Abstract
Breast cancer is the most frequent invasive tumor diagnosed in women, causing over 400 000 deaths yearly worldwide. Like other tumors, it is a disease with a complex, heterogeneous genetic and biochemical background. No single genomic or metabolic condition can be regarded as decisive for its formation and progression. However, a few key players can be pointed out and among them is the TP53 tumor suppressor gene, commonly mutated in breast cancer. In particular, TP53 mutations are exceptionally frequent and apparently among the key driving factors in triple negative breast cancer —the most aggressive breast cancer subgroup—whose management still represents a clinical challenge. The majority of TP53 mutations result in the substitution of single aminoacids in the central region of the p53 protein, generating a spectrum of variants (’mutant p53s’, for short). These mutants lose the normal p53 oncosuppressive functions to various extents but can also acquire oncogenic properties by gain-of-function mechanisms. This review discusses the molecular processes translating gene mutations to the pathologic consequences of mutant p53 tumorigenic activity, reconciling cell and animal models with clinical outcomes in breast cancer. Existing and speculative therapeutic methods targeting mutant p53 are also discussed, taking into account the overlap of mutant and wild-type p53 regulatory mechanisms and the crosstalk between mutant p53 and other oncogenic pathways in breast cancer. The studies described here concern breast cancer models and patients—unless it is indicated otherwise and justified by the importance of data obtained in other models.
Significance of TP53 mutations in breast cancer
TP53 gene and its mutations in spontaneous breast cancer
P53 protein, encoded by the TP53 tumor suppressor gene, is one of the main molecular decision makers of stress response in human cells (1). Embedded within a complex signaling pathway, p53 senses a plethora of stress signals originating from deregulated expression of oncogenes, DNA damage, metabolic deprivation or telomere erosion. Depending on the cellular context and on the type of stress, p53 elicits apoptosis, DNA repair, transient or permanent cell cycle arrest and, lately found as surprisingly crucial, metabolic homeostasis maintenance (2). P53 activation–inactivation upon stress depends on a repertoire of posttranslational modifications (PTMs) and interactions with proteins that induce p53 stabilization and subcellular relocalization, allowing it to induce appropriate sets of genes. The oncosuppressive functions of p53 may be inhibited by several mechanisms, but TP53 has come to researchers’ attention primarily due to its exceptional mutation frequency—higher than in any other tumor suppressor gene in humans overall. On average, TP53 is mutated in 31% of all tumors included in the Catalog of Somatic Mutations in Cancer (COSMIC) database (3), and is mutated in ~23% of breast cancer samples, where it is the second most frequently mutated gene after the PI3KCA protooncogene (26% in COSMIC). Mutations in TP53 occur more frequently in other types of tumors, in particular ovarian (50% of cases in COSMIC), large intestine (43%) and lung (36%) cancers . Although these sporadic cancers depend more heavily on TP53 mutations than breast cancer, the presence of mutated TP53 is still one of the main molecular characteristics of this type of tumor.
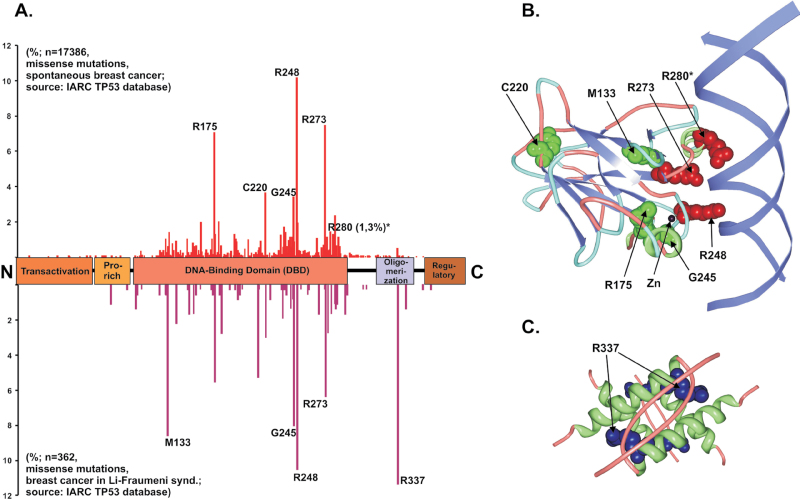
According to the current release of the International Agency for Research on Cancer (IARC) TP53 database (http://www-p53.iarc.fr/), included in COSMIC, ~70% of the breast cancer alterations in TP53 are missense mutations (4). This proportion, as well as the spectrum of mutated codons in the gene (the hotspots), reflect the p53 mutational pattern of other tumors (Figure 1 and Table 1). A noteworthy difference is codon 220, which is the fourth most frequent missense mutation in breast cancer (3.6%), whereas it ranks seventh in other cancers (2%). Another such overrepresentation is codon 163 (2% in breast cancer, 1% in other cancers) (5). Although no explanation of these differences has been provided, geographic or ethnical characteristics have been suggested to influence the occurrence of specific mutations, possibly due to the link to environmental mutagens (6,7). Associations of TP53 mutation with breast-cancer-predisposing BRCA1/2 germline mutations have been also found, probably favored by a bias in the dysfunctional DNA repair mechanisms (8,9). In sporadic breast cancers, high TP53 mutation frequencies have been significantly associated with two polymorphisms: the homozygous Arginine at codon 72 of p53 (10); and the presence of the highly active allelic variant G of glutathione-S-transferases (GSTs) (11). Importantly, differences have been found in the specific TP53 mutation occurrence in breast cancer types and grades, as well as in the survival of patients bearing particular hotspot mutations (discussed in part IV).
Fig. 1.
Frequency and structure of p53 missense alterations in breast cancer. (A) Human p53 domain structure with indicated frequency (percent bars) of missense changes in TP53 found in spontaneous (above) or Li–Fraumeni-associated (below) breast cancer. The five most frequently changed codons are indicated by numbers and residue names. The codon 280 marked with (*) is included due to the widespread use of the MDA-MB-231 cell line, bearing R280K p53, as a model for invasive breast cancer (see Table I). (B) Structural position of residues affected by most frequent missense-mutation-related changes in the DNA-binding domain (DBD) of human p53. DNA-contacting Arginine side chains are colored red (R280 is marked with * as in A), residues whose change affects DBD folding in p53 ‘structural mutants’ are colored green. The position of the DBD-stabilizing Zinc (Zn) atom is also indicated. Derived from Protein Data Bank (PDB) ID: 1TUP (33). (C) Structural position of the residues most frequently altered in Li–Fraumeni breast cancer—Arginine 337 (side chains colored blue)—in the oligomerization domains of human p53 tetramer. Derived from PDB ID: 1C26 (163).
Table I.
Oncogenic properties of most frequent TP53 missense mutations in breast cancer
| Codon | Frequency of missense mutations in breast cancer (# most frequent in breast cancer); in other tumors (# most frequent) | Ten-year mortality rate (/1000) in breast cancer patients, based on (17) | Human breast cancer cell lines with endogenous mutation, based on Handbook of p53 Mutations In Cell Lines, v. 1.0 (http://p53.free.fr/) | Human mammary epithelial cell characteristics associated with the presence of mutant p53 | Knockin mouse models with mutant TP53 expressed in mammary epithelium | Mammary carcinomas associated with mutant p53 in mouse models |
|---|---|---|---|---|---|---|
| 248 | 10.1 % (1); 8.8% (2) | 78.65 (R248Q: 69.06; R248W: 108.84) | H-31 (R248Q)HCC1143 (R248Q)HCC2157 (R248W)HCC70 (R248Q) | Altered growth and cell polarity in 3D cultures, EMT induction (MCF10A cells with introduced p53 R248 mutant) (104,121) | Mammary gland-specific expression (R245W) (166) | No increase in mammary carcinomas compared with parental strain (166,167) |
| HupKI – exons 4–9 of murine p53 replaced by the human sequence (R248W) (84) | No mammary carcinomas; increased genomic instability and altered tumor spectrum compared with p53 –/– (84) | |||||
| 273 | 7.5% (2); 8.9% (1) | 68.29 (R273H: 67.29; R273C: 64.86) b | HCC38 (R273L)MDA-MB-468 (R273H)R30T (R273C)SUM229PE (R273C) | Disordered growth in 3D cultures, induction of mevalonate pathway genes; induction of migration-related mutant p53 signature genes; inhibition of apoptosis (MDA-MB-468) (48,90,104);Altered growth and cell polarity in 3D cultures, EMT induction (MCF10A cells with introduced p53 R273H mutant) (104,121);Immortalization of normal mammary epithelial cells (95) | Germline knockin (R270H) (26) | Low incidence of mammary carcinoma; altered tumor spectrum compared with p53 –/– and +–- (26) |
| Mammary gland-specific expression (R270H) (30,51,167) | High frequency of mammary tumors; DN but not GOF of mutant p53 observed (30,51); Higher tumor grade compared with wt p53 in the presence of T antigen (167) | |||||
| HupKI – exons 4–9 of murine p53 replaced by the human sequence (R273H) (84) | No mammary carcinomas; Altered tumor spectrum compared with p53–/– (84) | |||||
| 175 | 7.0% (3); 6.5% (3) | 68.29 | HCC1395 (R175H)SK-BR-3 (R175H) | Increased growth rate, tumorigenic potential, chemoresistance; increased expression of pro-angiogenic genes, NF-Y and NF-κB targets; inhibition of p73; inhibition of apoptosis mediated by the vit. D receptor (SK-BR-3) (44,47,89,91,168,169); altered growth and cell polarity in 3D cultures, EMT induction (MCF10A cells with introduced p53 R273H mutant) (104,121) | Germline knockin (R172H)(26,27,57) | No mammary carcinomas; altered tumor spectrum compared with p53 –/– and +/– (26,27);Increased mutant p53 stability and tumorigenesis in mdm2 –/– or p16 –/– background (57) |
| Mammary gland-specific expression (R172H) (170) | Low level of spontaneous tumorigenesis, predisposition to the development of mammary tumors when treated with the chemical carcinogen DMBA; increased genomic instability (170) | |||||
| 220 | 3.6% (4); 2.0% (7) | 45.63 | HCC1419 (Y220C)MDA-MB-330 (Y220C) | Unknown | None | Unknown |
| 245 | 3.4% (5); 4.1% (4) | 37.68 | None | Altered growth and cell polarity in 3D cultures, weak GOF (MCF10A cells with introduced p53 G245S mutant) (104,121) | None | Unknown |
| 280a | 1.3% (16); 1.3% (13) | Unknown | CAMA-1 (R280T)MDA-MB-231 (R280K) | Disordered growth in 3D cultures, induction of mevalonate pathway genes; induction of migration-related mutant p53 signature genes and migratory phenotype; inhibition of p63 downstream transcriptional program and TGFβ-induced metastasis; inhibition of apoptosis mediated by the vit. D receptor, induction of chemokines and other inflammatory mediators (MDA-MB-231) (48,75,91,104,108) | None | Unknown |
a280 codon is included as in Figure 1; b Increased mortality from total codon 273 changes is due to other 273 variants, albeit very few in number (17)
Hereditary TP53 mutations and breast cancer
The significance of TP53 gene alterations in breast cancer is supported by the frequent occurrence of this cancer type in the Li–Fraumeni syndrome, a hereditary tumor-predisposing disorder associated with germline TP53 mutations (12). Taking into account the tissue and organ specificity of tumors, breast cancer is the single most frequent event in Li–Fraumeni syndrome, accounting for >25% of all tumors in affected families (13). The mutational spectrum of TP53 in Li–Fraumeni breast cancer resembles that in spontaneous breast cancer, with ~65% missense mutations, but differs in the hotspot distribution (Figure 1). This is mainly due to the unusually high frequency (up to 16%) of codon 337 mutation in Li–Fraumeni patients, which is 11% in syndrome-related breast cancers (IARC TP53 database). This bias probably reflects a founder effect in the Southern Brazilian population (14), due to the genetic background dependence of the mutation’s penetrance (15). The tumor spectrum associated with germline alterations at codon 337 is particular, with 67% of diagnosed adrenal tumors and breast cancers down to 11.6% (IARC TP53 database). This underlines that specific kinds of TP53 alterations may have a different impact on breast cancer development.
