL) was added in ZnBA buffer containing 1% BSA and 120 ng/μL salmon sperm DNA and incubated for 1 h at room temperature. The unpurified lysate with the empty vector was used as a control. After washing with water, 100 μL mouse anti-Mbp antibody (Sigma-Aldrich) in ZnBA with 1% BSA was added and incubated for 30 min at room temperature. After washing with water, 100 μL alkaline phosphatase-conjugated goat anti-mouse antibody (Sigma-Aldrich) in ZnBA with 1% BSA was added and incubated for 30 min at room temperature. After washing with water, the colorimetric ELISA was performed by adding 100 μL substrate pNPP solution and reactions were stopped after 3 h incubation with 50 μL 2N NaOH and the absorbance was measured at 405 nm with a plate reader.
Cloning of the DNA-guided nano-assembly biosensor constructs
The DNA-guided nano-assembly (DGnA) biosensor is a two-module construct comprising module 1 and module 2 (Supplementary Figs. S4 and S5). Each module was assembled using InFusion PCR kit (Clontech, Mountain View, CA) from a L1PA2-targeting zinc finger domain as described above and a half of the split intein-firefly luciderase fusion domain, and were placed into separate mammalian expression vectors DNA4/HisMax (Invitrogen, Carlsbad, CA). The domain organizations of each module is shown as the following: Module 1: ZF1_NLuc_NInt; Module 2: ZF2_CInt_CLuc. Both modules were His- and NLS-tagged at the N-termini. The original split intein-luciferase fusion constructs (NLuc_NInt and CInt_CLuc) were kind gifts from Dr. Edmund Schwartz and Dr. Tom Muir from the Rockefeller University.
Cell culture, transfection, and drug treatments
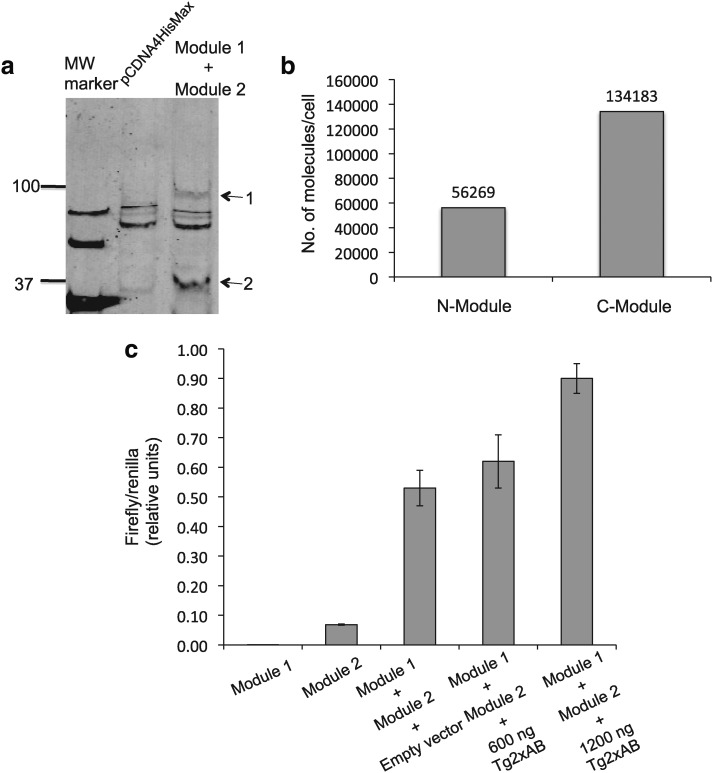
Human HeLa cells (ATCC, Manassas, VA) were grown in Eagle's minimum essential medium (ATCC) with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich) in plastic six-well tissue culture plates at 37°C in a humidified atmosphere containing 5% CO2. Transfection efficiency using Effectene (Invitrogen) and Fugene (Roche, Basel, Switzerland) was evaluated using replicate experiments involving transfection with 750 ng of plasmid DNA. Using real-time quantitative PCR, we estimate an average of 2000 internalized molecules per nucleus. By extrapolation, 600 ng of artificial target plasmid would result in internalization of ∼1600 molecules/nucleus, while 1200 ng of artificial target plasmid would result in internalization of ∼3200 molecules/nucleus. These numbers of internalized plasmid molecules are consistent with the published literature, where transfection of HeLa cells with lipofectamine has been quantified by Glover et al. (2010) using real-time PCR, yielding results similar to ours. Since the copy number of L1PA2 repetitive elements in the human genome is ∼4600 (or 9200 in a diploid genome), these experimental estimates of transfection efficiency indicate that we can internalize artificial plasmid DNA targets in HeLa cells (as performed to generate the data in Fig. 2c) in numbers representing roughly one-third of the number of L1PA2 elements in the genome.
