Abstract
The assembly of actin monomers into filaments and networks plays vital roles throughout eukaryotic biology, including intracellular transport, cell motility, cell division, determining cellular shape, and providing cells with mechanical strength. The regulation of actin assembly and modulation of filament mechanical properties are critical for proper actin function. It is well established that physiological salt concentrations promote actin assembly and alter the overall bending mechanics of assembled filaments and networks. However, the molecular origins of these salt-dependent effects, particularly if they involve nonspecific ionic strength effects or specific ion-binding interactions, are unknown. Here, we demonstrate that specific cation binding at two discrete sites situated between adjacent subunits along the long-pitch helix drive actin polymerization and determine the filament bending rigidity. We classify the two sites as “polymerization” and “stiffness” sites based on the effects that mutations at the sites have on salt-dependent filament assembly and bending mechanics, respectively. These results establish the existence and location of the cation-binding sites that confer salt dependence to the assembly and mechanics of actin filaments.
Keywords: ion-linkage, structural bioinformatics, persistence length, polyelectrolyte
The polymerization of the protein actin into double-stranded helical filaments powers many eukaryotic cell movements and provides cells with mechanical strength and integrity (1–4). Filament formation is favored when the total actin concentration exceeds the critical concentration (Cc) for assembly-defined as the monomer concentration at steady state for ATP-actin, or the dissociation constant for the reversible-equilibrium binding reaction of monomer binding to ADP-actin filament ends. Accordingly, the Cc of ADP-actin is linked to the filament subunit interaction free energy such that lower Cc values reflect greater thermodynamic stability (5).
The effects of solution ionic conditions on the assembly and stability of actin filaments have been investigated for several decades (6–12). The actin Cc and (monomer and filament) conformation depend on the nucleotide-associated divalent cation (Ca2+ or Mg2+) as well as the type and concentration of ions in solution (6, 7, 13–15), a behavior shared among characterized actins and their bacterial homologs (16). However, it is not firmly established if these salt effects on actin filament assembly and mechanics originate from nonspecific ion effects (e.g., electrostatic screening, counterion condensation, etc.) and/or specific ion binding interactions, potentially at discrete sites. Identification of saturable cation binding sites with different affinities favors specific and discrete binding sites on monomers (8–10, 17), but the location of these sites and their contributions to filament assembly and stiffness are unknown.
Here we identify distinct cation-binding sites at subunit interfaces that regulate actin filament assembly and rigidity. Site-specific substitution of a charged amino acid at one of the sites modulates the salt dependence of filament flexural rigidity, while substitution at the second site alters salt-dependent filament assembly. These studies provide a structural and thermodynamic basis for cation-linked actin filament assembly and bending mechanics.
Results and Discussion
Specific Cation-Binding Interactions Promote Actin Polymerization.
To determine if general (i.e., nonspecific) electrostatic screening or specific cation-binding interactions dominate the effects of salts on actin filament assembly, we evaluated how the cation dependence of the Cc compares to predictions made by general and specific ion-binding theories. We focused on the abundant intracellular cations K+ and Mg2+, as well as Ca2+ and Na+. We utilized ADP-actin to eliminate cation effects on ATP hydrolysis and phosphate release, and because filaments assembled from ATP-actin monomers are comprised of > 95% ADP subunits at steady state and therefore ADP-actin monomers dominate the mechanical behavior of actin filaments. For our Cc measurements, we assembled ADP-actin filaments from ADP-actin monomers to eliminate ATP hydrolysis and mixed nucleotide state effects at filament ends, which determine the Cc.
The ADP-actin Cc depends on the concentration and type of cations in solution (Figs. S1 and S2). This observation indicates that the actin filament thermodynamic stability depends on solution cations (18–20), since the Cc of ADP-actin reflects the free energy associated with filament subunit incorporation (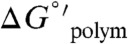 ) according to
) according to 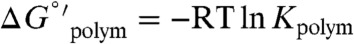 , where Kpolym is a macroscopic overall equilibrium constant for incorporation of monomers into filaments. We emphasize that Kpolym is an “observed” binding constant under given experimental conditions (e.g., salt concentration), defined only by the reaction between monomers and filament ends, and does not explicitly account for contributions from linked equilibria such as ion binding. We note that this relation holds only for ADP-actin since it assembles following a reversible equilibrium reaction, and therefore Cc reflects the (average) affinity of ADP-actin monomers for filament ends. Assembly of ATP-actin, on the other hand, has additional linked equilibria (e.g., ATP hydrolysis and Pi release), and the barbed and pointed filament ends vary in nucleotide composition (21, 22). The Cc of ATP-actin monomers is therefore more accurately described as the monomer concentration once polymerization has reached steady-state.
, where Kpolym is a macroscopic overall equilibrium constant for incorporation of monomers into filaments. We emphasize that Kpolym is an “observed” binding constant under given experimental conditions (e.g., salt concentration), defined only by the reaction between monomers and filament ends, and does not explicitly account for contributions from linked equilibria such as ion binding. We note that this relation holds only for ADP-actin since it assembles following a reversible equilibrium reaction, and therefore Cc reflects the (average) affinity of ADP-actin monomers for filament ends. Assembly of ATP-actin, on the other hand, has additional linked equilibria (e.g., ATP hydrolysis and Pi release), and the barbed and pointed filament ends vary in nucleotide composition (21, 22). The Cc of ATP-actin monomers is therefore more accurately described as the monomer concentration once polymerization has reached steady-state.
The value of Kpolym does not scale with the solution ionic strength independent of ion type (Fig. S3), indicating that the effects of various salts on actin filament thermodynamic stability are specific and not purely ionic-strength effects. Hence, salt effects on actin polymerization reflect differential binding—to discrete binding sites and/or condensation/polyelectrolyte effects (23, 24)—and coupled salt-dependent conformational changes. We therefore interpret the salt dependence of Cc according to established formalisms of linked binding equilibria (25).