The fact that most inborn TP53 mutations preferentially induce breast cancer may imply that p53 alterations are the early events in spontaneous mammary tumors also. The fact that missense mutations in TP53 are more frequent in high-grade spontaneous breast carcinomas (16,17) suggests that early p53 mutations might be one of the decisive events in the development of breast cancers on the ‘high-grade-like’ rather than ‘low-grade-like’ molecular pathway, taking into account their proposed early divergence (18,19). The rise of new approaches for monitoring genomes and global expression profiles of single cells (20) or tumor mass fragments (21) should allow to draw a more accurate picture of the timing and ‘topology’ of TP53 mutations in breast cancer.
Mutant TP53 in mouse models: importance of an oncogenic p53 gain-of-function
Although the idea that missense mutations can confer oncogenic properties to p53, in contrast to a mere loss-of-function, has been around for many years(22–24), only specific mouse models have proved that mutant p53 gain-of-function (GOF)—defined as the ability of p53 missense mutants to actively contribute to tumor progression and aggressiveness—indeed affects tumorigenesis in vivo.
The most accurate mouse models of Li–Fraumeni TP53 missense mutations were generated by gene knockin (Table 1). The mutant TP53 alleles used in crucial studies encoded R172H and R270H p53 variants, the murine counterparts of human R175H and R273H hotspot mutants (25–27). Tumor spectra in these mice differed, and metastasis frequency was increased, when compared with mice having TP53 +/– and –/– genotypes; there were also differences between R172H and R270H variants (26).
In contrast to Li–Fraumeni patients with the corresponding R175H or R273H mutations, these mice displayed low frequency of mammary carcinomas (26,27), similar to TP53 knockout mice (Table 1) (28,29). This apparent inconsistency may find an explanation in their particular genetic backgrounds, as well as in the fact that early appearance of other tumors in these animals would mask the formation of mammary carcinomas. Indeed, when a single TP53 R270H allele was expressed specifically in mammary epithelium, increased formation of breast cancer was observed (30).
Mouse models generated so far provided a great deal of important data (Table 1), but as illustrated by the Li–Fraumeni reconstruction attempts, possess limitations in simulating the phenotypic effects of human TP53 mutations. Nonetheless, after inclusion of more TP53 mutations, combined with alterations in other oncogenic pathways, careful dissection of the impact of genetic background and extensive use of cancer xenografts, mouse models may yet provide a tremendous contribution to the understanding of the role of p53 in breast cancer.
Molecular mechanism of mutant p53 action in breast cancer cells
Transcriptional activity of mutant p53
The role of wild-type (wt) TP53 in tumor suppression is strongly linked to the molecular properties of the p53 protein. Even though p53 has important transcription-independent roles (31), the protein works primarily as a tetrameric transcription factor (32). Solving the crystal structure of the DNA-binding domain (DBD) of wt p53, the largest and most structured of its domains, in complex with the target DNA sequence (Figure 1B) was crucial for understanding how oncogenic hotspot mutations affect p53 activity (33). The most commonly changed residues in breast cancer—R248Q and R273H—affect contact between p53 and DNA and hence have been dubbed ‘contact mutants’. In contrast, R175H and Y220C substitutions generate p53 ‘structural mutants’, with distorted DBD structure under physiological conditions. Careful biophysical studies in vitro uncovered a gradient in the extent of p53 DBD destabilization by the specific hotspot TP53 mutations (34), suggesting that different mutants may be functionally different proteins. Transactivation assays in yeast or human cultured cells have been designed to assess the activity of mutant p53s (35–37). In a study concerning TP53 mutants frequently expressed in breast cancer, many mutant p53 variants were shown to possess an altered promoter activation spectrum. Y220C, for instance, still transactivates the most sensitive wt p53 response element (from the promoter of the p21 gene—WAF1), whereas other response elements are not activated (38). The shift, rather than a full displacement in the transcriptional specificity of wt p53, is an element of mutant p53 GOF. Transcriptomic analysis of the activity of six different TP53 hotspot mutants in a p53 –/– background of H1299 lung carcinoma cells indicated that mutant p53s regulate predominantly a specific subset of genes whose promoters are also bound by wt p53 (39). Nevertheless, an accumulating body of evidence in different models, including breast cancer cells, suggests that mutant p53 also acquires distinct DNA-binding and transactivation properties because many loci lacking p53-responsive elements are direct transcriptional targets of hotspot p53 mutants (40–44). Mutant p53 can also directly activate transcription of specific micro–RNA (45) and attenuate micro-RNA processing, presumably affecting their general levels in cells (46). Using breast cancer cell lines, Blandino and colleagues found 40 promoters bound de novo by R175H p53 (47), whereas our group identified 10 novel genes controlled and bound by R280K p53 (48).
Despite increasing interest, no consensus target sequence of mutant p53 has been defined so far. However, an overrepresentation of nuclear factor-kappaB (NF-κB) target sites has been observed in promoters bound by the p53 R175H mutant (47), and a p63-binding consensus emerged as the main feature of sites bound by mutant p53s in H1299 lung carcinoma cells and HaCaT keratinocytes (39,49), suggesting that mutant p53 cooperation with different transcription factors may be an important route to execute its GOF activity.
Because p53 is a tetrameric transcription factor, hetero-oligomerization of wt and mutant monomers results in a dominant negative effect. Even though this dominant negative effect of mutant p53 has been demonstrated (50,51), its importance in tumorigenesis is controversial. In H1299 cells, mutations in the DBD of p53 have been shown to be relatively ineffective in the functional inactivation of heterotetramers of wt p53 (52), and the influence of heterozygous TP53 missense mutations in cell lines and tumor samples (including breast cancer) has been questioned (53,54). Taking into account different observations, it is conceivable that many mutant p53 variants can interfere with wt p53 to facilitate tumor formation by lowering the tumor suppressor barrier, but mutant p53 GOF provides an additional boost , such as increased invasiveness and chemoresistance, for tumorigenesis; this effect could be potentiated when the wt allele is lost (27,55). The cell/tumor type context is also an important variable to consider in further research—because the mutant p53’s GOF penetrance may be dependent on the particular molecular background.
Stability and PTMs of mutant p53
Complementing the altered transcriptional abilities, the oncogenic properties of most frequent p53 mutants also benefit from an increased p53 level. In normal cells, p53 is destabilized mainly by the action of Mdm2, an E3 ubiquitin ligase that is a direct p53 transcriptional target (56). In tumor cells, this negative feedback is frequently abrogated, resulting in increased p53 level. The importance of mutant p53 accumulation is underscored by the observation that Li–Fraumeni mouse models with R172H p53 knockin, despite different tumor spectra and metastasis incidence, have survival curves similar to p53 –/– mice (26,27). However, when they are crossed with MDM2 –/– mice, resulting in increased mutant p53 stability, survival of the animals is drastically shortened (57), indicating that R175H p53 GOF is strongly dependent on p53 stability. Even though mutant p53 variants can be ubiquitinated by MDM2 as well as other E3 ligases and degraded (58), several mechanisms may counteract this process in mammary tumor cells: for instance, p16INK4 upregulation(59) or Hsp90-mediated stabilization (60).