FIG. 2.
Luciferase activity of the DGnA biosensor in response to specific target DNA transfected into HeLa cells. (a) Immunoblot analysis of the His-tagged modules 1 and 2 of the biosensor expressed in HeLa cells. The empty vector is the expression vector pcDNA4/HisMax without biosensor inserts. HeLa cells transfected with empty vector (second lane) show several bands, probably representing cross-reactivity of the anti-His antibody with cellular proteins. Cells transfected with expression vectors harboring each of the two biosensor constructs show additional bands corresponding to the expected molecular weights for Module 1 (95.8 kd) and Module 2 (36.1 kd). (b) Quantitation of the protein expression of the two modules in transfected HeLa cells using a Western blot analyzed in a LiCOR instrument. The 50 kDa band with the known protein amount (15 ng) of the Smart His-tagged Protein Standard (SS) was used as the reference. (c) Measurement of luminescence in HeLa cells transiently transfected with the two modules of the biosensor along with increasing amounts of co-transfected target DNA. Firefly luciferase luminescent signal was normalized by Renilla luciferase luminescent signal as the transfection control. Tg2xAB, a puc19 vector carrying target DNA, comprising two copies of the target 1 and 2 sequences, separated by 2 bp.
To measure the DGnA biosensor response to epigenetic drug treatments, different concentrations of fresh 5-aza-2-deoxycytidine (Sigma-Aldrich) or 5-aza-cytidine (Sigma-Aldrich) were added to the cells every 24 h through change of medium. The drug treatments were maintained before and after the biosensor transfection throughout the desired protocol. No artificial DNA targets were used in these experiments. One day before biosensor transfection, 2.0×105 cells were seeded in 2 mL growth medium per well. Next day the plasmids of 100 ng module 1 and 50 ng module 2 were transfected using the transfection reagent FugeneHD (Roche) according to manufacturer's instructions. 10 ng Renilla luciferase plasmid pRL-TK (Promega, Madison, WI) was co-transfected as a control for transfection efficiency. After 20 h post-transfection growth, cells were harvested with ice cold PBS buffer and cell pellets were lysed in 40 μL 1× passive lysis buffer (Promega) containing 1× protease inhibitors (Roche, Madison, WI) and analyzed for protein expression and Luciferase activity.
Western blot analysis of the DGnA biosensor expression and Luciferase assay
For Western blot analysis, cell lysates were mixed with the same volume of 2× Laemmli sample buffer, boiled, and loaded onto 10% polyacrylamide gels containing sodium dodecyl sulfate. Proteins were transferred to polyvinylidene fluoride membranes Immobilon™FL (Millipore, Billerica, MA). We used a mouse anti-His antibody (Sigma-Aldrich) and a goat anti-mouse secondary antibody IRdye800CW (LI-COR, Lincoln, NE). The protein bands were visualized and quantitated using the Odyssey Infrared Imaging System (LI-COR). The protein marker of the Smart His-tagged Protein Standard (GenScript, Piscataway, NJ) was used as a reference for quantitation. For luciferase assay, cell lysates was analyzed using the dual-luciferase reporter assay kit according to the manufacturer's instruction (Promega) and the luminescence was read using a luminometer GloMax-20/20 (Promega).
L1PA2 DNA methylation assay
Genomic DNA samples were isolated from drug-treated and control HeLa cells using a DNA isolation kit for blood and cells (Qiagen, Valencia, CA). The primers used for L1PA2 bisulfate-PCR is: TTTTATTAGGGAGTGTTAGATAGTGGG and ACCCACTTAAAAAAACAATCTACCC. DNA methylation was measured by bisulfite-PCR, followed by mass spectrometry-based MassARRAY analysis using the Sequenom EpiTyper analysis platform at the Yale Center for Genome Analysis.
