Abstract
Metal-catalyzed oxidation may result in structural damage to proteins and has been implicated in aging and disease, including neurological disorders such as Alzheimer's disease and amyotrophic lateral sclerosis. The selective modification of specific amino acid residues with high metal ion affinity leads to subtle structural changes that are not easy to detect but may have dramatic consequences on physical and functional properties of the oxidized protein molecules. PrP contains a histidine-rich octarepeat domain that binds copper. Because copper-binding histidine residues are particularly prone to metal-catalyzed oxidation, we investigated the effect of this reaction on the recombinant prion protein SHaPrP(29–231). Using Cu2+/ascorbate, we oxidized SHaPrP(29–231) in vitro. Oxidation was demonstrated by liquid chromatography/mass spectrometry, which showed the appearance of protein species of higher mass, including increases in multiples of 16, characteristic of oxygen incorporation. Digestion studies using Lys C indicate that the 29–101 region, which includes the histidine-containing octarepeats, is particularly affected by oxidation. Oxidation was time- and copper concentration-dependent and was evident with copper concentrations as low as 1 μM. Concomitant with oxidation, SHaPrP(29–231) suffered aggregation and precipitation, which was nearly complete after 15 min, when the prion protein was incubated at 37°C with a 6-fold molar excess of Cu2+. These findings indicate that PrP, a copper-binding protein, may be particularly susceptible to metal-catalyzed oxidation and that oxidation triggers an extensive structural transition leading to aggregation.
Metal-catalyzed oxidation (MCO) of proteins has been implicated in a variety of diseases, including several neurodegenerative ailments such as Alzheimer's disease (1) and amyotrophic lateral sclerosis (2) as well as in aging (3, 4). The reaction involves reduction of Fe (III) or Cu (II) by a suitable electron donor such as NADH, NADPH, ascorbate, or mercaptane. Fe (II) and Cu (I) ions bound to specific metal-binding sites on proteins react with H2O2 to generate ⋅OH (5, 6), which immediately oxidizes neighboring amino acid residues. The reaction typically results in structural alterations and loss of enzymatic activity. MCO of proteins is a highly selective reaction that occurs primarily at protein sites with transition metal-binding capacity. Among such sites are those that include histidine residues, with affinity for Cu (II). Previous work has established that the histidine-rich octarepeat region of the prion protein constitutes a copper-binding site (7–9). At physiological pH, one molecule of Cu (II) can coordinate with each of the four octarepeats through interaction with one oxygen and three nitrogen atoms, one of the latter corresponding to the histidine imidazole nitrogen and two to glycine backbone amides (9). We hypothesized that copper binding would make the prion protein a relatively easy target of metal-catalyzed oxidation and that the ensuing damage might lead to structural alterations that could be relevant to the conversion of the soluble cellular isoform, PrPC, to the pathogenic and infectious “scrapie” isoform, PrPSc. Such conversion is the central event in the pathogenesis of the transmissible spongiform encephalopathies (TSEs). The ability of PrPSc to catalyze this self-perpetuating conformational change is the basis for prion infectivity. While the mechanism of the conversion process is not understood, explaining the inception of a first pool of PrPSc molecules poses a problem of its own. Generation of PrPSc de novo obviously is a necessary event in genetic and sporadic cases of TSE. Genetic cases are the consequence of point mutations located mainly in the carboxyl-terminal domain of PrP or insertions of additional octapeptide repeats in the flexible amino-terminal region (10). It seems reasonable to assume that mutations result in a PrP molecule of decreased structural stability, which is more prone than wild-type PrP to spontaneously acquire the PrPSc conformation (11). More difficult to explain is the genesis of PrPSc in sporadic cases of TSE. In humans, sporadic Creutzfeldt–Jakob disease accounts for ≈85% of all cases (12). It has been argued that somatic mutations or extremely rare, random conversion events could lead to the formation of an initial pool of PrPSc. However, certain covalent posttranslational modifications affecting a small fraction of PrP molecules also might introduce structural instability into them, making them more prone to the kind of conformational changes that affect mutant PrP.
Here, we show that in the presence of oxygen and of ascorbate as a reducing agent, copper binding to PrP results in a facile oxidation of the protein molecule, a reaction that is accompanied by extensive aggregation and precipitation of PrP.
Materials and Methods
Proteins and Reagents.
Unless otherwise specified, reagents were from Aldrich or Sigma, of the highest purity available. Syrian hamster recombinant SHa(29–231) and SHa(90–231) prion proteins were expressed in Escherichia coli, purified as described (13, 14), and stored as a 1 mg/ml solution in 8 M guanidine at −70°C. Small portions were thawed as required and refolded at room temperature for 1 h by 10-fold dilution in 50 mM Hepes buffer, pH 7.4. Protein concentration was determined by using the BCA assay (Pierce).