The following scheme defines the overall reaction for reversible ion-linked actin polymerization:
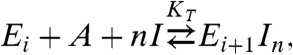 |
[1] |
where Ei and Ei+1 are the ends of filaments comprised of i or i + 1 subunits, A is a free actin monomer, n is the net number of ions (I) taken up per incorporated filament subunit, and KT is the overall thermodynamic equilibrium constant for subunit addition that accounts for the linked ion binding reaction, defined by:
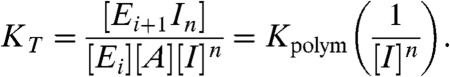 |
[2] |
Eq. 2 can be expressed in a linear form by taking the log of both sides and rearranging:
| [3] |
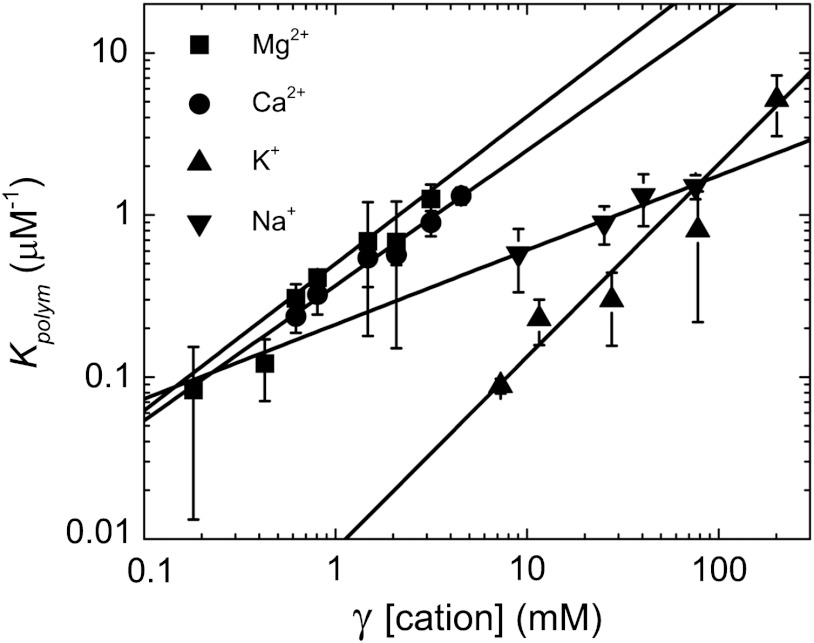
thereby permitting the net change in number of actin-associated ions (n) upon polymerization to be estimated from the slope of the double log plot of Kpolym versus cation activity, given by the product of [cation] and the ionic strength-dependent activity coefficient (γ) (26) (Fig. 1). Actin filaments are linear polymers with net negative charge, so we expect that the salt effects here are due to cation interaction with the polymer. Accordingly, we restrict analysis and discussion of the linkage of different cations in the presence of identical counterions.
Fig. 1.
Specific cation binding drives actin polymerization. Linear fits of the activity coefficient (γ) corrected cation concentration dependence of Kpolym (rabbit skeletal muscle actin, 5% pyrene labeled) yields slopes of 1.18 ± 0.02, 0.46 ± 0.10, 0.91 ± 0.09, and 0.83 ± 0.02, for K+, Na+, Mg2+, or Ca2+, respectively. Uncertainty bars represent the standard error (SEM).
The cation dependence of Cc indicates that approximately 1 net Mg2+, Ca2+, or K+ associates per actin subunit upon incorporation into filaments (Fig. 1). This value agrees with a previous study showing that binding of a single low-affinity Mg2+ is linked to actin polymerization induced by MgCl2 (15). Approximately half as many net Na+ are taken up per actin monomer during polymerization. The difference between K+ and Na+ is consistent with ionic species-specific effects that are not Coulombic. The intercept value of Kpolym extrapolated to 1M cation is related to the intrinsic binding free energy of an actin subunit and associated cation with a filament end (27, 28). These values are not identical for all cations evaluated, suggesting that filaments assembled with different cationic species have variable thermodynamic stabilities and salt-dependent conformational distribution(s) (6, 7, 20, 29, 30).
Cation Binding Stiffens Actin Filaments.
Electrostatic potential changes due to both screening and ion binding play a critical role in the structure and mechanical properties of charged biopolymers including DNA and RNA (25, 27, 28). For example, the partial neutralization of phosphates with monovalent cations lowers the rigidity of DNA (31). Considering that actin filaments are polyampholytes that behave like negatively charged linear polyelectrolytes (32, 33), it is conceivable that screening and/or cation binding play an important role in actin filament flexural rigidity. However, the molecular mechanism of cation effects on actin filament mechanics remains elusive.
