Abstract
The fluG gene is a member of a family of genes required for conidiation and sterigmatocystin production in Aspergillus nidulans. We examined the role of the Aspergillus flavus fluG orthologue in asexual development and aflatoxin biosynthesis. Deletion of fluG in A. flavus yielded strains with an approximately 3-fold reduction in conidiation but a 30-fold increase in sclerotial formation when grown on potato dextrose agar in the dark. The concurrent developmental changes suggest that A. flavus FluG exerts opposite effects on a mutual signaling pathway for both processes. The altered conidial development was in part attributable to delayed expression of brlA, a gene controlling conidiophore formation. Unlike the loss of sterigmatocystin production by A. nidulans fluG deletion strains, aflatoxin biosynthesis was not affected by the fluG deletion in A. flavus. In A. nidulans, FluG was recently found to be involved in the formation of dehydroaustinol, a component of a diffusible signal of conidiation. Coculturing experiments did not show a similar diffusible meroterpenoid secondary metabolite produced by A. flavus. These results suggest that the function of fluG and the signaling pathways related to conidiation are different in the two related aspergilli.
INTRODUCTION
Aspergillus nidulans has been the model system for studying asexual conidiation at the molecular level. The genes brlA (bristle) (1), abaA (abacus) (39), and wetA (wet-white conidia) (29) are expressed sequentially and form the central developmental pathway of conidiation (17). BrlA is a transcription factor that mediates the budding growth of conidiophores. Mutations in brlA prevent vesicle formation, yielding stalks that grow indeterminately and that are unable to bear conidia (1). BrlA activates the expression of abaA, a gene that controls phialide differentiation (39). AbaA further mediates transcriptional activation of wetA (31) and also expression of brlA (3, 4). Efforts to elucidate early events leading to activation of conidiation in A. nidulans have identified six genes, fluG (fluffy) (2) and flbA to flbE (fluffy with low brlA expression), that are required for normal activation of brlA (42). Mutations of these genes result in proliferation of undifferentiated vegetative hyphae that produce fluffy cotton-like colonies. A. nidulans fluG is not transcribed in dormant conidia and is significantly downregulated during early vegetative growth; its expression before and after induction of conidiation does not change significantly (2, 7). A positive role of fluG in brlA activation has been suggested in A. nidulans, although the genetic relationship of brlA and fluG is not well defined. A. nidulans fluG mutants do not produce the secondary metabolite sterigmatocystin (20), the penultimate precursor of the carcinogenic aflatoxins. Because contact of the wild-type A. nidulans strain with the fluG null mutant remediates phenotypic defects (26), it was postulated that FluG synthesizes a diffusible factor that initiates conidiation. The effector molecules have recently been determined to be dehydroaustinol and diorcinol, which form a diffusible complex (36).
Aspergillus flavus, a major producer of aflatoxins, is related to A. nidulans in that they share most of the aflatoxin biosynthesis pathway. A. flavus normally reproduces and disseminates asexually, mainly through production of conidia, but some strains produce melaninized hyphal aggregates called sclerotia, which serve as an alternative reproductive form and a survival structure (13). Sclerotia are considered to be a vestige of the cleistothecia produced by other sexual aspergilli, including A. nidulans (19). Only recently has sexual reproduction under laboratory conditions been demonstrated with A. flavus strains of different mating types (21). The relationship between regulatory factors controlling the formation of sclerotia and conidiation is not well understood. SclR, the sclerotial pathway-specific helix-loop-helix transcription factor, was reported to promote sclerotial formation and to regulate hyphal morphology in Aspergillus oryzae (23). Deletion of sclR yields strains with sparse sclerotia but produces dense conidia. Overexpression of sclR results in strains with extremely branched and intertwined aerial hyphae. Conserved global regulators, such as VeA and LaeA, also affect A. flavus asexual development. The veA deletion strains of A. flavus ATCC MYA384 are unable to produce sclerotia and secondary metabolites (16). The A. flavus laeA deletion strains also cannot produce sclerotia and exhibit developmental abnormalities, including increased production of conidiophores, reduced conidial chain elongation, and marked reduction in hydrophobicity (11). In A. nidulans, VeA acts as a negative regulator of conidiation and a positive regulator of sexual development (25, 32), and LaeA acts as a positive regulator of secondary metabolite production (6). VeA and LaeA of A. nidulans are able to form a complex with two other velvet family proteins, VelB and VosA (33), to coordinate light-dependent sexual and asexual development and secondary metabolism (5, 37).
