Abstract
Mosquitoes are insects of the Diptera, Nematocera, and Culicidae families, some species of which are important disease vectors. Identifying mosquito species based on morphological characteristics is difficult, particularly the identification of specimens collected in the field as part of disease surveillance programs. Because of this difficulty, we constructed DNA barcodes of the cytochrome c oxidase subunit 1, the COI gene, for the more common mosquito species in China, including the major disease vectors. A total of 404 mosquito specimens were collected and assigned to 15 genera and 122 species and subspecies on the basis of morphological characteristics. Individuals of the same species grouped closely together in a Neighborhood-Joining tree based on COI sequence similarity, regardless of collection site. COI gene sequence divergence was approximately 30 times higher for species in the same genus than for members of the same species. Divergence in over 98% of congeneric species ranged from 2.3% to 21.8%, whereas divergence in conspecific individuals ranged from 0% to 1.67%. Cryptic species may be common and a few pseudogenes were detected.
Introduction
Approximately 41 genera and 3500 species and subspecies of mosquito exist worldwide. Although mosquitoes have been studied more extensively than most other insect groups because of their role as vectors of disease, our taxonomic knowledge of these insects is far from complete. Numerous Chinese taxonomists have worked on mosquito classification since 1932, particularly since Edwards provided the modern mosquito classification system [1]. Feng Lan-Zhou reported 100 Chinese mosquito species in 1938 [2]. This number has since then increased to approximately 390 described species and new species are still being identified, particularly within the genera Armigeres, Heizmannia, Topomyia and Uranotaenia.
Some species are vectors of medically important pathogens, such as malaria, Dengue fever and Japanese B encephalitis. Species identification therefore constitutes the first step in the surveillance and control of mosquito-borne diseases. The identification of mosquito species is mainly done on the basis of morphological characteristics. This can be problematic because diagnostic morphological features are often damaged during collection or storage, or are not present in all developmental stages. Moreover, the morphological characteristics used to identify intact adult specimens often vary so little between species that usually only experienced mosquito taxonomists are able to distinguish mosquito species reliably [3].
DNA analysis provides a more accurate way of identifying species and the use of molecular data, in combination to morphological methods, has resolved some long-standing taxonomic questions [4], [5]. The increase in the number of available molecular markers has facilitated the accurate identification of mosquito species, particularly within groups of sibling species. For instance, Anopheles anthropophagus and Anopheles sinensis can be identified more simply, rapidly, and accurately using the ITS2 sequence than on the basis of morphology [6], [7].
After Tautz proposed using DNA sequences as the main basis of biological classification in 2002 [8], [9] Paul Hebert suggested that sequencing the COI gene could allow DNA barcoding that would facilitate such classification [10]–[12]. Many studies have since then demonstrated that the COI gene is a valid molecular tool for identifying mosquito species [13], [14] and revealing cryptic species [15]–[18].
Although several studies on the distribution of Chinese mosquito species have been conducted using classical morphology identifying sibling and cryptic species remains problematic. Here we provide an updated classification of nearly one-third of China’s mosquito species based on a combination of molecular and morphological methods.
Results
Specimen Collection
A total of 122 mosquito species belonging to 15 genera and three subfamilies were collected from sampling sites in eight Chinese provinces (Figure 1, Table 1). We identified mosquitoes on the basis of diagnostic morphological characteristics of their adult and larval stages and cercopoda [19], and by using molecular methods to distinguish sibling species [6], [7].
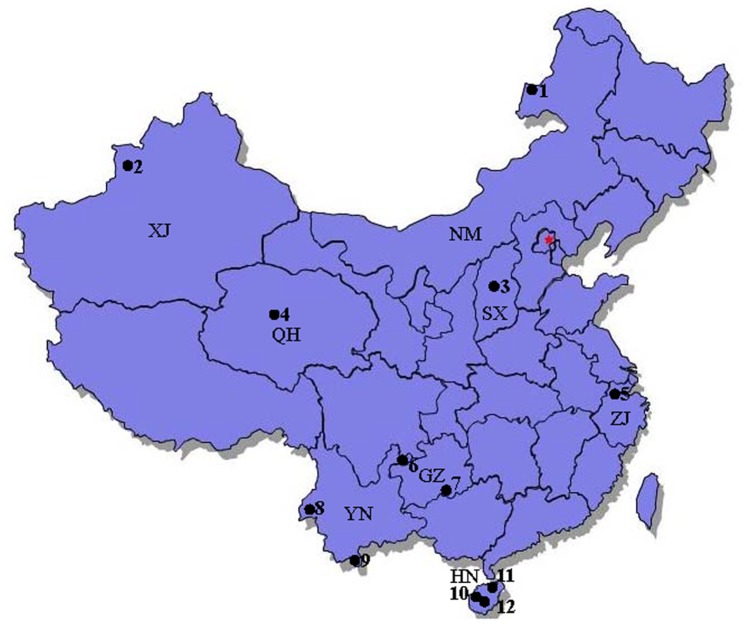
Figure 1. Map of the study area showing the sampling sites of mosquitoes collected in this study.
Site 1: Manzhouli City, Neimeng ProvinceXinjiang; Site 2: Yili, Kazakh Autonomous Prefecture, Xinjiang Province; Site 3: Taiyuan City, Shanxi Province; Site 4: Golmud River, QinghaiQinghai Province; Site 5: Tianmu Mountain, Zhejiang Province; Site 6: Zhenxiong County, Yunnan Province; Site 7: Maolan Natural Reserve, Guizhou Province; Site 8: Ruili City, Yunnan Province; Site 9: Mengla County, Yunnan Province; Site 10: Changjiang County, Hainan Province; Site 11: Limushan Nature Reserve, Hainan Province; Site 12: Mangrove Nature Reserve, Hainan Province.