Results
Polydactyl zinc finger (PZF) proteins bind double-stranded DNA based on sequence-specific molecular recognition of DNA bases by helical peptide domains, which penetrate the major groove of B-DNA (Klug, 2010). Each PZF recognizes 3 bp of DNA in a quasi-modular manner, and thereby the rational design of artificial PZFs is possible (Blancafort et al., 2004; Bhakta and Segal, 2010). Just as the PCR achieves single-locus specificity based on the use of two different primers, it is possible to target a genomic locus with high specificity employing a set of two contiguous five-finger PZF modules, which together recognize 15+15=30 bp. The use of separate modules of PZF proteins to recognize specific DNA sequences, resulting in the assembly of a green fluorescent protein reporter has been described (Stains et al., 2006; Furman et al., 2009). We modified this system to be under the control of a conditional protein splicing system, in which fragments of an artificially split Saccharomyces cerevisiae vacuolar membrane ATPase (VMA) intein complement through rapamycin-induced heterodimerization of the FK506 binding protein (FKBP) and FKB12-Rapamycin binding domain of mTor (FBR) Proteins (Mootz and Muir, 2002). This intein-based biosensor system was able to report the presence of the drug rapamycin in a living animal, in real time (Schwartz et al., 2006) meeting one of our key design requirements.
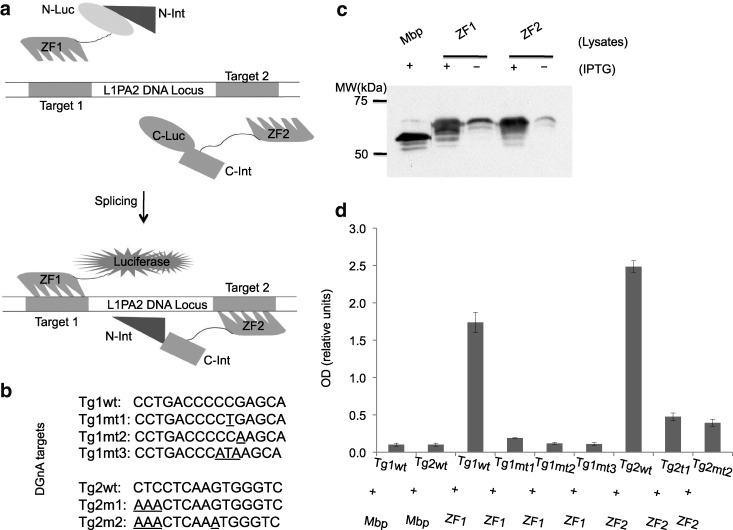
In our biosensor design, specific DGnA of a luminescent reporter is driven by two independent protein fusions each comprising a PZF module with five PZF domains, a half-intein domain, and a half-luciferase-enzyme domain. Figure 1a illustrates the two zinc finger biosensor structures ZF1 and ZF2, and the resulting DGnA process, in which each of the modules specifically binds at adjacent sites in the genome, followed by intein-catalyzed splicing and the generation of covalently joined and active luciferase. We targeted specific DNA sequences representing the consensus sequence (see Supplementary Fig. S1) of the 5′-UTR of Line-1 elements belonging to the L1PA2 subfamily, a group of sequences that numbers ∼4600 interspersed copies, originating in Hominoidea, and largely absent in older species of monkeys such as Rhesus macaque. To evaluate DNA-recognition specificity, we expressed the artificial zinc finger proteins in Escherichia coli (Fig. 1c), and confirmed by means of ELISA assays that each of the two PZF modules specifically binds to its cognate sequence, and not to other DNA sequences (Fig. 1b, d). Note that the two strongest binding signals in Figure 1d correspond to the perfect L1PA2 genomic target sequences Tg1wt and Tg2wt.
FIG. 1.
The two-module design of the DGnA biosensor and characterization of DNA binding of the zinc finger protein in each module. (a) schematic diagram showing His- and NLS-tagged module 1 (ZF1_NLuc_NInt) and module 2 (ZF2_CInt_CLuc). Each module contains a pentadactyl zinc finger domain linked with one half of the split VMA intein and a split firefly luciferase fusion domain. Binding of both modules via their respective zinc finger domains to their specific L1PA2 target sites induces intein-mediated protein splicing. This reaction results in covalent reconstitution of a functional luciferase capable of generating luminescence in the presence of luciferin. NLS, nuclear localization signal. ZF1 and ZF2, zinc finger domain 1 and 2. NLuc and NInt, N-terminal half of the firefly lucifease and VMA intein fusion domain. CLuc and CInt, C-terminal half of the VMA intein and firefly fusion domain. (b) DNA target sequences used to evaluate artificial zinc finger binding. Underlined bases represent changes from L1PA2 consensus sequence. (c) Immunoblot analysis of Mbp-tagged zinc finger domains in bacterial lysates following IPTG induction. (d) ELISA analysis of bacterially expressed zinc finger protein binding to the wild-type and mutant DNA targets. Bacterial lysates containing expressed zinc finger domains were used without further purification (see Materials and Methods section), and the control lysate was generated by an empty vector expressing only the Mbp protein. Tg, target; wt, wild-type; mt, mutant; DGnA, DNA-guided nano-assembly; VMA, vacuolar membrane ATPase; Mbp, maltose binding protein; IPTG, isopropyl-beta-D-1-thiogalactopyranoside.