Mutant p53 stabilization and activity can depend also on PTMs other than ubiquitination, although not many studies have addressed their role (61,62). In breast cancer cell lines and tumor samples, p53 phosphorylation is detected regardless of TP53 mutational status (63), indicating that oncogenic stress can modify mutant p53 on the same regulatory sites as it does on wt p53 (48). Expression of a non-phosphorylatable variant (S392A) enhances the oncogenic potential of p53 R175H in cultured cells, suggesting that Ser 392 phosphorylation might negatively affect the GOF. Accordingly, in breast cancer samples with high levels of mutant p53, the phosphorylation at S392 was reduced (64).
Despite the massive amount of knowledge on wt p53 PTMs, the same aspect of mutant p53 still holds many unknowns. In particular, next to nothing is known about acetylation, methylation or sumoylation specific to mutant p53 in breast cancer, whereas acetylation by p300/CBP-associated factor (PCAF) was found to reactivate selected p53 mutants in H1299 cells (65). This field of research deserves greater efforts, because pharmacological intervention on enzymes that apply regulatory PTMs on mutant p53 might become an approach of antitumor treatment.
Effect of TP53 mutations on p53 paralogs and isoforms in breast cancer
Human p53 possesses two main paralogs—p63 and p73—expressed as multiple isoforms sharing a significant similarity to p53 (66,67). Though not being classical tumor suppressors, both possess antitumorigenic functions (68,69). P53 does not hetero-oligomerize with its paralogs (70), but p73 and p63 were found to bind mutant p53 via its DBD (71,72). As a consequence, p73 and p63 can be drawn into mutant p53 aggregates in osteosarcoma cells, presumably blocking their normal function (73). Such interactions are involved in the direct negative effect of mutant p53 on the antitumor activity of its paralogs in different tumor models (26,27,70,74). In breast cancer cells, p53 mutant variants were shown to repress p73- and p63-driven transcription of target genes (71,72). In response to transforming growth factor (TGF)-beta signaling, mutant p53 binds p63 in a complex with Smad2, preventing transcription of Sharp-1 and Cyclin G2, two crucial p63 target genes that suppress metastatic behavior of breast cancer cells (48,75). Similarly, mutant p53 variants introduced into epithelial lung H1299 and mammary MCF-10A cells were found to promote an invasive phenotype by inhibiting p63-dependent regulation of integrin and epidermal growth factor receptor (EGFR) recycling (76). Finally, high-throughput microarray and ChIP-seq data from lung and skin cancer cells (39,49) revealed that p63-promoter-binding sites are frequently targeted by mutant p53, indicating that direct influence on p63 is one of the distinctive mechanisms of mutant p53 GOF.
The last 10 years have brought significant advances in our knowledge of p53 isoforms (77). Indeed, p53 can be expressed in various alternative N-terminal and C-terminal isoforms. Notably, they were reported to be differentially expressed between normal and breast cancer tissues (78). In normal breast tissue, the C-terminally truncated and modified β and γ isoforms were both present, whereas p53β was only detected in 33% of tested tumors, and p53γ in none. In contrast, although N-terminally truncated Δ133p53 was not detectable in healthy controls, it was found in 80% of all tested breast tumors. Successive studies demonstrated that expression of the p53γ isoform significantly improves the outcome of breast cancer patients bearing mutant p53 (79). The mutations were present in both full-length p53 and p53γ, leading to a conclusion that oncogenic properties induced by a mutation in TP53 are not simply transferred to a shorter p53 isoform. It will be interesting to address in further research whether there is an influence of specific hotspot mutations on the function of p53 isoforms, and whether this function extends beyond the influence on full-length p53.
Mutant p53 involvement in mechanisms of breast cancer development
Mutant p53 in early tumorigenesis of breast cancer: DNA damage response, genomic instability and apoptosis avoidance
Mutant p53 influence in breast cancer may stretch from early events, raising the probability of tumor development (as in Li–Fraumeni patients), to processes characteristic of advanced stages of cancer (Figure 2). The loss of p53 function as the ‘guardian of the genome’ has been classically associated with a deregulation of its downstream targets and impairment of DNA repair, cell cycle arrest or apoptosis (80,81). However, the presence of p53 mutant variants introduces additional oncogenic layers to the cell stress response.
Fig. 2.
Mutant p53 involvement in processes associated with breast cancer development. Mutant p53 is known to affect multiple oncogenic processes (164,165). Although different oncogenic mechanisms overlap during tumorigenesis, here they are arbitrarily divided into mechanisms indispensable for early tumorigenesis at the level of single-cell biochemistry (green), mechanisms supporting multicellular tumor mass growth (orange), and features necessary for metastasis to secondary sites (red). The asterisks (*) indicate oncogenic mechanisms known to be important for breast cancer, linked to p53 gain-of-function in other tumors, but not yet directly tested for mutant p53 dependence in mammary carcinoma cells or mouse models. See text for detailed information and references.
First, signaling pathways and mechanisms normally controlling wt p53 in response to genotoxic stress may similarly activate and stabilize mutant p53 (82,83). Second, one of the characteristics of mutant p53 GOF in various models, involving transcription-independent mechanisms, is the contribution to genomic instability (84,85). In breast cancer samples, mutations in TP53 have been associated with increased incidence of chromosomal abnormalities in patients (86,87). In mouse mammary epithelial cells, introduction of R172H p53 induced centrosome amplification and increased the frequency of aberrant mitoses, leading to altered chromosome numbers (88). Third, mutant p53 reduces sensitivity to both spontaneous and DNA-damage-induced apoptosis (88). This effect was present in different breast carcinoma cell lines and was found to depend on mutant p53, as opposed to the loss of p53 (89,90). Several studies have explored the mechanism of these anti-apoptotic effects: mutant p53 variants were found to convert Vitamin D3 from a pro- to an anti-apoptotic factor, shifting the transactivation spectrum of Vitamin D receptor toward a prosurvival profile (91). Positive association in breast cancer has been found between mutant p53 and expression of the anti-apoptotic splice variant of Survivin (92). Mutant p53s have been also found to increase the levels of Bcl-2 (93). Recently, Bcl-2 has been described to be frequently overproduced in triple-negative breast cancers (TNBCs) and, in contrast to other mammary tumor types, to correlate with poor patient survival (94). Fourth, mutant p53 may affect cellular senescence. In this respect, specific studies are needed to understand the contribution of mutant p53 variants to overcoming cellular senescence in breast cancer, because this is an effect already observed in mammary epithelial cells (95) and in other cancers (96,97).
Mutant p53 in breast cancer growth: effect on metabolism, inflammation and angiogenesis
A large fraction of breast cancers expresses estrogen (ER) and progesterone receptors (PR), and their proliferation depends on hormonal stimulation. Despite prognostic associations discussed in the following sections, there is little clarity in the functional interactions between ER and PR and mutant p53. Block of estrogen-dependent signaling was shown to reduce wt and mutant p53 protein levels in ER-positive mammary tumor cells (98,99). ER-alpha and mutant p53 may also potentially cooperate to transcribe selected target genes by cooperative docking to non-canonical promoter-binding sites (100). Despite this potential cooperation, ER-alpha was shown to inhibit wt p53-dependent transcription in breast tumor cell lines via direct binding and recruitment of transcriptional repressors (101–103). In theory, such a complex can form also with some p53 mutants, and therefore ER-alpha may potentially influence the transcriptional landscape of cells expressing mutant p53. Additional studies are urgently needed to clarify the functional interaction between ER and mutant p53 in breast cancer, especially in view of the clinical use of anti-estrogenic drugs.
One of the earliest discoveries regarding the pro-oncogenic activities of mutant p53 was its ability to sustain tumor growth of fibroblast cells injected subcutaneously in immunocompromised mice (24). Recently, mutant p53s were shown to promote breast cell proliferation in both two- (2D) and three-dimensional (3D) cultures by inducing the expression of genes involved in the mevalonate pathway (104). Indeed, either mutant p53 depletion or treatment with inhibitors of this pathway could reduce the growth and tumorigenic potential of mutant p53-harboring cells (104). The involvement of mutant p53 in tumor-associated inflammation may also be relevant, because cases of inflammatory breast cancer display increased NF-κB signaling and frequency of p53 mutation (105,106). Tissue culture experiments indicate that mutant p53 promotes NF-κB activity in response to tumor necrosis factor (TNF)-alpha (107) and sustains expression of inflammatory cytokines and receptors in breast cancer cell lines (108).
In different experimental systems, mutant p53 was also reported to affect numerous other proliferative signals and metabolic pathways potentially important for development of breast cancer, although no specific experiments with mammary cells were performed (see also Figure 2). Mutant p53 has been demonstrated to promote the expression levels of 15-lypoxygenase (109), an enzyme that was found to be associated with mammary tumor progression and survival of breast cancer cells (110). Mutant p53 was also shown to induce the expression levels of the insulin-like growth factor I receptor gene (IGF1R (111)), which once activated is able to trigger both phosphatidylinositol 3-kinase (PI3K)/AKT and Ras/mitogen-activated protein kinase (MAPK) prosurvival pathways, often deregulated in breast cancer (112). Mutant p53 was also demonstrated to inhibit autophagy (113), a process shown to modulate breast tumor formation and development (114).
In order to sustain faster growth and match the increased requirement of oxygen and nutrients, the tumor mass needs to be vascularized. Mutant p53 was reported to promote vascular endothelial growth factor (VEGF) expression in NIH 3T3 cells (115) and to trigger E2F1-dependent induction of ID4, which in turn contributes to stabilization of the mRNAs of the pro-angiogenic factors IL8 and GRO-α in breast cancer cells (44). In accordance, increased levels of mutant p53 and VEGF have been described to be correlated with poor clinical outcome of breast carcinoma patients (116).