Table 1. List of mosquito species, collection sites and GenBank accession numbers.
| Mosquito species | Collection site | GenBank accession number |
| An. lindesayi | Site 6, Yunnan | JQ728147; JQ728148;JQ728149 |
| Site 7, Guizhou | JQ728370 | |
| Site 5, Zhejiang | JQ728076 | |
| An. gigas baileyi | Site 6, Yunnan | JQ728161;JQ728162;JQ728163 |
| An. barbirostris | Site 8, Site 9, Yunnan | JQ728025;JQ728220 |
| Site 10–12, Hainan | JQ728403;JQ728404;JQ728405 | |
| An. barbumbrosus | Site 9, Yunnan | JQ728212 |
| An. jamesii | Site 9, Yunnan | JQ728209 |
| An. messeae | Site 1, Neimeng | JQ728113; JQ728114; JQ728115; JQ728116; JQ728077 |
| Site 2, Xinjiang | JQ728279; JQ728280 | |
| An. sinensis | Site 6, Site 8, Site 9, Yunnan | JQ728141;JQ728388;JQ728389; JQ728390; JQ728391;JQ728343; JQ728233 |
| Site 10–12, Hainan | JQ728409; JQ728410;JQ728411 | |
| Lab | JQ728020 | |
| An. yatsushiroensis | Site 3, Shanxi | JQ728372; JQ728373 |
| An. hyrcanus | Site 2, Xinjiang | JQ728293; JQ728294;JQ728295 |
| An. claviger | Site 2, Xinjiang | JQ728274 |
| An. kweiyangensis | Site 6, Yunnan | JQ728386 |
| Site 5, Zhejiang | JQ728378 | |
| An.sawadwongpormi | Site 12, Hainan | JQ728407; JQ728408 |
| An. peditaeniatus | Site 8, Site 9, Yunnan | JQ728088; JQ728089;JQ728090; JQ728342; JQ728230; JQ728231 |
| An. maculatus | Site 9, Yunnan | JQ728164 |
| An. xui | Site 9, Yunnan | JQ728232; JQ728203 |
| An. tessellatus | Site 8, Site 9, Yunnan | JQ728102; JQ728103 |
| Site 10–12, Hainan | JQ728050; JQ728051; JQ728052; JQ728053;JQ728054 | |
| An. kochi | Site 8, Site 9, Yunnan | JQ728307; JQ728242;JQ728243; JQ728290; JQ728291; JQ728292 |
| An. aitkenii | Site 9, Yunnan | JQ728268;JQ728269; JQ728270 |
| An. pseudowillmori | Site 9, Yunnan | JQ728241 |
| An. vagus | Site 8, Site 9, Yunnan | JQ728070; JQ728042 |
| Site 10–12, Hainan | JQ728305; JQ728045; JQ728044; JQ728043 | |
| An. minimus | Site 9, Yunnan | JQ728026; JQ728027; JQ728028; JQ728029 |
| Site 10, Hainan | JQ728406; JQ728030 | |
| An. aconitus | Site 9, Yunnan | JQ728412; JQ728413; JQ728414; JQ728415; JQ728416 |
| Site 10, Hainan | JQ728306; JQ728417; JQ728418; JQ728419 | |
| An. jeyporiensis | Site 9, Yunnan | JQ728235; JQ728236; JQ728218 |
| An. dirus | Site 12, Hainan | JQ728302; JQ728303 |
| An. splendidus | Site 8, Yunnan | JQ728261 |
| Cx. halifaxia | Site 9, Yunnan | JQ728180; JQ728387; JQ728333 |
| Site 10, Hainan | JQ728073; JQ728074; JQ728075 | |
| Cx. brevipalpis | Site 8, Site 9, Yunnan | JQ728158; JQ728159; JQ728160; JQ728336 |
| Site 7, Guizhou | JQ728358; JQ728359 | |
| Cx. foliatus | Site 9, Yunnan | JQ728234 |
| Cx. minor | Site 9, Yunnan | JQ728188; JQ728189 |
| Site 12, Hainan | JQ728374 | |
| Cx. infantulus | Site 8, Yunnan | JQ728267 |
| Cx. malayi | Site 5, Zhejiang | JQ728092 |
| Cx. richei | Site 5, Zhejiang | JQ728091; JQ728265 |
| Cx. peytoni | Site 9,Yunnan | JQ728379; JQ728380 |
| Cx. spiculosus | Site 8, Site 9, Yunnan | JQ728022; JQ728023; JQ728024 |
| Cx. bicornutus | Site 9, Yunnan | JQ728205 |
| Cx. fuscocephala | Site 8, Site 9, Yunnan | JQ728383; JQ728338; JQ728339; JQ728237; JQ728354 |
| Cx. hayashii | Site 9, Yunnan | JQ728264; JQ728266 |
| Cx. fuscanus | Site 9, Yunnan | JQ728037 |
| Cx. rubithoracis | Site 9, Yunnan | JQ728155 |
| Cx. infula | Site 9, Yunnan | JQ728199 |
