Abstract
Protein that interacts with C kinase 1 (PICK1) is a critical mediator of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR) trafficking in neural synapses. However, its ubiquitous expression suggests that it may have other non-neural functions. Here we show that PICK1 antagonizes transforming growth factor beta (TGF-β) signaling by targeting TGF-β type I receptor (TβRI) for degradation. Biochemical analyses reveal that PICK1 directly interacts with the C-terminus of TβRI via its PDZ domain and acts as a scaffold protein to enhance the interaction between TβRI and caveolin-1, leading to enhanced lipid raft/caveolae localization. Therefore, PICK1 increases caveolin-mediated endocytosis, ubiquitination and degradation of TβRI. Moreover, a negative correlation between PICK1 expression and TβRI or phospho-Smad2 levels is observed in human breast tumors, indicating that PICK1 may participate in breast cancer development through inhibition of TGF-β signaling. Our findings reveal a non-neural function of PICK1 as an important negative regulator of TGF-β signaling.
Keywords: PICK1, TGF-β signaling, receptor degradation, caveolin, endocytosis
Introduction
Transforming growth factor beta (TGF-β) regulates cell proliferation, differentiation, migration and death, and its abnormity has been closely related to pathology, including fibrosis, vascular disorders and cancer development1,2,3,4,5. Through a phosphorylation cascade, TGF-β type I receptor (TβRI), which is activated by the type II receptor (TβRII) in the ligand-bound receptor complex, transduces signaling to intracellular molecules, Smad proteins. Then, the activated Smad2/3-Smad4 complex translocates from the cytoplasm into the nucleus, and regulates the expression of target genes6. As TGF-β plays vital roles in many physiological processes, its duration and intensity are under tight controls7.
TGF-β receptors link extracellular stimulation to intracellular responses, so their distributions and stabilities are critical for TGF-β signal transduction8,9. Like many other receptors on cell surface, TGF-β receptors undergo constant internalization, which is mediated by two major endocytic pathways: clathrin-mediated endocytosis and caveolin-mediated endocytosis10. The clathrin-mediated endocytosis of TGF-β receptors facilitates phosphorylation of downstream R-Smad and results in enhanced signaling. In contrast, caveolin-mediated endocytosis, which mainly occurs in lipid raft region, inhibits signaling through promoting receptors' ubiquitination followed by degradation10,11. The dynamic balance between these two pathways maintains appropriate response of a cell to TGF-β stimulation.
Protein that interacts with C kinase 1 (PICK1) is a PDZ (PSD-95/Dlg/ZO-1) domain and BAR (Bin/amphiphysin/Rvs) domain containing protein first cloned as a PKC-binding partner through yeast two hybrid system12. As a peripheral protein, PICK1 has been reported to interact with a series of membrane proteins including α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR)13,14,15,16, acid-sensing ion channel (ASICs)17 and ErbB2/Her-218. In most of the cases, PICK1 acts as a critical regulator of membrane receptors' subcellular trafficking to modulate neural processes such as learning and memory19,20. PICK1 is widely expressed in brain, testis, heart, lung, liver, kidney and muscle12. Its abnormal expression in brain has been linked to schizophrenia and other mental disorders21,22,23. However, the significance of PICK1 in other organs has not been well characterized. Its potential effects on signal transduction and related diseases remain unclear. Recent evidences suggest that PICK1 may affect human cancer development18,24,25,26, in which TGF-β signaling has been reported to play significant roles5.
A PDZ domain-containing protein GIPC (GAIP-interacting protein, C terminus) has been shown to interact with the TGF-β type III receptor and regulate TGF-β signaling27, but no PDZ proteins have been reported to interact with TβRI. In this study, we demonstrated that PICK1 antagonized TGF-β signaling and released its inhibition of cell growth. We found that PICK1 bound to TβRI directly and facilitated its internalization through caveolin-mediated endocytic pathway, which led to degradation of TβRI and turnoff of the signal. Moreover, the restriction of PICK1 on TGF-β signaling was observed in human breast cancer, which indicated that PICK1 might affect breast cancer development in a TGF-β-dependent manner. Not only did this study reveal the relationship between PICK1 and TGF-β signal transduction, but also it provided new insight into physiological and pathological functions of PICK1, especially its effects on tumor development.
Results
PICK1 attenuates TGF-β/Smad signaling
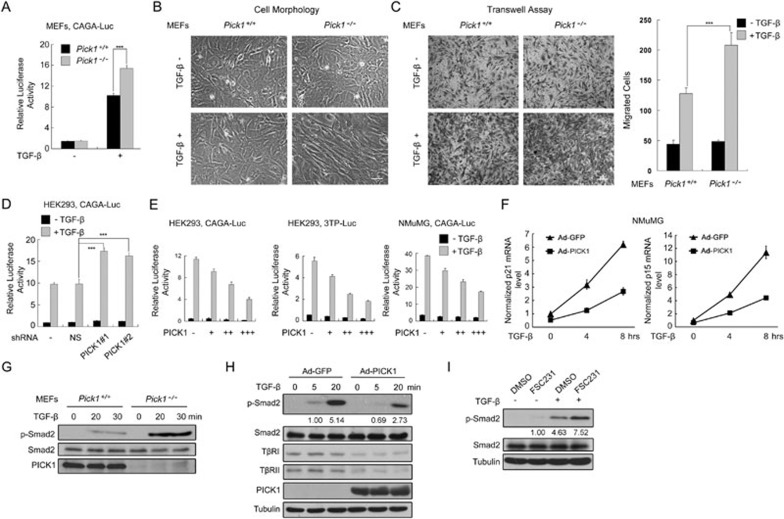
Sequence analysis predicted that the C-terminus of TβRI (QQEGIKM) might function as a potential PICK1 PDZ-binding motif (-X-Φ-X-Φ, where X stands for any residues and Φ stands for hydrophobic residues), thus suggesting that PICK1 may regulate trafficking and function of TβRI. To assess this possibility, we investigated the effect of PICK1 on TGF-β signaling by comparing TGF-β responses between Pick1+/+ and Pick1−/− mouse embryonic fibroblast (MEF) cells. Reporter assay using CAGA-luciferase revealed that TGF-β treatment increased the Smad activation in Pick1+/+ MEFs, and TGF-β response was markedly enhanced in Pick1−/− MEFs (Figure 1A). Consistently, Pick1−/− MEFs were more sensitive to TGF-β in the induction of the morphologic change to an elongate shape than Pick1+/+ MEFs (Figure 1B). Besides, loss of PICK1 promoted the mobility of MEFs upon TGF-β stimulation (Figure 1C). These data indicate that deletion of PICK1 enhances TGF-β response. To verify this, we employed RNA interference to knock down PICK1 expression (Supplementary information, Figure S1) and found that knockdown of PICK1 by shRNAs led to enhanced TGF-β response (Figure 1D). These data further support that disruption of PICK1 expression sensitizes cells to TGF-β responses.
Figure 1.