Reactions.
A 25-μl volume of 0.1 μg/μl refolded SHaPrP(29–231) or 0.072 μg/μl SHaPrP(90–231) was supplemented with 3 μl of a freshly prepared mixture of ascorbate and CuCl2 (pH 7.2) to achieve final concentrations of 4 μM PrP, 25 mM ascorbate, and the desired Cu2+ concentration. This relatively high concentration of ascorbate was used for experimental convenience, but much lower concentrations support vigorous metal-catalyzed oxidation (15). Control samples were supplemented with 3 μl of 50 mM Hepes buffer, pH 7.2. The mixtures were incubated at 37°C for 15 min, and the reaction was stopped by adding 2 μl of neutralized EDTA to a final concentration of 2 mM. Samples were spun at 10,000 × g for 10 min in a tabletop centrifuge, and supernatants were analyzed by HPLC. For anaerobic incubations, PrP stock solution and all other reagents were sparged with argon and transferred to an anaerobic chamber (Forma Scientific, Marietta, OH) equipped with manipulation gloves. PrP was refolded and oxidized as described inside the chamber. Cu+ solutions were freshly prepared before use from CuCl, treated as described (16) to eliminate any traces of Cu2+ present in the solid stock.
Lys C Digestion.
Control and oxidized samples were prepared as described, except that 100-μl volumes of the refolded PrP solutions and proportional volumes of other reagents were used. Samples were digested overnight at 37°C in a shaking bath with Achromobacter LysC (Wako) at a 1:17 Lys C to PrP mass ratio. The pH of digests was adjusted to ≈9 by addition of a half-volume of 200 mM borate buffer, pH 9.2, and samples were reduced with DTT at a final concentration of 10 mM for 1 h at 37°C. Samples were then spun at 10,000 × g for 10 min, and supernatants were analyzed by HPLC.
HPLC Analysis of SHaPrP(29–231) and Its Lys C Digestion Products.
Reverse-phase HPLC was performed on a Hewlett–Packard Series 1100 liquid chromatograph (Agilent) equipped with a Vydac C18 column (218TP5205; Vydac, Hesperia, CA). PrP was eluted with a gradient beginning with solvent A (0.05% trifluoroacetic acid) and an increasing percentage of solvent B (0.05% trifluoroacetic acid/acetonitrile): 0–15% in 3 min, 15–65% in 30 min, and return to 0% in 5.5 min. For elution of PrP Lys C digests, a shallower gradient was used, with the percentage of solvent B increased from 0% to 50% in 50 min and returned to 0% in 5.5 min. The flow was maintained at 0.2 ml/min. The effluent was fed to the flow cell of a Hewlett–Packard Series 1100 diode array detector, whose outlet was connected to a mixing tee receiving 0.1 ml/min glacial acetic acid pumped by an auxiliary pump (17). The mixture then entered the source of a Hewlett-Packard Series 1100 electrospray mass selective detector. Absorbance and mass data were recorded simultaneously.
Amino Acid Analysis.
Control samples and precipitates of oxidized samples were hydrolyzed in 6 M HCl at 155°C for 30 min in the presence of 2 mM DTT/2 μg/μl phenol/2 μg/μl benzoic acid. Hydrolysates were dried in vacuo (SpeedVac; Savant) and dissolved in deionized water. Amino acid analysis was performed after automated o-phthalaldehyde derivatization by reverse-phase HPLC. Methionine oxidation was assessed separately in samples treated with CNBr before hydrolysis (18). This treatment converts methionine to homoserine, which is detected partially as such and partially as homoserine lactone after acid hydrolysis, whereas methionine sulfoxide and other products of methionine oxidation are not affected by the reaction, reverting to methionine during acid hydrolysis, and are detected as such. Oxohistidine, unstable under typical analytical conditions, was detected in hydrolysates prepared in the presence of 100 mM DTT as described (19).
Transmission Electron Microscopy (TEM).
TEM studies were performed by Yuhui Xu at the National Heart, Lung, and Blood Institute Core Electron Microscopy Facility. SHaPrP(29–231) (0.1 μg/μl) was oxidized by 25 μM Cu2+/25 mM ascorbate as described; 10-μl portions were taken after vigorous vortexing to achieve homogenization, deposited on a Formvar and carbon-coated grid and negatively stained with 2% uranyl acetate. Samples were examined with a JEOL 1200 EM II transmission electron microscope at an accelerating voltage of 80 kV.