| Cx. nigropunctatus | Site 8, Site 9, Yunnan | JQ728087;JQ728347;JQ728348; JQ728206; JQ728207; JQ728208 JQ728071; JQ728072 |
| Site 10, Hainan | JQ728298 | |
| Cx. pipiens | Site 2, Xinjiang | JQ728284; JQ728285; JQ728286 |
| Lab | JQ728036; JQ728035 | |
| Cx.pipiens quinquefasciatus | Site 6, Site 8–9, Yunnan | JQ728381;JQ728382;JQ728327 |
| Lab | JQ728021 | |
| Cx. pipiens pallens | Lab | JQ728040 |
| Cx. pallidothorax | Site 10, Hainan | JQ728057; JQ728058 |
| Cx. whitmorei | Site 9, Yunnan | JQ728304 |
| Cx.bitaeniorhynchus | Site 8–9, Yunnan | JQ728034; JQ728349; JQ728200 |
| Cx. sitiens | Site 10, Hainan | JQ728396; JQ728397; JQ728398; JQ728399; JQ728400; JQ728401; JQ728402 |
| Cx. mimulus | Site 9, Yunnan | JQ728244; JQ728245; JQ728246; JQ728247 |
| Site 5, Zhejiang | JQ728082; JQ728083; JQ728084; JQ728085; JQ728086 | |
| Cx. mimeticus | Site 9, Yunnan | JQ728150; JQ728151; JQ728152 |
| Site 5, Zhejiang | JQ728078 | |
| Cx. murrelli | Site 5, Zhejiang | JQ728079; JQ728080; JQ728081; JQ728017 |
| Cx. vagans | Site 1, Neimeng | JQ728101 |
| Cx. modestus | Site 1, Neimeng | JQ728108; JQ728109; JQ728110; JQ728111; JQ728112 |
| Site 3, Shanxi | JQ728375; JQ728376 | |
| Site 2, Xinjiang | JQ728296 | |
| Cx. tritaeniorhynchus | Site 6, Site 8–9, Yunnan | JQ728031; JQ728350; JQ728346; JQ728238 |
| Site 10–12, Hainan | JQ728059; JQ728060;JQ728061; JQ728062 | |
| Cx. gelidus | Site 9, Yunnan | JQ728366 |
| Ae. prominens | Site 9, Yunnan | JQ728239;JQ728240;JQ728145; JQ728146 |
| Ae. flavescens | Site 1, Neimeng | JQ728104; JQ728105; JQ728106; JQ728107 |
| Ae. dorsalis | Site 1, Neimeng | JQ728117; JQ728118; JQ728119; JQ728120 |
| Site 4, Qinghai | JQ728317 | |
| Site 2, Xinjiang | JQ728281; JQ728282; JQ728283 | |
| Ae. omorii | Site 9, Yunnan | JQ728272 |
| Ae. fengi | Site 5, Zhejiang | JQ728015 |
| Ae. albolateralis | Site 10, Hainan | JQ728394; JQ728395 |
| Site 7, Guizhou | JQ728365 | |
| Site 9, Yunnan | JQ728289 | |
| Ae. khazani | Site 7, Guizhou | JQ728364 |
| Ae. desmotes | Site 7, Guizhou | JQ728361 |
| Ae. tonkinensis | Site 7, Guizhou | JQ728360 |
| Ae. japonicus | Site 6, Yunnan | JQ728181 |
| Site 5, Zhejiang | JQ728068; JQ728069 | |
| Ae. albolineatus | Site 10, Hainan | JQ728308 |
| Ae. chrysolineatus | Site 9, Yunnan | JQ728271 |
| Ae. formosensis | Site 7, Guizhou | JQ728362; JQ728363 |
| Site 9, Yunnan | JQ728260; JQ728153 | |
| Ae. elsiae | Site 9, Yunnan | JQ728332 |
| Site 5, Zhejiang | JQ728093; JQ728094 | |
| Ae. togoi | Lab | JQ728038; JQ728039 |
| Ae. vexans | Site 11, Hainan | JQ728135; JQ728136; JQ728137; JQ728049 |
| Site 1, Neimeng | JQ728095; JQ728096;JQ728097; JQ728098; JQ728099 | |
| Site 9, Yunnan | JQ728392; JQ728393 | |
| Site 2, Xinjiang | JQ728287; JQ728288 | |
| Ae. kasachstanicus | Site 2, Xinjiang | JQ728276; JQ728277; JQ728278 |
| Ae. aegypti | Site 8, Yunnan | JQ728344; JQ728345 |
| Lab | JQ728041 | |
| Ae. novoniveus | Site 7, Guizhou | JQ728368; JQ728369 |
| Ae. dissimilis | Site 9, Yunnan | JQ728018; JQ728385; JQ728384; JQ728259; JQ728258 |
| Ae. craggi | Site 5, Zhejiang | JQ728142; JQ728143 |
| Ae. niveoides | Site 8, Yunnan | JQ728201 |
| Ae. annandalei | Site 8–9, Yunnan | JQ728202; JQ728227 |
| Ae. subsimilis | Site 8, Yunnan | JQ728226 |
| Ae. aureostriatus kanaranus | Site 9, Yunnan | JQ728225 |
| Ae. gilli | Site 9, Yunnan | JQ728215; JQ728216 |
