Abstract
Microbial eukaryotes may extinguish much of their nuclear phylogenetic history due to endosymbiotic/horizontal gene transfer (E/HGT). We studied E/HGT in 32,110 contigs of expressed sequence tags (ESTs) from the dinoflagellate Alexandrium tamarense (Dinophyceae) using a conservative phylogenomic approach. The vast majority of predicted proteins (86.4%) in this alga are novel or dinoflagellate-specific. We searched for putative homologs of these predicted proteins against a taxonomically broadly sampled protein database that includes all currently available data from algae and protists and reconstructed a phylogeny from each of the putative homologous protein sets. Of the 2,523 resulting phylogenies, 14-17% are potentially impacted by E/HGT involving both prokaryote and eukaryote lineages, with 2-4% showing clear evidence of reticulate evolution. The complex evolutionary histories of the remaining proteins, many of which may also have been affected by E/HGT, cannot be interpreted using our approach with currently available gene data. We present empirical evidence of reticulate genome evolution that combined with inadequate or highly complex phylogenetic signal in many proteins may impede genome-wide approaches to infer the tree of microbial eukaryotes.
Keywords: dinoflagellates, endosymbiosis, eukaryote evolution, horizontal gene transfer, phylogenomics
Introduction
Numerous attempts have been made in recent years to erect a comprehensive phylogeny of eukaryotes (Burki et al. 2007, Hackett et al. 2007, Yoon et al. 2008, Parfrey et al. 2010). In spite of an expanding database of gene and genome data, many nodes in the tree have proven refractory to resolution using multi-gene phylogenetic methods. This is in part explained by plastid endosymbiosis, whereby a foreign cell (e.g., a cyanobacterium or a red alga) is captured and retained by an ancient eukaryotic lineage as a photosynthetic organelle (Reyes-Prieto et al. 2007). Endosymbiosis results in outright gene loss (owing to loss of gene function) from the captured cell and more importantly for phylogenetic analysis, the movement of hundreds of its genes to the host nuclear genome via endosymbiotic gene transfer (EGT), a specific instance of horizontal gene transfer (HGT; Martin & Herrmann 1998, Reyes-Prieto et al. 2006), yielding chimeric nuclear genomes. These forces are particularly prominent in taxa that have undergone serial endosymbioses (Yoon et al. 2005, Patron et al. 2007, Moustafa et al. 2009). However, the contribution by E/HGT to reticulate genome evolution in photosynthetic lineages (or in taxa that have secondarily lost the plastid; Reyes-Prieto et al. 2008) and its impact on phylogeny reconstruction among eukaryotes are poorly understood. It is conceivable that a patchy distribution of genes impacted by E/HGT explains the inability to unambiguously resolve the interrelationships of algal phyla such as Rhodophyta, Viridiplantae (green algae and plants), Glaucophyta (together, the Plantae; Rodríguez-Ezpeleta et al. 2005, Price et al. 2012), Cryptophyta, and Haptophyta (Stiller 2007, Yoon et al. 2008, Baurain et al. 2010, Parfrey et al. 2010, Chan et al. 2011c). These taxa are either donors (e.g., red and green algae) or recipients (e.g., Cryptophyta, Haptophyta) of genes implicated in EGT.
In contrast to eukaryotes, much more is known about HGT in prokaryotes where the non-lineal movement of genes between taxa is so extensive (Beiko et al. 2005, Lerat et al. 2005, Puigbo et al. 2010) that the ability to infer a bacterial tree of life (TOL) has been called into question (Doolittle & Bapteste 2007, Lawrence & Retchless 2010). Although a more-complex cellular organization and gene (e.g., exon-intron) structure may hinder HGT in eukaryotes (Keeling & Palmer 2008), there is no a priori reason to believe that over evolutionary time scales (i.e., hundreds of millions of years) the genomes of microbial eukaryotes would be immune to HGT. Like prokaryotes, many of these taxa are unicellular (i.e., each cell comprises a potential germ line), free-living, and often phagotrophic with an unknown or poorly understood history of plastid endosymbiosis. Furthermore, opportunities presumably exist for HGT due to long-term associations with a multitude of foreign DNA from prey (Doolittle 1998), symbionts, and pathogens (Bowler et al. 2008, Worden et al. 2009). Therefore, unlike most plants (Bock 2010) and animals, it is important to study genome data from microbial eukaryotes with the expectation of reticulate evolution for a subset of genes rather than attempting to fit a model of strict vertical gene ancestry, and non-complying data is excluded as “noise”. The question remains however, does E/HGT impart a negligible signal to protist gene evolution or can these forces dominate the history of their genomes? If the latter holds, then it is crucial to detect E/HGT on a genome-wide basis in microbial eukaryotes using a gene-by-gene approach that attempts to rise above biases introduced by taxon sampling or phylogenetic artifacts (Bodył et al. 2009, Stiller 2011).
Among microbial eukaryotes, dinoflagellates are an important group of primary producers and grazers that have one of the most complex evolutionary histories known (Hackett et al. 2004). The widespread peridinin-containing plastid in these taxa is of red algal origin via secondary (i.e., eukaryotic) endosymbiosis (Yoon et al. 2005). However some fucoxanthin-containing dinoflagellates have undergone an additional (tertiary) endosymbiosis resulting in a haptophyte-derived plastid (Ishida & Green 2002). As shown in the schematic tree in Fig. 1A, the interrelationships between various eukaryote phyla are currently poorly resolved. This is therefore a working hypothesis that summarizes the results of recent studies that, taken separately, often conflict depending on the nature of the data and chosen analysis. In particular, the grouping of Alveolata with the cryptophytes, haptophytes, and stramenopiles (which includes the ubiquitous diatoms) under the supergroup “Chromalveolata” is highly contentious (Baurain et al. 2010); e.g., in some analyses either or both the Cryptophyta and Haptophyta are sister to Plantae (Burki et al. 2008, Parfrey et al. 2010, Parfrey et al. 2011).
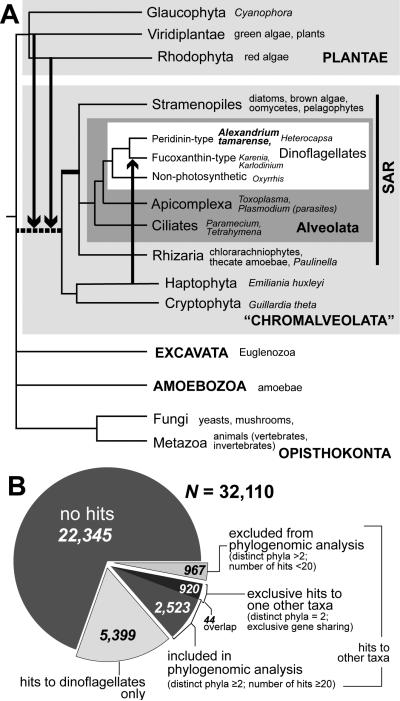
FIGURE 1.
The putative eukaryote tree of life (TOL). (A) Schematic tree showing major endosymbiotic/horizontal gene transfer (E/HGT) events that have occurred as a result of plastid evolution. The contribution of genes from lineages of the red and green algae is thought to be prominent in most chromalveolate (including dinoflagellates), whereas haptophyte-derived genes are known to be present in fucoxanthin-type dinoflagellates. Other dinoflagellates have undergone tertiary endosymbiosis with different algae (Hackett et al. 2004). The arrows represent instances of gene transfer/sharing. The grouping of “Chromalveolata”, in which the ancestral branch is shown as a dashed line remains controversial in the literature. (B) The number of Alexandrium tamarense genes used in this study, breakdown by those with no hits in the current database, those with hits only in dinoflagellates, and those with hits with other taxa.