We now report that in A. flavus deletion of fluG delays and decreases conidiation but elevates sclerotial production, depending on the growth medium. These developmental changes were not remediated by coculturing with fluG-positive strains. Surprisingly, unlike the loss of the ability to produce sterigmatocystin in A. nidulans, the fluG deletion strains still retained their ability to produce aflatoxin. These results show that fluG functions distinctly in A. flavus.
MATERIALS AND METHODS
Fungal strains and media.
The A. flavus strain CA14 Δku70 ΔpyrG was the transformation recipient. This mutant was derived from KuPG no. 1, a strain originally derived from aflatoxigenic wild-type CA14 (10). The ku70 gene of the nonhomologous end-joining pathway was deleted in the recipient strain to increase the gene-targeting frequency. The ΔwA strain used in coculture studies produces white conidia due to the deletion of the polyketide synthase gene, wA, involved in conidial pigment synthesis (12). Growth media used for observing morphological changes, such as conidiation and sclerotium formation, were potato dextrose agar (PDA) (EMD, Damstadt, Germany) and Wickerham medium (34) with a minor modification. The medium contained 2.0 g yeast extract, 3.0 g peptone, 5.0 g corn steep solids, 2.0 g dextrose, 30.0 g sucrose, 2.0 g NaNO3, 1.0 g K2HPO4 · 3H2O, 0.5 g MgSO4 · 7H2O, 0.2 g KCl, 0.1 g FeSO4 · 7H2O (10-fold the original recipe), and 15.0 g agar per liter (pH 5.5).
Deletion of fluG and complementation of the fluG deletion strain.
For the deletion of the fluG gene in A. flavus, a double-crossover gene knockout strategy was used (12). The complete fluG gene sequence of A. flavus NRRL3357 (AFL2G_11076.2) was obtained from the Aspergillus Comparative Database at Broad Institute (http://www.broadinstitute.org/annotation/genome/aspergillus_group/MultiHome.html). The primer set fluG5Sc, ACAGAGCTCGCCGCGCTCTAC, and fluG5B, CTGTGGATCCCGGCTCACCATA, was used to amplify a 5′ region. The set fluG3S, CGCCTCGTCGACCGCTACTCTC, and fluG3XH, ATTAAGCTTCGCCGATGATGCTCTGCA, was used to amplify a 3′ region. The two PCR fragments, after digestion with appropriate restriction enzymes, were cloned sequentially into the vector pPG15-3, which contains a pyrG selection marker. The fluG deletion vector was linearized with SacI and XbaI prior to transformation. Putative fluG deletion strains were verified by PCR analyses based on the genomic patterns expected from the fluG disruption. A confirmed fluG deletion strain was used in the fluG complementation experiment. For complementation of the fluG deletion, the full-length genomic fragment from the start to the stop codons was amplified using AccuPrime Pfx PCR Supermix (Invitrogen, Carlsbad, CA) with primers fluGNot, ACTATAGCGGCCGCATGGATCTTACTTCCCTCCAATCC, and fluGRsr, ACTAGACGGTCCGCTAATAACGCTCAAGAAGCCACGTTC. The amplified gene was digested with NotI and ligated into NotI-SmaI-digested pTR1-GPD-TRPC, which contains the A. nidulans glyceraldehyde-3-phosphate dehydrogenase gene (gpdA) promoter and trpC terminator and the A. oryzae pyrithiamine resistance gene (ptrA) as the selection marker.
Generation of laeA and fluG double-deletion strains.
The laeA deletion strain was created from A. flavus Δku70 ΔpyrG, and the uracil auxotrophy was regenerated by growing spores (107) of the laeA deletion strain on PDA plates supplemented with 2 mg uracil/ml and 2 mg fluoroorotic acid (FOA)/ml to force out the reusable pyrG marker (12). The aforementioned fluG deletion vector was used to disrupt fluG in the resulting laeA deletion strain to generate laeA fluG double-deletion strains, using pyrG as the selection marker.
Conidial and sclerotial production.