| Ae. albopictus | Site 10–12, Hainan | JQ728063; JQ728064; JQ728065; JQ728066; JQ728067;JQ728299 JQ728300; JQ728301 |
| Site 7, Guizhou | JQ728192; JQ728193; JQ728194 | |
| Lab | JQ728019 | |
| Ae. subalbopictus | Site 7, Guizhou | JQ728198 |
| Ae. pseudalbopictus | Site 7, Guizhou | JQ728197 |
| Ae. albotaeniatus mikiranus | Site 9, Yunnan | JQ728248; JQ728249; JQ728250; JQ728251; JQ728154 |
| Ae. assamensis | Site 9, Yunnan | JQ728190; JQ728191 |
| Site 7, Guizhou | JQ728355; JQ728356 | |
| Ae. Vittatus | Site 10, Hainan | JQ728328 |
| Ae. mediolineatus | Site 12, Hainan | JQ728297 |
| Ae. malikuli | Site 9, Yunnan | JQ728324; JQ728325; JQ728326 |
| Ae. harveyi | Site 8–9, Yunnan | JQ728211; JQ728351; JQ728352; JQ728353 |
| Ar. flavus | Site 9, Yunnan | JQ728321; JQ728322; JQ728323 |
| Ar. durhami | Site 9, Yunnan | JQ728171; JQ728172; JQ728173; JQ728174; JQ728175; JQ728331 |
| Ar. subalbatus | Site 6, Yunnan | JQ728219 |
| Lab | JQ728033 | |
| Hz. proxima | Site 9, Yunnan | JQ728213; JQ728214 |
| Hz. menglianensis | Site 9, Yunnan | JQ728377 |
| Hz. lii | Site 9, Yunnan | JQ728252; JQ728253 |
| Hz. chengi | Site 9, Yunnan | JQ728255; JQ728257 |
| Hz. reidi | Site 8–9, Yunnan | JQ728182; JQ728183; JQ728184; JQ728254; JQ728256; JQ728217 |
| Ur. nivipleura | Site 9, Yunnan | JQ728221; JQ728222 |
| Ur. macfarlanei | Site 11, Hainan | JQ728128; JQ728129; JQ728130; JQ728131; JQ728132; JQ728133; JQ728134; JQ728016 |
| Site 2, Xinjiang | JQ728311 | |
| Ur. lutescens | Site 9, Yunnan | JQ728165; JQ728335; JQ728334 |
| Ur.bicolor | Site 9, Yunnan | JQ728223; JQ728224 |
| Ur. novobscura | Site 8, Yunnan | JQ728357 |
| Ur. jinhongensis | Site 9, Yunnan | JQ728228; JQ728229 |
| Tx. gravelyi | Site 8–9, Yunnan | JQ728144; JQ728341; JQ728330; JQ728210 |
| Tx. edwardsi | Site 9, Yunnan | JQ728337 |
| Tx. splendens | Site 8, Yunnan | JQ728340; JQ728126; JQ728127 |
| Tx. kempi | Site 8, Yunnan | JQ728329 |
| Tx. aurifluus | Site 9, Yunnan | JQ728204 |
| Tr. aranoides | Site 8–9, Yunnan | JQ728166; JQ728167; JQ728168; JQ728169; JQ728170; JQ728262; JQ728263 |
| Tr. tarsalis | Site 5, Zhejiang | JQ728014 |
| Site 7, Guizhou | JQ728371 | |
| Tr. similis | Site 7, Guizhou | JQ728367; JQ728320 |
| Ml. jacobsoni | Site 9, Yunnan | JQ728185; JQ728186; JQ728187; JQ728273 |
| Ml. genurostris | Site 10, Hainan | JQ728046 |
| Cq. crassipes | Site 9, Yunnan | JQ728179 |
| Site 10–11, Hainan | JQ728121; JQ728122; JQ728123; JQ728124; JQ728125; JQ728319 | |
| Cq. richiardii | Site 2, Xinjiang | JQ728309; JQ728310 |
| Cs. nipponica | Site 2, Xinjiang | JQ728316 |
| Site 1, Neimeng | JQ728100 | |
| Cs. annulata | Site 2, Xinjiang | JQ728312; JQ728313; JQ728314; JQ728315 |
| Ma. uniformis | Site 8–9, Yunnan | JQ728176; JQ728177; JQ728178 |
| Site 10–12, Hainan | JQ728055; JQ728056; JQ728047; JQ728048; JQ728318 | |
| Mi. luzonensis | Site 9, Yunnan | JQ728156; JQ728157 |
| Or. anopheloides | Site 12, Hainan | JQ728138; JQ728139; JQ728140 |
| To. houghtoni | Site 9, Yunnan | JQ728195; JQ728196; JQ728275 |
Sequence Analysis