PICK1 attenuates TGF-β signaling. (A) MEFs transfected with CAGA-luciferase were treated with 100 pM TGF-β1 for 16 h and harvested for luciferase measurement. (B) MEFs were treated with 200 pM TGF-β1 for 24 h. Scale bar, 50 μm. (C) MEFs in the transwell were treated with 200 pM TGF-β1 for 16 h and fixed with methanol. After crystal violet staining, the migrated MEFs were quantitated (right panel). Scale bar, 100 μm. (D) HEK293 cells transfected with CAGA-luciferase and shRNA were treated with TGF-β1. Nonspecific (NS) shRNA was used as a negative control. (E) Cells transfected with different amounts of PICK1 plasmid were treated with TGF-β1 for 16 h. (F) NMuMG cells infected with adenovirus expressing GFP or PICK1 were harvested to examine expression of p21 and p15 using quantitative RT-PCR. (G) MEFs were treated with 100 pM TGF-β1 for various time and harvested for immunoblotting with the indicated antibodies. (H) NMuMG cells infected with GFP or PICK1 adenovirus were treated with TGF-β1 for the indicated time, and harvested for immunoblotting. The band intensity was quantitated with BandScan 5.0. (I) NMuMG cells were treated with DMSO or FSC231 for 4 h followed by TGF-β1 treatment for 30 min. Then, the cells were harvested for immunoblotting. Reporter activity was normalized with co-transfected Renilla and the data represent the mean ± S.D. (n = 3). ***P < 0.001.
To confirm the negative effect of PICK1 on TGF-β signaling, we overexpressed PICK1 and examined its effect on TGF-β-induced expression of the reporters CAGA-luciferase and 3TP-luciferase. Overexpression of PICK1 inhibited the transcriptional activity of TGF-β in HEK293, NMuMG and HaCaT cells in a dose-dependent manner (Figure 1E and Supplementary information, Figure S2). TGF-β upregulates the expression of p21 and p15 via Smad proteins28,29. The TGF-β-induced expression of p21 and p15 was also attenuated by PICK1 in NMuMG cells, as shown by qRT-PCR (Figure 1F). In agreement with this, the antiproliferative effect of TGF-β was abolished by PICK1 overexpression in NMuMG cells (Supplementary information, Figure S3).
As Smad2/3 proteins are the main mediators of TGF-β signaling and activated by TGF-β receptors via C-terminal serine phosphorylation, we then assessed the effect of PICK1 on Smad phosphorylation. Although the Smad2 level was lower in Pick1+/+ MEFs, stronger phosphorylation was observed in Pick1−/− MEFs upon TGF-β treatment (Figure 1G), while overexpression of PICK1 decreased TGF-β-induced Smad2 phosphorylation in NMuMG cells (Figure 1H). FSC231 is a small-molecule inhibitor of PICK1, which abolishes the interaction of PICK1 PDZ domain with other proteins30. As shown in Figure 1I, FSC231 enhanced TGF-β-induced Smad2 phosphorylation. Taken together, these data suggest that PICK1 negatively modulates TGF-β/Smad signaling.
PICK1 promotes TβRI degradation
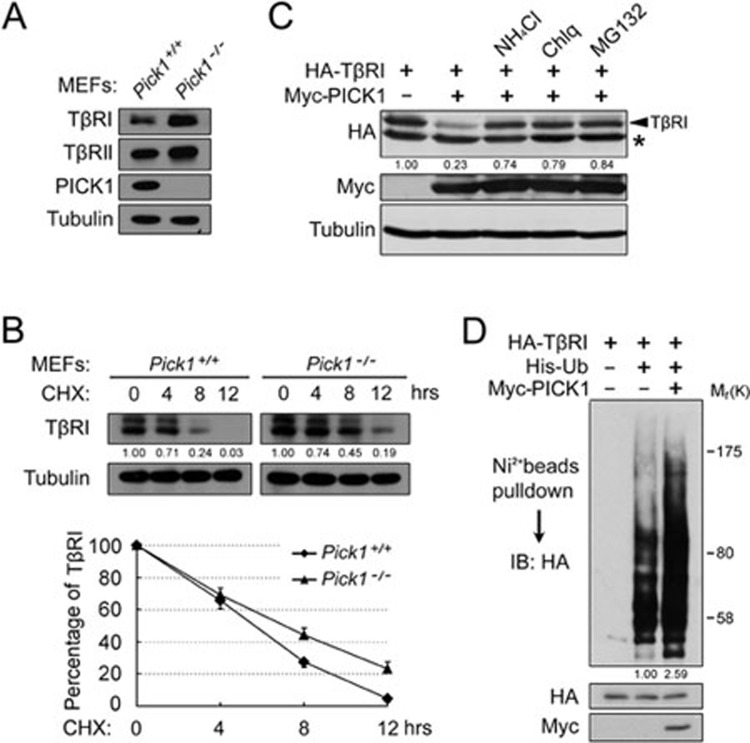
As PICK1 attenuates TGF-β/Smad signaling, we then assessed whether PICK1 regulates TGF-β receptor stability. Immunblotting analysis revealed that the TβRI protein level was higher in Pick1−/− MEFs than in Pick1+/+ MEFs (Figure 2A). However, the TβRI mRNA level was similar in these cells (data not shown). These data suggest that PICK1 may modulate TβRI stability. To verify this, we followed TβRI turnover after proteins synthesis was blocked by cycloheximide (CHX) and found that loss of PICK1 resulted in a prolonged half-life of TβRI in MEFs (Figure 2B).
Figure 2.
PICK1 enhances TβRI degradation. (A) MEFs were harvested for immunoblotting. Tubulin served as a loading control. (B) MEFs were treated with cycloheximide (CHX) for the indicated time and harvested for immunoblotting. The band intensity was quantitated and the statistical analysis of 3 independent experiments was provided. (C) HEK293T cells transfected with TβRI and PICK1 as indicated were treated with NH4Cl (25 mM), chloroquine (100 μM) or MG132 (25 μM) for 4 h before harvested for immunoblotting. * indicates a nonspecific band. (D) HEK293T cells transfected with the indicated constructs were treated with chloroquine and MG132 and harvested for Ni-nitrilotriacetate bead precipitation, followed by immunoblotting.
Consistently, overexpression of PICK1 destabilized TβRI in HEK293T cells, which could be rescued by the lysosome inhibitors NH4Cl and chloroquine or the proteasome inhibitor MG132 (Figure 2C), indicating that both the lysosome and proteasome degradation pathways are involved in PICK1-mediated degradation of TβRI. As Smurf-mediated ubiquitination of TβRI is required for its degradation31,32, we examined whether PICK1 affected TβRI ubiquitination. Indeed, PICK1 enhanced TβRI ubiquitination (Figure 2D).
Protein kinase C is important in regulating cellular functions of PICK133, however, PICK1-mediated degradation of TβRI was not markedly influenced by it (Supplementary information, Figure S4).