We then showed that each of the two fusion proteins could be expressed in HeLa cells (Fig. 2a), and optimized protein expression levels by adjusting the amount of transfected DNA until an expression level ranging between 56,000 and 134,000 molecules per cell (Fig. 2b) was attained for each of the protein modules. The feasibility of the DGnA process was evaluated by co-transfecting HeLa cells with an artificial DNA plasmid that harbors the entire target region corresponding to the L1PA2 consensus sequence. As shown in Figure 2c, transfection was performed with different amounts of target DNA or with an irrelevant DNA target, and biosensor performance was assessed as a ratio of firefly luciferase (biosensor) to Renilla luciferase (transfection control) in a dual luciferase assay. The results, as indicated by increasing values for the luminescence ratio of firefly luciferase to Renilla luciferase, are consistent with the two genetically encoded biosensor modules being able to bind the artificial L1PA2 sequence (denominated Tg2xAB) in the transfected targets, and undergoing conditional protein splicing to generate, via the DGnA process, molecules of covalently joined and active luciferase. We also observed a significant amount of luminescence signal for the empty (target-free) vector DNA input measurement, most likely arising from background (nonconditional) intein splicing as reported by others (Schwart et al., 2006) that can occur in the absence of DNA binding. Since the largest input used in this experiment (1200 ng of transfected plasmid) has been determined to correspond to ∼3200 molecules of artificial target internalized in cell nuclei (see Materials and Methods section), and this number of artificial targets is smaller than the number of L1PA2 elements in a diploid genome, it seems unlikely that background signal could be generated by a small subcomponent of undermethylated L1PA2 genomic DNA. Despite the nonconditional intein splicing background issue, the experiment shown in Figure 2c demonstrates a strong luminescence response in the presence of artificial DNA targets.
To demonstrate the utility of the genetically encoded biosensors in a relevant biological context we performed several experiments where cells are exposed to drugs capable of inducing DNA demethylation. The responses of mammalian cells to the demethylating drugs decitabine and 5-azacitidine have been extensively documented in the literature. To illustrate the effects of decitabine on the DNA methylation levels of L1PA2 repetitive elements in HeLa cells, in which Line-1 elements have been reported to be predominantly methylated (Teneng et al., 2011), we treated a HeLa cell culture with decitabine, at a concentration of 2.5 μM, for a period of 4 or 5 days. The drug induced a loss of methylation corresponding to values lower than 70% of the control HeLa cell culture methylation levels in 8 out of 21 CpG dinucleotides. The lowest observed levels of relative DNA methylation compared to controls, after decitabine treatment, were 51%, 54%, and 55% for three of the 21 CpG dinucleotides assayed (Table 1). The drug concentrations used in these and subsequent experiments were chosen to be roughly comparable to the maximum concentrations (Cmax) achieved in human plasma at clinically used dosages and schedules of administration. Human plasma Cmax values are 3 to 11 μM 5-azacytidine and 0.3 to 1.6 μM decitabine (Marcucci et al., 2005; Blum et al., 2008; Cashen et al., 2008). Similar drug concentrations to those used in our study have been utilized in recent cell-line based studies of these drugs (Hollenbach et al., 2010).
Table 1.