Results
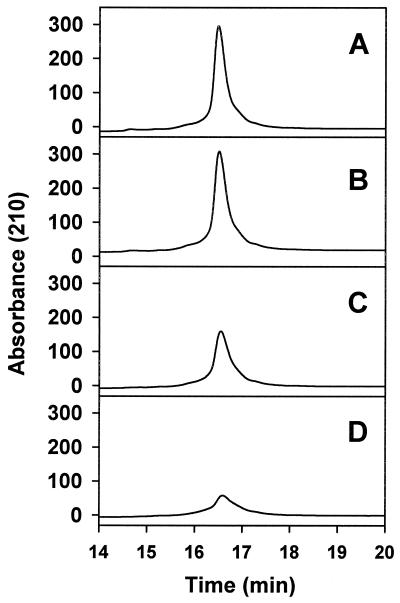
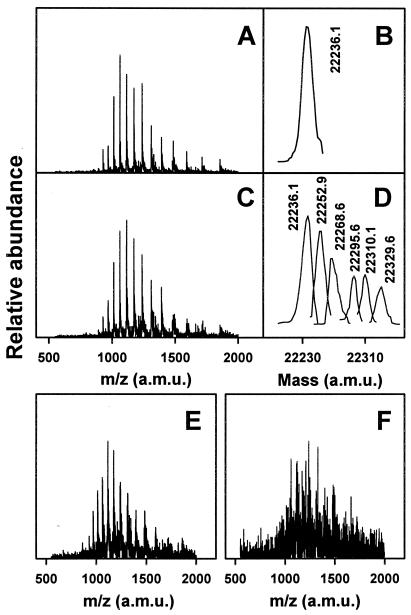
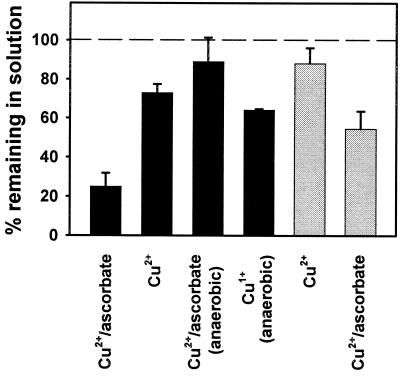
SHaPrP(29–231) proved to be very susceptible to oxidation by the Cu2+/ascorbate system. Oxidation induced aggregation and precipitation of the prion protein in a Cu2+ concentration-dependent (Fig. 1) and time-dependent (data not shown) manner. The protein remaining in solution shows increased mass heterogeneity, including species with mass increments in multiples of 16, characteristic of oxygen incorporation to form oxohistidine or methionine sulfoxide (Fig. 2). This mass heterogeneity was clearly noticeable at a Cu2+ concentration as low as 1 μM and increased dramatically with higher Cu2+ concentration to a point at which deconvolution of the masses was no longer possible. The complete, metal-catalyzed oxidation system (Cu2+, ascorbate, air) was necessary to obtain the described results. Cu (II) alone, at the maximum concentration used in the metal-catalyzed oxidation system (25 μM), induced a modest precipitation of PrP (Fig. 3) that was not accompanied by oxidative modification as assessed by mass spectrometry. Samples incubated with ascorbate alone were identical to controls. Oxygen is also a requirement for the reaction, as incubations carried out under anaerobic conditions resulted in almost no oxidative modification or precipitation of PrP (Fig. 3). Incubation of PrP with 25 μM Cu+ under anaerobic conditions resulted in a degree of precipitation of the protein that was slightly higher than that induced by Cu2+.
Figure 1.
Oxidation-induced aggregation and precipitation of SHaPrP(29–231). SHaPrP(29–231), 0.1 μg/μl, was incubated for 15 min at 37°C, in the absence (control) or presence of CuCl2 and 25 mM ascorbate acid. Samples were centrifuged at 15,000 × g for 10 min, and supernatants were analyzed by HPLC as described in Materials and Methods. (A) Control; (B) CuCl2 (1 μM); (C) CuCl2 (5 μM); (D) CuCl2 (25 μM).
Figure 2.
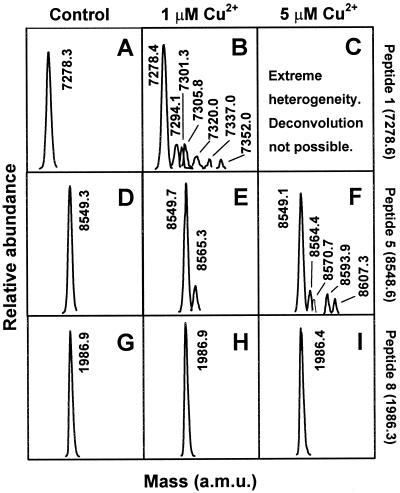
Mass spectral analysis of copper-oxidized SHaPrP(29–231). Mass spectra were obtained from peaks shown in Fig. 1. (A) Raw spectrum of control sample. (B) Deconvoluted spectrum of control sample. (C) Raw spectrum of sample treated with 1 μM CuCl2/25 mM ascorbate. (D) Deconvoluted spectrum of sample treated with 1 μM CuCl2/25 mM ascorbate. (E) Raw spectrum of sample treated with 5 μM CuCl2/25 mM ascorbate. (F) Raw spectrum of sample treated with 25 μM CuCl2/25 mM ascorbate. Raw spectra shown in E and F could not be deconvoluted.