PICK1 interacts with TβRI via its PDZ domain
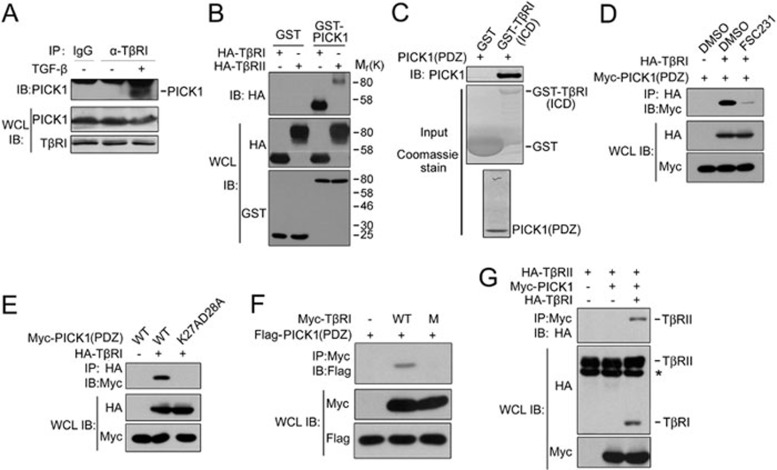
To address how PICK1 controls TβRI turnover, we examined whether PICK1 interacts with TGF-β receptors. In HEK293 cells, co-immunoprecipitation revealed that TβRI associated with PICK1 at the endogenous protein level upon TGF-β stimulation (Figure 3A). When they were overexpressed, the interaction between PICK1 and TβRI could be detected without TGF-β treatment (Figure 3B).
Figure 3.
PICK1 directly interacts with TβRI via its PDZ domain. (A) HEK293 cells were treated with 100 pM TGF-β1 for 30 min before harvested for immunoprecipitation (IP) and immunoblotting (IB) analysis. WCL, whole-cell lysate. (B) HEK293T cells transfected with GST-PICK1 and HA-tagged receptors were subjected for GST pull-down and immunoblotting. (C) Bacterially expressed recombinant GST-TβRI(ICD) and PICK1(PDZ) were subjected for GST pull-down and immunoblotting. GST and GST-TβRI(ICD) inputs were shown by Coomassie staining. (D) HEK293T cells transfected with HA-TβRI and Myc-PICK1(PDZ) were treated with FSC231 for 4 h and harvested for anti-HA immunoprecipitation and anti-Myc immunoblotting. (E-G) HEK293T cells transfected with the indicated constructs were harvested for immunoprecipitation and immunoblotting. M: mutation of Ile501 and Met503 to Asp at the C-tail of TβRI. * indicates a nonspecific band.
It has been shown that the PDZ domain of PICK1 mediates its interaction with other proteins19. To test whether the PICK1-TβRI interaction was direct, we generated recombinant PICK1 PDZ domain and GST-fused intracellular domain of TβRI(ICD). GST pull-down demonstrated that the PDZ domain could directly bind to TβRI(ICD) (Figure 3C). FSC231 interfered with the interaction between TβRI and PICK1(PDZ) (Figure 3D), further supporting that PICK1 directly interacts with TβRI via its PDZ domain. It has been reported that mutation of Lys27 and Asp28 to Ala can completely disrupt the binding of the PICK1 PDZ domain to other proteins13,34. Consistently, only wild-type PICK1(PDZ), but not the mutant K27A,D28A, was co-immunoprecipitated by TβRI (Figure 3E). We further identified the C-terminal tail of TβRI as a PDZ-binding motif as mutation of the residues Ile501 and Met503 to Asp at the TβRI tail abolished its interaction with the PICK1 PDZ domain (Figure 3F).
The data in Figure 3B showed that PICK1 also weakly interacted with TβRII. To explore whether this interaction requires the presence of TβRI, we tested whether PICK1 interacts with TβRII in L17 cells that do not express TβRI35. As shown in Figure 3G, PICK1 interacted with TβRII only when TβRI was co-expressed. As overexpressed receptors can form a complex in the absence of TGF-β36,37, these results implicate that PICK1 associates with TβRII through TβRI.
Caveolin-1 is essential for PICK1-mediated degradation of TβRI
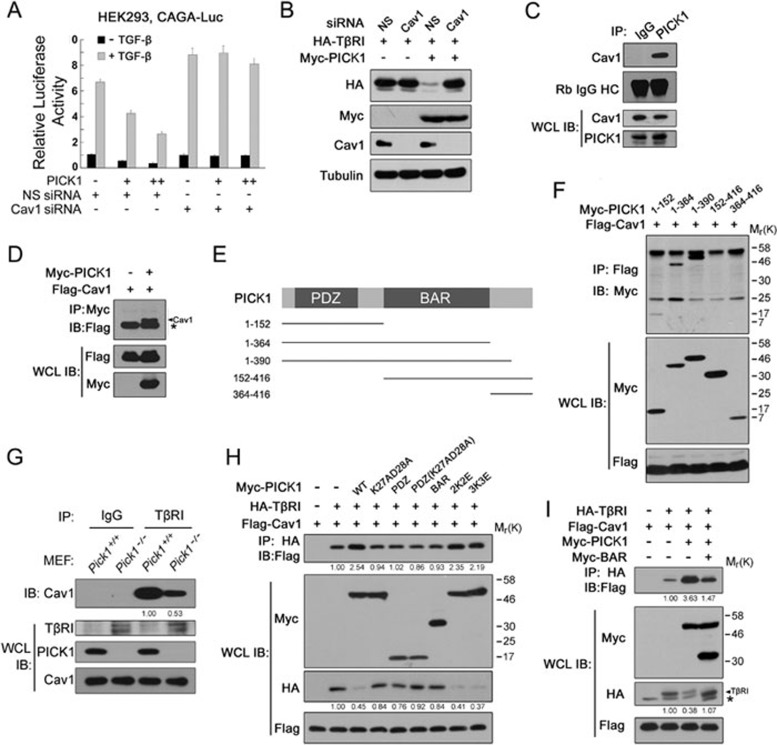
PICK1 acts as a scaffold protein regulating membrane trafficking of AMPK receptors19,20. It has been suggested that caveolin-1-mediated endocytosis is critical for TβRI degradation10,38. Therefore, we tested whether caveolin-1 is involved in PICK1-mediated TβRI degradation. First, we examined the effect of caveolin-1 on PICK1 inhibition of TGF-β transcriptional responses. As shown in Figure 4A, caveolin-1 knockdown with siRNA abolished the inhibitory effect of PICK1 on TGF-β-induced expression of CAGA-luciferase. Consistent with this, PICK1-mediated degradation of TβRI was prevented when caveolin-1 was depleted (Figure 4B). These data suggest that caveolin-1 is required for PICK1-mediated inhibition of TGF-β signaling and TβRI degradation.
Figure 4.
PICK1 promotes the TβRI-caveolin-1 interaction. (A) HEK293 cells transfected with CAGA-luciferase and caveolin-1 siRNA were treated with 100 pM TGF-β1 for 16 h before harvested for luciferase assay. (B) HEK293T cells transfected with the indicated constructs or siRNA were harvested for immunoblotting. Tubulin was used as a loading control. (C, D) Interaction between PICK1 and caveolin-1. * indicates IgG light chain. (E) PICK1 deletion constructs for domain mapping. (F) HEK293T cells transfected with the indicated constructs were harvested for immunoprecipitation and immunoblotting. (G) The TβRI-caveolin-1 interaction was examined in MEFs with immunoprecipitation and immunoblotting. (H, I) HEK293T cells transfected with the indicated constructs were harvested for immunoprecipitation and immunoblotting. * indicates a nonspecific band.