Measurements of Line-1 5-UTR DNA Methylation in HeLa Cells Treated with 2.5 μM Decytabine for 4 or 5 days
| Line-1 5′UTR CpGs | Control methylated fraction | DAC-4 days methylated fraction | DAC-5 days methylated fraction | DAC-4 days ratio relative to control | DAC-5 days ratio relative to control |
|---|---|---|---|---|---|
| I1PA2-1_CpG_1 | 0.28 | 0.23 | 0.20 | 0.82 | 0.72 |
| I1PA2-1_CpG_4.5.6 | 0.47 | 0.41 | 0.39 | 0.87 | 0.83 |
| I1PA2-1_CpG_7 | 0.54 | 0.63 | 0.48 | 1.16 | 0.89 |
| I1PA2-1_CpG_8 | 0.16 | 0.17 | 0.19 | 1.06 | 1.23 |
| I1PA2-1_CpG_9 | 0.54 | 0.63 | 0.48 | 1.16 | 0.89 |
| I1PA2-1_CpG_10 | 0.61 | 0.42 | 0.36 | 0.69 | 0.59 |
| I1PA2-1_CpG_12 | 0.54 | 0.63 | 0.48 | 1.16 | 0.89 |
| I1PA2-1_CpG_13 | 0.39 | 0.32 | 0.26 | 0.82 | 0.67 |
| I1PA2-1_CpG_14 | 0.71 | 0.51 | 0.57 | 0.72 | 0.80 |
| I1PA2-1_CpG_15.16 | 0.59 | 0.40 | 0.40 | 0.68 | 0.67 |
| I1PA2-1_CpG_17 | 0.44 | 0.37 | 0.27 | 0.84 | 0.63 |
| I1PA2-1_CpG_18.19 | 0.53 | 0.45 | 0.37 | 0.85 | 0.69 |
| I1PA2-1_CpG_21 | 0.44 | 0.44 | 0.32 | 1.00 | 0.72 |
| I1PA2-1_CpG_22 | 0.43 | 0.36 | 0.31 | 0.82 | 0.72 |
| I1PA2-1_CpG_23 | 0.62 | 0.57 | 0.34 | 0.92 | 0.55 |
| I1PA2-1_CpG_25 | 0.42 | 0.37 | 0.30 | 0.87 | 0.71 |
| I1PA2-1_CpG_26 | 0.26 | 0.21 | 0.19 | 0.80 | 0.71 |
| I1PA2-1_CpG_28.29 | 0.36 | 0.32 | 0.28 | 0.90 | 0.79 |
| I1PA2-1_CpG_30 | 0.26 | 0.21 | 0.19 | 0.80 | 0.71 |
| I1PA2-1_CpG_31 | 0.28 | 0.32 | 0.14 | 1.12 | 0.51 |
| I1PA2-1_CpG_33 | 0.32 | 0.22 | 0.17 | 0.69 | 0.54 |
DNA methylation was measured by bisulfite-polymerase chain reaction, followed by mass spectrometry analysis using the Sequenom EpiTyper analysis platform. Data in the Ratio columns are presented as percentage of methylation at a given CpG site in the sequence obtained from three separate experiments. The Ratio is the post-treatment methylation value divided by the value of the pretreated control. Bold text indicates those ratios at or below 0.70.
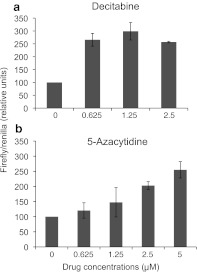
We performed an experiment, using triplicate cell cultures, where the biosensors were transfected into HeLa cells 3 days after the start of drug treatment. The results, shown in Figure 3a, demonstrate that incremental doses of decitabine lead to significant increases in luminescent output of the DGnA biosensors, demonstrating the ability of the system to report changes in accessibility of the genomic DNA harboring L1PA2 elements, as they become demethylated. We performed a similar experiment using the demethylating drug 5-azacytidine, and the results are shown in Figure 3b. In this second experiment, also performed in triplicate, the DGnA biosensors report the increased DNA accessibility in L1PA2 genomic DNA, resulting from 5-azacytidine treatment, showing a drug concentration-dependent increase in relative luminescent output.
FIG. 3.
Luciferase activity of the L1PA2 DGnA biosensor in response to decitabine or 5-azacytidine treatment in HeLa cells. (a) Measurements of luminescence in HeLa cells co-transfected with the two modules of the L1PA2 biosensor after a 4-days decitabine treatment. The values shown are the average of experiments performed in triplicate, and the bars represent the Standard Error. (b) Measurements of luminescence in HeLa cells co-transfected with the two modules of the L1PA2 biosensor following 4-days of 5-azacytidine treatment. The values shown are the average of experiments performed in triplicate, and the bars represent the Standard Error. Firefly luciferase luminescent signal was normalized by Renilla luciferase luminescent signal as the transfection control.
Discussion
The potential for future imaging applications of zinc-finger based biosensors is supported by the success of recent efforts to image repetitive DNA elements using PZF fusions coupled to Green Fluorescent protein (Lindhout et al., 2007, 2010). These simple fusions have enabled the detection of centromere repeats in Arabidopsis thaliana and in mouse cells. Our initial demonstration of the utility of the more complex splicing-based DGnA biosensor technology illustrates some of the limitations of the system as currently implemented using luciferase as a signal-generating moiety. The signal to noise ratio is not yet high enough to enable the detection of DNA accessibility changes at single-copy gene loci. The major cause of noise in the system is the occurrence of background intein splicing in the absence of target DNA. It might be possible to engineer modifications in the structure of the intein to reduce the rate of the spontaneous splicing reaction. Since the binding of the zinc finger modules to DNA is of very high affinity, the modified inteins will linger long enough when bound to DNA to permit splicing even when the engineered fragments have lower affinity for each other. Other modifications that could enhance the performance of the system include the utilization of natural or engineered luciferases with higher photon yields.