Individual species were represented by one to eight individuals giving a total of 404 COI sequences, representing 122 species and subspecies. We identified and excluded 3 pseudogenes from further analyses by only selecting sequences without insertions, deletions and stop codons. COI sequences contain a large number of A+T pairs (average of 69% for all codons), particularly at the third codon position (93.4%) (Table S1). There was, however, no G content in Orthopodomyia anopheloides and Topomyia houghtoni at the third codon. As in the case of Drosophila [20], [21], this quite strong bias is apparently caused by the relative abundance of iso-accepting tRNA. All sequences contained less T in the first codon compared to the second. However, the A content of the first codon was higher than that of the second. The average R-value (transitions/transversions) was 0.7.
Neighbor-Joining (NJ) Tree
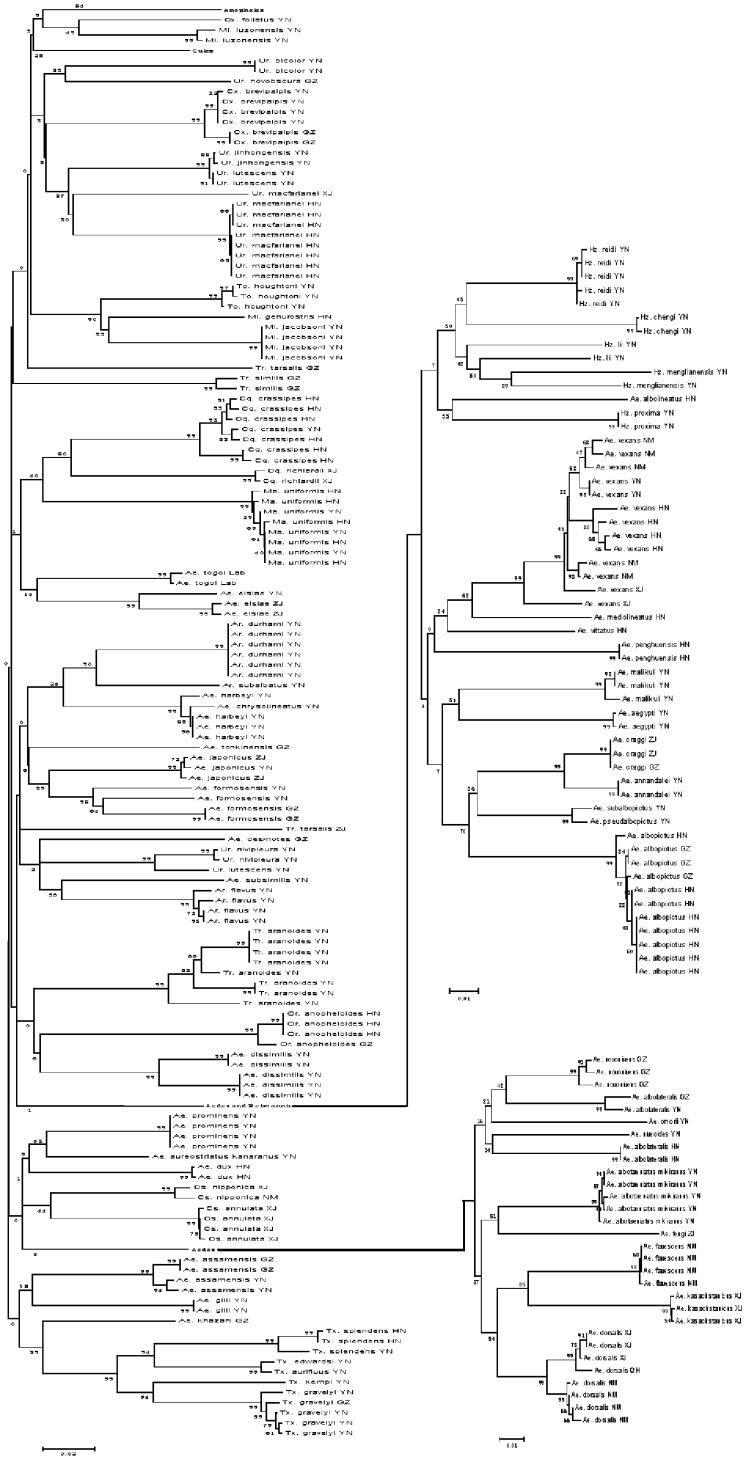
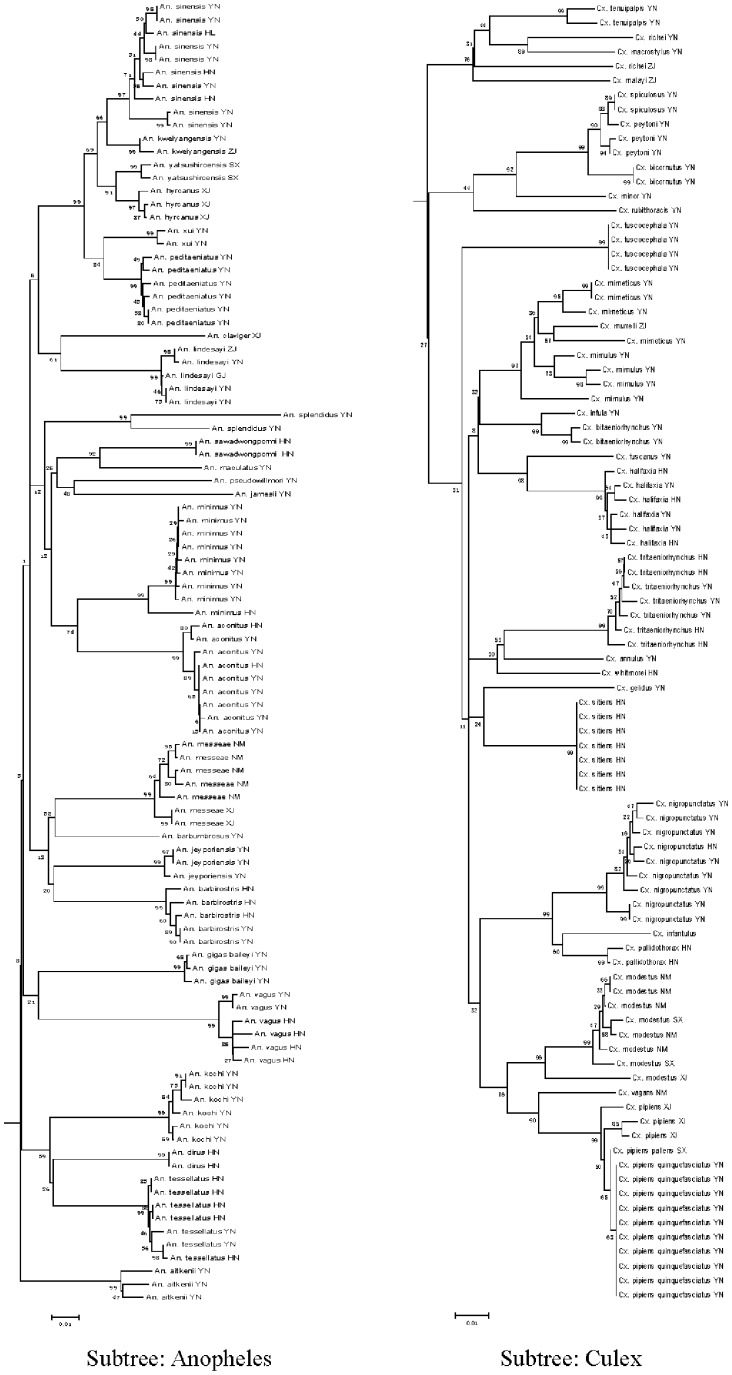
The Neighbor-Joining (NJ) tree method is conceptually related to clustering, but without the assumption of clock-like behavior [22]. COI gene fragments accurately revealed species boundaries and provided a clear phylogenetic signal (Figs. 2 and 3). Most of the major branches on the tree represent distinct taxonomic groups, including all genera and subgenera. Moreover, specimens of the same species always grouped closely together, regardless of collection site, and, except for some specimens from Hainan Island, no obvious geographic differences in sequences within the same species were found.
Figure 2. NJ phylogenetic tree based on Kimura two-parameter genetic distances of COI gene sequences of mosquitoes prevalent in China.
Sequence analysis was conducted using MEGA version 4.0 software with 1000 replications. Most major branches on the tree represent recognized groups, including all genera and subgenera except Anopheles and Culex which comprise separate subtrees and are shown in detail in Fig.3.
Figure 3. Two distinct sub-trees comprised of Anopheles and Culex in the NJ phylogenetic tree (Fig. 2).
Combining NJ tree and bootstrap analysis is the most appropriate method for evaluating phylogenetic trees using distance methods [23]. Nodes linking sequences of individuals of the same species had a high bootstrap value (98%–99%) whereas some linking sequences of geographically different individuals had low bootstrap values (6%–99%).
Species Boundaries
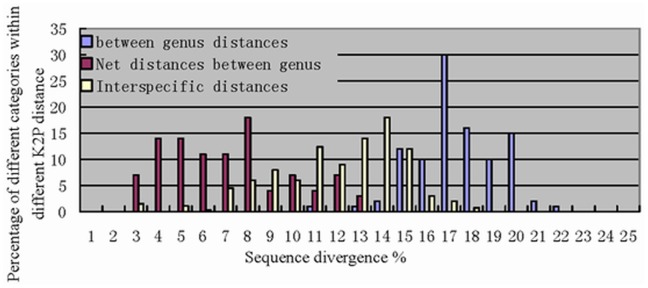
All species had a distinct set of COI sequences. Excluding the Culex mirneticus subgroup and the species listed in Table 2 (see Discussion section), most (98%) conspecific sequences showed <2% (range = 0% to 1.67%), whereas >98% of interspecific divergence was in specimens with >2% K2P divergence (range = 2.3% to 21.8%). Sequence divergence was even higher among species in different genera, ranging from 10.9% to 21.8% (Fig. 4).
Table 2. Intraspecific K2P distance, transversion distance, and morphological characteristics of some mosquitoes.
| Species | K2P distance (%) | Transversion distance (%) | Variation in morphological characters |
| A. dorsalis | 2.98 | 1.11 | stripe shape and color of metascutellum |
| A. vexans | 4.71 | 1.86 | mesopleuron and urotergite |
| T. aranoides | 5.72 | 1.29 | stable |
| T. splendens | 2.79 | 1.29 | stable |
| C. modestus | 4.71 | 1.67 | larvae chest hair and male terminalia |
| C. crassipes | 3.57 | 0.37 | stable |
| A. sinensis | 2.61 | 0.18 | stable |
Figure 4. Pairwise comparisons between COI sequences among mosquito species separated into three categories; interspecific distances, between gene distances and net distances between genera.