To elucidate the relationship of PICK1 and caveolin-1 in regulation of TGF-β signaling, we investigated whether PICK1 interacts with caveolin-1. Co-immunoprecipitation showed that PICK1 interacted with caveolin-1 at the endogenous protein levels in NMuMG cells (Figure 4C) and at the ectopically expressed protein levels in HEK293T cells (Figure 4D). Domain mapping revealed that the 1-152 residues of PICK1 containing the PDZ domain was essential for its interaction with caveolin-1 (Figure 4E and 4F). However, the binding ability of the 1-152 fragment to caveolin-1 was weaker than that of full-length PICK1, suggesting that other domains may also contribute to this interaction.
As PICK1 interacts with both TβRI and caveolin-1, it might serve as a scaffold protein promoting the TβRI-caveolin-1 association. Indeed, we observed that the interaction between TβRI and caveolin-1 was greatly attenuated in Pick1−/− MEFs (Figure 4G). Consistently, the TβRI-caveolin-1 association was markedly enhanced by wild-type PICK1 although the total TβRI level was lower as the result of PICK1-mediated degradation (Figure 4H). However, none of the PDZ domain, the BAR domain or K27A,D28A mutant had effect on the TβRI-caveolin-1 interaction and TβRI degradation. PICK1 K266E,K268E (2K2E) and K251E,K252E,K257E (3K3E) mutants, which lose the lipid-binding ability39, could not disrupt the PICK1 activity to interact with TβRI and promote its degradation.
The BAR domain of PICK1 is critical for PICK1 dimerization40. Overexpression of the BAR domain could bind to wild-type PICK1 and then disrupt the wild-type PICK1 dimerization efficiently (Supplementary information, Figure S5). To test whether dimerization is important for PICK1 to promote the interaction between TβRI and caveolin-1, we co-expressed the BAR domain and found that it decreased the PICK1-enhanced TβRI-caveolin-1 interaction (Figure 4I), implicating that PICK1 dimerization is critical for its enhancement on the TβRI-caveolin-1 interaction.
PICK1 facilitates lipid raft localization and caveolin-mediated endocytosis of TβRI
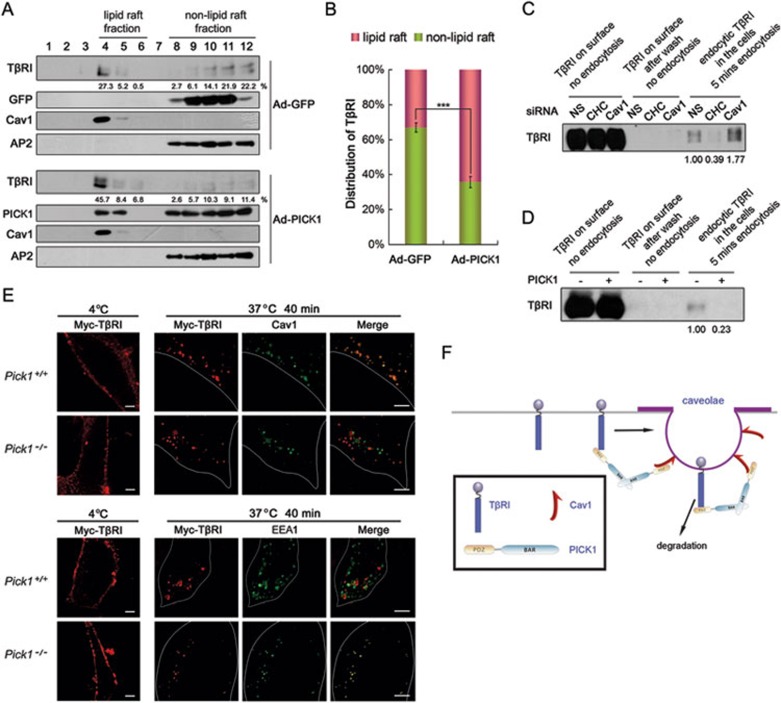
As PICK1-enhanced TβRI degradation is dependent on caveolin-1, we assessed whether localization of TβRI in lipid raft is affected by PICK1 in NMuMG cells. In GFP-expressing control cells, TβRI was distributed in both lipid raft (33%) and non-lipid raft fractions, while in the cells expressing PICK1, more TβRI was found in lipid raft fractions (60%) (Figure 5A and 5B).
Figure 5.
PICK1 facilitates lipid raft localization and caveolin-mediated endocytosis of TβRI. (A, B) NMuMG cells infected with GFP or PICK1 adenovirus were harvested for lipid raft purification using sucrose density gradient centrifugation. Fraction aliquots were immunoblotted with the indicated antibodies. Caveolin-1 and AP2 indicate lipid raft/caveolae and non-lipid raft fractions, respectively. Density of TβRI was quantitated and the statistical analysis of 3 independent experiments was shown in B. The data represent the mean ± S.D. (n = 3). ***P < 0.001. (C, D) HEK293 cells were transfected with clathrin heavy chain (CHC) or caveolin-1 siRNA (C) or PICK1 construct (D), and the cell surface was labeled with biotin. After precipitation with streptavidin beads, TβRI on cell surface and endocytic TβRI were detected with anti-TβRI antibody. (E) MEFs transfected with Myc-TβRI were labeled with anti-Myc antibody at 4 °C for 6 h. Then the cells were washed with cold PBS twice, followed by incubation at 37 °C for 40 min. After fixation and permeabilization, the cells were visualized by immunofluorescence with anti-Myc (red) and anti-caveolin-1 (green) or with anti-Myc (red) and anti-EEA1 (green). Scale bar, 5 μm. (F) A working model showing that PICK1 acts as a scaffold protein to enhance the TβRI-caveolin-1 interaction and caveolin-mediated endocytosis and degradation of the receptor.
TβRI can be internalized via both clathrin-mediated and caveolin-mediated endocytosis10, and caveolin-mediated endocytosis turns off TGF-β signaling by promoting TβRI degradation. These two endocytosis pathways are maintained at a dynamic balance, and blockage of one pathway would lead to amplification of the other38. Endocytosis of biotin-labeled TβRI was employed to study the effect of PICK1 on this dynamic balance of the receptor41. Clathrin heavy chain siRNA and caveolin-1 siRNA were used to block clathrin-mediated endocytosis and caveolin-mediated endocytosis, respectively (Figure 5C and Supplementary information, Figure S6). We found that most TβRI internalization into the cell within 5 min was mainly through a rapid clathrin-mediated endocytosis (Figure 5C), which was blocked by PICK1 (Figure 5D), suggesting that PICK1 can prevent TβRI from the fast internalization through the clathrin-dependent pathway. In agreement with this, the majority of internalized TβRI was co-stained with the caveolin-1-positive vesicles in Pick1+/+ MEFs, while the proportion of TβRI-caveolin-1 co-localization was dramatically decreased in Pick1−/− MEFs (Figure 5E). Consistently, the co-localization between internalized TβRI and EEA1 was increased in Pick1−/− cells (Figure 5E). These data suggest that PICK1 can shift the internalization of TβRI from the clathrin-dependent pathway to the caveolin-dependent pathway.