Similarly, although a red algal secondary endosymbiosis has clearly occurred in many of these phyla due to the presence of a plastid derived from this lineage (McFadden 2001, Yoon et al. 2002, McFadden & van Dooren 2004), a possible cryptic green algal endosymbiosis that has been postulated to predate the red algal capture (Moustafa et al. 2009) warrants further investigation. A recent study of diatom membrane transporters (Chan et al. 2011b) demonstrates red and/or green algal origins of these genes, implying that E/HGT could play a crucial role in environmental adaptation among microbial eukaryotes. In most of these phylogenetic analyses, the simplest explanation, with the invocation of the least number of evolutionary events, is assumed to be a plausible explanation of the data. Nevertheless, as with any phylogenetic analysis, one cannot definitively dismiss the possibility of data biases, such as stochastic sequence variation, rate heterogeneity, convergent evolution, or simply, inadequate taxon sampling, which would mislead the interpretation of evolutionary history (Gruenheit et al. 2008, Bodył et al. 2009, Stiller et al. 2009, Christin et al. 2010, Stiller 2011). Whereas the impact of plastid endosymbiosis on the interrelationships among eukaryote lineages is not yet clearly understood, the clade defined by stramenopiles, Alveolata, and Rhizaria (SAR) has been recovered in several multi-gene phylogenetic analyses (Burki et al. 2007, Hackett et al. 2007) and provides a provisional phylogenetic affiliation of dinoflagellates/alveolates to other eukaryotes in the TOL (Fig. 1A).
Given this current state of understanding, the nuclear genomes of peridinin dinoflagellates are expected to show a history of EGT involving at least the canonical red algal endosymbiosis (Li et al. 2006) although other cryptic endosymbiosis are also likely to have occurred during the long evolutionary history of dinoflagellates and sister phyla such as stramenopiles (Moustafa et al. 2009, Baurain et al. 2010) that stretches back hundreds of millions of years (Yoon et al. 2004). In addition, HGT between dinoflagellates and various bacterial sources has been demonstrated in recent studies (Nosenko & Bhattacharya 2007, Keeling 2009). Intriguingly, this complex history of gene recruitment in dinoflagellates occurs in the backdrop of some of the most peculiar properties known with respect to cell and DNA biology (Wisecaver & Hackett 2011): e.g., permanently condensed chromosomes, highly reduced and fragmented organelle DNA (Koumandou et al. 2004), and nuclear genomes of immense size, i.e., estimate ranging from 1.5 to 220 Gbp (LaJeunesse et al. 2005, Hackett & Bhattacharya 2008). The source of evolutionary pressure on dinoflagellates to maintain such massive nuclear genomes, given the high energetic costs of DNA replication is currently unknown. However, an outcome of large genome size may be the ability to incorporate foreign genes, which may ultimately provide selective advantages in variable environmental conditions. Therefore, the dinoflagellates provide an interesting target for studying genome evolution in a system that may be considered a “worst-case” scenario in terms of the complexity of evolutionary history among microbial eukaryotes. Here we analyze a comprehensive expressed sequence tag (EST) dataset from the ecologically and economically important, toxic dinoflagellate Alexandrium tamarense (Lebour) E. Balech to assess the extent of “gene sharing” in microbial eukaryotes. Here we use the phrase gene sharing to describe non-lineal gene transfer (and/or exchange), regardless of the direction of transfer.
Materials and Methods
Generation of expressed sequence tag (EST) data
The ESTs were derived from the vegetative (haploid) phase of A. tamarense CCMP 1598 (Hackett et al. 2005). A pooled set of transcripts was generated by combining mRNAs isolated from cells grown under six different culture conditions (f/2 semi-continuous, G1 pooled, -N semi-continuous, glutamine semi-continuous, -P semi-continuous, and xenic semi-continuous) to maximize gene discovery across a variety of environmental conditions. The xenic culture was prepared using the bacterized clone CCMP1493; i.e., a culture of A. tamarense that contained naturally occurring bacteria. Details of these culture conditions and the cDNA isolation procedure can be found in Moustafa et al. (2010). For 454 sequencing, first strand synthesis of A. tamarense cDNA was performed using the Superscript® III First-Strand Synthesis System (Invitrogen, USA). The first-strand reaction included 5 μL of total RNA and 50 pmol modified oligo-dT primer with PIIA tag (5’ AAG CAG TGG TAT CAA CGC AGA GTT TGT TTT TTT TTC TTT TTT TTT TVN 3’). The reaction was incubated at 50°C for 90 min The PIIA tag was annealed to the 5’ end of full-length dinoflagellate transcripts using the Advantage® 2 PCR kit (Clontech, USA), taking advantage of the trans-spliced leader sequence present on mature dinoflagellate transcripts (Zhang et al. 2007, Zhang et al. 2009). The reaction included 20 μL of first-strand cDNA and 20 pmol of the spliced leader-annealed PIIA primer (PIIA primer: 5’AAG CAG TGG TAT CAA CGC AGA GTC CGT AGC CAT TTT GGC TCA AG 3’). One PCR cycle was performed (95°C for 1 min, 68°C for 6 min). A PCR cleanup was performed using the QIAquick® PCR purification kit (Qiagen, USA) to remove unincorporated primers and dNTPs. PCR amplification of the resulting double-stranded cDNA was performed with the Advantage® 2 PCR kit (Clontech, USA) using 10 pmol PIIA PCR primer (5’ AAG CAG TGG TAT CAA CGC AGA GT 3’). Cycling parameters included an initial denaturation step at 95°C for 1 min followed by 18 cycles of 95°C for 30 s, 58°C for 30 s, 68°C for 6 min PCR products were visualized on an agarose gel to confirm expected cDNA size range and cleaned using the CHROMA SPIN™ columns (400 size selection; Clontech, USA). Multiple cDNA synthesis/amplification reactions were pooled to generate the 5μg DNA needed for 454 sequencing and reduce the stochastic bias of PCR amplification. The cDNA was sequenced with a 454 FLX-Titanium pyrosequencing machine at the Arizona Genomics Institute (Tucson, AZ, USA). Combination and clustering of all A. tamarense data using CAP3 (Huang & Madan 1999) resulted in 32,716 distinct EST contigs for downstream analysis. Excluding short ESTs (length <150 nt), we used 32,110 ESTs with lengths ranging from 150 to 3,418 nt (average length 506 nt) for subsequent analysis of HGT/EGT. The ESTs generated from the subtracted libraries are available from http://dbdata.rutgers.edu/alexbase/. The EST assembly used in this study is available from http://dbdata.rutgers.edu/data/dino/. An earlier study (Moustafa et al. 2010) has demonstrated that A. tamarense encodes ca. 40,000 unique cDNA signatures (i.e., determined using massively parallel signature sequencing, MPSS). Therefore the EST unigene set used in the current research represents ca. 80% of the expressed genes in the dinoflagellate.
Because the conserved 5' spliced leader sequence in dinoflagellates (Zhang et al. 2007) was used to target cDNAs for EST library construction, we determined the proportion of assembled ESTs that actually encoded the complete 5' terminus. Here we used the spliced leader sequence 5'-DCCGUAGCCAUUUUGGCUCAAG-3' as a query against the assembled unigenes and found that at the identify level of 21 nt, a total of 6,492 contigs contain this sequence. By significantly relaxing the threshold to 10 nt identity, we found 8,158 matching EST contigs, suggesting that ca. 25% of the 32,110 contigs contain the 5' terminus of the A. tamarense coding regions.