Mutant p53 in metastasis, epithelial to mesenchymal transition and stemness
The majority of deaths in breast cancer patients are associated with metastasis. Mutant p53 has been recently reported to boost the metastatic potential of breast cancer cells. As discussed earlier, this process is in part linked to mutant p53-dependent inhibition of its paralog p63. However, mutant p53 upon phosphorylation-dependent prolyl isomerization by Pin1 can promote tumor cell migration and invasion by directly inducing a transcriptional program sustaining the aggressive potential of breast cancer (48).
Metastasis formation is a complex process intimately connected to cell plasticity and epithelial to mesenchymal transition within the primary tumor (117). In breast cancer cells, mutant p53 is able to both repress epithelial markers such as E-cadherin (118) and induce some crucial transcription factors regulating mesenchymal phenotypes such as Twist (119), ZEB-1 and ZEB-2 (120). Accordingly, MCF-10A-immortalized mammary epithelial cells ectopically expressing mutant p53 undergo epithelial to mesenchymal transition, losing cell polarity and the capability to grow as spheroids in 3D cultures, an effect that can be only partially achieved by the endogenous wt p53 knockdown (121).
Enhanced plasticity due to epithelial to mesenchymal transition can also generate cancer stem-like cells, ultimately responsible of tumor growth and aggressiveness (122). In this regard, wt p53 has recently been shown to act as a crucial safeguard of stem cell self-renewal. Its loss promotes symmetric cell division, favoring expansion of cancer stem cells, breast tumor growth and dissemination (123). Taking into account these observations, it is conceivable to hypothesize that mutant p53 could affect many aspects of cancer stem cell formation and expansion. Supporting this idea, the presence of mutant p53 in primary breast tumors correlates with stem cell transcriptional signatures (124), whereas poorly differentiated and grade 3 breast carcinomas displaying the highest stem cell content (125) are characterized by high frequency of TP53 alterations (17). Recently, mutant p53 was shown to facilitate somatic cell reprogramming, thus increasing the malignant phenotype of such cells (126). This evidence further underlines the multifaceted role of mutant p53 in controlling stem cell phenotypes and tumor aggressiveness.
Diagnostic implications of mutant p53 in breast cancer
Mutant p53 variants in breast cancer prognosis and heterogeneity
Increasing evidence regarding the oncogenic activities of mutant p53 prompted many groups to analyze TP53 mutations as a prognostic or predictive marker (127). In an important study analyzing 1794 breast cancer patients, the stratification of different kinds of TP53 mutations showed that all loss-of-function p53 mutants correlated with poor prognosis compared with wt p53 cases (17). However, mutant p53 GOF and dominant negative indications were also found: among the most frequent missense mutations, codon 248 alterations were linked to reduced survival, whereas mutations at codon 220 and 245 were associated with a better prognosis compared with any other missense variants (Table 1) (17). Interestingly, a difference was found between R248Q and R248W variants, where the latter, though less frequent, correlated with significantly higher patient mortality. This indicates that the two mutant proteins might be functionally different; this observation, together with the mouse model studies (26), underlines that distinguishing the tumorigenic properties of specific p53 hotspot mutants is a potentially critical and still open problem in oncology. Currently, specific transcriptional programs or cell phenotypes cannot be correlated with selected missense p53 mutations or even with the broad distinction between contact and structural mutants (39) (Table 1). However, we suggest that future research on breast tumor progression and its clinical applications will have to take into account the properties of the specific mutants. This will require extensive application of DNA sequencing to biological samples along with the analysis of p53 expression levels.
TP53 mutations were found to be more frequent in high-grade, large-size, node-positive cases and in estrogen- and progesterone- receptor-negative (ER-, PR-) tumors (17). Considering division of breast tumors according to the current molecular subtype classification (128,129), TP53 mutation numbers are usually low in well-differentiated and hormone–receptor-positive luminal subtype A and significantly higher in human epidermal growth factor receptor 2 (HER2) positive and basal-like tumors. In a recent study involving a detailed, genome-scale analysis of nearly 2000 breast cancer cases, TP53 mutations were found in 34% basal-like, 22% HER2, 13% luminal B and 5% luminal A molecular subtypes (130). Taking into account the hormone receptors, TP53 mutations were detected in 8% cases of ER+ and/or PR+ tumors and in 29% of hormone-receptor-negative ones (130). Interestingly, in ER+/PR+ tumors, despite the reduced frequency, the prognostic value of TP53 mutation remains strong (17). P53 alterations correlate with poor clinical outcome also in HER2-positive cancers (131,132). TNBCs (ER-, PR- and HER2-), which mainly belong to the molecular basal-like subtype (129), are more likely to be grade 3 tumors (16). A recent study conducted with high- throughput genomic approaches confirmed that in TNBCs, mutations in TP53 are more frequent than in any other oncogene or tumor suppressor, reaching 54% of samples (133). The increase in TP53 mutations in hormone-receptor-negative tumors and their exceptionally frequent occurrence in TNBC suggests that in the absence of hormone-related stimulatory signaling characteristic to the mammary epithelium, mutant p53 may become increasingly critical for breast cancer progression. Moreover, once established, mutant p53 dependence seems to provide a more severe pro-oncogenic activity compared with hormone dependence. This is supported by the fact that taking into account all the heterogeneity of breast cancer, whenever present, TP53 mutations are more frequent in high-grade, large-size, node-positive cases, thus possessing a prognosis-worsening, driving role (17,130).
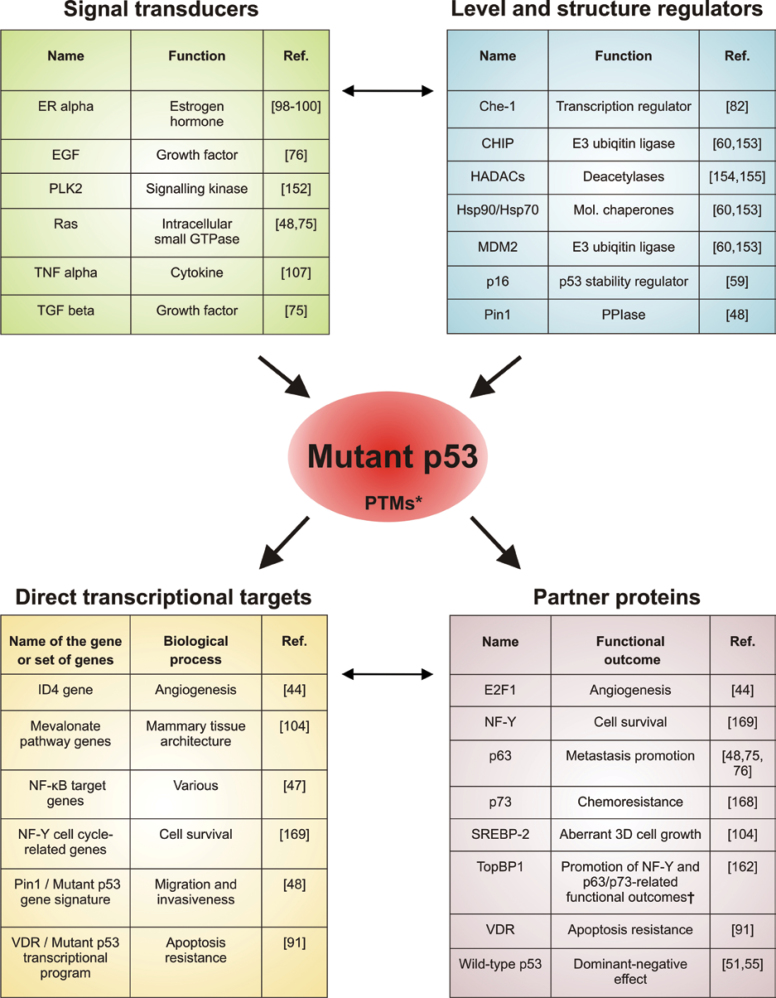
For clinical applications, it would be important to improve the reliability of p53 mutation as the prognostic marker. Different studies have shown that this can be achieved by combining the analysis of TP53 mutational status with the assessment of other factors relevant for either mutant p53 stability or pro-oncogenic functions (Figure 3). Examples of these are the Polo-like kinase 1 and the phoshorylation-dependent prolyl isomerase Pin1 (48,134). In particular, combined analysis of Pin1 protein levels and TP53 missense mutations has been shown to outperform the mere determination of TP53 mutational status as the predictor of clinical outcome (48). Further research is needed to identify other molecular parameters to improve the prognostic value of TP53 mutational status.
Fig. 3.
Mutant p53 as the hub of oncogenic pathways in breast cancer. The activity of mutant p53 is regulated through upstream signal transducers as well as regulators of transcription, stability and structure. Its biological effects are mediated by direct transcriptional activity and through association with downstream protein effectors. These mechanisms are interconnected (small arrows), especially downstream of mutant p53, as most proteins bound directly are transcription regulators. The figure includes only the factors/processes that were found to affect the tumorigenic features of breast cancer models or patients and target genes found to be directly regulated by mutant p53. *PTMs—Posttranslational Modifications may be affected by upstream factors and may influence downstream effects of mutant p53. †TopBP1 is implicated as a coordinator protein of mutant p53 GOF exerted via NF-Y, p63 and p73.
Predictive value of mutant p53 in breast cancer
Despite increasing evidence supporting the prognostic significance of TP53 mutations, few studies have been performed in order to assess its predictive value in patient response to therapy. Among the most commonly used chemotherapeutic regimens, the clearest available results concern treatment with anthracyclines, as several preclinical and clinical reports have indicated that mutant TP53-bearing breast cancer cells and tissues are more resistant to these drugs, compared with wt TP53-bearing ones(37,135,136). A more complex scenario arises when taxane-based chemotherapy is considered. Despite promising early results (137), a large phase-3 clinical trial has recently demonstrated that mutant TP53-harboring breast carcinomas have similar sensitivity to taxanes as those with wt TP53, and TP53 status seems not to be decisive in selecting patients for such chemotherapy (138).