To address whether dimerization is important for PICK1 to mediate caveolin-dependent endocytosis of TβRI, we expressed the BAR domain in HeLa cells and found that disruption of PICK1 dimerization markedly decreased the co-localization between endocytic TβRI and caveolin-1 (Supplementary information, Figure S7).
These results together suggest that PICK1 facilitates lipid raft localization and caveolin-mediated endocytosis of TβRI and thus accelerates TβRI degradation.
PICK1 expression is negatively correlated with TGF-β signaling in human breast cancer
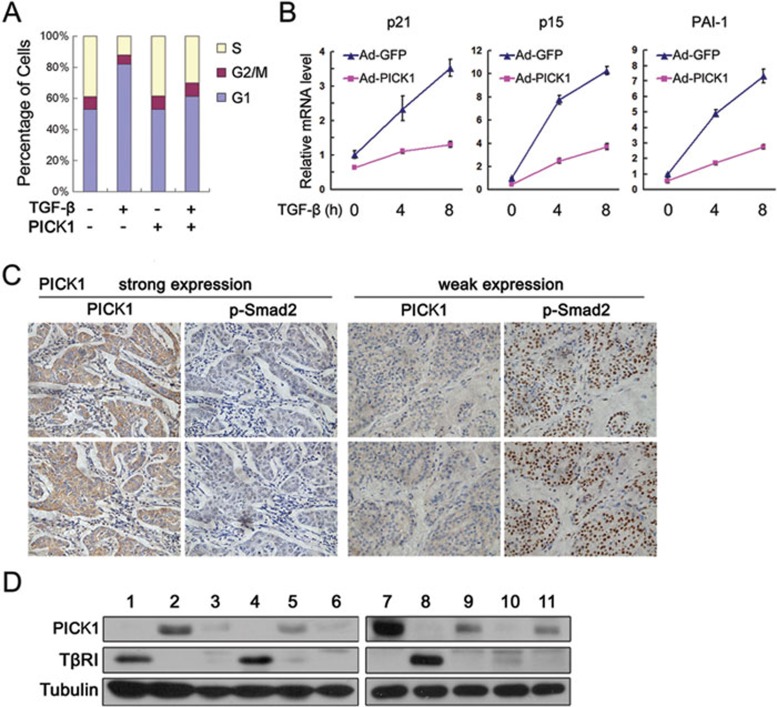
Deregulation of TGF-β signaling contributes to tumorigenesis. It has been reported that PICK1 is highly expressed in breast cancer cells26. To investigate whether the negative regulation of PICK1 on TGF-β signaling contributes to breast cancer development, we first examined the effect of PICK1 on the antiproliferative activity of TGF-β in breast epithelial MCF10A cells. As shown in Figure 6A and 6B, overexpression of PICK1 not only attenuated G1 phase arrest induced by TGF-β, but also remarkably decreased mRNA levels of PAI1, p21 and p15, the latter two genes playing pivotal roles in cell proliferation. Immunohistochemistry analysis revealed that PICK1 expression and phosphor-Smad2 level, a TGF-β signaling indicator, showed a significant negative correlation (P = 4.3e-12; correlation coefficient = 0.54) in 141 breast cancer samples (Figure 6C). Furthermore, this negative correlation was also observed in 11 fresh breast tumors (Figure 6D). These results suggest that PICK1 may promote tumorigenesis by inhibiting TGF-β signaling.
Figure 6.
PICK1 protein levels are negatively correlated with TGF-β signaling activities in human breast cancer. (A) MCF10A cells were infected with GFP or PICK1 adenovirus and treated with TGF-β1 for 20 h, followed by FACS analysis. (B) Expression analysis of TGF-β target genes by quantitative RT-PCR in MCF10A cells infected with GFP or PICK1 adenovirus. (C) Representative images from immunohistochemistry staining of breast cancer tissues with anti-PICK1 and anti-p-Smad2 antibodies in consecutive sections. (D) Immunoblotting of 11 fresh human breast cancer samples using the indicated antibodies.
Discussion
TGF-β signaling is tightly regulated to ensure its appropriate activity in embryo development and tissue homeostasis. PICK1 is a scaffold protein to control trafficking of several membrane receptors, including AMPA receptor and ASIC, and has been implicated to function in nervous system and male fertility17,19,20,42. Here, we identify PICK1 as a novel negative regulator of TGF-β signaling by modulating subcellular localization and trafficking of TβRI. By promoting the interaction between TβRI and caveolin-1, PICK1 facilitates caveolin-mediated endocytosis of cell surface TβRI, resulting in TβRI degradation and signaling termination (Figure 5F).
TGF-β receptors are dynamically distributed on the plasma membrane10. Interleukin-643, ADAM1244 and cholesterol depletion45 can partition TβRI from the lipid raft microdomains into non-lipid raft regions, while hyaluronan46 and the polysaccharide heparin sulfate47 promote lipid raft localization of TβRI. Several lines of evidence showed that caveolin, which is enriched in lipid rafts, is required for TβRI turnover38,46,48,49. In the current study, we found that PICK1 could promote caveolae localization of TβRI. This is achieved for PICK1 to function as a scaffold protein to bridge TβRI and caveolin-1. Consistently, less TβRI was found in Pick1−/− cells. Interestingly, unlike AMPA receptor, PICK1-mediated regulation of membrane distribution of TβRI is independent of protein kinase C activity. Of note, our previous work demonstrated that lipid raft location of TβRI is also required for TGF-β-induced activation of MAPKs49. We observed that loss of PICK1 or knockdown of caveolin-1 led to enhanced phosphorylation level of p38 (data not shown), supporting the notion that TβRI localization in caveolae mainly modulates receptor turnover while its distribution in non-caveolae lipid rafts is important for MAPK activation.
TβRI enters the cell via both clathrin- or lipid raft-mediated endocytosis10,50. These two distinct endocytic pathways predetermine whether TβRI promotes a signaling response or is degraded9. Clathrin-mediated endocytosis of TβRI into early endosomes enhances TGF-β signaling while lipid raft/caveolin-mediated endocytosis enhances ubiquitination and degradation of TβRI. Consistent with this, our biotin-labeled TβRI endocytosis assay revealed that rapid internalization of TβRI is mainly through clathrin-mediated endocytosis, indicating that clathrin-mediated endocytosis may be important for immediate cell response to TGF-β stimulation. Furthermore, PICK1 inhibits rapid clathrin-mediated internalization of TβRI, suggesting that PICK1-enhanced caveolae localization of TβRI leads to signal turnoff not only by promoting TβRI degradation, but also by shielding the receptor from entering early endosomes.