Figure 3.
Aggregation and precipitation of SHaPrP(29–231) and SHaPrP(90–231). Solid bars, SHaPrP(29–231); shaded bars, SHaPrP(90–231). Percentage of protein remaining in solution was calculated with respect to control samples incubated and treated under identical conditions, using the areas of absorbance at 210 nm. Concentrations of metal ion and ascorbate were 25 μM and 25 mM, respectively. Data represent means of three separate experiments; bars, SD.
Mapping of Oxidative Modifications by Using Lys C and Liquid Chromatography/Mass Spectrometry.
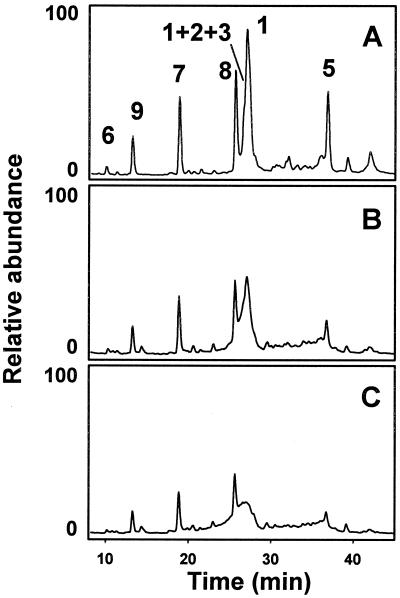
Lys C digestion generated a mixture of peptides that were resolved by HPLC and unequivocally identified by mass spectral analysis. Table 1 shows the theoretical Lys C digestion peptide list of PrP with calculated average masses. Fig. 4A shows the total ion current signal profile of a control PrP digest. All expected peptides were identified except for the small hydrophilic peptide (107), TNMK, which probably elutes very early. Two other very small peptides (102–104 and 105–106), PSK and PK, both with proline in their amino terminus, were not fully cleaved by Lys C and appeared as part of a 29–106 peptide that was not completely resolved by HPLC from peptide 29–101. Oxidized PrP was digested less efficiently by Lys C, and peptide recovery was lower, probably a consequence of decreased accessibility of aggregated PrP. The chromatographic profile was, nevertheless, very similar to that of control PrP (Fig. 4 B and C). Mass analysis shows that peptides 195–204, 205–220, and 221–231, all corresponding to the carboxyl terminus, were unchanged. As an example, the total ion mass signals corresponding to peptide 205–220 is shown in Fig. 5. Alternately, extensive mass heterogeneity was seen in peptide 111–185 and especially in peptide 29–101, corresponding to the amino terminus of PrP (Fig. 6). The chromatographic peaks corresponding to these peptides broadened with increasing Cu2+ concentration, as a more complex mixture of close-eluting oxidation products formed. Peptide 29–101 could not be deconvoluted in the 5 μM and higher Cu2+ concentration samples. Although integration of the broad peaks corresponding to these peptides is problematic, it appears that their yield is lower than that of the unmodified peptides, perhaps indicative of an even more restricted accessibility to Lys C of PrP amino-terminal domains in the aggregates.
Table 1.
SHa PrP(29–231) Lys C digestion peptides
| n | From–to | Average calculated mass | Observed mass |
|---|---|---|---|
| 1 | 29–101 | 7278.59 | 7278.0 |
| 2 | 102–104 | 330.38 | * |
| 3 | 105–106 | 243.31 | * |
| 4 | 107–110 | 492.60 | Not detected |
| 5 | 111–185 | 8548.60 | 8549.1 |
| 6 | 186–194 | 1016.12 | 1015.5 |
| 7 | 195–204 | 1153.21 | 1152.7 |
| 8 | 205–220 | 1986.34 | 1986.8 |
| 9 | 221–231 | 1331.36 | 1330.9 |
29 GGWNTGGSRYPGQGSPGGNRYPPQGGGTWGQPHGGGWGQPHGGGWGQPHGGGWGQPHGGGWGQGGGTHNQWNKPSKPKTNMKHMAGAAAAGAVVGGLGGYMLGSAMSRPMMHFGNDWEDRYYRENMNRYPNQVYYRPVDQYNNQNNFVHDCVNITIKQHTVTTTTKGENFTETDIKIMERVVEQMCTTQYQKESQAYYDGRRS
231
Lysine residues are shown in bold characters. Histidine residues are underlined.