All sequences were grouped with MEGA software, each group includes all species of a particular genus.
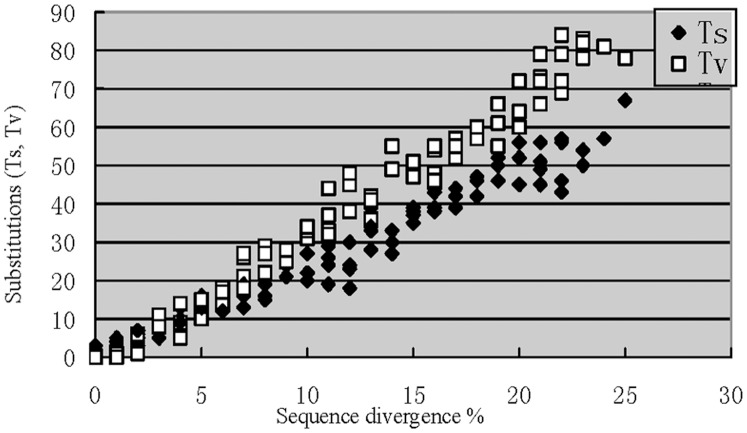
Transition and transversion distances varied consistently with sequence divergence (Fig. 5). Transition distance was significantly greater than transversion distance when sequence divergence was <2%. However, transversion distances increased slowly with sequence divergence to eventually exceed transition distances at K2P divergence of ≥6%. Both transition and transversion distances then decreased until K2P divergence reached about 15%. The relationship between the transversion distance, sequence divergence, and morphological characteristics are shown in Tables 2 and 3.
Figure 5. The numbers of COI transitions (ts) and transversions (tv) plotted against sequence divergence.
Table 3. Interspecific K2P distance, transversion distance, and morphological characters of some mosquitoes.
| Species | K2P distance (%) | Transversiondistance (%) | Variation in morphological characters |
| Ae. craggi and Ae. annandalei | 2.99 | 0.37 | male terminalia |
| Cx. minor and Cx. spiculosus | 1.86 | 0.37 | male antenna and terminalia |
Discussion
Accuracy of COI
The primary function of DNA barcoding is accurate species identification. We found that COI sequence differences among congeneric mosquito species were approximately 30 times higher than the average differences within species. Moreover, more than 98% of COI fragments had clear interspecific boundaries, a result consistent with the results of other authors [13]. The average conspecific K2P divergence in this study, 0.39%, is similar to values reported for fish species in Australia [24] and slightly higher than those reported for North American birds (0.27%) [25] and moths (0.25%) [10]. It is slightly less than the K2P divergence value reported for Canadian mosquitoes (0.55%) [13].
Transversion Distance and Speciation
Mitochondrial DNA (mtDNA) functions as a molecular clock in that transversions accumulate in a linear fashion over time [26], [27]. Comparison of the molecular and morphological data indicates that the number of transversions may raise to about 7 value without apparent or detectable changes in morphology. (Fig. 5). Transition distance was significantly greater than transversion distance when sequence divergence was below 2% at which level there were almost no morphological differences between specimens. At higher levels of sequence divergence transversion distances slowly increased, eventually exceeding transition distances when sequence divergence reached 6%. Morphological differences were undetectable when sequence divergence was about 2% but were distinct when this reached 6%. Transversion distances increased steadily at sequence divergence levels of 6% to 15% at which level plesiomorphy also first became evident. Plesiomorphy stabilized at sequence divergence of 15%. In addition, the vast majority of intraspecific distances occurred between sequence divergence levels of 6% and 15% whereas most intergeneric distances occurred from 15% to 20% (Fig. 4). Very few intraspecific, and no intergeneric, distances occurred between sequence divergence levels of 2% and 6%.
We found that transversion distances indicated a clear boundary between species. The transversion distance between most species was <1.1% at sequences divergence values of less than 2%. There were, however, some exceptions; although the transversion distance between two plesiomorphous species was usually <1.1% (Table 3), some species with anomalous intraspecific COI sequences divergences >2% (Table 2) had intraspecific transversion distances >1.1%. This suggests the presence of cryptic species, which, if confirmed, in turn suggests that transversion distances may be a useful supplement to barcoding information in species identification. Further research on the use of transversion as an additional index of taxonomic similarity is recommended.
Molecular Data Versus Morphology
Sequence divergence values of 14% to 16% were indicative of either interspecific or intergeneric differences. There are two possible reasons for this; temporary substitution saturation of the COI fragment and the limitations of morphological identification.
We found some cases of high intraspecific sequence divergence among Aedes dorsalis, Aedes vexans, Culex modestus, Tripteroides aranoides, and Toxorhynchites splendens (Table 2). Although the degree of niche separation within these species remains unclear, this result suggests the existence of cryptic species. We also detected intraspecific sequence divergence slightly greater than the 2% threshold within Coquillettidia crassipes and Anopheles sinensis (Table 2). Although no morphological differences within these species were observed, differences in feeding habits and habitat have been documented within Anopheles sinensis populations[19]. This, together with the >2% level of COI sequence divergence, suggests the presence of cryptic species [28]. Some cases of low interspecific sequence divergence were found among some pairs of species (Table 3), including Aedes craggi and Aedes annandalei, as well as Culex spiculosus and Culex minor. Although there is no evidence of niche separation between these species, slight morphological differences were observed. This suggests that the taxonomic status of these species should be re-confirmed. Although few doubt that mtDNA barcodes are a valuable molecular tool for matching unidentified specimens to described taxa, there has been relatively little use of barcodes to delimit species [29]. More research on rDNA, morphology, biogeography and ethology are required to improve the applicability of barcoding to species-level taxonomy.
Culex neomimulus was previously classified as Culex mimulus in the Culex mirneticus group [30]. Although our COI data supports the previous view, we found that anomalous COI sequence divergence values were relatively common in the Culex mirneticus group with some morphologically distinct specimens having similar barcodes. This could be due to infection with the Wolbachia bacteria. The maternally inherited Wolbachia bacteria causes a loss of haplotype diversity in populations by inducing a selective sweep of the initially infected individual's haplotype through a population. We detected Wolbachia infection in Culex mimulus so it’s possible that this may also occur in this species. Although Smith et.al concluded that the presence of Wolbachia DNA in total genomic extracts is unlikely to compromise the accuracy of the DNA barcode library, this is a complex problem that requires further investigation [31].