Analysis of exclusive gene sharing
Within the context of this study, we define a taxon as each individual terminal node of a phylogenetic tree, and a phylum as the group of such closely related taxa, such as dinoflagellates, apicomplexans, stramenopiles, Rhodophyta, and Viridiplantae. For this and the following phylogenomic analyses, we used an in-house database that consists of all annotated protein sequences from RefSeq release 43 at GenBank (http://ncbi.nih.nlm.gov/RefSeq/), predicted protein models available from the Joint Genome Institute (ftp://ftp.jgi-psf.org/pub/JGI_data/), and six-frame translated proteins from EST datasets of all publicly available algal and unicellular eukaryote sources, i.e., dbEST at GenBank (http://ncbi.nih.nlm.gov/dbEST/) and TBestDB (http://tbestdb.bcm.umontreal.ca/), as well as data from Porphyridium cruentum (S. F. Gray) Nägeli (Porphyridiophyceae) and Calliarthron tuberculosum (Postels and Ruprecht) E. Y. Dawson (Florideophyceae) (Chan et al. 2011c), totaling 14,029,220 sequences (Table S1 in the supplementary material). Using 32,110 unigenes of A. tamarense (each was translated into proteins in six frames) as a query platform against the database (BLASTP, e-value ≤10-10), we adopted a simplified reciprocal BLAST approach (Chan et al. 2011c) to identify the homologous protein sequence set for each of these genes at high confidence. The frame in which the encoded protein has the most hits among the six frames for a unigene is considered the correct frame. For each of the top five BLASTP hits (or less if there are <5 hits) for an A. tamarense protein, we generated a list of hits via BLASTP searches against our database. The sequence hits that are found in all of these lists (including the A. tamarense protein) are grouped into a set. A protein set that consists of only the dinoflagellates and one other phylum represents putative cases of exclusive gene sharing between the two phyla.
Inference of E/HGT
For each of the remaining 4,366 ESTs (14% of 32,110) that have significant matches in the database, gene (i.e., protein) sets were identified (maximum size = 100 with no single species represented more than four times). No more than five bacterial groupings (e.g., Actinobacteria, Proteobacteria, Cyanobacteria, according to the NCBI Taxonomy) are represented in a gene set. Given that the current database is data-rich in the groups of Metazoa and Fungi, ≤15 species (for each of these two groups) were represented in a gene set. Gene sets with <4 sequences were excluded from analysis because they are not phylogenetically meaningful. Multiple sequence alignments for each gene set were performed using MUSCLE (Edgar 2004). Phylogenetically non-informative sites (i.e., fast evolving, divergent, ambiguously aligned blocks) in the alignments were removed using GBLOCKS (Talavera & Castresana 2007) with parameters b3 = 200, b4 = 2, and b5 = a. Sequence alignments of length <75 amino acid positions were excluded from analysis. We did not specifically test for rate heterogeneity, compositional bias, or stochastic sequence variation in our dataset, but this alignment “cleaning” step acts as a precautionary measure to reduce potential phylogenetic artifacts in the subsequent tree inference. The phylogeny for each alignment was reconstructed using a maximum likelihood approach (Stamatakis 2006) with the WAG amino acid substitution model (Whelan & Goldman 2001) with a discrete gamma distribution (Yang 1994) and non-parametric bootstrap of 100 replicates. The EST unigenes and their encoded proteins, as well as the alignments and trees generated from this work are available from http://dbdata.rutgers.edu/data/dino/. We adopted a two-step approach, as modified from Chan et al. (2011b), for phylogeny sorting to examine strongly supported monophyletic relationship (based on non-parametric bootstrap support at ≥90%, ≥70%, and ≥50%) between dinoflagellates and one other phylum. First, we used a simple computational text-parsing tool (PERL script available upon request) to rapidly identify trees with potentially (interpretable) topologies among phylogenies (in NEWICK format) that contain ≥2 distinct phyla and ≥20 terminal taxa (number of branch tips). To minimize the effect of missing data (another common bias of phylogenetic artifact), we required dinoflagellates and the second phylum to each have ≥2 sequences within the monophyletic clade. After that, each of these sorted trees was manually inspected by eye to identify putative instances of E/HGT.
Functional annotation of EST
Putative functions of the EST unigenes were annotated using Blast2GO, based on sequence similarity searches (BLASTP) against the GenBank non-redundant (NR) protein database (e-value ≤10-5). See Appendix S1 in the supplementary material for the complete list of annotations for the A. tamarense genes used in this study.
Results and Discussion
Tracing the evolutionary origins of dinoflagellate proteins
Assembly of the A. tamarense ESTs from Sanger and Roche 454 pyrosequencing data resulted in 32,110 unigenes, which is 80% of the ca. 40K unique genes in this species. This latter number was previously estimated by counting unique, high quality sequence tags generated using massively parallel signature sequencing (MPSS) of RNA derived from cells grown under different culture conditions (Moustafa et al. 2010). For each of the unigenes, we used a simplified reciprocal BLAST approach (Chan et al. 2011c; see Methods) to identify putative homologs within a broadly sampled in-house protein sequence database (ca. 14 million sequences obtained from public sources; Table S1). This study included 23,654 predicted protein sequences from the parasitic flagellate, Perkinsus marinus that forms a basal sister lineage to dinoflagellates (i.e., including the early-branching heterotrophic dinoflagellate, Oxyrrhis marina) (Saldarriaga et al. 2003). Here we considered Perkinsus (Perkinsea) and the dinoflagellates (Dinophyceae) to comprise a single phylogenetic entity. Using our conservative approach, only 9,765 (30.4%) predicted proteins had significant BLAST hits (e-value ≤ 10-10) to the database. This suggests that most A. tamarense genes are novel or alternatively, too divergent when compared to existing data to be identified based on sequence similarity. In addition to 22,345 unigenes without hits, 5,399 had hits only to other dinoflagellates. These 27,744 (86.4%) genes, putatively dinoflagellate-specific (Fig. 1B; some of these include Perkinsus), are likely to contribute to the many unique characteristics shared by these taxa. The remaining 4,366 genes that have hits to other taxa in the database (13.6% of 32,110) provided the input data for phylogenomic analysis.
To assess the phylogenetic history of dinoflagellate genes, we relied on the well-supported affiliation of these taxa with other members of the phylum Alveolata that also includes the heterotrophic ciliates and parasitic apicomplexans (Cavalier-Smith 1991, Gajadhar et al. 1991). Given these existing data and our limited understanding of eukaryote evolution (Figure 1A), we postulated that, ignoring intra-phylum E/HGT, dinoflagellate genes of vertical descent would be most closely related to other alveolates with strong bootstrap support or, in the absence of other alveolate homologs (e.g., photosynthetic genes absent in ciliates and apicomplexans), be sister to stramenopiles and/or Rhizaria (i.e., the SAR clade (Burki et al. 2007)).