Observed as part of an unresolved peak fronting peak 1, with mass: 7815.7 (average calculated mass of 1 + 2 + 3 − 2H2O = 7816.28).
Figure 4.
Lys C digestion of control and oxidized SHaPrP(29–231). SHaPrP(29–231) control and oxidized samples were digested with Lys C and reduced with 10 mM DTT at pH 9. Digests were analyzed by HPLC. Chromatograms represent the total ion current signal from the mass selective detector. Peptides are identified according to Table 1. (A) Control sample. (B) Sample treated with 1 μM CuCl2/25 mM. (C) Sample treated with 5 μM CuCl2/25 mM.
Figure 5.
Deconvoluted mass spectra of individual control and oxidized SHaPrP(29–231) Lys C digests. (A–C) Peptide 1 (29). (D–F) Peptide 5 (111). (G–I) Peptide 8 (205). Samples are identified at the top of each image: control sample, left column; sample treated with 1 μM CuCl2/25 mM, center column; sample treated with 5 μM CuCl2/25 mM, right column.
Figure 6.
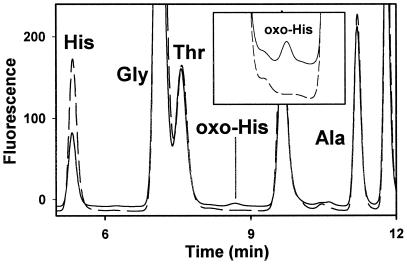
Detection of oxohistidine in oxidized SHaPrP(29–231). SHaPrP(29–231), 0.1 μg/μl, was oxidized with 25 μM CuCl2 and 25 mM ascorbate. The sample was centrifuged at 15,000 rpm for 10 min. The precipitate was hydrolyzed in the presence of 100 mM DTT, and the hydrolysate was analyzed by reverse-phase HPLC after o-phthalaldehyde derivatization as described (19). A standard of pure benzoyl-oxohistidine was used as control.
Amino Acid Composition of Oxidized PrP.
Amino acid analysis of the oxidized PrP precipitate (25 μM Cu2+-treated sample) showed a large decrease in histidine content (45% loss) and a lesser loss of tyrosine and phenylalanine (Table 2). Analysis of whole oxidized samples, including the ≈25% portion of oxidized PrP remaining in the supernatant, was performed after protein precipitation with 85% methanol. Results similar to those obtained with the precipitate were seen. Lower losses of histidine were seen in samples oxidized with lower concentrations of Cu2+ (data not shown). Oxidation of histidine is known to result in a complex mixture of products including oxohistidine and aspartate. Oxohistidine was found in the oxidized PrP precipitate, whereas no oxohistidine was present in control samples (Fig. 6). Oxohistidine also was detected in Lys C digests of oxidized PrP (25 μM Cu2+-treated samples) by simultaneous sequencing (data not shown; ref. 20).
Table 2.
Relative amino acid composition of aggregate, oxidized, and control SHa PrP(29–231)
| Amino acid | Oxidized/control |
|---|---|
| Aspartate | 1.10 |
| Glutamate | 1.06 |
| Serine | 1.00 |
| Histidine | 0.56 |
| Glycine | 0.91 |
| Threonine | 0.97 |
| Arginine | 0.97 |
| Alanine | 1.01 |
| Tyrosine | 0.76 |
| Methionine | 1.07 |
| Valine | 1.03 |
| Phenylalanine | 0.83 |
| Isoleucine | 1.01 |
| Leucine | 1.10 |
| Lysine | 0.95 |
| 95% Confidence interval | 0.86–1.14 |
Data for residual methionine and homoserine, obtained in separate analyses after CNBr treatment, were 1.01 and 0.97, respectively. Oxidation by 25 uM Cu++/ascorbate. Results are means of three samples.
Methionine is another possible target of oxidation, and a previous study showed evidence of methionine destruction in PrP refolded in the presence of millimolar concentrations of Cu2+ (21). However, analysis of our oxidized samples showed no decrease in methionine or increase in methionine sulfoxide.
TEM Studies.
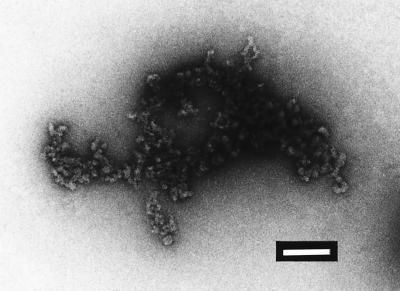
Oxidized PrP was studied by TEM after negative staining. Very conspicuous globular aggregates but no fibrillar structures were visualized (Fig. 7). Samples were allowed to stand at 4°C for 3 days, resulting in the appearance of turbidity apparent to the naked eye. These samples were studied again by using TEM, but still no fibrillar structures were seen.
Figure 7.