As reported here, the Line-1PA2 epigenomic biosensor can be used to monitor the cellular response to demethylating drugs that modulate epigenetic states in chromatin. We emphasize that the system is currently capable of reporting changes in DNA accessibility, which may or may not reflect changes in DNA methylation. Nonetheless, future improvements in the molecular design of the biosensors could involve incorporating modules with the capability of recognizing methylated DNA. It has been reported that the human zinc-finger proteins Kaiso, ZBTB4, and ZBTB38 can bind methylated DNA in a sequence-specific manner (Sasai et al., 2010). As we begin to more fully understand the molecular recognition contacts of Kaiso and Kaiso-like zinc fingers, new possibilities will emerge for re-engineering biosensors with more powerful recognition capabilities.
With a potentially more gentle system for recombinant vector delivery, such as exosomes (Lakhal and Wood, 2011), the biosensors could be used to measure drug responses in epithelial cells from individual patients, or even in proxy tissues derived from different individuals using iPS cell technology. In this last application, the biosensors would enable research in the individualized pharmacogenetics and pharmacokinetics of demethylating drug responses. Notably, it has been reported (Issa et al., 2005) as part of phase II clinical trail studies with demethylating drugs, that there are marked decreases in the methylation of Line-1 elements in patients with leukemia undergoing treatment with decitabine. It would be exciting if improved versions of the epigenomic biosensors could be utilized to report on the status of Line-1 DNA accessibility in live blood cells derived from patients undergoing treatment with demethylating drugs. It must be emphasized that the DGnA technology is modular, and that the biosensors can be readily modified to target other genomic loci of interest through the use of appropriately modified zinc finger modules. One could, for example, redesign the two zinc finger modules of the biosensor to target other Line-1 subfamilies, such as the human-specific Line-1HS elements. Other retroelements, such as Alu, SVA, or HERV subfamilies could also be specifically targeted. Different kinds of centromeric repeat sequences could be targeted to report on the silencing status of specific centromere domains. A recent publication reports that Satellite 2 demethylation induced by 5-azacytidine is associated with missegregation of chromosomes 1 and 2 in human somatic cells (Prada et al., 2012). The ability to observe in living cells the loss of silencing of satellite DNA in the context of demethylating drug treatment can enable new experimental approaches for the study of drug-induced genome instability.
The modularity of the biosensor can be extended to other designs driving the assembly of split modules comprising a variety of enzymes suitable for other imaging modalities. Notably, a recent report described a molecularly engineered split thymidine kinase reporter for imaging protein-protein interactions with positron emission tomography (Massoud et al., 2010), which enabled imaging in live animal models. In the case of the DGnA system, thymidine kinase or any monomeric protein can in principle be split to generate inactive fragments, and rejoined by splicing to generate active enzyme under the specific control of a genomic guide (target) sequence. The flexibility in possible imaging modalities should make the Line-1 biosensors we have described excellent tools to study the loss of silencing of these retroelements, possibly associated with Line-1 sense and antisense promoter activation, in a variety of experimental settings. We expect that DGnA biosensors will find many applications in basic research, including systems biology, and we also envision exciting possibilities in advanced screens for discovery or improvement of epigenetic modulation drugs. We also envision a novel and ambitious therapeutic strategy, where an abnormal genome becomes a drug target, and the DGnA process operates in the cell nucleus as a factory of therapeutic proteins. For example, a pro-apoptotic protein could be assembled via DGnA, upon detection of DNA structural abnormalities (such as ERBB2 or Myc amplification) or abnormal loss of epigenetic silencing of Line-1 retroelement sequences.
Supplementary Material
Acknowledgments
We are indebted to Dr. Tom Muir at the Rockefeller University for advice on the use of intein splicing systems and for the gift of intein-luciferase plasmid constructs. We thank the Yale Liver Center for use of a LICOR instrument purchased with Center grant # USPHS DK P30-34989 and the Yale Center for Genome Analysis for the DNA methylation analysis. This work was supported by Grant 5R01GM080242-03 from the National Institutes of Health and by a gift from the Virginia Alden Wright Family Fund to the Yale New Haven Hospital Department of Surgery.
Disclosure Statement
No competing financial interests exist.