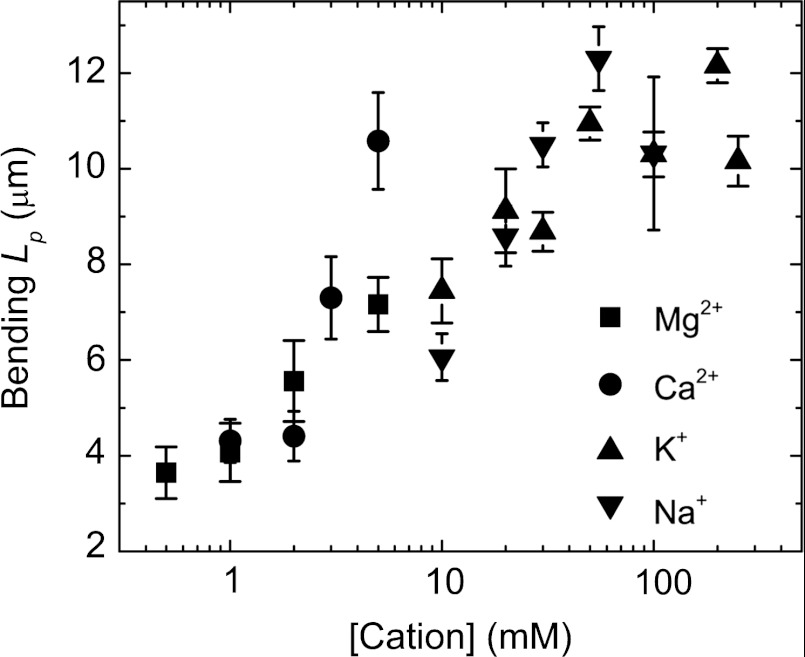
We evaluated the salt dependence of actin filament flexural rigidity by directly visualizing filaments undergoing thermally driven fluctuations in shape and calculating their bending persistence lengths (Lp) from the average angular correlation along their contour length (34, 35). Because filament contour lengths are comparable to Lp, small changes in filament rigidity yield readily detectable changes in filament shape (Fig. S4). The actin filament-bending Lp increases approximately 4-fold from 3.5 μm to 12.7 μm over the range of salt concentrations evaluated (0.5–5 mM for divalent cations, and 10–250 mM for monovalent cations; Fig. 2), indicating that cations stiffen actin filaments. The stiffening effects of divalent cations continue to increase at concentrations higher than those needed for polymerization (Fig. S5). The two distinct [cation] regimes over which Lp and Cc vary suggest that additional cation interactions at sites distinct from those required for polymerization modulate the filament bending rigidity.
Fig. 2.
Cation binding stiffens actin filaments. Bending persistence length (Lp) of actin filaments (rabbit skeletal muscle actin, Alexa 488 labeled) in K+, Na+, Mg2+, or Ca2+. Uncertainty bars represent SEM.
The effects of salt on actin filament Lp could result from electrostatic screening as addressed by the Odijk-Skolnick-Fixman (OSF) (36, 37) and Manning theories (38) and/or specific cation binding. The Lp of polyelectrolytes, including DNA (38–40), diminishes with salt (< 10 mM Na+ or < 1 mM Mg2+). In marked contrast, the Lp of actin filaments increases with [salt] (Fig. 2), indicating that actin filaments behave differently from DNA. It is possible that the salt-dependent increase in actin filament Lp arises from neutralization or screening of repulsive interactions, thereby increasing the subunit interface area and/or intersubunit interaction energy (41). However, data presented below support a mechanism in which cation binding to a discrete site helps regulate the bending stiffness of actin filaments.
Structural Bioinformatics Predicts Two Discrete Actin Filament-Specific Cation-Binding Sites.
The salt dependence of actin filament thermodynamic stability (Cc) and bending rigidity (Lp) is best described by specific cation-binding interactions rather than general electrostatic screening effects. We therefore utilized a structural bioinformatics approach (SI Text) to predict the location of actin filament-specific cation-binding sites. We tested our predictions with experiments where site-specific mutations were engineered within the most highly ranked predicted sites (SI Text).
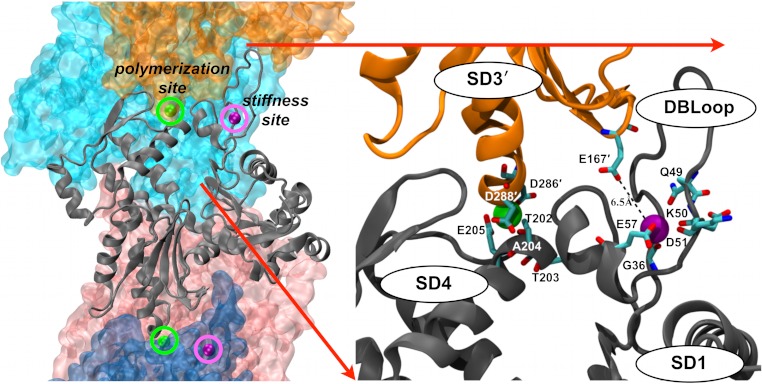
We employed WebFEATURE (42) with our own customized scripts (Materials and Methods and SI Text) to identify potential filament-specific cation binding sites distinct from the low affinity cation binding sites of monomers (8). Our procedure (SI Text) predicts two filament-specific cation binding sites on actin. Both sites are formed by residues from two adjacent filament subunits, such that coordinated cations are specifically positioned between neighboring subunits along the long-pitch helix (Fig. 3).
Fig. 3.
Structural bioinformatics predicts two classes of discrete actin filament-specific cation-binding sites. The actin filament on the left (PDB ID 3MFP “biological assembly”) is oriented with the barbed end at the bottom, and is colored by subunit. The central subunit is rendered as a cartoon showing the location of the predicted cation-binding sites. “Polymerization” sites (green spheres) have the highest prediction score from comparing WebFEATURE cation-binding site prediction results between the F-actin monomer (3MFP) and F-actin polymer (3MFP “biological assembly,” Materials and Methods). “Stiffness” sites (purple spheres) have the highest prediction score from comparing WebFEATURE cation-binding site prediction results between the G-actin monomer (PDB ID 1J6Z) and F-actin monomer (3MFP).
We refer to the two predicted actin filament-specific cation binding sites as “polymerization” and “stiffness” sites based on the following observations. The polymerization site is located between subdomains 3 and 4 of adjacent filament subunits. Residues comprising our predicted polymerization site (rendered as ball and stick in Fig. 3) are conserved among the vast majority of actins (43). Mutations within this predicted site can render cytoplasmic actin nonpolymerizable (44) and can be lethal in S. cerevisiae (herein referred to as yeast) (45). We note that an acidic residue within 3 Å of the polymerization site (Asp288) is predicted to have a pKa shifted from approximately 4 to 7.1 [using PROPKA software (46)] when incorporated within a filament (SI Text). Consequently, protonation at this site may account for the filament stabilizing effects of decreasing the solution pH (47). In this manner, protonation behaves analogous to cation binding.