As suggested for the prognostic value of TP53 status, the combination of p53 assessment with the analysis of additional pathways or molecular markers—primarily those controlling mutant p53 stability or activity—should be considered in order to strengthen its predictive properties. Furthermore, it would be important to verify whether the combination of chemotherapeutic drugs with molecules directly or indirectly impinging on mutant p53 activity could be useful as a therapeutic strategy. A striking example is provided by the recent results obtained by treating TNBC tumors implanted in mice with a combination of Chk 1 inhibitors and the DNA-damaging drug irinotecan (139). Under these conditions, tumors with mutant p53 responded to a significantly better extent, indicating that it may be a key determinant influencing tumor response to therapy. Another recent intriguing example has been provided by the observation that Wnt-induced mouse mammary tumors expressing mutant p53 show a superior clinical response to doxorubicin treatment when compared to similar tumors with wild-type p53 (140).
Mutant p53 as the drug target in breast cancer
Direct targeting of mutant p53
Mutant p53 has been considered a potential direct target of therapy and structural information on p53 has been used to rationally identify molecules reactivating wt transcriptional function of mutant p53 or restoring its native structure (141,142). Peptides designed to interfere with p53/p73 binding were able to sensitize breast cancer cells to chemotherapeutic drugs (143). Another noteworthy study concerns a rational design of small compounds predicted to bind and stabilize in silico the structure of one mutant p53 variant distinctive in breast cancer—Y220C (144). Although these are elegant examples of how the detailed structural information on p53 could be potentially transferred to clinics, all these above-mentioned compounds have not yet been tested to confirm their effects in vivo.
Instead, a random screening approach allowed the identification of short peptide aptamers that bind specifically to mutant p53 and, importantly, trigger cell death in breast cancer cells expressing mutant, but not wt, p53 (145). Other interesting molecules of potential therapeutic impact identified by screening approaches are CP-31398 (146), P53R3 (147), RETRA (148) and PRIMA-1 (149). This last compound was one of the first examples of drugs capable of restoring wt p53 conformation, thereby allowing sequence-specific DNA binding and induction of p53 target genes (149). In particular, PRIMA-1 induces apoptosis in tumor cells (other than breast cancer) and inhibits human xenograft tumor growth in SCID mice (149). PRIMA-1 derivatives have been produced to increase its efficacy and the most powerful one is its methylated form, PRIMA-1MET/APR-246 (150), which is currently a promising drug candidate and is being tested in a clinical phase I/II trial, though not in breast cancer (151).
Indirect targeting of mutant p53–challenging mechanisms common to wt and mutant p53
The alternative to targeting mutant p53 itself is to block its upstream activators. Results obtained in several models have suggested candidates, such as the signaling pathway components EGFR (76), transforming growth factor TGFBR1 (75), Ras-activated kinases like p38, JNK1-2, MEK/extracellular signal-regulated kinase ERK and CK1ε/δ (48,75), and the Polo-like kinase family members 1 (151) and 2 (152), for pharmacological inhibition in breast cancer. Other potential targets include proteins affecting mutant p53 levels and structure, such as prolyl isomerase Pin1 (48), Hsp90 (153) or the histone deacetylases (154,155).
There are, however, risks associated with inhibiting some of these factors because of the numerous regulatory mechanisms shared by wt and mutant p53. This includes p53 degradation, which in the case of both wt and mutant depends at least partially on Mdm2 and p16INK4 (57,58). Wt p53 folding, stability and activity are supported by Hsp70–Hsp90 chaperone machinery (156) and prolyl isomerase Pin1 (157), whereas the same proteins are found to support mutant p53’s oncogenic potential (48,158). Therefore, an important prerequisite to their clinical applications is to know the status of TP53 sequence and expression in the tumor; Mdm2 and Hsp90 inhibitors, such as Nutlin and 17AAG, respectively, are being considered candidate drugs for breast cancer (159, 171). A more appealing possibility is to target specific inducers of mutant p53. One example is topoisomerase IIβ-binding protein (TopBP1), which is aberrantly expressed in breast cancer (160), where it is associated with high tumor grade and shorter patient survival (161). TopBP1 in breast cancer cells inhibits wt p53 function, whereas it promotes GOF of mutant p53 variants (161,162). This makes TopBP1 a very interesting potential drug target in breast cancers bearing either wt or mutant p53.
Conclusions
The TP53 gene is mutated less frequently in breast cancer than in other tumors, yet it is the second most frequent genetic alteration in this type of cancer. TP53 mutation frequency and its prognostic value differ between breast cancer subtypes, where, like in other tumors, p53 has to be considered within a web of highly interconnected tumorigenic pathways (Figure 3). In some cases, like TNBC, mutant p53 probably acts as a balance-shifting ‘driver’, whereas in others, it may just be a contingent ‘passenger’. However, in overall majority of cases, missense mutant p53 seems associated with an aggressive tumor phenotype, suggesting that mutant p53 is intrinsically predisposed to overtake the driver role.
The tumor-driving potential of mutant p53 is exerted at multiple levels: dampening oncosuppressive pathways (p63 and other growth suppressors); enhancing multiple oncogenic pathways relevant in breast cancer (such as PI3K/AKT, Ras/mitogen-activated protein kinase (MAPK) and NF-κB) both directly and indirectly; and altering the cellular physiology at posttranscriptional level via non-coding RNAs.
Additional functional intersections of mutant p53 with cellular pathways will certainly emerge in the near future due to new technologies and high-throughput approaches. Following validation in breast cancer models, definition of these cross talks will provide essential conceptual tools for basic and clinical research, granting a better understanding of breast cancer biology and allowing a more accurate stratification of patients for personalized therapy. This knowledge will be critical not only for the development of novel anticancer approaches, but also—and perhaps more importantly— to allow more efficient use of currently available drugs.
Funding
Associazione Italiana per la Ricerca sul Cancro (AIRC) Special Program Molecular Clinical Oncology “5 per mille” (Grant n. 10016) to G.D.S. and L.C.; FEBS postdoctoral fellowship to D.W.
Acknowledgements
We thank A. Testa for reading and editing the manuscript, A. Rustighi and A. Bisso for helpful discussions and reading of the manuscript. We apologize to the colleagues whose contributions have not been cited due to space constraints.
Conflict of Interest Statement: None declared.
Abbreviations
- COSMIC
Catalog of Somatic Mutations in Cancer
- DBD
DNA-binding domain
- ER
estrogen receptor
- GOF
gain-of-function
- HER2
human epidermal growth factor receptor 2
- IARC
International Agency for Research on Cancer
- NF-κB
nuclear factor-kappaB
- PR
progesterone receptor
- PTM
posttranslational modification
- TNBC
triple-negative breast cancer
- TopBP1
topoisomerase IIβ-binding protein
References
- 1. Horn H.F., et al. (2007). Coping with stress: multiple ways to activate p53. Oncogene, 26, 1306––1316 [DOI] [PubMed] [Google Scholar]
- 2. Li T., et al. (2012). Tumor Suppression in the Absence of p53-Mediated Cell-Cycle Arrest, Apoptosis, and Senescence. Cell, 149, 1269––1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Forbes S.A., et al. (2011). COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res., 39, D945––50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Petitjean A., et al. (2007). Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat., 28, 622––629 [DOI] [PubMed] [Google Scholar]
- 5. Feki A., et al. (2004). Mutational spectrum of p53 mutations in primary breast and ovarian tumors. Crit. Rev. Oncol. Hematol., 52, 103––116 [DOI] [PubMed] [Google Scholar]
- 6. Olivier M., et al. (2001). TP53 mutation patterns in breast cancers: searching for clues of environmental carcinogenesis. Semin. Cancer Biol., 11, 353––360 [DOI] [PubMed] [Google Scholar]
- 7. Hill K.A., et al. (2002). p53 as a mutagen test in breast cancer. Environ. Mol. Mutagen., 39, 216––227 [DOI] [PubMed] [Google Scholar]
- 8. Greenblatt M.S., et al. (2001). TP53 mutations in breast cancer associated with BRCA1 or BRCA2 germ-line mutations: distinctive spectrum and structural distribution. Cancer Res., 61, 4092––4097 [PubMed] [Google Scholar]
- 9. Manié E., et al. (2009). High frequency of TP53 mutation in BRCA1 and sporadic basal-like carcinomas but not in BRCA1 luminal breast tumors. Cancer Res., 69, 663––671 [DOI] [PubMed] [Google Scholar]
- 10. Langerød A., et al. (2002). The TP53 codon 72 polymorphism may affect the function of TP53 mutations in breast carcinomas but not in colorectal carcinomas. Cancer Epidemiol. Biomarkers Prev., 11, 1684––1688 [PubMed] [Google Scholar]
- 11. Nedelcheva Kristensen V., et al. (1998). Single tube multiplex polymerase chain reaction genotype analysis of GSTM1, GSTT1 and GSTP1: relation of genotypes to TP53 tumor status and clinicopathological variables in breast cancer patients. Pharmacogenetics, 8, 441––447 [DOI] [PubMed] [Google Scholar]
- 12. Malkin D. (2011). Li-fraumeni syndrome. Genes Cancer, 2, 475––484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Olivier M., et al. (2010). TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol., 2, a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garritano S., et al. (2010). Detailed haplotype analysis at the TP53 locus in p.R337H mutation carriers in the population of Southern Brazil: evidence for a founder effect. Hum. Mutat., 31, 143––150 [DOI] [PubMed] [Google Scholar]