Negative-stain TEM of oxidized SHaPrP(29–231). SHaPrP(29–231), 0.1 μg/μl, was oxidized with 25 μM Cu2+/25 mM ascorbate. The sample was prepared for TEM as described in Materials and Methods. (Bar, 200 nm.)
Copper-Catalyzed Oxidation of SHaPrP(90–231).
The shorter SHaPrP(90–231) was subjected to Cu2+/ascorbate-catalyzed oxidation at the same molar concentration as SHaPrP(29–231). SHaPrP(90–231) also underwent oxidation, although it was less sensitive than SHaPrP(29–231). Oxidation also was accompanied by aggregation and precipitation (Fig. 3). Mass spectral analysis, including analysis of Lys C digestion, was performed. Qualitatively, the results were very similar to those obtained with SHaPrP(29–231), but the extent of oxidative modification was lower. The (90) amino-terminal region, GQGGGTHNQWNK, and the peptide 111–185, which is the same as peptide 5 in SHaPrP(29–231) PrP, suffered considerable modification.
Discussion
Copper binding is an intriguing property of PrP. Well characterized in vitro (7–9, 14), it is still not known to what extent, under what circumstances, and with which consequences it occurs in vivo. The reported affinity constant is very low as compared with those exhibited by well established copper-binding proteins, such as superoxide dismutase (SOD). Although this low affinity casts doubts on a copper-dependent enzymatic activity of PrP, it remains compatible with PrP acting as a copper carrier, moving Cu2+ molecules from the extracellular milieu into the cell (14). Alternatively, PrP has been viewed as an antioxidant defense molecule. In this line, several reports have suggested that PrP, particularly if refolded in the presence of millimolar concentrations of Cu2+, has SOD activity (22, 23). However, these findings remain controversial, because brains from PrP-deficient mice exhibit unchanged SOD activity (24). Whatever the involvement of copper in prion biology might be, the present studies show that SHaPrP(29–231) is highly susceptible to MCO in the presence of this metal ion and a suitable reducing agent. The histidine residues in the octarepeats are especially susceptible. To a lesser degree, all other histidines are oxidizable, perhaps with the exception of His187, which is relatively buried (25). This is no surprise, given that octarepeat histidines are involved in Cu2+ binding, creating a site-specific target for MCO. Further, the accessibility of the flexible amino-terminal tail of PrP could facilitate the access of reducing agents (ascorbate in our model) necessary for in situ redox cycling of the transition metal. Of note, all of the histidines present in the carboxyl, well structured domain of PrP are also very solvent-exposed, which would explain their high susceptibility to oxidation. The concentrations of Cu2+ used in our experiments are relevant to the known affinity of PrP for this cation; Stockel et al. (7) have shown that the dissociation constant for SHaPrP(29–231)-Cu2+ is ≈14 μM at pH 6, whereas studies conducted at pH 7.4 with SHaPrP(23–98) showed a maximum binding capacity of 5 mol/mol with dissociation constants ranging from ≈0.1 to ≈5 μM (14).
Our studies show that oxidation induces extensive aggregation and precipitation of PrP. Metal-catalyzed oxidation previously has been shown to induce aggregation of other proteins such as relaxin (26) and synuclein (27). In the case of relaxin, aggregation induced by an oxidative model system similar to ours is mediated by oxidation of histidine residues (26). The nature of the chemical modifications responsible for aggregation after MCO remains unknown, but it is worth noting that oxidation of histidine converts it from a basic to neutral (oxohistidine) or acidic (aspartate) residue. Our data indicate that Cu+, which is generated during the reaction, has a certain capacity to induce aggregation of PrP, although it accounts only for a fraction of it.
It is possible that certain nonenzymatic, posttranslational covalent modifications, particularly oxidation, introduce structural instability to the prion molecule, generating a pool of PrP molecules more prone to conformational change. These altered PrP molecules could play a role in formation of the initial cohort of PrPSc, which is formed de novo in sporadic TSE (Fig. 8). This proposal does not challenge the possibility of spontaneous conformational changes leading to generation of PrPSc de novo but, rather, offers another mechanism that could facilitate such conformational changes. A relevant example of this concept is provided by rat phosphoglycerate kinase (PGK). This enzyme can be “aged” by prolonged incubation in vitro. Artificially aged PGK exhibits characteristics that are typical of PGK molecules purified from old animals, such as increased resistance to heat inactivation. It was thought initially that the only cause of such “aged” phenotype is a conformational change, as denaturation and refolding of the “aged” enzyme results in a molecule of properties identical to those of “young” enzyme treated in a similar way (28). However, it was discovered later that the conformational change is triggered by oxidation of certain cysteine residues, which can be re-reduced later without reversal of the conformational changes (28).
Figure 8.