Exclusive gene sharing by A. tamarense
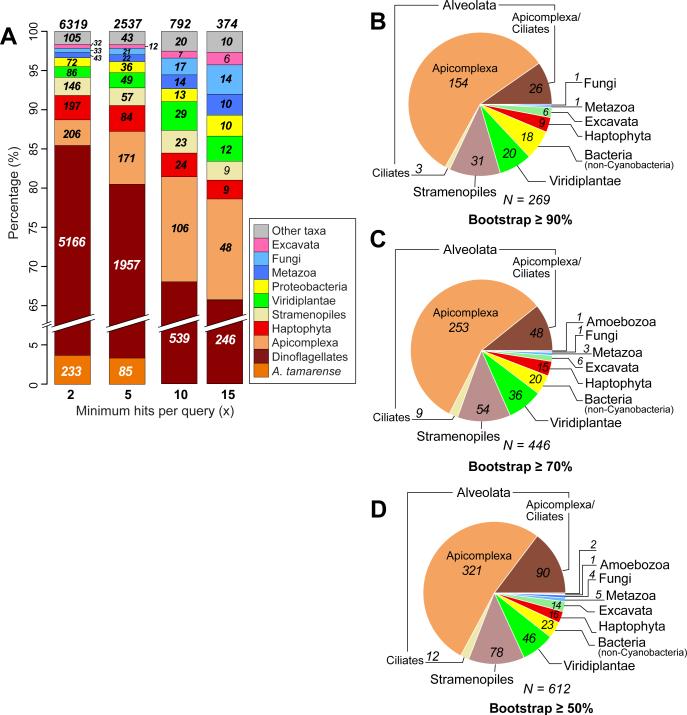
Under the assumption that the signal of gene sharing (or apparent signal, due to the limits of detection using BLAST or potentially convergence (Sanderson & Donoghue 1989, Brandley et al. 2009)) is correlated with evolutionary distance, we studied the distribution of hits to the dinoflagellate data. Of the 9,765 A. tamarense proteins with hits, 920 have hits only to dinoflagellates and one other taxon (Fig. 1B; see also Table S2 in the supplementary material). Figure 2A shows the proportion of A. tamarense genes for which hits are found solely among other dinoflagellates, or between dinoflagellates and one other taxon. Assuming that inadequate sampling is less of a concern as the number of total hits (i.e., taxonomic breadth) increases, we examined the two-taxon associations by requiring an increasing minimum number of hits per query (x) from both taxa (x ≥2, ≥5, ≥10, and ≥15). At x ≥2, the four most abundant foreign taxa that contain genes with hits exclusively to dinoflagellates are Apicomplexa (206 proteins), Haptophyta (197), stramenopiles (146), and Viridiplantae (86; Fig. 2A). As x increases (across the bars from left to right in Fig. 2A), the proportion of hits that showed a dinoflagellate-Apicomplexa association generally increased; i.e., 206 (3%) → 171 (7%) → 106 (13%) → 48 (13%). In comparison, the proportion of proteins showing hits only to dinoflagellates decreased under the same condition; i.e., 5,399 (85%) → 2,042 (80%) → 539 (68%) → 246 (66%). Our approach therefore provided a signal of evolutionary association for dinoflagellate proteins (independent of phylogeny) that, as would be predicted, shows a close association with Apicomplexa (i.e., both groups are alveolates; likely explained by vertical inheritance), to a lesser extent with stramenopiles (i.e., the SAR hypothesis), as well as with the more distantly related haptophytes (197 [3%] → 84 [3%] → 24 [3%] → 9 [2%]) for which a complete genome sequence is available from Emiliania huxleyi (i.e., these are probable instances of exclusive gene sharing). There are currently no completed genomes among cryptophytes and Rhizaria, likely explaining their absence from this list of exclusive hits.
FIGURE 2.
Evolutionary origins of dinoflagellate protein families. (A) The distribution of phyla with exclusive BLASTP hits to A. tamarense proteins, across the minimum number of hits per query, x ≥2, ≥5, ≥10, and ≥15. The different phyla that share proteins exclusively with A. tamarense are shown. For each bar, the number of proteins for each phylum is shown, with the total on top. (B) Distribution of phyla that are found to share genes with dinoflagellates, based on the number of protein phylogenies in which a strongly supported (bootstrap ≥90%) monophyly between these phyla and dinoflagellates was recovered. This distribution is also shown for (C) bootstrap ≥70% and (D) bootstrap ≥50%. In these cases, at least two dinoflagellate sequences and two from the sister taxon are required to be counted as a monophyletic lineage at the prescribed bootstrap cut-off value.
Phylogenies of dinoflagellate protein families
Using an automated pipeline, we generated a maximum likelihood tree for 4,366 A. tamarense protein alignments. Using a similar approach adopted from an earlier study of red algae (Chan et al. 2011c) but using less stringent conditions given the large number of unknown genes in dinoflagellates, we focused on phylogenies that contain ≥2 distinct phyla and ≥20 terminal taxa (2,523 proteins); i.e., those with a sufficiently broad sampling of taxa to infer protein (thus gene) history. Using the initially stringent bootstrap cut-off of ≥90% as evidence of a monophyletic clade in individual trees combined with the requirement of the clade comprising sequences from ≥2 dinoflagellate species and ≥2 species of another taxon, we found 251 protein families (10% of 2,523 trees) to have a eukaryote origin and 18 (0.7%) to have arisen from prokaryotes, totaling 269 trees (Fig. 2B). These numbers rose to 426 (17%) and 20 (0.8%) totaling 446 trees and to 589 (23%) and 23 (0.9%) totaling 612 trees at bootstrap ≥70% and ≥50%, respectively (Fig. 2, C and D). The percentage of trees showing an apicomplexan affiliation for A. tamarense protein families across each of these analyses did not change significantly: i.e., 57% (bootstrap ≥90%), 57% (bootstrap ≥70%), and 52% (bootstrap ≥50%). In summary, among the 269 trees recovered using the most conservative bootstrap cut-off ≥90%, dinoflagellates were most often positioned as sister to chromalveolate lineages (223 trees); i.e., Alveolata (Apicomplexa and/or Ciliates; 183), stramenopiles (31), and Haptophyta (9). The remaining 46 trees showed a sister relationship between the dinoflagellates and other lineages: i.e., Viridiplantae (20), bacteria that are not Cyanobacteria (18), Excavata (6), Fungi (1), and Metazoa (1). The 214 trees that unite the Alveolata and stramenopiles putatively reflect vertical inheritance consistent with the SAR hypothesis, assuming high divergence and/or loss of genes (or the inability to identify homologs) in other taxa. Therefore our analysis returned several hundred genes (e.g., 321 at cut-off ≥50%) that provide the expected result of dinoflagellate-apicomplexan monophyly. These potentially comprise a set of proteins that may be useful for reconstructing the TOL for chromalveolates and other eukaryote taxa. Despite its initial appeal, this straightforward interpretation did not stand up to analysis of individual trees as described below. The sporadic distribution of diverse taxa that are sister to the dinoflagellates in the other 55, 82, and 111 proteins out of the 2,523 that were analyzed (ca. 2% at bootstrap ≥90%, 3% at bootstrap ≥70%, and 4% at bootstrap ≥50%) likely reflects the convoluted evolutionary history of these genes. These data included two classes of topologies that offer different insights into gene history.
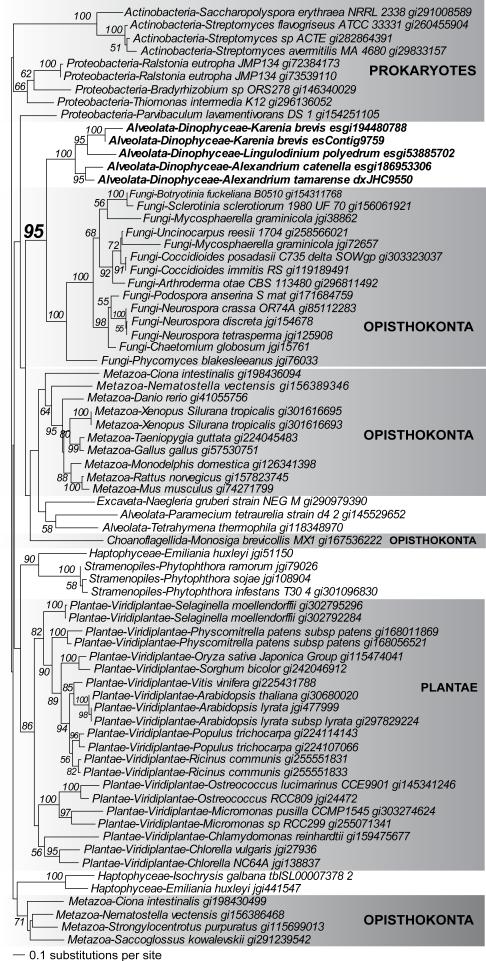
In the first class, dinoflagellates and a non-SAR phylum formed a strongly supported clade, in which each of these phyla were themselves (strongly supported) monophyletic groups with no homologs present elsewhere in the tree. This type of phylogeny could be interpreted as cases of vertical inheritance with massive gene loss (e.g., in other alveolate or stramenopile lineages to which dinoflagellates are most closely related), rather than E/HGT. An alternative explanation is inadequate taxon sampling (missing data) in our current database. An example of such a tree is shown in Fig. 3. This phylogeny of acyl-CoA dehydrogenases supports a specific affiliation between dinoflagellates and fungi that is most easily explained by an ancient HGT event between these taxa. This interpretation must however be tempered by the fact that public databases currently have a strong imbalance in data sampling among eukaryotes (i.e., Fungi- and plant-rich, protist-poor) and the observation that dinoflagellates are sister to, and not nested within, Fungi. It is therefore conceivable that the addition of more chromalveolate genome data could lead to the growth of the dinoflagellate clade so that it encompasses a variety of other chromalveolate taxa and the affiliation to fungi is weakened. In addition to convergent evolution and data biases, ancient gene duplications and losses among eukaryotes could also potentially result in a tree such as Fig. 3, in which relationships reflect paralog rather than ortholog gene history.