The stiffness site is comprised largely by residues within the DNAse I binding loop (DB-loop) of subdomain 2 but also includes Glu167 within subdomain 3 of an adjacent subunit. Most actins have an acidic residue at position 167 in the stiffness site (43). However, Ala occupies this site in yeast actin and yeast actin filaments are more compliant in bending than their vertebrate counterparts (35).
Substitutions at Predicted Sites Modulate Cation-Dependent Rigidity and Polymerization.
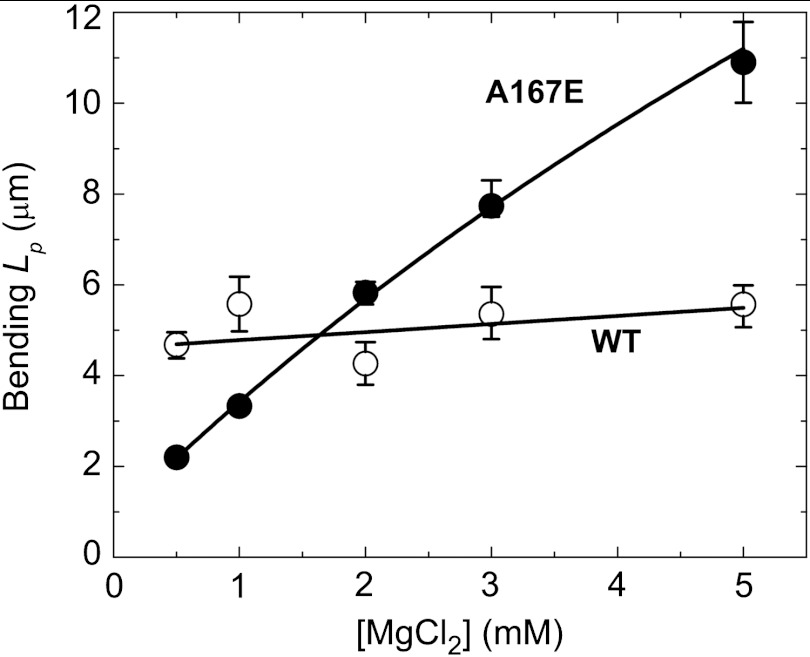
The lack of an acidic residue (Glu167) in the stiffness site of yeast actin filaments suggests that weak cation binding and occupancy at this site render them more flexible than vertebrate filaments. Consistent with this hypothesis, the bending rigidity of wild-type (wt) yeast actin filaments depends weakly, if at all, on [Mg2+] (Fig. 4). In contrast, yeast actin filaments engineered with Glu167 at the stiffness site (A167E) display a strong Mg2+-dependent rigidity (Fig. 4) similar to vertebrate actin filaments (Fig. 2), without affecting the Cc for polymerization (30). We note that despite sharing salt-dependent rigidity, the Lp values of vertebrate and A167E yeast actin filaments differ at any given salt concentration. This behavior is not surprising given that contacts between the two long-pitch helical strands of yeast actin filaments are less extensive than those of muscle actin (48) and that filament mechanical properties (e.g., flexural rigidity) are influenced greatly by the filament subunit interaction energies and interface areas (41, 49–51).
Fig. 4.
The “stiffness site” controls the cation dependence of actin filament rigidity. Bending persistence length (Lp) of A167E mutant yeast actin (Alexa 488 labeled) filaments increases with Mg2+-binding, whereas wt filaments shows no [Mg2+] dependence of Lp. Uncertainty bars represent SEM.
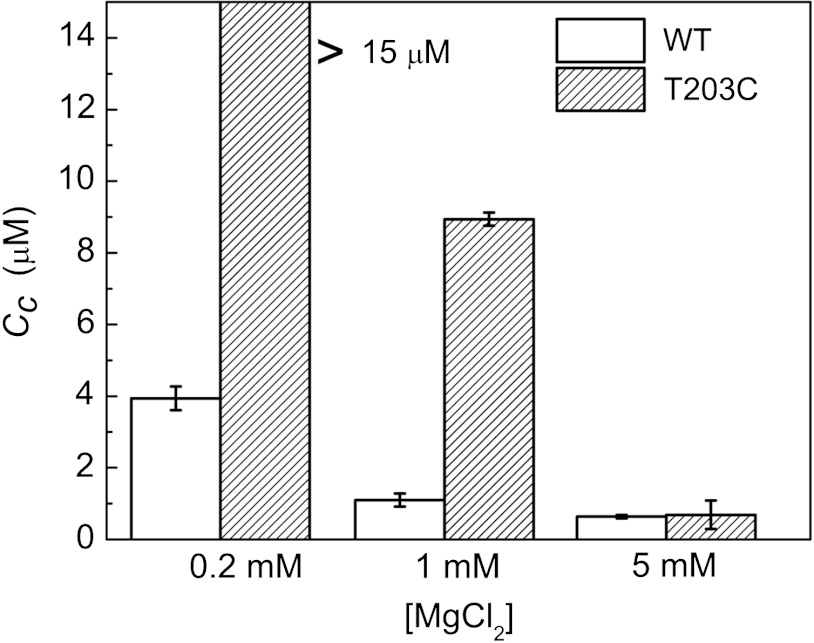
Similar to vertebrate actin (Fig. S2), the Cc of wt yeast actin depends on the cation concentration and approaches a minimum value at approximately 1–5 mM Mg2+ (Fig. 5). Such comparable behavior is expected given the strict conservation of polymerization site residues between both actin isoforms. High-resolution structures of actin dimers reveal that T203 and D288 can be within hydrogen-bonding distance (approximately 2.9 Å) (52, 53). Therefore, we mutated the highly conserved T203 at the polymerization site, which could potentially participate in direct cation binding and/or orienting D288 through H-bonding for proper coordination geometry of the bound cation. Disruption of the yeast actin polymerization site with the mutation (T203C) shifts the Mg2+-dependence of Cc (Fig. 5), in accord with a reduction in the Mg2+ binding affinity. This behavior strongly supports the existence and location of the cation binding site that drives actin polymerization.