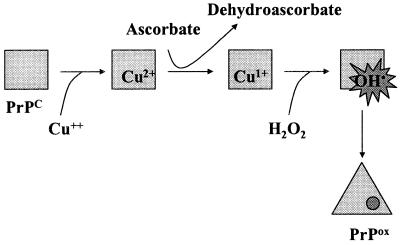
Scheme showing oxidation of PrP by the Cu2+/ascorbate system. Cu2+ bound to PrP is reduced to Cu+ by ascorbate. Alternatively, Cu+ generated by reduction of Cu2+ by ascorbate in solution may bind to PrP. Reaction of Cu+ with H2O2, generated under aerobic conditions by oxidation of ascorbate, generates ⋅OH, which reacts in situ with histidine and other residues to generate PrPox. Oxidized PrP undergoes a rapid conformational change and aggregates.
A necessary, although not sufficient characteristic of oxidized PrP therefore must be structural instability, evidenced by the tendency to aggregation of oxidized PrP observed in our experiments. Other in vitro manipulations also have resulted in increased aggregability of PrP (29–31). Some of them involve covalent modification of PrP, in particular, reduction of its only disulfide bridge (30, 31). Manipulations that involve physiologically relevant procedures obviously are of particular interest. In general, aggregation has been reported to result in formation of mixtures of globular and fibrillar structures, the former sometimes evolving to the latter upon extended incubation. In our hands, oxidized PrP aggregated, forming unstructured globules that grew larger with time. Fibrils were not observed. It should be noted that all fibrils formed in vitro thus far lack infectivity and exhibit only a modest increase in resistance to proteinase K digestion, which clearly differs from the proteinase K resistance of PrPSc (30). Thus, generation of fibrils in vitro does not necessarily imply a material that is more relevant to the formation of PrPSc. Rather, the increased tendency of covalently modified PrP to undergo conformational change is key. In vivo, additional cellular factors such as chaperones or protein X may prevent immediate aggregation and trap the intermediate, partially unfolded forms, which then may be degraded or, alternatively, interact in ways relevant to the formation of PrPSc. Addition of chaperones greatly facilitates PrPSc-assisted conversion of PrPC to PrPSc in vitro (32). A role for oxidatively modified PrP may be considered an extension of previous proposals, implicating partially unfolded intermediates in conversion of PrPC to PrPSc (10).
In summary, we have shown that PrP is a target of copper-catalyzed oxidation and that this reaction leads to profound structural changes in the protein molecule. Oxidation therefore must be taken into account as a potential side reaction when considering the role of copper in prion biology.
Acknowledgments
We thank Dr. Yuhui Xu from the National Heart, Lung, and Blood Institute Core Electron Microscopy Facility for performing TEM studies.
Abbreviations
- MCO
metal-catalyzed oxidation
- PGK
phosphoglycerate kinase
- PrP
prion protein
- PrPC
prion protein, cellular isoform
- PrPSc
prion protein, scrapie isoform
- TEM
transmission electron microscopy
- TSE
transmissible spongiform encephalopathy
References
- 1.Smith C D, Carney J M, Starke-Reed P E, Oliver C N, Stadtman E R, Floyd R A, Markesbery W R. Proc Natl Acad Sci USA. 1991;88:10540–10543. doi: 10.1073/pnas.88.23.10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowling A C, Schultz J B, Brown R H, Jr, Beal M F. J Neurochem. 1993;61:2322–2325. doi: 10.1111/j.1471-4159.1993.tb07478.x. [DOI] [PubMed] [Google Scholar]
- 3.Oliver C N, Ahn B-W, Moerman E J, Goldstein S, Stadtman E R. J Biol Chem. 1987;262:5488–5491. [PubMed] [Google Scholar]
- 4.Starke-Reed P E, Oliver C N. Arch Biochem Biophys. 1989;275:559–567. doi: 10.1016/0003-9861(89)90402-5. [DOI] [PubMed] [Google Scholar]
- 5.Stadtman E R, Berlett B S. Drug Metab Rev. 1998;30:225–243. doi: 10.3109/03602539808996310. [DOI] [PubMed] [Google Scholar]
- 6.Berlett B S, Stadtman E R. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 7.Stöckel J, Safar J, Wallace A C, Cohen F E, Prusiner S B. Biochemistry. 1998;37:7185–7193. doi: 10.1021/bi972827k. [DOI] [PubMed] [Google Scholar]