FIGURE 3.
Phylogeny of acyl-CoA dehydrogenase that provides evidence of an HGT event involving dinoflagellates and fungi. Non-parametric bootstrap support values ≥50% are shown at the nodes of the tree. Dinoflagellates are highlighted in boldface. The unit of branch length is in the number of substitutions per site.
In the second class of trees, dinoflagellates were nested with strong bootstrap support within another phylum, likely supporting an origin via E/HGT. Although we implemented a number of precautionary measures to reduce potential artifacts during phylogeny reconstruction (see Materials and Methods), we cannot dismiss the possibility that some of these trees could also be a byproduct of phylogenetic artifacts. Therefore, without further experimental verification, these putative E/HGT instances could alternatively be explained by convergent evolution, sampling biases, or other unknown evolutionary aspects, which result in a strongly supported clade of distantly related lineages (Bodył et al. 2009, Christin et al. 2010, Stiller 2011). Two examples of the second class of putative E/HGT candidates in A. tamarense returned by our analysis are shown in Fig. 4. The first is the previously recognized proteobacterial origin of histone-like DNA binding proteins (major basic nuclear proteins) in dinoflagellates (Hackett et al. 2005; Fig. 4A) that have undergone multiple gene duplications in different dinoflagellate lineages and are usually highly represented in EST libraries (Hackett et al. 2005). The second HGT candidate is a GTP-binding protein of the YchF family that is broadly distributed among eukaryotes (Fig. 4B). This tree shows a shared origin of the gene specifically in dinoflagellates and picoprasinophytes (bacterium-sized green algae, e.g., Ostreococcus and Micromonas). It is important to note here that the dinoflagellate-picoprasinophyte clade is nested within Viridiplantae (with bootstrap support 93%), thereby supporting gene origin in dinoflagellates E/HGT from a green algal source. An alternative explanation is that dinoflagellates and green algae are specifically related to each other independent of other Viridiplantae and alveolates, a result that is not supported by other vertically inherited gene markers. The YchF phylogeny however combines examples of vertical inheritance (e.g., in Fungi and in Alveolata excluding dinoflagellates) and HGT with the gene in the choanoflagellate (Monosiga ovata) and the photosynthetic stramenopiles (e.g., Phaeodactylum tricornutum) having a green algal origin from a putative ancient gene duplication product shared by Viridiplantae. This tree demonstrates a key point we wish to make, that vertical inheritance is a relative characteristic of eukaryote protein families. Lineages with a history of phagotrophy such as chromalveolates and choanoflagellates are much more likely to show evidence of E/HGT (again, notwithstanding other explanations due to phylogenetic artifacts) than most plant, fungal, and animal lineages that have often been used as models to elucidate gene history and function. Therefore, unusual (but phylogenetically, highly misleading) examples of E/HGT (or simply an unusual association of distantly related phyla) need to be identified in protists if genome data, such as that shown in Figure 4B are to be used as markers of eukaryote evolution (see also discussion of eukaryotic translation elongation factor 2 in Hackett et al. 2007).
FIGURE 4.
Examples of proteins that have an origin in dinoflagellates via E/HGT. (A) Phylogeny of genes encoding the DNA-binding major basic nuclear proteins in dinoflagellates that has a proteobacterial HGT origin prior to the diversification of these algal taxa. (B) Phylogeny of a gene encoding a GTP-binding protein of the YchF family. This tree shows strong bootstrap support (97%) for a shared origin of the gene in picoprasinophytes and dinoflagellates. For both trees, non-parametric bootstrap support values ≥50% are shown at the nodes. Dinoflagellates are highlighted in boldface. The unit of branch length is in the number of substitutions per site.
Complicating factors in phylogenetic inference
Thus far, we have identified putative cases of E/HGT based on the presence of a strongly supported clade within a phylogeny that contains lineages of only dinoflagellates and one sister phylum. However, examples of E/HGT from Plantae into lineages basal to dinoflagellates (i.e., endosymbiotic transfer of algal genes into the ancestral alveolate or SAR lineage) could also indicate non-lineal sources of genes in these taxa. These cases, alternatively explained by convergent evolution, would not be detected by the approach described above. To evaluate this important aspect of gene transfer, we implemented a less-stringent approach for phylogeny sorting in which we allowed the presence of interrupting phyla (other than dinoflagellates and the Plantae) within the clade, whereby they constitute ≤30% of the total number of lineages within the clade. That is, dinoflagellates and the Plantae taxa (each with ≥2 sequences) constitute >70% of the total lineages within a strongly supported clade. These sorted trees are available at http://dbdata.rutgers.edu/data/dino/. Many of these phylogenies document a history of non-exclusive gene sharing involving dinoflagellates that could be explained by EGT events from Plantae to photosynthetic chromalveolates and Euglenozoa (e.g., phylogeny of the cytochrome b6-f complex iron-sulfur subunit, shown in Fig. S1). This predicted migration of endosymbiont genes to the nucleus of A. tamarense and other dinoflagellates accounts for 4, 147, and 142 genes of rhodophyte, Viridiplantae, and rhodophyte/Viridiplantae/glaucophyte origin, respectively at bootstrap ≥90% (totaling 293). At bootstrap ≥70%, the total number became 353. Together with the number of putatively E/HGT-implicated genes we observed in Fig. 2, we estimate as many as 348 (at bootstrap ≥90%) to 435 (at bootstrap ≥70%) of dinoflagellate genes (14-17% of the examined phylogenies in this study) are implicated in non-lineal gene sharing during their evolutionary history. These numbers, albeit speculative, represent conservative estimates that are dependent on the criteria used in phylogeny sorting.
Another example of a complex protein family history in dinoflagellates and other chromalveolates is shown in Fig. S2 (see supplementary material). This is the phylogeny of a putative triose-phosphate isomerase, an enzyme that in plants is involved in multiple metabolic pathways including glycolysis, gluconeogenesis, and the Calvin cycle. The tree shows a well-supported haptophyte-dinoflagellate association: i.e., monophyly of the haptophyte Isochrysis galbana and dinoflagellates at bootstrap 98%. Other gene copies from the dinoflagellates are distributed outside the clade, associated with the other haptophyte in the analysis, Emiliania huxleyi, and the stramenopile Aureococcus anophagefferens (at bootstrap 100%), whereas the parasitic alveolate, Perkinsus marinus is positioned elsewhere in the tree. This phylogeny generally shows weak support (bootstrap <50%) at the deeper splits, particularly the ambiguous separation among lineages of chromalveolates (e.g., the resolution of alveolates and stramenopiles) and excavates. The origin of this gene in the dinoflagellates remains unclear due to the patchy distribution of diverse taxa (except for Viridiplantae and Opisthokonta) within the tree, even though a strong haptophyte-dinoflagellate association is recovered. The phylogeny again combines vertical inheritance in groups such as Metazoa and Viridiplantae with a highly complex pattern likely explained by serial endosymbiosis, HGT, gene duplication and loss, and stochastic variation of sequences among chromalveolates that is currently impossible to decipher. Many dinoflagellate protein families display this type of topology (see http://dbdata.rutgers.edu/data/dino/). These results provide empirical evidence for a complex evolutionary history of dinoflagellate genes and the existence of complicating factors other than E/HGT (e.g., gene duplication and loss) that would significantly attenuate, if not invalidate, the use of these genes in inferring phylogenetic relationships, particularly among the chromalveolates.