PICK1 is ubiquitously expressed in many tissues outside the nervous system12, however, its non-neural functions remains largely unclear. Increased expression of PICK1 was reported in breast cancer26. We observed a negative correlation between PICK1 and p-Smad2 levels in breast cancer samples, indicating that PICK1 may promote breast cancer formation by restricting TGF-β signaling. In agreement with this, our results revealed that overexpression of PICK1 in breast epithelial cells abolished TGF-β-induced growth inhibition.
In summary, this study provides the evidence that PICK1 facilitates TGF-β receptor degradation in a caveolin-dependent manner. Our findings extend the understanding of the dynamic regulation of TGF-β receptors and offer new insight into the cellular and pathological functions of PICK1.
Materials and Methods
Plasmids and RNA interference
Human PICK1 was cloned into pCMV-Myc, Flag-pCDNA3.1 and PEBG1 for eukaryotic expression. MBP-PICK1 was generated by subcloning into the EcoRI and SalI sites of pMal-s (a gift from Jingming Yuan) for prokaryotic expression. pCMV-Myc-PICK1 K27A, D28A, 2K2E and 3K3E were constructed by the mutagenesis of pCMV-Myc-PICK1 using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) and confirmed by sequencing. For RNA interference, two PICK1 shRNA constructs were generated using pll3.7. The targeted sequences are as follows: 5′-GAAGTACCTGGACGTGAAG-3′ and 5′-GAGGAGCTGGAGCGGACCG-3′. siRNAs targeting caveolin-1 and clathrin heavy chain were purchased from GenePharma. The sequences of siRNAs were as follows: negative control, sense 5′-UUCUCCGAACGUGUCACGUTT-3′, anti-sense 5′-ACGUGACACGUUCGGAGAATT-3′ caveolin-1, sense 5′-GGGACAUCUCUACACCGUUCC-3′, anti-sense 5′-GGAACGGUGUAGAGAUGUCCC-3′ clathrin heavy chain, sense 5′-GUAAUCCAAUUCGAAGACCTT-3′, anti-sense 5′-GGUCUUCGAAUUGGAUUACTT-3′.
Reagents and antibodies
FSC231 was a generous gift from Dr Ulrik Gether. Propidium iodide, NH4Cl, chloroquine and cycloheximide (CHX) were obtained from Sigma, MG132 from Calbiochem, TGF-β1 from R&D Systems, HRV3C from Novagen, Transferrin conjugates (Fluor 488) from Invitrogen. Antibodies were purchased from ABR (α-Adaptin2 and α-PICK1), BD Biosciences (α-caveolin-1), Cell Signaling (α-Myc tag), Millipore (α-Myc tag and α-p-Smad2), Proteintech Group (α-PICK1), Santa Cruz (α-GST tag, α-HA tag, α-PICK1, α-Smad2, α-TβRI, α-TβRII and α-tubulin), Sigma (α-Flag tag M2), eBioscience (Rabbit TrueBlot HRP-conjugated anti-rabbit lgG), GE Healthcare (ECL HRP-linked anti-mouse lgG and ECL HRP-linked anti-rabbit lgG) and Jackson ImmunoResearch (fluorescein isothiocyanate (FITC)-conjugated anti-mouse and tetramethylrhodamine β-isothiocyanate (TRITC)-conjugated anti-rabbit).
Cell lines and transfection
HEK293, HEK293T, HeLa, HaCaT, NMuMG cells and MEFs were maintained in DMEM medium supplemented with 10% FBS (Hyclone) in a 37 °C humidified incubator containing 5% CO2. L17 cells were maintained in MEM medium with 10% FBS and MCF10A was maintained in MCF10A specific culture medium. Transfection was performed with VigoFect (Vigorous) and Lipofectamine 2000 (Invitrogen) following the manufacturer's recommendations.
Pick1−/− mice and MEF isolation
Pick1−/− mice have been reported42. MEFs were isolated and cultured as previously described51.
Adenoviral expression
Ad-EGFP and Ad-PICK1 were generated with AdEasy™ Adenoviral Vector System (Stratagene). PICK1 was cloned into the XhoI and HindIII sites of pShuttle-CMV, and then transfered to pAdEasy-1 vector using homologous recombination in vivo in BJ5183 cells. Linearized pAdEasy-1-PICK1 produced viral stocks after transfection into 293A cells.
Reporter assay, immunoblotting, immunofluorescence, immunoprecipitation and flow cytometry
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted with Trizol reagent (Invitrogen) and cDNA was synthesized with Revertra Ace (Toyobo). A Mx3000p Quantitative PCR system (Stratagene) was employed to perform qRT-PCR using EvaGreen dye (Biotium). The primers were: mouse p21 (5′-GTGATTGCGATGCGCTCATG-3′ and 5′-TCTCTTGCAGAAGACCAATC-3′), mouse p15 (5′-CCCTGCCACCCTTACCAGA-3′ and 5′-CAGATACCTCGCAATGTCACG-3′), mouse GAPDH (5′-AAGAAGGTGGTGAAGCAG-3′ and 5′-TCATACCAGGAAATGAGC-3′), human p21 (5′-TGGAGACTCTCAGGGTCGAAAA-3′ and 5′-GCGTTTGGAGTGGTAGAAATCTG-3′), human p15 (5′-CACCGTTGGCCGTAAACTTAAC-3′ and 5′-TAATGAAGCTGAGCCCAGGTCT-3′), human PAI1 (5′-GAGACAGGCAGCTCGGATTC-3′ and 5′-GGCCTCCCAAAGTGCATTAC-3′) and human GAPDH (5′-GAAGGTGAAGGTCGGAGTC-3′ and 5′-GAAGATGGTGATGGGATTTC-3′).
In vitro pull-down assay
MBP-PICK1 and GST-TβRI (intracellular domain, ICD) fusion proteins were expressed in E. coli and purified with glutathione-sepharose (Amersham Pharmacia Biotech) or amylose resin (New England Biolabs). PDZ domain of PICK1 was obtained after on-beads-cleavage of MBP-PDZ with HRV3C. The beads binding MBP-PICK1 or GST-TβRI(ICD) were washed extensively with binding buffer (50 mM Tris-HCl pH 8.0, 250 mM NaCl) and were incubated with purified TβRI(ICD) or PICK1(PDZ) for 1 h, respectively. Bound proteins were extracted with loading buffer and analyzed by immunoblotting.
Receptor endocytosis assay and quantitative analysis of endogenous TβRI endocytosis by biotinylation
HEK293 cells were placed on ice, washed twice with ice-cold PBS and then labeled with 0.2 mM Sulfo-NHS-SS-biotin (Thermo) in PBS for 30 min. After washing twice with ice-cold PBS and replacing with pre-warmed DMEM, dishes were placed in 37 °C incubator for 5 min, allowing for TβRI endocytosis. All dishes were then returned to ice and washed once with ice-cold PBS to stop membrane trafficking. To strip biotin remaining on cell surface, cells were treated twice with stripping buffer (50 mM cold L-Glutathione reduced, 75 mM NaCl, 75 mM NaOH, and 10% FBS) at 4 °C for 20 min. The cells are lysed in lysis buffer (10 mM EDTA, 1% Triton X-100, 0.1% SDS and protease inhibitors in PBS) and incubated with streptavidin beads (Thermo) for 1 h at 4 °C. Strptavidin-precipitated TβRI protein was detected with immunoblotting. The biotinylated TβRI remaining within the cells should be compared to the total TβRI level on cell surface before stripping.