References
- Aravin A.A. Sachidanandam R. Bourc'his D. Schaefer C. Pezic D. Toth K.F. Bestor T. Hannon G.J. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio V.P. Deininger P.L. Roy-Engel A.M. LINE dancing in the human genome: transposable elements and disease. Genome Med. 2009;1:97.1–97.8. doi: 10.1186/gm97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran A. Parikh S. Liu Y. Cuevas B.D. Johnson G.L. Futscher B.W. Blancafort P. Re-activation of a dormant tumor suppressor gene maspin by designed transcription factors. Oncogene. 2007;26:2791–2798. doi: 10.1038/sj.onc.1210072. [DOI] [PubMed] [Google Scholar]
- Bhakta M.S. Segal D.J. The generation of zinc finger proteins by modular assembly. Methods Mol Biol. 2010;649:3–30. doi: 10.1007/978-1-60761-753-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancafort P. Segal D.J. Barbas C.F., 3rd Designing transcription factor architectures for drug discovery. Mol Pharmacol. 2004;66:1361–1371. doi: 10.1124/mol.104.002758. [DOI] [PubMed] [Google Scholar]
- Blum W. Klisovic R.B. Hackanson B. Liu Z. Liu S., et al. Phase I study ofn decitabine alone or in combination with valproic acid in acute myeloid leukemia. J Clin Oncol. 2008;25:3884–3891. doi: 10.1200/JCO.2006.09.4169. [DOI] [PubMed] [Google Scholar]
- Bratthauer G.L. Fanning T.G. Active LINE-1 retrotransposons in human testicular cancer. Oncogene. 1992;7:507–510. [PubMed] [Google Scholar]
- Bratthauer G.L. Fanning T.G. LINE-1 retrotransposon expression in pediatric germ cell tumors. Cancer. 1993;71:2383–2386. doi: 10.1002/1097-0142(19930401)71:7<2383::aid-cncr2820710733>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Cashen A.F. Shah A.K. Todt L. Fisher N. DiPersio J. Pharmacokinetics of decitabine administered as a 3-h infusion to patients with acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS) Cancer Chemother Pharmacol. 2008;61:759–766. doi: 10.1007/s00280-007-0531-7. [DOI] [PubMed] [Google Scholar]
- Furman J.L. Badran A.H. Shen S. Stains C.I. Hannallah J. Segal D.J. Ghosh I. Systematic evaluation of split-fluorescent proteins for the direct detection of native and methylated DNA. Bioorg Med Chem Lett. 2009;19:3748–3751. doi: 10.1016/j.bmcl.2009.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasche J.A. Hoffmann J. Boland C.R. Goel A. Interleukin-6 promotes tumorigenesis by altering DNA methylation in oral cancer cells. Int J Cancer. 2011;129:1053–1063. doi: 10.1002/ijc.25764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gius D. Cui H. Bradbury C.M. Cook J. Smart D.K. Zhao S. Young L. Brandenburg S.A. Hu Y. Bisht K.S. Ho A.S. Mattson D. Sun L. Munson P.J. Chuang E.Y. Mitchell J.B. Feinberg A.P. Distinct effects on gene expression of chemical and genetic manipulation of the cancer epigenome revealed by a multimodality approach. Cancer Cell. 2004;6:361–371. doi: 10.1016/j.ccr.2004.08.029. [DOI] [PubMed] [Google Scholar]
- Glover D.J. Leyton D.L. Moseley G.W. Jans D.A. The efficiency of nuclear plasmid DNA delivery is a critical determinant of transgene expression at the single cell level. J Gene Med. 2010;12:77–85. doi: 10.1002/jgm.1406. [DOI] [PubMed] [Google Scholar]
- Hagemann S. Heil O. Lyko F. Brueckner B. Azacytidine and decitabine induce gene-specific and non-random DNA demethylation in human cancer cell lines. PLoS One. 2011;6:e17388. doi: 10.1371/journal.pone.0017388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M. Moazed D. Transposon silencing by piRNAs. Cell. 2009;138:1058–1060. doi: 10.1016/j.cell.2009.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins R.D. Hon G.C. Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet. 2010;11:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbach P.W. Nguyen A.N. Brady H. Williams M. Ning Y. Richard N. Krushel L. Aukerman S.L. Heise C. MacBeth K.J. A comparison of azacitidine and decitabine activities in acute myeloid leukemia cell lines. PLoS One. 2010;5:e9001. doi: 10.1371/journal.pone.0009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S. Watanabe T. Gotoh K. Totoki Y. Toyoda A. Ikawa M. Asada N. Kojima K. Yamaguchi Y. Ijiri T.W. Hata K. Li E. Matsuda Y. Kimura T. Okabe M. Sakaki Y. Sasaki H. Nakano T. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhal S. Wood M.J. Exosome nanotechnology: an emerging paradigm shift in drug delivery: exploitation of exosome nanovesicles for systemic in vivo delivery of RNAi heralds new horizons for drug delivery across biological barriers. Bioessays. 2011;33:737–741. doi: 10.1002/bies.201100076. [DOI] [PubMed] [Google Scholar]