- 8.Brown D R, Qin K, Herms J W, Madlung A, Manson J, Strome R, Fraser P E, Kruck T, von Bohlen A, Schulz-Schaeffer W, et al. Nature (London) 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 9.Aronoff-Spencer E, Burns C S, Avdievich N I, Gerfen G J, Peisach J, Antholine W E, Ball H L, Cohen F E, Prusiner S B, Millhauser G L. Biochemistry. 2000;39:13760–13771. doi: 10.1021/bi001472t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prusiner S B. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liemann S, Glockshuber R. Biochemistry. 1999;38:3258–3267. doi: 10.1021/bi982714g. [DOI] [PubMed] [Google Scholar]
- 12.Ironside J. J Pathol. 1998;186:227–234. doi: 10.1002/(SICI)1096-9896(1998110)186:3<227::AID-PATH174>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 13.Viles J H, Cohen F E, Prusiner S B, Goodin D B, Wright P E, Dyson H J. Proc Natl Acad Sci USA. 1999;96:2042–2047. doi: 10.1073/pnas.96.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whittal R M, Ball H L, Cohen F E, Burlingame A L, Prusiner S B, Baldwin M A. Protein Sci. 2000;9:332–343. doi: 10.1110/ps.9.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine R L. J Biol Chem. 1983;258:11828–11833. [PubMed] [Google Scholar]
- 16.Davis D A, Branca A A, Pallenberg A J, Marschner T M, Patt L M, Chatlynne L G, Humphrey R W, Yarchoan R, Levine R L. Arch Biochem Biophys. 1995;322:127–134. doi: 10.1006/abbi.1995.1444. [DOI] [PubMed] [Google Scholar]
- 17.Apffel A, Fischer S, Goldberg G, Goodley P C, Kuhlmann F E. J Chromatogr. 1995;712:177–190. doi: 10.1016/0021-9673(95)00175-m. [DOI] [PubMed] [Google Scholar]
- 18.Levine R L, Mosoni L, Berlett B S, Stadtman E R. Proc Natl Acad Sci USA. 1996;93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewisch S A, Levine R L. Anal Biochem. 1995;231:440–446. doi: 10.1006/abio.1995.9974. [DOI] [PubMed] [Google Scholar]
- 20.Taggart C, Cervantes-Laurean D, Kim G, McElvaney N G, Wehr N, Moss J, Levine R L. J Biol Chem. 2000;275:27258–27265. doi: 10.1074/jbc.M004850200. [DOI] [PubMed] [Google Scholar]
- 21.Wong B S, Wang H, Brown D R, Jones I M. Biochem Biophys Res Commun. 1999;259:352–355. doi: 10.1006/bbrc.1999.0802. [DOI] [PubMed] [Google Scholar]
- 22.Brown D R, Wong B S, Hafiz F, Clive C, Haswell S J, Jones I M. Biochem J. 1999;344:1–5. [PMC free article] [PubMed] [Google Scholar]
- 23.Brown D R, Clive C, Haswell S J. J Neurochem. 2001;76:69–76. doi: 10.1046/j.1471-4159.2001.00009.x. [DOI] [PubMed] [Google Scholar]
- 24.Waggoner D J, Drisaldi B, Bartnikas T B, Casareno R L B, Prohaska J R, Gitlin J D, Harris D A. J Biol Chem. 2000;275:7455–7458. doi: 10.1074/jbc.275.11.7455. [DOI] [PubMed] [Google Scholar]
- 25.James T L, Liu H, Ulyanov N B, Farr-Jones S, Zhang H, Donne D G, Kaneko K, Groth D, Mehlhorn I, Prusiner S B, Cohen F E. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khossravi M, Shire S J, Borchardt R T. Biochemistry. 2000;39:5876–5885. doi: 10.1021/bi9924720. [DOI] [PubMed] [Google Scholar]
- 27.Paik S R, Shin H-J, Lee J-H. Arch Biochem Biophys. 2000;378:269–277. doi: 10.1006/abbi.2000.1822. [DOI] [PubMed] [Google Scholar]
- 28.Cook L, Gafni A. J Biol Chem. 1998;263:13991–13993. [PubMed] [Google Scholar]
- 29.Swietnicki W, Morillas M, Chen S G, Gambetti P, Surewicz W K. Biochemistry. 2000;39:424–431. doi: 10.1021/bi991967m. [DOI] [PubMed] [Google Scholar]
- 30.Jackson G S, Hosszu L L P, Power A, Hill A F, Kenney J, Saibil H, Craven J, Waltho J P, Clarke A R, Collinge J. Science. 1999;283:1935–1937. doi: 10.1126/science.283.5409.1935. [DOI] [PubMed] [Google Scholar]
- 31.Requena J R, Levine R L. Free Radical Biol Med. 2001;30:141–147. doi: 10.1016/s0891-5849(00)00430-5. [DOI] [PubMed] [Google Scholar]
- 32.DebBurman S K, Raymond G J, Caughey B, Lindquist S. Proc Natl Acad Sci USA. 1997;94:13938–13943. doi: 10.1073/pnas.94.25.13938. [DOI] [PMC free article] [PubMed] [Google Scholar]