Fig. 5.
The “polymerization site” modulates the cation dependence of the critical concentration. T203C yeast actin shows little or no polymerization at 0.2 mM Mg2+ (Cc > 15 μM), Cc at 1 mM Mg2+ that is higher (Cc = 8.9 μM) than that of wt yeast actin, but a Cc value comparable to that of wt is achieved at 5 mM Mg2+. Uncertainty bars represent SEM.
Conclusion
We predict the existence and identify the locations of two distinct, filament-specific classes of cation-binding sites on actin. We refer to these as “polymerization” and “stiffness” sites given the effects that mutations in the sites have on salt-dependent assembly and bending rigidity. Occupancy of the polymerization site drives actin filament assembly, while occupancy of the stiffness site modulates filament bending rigidity. Stiffness sites are located at the interface between the DB-loop and SD3 of adjacent subunits, consistent with DB-loop conformation and remodeling playing an important role in determining overall actin filament mechanics and structural dynamics (6, 7). Mutations adjacent to the stiffness site have been linked to hypertrophic cardiomyopathy (54), consistent with the stiffness site being critical for actin function. Binding of the actin filament regulatory protein, cofilin (55), is coupled to stiffness site reorganization (56), cation release (33), and enhanced actin filament bending (34, 35, 50) and twisting (57), suggesting that displacement of stiffness site cations contributes to the effects of cofilin on actin filament mechanics. The work presented here favors a general mechanism in which cation binding to discrete filament-specific sites is the dominant effect underlying salt-dependent actin polymerization and filament bending mechanics.
Materials and Methods
Proteins.
Actin was isolated from rabbit skeletal muscle acetone powder, gel filtered over Sephacryl S-300 equilibrated in buffer A (0.2 mM ATP, 0.1 mM CaCl2, 0.5 mM DTT, 1 mM NaN3, 2 mM Tris-HCl, pH 8.0), and fluorescently labeled with pyrenenyl-iodoacetamide or Alexa 488-succimidyl ester (Molecular Probes, Eugene, OR) as described (34, 58). The labeling efficiency was 0.85–0.90 fluorophore per actin. Yeast actins were purified using DNAse I affinity chromatography with the previously described modifications (59) and labeled with Alexa 488-succimidyl ester (Molecular Probes) as described (34, 58), where indicated.
Ca-ATP-G-actin was converted into Mg-ATP-G-actin with 200 μM EGTA and MgCl2 equal to the [G-actin] plus 10 μM. Mg-ADP actin monomers were prepared by depleting free ATP from a solution of 50 μM Mg-ATP-actin monomers with Dowex-1 beads, followed by addition of 20 U/mL of hexokinase, 200 μM ADP, and 1 mM glucose, and incubated for 3 h on ice (60–62). K+-ADP actin monomers were prepared by adding 1 mM K+-ADP, 1 mM K+–EGTA, 1 mM K+–EDTA, 20 U/mL hexokinase, and 1 mM glucose to Ca-ATP actin monomers in buffer A. Ca2+-ADP- and Na+-ADP- actin were prepared using the respective salts.
Determination of the Critical Concentration for Actin Polymerization.
Actin monomers were polymerized at room temperature with 0.1 volume of 10× polymerization buffer containing 10 mM Tris (pH 7.5 at 25 °C), 1 mM DTT, 1 mM ADP (ATP for yeast actin), and a range of the indicated salt concentrations. Polymerized samples were briefly sonicated in a water bath, diluted to a series of desired final concentrations (0.05–25 μM), and equilibrated for 4–6 h. The fluorescence emission (λex = 365 nm; λem = 400–420 nm) of equilibrated rabbit skeletal actin (5% pyrene labeled) samples were measured at 25 °C with a Varian Cary spectrofluorometer (Agilent Technologies, Palo Alto, CA). The critical concentration (Cc) was determined from the x intercept of the best fit of the [actin] dependence of the integrated emission peak intensities to a linear function after subtraction of actin monomer background fluorescence. The Cc of unlabeled wild-type and T203C yeast actin was measured by light scattering at 400 nm (63, 64).
Determination of the Actin Filament Bending Persistence Length.
Images of Alexa-488 labeled actin filaments that were thermally fluctuating in 2D were acquired for 100 ms using a Nikon Eclipse TE300 microscope equipped with a Coolsnap HQ cooled CCD camera (Roper Scientific, Tucson, AZ) and μManager software (National Institutes of Health). The depth of the sample was estimated to be < 3 μm, which was necessary to ensure the 2D motion and thus prevent the actin filament from rotating axially (34). Digital images were processed and skeletonized using ImageJ software (NIH). Single actin filaments longer than four pixels (0.18 μm/pixel) were automatically detected using a custom Matlab script, but any bundles were excluded. Detected filaments were reconstructed with an average third-order Bezier spline to minimize measurement errors (34, 35). The bending persistence lengths (Lp) of actin filaments were determined from digitized images (20 images, n≥200 filaments for each data set), by fitting the average angular correlation of segment lengths (s) to the following 2D correlation function (34, 35):
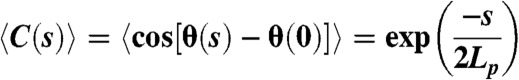 |
[4] |
Actin Structures and Prediction of Cation-Binding Sites.