Antibody-labeled TβRI endocytosis
Pick1+/+ and Pick1−/− MEFs were placed on glass coverslips and transfected with TβRI tagged with Myc right after signal peptide. Two days later, cells were washed twice with ice-cold PBS, followed by incubation with α-Myc tag antibody (1:100 in PBS containing 10% FBS) for 6 h at 4 °C. Then cells were washed with cold PBS twice and returned to 37 °C for 40 min, allowing endocytosis to occur. The cells were fixed and prepared for immunofluorescence analysis.
Lipid raft fractionation
Lipid raft fractionation was performed as previously described49.
Immunohistochemistry and tissue array
Immunohistochemistry was performed with human breast tumor tissue array (grades I-III with normal controls) by Cybrdi Inc (Xi'an, China). A spearman's correlation test was used to analyze the relationship between PICK1 and p-Smad2.
Tumor samples
Fresh breast cancer tumor samples were obtained from the First Affiliated Hospital of Nanchang University by following the Ethics Committee guideline and stored at −80 °C. The frozen tissue was weighed, cut into small pieces and homogenized in lysis buffer (20 mM Tris-HCl, 150 mM NaCl, 0.5% NP40, 1 mM EDTA, 1% SDS) with Complete Protease Inhibitor cocktail (Roche) and PhosSTOP phosphatase inhibitor (Roche).
Statistic analysis
The band intensity was quantitated with BandScan 5.0. Student's t-test was performed in statistic analysis.
Acknowledgments
We are grateful to Ulrik Gether (Department of Neuroscience and Pharmacology, University of Copenhagen, Denmark) for FSC231 and Jingming Yuan (Institute of Biotechnology, Shanxi University, China) for pMal-s. This work was supported by grants from the National Basic Research Program of China (973 Program; 2010CB833706, 2011CB943803), the National Natural Science Foundation of China (30930050, 30921004), and Tsinghua University Initiative Scientific Research Program (2010THZ0) to YGC.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Knockdown efficiency of PICK1 shRNAs.
Overexpression of PICK1 inhibits the TGF-β-induced expression of CAGA-luciferase in HaCaT cells.
PICK1 attenuates TGF-β-induced cell cycle arrest in NMuMG cells.
PICK1-mediated degradation of TβRI was not apparently influenced by PKC.
Co-expression of Myc-BAR domain disrupted dimerization between GST-PICK1 and Flag-PICK1.
Depletion of clathrin heavy chain with siRNA effectively blocks clathrin-mediated endocytosis.
Disruption of PICK1 dimerization by the BAR domain markedly decrease the co-localization between endocytic TβRI and caveolin-1.
References
- Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Massague J, Chen YG. Controlling TGF-beta signaling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- Itoh S, ten Dijke P. Negative regulation of TGF-beta receptor/Smad signal transduction. Curr Opin Cell Biol. 2007;19:176–184. doi: 10.1016/j.ceb.2007.02.015. [DOI] [PubMed] [Google Scholar]
- Kang JS, Liu C, Derynck R. New regulatory mechanisms of TGF-beta receptor function. Trends Cell Biol. 2009;19:385–394. doi: 10.1016/j.tcb.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Chen YG. Endocytic regulation of TGF-beta signaling. Cell Res. 2009;19:58–70. doi: 10.1038/cr.2008.315. [DOI] [PubMed] [Google Scholar]
- Lonn P, Moren A, Raja E, Dahl M, Moustakas A. Regulating the stability of TGFbeta receptors and Smads. Cell Res. 2009;19:21–35. doi: 10.1038/cr.2008.308. [DOI] [PubMed] [Google Scholar]
- Staudinger J, Zhou J, Burgess R, Elledge SJ, Olson EN. PICK1: a perinuclear binding protein and substrate for protein kinase C isolated by the yeast two-hybrid system. J Cell Biol. 1995;128:263–271. doi: 10.1083/jcb.128.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Zhang X, Staudinger J, Huganir RL. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- Rocca DL, Martin S, Jenkins EL, Hanley JG. Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat Cell Biol. 2008;10:259–271. doi: 10.1038/ncb1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Ziff EB. PICK1 interacts with ABP/GRIP to regulate AMPA receptor trafficking. Neuron. 2005;47:407–421. doi: 10.1016/j.neuron.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Hanley JG, Henley JM. PICK1 is a calcium-sensor for NMDA-induced AMPA receptor trafficking. EMBO J. 2005;24:3266–3278. doi: 10.1038/sj.emboj.7600801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron A, Deval E, Salinas M, et al. Protein kinase C stimulates the acid-sensing ion channel ASIC2a via the PDZ domain-containing protein PICK1. J Biol Chem. 2002;277:50463–50468. doi: 10.1074/jbc.M208848200. [DOI] [PubMed] [Google Scholar]
- Jaulin-Bastard F, Saito H, Le Bivic A, et al. The ERBB2/HER2 receptor differentially interacts with ERBIN and PICK1 PSD-95/DLG/ZO-1 domain proteins. J Biol Chem. 2001;276:15256–15263. doi: 10.1074/jbc.M010032200. [DOI] [PubMed] [Google Scholar]
- Xu J, Xia J. Structure and function of PICK1. Neurosignals. 2006;15:190–201. doi: 10.1159/000098482. [DOI] [PubMed] [Google Scholar]
- Hanley JG. PICK1: a multi-talented modulator of AMPA receptor trafficking. Pharmacol Ther. 2008;118:152–160. doi: 10.1016/j.pharmthera.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Hong CJ, Liao DL, Shih HL, Tsai SJ. Association study of PICK1 rs3952 polymorphism and schizophrenia. Neuroreport. 2004;15:1965–1967. doi: 10.1097/00001756-200408260-00026. [DOI] [PubMed] [Google Scholar]
- Dev KK, Henley JM. The schizophrenic faces of PICK1. Trends Pharmacol Sci. 2006;27:574–579. doi: 10.1016/j.tips.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikida T, Mustafa AK, Maeda K, et al. Modulation of D-serine levels in brains of mice lacking PICK1. Biol Psychiatry. 2008;63:997–1000. doi: 10.1016/j.biopsych.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WJ, Chang YF, Wang WL, Huang CY. Mitogen-stimulated TIS21 protein interacts with a protein-kinase-Calpha-binding protein rPICK1. Biochem J. 2001;354:635–643. doi: 10.1042/0264-6021:3540635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffon KJ, Hruska-Hageman A, Klotz M, Traver GL, Zabner J. A role for the PDZ-binding domain of the coxsackie B virus and adenovirus receptor (CAR) in cell adhesion and growth. J Cell Sci. 2004;117:4401–4409. doi: 10.1242/jcs.01300. [DOI] [PubMed] [Google Scholar]