An aliquot of conidial suspension (103) was seeded at the center of each petri dish (Falcon; 60 by 15 mm) of PDA or Wickerham medium agar. For quantitative comparison of the production of conidia and sclerotia, cultures in triplicate were grown in the dark or under white light at 30°C for 1 week. At the end of the growth period, conidia were washed off the agar plates using 0.01% Triton X-100 solution and counted on a hemocytometer. Sclerotia, if present, were counted manually.
qRT-PCR analysis.
Cultures for quantitative reverse transcription-PCR (qRT-PCR) analyses were grown on duplicate PDA plates (100 by 15 mm) with single-point inoculation at 30°C for 1 week in the dark. Two 5-mm agar plugs, 1.5 cm from the inoculation site, were cored from each plate with a plastic transfer pipette (Transfertubes; Spectrum, Houston, TX) on day 3, day 5, and day 7. The plugs were placed in a 2-ml microcentrifuge tube on dry ice and stored at −80°C overnight. Total RNA was prepared from the plugs with mycelia using a ZR Fungal/Bacterial RNA MiniPrep kit (Zymo Research, Orange County, CA). Mycelia disrupted with ZR bashing beads were processed in a Mini-Beadbeater-8 cell disrupter (Biospec Products, Bartlesville, OK). Treatment of total RNA with DNase 1 (Ambion, Austin, TX) was performed on the column before cleanup and eluted in RNase- and DNase-free water. qRT-PCR was carried out in a 20-μl reaction volume with the LuminoCT SYBR green qPCR ReadyMix (Sigma-Aldrich, St. Louis, MO) and the Reverse Transcriptase Enzyme Mix (Applied Biosystems, Foster City, CA) in an Applied Biosystems StepOne thermal cycler. The amplification conditions were as follows: an initial step of 48°C for 30 min for reverse transcription, followed by 40 cycles, with each cycle consisting of 95°C for 5 s and 60°C for 20 s. The gene-specific primers were 18S-F (TTCCTAGCGAGCCCAACCT) and 18S-R (CCCGCCGAAGCAACTAAG), brlA-F (TATCCAGACATTCAAGACGCACAG) and brlA-R (GATAATAGAGGGCAAGTTCTCCAAAG), abaA-F (GAGTGGCAGACCGAATGTATGTTG) and abaA-R (TAGTGGTAGGCATTGGGTGAGTTG), wetA-F (CCACAGCAGCCGATCCA) and wetA-R (CCCCTTGCAGGATGTCATG), and sclR-F (TGCCGCACACAACATCATT) and sclR-R (TTCTCCAAGGCCACGAACTT).
Examination of aflatoxin production by semiquantitative TLC.
Cultures of PDA plugs for thin-layer chromatography (TLC) analysis were sampled on day 3, day 5, and day 7 and removed at the same time as those used for qRT-PCR analysis. Two plugs in each 2-ml microcentrifuge tube were extracted with 300 μl of methanol for 1 h. The tubes were spun at the maximum speed for 5 min, and 200 μl was drawn off in a 0.5-ml microcentrifuge tube and evaporated to dryness for TLC. The metabolite extracts were redissolved in 20 μl methanol, and 10 μl was spotted and developed on a Si250 silica gel plate (J. T. Baker, Phillipsburg, NJ) with a solvent system of toluene-ethyl acetate-acetic acid (65:35:10 [vol/vol/vol]).
RESULTS
Role of FluG in A. flavus conidiation and sclerotial production.