We employed a structural bioinformatics approach to predict potential cation binding sites on actin. We utilized the Ca2+ binding site model implemented within WebFEATURE (42, 65, 66) to compare predicted sites on the G-actin monomer [Protein Data Bank (PDB) ID 1J6Z (67)], F-actin (conformation) monomer [PDB ID 3MFP (68)], and F-actin polymer (3MFP “biological assembly” file comprised of 5 subunits). While the particular WebFEATURE model we chose was trained using the characteristics of Ca2+ binding sites from the Protein Data Bank (66), we utilized the resulting HITs as predictions of general cation-binding sites with a range of possible affinities for specific cations. Individual .pdb files were edited (using a text editor) to include only protein and ADP atoms, and the 5-mer F-actin model (3MFP “biological assembly”) was edited to have consecutive chain identifiers (A,B,C,D,E) to better keep track of individual site predictions and for visualization software (VMD) (69).
We compared WebFEATURE Ca2+ binding site predictions (HITs) between the G and F conformations of the actin monomer and between the F conformation of the monomer with the F-actin polymer. For the first comparison, the G-actin monomer predictions were subtracted from F-actin monomer predictions. For the second comparison, F-actin monomer predictions were subtracted from F-actin polymer predictions (SI Text).
Identification of Discrete Actin Filament-Specific Cation-Binding Sites.
Following the above subtraction process, we grouped clusters of HITs in order to generate individual cation-binding site predictions. The highest-scoring WebFEATURE HITs tend to occur in clusters of points on a regular grid pattern (WebFEATURE uses a 1.5-Å grid). Cation-binding sites in proteins typically include several coordinating atoms about 2.1–4.8 Å from the metal center (70). To capture this behavior in our prediction of discrete cation-binding sites on actin, we grouped individual WebFEATURE HITs based on series of steps described in SI Text.
We ranked the predicted discrete cation-binding sites according to the total prediction score of each cluster of HITs from WebFEATURE. We focused on the highest-scoring predicted cation-binding sites generated for each of the two comparisons: F-actin conformation monomer vs. G-actin, and F-actin polymer vs. F-actin conformation monomer.
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by National Institutes of Health Grant RO1-GM097348 and American Heart Association Established Investigator Award 0655849T awarded to E.M.D.L.C. and National Institutes of Health Grant RO1-GM077190 awarded to E.R. We thank Dr. Nathan Baker for recommending cation prediction software, Dr. Peter Rubenstein for the yeast cell line-expressing actin mutant A167E, and Dr. Thomas D. Pollard for valuable suggestions on an earlier version of the manuscript. We are indebted to Drs. Timothy M. Lohman and M. Thomas Record for discussions and clarifications regarding salt effects on macromolecular interactions and critical reading of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211078109/-/DCSupplemental.
References
- 1.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 2.Ariel G, Andelman D. Polyelectrolyte persistence length: Attractive effect of counterion correlations and fluctuations. Europhys Lett. 2003;61:67–73. [Google Scholar]
- 3.Pollard TD, Cooper J. Actin and actin-binding proteins: A critical evaluation of mechanisms and functions. Annu Rev Biochem. 1986;55:987–1035. doi: 10.1146/annurev.bi.55.070186.005011. [DOI] [PubMed] [Google Scholar]
- 4.Korn ED, Carlier MF, Pantaloni D. Actin polymerization and ATP hydrolysis. Science. 1987;238:638–644. doi: 10.1126/science.3672117. [DOI] [PubMed] [Google Scholar]
- 5.Oosawa F, Kasai M. Theory of linear and helical aggregations of macromolecules. J Mol Biol. 1962;4:10–21. doi: 10.1016/s0022-2836(62)80112-0. [DOI] [PubMed] [Google Scholar]
- 6.Orlova A, Egelman EH. A conformational change in the actin subunit can change the flexibility of the actin filament. J Mol Biol. 1993;232:334–341. doi: 10.1006/jmbi.1993.1393. [DOI] [PubMed] [Google Scholar]
- 7.Strzelecka-Golaszewska H, Wozniak A, Hult T, Lindberg U. Effects of the type of divalent cation, Ca2+ or Mg2+, bound at the high-affinity site and of the ionic composition of the solution on the structure of F-actin. Biochem J. 1996;316:713–721. doi: 10.1042/bj3160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strzelecka-Golaszewska H, Prochniewicz E, Drabikowski W. Interaction of actin with divalent cations: 1: The effect of various cations on the physical state of actin. Eur J Biochem. 1978;88:219–227. doi: 10.1111/j.1432-1033.1978.tb12441.x. [DOI] [PubMed] [Google Scholar]
- 9.Estes JE, Selden LA, Kinosian HJ, Gershman LC. Tightly-bound divalent cation of actin. J Muscle Res Cell Motil. 1992;13:272–284. doi: 10.1007/BF01766455. [DOI] [PubMed] [Google Scholar]
- 10.Zimmerle CT, Patane K, Frieden C. Divalent cation binding to the high-and low-affinity sites on G-actin. Biochemistry. 1987;26:6545–6552. doi: 10.1021/bi00394a039. [DOI] [PubMed] [Google Scholar]
- 11.Frieden C. The Mg2+-induced conformational change in rabbit skeletal muscle G-actin. J Biol Chem. 1982;257:2882–2886. [PubMed] [Google Scholar]
- 12.Martonosi A, Molino CM, Gergely J. Binding of divalent cations to actin. J Biol Chem. 1964;239:1057–1064. [PubMed] [Google Scholar]
- 13.Carlier MF. Actin: Protein structure and filament dynamics. J Biol Chem. 1991;266:1–4. [PubMed] [Google Scholar]