It should also be noted that much of the genome data that are currently available from microbial eukaryotes (including the dinoflagellates in this study) comprise EST sequences that usually contain incomplete/partial gene transcripts and may include unidentified contaminants or potentially be misidentified due to sample mislabeling. These sequences, when included in an alignment can introduce systematic biases (e.g., long-branch attraction, (Bergsten 2005) resulting in inaccuracies in phylogenetic inference. Our simplified reciprocal BLAST approach was designed to reduce the inclusion of short, partial transcripts in a sequence alignment to minimize (not exclude) these biases. Nevertheless, our gene-by-gene approach did not consider other complicating factors that limit the ability to detect E/HGT, such as transfer of genetic fragments irrespective of gene boundaries (Chan et al. 2009), as demonstrated in prokaryotes (Inagaki et al. 2006, Chan et al. 2011a) and eukaryotes (Nikoh & Nakabachi 2009), as well as genome amelioration (Lawrence & Ochman 1997, Chan et al. 2006). These complicating issues with regard to detecting HGT are described in a recent review (Ragan & Beiko 2009). Given the existence of taxon (gene) sampling biases, the numbers reported here should be considered as conservative estimates of E/HGT because they were inferred using relatively stringent phylogenetic criteria (e.g., imposing a minimum threshold for the number of distinct phyla and terminal taxa in trees). The expectation was that this approach played to the strengths of phylogenetic inference methods and therefore was more likely to have few significant artifacts.
Conclusions
We used a rich collection of ESTs from a microbial eukaryote (Hackett et al. 2004) to study genome evolution. As found for many previously unexplored protist lineages, most of the expressed genes in A. tamarense are unique to this species or to dinoflagellates in general, removing 86.4% of the genome data from our comparative analyses. Of the remaining genes with BLASTP hits to our database, only 7.9% were present in ≥2 distinct phyla with ≥20 terminal taxa and therefore broadly enough distributed to be considered as a potential phylogenetic marker of the TOL. Here we defined a phylogenetic marker to be any gene/protein that is broadly distributed across the eukaryote TOL and provides moderate to strong bootstrap support for vertical inheritance; i.e., no detectable evidence for non-lineal gene sharing (due to E/HGT). Therefore, although the phylogenomic analysis provides clear evidence for the expected apicomplexan affiliation for A. tamarense proteins, this small piece of the “genome pie” in the dinoflagellate also contains clear examples of E/HGT or an evolutionary history that is too complex to yet be interpreted with currently available gene and genome data.
The major implication of our work is obvious: we have much to learn about protist genome evolution. It is clear that analysis of genome data with readily identifiable homologs in other lineages addresses only a small fraction of the encoded information in dinoflagellates; the assumption of vertical gene inheritance reduces this number further. When these taxa are included in multi-gene analyses, extreme care must be taken with each gene in each genome to assure that a common set of vertically inherited genes are compared across diverse taxa. In the extreme case, one could argue that there are potentially no vertically inherited phylogenetic markers that can be applied across the entire eukaryote TOL if the requirement is that the orthologs in each taxon are readily identifiable and free of E/HGT. TOL inference is therefore by necessity a compromise between the choice of appropriate taxa and marker genes. Ultimately far more genome data are needed from dinoflagellates and other alveolates to fully understand the phylum- and species-specific patterns of E/HGT and vertical inheritance. The failure to go beyond a select set of conserved gene trees (e.g., rDNA, heat shock proteins) to represent dinoflagellate evolution is problematic, and in some ways analogous to what has been found in comparisons of rDNA in phytoplankton, whereby significantly different genome structures and protein divergence are often masked by nearly identical rDNA sequences (Palenik et al. 2007, Worden et al. 2009, Cuvelier et al. 2010). These results also suggest the possibility that the biogeographic distribution of microbes (irrespective of phylogenetic relatedness, (Gogarten et al. 2002) may facilitate gene sharing among taxa living in similar environmental niches (Beiko et al. 2005). Conversely, related taxa living in different environments may show inconsistent patterns of gene sharing based on local circumstances. Both of these exciting prospects would further complicate the evolutionary history of protist genomes. Therefore, it may turn out that E/HGT has a far more complex distribution on the tree of eukaryotes than can be explained by fitting available data to the most parsimonious explanation (e.g., red algal-derived plastid distribution in support of the chromalveolate hypothesis, Cavalier-Smith 1998, Yoon et al. 2002).
Finally, the extent of gene sharing among prokaryotes has previously been estimated to be 2% (Ge et al. 2005), 13% (Beiko et al. 2005), 60% (Lerat et al. 2005), and 90% (Mirkin et al. 2003) of genes or bipartitions based on phylogenetic approaches. The findings of a previous study based on the characteristics of genome sizes (Dagan & Martin 2007) suggest that all prokaryote genes have undergone HGT. Here we find the levels of E/HGT in dinoflagellates are comparable to prokaryotes. A comprehensive understanding of the eukaryote TOL will therefore necessitate similar care as has been used to assess prokaryote phylogeny when taxa such as alveolates are included in genome-wide multi-gene datasets that include their major partners of gene sharing (e.g., stramenopiles, red, and green algae).
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (R01-ES013679-01A2 to DB, MBS, DLE, and DMA), and by the National Institute of Environmental Health Sciences (1-P50-ES012742) and the National Science Foundation (OCE-0430724) through the Woods Hole Center for Oceans and Human Health (to DMA and DLE). We are grateful to two anonymous reviewers for constructive comments on the manuscript.
Abbreviations
- EGT
endosymbiotic gene transfer
- EST
expressed sequence tag
- GTP
guanosine triphosphate
- HGT
horizontal gene transfer
- SAR
Stramenopiles-Alveolata-Rhizara
- TOL
tree of life
Footnotes
Received _________. Accepted _________.
Contributor Information
Cheong Xin Chan, Department of Ecology, Evolution and Natural Resources, and Institute of Marine and Coastal Sciences, Rutgers University, New Brunswick, NJ 08901, USA.
Marcelo B. Soares, Northwestern University, Children's Memorial Research Center, Chicago, IL 60614, USA
Maria F. Bonaldo, Northwestern University, Children's Memorial Research Center, Chicago, IL 60614, USA
Jennifer H. Wisecaver, Department of Ecology and Evolutionary Biology, The University of Arizona, Tucson, AZ 85721, USA
Jeremiah D. Hackett, Department of Ecology and Evolutionary Biology, The University of Arizona, Tucson, AZ 85721, USA
Donald M. Anderson, Woods Hole Oceanographic Institution, Woods Hole, MA 02543, USA
Deana L. Erdner, Marine Science Institute, University of Texas, Port Aransas, TX 78373, USA
Debashish Bhattacharya, Department of Ecology, Evolution and Natural Resources, and Institute of Marine and Coastal Sciences, Rutgers University, New Brunswick, NJ 08901, USA.