- Lander E.S. Linton L.M. Birren B. Nusbaum C. Zody M.C. Baldwin J., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lindhout B.I. Fransz P. Tessadori F. Meckel T. Hooykaas P.J. van der Zaal B.J. Live cell imaging of repetitive DNA sequences via GFP-tagged polydactyl zinc finger proteins. Nucleic Acids Res. 2007;35:e107. doi: 10.1093/nar/gkm618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindhout B.I. Meckel T. van der Zaal B.J. Zinc finger-mediated live cell imaging in Arabidopsis roots. Methods Mol Biol. 2010;649:383–398. doi: 10.1007/978-1-60761-753-2_24. [DOI] [PubMed] [Google Scholar]
- Lizardi P.M. As we bring demethylating drugs to the clinic, we better know the DICE being cast. Oncogene. 2010;29:5772–5774. doi: 10.1038/onc.2010.372. [DOI] [PubMed] [Google Scholar]
- Marcucci G. Silverman L. Eller M. Lintz L. Beach C.L. Bioavailability of azacitidine subcutaneous versus intravenous in patients with the myelodysplastic syndromes. J Clin Pharmacol. 2005;45:597–602. doi: 10.1177/0091270004271947. [DOI] [PubMed] [Google Scholar]
- Massoud T.F. Paulmurugan R. Gambhir S.S. A molecularly engineered split reporter for imaging protein-protein interactions with positron emission tomography. Nat Med. 2010;16:921–926. doi: 10.1038/nm.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz H.D. Muir T.W. Protein splicing triggered by a small molecule. J Am Chem Soc. 2002;124:9044–9045. doi: 10.1021/ja026769o. [DOI] [PubMed] [Google Scholar]
- Park P.J. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prada D. González R. Sánchez L. Castro C. Fabián E. Herrera L.A. Satellite 2 demethylation induced by 5-azacytidine is associated with missegregation of chromosomes 1 and 16 in human somatic cells. Mutat Res. 2012;729:100–105. doi: 10.1016/j.mrfmmm.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Sasai N. Nakao M. Defossez P.A. Sequence-specific recognition of methylated DNA by human zinc-finger proteins. Nucleic Acids Res. 2010;38:5015–5022. doi: 10.1093/nar/gkq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz W.A. Steinhoff C. Florl A.R. Methylation of endogenous human retroelements in health and disease. Curr Top Microbiol Immunol. 2006;310:211–250. doi: 10.1007/3-540-31181-5_11. [DOI] [PubMed] [Google Scholar]
- Schwartz E.C. Saez L. Young M.W. Muir T.W. Post-translational enzyme activation in an animal via optimized conditional protein splicing. Nat Chem Biol. 2006;3:50–54. doi: 10.1038/nchembio832. [DOI] [PubMed] [Google Scholar]
- Stains C.I. Furman J.L. Segal D.J. Ghosh I. Site-specific detection of DNA methylation utilizing mCpG-SEER. J Am Chem Soc. 2006;128:9761–9765. doi: 10.1021/ja060681j. [DOI] [PubMed] [Google Scholar]
- Teneng I. Montoya-Durango D.E. Quertermous J.L. Lacy M.E. Ramos K.S. Reactivation of L1 retrotransposon by benzo(a)pyrene involves complex genetic and epigenetic regulation. Epigenetics. 2011;6:355–367. doi: 10.4161/epi.6.3.14282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B. Kimhi S. Howard G. Eden A. Lyko F. Demethylation of a LINE-1 antisense promoter in the cMet locus impairs Met signalling through induction of illegitimate transcription. Oncogene. 2010;29:5775–5784. doi: 10.1038/onc.2010.227. [DOI] [PubMed] [Google Scholar]
- Wolff E.M. Byun H.M. Han H.F. Sharma S. Nichols P.W. Siegmund K.D. Yang A.S. Jones P.A. Liang G. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet. 2010;6:e1000917. doi: 10.1371/journal.pgen.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.