- 14.Rich SA, Estes JE. Detection of conformational changes in actin by proteolytic digestion: Evidence for a new monomeric species. J Mol Biol. 1976;104:777–792. doi: 10.1016/0022-2836(76)90181-9. [DOI] [PubMed] [Google Scholar]
- 15.Frieden C. Polymerization of actin: Mechanism of the Mg2+-induced process at pH 8 and 20 degrees C. Proc Natl Acad Sci USA. 1983;80:6513–6517. doi: 10.1073/pnas.80.21.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonkaria S, et al. Insight into the assembly properties and functional organisation of the magnetotactic bacterial actin-like homolog, MamK. PLoS One. 2012;7:e34189. doi: 10.1371/journal.pone.0034189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strzelecka-Golaszewska H, Prochniewicz E, Drabikowski W. Interaction of actin with divalent cations: 2: Characterization of protein-metal complexes. Eur J Biochem. 1978;88:229–237. doi: 10.1111/j.1432-1033.1978.tb12442.x. [DOI] [PubMed] [Google Scholar]
- 18.Rouayrenc JF, Travers F. The 1st step in the polymerization of actin. Eur J Biochem. 1981;116:73–77. doi: 10.1111/j.1432-1033.1981.tb05302.x. [DOI] [PubMed] [Google Scholar]
- 19.Tellam R. Mechanism of CaCl2-induced actin polymerization. Biochemistry. 1985;24:4455–4460. doi: 10.1021/bi00337a029. [DOI] [PubMed] [Google Scholar]
- 20.Oda T, Makino K, Yamashita I, Namba K, Maeda Y. Distinct structural changes detected by X-ray fiber diffraction in stabilization of F-actin by lowering pH and increasing ionic strength. Biophys J. 2001;80:841–851. doi: 10.1016/S0006-3495(01)76063-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujiwara I, Vavylonis D, Pollard TD. Polymerization kinetics of ADP-and ADP-P-i-actin determined by fluorescence microscopy. Proc Natl Acad Sci USA. 2007;104:8827–8832. doi: 10.1073/pnas.0702510104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yarmola EG, Bubb MR. Profilin: Emerging concepts and lingering misconceptions. Trends Biochem Sci. 2006;31:197–205. doi: 10.1016/j.tibs.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Manning GS. Counterion binding in polyelectrolyte theory. Acc Chem Res. 1979;12:443–449. [Google Scholar]
- 24.Draper DE. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Record MT, Anderson CF, Lohman TM. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: The roles of ion association or release, screening and ion effects. Q Rev Biophys. 1978;11:103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- 26.Robinson RA, Stokes RH. Electrolyte Solutions. 2nd Ed. London, UK: Butterworths; 1959. [Google Scholar]
- 27.Record MT, Lohman TM, deHaseth PL. Ion effects on ligand-nucleic acid interactions. J Mol Biol. 1976;107:145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- 28.Lohman TM, deHaseth PL, Record MT. Pentalysine-deoxyribonucleic acid interactions: A model for the general effects of ion concentrations on the interactions of proteins with nucleic acids. Biochemistry. 1980;19:3522–3530. doi: 10.1021/bi00556a017. [DOI] [PubMed] [Google Scholar]
- 29.Vahdat A, Miller C, Phillips M, Muhlrad A, Reisler E. A novel 27/16 kDa form of subtilisin cleaved actin: Structural and functional consequences of cleavage between Ser234 and Ser235. FEBS Lett. 1995;365:149–151. doi: 10.1016/0014-5793(95)00446-g. [DOI] [PubMed] [Google Scholar]
- 30.Stokasimov E, McKane M, Rubenstein PA. Role of intermonomer ionic bridges in the stabilization of the actin filament. J Biol Chem. 2008;283:34844–34854. doi: 10.1074/jbc.M804419200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirzabekov AD, Rich A. Asymmetric lateral distribution of unshielded phosphate groups in nucleosomal DNA and its role in DNA bending. Proc Natl Acad Sci USA. 1979;76:1118–1121. doi: 10.1073/pnas.76.3.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang JX, Janmey PA. The polyelectrolyte nature of F-actin and the mechanism of actin bundle formation. J Biol Chem. 1996;271:8556–8563. doi: 10.1074/jbc.271.15.8556. [DOI] [PubMed] [Google Scholar]
- 33.Cao WX, Goodarzi JP, De La Cruz EM. Energetics and kinetics of cooperative cofilin-actin filament interactions. J Mol Biol. 2006;361:257–267. doi: 10.1016/j.jmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 34.McCullough BR, Blanchoin L, Martiel JL, De La Cruz EM. Cofilin increases the bending flexibility of actin filaments: Implications for severing and cell mechanics. J Mol Biol. 2008;381:550–558. doi: 10.1016/j.jmb.2008.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCullough BR, et al. Cofilin-linked changes in actin filament flexibility promote severing. Biophys J. 2011;101:151–159. doi: 10.1016/j.bpj.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Odijk T. Polyelectrolytes near rod limit. J Polym Sci Polym Phys. 1977;15:477–483. [Google Scholar]
- 37.Skolnick J, Fixman M. Electrostatic persistence length of a wormlike polyelectrolyte. Macromolecules. 1977;10:944–948. [Google Scholar]
- 38.Manning G. The persistence length of DNA is reached from the persistence length of its null isomer through an internal electrostatic stretching force. Biophys J. 2006;91:3607–3616. doi: 10.1529/biophysj.106.089029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Ionic effects on the elasticity of single DNA molecules. Proc Natl Acad Sci USA. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hagerman PJ. Flexibility of DNA. Annu Rev Biophys Biophys Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- 41.De La Cruz EM, Roland J, McCullough BR, Blanchoin L, Martiel JL. Origin of twist-bend coupling in actin filaments. Biophys J. 2010;99:1852–1860. doi: 10.1016/j.bpj.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang MP, Banatao DR, Klein TE, Brutlag DL, Altman RB. WebFEATURE: An interactive web tool for identifying and visualizing functional sites on macromolecular structures. Nucleic Acids Res. 2003;31:3324–3327. doi: 10.1093/nar/gkg553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheterline P, Clayton J, Sparrow J. Actins. 3rd Ed. London: Academic Press; 1996. [Google Scholar]