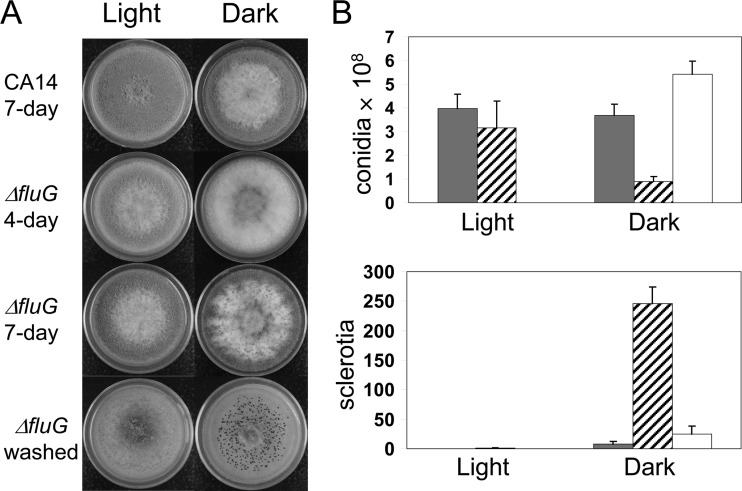
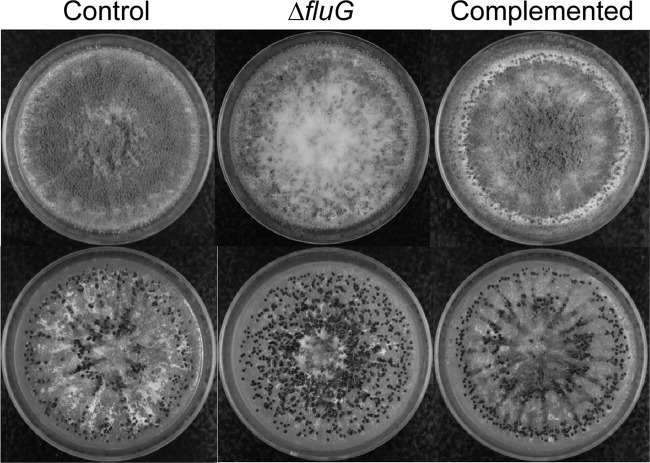
When grown on PDA plates in the dark, the fluG deletion (ΔfluG) strains, compared to the wild-type strain, showed delayed and decreased conidiation but had increased sclerotial production (Fig. 1). The ΔfluG strains produced about two-thirds fewer conidia than the wild-type strain and, concomitantly, about 30-fold more sclerotia. The fluG complemented strains on PDA exhibited two types of colony morphology, one similar to the wild type and another having much denser conidiation (Fig. 1A; see Fig. S1 in the supplemental material). PCR analyses with location-specific primers suggested that each morphotype corresponded to a homologous integration at a specific fluG gene region in the ΔfluG recipient strain, which is defective in the nonhomologous end-joining pathway. The two recombinational events resulted in fluG being under the control of either the native fluG promoter or the stronger A. nidulans gpdA promoter, respectively (see Fig. S1 in the supplemental material). Alterations in production of conidia and sclerotia in the dark were remediated in the fluG complemented strains (Fig. 1B). The complemented strains with fluG under the gpdA promoter produced twice the number of conidia produced by the ones with the native fluG promoter (1.18 × 109 ± 0.17 × 109 versus 5.43 × 108 ± 0.54 × 108 per plate) and did not produce sclerotia on PDA. Conidiation of the ΔfluG strains was not affected compared to the wild-type strain when the cultures were grown under light (Fig. 1), and sclerotia were not produced by either the wild-type or the ΔfluG strains. Wickerham medium was highly conducive to sclerotial formation and conidiation; 30-fold more sclerotia and 4-fold more conidia were produced by the control strain, KuPG, on this corn steep-containing medium than when grown on PDA. When grown in the dark on Wickerham medium, the ΔfluG strains showed a 6-fold decrease in conidiation (2.35 × 108 ± 0.15 × 108 versus 1.53 × 109 ± 0.21 × 109) and an approximate 2-fold increase in sclerotial production (916 ± 28 versus 503 ± 125) (Fig. 2). Complemented strains showed almost wild-type levels of conidiation (1.35 × 109 ± 0.18 × 109) and sclerotial production (512 ± 38).
Fig 1.
Morphology and production of conidia and sclerotia by A. flavus ΔfluG strains on PDA. (A) Cultures were grown at 30°C under white light and in the dark for the specified numbers of days. Washed, conidia of the 7-day-old cultures were washed off to expose sclerotia (black aggregates). (B) Quantification of conidia and sclerotia. Gray bars, wild-type CA14; crosshatched bars, ΔfluG strains; open bars, fluG complemented strains. Conidiation and sclerotial production under light by the fluG complemented strains were not determined. The complemented strains shown are those putatively having the native fluG promoter (see Fig. S1 in the supplemental material). The numbers of conidia and sclerotia are the total count per plate. The error bars indicate standard deviations.
Fig 2.