References
- Baurain D, Brinkmann H, Petersen J, Rodríguez-Ezpeleta N, Stechmann A, Demoulin V, Roger AJ, Burger G, Lang BF, Philippe H. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol. Biol. Evol. 2010;27:1698–709. doi: 10.1093/molbev/msq059. [DOI] [PubMed] [Google Scholar]
- Beiko RG, Harlow TJ, Ragan MA. Highways of gene sharing in prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14332–37. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsten J. A review of long-branch attraction. Cladistics. 2005;21:163–93. doi: 10.1111/j.1096-0031.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- Bock R. The give-and-take of DNA: horizontal gene transfer in plants. Trends Plant Sci. 2010;15:11–22. doi: 10.1016/j.tplants.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Bodył A, Mackiewicz P, Stiller JW. Early steps in plastid evolution: current ideas and controversies. BioEssays. 2009;31:1219–32. doi: 10.1002/bies.200900073. [DOI] [PubMed] [Google Scholar]
- Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, Rayko E, Salamov A, Vandepoele K, Beszteri B, Gruber A, Heijde M, Katinka M, Mock T, Valentin K, Verret F, Berges JA, Brownlee C, Cadoret JP, Chiovitti A, Choi CJ, Coesel S, De Martino A, Detter JC, Durkin C, Falciatore A, Fournet J, Haruta M, Huysman MJ, Jenkins BD, Jiroutova K, Jorgensen RE, Joubert Y, Kaplan A, Kroger N, Kroth PG, La Roche J, Lindquist E, Lommer M, Martin-Jezequel V, Lopez PJ, Lucas S, Mangogna M, McGinnis K, Medlin LK, Montsant A, Oudot-Le Secq MP, Napoli C, Obornik M, Parker MS, Petit JL, Porcel BM, Poulsen N, Robison M, Rychlewski L, Rynearson TA, Schmutz J, Shapiro H, Siaut M, Stanley M, Sussman MR, Taylor AR, Vardi A, von Dassow P, Vyverman W, Willis A, Wyrwicz LS, Rokhsar DS, Weissenbach J, Armbrust EV, Green BR, Van de Peer Y, Grigoriev IV. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–44. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Brandley MC, Warren DL, Leaché AD, McGuire JA. Homoplasy and clade support. Syst. Biol. 2009;58:184–98. doi: 10.1093/sysbio/syp019. [DOI] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Minge M, Skjæveland Å, Nikolaev SI, Jakobsen KS, Pawlowski J. Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE. 2007;2:e790. doi: 10.1371/journal.pone.0000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Pawlowski J. Phylogenomics reveals a new “megagroup” including most photosynthetic eukaryotes. Biol. Lett. 2008;4:366–69. doi: 10.1098/rsbl.2008.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Cell diversification in heterotrophic flagellates. In: Patterson DJ, Larsen J, editors. The Biology of Free-Living Heterotrophic Flagellates. Oxford University Press; Oxford: 1991. pp. 113–31. [Google Scholar]
- Cavalier-Smith T. A revised six-kingdom system of life. Biological Reviews. 1998;73:203–66. doi: 10.1017/s0006323198005167. [DOI] [PubMed] [Google Scholar]
- Chan CX, Beiko RG, Ragan MA. Detecting recombination in evolving nucleotide sequences. BMC Bioinformatics. 2006;7:412. doi: 10.1186/1471-2105-7-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CX, Beiko RG, Ragan MA. Lateral transfer of genes and gene fragments in Staphylococcus extends beyond mobile elements. J. Bacteriol. 2011a;193:3964–77. doi: 10.1128/JB.01524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CX, Darling AE, Beiko RG, Ragan MA. Are protein domains modules of lateral genetic transfer? PLoS ONE. 2009;4:e4524. doi: 10.1371/journal.pone.0004524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CX, Reyes-Prieto A, Bhattacharya D. Red and green algal origin of diatom membrane transporters: insights into environmental adaptation and cell evolution. PLoS ONE. 2011b;6:e29138. doi: 10.1371/journal.pone.0029138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CX, Yang EC, Banerjee T, Yoon HS, Martone PT, Estevez JM, Bhattacharya D. Red and green algal monophyly and extensive gene sharing found in a rich repertoire of red algal genes. Curr. Biol. 2011c;21:328–33. doi: 10.1016/j.cub.2011.01.037. [DOI] [PubMed] [Google Scholar]
- Christin PA, Weinreich DM, Besnard G. Causes and evolutionary significance of genetic convergence. Trends Genet. 2010;26:400–05. doi: 10.1016/j.tig.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Cuvelier ML, Allen AE, Monier A, McCrow JP, Messié M, Tringe SG, Woyke T, Welsh RM, Ishoey T, Lee JH, Binder BJ, DuPont CL, Latasa M, Guigand C, Buck KR, Hilton J, Thiagarajan M, Caler E, Read B, Lasken RS, Chavez FP, Worden AZ. Targeted metagenomics and ecology of globally important uncultured eukaryotic phytoplankton. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14679–84. doi: 10.1073/pnas.1001665107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Martin W. Ancestral genome sizes specify the minimum rate of lateral gene transfer during prokaryote evolution. Proc. Natl. Acad. Sci. U. S. A. 2007;104:870–75. doi: 10.1073/pnas.0606318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF. You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 1998;14:307–11. doi: 10.1016/s0168-9525(98)01494-2. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Bapteste E. Pattern pluralism and the Tree of Life hypothesis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2043–49. doi: 10.1073/pnas.0610699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–97. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajadhar AA, Marquardt WC, Hall R, Gunderson J, Ariztia-Carmona EV, Sogin ML. Ribosomal RNA sequences of Sarcocystis muris, Theileria annulata and Crypthecodinium cohnii reveal evolutionary relationships among apicomplexans, dinoflagellates, and ciliates. Mol. Biochem. Parasitol. 1991;45:147–54. doi: 10.1016/0166-6851(91)90036-6. [DOI] [PubMed] [Google Scholar]
- Ge F, Wang LS, Kim J. The cobweb of life revealed by genome-scale estimates of horizontal gene transfer. PLoS Biol. 2005;3:e316. doi: 10.1371/journal.pbio.0030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 2002;19:2226–38. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- Gruenheit N, Lockhart PJ, Steel M, Martin W. Difficulties in testing for covarion-like properties of sequences under the confounding influence of changing proportions of variable sites. Mol. Biol. Evol. 2008;25:1512–20. doi: 10.1093/molbev/msn098. [DOI] [PubMed] [Google Scholar]
- Hackett JD, Anderson DM, Erdner DL, Bhattacharya D. Dinoflagellates: a remarkable evolutionary experiment. Am. J. Bot. 2004;91:1523–34. doi: 10.3732/ajb.91.10.1523. [DOI] [PubMed] [Google Scholar]
- Hackett JD, Bhattacharya D. The genomes of dinoflagellates. In: Katz LA, Bhattacharya D, editors. Genomics and Evolution of Microbial Eukaryotes. Oxford University Press; New York: 2008. pp. 48–63. [Google Scholar]
- Hackett JD, Scheetz TE, Yoon HS, Soares MB, Bonaldo MF, Casavant TL, Bhattacharya D. Insights into a dinoflagellate genome through expressed sequence tag analysis. BMC Genomics. 2005;6:80. doi: 10.1186/1471-2164-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JD, Yoon HS, Li S, Reyes-Prieto A, Rummele SE, Bhattacharya D. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of Rhizaria with chromalveolates. Mol. Biol. Evol. 2007;24:1702–13. doi: 10.1093/molbev/msm089. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–77. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki Y, Susko E, Roger AJ. Recombination between elongation factor 1 alpha genes from distantly related archaeal lineages. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4528–33. doi: 10.1073/pnas.0600744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida K, Green BR. Second- and third-hand chloroplasts in dinoflagellates: phylogeny of oxygen-evolving enhancer 1 (PsbO) protein reveals replacement of a nuclear-encoded plastid gene by that of a haptophyte tertiary endosymbiont. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9294–99. doi: 10.1073/pnas.142091799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. Chromalveolates and the evolution of plastids by secondary endosymbiosis. J. Eukaryot. Microbiol. 2009;56:1–8. doi: 10.1111/j.1550-7408.2008.00371.x. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 2008;9:605–18. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- Koumandou VL, Nisbet RER, Barbrook AC, Howe CJ. Dinoflagellate chloroplasts - where have all the genes gone? Trends Genet. 2004;20:261–67. doi: 10.1016/j.tig.2004.03.008. [DOI] [PubMed] [Google Scholar]