- 15. Assumpção J.G. et al, et al. (2008). Association of the germline TP53 R337H mutation with breast cancer in southern Brazil. BMC Cancer, 8, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bull S.B. et al, et al. (2004). The combination of p53 mutation and neu/erbB-2 amplification is associated with poor survival in node-negative breast cancer. J. Clin. Oncol., 22, 86––96 [DOI] [PubMed] [Google Scholar]
- 17. Olivier M. et al, et al. (2006). The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin. Cancer Res., 12, 1157––1167 [DOI] [PubMed] [Google Scholar]
- 18. Wiechmann L. et al, et al. (2008). The molecular journey from ductal carcinoma in situ to invasive breast cancer. Cancer, 112, 2130––2142 [DOI] [PubMed] [Google Scholar]
- 19. Bombonati A. et al, et al. (2011). The molecular pathology of breast cancer progression. J. Pathol., 223, 307––317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Navin N. et al, et al. (2011). Tumour evolution inferred by single-cell sequencing. Nature, 472, 90––94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gerlinger M. et al, et al. (2012). Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med., 366, 883––892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wolf D. et al, et al. (1984). Reconstitution of p53 expression in a nonproducer Ab-MuLV-transformed cell line by transfection of a functional p53 gene. Cell, 38, 119––126 [DOI] [PubMed] [Google Scholar]
- 23. Halevy O. et al, et al. (1990). Different tumor-derived p53 mutants exhibit distinct biological activities. Science, 250, 113––116 [DOI] [PubMed] [Google Scholar]
- 24. Dittmer D. et al, et al. (1993). Gain of function mutations in p53. Nat. Genet., 4, 42––46 [DOI] [PubMed] [Google Scholar]
- 25. Liu G. et al, et al. (2000). High metastatic potential in mice inheriting a targeted p53 missense mutation. Proc. Natl. Acad. Sci. U.S.A., 97, 4174––4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Olive K.P. et al, et al. (2004). Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell, 119, 847––860 [DOI] [PubMed] [Google Scholar]
- 27. Lang G.A. et al, et al. (2004). Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell, 119, 861––872 [DOI] [PubMed] [Google Scholar]
- 28. Donehower L.A. et al, et al. (1992). Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature, 356, 215––221 [DOI] [PubMed] [Google Scholar]
- 29. Jacks T. et al, et al. (1994). Tumor spectrum analysis in p53-mutant mice. Curr. Biol., 4, 1––7 [DOI] [PubMed] [Google Scholar]
- 30. Wijnhoven S.W. et al, et al. (2005). Mice expressing a mammary gland-specific R270H mutation in the p53 tumor suppressor gene mimic human breast cancer development. Cancer Res., 65, 8166––8173 [DOI] [PubMed] [Google Scholar]
- 31. Speidel D. et al, et al. (2006). Dissection of transcriptional and non- transcriptional p53 activities in the response to genotoxic stress. Oncogene, 25, 940––53 [DOI] [PubMed] [Google Scholar]
- 32. Clore G.M. et al, et al. (1994). High-resolution structure of the oligomerization domain of p53 by multidimensional NMR. Science, 265, 386––391 [DOI] [PubMed] [Google Scholar]
- 33. Cho Y. et al, et al. (1994). Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science, 265, 346––355 [DOI] [PubMed] [Google Scholar]
- 34. Bullock A.N. et al, et al. (2000). Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: definition of mutant states for rescue in cancer therapy. Oncogene, 19, 1245––1256 [DOI] [PubMed] [Google Scholar]
- 35. Kato S. et al, et al. (2003). Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc. Natl. Acad. Sci. U.S.A., 100, 8424––8429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Deissler H. et al, et al. (2004). Spectrum of p53 mutations in biopsies from breast cancer patients selected for preoperative chemotherapy analysed by the functional yeast assay to predict therapeutic response. Oncol. Rep., 11, 1281––1286 [PubMed] [Google Scholar]
- 37. Bertheau P. et al, et al. (2007). Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLoS Med., 4, e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jordan J.J. et al, et al. (2010). Altered-function p53 missense mutations identified in breast cancers can have subtle effects on transactivation. Mol. Cancer Res., 8, 701––716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Neilsen P.M. et al, et al. (2011). Mutant p53 uses p63 as a molecular chaperone to alter gene expression and induce a pro-invasive secretome. Oncotarget., 2, 1203––1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chicas A. et al, et al. (2000). Mutant p53 forms a complex with Sp1 on HIV-LTR DNA. Biochem. Biophys. Res. Commun., 279, 383––390 [DOI] [PubMed] [Google Scholar]
- 41. Scian M.J. et al, et al. (2004). Tumor-derived p53 mutants induce oncogenesis by transactivating growth-promoting genes. Oncogene, 23, 4430––4443 [DOI] [PubMed] [Google Scholar]
- 42. Weisz L. et al, et al. (2004). Transactivation of the EGR1 gene contributes to mutant p53 gain of function. Cancer Res., 64, 8318––8327 [DOI] [PubMed] [Google Scholar]
- 43. Yan W. et al, et al. (2009). Identification of GRO1 as a critical determinant for mutant p53 gain of function. J. Biol. Chem., 284, 12178––12187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fontemaggi G. et al, et al. (2009). The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis. Nat. Struct. Mol. Biol., 16, 1086––1093 [DOI] [PubMed] [Google Scholar]
- 45. Donzelli S. et al, et al. (2011). MicroRNA-128-2 targets the transcriptional repressor E2F5 enhancing mutant p53 gain of function. Cell Death Differ., 19, 1038––1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suzuki H.I. et al, et al. (2009). Modulation of microRNA processing by p53. Nature., 460, 529––533 [DOI] [PubMed] [Google Scholar]
- 47. Dell’Orso S. et al, et al. (2011). ChIP-on-chip analysis of in vivo mutant p53 binding to selected gene promoters. OMICS., 15, 305––312 [DOI] [PubMed] [Google Scholar]
- 48. Girardini J.E. et al. (2011). A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell., 20, 79––91 [DOI] [PubMed] [Google Scholar]
- 49. Martynova E. et al. (2012). Gain-of-function p53 mutants have widespread genomic locations partially overlapping with p63. Oncotarget, 3, 132––143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chène P. (1998). In vitro analysis of the dominant negative effect of p53 mutants. J. Mol. Biol., 281, 205––209 [DOI] [PubMed] [Google Scholar]
- 51. Wijnhoven S.W. et al, et al. (2007). Dominant-negative but not gain-of-function effects of a p53.R270H mutation in mouse epithelium tissue after DNA damage. Cancer Res., 67, 4648––4656 [DOI] [PubMed] [Google Scholar]
- 52. Chan W.M. et al, et al. (2004). How many mutant p53 molecules are needed to inactivate a tetramer? Mol. Cell. Biol., 24, 3536––3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Monti P. et al, et al. (2011). Dominant-negative features of mutant TP53 in germline carriers have limited impact on cancer outcomes. Mol. Cancer Res., 9, 271––279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stoczynska-Fidelus E., et al. (2011). Limited importance of the dominant-negative effect of TP53 missense mutations. BMC Cancer, 11, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Junk D.J. et al, et al. (2008). Different mutant/wild-type p53 combinations cause a spectrum of increased invasive potential in nonmalignant immortalized human mammary epithelial cells. Neoplasia, 10, 450––461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bond G.L. et al, et al. (2005). MDM2 is a central node in the p53 pathway: 12 years and counting. Curr. Cancer Drug Targets, 5, 3––8 [DOI] [PubMed] [Google Scholar]
- 57. Terzian T. et al, et al. (2008). The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev., 22, 1337––1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lukashchuk N. et al, et al. (2007). Ubiquitination and degradation of mutant p53. Mol. Cell. Biol., 27, 8284––8295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang J. et al, et al. (2006). p16INK4a modulates p53 in primary human mammary epithelial cells. Cancer Res., 66, 10325––10331 [DOI] [PubMed] [Google Scholar]
- 60. Muller P. et al, et al. (2008). Chaperone-dependent stabilization and degradation of p53 mutants. Oncogene, 27, 3371––3383 [DOI] [PubMed] [Google Scholar]
- 61. Matsumoto M. et al, et al. (2006). Posttranslational phosphorylation of mutant p53 protein in tumor development. Med. Mol. Morphol., 39, 79––87 [DOI] [PubMed] [Google Scholar]
- 62. Dai C. et al, et al. (2010). p53 post-translational modification: deregulated in tumorigenesis. Trends Mol. Med., 16, 528––536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Minamoto T. et al, et al. (2001). Distinct pattern of p53 phosphorylation in human tumors. Oncogene, 20, 3341––3347 [DOI] [PubMed] [Google Scholar]
- 64. Yap D.B. et al, et al. (2004). Ser392 phosphorylation regulates the oncogenic function of mutant p53. Cancer Res., 64, 4749––4754 [DOI] [PubMed] [Google Scholar]
- 65. Perez R.E. et al, et al. (2010). Restoration of DNA-binding and growth-suppressive activity of mutant forms of p53 via a PCAF-mediated acetylation pathway. J. Cell. Physiol., 225, 394––405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dötsch V. et al, et al. (2010). p63 and p73, the ancestors of p53. Cold Spring Harb. Perspect. Biol., 2, a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Collavin L. et al, et al. (2010). p53-family proteins and their regulators: hubs and spokes in tumor suppression. Cell Death Differ., 17, 901––911 [DOI] [PubMed] [Google Scholar]
- 68. Melino G. (2011). p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ., 18, 1487––1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zawacka-Pankau J. et al, et al. (2010). p73 tumor suppressor protein: a close relative of p53 not only in structure but also in anti-cancer approach? Cell Cycle, 9, 720––728 [DOI] [PubMed] [Google Scholar]