- 44.Rould MA, Wan Q, Joel PB, Lowey S, Trybus KM. Crystal structures of expressed nonpolymerizable monomeric actin in the ADP and ATP states. J Biol Chem. 2006;281:31909–31919. doi: 10.1074/jbc.M601973200. [DOI] [PubMed] [Google Scholar]
- 45.Wertman KF, Drubin DG, Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bas DC, Rogers DM, Jensen JH. Very fast prediction and rationalization of pK(a) values for protein-ligand complexes. Proteins. 2008;73:765–783. doi: 10.1002/prot.22102. [DOI] [PubMed] [Google Scholar]
- 47.Wang F, Sampogna RV, Ware BR. pH dependence of actin self-assembly. Biophys J. 1989;55:293–298. doi: 10.1016/S0006-3495(89)82804-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orlova A, Chen X, Rubenstein PA, Egelman EH. Modulation of yeast F-actin structure by a mutation in the nucleotide-binding cleft. J Mol Biol. 1997;271:235–243. doi: 10.1006/jmbi.1997.1163. [DOI] [PubMed] [Google Scholar]
- 49.Chu JW, Voth GA. Coarse-grained modeling of the actin filament derived from atomistic-scale simulations. Biophys J. 2006;90:1572–1582. doi: 10.1529/biophysj.105.073924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfaendtner J, De La Cruz EM, Voth GA. Actin filament remodeling by actin depolymerization factor/cofilin. Proc Natl Acad Sci USA. 2010;107:7299–7304. doi: 10.1073/pnas.0911675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saunders MG, Voth GA. Comparison between actin filament models: Coarse-graining reveals essential differences. Structure. 2012;20:641–653. doi: 10.1016/j.str.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 52.Kudryashov DS, et al. The crystal structure of a cross-linked actin dimer suggests a detailed molecular interface in F-actin. Proc Natl Acad Sci USA. 2005;102:13105–13110. doi: 10.1073/pnas.0506429102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Otomo T, et al. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 54.Müller M, et al. Functional characterization of the human α-cardiac actin mutations Y166C and M305L involved in hypertrophic cardiomyopathy. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-1030-5. doi: 10.1007/s00018-012-1030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De La Cruz EM. How cofilin severs an actin filament. Biophys Rev. 2009;1:51–59. doi: 10.1007/s12551-009-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galkin VE, et al. Remodeling of actin filaments by ADF/cofilin proteins. Proc Natl Acad Sci USA. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prochniewicz E, Janson N, Thomas DD, De La Cruz EM. Cofilin increases the torsional flexibility and dynamics of actin filaments. J Mol Biol. 2005;353:990–1000. doi: 10.1016/j.jmb.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 58.De La Cruz EM. Cofilin binding to muscle and non-muscle actin filaments: Isoform-dependent cooperative interactions. J Mol Biol. 2005;346:557–564. doi: 10.1016/j.jmb.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 59.Grintsevich EE, et al. Mapping the cofilin binding site on yeast G-actin by chemical cross-linking. J Mol Biol. 2008;377:395–409. doi: 10.1016/j.jmb.2007.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De La Cruz EM, et al. Polymerization and structure of nucleotide-free actin filaments. J Mol Biol. 2000;295:517–526. doi: 10.1006/jmbi.1999.3390. [DOI] [PubMed] [Google Scholar]
- 61.De La Cruz EM, Pollard TD. Nucleotide-free actin: Stabilization by sucrose and nucleotide-binding kinetics. Biochemistry. 1995;34:5452–5461. doi: 10.1021/bi00016a016. [DOI] [PubMed] [Google Scholar]
- 62.Frederick KB, Sept D, De La Cruz EM. Effects of solution crowding on actin polymerization reveal the energetic basis for nucleotide-dependent filament stability. J Mol Biol. 2008;378:540–550. doi: 10.1016/j.jmb.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wegner A, Engel J. Kinetics of cooperative association of actin to actin-filaments. Biophys Chem. 1975;3:215–225. doi: 10.1016/0301-4622(75)80013-5. [DOI] [PubMed] [Google Scholar]
- 64.Cooper JA, Pollard TD. Methods to measure actin polymerization. Methods Enzymol. 1982;85:182–210. doi: 10.1016/0076-6879(82)85021-0. [DOI] [PubMed] [Google Scholar]
- 65.Wei L, Altman RB. Recognizing protein binding sites using statistical descriptions of their 3D environments. Pac Symp Biocomput. 1998:497–508. [PubMed] [Google Scholar]
- 66.Liu T, Altman RB. Prediction of calcium-binding sites by combining loop-modeling with machine learning. BMC Struct Biol. 2009;9:72. doi: 10.1186/1472-6807-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Otterbein LR, Graceffa P, Dominguez R. The crystal structure of uncomplexed actin in the ADP state. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- 68.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 69.Humphrey W, Dalke A, Schulten K. VMD:Visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 70.Nayal M, Dicera E. Predicting Ca2+-binding sites in proteins. Proc Natl Acad Sci USA. 1994;91:817–821. doi: 10.1073/pnas.91.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.