- Zhang B, Cao W, Zhang F, et al. Protein interacting with C alpha kinase 1 (PICK1) is involved in promoting tumor growth and correlates with poor prognosis of human breast cancer. Cancer Sci. 2010;101:1536–1542. doi: 10.1111/j.1349-7006.2010.01566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobe GC, Liu X, Fang SJ, How T, Lodish HF. A novel mechanism for regulating transforming growth factor beta (TGF-beta) signaling. Functional modulation of type III TGF-beta receptor expression through interaction with the PDZ domain protein, GIPC. J Biol Chem. 2001;276:39608–39617. doi: 10.1074/jbc.M106831200. [DOI] [PubMed] [Google Scholar]
- Feng XH, Lin X, Derynck R. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-beta. EMBO J. 2000;19:5178–5193. doi: 10.1093/emboj/19.19.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardali K, Kurisaki A, Moren A, et al. Role of Smad proteins and transcription factor Sp1 in p21(Waf1/Cip1) regulation by transforming growth factor-beta. J Biol Chem. 2000;275:29244–29256. doi: 10.1074/jbc.M909467199. [DOI] [PubMed] [Google Scholar]
- Thorsen TS, Madsen KL, Rebola N, et al. Identification of a small-molecule inhibitor of the PICK1 PDZ domain that inhibits hippocampal LTP and LTD. Proc Natl Acad Sci USA. 2010;107:413–418. doi: 10.1073/pnas.0902225107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisawa T, Fukuchi M, Murakami G, et al. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- Kavsak P, Rasmussen RK, Causing CG, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- Perez JL, Khatri L, Chang C, et al. PICK1 targets activated protein kinase Calpha to AMPA receptor clusters in spines of hippocampal neurons and reduces surface levels of the AMPA-type glutamate receptor subunit 2. J Neurosci. 2001;21:5417–5428. doi: 10.1523/JNEUROSCI.21-15-05417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger J, Lu J, Olson EN. Specific interaction of the PDZ domain protein PICK1 with the COOH terminus of protein kinase C-alpha. J Biol Chem. 1997;272:32019–32024. doi: 10.1074/jbc.272.51.32019. [DOI] [PubMed] [Google Scholar]
- Boyd FT, Massague J. Transforming growth factor-beta inhibition of epithelial cell proliferation linked to the expression of a 53-kDa membrane receptor. J Biol Chem. 1989;264:2272–2278. [PubMed] [Google Scholar]
- Nohe A, Hassel S, Ehrlich M, et al. The mode of bone morphogenetic protein (BMP) receptor oligomerization determines different BMP-2 signaling pathways. J Biol Chem. 2002;277:5330–5338. doi: 10.1074/jbc.M102750200. [DOI] [PubMed] [Google Scholar]
- Thompson TB, Woodruff TK, Jardetzky TS. Structures of an ActRIIB:activin A complex reveal a novel binding mode for TGF-beta ligand:receptor interactions. EMBO J. 2003;22:1555–1566. doi: 10.1093/emboj/cdg156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL. Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat Cell Biol. 2003;5:410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- Jin W, Ge WP, Xu J, et al. Lipid binding regulates synaptic targeting of PICK1, AMPA receptor trafficking, and synaptic plasticity. J Neurosci. 2006;26:2380–2390. doi: 10.1523/JNEUROSCI.3503-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Lu Z, Murray JT, Luo W, et al. Transforming growth factor beta activates Smad2 in the absence of receptor endocytosis. J Biol Chem. 2002;277:29363–29368. doi: 10.1074/jbc.M203495200. [DOI] [PubMed] [Google Scholar]
- Xiao N, Kam C, Shen C, et al. PICK1 deficiency causes male infertility in mice by disrupting acrosome formation. J Clin Invest. 2009;119:802–812. doi: 10.1172/JCI36230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XL, Topley N, Ito T, Phillips A. Interleukin-6 regulation of transforming growth factor (TGF)-beta receptor compartmentalization and turnover enhances TGF-beta1 signaling. J Biol Chem. 2005;280:12239–12245. doi: 10.1074/jbc.M413284200. [DOI] [PubMed] [Google Scholar]
- Atfi A, Dumont E, Colland F, et al. The disintegrin and metalloproteinase ADAM12 contributes to TGF-beta signaling through interaction with the type II receptor. J Cell Biol. 2007;178:201–208. doi: 10.1083/jcb.200612046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Liu IH, Fliesler SJ, et al. Cholesterol suppresses cellular TGF-beta responsiveness: implications in atherogenesis. J Cell Sci. 2007;120:3509–3521. doi: 10.1242/jcs.006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Williams JD, Fraser DJ, Phillips AO. Hyaluronan regulates transforming growth factor-beta1 receptor compartmentalization. J Biol Chem. 2004;279:25326–25332. doi: 10.1074/jbc.M403135200. [DOI] [PubMed] [Google Scholar]
- Chen CL, Huang SS, Huang JS. Cellular heparan sulfate negatively modulates transforming growth factor-beta1 (TGF-beta1) responsiveness in epithelial cells. J Biol Chem. 2006;281:11506–11514. doi: 10.1074/jbc.M512821200. [DOI] [PubMed] [Google Scholar]
- Bizet AA, Liu K, Tran-Khanh N, et al. The TGF-β co-receptor, CD109, promotes internalization and degradation of TGF-β receptors. Biochim Biophys Acta. 2011;1813:742–753. doi: 10.1016/j.bbamcr.2011.01.028. [DOI] [PubMed] [Google Scholar]
- Zuo W, Chen YG. Specific activation of mitogen-activated protein kinase by transforming growth factor-beta receptors in lipid rafts is required for epithelial cell plasticity. Mol Biol Cell. 2009;20:1020–1029. doi: 10.1091/mbc.E08-09-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roy C, Wrana JL. Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat Rev Mol Cell Biol. 2005;6:112–126. doi: 10.1038/nrm1571. [DOI] [PubMed] [Google Scholar]
- Wen J, Chiang YJ, Gao C, et al. Loss of Dact1 disrupts planar cell polarity signaling by altering dishevelled activity and leads to posterior malformation in mice. J Biol Chem. 2010;285:11023–11030. doi: 10.1074/jbc.M109.085381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Lin Z, Chen F, et al. Human BAMBI cooperates with Smad7 to inhibit transforming growth factor-beta signaling. J Biol Chem. 2009;284:30097–30104. doi: 10.1074/jbc.M109.049304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Cao W, Bao L, et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12:781–790. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Knockdown efficiency of PICK1 shRNAs.
Overexpression of PICK1 inhibits the TGF-β-induced expression of CAGA-luciferase in HaCaT cells.
PICK1 attenuates TGF-β-induced cell cycle arrest in NMuMG cells.
PICK1-mediated degradation of TβRI was not apparently influenced by PKC.
Co-expression of Myc-BAR domain disrupted dimerization between GST-PICK1 and Flag-PICK1.
Depletion of clathrin heavy chain with siRNA effectively blocks clathrin-mediated endocytosis.
Disruption of PICK1 dimerization by the BAR domain markedly decrease the co-localization between endocytic TβRI and caveolin-1.