- 70. Gaiddon C. et al, et al. (2001). A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol., 21, 1874––1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Strano S. et al, et al. (2000). Physical and functional interaction between p53 mutants and different isoforms of p73. J. Biol. Chem., 275, 29503––29512 [DOI] [PubMed] [Google Scholar]
- 72. Strano S. et al, et al. (2002). Physical interaction with human tumor-derived p53 mutants inhibits p63 activities. J. Biol. Chem., 277, 18817––18826 [DOI] [PubMed] [Google Scholar]
- 73. Xu J. et al, et al. (2011). Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat. Chem. Biol., 7, 285––295 [DOI] [PubMed] [Google Scholar]
- 74. Di Como C.J. et al, et al. (1999). p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol. Cell. Biol., 19, 1438––1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Adorno M. et al, et al. (2009). A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell, 137, 87––98 [DOI] [PubMed] [Google Scholar]
- 76. Muller P.A. et al, et al. (2009). Mutant p53 drives invasion by promoting integrin recycling. Cell, 139, 1327––1341 [DOI] [PubMed] [Google Scholar]
- 77. Khoury M.P. et al, et al. (2011). p53 Isoforms: An Intracellular Microprocessor? Genes Cancer, 2, 453––465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bourdon J.C. et al, et al. (2005). p53 isoforms can regulate p53 transcriptional activity. Genes Dev., 19, 2122––2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bourdon J.C. et al, et al. (2011). p53 mutant breast cancer patients expressing p53γ have as good a prognosis as wild-type p53 breast cancer patients. Breast Cancer Res., 13, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ellis P.A. et al, et al. (1997). Absence of p21 expression is associated with abnormal p53 in human breast carcinomas. Br. J. Cancer, 76, 480––485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhan Q. et al, et al. (1996). Abrogation of p53 function affects gadd gene responses to DNA base-damaging agents and starvation. DNA Cell Biol., 15, 805––815 [DOI] [PubMed] [Google Scholar]
- 82. Bruno T. et al, et al. (2010). Che-1 promotes tumor cell survival by sustaining mutant p53 transcription and inhibiting DNA damage response activation. Cancer Cell, 18, 122––134 [DOI] [PubMed] [Google Scholar]
- 83. Suh Y.A. et al, et al. (2011). Multiple stress signals activate mutant p53 in vivo. Cancer Res., 71, 7168––7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Song H. et al, et al. (2007). p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat. Cell Biol., 9, 573––580 [DOI] [PubMed] [Google Scholar]
- 85. Hanel W. et al, et al. (2012). Links between mutant p53 and genomic instability. J. Cell. Biochem., 113, 433––439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jain A.N. et al, et al. (2001). Quantitative analysis of chromosomal CGH in human breast tumors associates copy number abnormalities with p53 status and patient survival. Proc. Natl. Acad. Sci. U.S.A., 98, 7952––7957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Jong Y.J. et al, et al. (2004). Chromosomal comparative genomic hybridization abnormalities in early- and late-onset human breast cancers: correlation with disease progression and TP53 mutations. Cancer Genet. Cytogenet., 148, 55––65 [DOI] [PubMed] [Google Scholar]
- 88. Murphy K.L. et al, et al. (2000). A gain of function p53 mutant promotes both genomic instability and cell survival in a novel p53-null mammary epithelial cell model. FASEB J., 14, 2291––2302 [DOI] [PubMed] [Google Scholar]
- 89. Bossi G. et al, et al. (2006). Mutant p53 gain of function: reduction of tumor malignancy of human cancer cell lines through abrogation of mutant p53 expression. Oncogene, 25, 304––309 [DOI] [PubMed] [Google Scholar]
- 90. Lim L.Y. et al, et al. (2009). Mutant p53 mediates survival of breast cancer cells. Br. J. Cancer, 101, 1606––1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Stambolsky P. , et al. (2010). Modulation of the vitamin D3 response by cancer-associated mutant p53. Cancer Cell, 17, 273––285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Végran F., et al. (2007). Association of p53 gene alterations with the expression of antiapoptotic survivin splice variants in breast cancer. Oncogene, 26, 290––297 [DOI] [PubMed] [Google Scholar]
- 93. Pratt M.A. et al, et al. (2007). Cytoplasmic mutant p53 increases Bcl-2 expression in estrogen receptor-positive breast cancer cells. Apoptosis, 12, 657––669 [DOI] [PubMed] [Google Scholar]
- 94. Tawfik K. et al, et al. (2012). Prognostic significance of Bcl-2 in invasive mammary carcinomas: a comparative clinicopathologic study between “triple-negative” and non-“triple-negative” tumors. Hum. Pathol., 43, 23––30 [DOI] [PubMed] [Google Scholar]
- 95. Gollahon L.S. et al, et al. (1996). Immortalization of human mammary epithelial cells transfected with mutant p53 (273his). Oncogene, 12, 715––725 [PubMed] [Google Scholar]
- 96. Morton J.P. et al, et al. (2010). Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc. Natl. Acad. Sci. U.S.A., 107, 246––251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Skinner H.D. et al, et al. (2012). TP53 disruptive mutations lead to head and neck cancer treatment failure through inhibition of radiation-induced senescence. Clin. Cancer Res., 18, 290––300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Liu G. et al, et al. (2000). Estrogen receptor protects p53 from deactivation by human double minute-2. Cancer Res., 60, 1810––1814 [PubMed] [Google Scholar]
- 99. Fernández-Cuesta L. et al, et al. (2011). Estrogen levels act as a rheostat on p53 levels and modulate p53-dependent responses in breast cancer cell lines. Breast Cancer Res. Treat., 125, 35––42 [DOI] [PubMed] [Google Scholar]
- 100. Menendez D. et al, et al. (2010). Estrogen receptor acting in cis enhances WT and mutant p53 transactivation at canonical and noncanonical p53 target sequences. Proc. Natl. Acad. Sci. U.S.A., 107, 1500––1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Konduri S.D. et al, et al. (2010). Mechanisms of estrogen receptor antagonism toward p53 and its implications in breast cancer therapeutic response and stem cell regulation. Proc. Natl. Acad. Sci. U.S.A., 107, 15081––15086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Liu W. et al, et al. (2006). Estrogen receptor-alpha binds p53 tumor suppressor protein directly and represses its function. J. Biol. Chem., 281, 9837––9840 [DOI] [PubMed] [Google Scholar]
- 103. Sayeed A. et al, et al. (2007). Estrogen receptor alpha inhibits p53-mediated transcriptional repression: implications for the regulation of apoptosis. Cancer Res., 67, 7746––7755 [DOI] [PubMed] [Google Scholar]
- 104. Freed-Pastor W.A. et al, et al. (2012). Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell, 148, 244––258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Aziz S.A. et al, et al. (2001). Case control study of prognostic markers and disease outcome in inflammatory carcinoma breast: a unique clinical experience. Breast J., 7, 398––404 [DOI] [PubMed] [Google Scholar]
- 106. Dawood S. et al, et al. (2011). Inflammatory breast cancer: what progress have we made? Oncology (Williston Park, N.Y.), 25, 264––70, 273 [PubMed] [Google Scholar]
- 107. Weisz L. et al, et al. (2007). Mutant p53 enhances nuclear factor kappaB activation by tumor necrosis factor alpha in cancer cells. Cancer Res., 67, 2396––2401 [DOI] [PubMed] [Google Scholar]
- 108. Yeudall W.A. et al, et al. (2012). Gain-of-function mutant p53 upregulates CXC chemokines and enhances cell migration. Carcinogenesis, 33, 442––451 [DOI] [PubMed] [Google Scholar]
- 109. Kelavkar U.P. et al, et al. (1999). Effects of mutant p53 expression on human 15-lipoxygenase-promoter activity and murine 12/15-lipoxygenase gene expression: evidence that 15-lipoxygenase is a mutator gene. Proc. Natl. Acad. Sci. U.S.A., 96, 4378––4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Reddy N. et al, et al. (1997). Characterization of a 15-lipoxygenase in human breast carcinoma BT-20 cells: stimulation of 13-HODE formation by TGF alpha/EGF. Biochem. Biophys. Res. Commun., 231, 111––116 [DOI] [PubMed] [Google Scholar]
- 111. Werner H. et al, et al. (1996). Wild-type and mutant p53 differentially regulate transcription of the insulin-like growth factor I receptor gene. Proc. Natl. Acad. Sci. U.S.A., 93, 8318––8323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Gallagher E.J. et al, et al. (2011). Minireview: IGF, Insulin, and Cancer. Endocrinology, 152, 2546––2551 [DOI] [PubMed] [Google Scholar]
- 113. Morselli E. et al, et al. (2008). Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle, 7, 3056––3061 [DOI] [PubMed] [Google Scholar]
- 114. Debnath J. (2011). The multifaceted roles of autophagy in tumors-implications for breast cancer. J. Mammary Gland Biol. Neoplasia, 16, 173––187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kieser A. et al, et al. (1994). Mutant p53 potentiates protein kinase C induction of vascular endothelial growth factor expression. Oncogene, 9, 963––969 [PubMed] [Google Scholar]
- 116. Linderholm B.K. et al, et al. (2001). The expression of vascular endothelial growth factor correlates with mutant p53 and poor prognosis in human breast cancer. Cancer Res., 61, 2256––2260 [PubMed] [Google Scholar]
- 117. Kalluri R. et al, et al. (2009). The basics of epithelial-mesenchymal transition. J. Clin. Invest., 119, 1420––1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Roger L. et al, et al. (2010). Gain of oncogenic function of p53 mutants regulates E-cadherin expression uncoupled from cell invasion in colon cancer cells. J. Cell. Sci., 123(Pt 8), 1295––1305 [DOI] [PubMed] [Google Scholar]