Morphologies of A. flavus ΔfluG and fluG complemented strains on Wickerham medium. (Top) Cultures after 7 days of growth at 30°C in the dark. (Bottom) Conidia of the colonies were washed off to expose sclerotia (black aggregates). The control strain is A. flavus KuPG no. 1.
Deletion of fluG affects conidial and sclerotial gene expression and aflatoxin production.
Transcript levels for the conidiation genes, brlA, abaA, and wetA, and the sclerotial regulatory gene, sclR, were measured on the control strain KuPG, two ΔfluG strains, and two fluG complemented strains at 3, 5, and 7 days after growth on PDA plates. qPCR analyses indicated that in the control strain, brlA expression (normalized to 18S rRNA) was the highest on day 3, decreased more than 99.5% (equivalent to an increase of more than 8 threshold cycles [Ct]) on day 5, and remained low thereafter. During the same period, brlA expression of the ΔfluG strains increased approximately 2-fold (ranging from 1.46 to 3.33) on day 5 and decreased more than 99.9% on day 7; the extent of decrease was similar to that of the control strain from day 3 to day 5. Expression of abaA and wetA of the ΔfluG strains also increased on day 5. On day 7, abaA transcript levels decreased moderately but wetA transcript levels remained the same (data not shown). Table 1 shows that when the ΔfluG strains on day 5 were compared to the control strain on day 5, they had an approximately 250-fold increase in brlA expression. Because of the delayed conidiation by the ΔfluG strains (Fig. 1A), a better comparison is brlA expression of the ΔfluG strains on day 5 to that of the control strain on day 3. Based on this comparison, the brlA transcript levels of the control and ΔfluG strains were found to be comparable (Table 1, data in parentheses). Overall, sclR expression based on same-day comparison was constant for all strains except the ΔfluG strains on day 5, a time corresponding to initiation of conidiation in the ΔfluG strains, which had substantially lower levels of expression than on day 3 and day 7. TLC analysis indicated that both ΔfluG and fluG complemented strains produced aflatoxin B1, although the amounts varied over the course of 7 days (Fig. 3).
Table 1.
Transcript levels relative to the control strain level for conidiation and sclerotium regulatory genes in fluG deletion and complemented strainsa
| Strain | Expression level on day: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
brlA |
abaA |
wetA |
sclR |
|||||||||
| 3 | 5 | 7 | 3 | 5 | 7 | 3 | 5 | 7 | 3 | 5 | 7 | |
| ΔfluG1 | 0.25 | 243.88b (0.84)c | 0.37 (0.80) | 0.61 | 2.46 (0.67) | 0.51 (0.82) | 0.74 | 0.41 (2.11) | 0.83 (0.38) | 0.73 | 0.14 | 1.31 |
| ΔfluG2 | 0.70 | 298.35 (1.02) | 0.16 (0.35) | 0.14 | 13.65 (3.70) | 0.51 (0.83) | 0.85 | 0.51 (2.64) | 1.03 (0.47) | 1.15 | 0.01 | 1.16 |
| Comp1 | 0.68 | 1.64 | 0.19 | 0.82 | 5.02 | 0.50 | 2.31 | 0.27 | 0.81 | 0.79 | 0.64 | 1.00 |
| Comp2 | 1.11 | 2.45 | 0.01 | 1.06 | 5.33 | 0.05 | 2.16 | 0.45 | 0.92 | 0.88 | 0.73 | 1.06 |
Independent deletion and complemented strains. The expression of the control strain, KuPG, is 1.00.
Values are expression levels compared to those of the control strain sampled on the same day, that is, 3 day versus 3 day, 5 day versus 5 day, and 7 day versus 7 day.
Values in parentheses are expression levels compared to those of the control strain sampled 2 days earlier, that is, 5 day versus 3 day and 7 day versus 5 day.
Fig 3.