- LaJeunesse TC, Lambert G, Andersen RA, Coffroth MA, Galbraith DW. Symbiodinium (Pyrrhophyta) genome sizes (DNA content) are smallest among dinoflagellates. J. Phycol. 2005;41:880–86. [Google Scholar]
- Lawrence JG, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 1997;44:383–97. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Retchless AC. The myth of bacterial species and speciation. Biol. Philos. 2010;25:569–88. [Google Scholar]
- Lerat E, Daubin V, Ochman H, Moran NA. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 2005;3:e130. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Nosenko T, Hackett JD, Bhattacharya D. Phylogenomic analysis identifies red algal genes of endosymbiotic origin in the chromalveolates. Mol. Biol. Evol. 2006;23:663–74. doi: 10.1093/molbev/msj075. [DOI] [PubMed] [Google Scholar]
- Martin W, Herrmann RG. Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol. 1998;118:9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI. Primary and secondary endosymbiosis and the origin of plastids. J. Phycol. 2001;37:951–59. [Google Scholar]
- McFadden GI, van Dooren GG. Evolution: red algal genome affirms a common origin of all plastids. Curr. Biol. 2004;14:R514–R16. doi: 10.1016/j.cub.2004.06.041. [DOI] [PubMed] [Google Scholar]
- Mirkin BG, Fenner TI, Galperin MY, Koonin EV. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol. Biol. 2003;3:2. doi: 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science. 2009;324:1724–26. doi: 10.1126/science.1172983. [DOI] [PubMed] [Google Scholar]
- Moustafa A, Evans AN, Kulis DM, Hackett JD, Erdner DL, Anderson DM, Bhattacharya D. Transcriptome profiling of a toxic dinoflagellate reveals a gene-rich protist and a potential impact on gene expression due to bacterial presence. PLoS ONE. 2010;5:e9688. doi: 10.1371/journal.pone.0009688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh N, Nakabachi A. Aphids acquired symbiotic genes via lateral gene transfer. BMC Biol. 2009;7:12. doi: 10.1186/1741-7007-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosenko T, Bhattacharya D. Horizontal gene transfer in chromalveolates. BMC Evol. Biol. 2007;7:173. doi: 10.1186/1471-2148-7-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenik B, Grimwood J, Aerts A, Rouze P, Salamov A, Putnam N, Dupont C, Jorgensen R, Derelle E, Rombauts S, Zhou KM, Otillar R, Merchant SS, Podell S, Gaasterland T, Napoli C, Gendler K, Manuell A, Tai V, Vallon O, Piganeau G, Jancek S, Heijde M, Jabbari K, Bowler C, Lohr M, Robbens S, Werner G, Dubchak I, Pazour GJ, Ren QH, Paulsen I, Delwiche C, Schmutz J, Rokhsar D, Van de Peer Y, Moreau H, Grigoriev IV. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:7705–10. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Grant J, Tekle YI, Lasek-Nesselquist E, Morrison HG, Sogin ML, Patterson DJ, Katz LA. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst. Biol. 2010;59:518–33. doi: 10.1093/sysbio/syq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJG, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13624–29. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patron NJ, Inagaki Y, Keeling PJ. Multiple gene phylogenies support the monophyly of cryptomonad and haptophyte host lineages. Curr. Biol. 2007;17:887–91. doi: 10.1016/j.cub.2007.03.069. [DOI] [PubMed] [Google Scholar]
- Price DC, Chan CX, Yoon HS, Yang EC, Qiu H, Weber AP, Schwacke R, Gross J, Blouin NA, Lane C, Reyes-Prieto A, Durnford DG, Neilson JAD, Lang BF, Burger G, Steiner JM, Löffelhardt W, Meuser JE, Posewitz MC, Ball S, Arias MC, Henrissat B, Coutinho PM, Rensing SA, Symeonidi A, Doddapaneni H, Green BR, Rajah VD, Boore J, Bhattacharya D. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science. 2012 doi: 10.1126/science.1213561. in press. [DOI] [PubMed] [Google Scholar]
- Puigbo P, Wolf YI, Koonin EV. The tree and net components of prokaryote evolution. Genome Biol. Evol. 2010;2:745–56. doi: 10.1093/gbe/evq062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan MA, Beiko RG. Lateral genetic transfer: open issues. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009;364:2241–51. doi: 10.1098/rstb.2009.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Prieto A, Hackett JD, Soares MB, Bonaldo MF, Bhattacharya D. Cyanobacterial contribution to algal nuclear genomes is primarily limited to plastid functions. Curr. Biol. 2006;16:2320–5. doi: 10.1016/j.cub.2006.09.063. [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A, Moustafa A, Bhattacharya D. Multiple genes of apparent algal origin suggest ciliates may once have been photosynthetic. Curr. Biol. 2008;18:956–62. doi: 10.1016/j.cub.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Prieto A, Weber AP, Bhattacharya D. The origin and establishment of the plastid in algae and plants. Annu. Rev. Genet. 2007;41:147–68. doi: 10.1146/annurev.genet.41.110306.130134. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Löffelhardt W, Bohnert HJ, Philippe H, Lang BF. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 2005;15:1325–30. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Saldarriaga JF, McEwan ML, Fast NM, Taylor FJ, Keeling PJ. Multiple protein phylogenies show that Oxyrrhis marina and Perkinsus marinus are early branches of the dinoflagellate lineage. Int. J. Syst. Evol. Microbiol. 2003;53:355–65. doi: 10.1099/ijs.0.02328-0. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ, Donoghue MJ. Patterns of variation in levels of homoplasy. Evolution. 1989;43:1781–95. doi: 10.1111/j.1558-5646.1989.tb02626.x. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stiller JW. Plastid endosymbiosis, genome evolution and the origin of green plants. Trends Plant Sci. 2007;12:391–6. doi: 10.1016/j.tplants.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Stiller JW. Experimental design and statistical rigor in phylogenomics of horizontal and endosymbiotic gene transfer. BMC Evol. Biol. 2011;11:259. doi: 10.1186/1471-2148-11-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller JW, Huang JL, Ding Q, Tian J, Goodwillie C. Are algal genes in nonphotosynthetic protists evidence of historical plastid endosymbioses? BMC Genomics. 2009;10 doi: 10.1186/1471-2164-10-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–77. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–99. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Wisecaver JH, Hackett JD. Dinoflagellate genome evolution. Annu. Rev. Microbiol. 2011;65:369–87. doi: 10.1146/annurev-micro-090110-102841. [DOI] [PubMed] [Google Scholar]
- Worden AZ, Lee J-H, Mock T, Rouzé P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, Foulon E, Grimwood J, Gundlach H, Henrissat B, Napoli C, McDonald SM, Parker MS, Rombauts S, Salamov A, Von Dassow P, Badger JH, Coutinho PM, Demir E, Dubchak I, Gentemann C, Eikrem W, Gready JE, John U, Lanier W, Lindquist EA, Lucas S, Mayer KFX, Moreau H, Not F, Otillar R, Panaud O, Pangilinan J, Paulsen I, Piegu B, Poliakov A, Robbens S, Schmutz J, Toulza E, Wyss T, Zelensky A, Zhou K, Armbrust EV, Bhattacharya D, Goodenough UW, Van de Peer Y, Grigoriev IV. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–72. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 1994;39:306–14. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Grant J, Tekle YI, Wu M, Chaon BC, Cole JC, Logsdon JM, Patterson DJ, Bhattacharya D, Katz LA. Broadly sampled multigene trees of eukaryotes. BMC Evol. Biol. 2008;8:14. doi: 10.1186/1471-2148-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 2004;21:809–18. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Pinto G, Bhattacharya D. The single, ancient origin of chromist plastids. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15507–12. doi: 10.1073/pnas.242379899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Van Dolah FM, Nosenko T, Lidie L, Bhattacharya D. Tertiary endosymbiosis driven genome evolution in dinoflagellate algae. Mol. Biol. Evol. 2005;22:1299–308. doi: 10.1093/molbev/msi118. [DOI] [PubMed] [Google Scholar]
- Zhang H, Campbell DA, Sturm NR, Lin SJ. Dinoflagellate spliced leader RNA genes display a variety of sequences and genomic arrangements. Mol. Biol. Evol. 2009;26:1757–71. doi: 10.1093/molbev/msp083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hou Y, Miranda L, Campbell DA, Sturm NR, Gaasterland T, Lin S. Spliced leader RNA trans-splicing in dinoflagellates. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4618–23. doi: 10.1073/pnas.0700258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.