- 119. Kogan-Sakin I. et al, et al. (2011). Mutant p53(R175H) upregulates Twist1 expression and promotes epithelial-mesenchymal transition in immortalized prostate cells. Cell Death Differ., 18, 271––281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ohashi S. et al, et al. (2010). Epidermal growth factor receptor and mutant p53 expand an esophageal cellular subpopulation capable of epithelial-to-mesenchymal transition through ZEB transcription factors. Cancer Res., 70, 4174––4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zhang Y. et al, et al. (2011). Mutant p53 disrupts MCF-10A cell polarity in three-dimensional culture via epithelial-to-mesenchymal transitions. J. Biol. Chem., 286, 16218––16228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Mani S.A. et al, et al. (2008). The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell, 133, 704––715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Cicalese A. et al, et al. (2009). The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell, 138, 1083––1095 [DOI] [PubMed] [Google Scholar]
- 124. Mizuno H. et al, et al. (2010). Inactivation of p53 in breast cancers correlates with stem cell transcriptional signatures. Proc. Natl. Acad. Sci. U.S.A., 107, 22745––22750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Pece S. et al, et al. (2010). Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell, 140, 62––73 [DOI] [PubMed] [Google Scholar]
- 126. Sarig R. et al, et al. (2010). Mutant p53 facilitates somatic cell reprogramming and augments the malignant potential of reprogrammed cells. J. Exp. Med., 207, 2127––2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Brosh R. et al, et al. (2009). When mutants gain new powers: news from the mutant p53 field. Nat. Rev. Cancer, 9, 701––713 [DOI] [PubMed] [Google Scholar]
- 128. Perou C.M. et al, et al. (2000). Molecular portraits of human breast tumours. Nature, 406, 747––52 [DOI] [PubMed] [Google Scholar]
- 129. Sorlie T. et al, et al. (2001). Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U. S. A., 98, 10869––74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Curtis C. et al, et al. (2012). The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 486, 346––352–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Rahko E. et al, et al. (2003). A mutant TP53 gene status is associated with a poor prognosis and anthracycline-resistance in breast cancer patients. Eur. J. Cancer, 39, 447––453 [DOI] [PubMed] [Google Scholar]
- 132. Yamashita H. et al, et al. (2004). Coexistence of HER2 over-expression and p53 protein accumulation is a strong prognostic molecular marker in breast cancer. Breast Cancer Res., 6, R24––R30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Shah S.P. et al, et al. (2012). The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature., 486, 395––399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. King S.I. et al, et al. (2012). Immunohistochemical detection of Polo-like kinase-1 (PLK1) in primary breast cancer is associated with TP53 mutation and poor clinical outcome. Breast Cancer Res., 14, E-pub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lowe S.W. et al, et al. (1994). p53 status and the efficacy of cancer therapy in vivo. Science, 266, 807––810 [DOI] [PubMed] [Google Scholar]
- 136. O’Connor P.M. et al, et al. (1997). Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res., 57, 4285––4300 [PubMed] [Google Scholar]
- 137. Wahl A.F. et al, et al. (1996). Loss of normal p53 function confers sensitization to Taxol by increasing G2/M arrest and apoptosis. Nat. Med., 2, 72––79 [DOI] [PubMed] [Google Scholar]
- 138. Bonnefoi H. et al, et al. (2011). TP53 status for prediction of sensitivity to taxane versus non-taxane neoadjuvant chemotherapy in breast cancer (EORTC 10994/BIG 1-00): a randomised phase 3 trial. Lancet Oncol., 12, 527––539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Ma C.X. et al, et al. (2012). Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J. Clin. Invest., 122, 1541––1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Jackson J.G. et al, et al. (2012). p53-Mediated Senescence Impairs the Apoptotic Response to Chemotherapy and Clinical Outcome in Breast Cancer. Cancer Cell, 21, 793––806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Selivanova G. et al, et al. (1997). Restoration of the growth suppression function of mutant p53 by a synthetic peptide derived from the p53 C-terminal domain. Nat. Med., 3, 632––638 [DOI] [PubMed] [Google Scholar]
- 142. Friedler A., et al. (2002). A peptide that binds and stabilizes p53 core domain: chaperone strategy for rescue of oncogenic mutants. Proc. Natl. Acad. Sci. U.S.A., 99, 937––942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Martynova E. et al, et al. (2012). Gain-of-function p53 mutants have widespread genomic locations partially overlapping with p63. Oncotarget, 3, 132––143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Boeckler F.M. et al, et al. (2008). Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc. Natl. Acad. Sci. U.S.A., 105, 10360––10365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Guida E. et al, et al. (2008). Peptide aptamers targeting mutant p53 induce apoptosis in tumor cells. Cancer Res., 68, 6550––6558 [DOI] [PubMed] [Google Scholar]
- 146. Foster B.A. et al, et al. (1999). Pharmacological rescue of mutant p53 conformation and function. Science, 286, 2507––2510 [DOI] [PubMed] [Google Scholar]
- 147. Weinmann L. et al, et al. (2008). A novel p53 rescue compound induces p53-dependent growth arrest and sensitises glioma cells to Apo2L/TRAIL-induced apoptosis. Cell Death Differ., 15, 718––729 [DOI] [PubMed] [Google Scholar]
- 148. Kravchenko J.E. et al, et al. (2008). Small-molecule RETRA suppresses mutant p53-bearing cancer cells through a p73-dependent salvage pathway. Proc. Natl. Acad. Sci. U.S.A., 105, 6302––6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Bykov V.J. et al, et al. (2002). Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med., 8, 282––288 [DOI] [PubMed] [Google Scholar]
- 150. Bykov V.J. et al, et al. (2005). PRIMA-1(MET) synergizes with cisplatin to induce tumor cell apoptosis. Oncogene, 24, 3484––3491 [DOI] [PubMed] [Google Scholar]
- 151. Wiman K.G. (2010). Pharmacological reactivation of mutant p53: from protein structure to the cancer patient. Oncogene, 29, 4245––4252 [DOI] [PubMed] [Google Scholar]
- 152. Valenti F. et al, et al. (2011). Mutant p53 oncogenic functions are sustained by Plk2 kinase through an autoregulatory feedback loop. Cell Cycle, 10, 4330––4340 [DOI] [PubMed] [Google Scholar]
- 153. Li D. et al, et al. (2011). Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol. Cancer Res., 9, 577––588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Li D. et al, et al. (2011). SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ., 18, 1904––1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Yan W. et al, et al. (2012). Histone deacetylase inhibitors suppress mutant p53 transcription via histone deacetylase 8. Oncogene. doi: 10.1038/onc.2012.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Walerych D. et al, et al. (2009). Hsp70 molecular chaperones are required to support p53 tumor suppressor activity under stress conditions. Oncogene, 28, 4284––4294 [DOI] [PubMed] [Google Scholar]
- 157. Mantovani F. et al, et al. (2007). The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nat. Struct. Mol. Biol., 14, 912––920 [DOI] [PubMed] [Google Scholar]
- 158. Blagosklonny M.V. et al, et al. (1995). Geldanamycin selectively destabilizes and conformationally alters mutated p53. Oncogene, 11, 933––939 [PubMed] [Google Scholar]
- 159. Zhang F. et al, et al. (2011). MDM2 antagonist nutlin-3a reverses mitoxantrone resistance by inhibiting breast cancer resistance protein mediated drug transport. Biochem. Pharmacol., 82, 24––34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Going J.J. et al, et al. (2007). Aberrant expression of TopBP1 in breast cancer. Histopathology, 50, 418––424 [DOI] [PubMed] [Google Scholar]
- 161. Liu K., et al. (2009). Regulation of p53 by TopBP1: a potential mechanism for p53 inactivation in cancer. Mol. Cell. Biol., 29, 2673––2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Liu K., et al. (2011). TopBP1 mediates mutant p53 gain of function through NF-Y and p63/p73. Mol. Cell. Biol., 31, 4464––4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Jeffrey P.D., et al. (1995). Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science, 267, 1498––1502 [DOI] [PubMed] [Google Scholar]
- 164. Hanahan D., et al. (2011). Hallmarks of cancer: the next generation. Cell, 144, 646––674 [DOI] [PubMed] [Google Scholar]
- 165. Solomon H., et al. (2011). Mutant p53 gain of function is interwoven into the hallmarks of cancer. J. Pathol., 225, 475––478 [DOI] [PubMed] [Google Scholar]
- 166. Krepulat F., et al. (2005). Epigenetic mechanisms affect mutant p53 transgene expression in WAP-mutp53 transgenic mice. Oncogene, 24, 4645––4659 [DOI] [PubMed] [Google Scholar]
- 167. Heinlein C., et al. (2008). Mutant p53(R270H) gain of function phenotype in a mouse model for oncogene-induced mammary carcinogenesis. Int. J. Cancer, 122, 1701––1709 [DOI] [PubMed] [Google Scholar]
- 168. Di Agostino S., et al. (2008). The disruption of the protein complex mutantp53/p73 increases selectively the response of tumor cells to anticancer drugs. Cell Cycle, 7, 3440––3447 [DOI] [PubMed] [Google Scholar]
- 169. Di Agostino S., et al. (2006). Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell, 10, 191––202 [DOI] [PubMed] [Google Scholar]
- 170. Li B., et al. (1998). A transgenic mouse model for mammary carcinogenesis. Oncogene, 16, 997––1007 [DOI] [PubMed] [Google Scholar]
- 171. Modi S., et al. (2011). HSP90 inhibition is effective in breast cancer: a phase II trial of tanespimycin (17-AAG) plus trastuzumab in patients with HER2-positive metastatic breast cancer progressing on trastuzumab. Clin. Cancer. Res., 17, 5132––9 [DOI] [PubMed] [Google Scholar]