Pseudogenes
The presence of pseudogenes can affect the accuracy of barcoding identification but, since their incidence was <1%, their influence on our data was presumably small. The distinctive characteristics of the COI gene (no insertions, deletions and stop codons) allowed pseudogenes to be easily identified and excluded from the sequences we obtained. Although the leakage of paternal mtDNA may influence the results of barcoding this phenomenon is only occasionally (<0.004%) found in higher animals.
A total of three pseudogenes were detected. For instance, one of the samples of Aedes dissimilis collected from the same area exhibited high interspecific sequence (3.74%) and transversion divergence (3.00%). A total of 12 different protein sequence sites were observed, which is very rare in the Culicidae. The substitution rate at nucleotide codons 1, 2, and 3 was 1∶2:2, very different to the average of 5∶1:18. We also amplified the pseudogenes of Uranotaenia lutescens and Culex halifaxia, which have insertions and deletions, respectively. The sequence divergence between pseudogenes and COI fragments in Culex halifaxia was 10.93% and the substitution rate at nucleotide codons 1, 2, and 3 was 5∶4:11. The divergence time formula of mtDNA and pseudogenes [32] suggests that the nuclear transfer event occurred 500 million years ago in Culex halifaxia and 170 million in Aedes dissimilis. We found an insertion site at 54 bp in the sequence of Uranotaenia lutescens, with a substitution rate at nucleotide codons 1, 2, and 3 of 7∶1:18. Two different protein sequence sites were also observed. These abnormal phenomena disappeared when the inserted site was deleted manually. Therefore, these anomalous sequences likely caused by the frameshift mutations of PCR.
Overall, DNA-based species identification systems depend on the ability to distinguish intraspecific from interspecific variation. This analysis of 404 COI sequences from 15 mosquito genera and 122 species and subspecies indicates that >98% of specimens formed distinctive clusters and that barcode divergence was relatively large between these groupings. Although it has limitations, DNA barcode technology has several advantages over traditional taxonomic methods as a tool for species identification. For example, it is unaffected by morphological variation between different life cycle stages. Another benefit is that it allows the homogenization, or calibration, of the taxonomic units identified in different areas. DNA barcode technology generally produces accurate results thereby greatly reducing the need for experienced taxonomists.
In summary, this study provides the first COI barcodes for mosquitoes in China and provides further evidence of the effectiveness of DNA barcoding in identifying recognized species. An insufficient number of specimens prevented in-depth investigation of sibling species complexes but we plan to address this area in the future. Care must be taken to exclude pseudogenes from COI databases to ensure the accuracy of molecular identification. COI databases also need to include specimens of the same species collected from different geographical locations in order to determine the extent of intraspecific variation. A complete evaluation of the effectiveness of DNA barcoding for the Culicidae can be achieved through multinational research.
Materials and Methods
Ethics Statement
No specific permits were required for this study. All experiments were conducted within state-owned land in China. Therefore, the local ethics committee deemed that approval was unnecessary.
Mosquito Collections
Mosquito specimens used for constructing DNA barcodes were collected from different Chinese Provinces in 2009 and 2010. Details on specimens collected are provided on Fig. 1 and Table. 1. Larval and adult mosquitoes were collected in the field. Adults were sampled with CO2-baited miniature light traps. Larvae were reared individually and associated larval and pupal skins were mounted. All specimens were identified using standard taxonomic keys [19].
Target Gene Preparation
Total DNA (100 µL to 150 µL) was extracted from each specimen using the Universal Genomic DNA Extration Kit (Invitrogen). PCR was performed to amplify the 5′ COI region of mtDNA using the following cycle: An initial denaturation of 1 min (94°C) followed by five cycles of 94°C for 40 s (denaturation), 45°C for 40 s (annealing), and 72°C for 1 min (extension); 30 cycles of 94°C for 40 s (denaturation), 51°C for 40 s (annealing), 72°C for 1 min (extension) and a final extension at 72°C for 5 min. PCR cocktails were made as follows: A 50 µL solution comprised of 0.3 µL Taq DNA polymerase (5 U/µL), 5 µL of 10×PCR buffer, 5 µL of 2 mmol/L dNTP, 2 µL of 10 µmol/L each of the forward and reverse primers, 5 µL of template DNA and sufficient ddH2O to make up to 50 µL. The primer pairs LCO1490 and HCO2198 [33] were used to amplify a 650 bp fragment of COI. The amplified fragments were run on a 1% agarose gel to check the integrity of the fragments after which the PCR product was purified with a normal PCR purification kit (Tiangen). Both reads (forward as well as reverse primer) were done.
Data Analysis
DNA sequences were aligned using Clustal X [34]. Sequence analysis and Ts/Tv calculation was conducted using MEGA version 4.0 software [14]. Sequence divergence and Ts, Tv distance among individuals was quantified using the Kimura two-parameter distance model [35]. An NJ tree of K2P distances was created to provide a graphic representation of the clustering pattern among different species [36].
Supporting Information
Sequence divergence and nucleotide composition for the mosquito genera. The frequencies of nucleotides in sequence are presented as the total average values for all Condon positions and for each condon position separately with the accuracy to tenths of a percent. (*) Figures in brackets are the number of mosquito species used to estimates of sequence divergence for the genus
(PDF)
Funding Statement
This work was supported by the Infective Diseases Prevention and Cure Project of China (NO: 2008ZX10402) and (NO: 2012ZX10004219).
References
- 1.Edwards FW (1932) Genera insectorum: Diptera : Fam. Culicidae: Verteneuil & Desmet.
- 2.Feng LC (1938a) A critical review of literature regarding the records of mosquito in China. Part I. Subfamily Culicinae, tribe Anophelini; Part II. Subfamily Culicinae, tribe Megarhinni and Culicini. Pek nat Hist Bull 12: 169–181; 285–318.
- 3. Bortolus A (2008) Error cascades in the biological sciences:the unwanted consequences of using bad taxonomy in Ecology. Ambio 37: 114–118. [DOI] [PubMed] [Google Scholar]
- 4. Hanel R, Sturmbauer C (2000) Multiple recurrent evolution of trophic types in northeastern atlantic and mediterranean seabreams (Sparidae, Percoidei). J Mol Evol 50: 276–283. [DOI] [PubMed] [Google Scholar]
- 5. Herran de la R, Rejon C, Rejon M, Garrido-Ramos M (2001) The molecular phylogeny of the Sparidae (Pisces, perciformes) based on two satellite DNA families. Heredity 87: 691–697. [DOI] [PubMed] [Google Scholar]
- 6. Phuc H, Ball A, Son L, Hanh N, Tu N, et al. (2003) Multiplex PCR assay for malaria vector Anopheles minimus and four related species in the Myzomyia Series from Southeast Asia. MED VET ENTOMOL 17: 423–428. [DOI] [PubMed] [Google Scholar]
- 7. Gao Q, Beebe N, Cooper R (2004) Molecular identification of the malaria vectors Anopheles anthropophagus and Anopheles sinensis (Diptera: Culicidae) in central China using polymerase chain reaction and appraisal of their position within the Hyrcanus Group . J Med Entomol 41: 5–11. [DOI] [PubMed] [Google Scholar]
- 8. Tautz D, Arctander P (2003) A plea for DNA taxonomy. TRENDS ECOL EVOL 18: 70–74. [Google Scholar]
- 9. Tautz D, Arctander P (2002) DNA points the way ahead in taxonomy. Nature 418: 479. [DOI] [PubMed] [Google Scholar]
- 10. Hebert P, Cywinska A, Ball S, deWaard J (2003a) Biological identifications through DNA barcodes. ProcR Soc Lond B 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hebert P, Ratnasingham S, deWaard J (2003b) Barcoding animal life: cytochrome c oxidase subunit 1 divergence,among closely related species. Proc R Soc Lond B (Suppl) 270: S96–S99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Remigio E, Hebert P (2003) Testing the utility of partial COI sequences for phylogenetic estimates of gastropod relationships. Mol Phylogenet 29: 641–647. [DOI] [PubMed] [Google Scholar]
- 13. Cywinska A, Hunter FF, Hebert PDN (2006) Identifying Canadian mosquito species through DNA barcodes. MED VET ENTOMOL 20: 413–424. [DOI] [PubMed] [Google Scholar]
- 14. Kumar NP, Rajavel AR, Natarajan R, Jambulingam P (2007) DNA Barcodes can distinguish species of Indian mosquitoes(Diptera:Culicidae). J Med Entomol 44: 1–7. [DOI] [PubMed] [Google Scholar]
- 15.Monaghan M, Balke M, Gregory T, Vogler A (2005) DNA-based species delineation in tropical beetles using mitochondrial and nuclear markers. Phil Trams R SocB 360: 1 925–921 933. [DOI] [PMC free article] [PubMed]
- 16. Brown JS (2003) Miller (2003) Studies on new Guinea moths description of a new species of Xenothctis meyrick(Lepidoptera:Torthricidae:Archipini). Proc Entomol Soc Wash 105: 1043–1050. [Google Scholar]
- 17. Hebert P (2004b) Penton (2004b) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator . Proc Natl Acad Sci 101: 14812–14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Burns JM, Janzen DH, Hajibabaei M, Hallwachs W, Hebert PDN (2007) DNA barcodes and cryptic species of skipper butterflies in the genus Perichares in Area de Conservació n Guanacaste, Costa Rica. PNAS 105: 6350–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baolin L (1997) FAUNA SINICA, INSECTA Vol.9, Diptera: Culicidae I, II. Beijing: Science Press.
- 20. Akashi H (1994) Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics 136: 927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moriyama EN, Powell JR (1997) Codon usage bias and tRNA abundance in Drosophila . Mol Bilo Evol 45: 514–523. [DOI] [PubMed] [Google Scholar]
- 22. Moftah M, Aziz SHA, Elramah S, Favereaux A (2011) Classification of sharks in the Egyptian Mediterranean waters using morphological and DNA Barcoding approaches. PLos ONE 6: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evol 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 24.Ward R, Zemlak T (2005) DNA barcoding Australia's fish species. Plut trans R Soc B: 1–11. [DOI] [PMC free article] [PubMed]
- 25. Hebert P, Stoeckle M, Zemlak T, Francis C (2004a) Identification of Birds through DNA Barcodes. PLos Biology 2: e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Papadopoulou A, Anastasiou I, Vogler AP (2010) Revisiting the insect mitochondrial molecular clock: the mid-aegean trench calibration. Mol Bilo Evol 27: 1659–1672. [DOI] [PubMed] [Google Scholar]
- 27. Marko PB, Moran AL (2002) Correlated evolutionary divergence of egg size and a mitochondrial protein across the isthmus of panama. Evolution 56: 1303–1309. [DOI] [PubMed] [Google Scholar]
- 28. Baker RJ, Bradley RD (2006) Speciation in mammals and the genetic species concept. J MAMMAL 87: 643–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lohse K (2009) Can mtDNA barcodes be used to delimit species? a response to Pons, et al. (2006). SYSTEMATIC BIOL 58: 439–442. [DOI] [PubMed] [Google Scholar]
- 30. Sirivanakarn S (1976) A revision of the subgenus Culex on the oriental region (Diptera:Culicidae). ContribAmentInst 12: 1–272. [Google Scholar]
- 31. Smith MA, Bertrand C, Crosby K, Eveleigh ES, Fernandez-Triana J, et al. (2012) Wolbachia and DNA Barcoding insects: patterns, potential, and problems. PLos ONE 7: e36514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li W, Gojobori T, Nei M (1981) Pseudogenes as a paradigm of neutral evolution. Nature 292: 237–239. [DOI] [PubMed] [Google Scholar]
- 33. Flomer O, Black M (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech 3: 294–299. [PubMed] [Google Scholar]
- 34. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kimura M (1980) A simple method for estimating evolutionary rates of base substitution through comparative studies of nucleotide sequence. J MOL EVOL 16: 111–120. [DOI] [PubMed] [Google Scholar]
- 36. Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic tree. MOL BIOL EVOL 4: 406–425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence divergence and nucleotide composition for the mosquito genera. The frequencies of nucleotides in sequence are presented as the total average values for all Condon positions and for each condon position separately with the accuracy to tenths of a percent. (*) Figures in brackets are the number of mosquito species used to estimates of sequence divergence for the genus
(PDF)