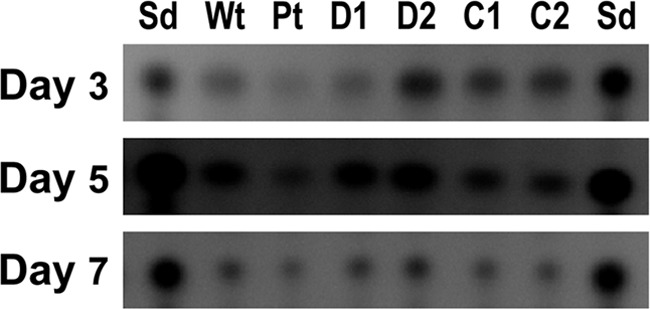
Time course semiquantitative TLC analyses of aflatoxin B1 production by A. flavus ΔfluG and fluG complemented strains. Wt, wild-type A. flavus CA14; Pt, control A. flavus KuPG no. 1; D, ΔfluG strains; C, fluG complemented strains with fluG under the gpdA (C1) and the fluG (C2) promoter, respectively. The numbers indicate independent isolates. Sd, aflatoxin B1 standard.
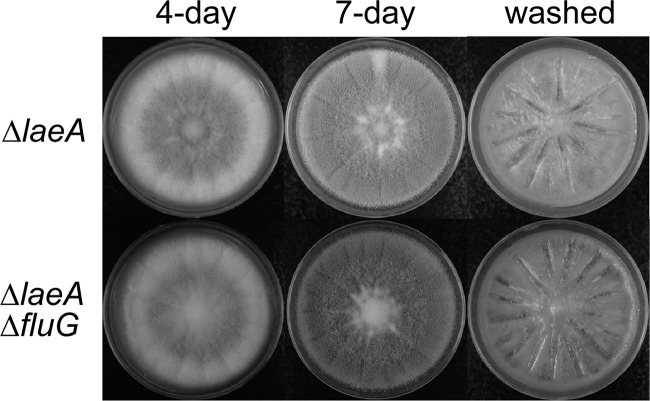
Production of conidia and sclerotia by ΔlaeA and ΔlaeA ΔfluG strains.
A previous study found that A. flavus ΔlaeA strains had developmental abnormalities, including increased conidiophore production and reduced conidial chain elongation when grown on PDA. These alterations gave dense, velvet-like colonies (11). To examine if fluG deletion affects development in ΔlaeA strains, we grew ΔlaeA ΔfluG strains on Wickerham agar plates. The ΔlaeA strains exhibited the same colony morphology and produced amounts of conidia comparable to those produced by the control strain (1.68 × 109 ± 0.09 × 109 versus 1.53 × 109 ± 0.21 × 109), but they did not produce sclerotia (Fig. 4). Deletion of fluG in the ΔlaeA strain caused only a moderate further decrease in conidiation (1.30 × 109 ± 0.04 × 109) compared to that of the ΔfluG strains. Like the ΔlaeA strains, ΔlaeA ΔfluG strains lacked the ability to produce sclerotia (Fig. 4).
Fig 4.
Morphologies of A. flavus ΔlaeA and ΔlaeA ΔfluG strains on Wickerham medium grown at 30°C in the dark for the specified numbers of days.
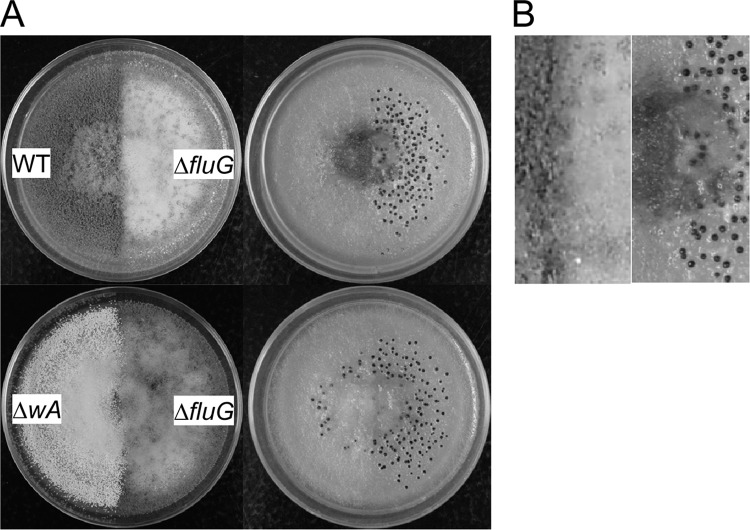
Coculture of fluG-intact and ΔfluG strains has no effect on resulting colony morphology.
Coculture of the ΔfluG strain with wild-type CA14 or with the CA14-derived white ΔwA strain, a strain that produces only a few sclerotia (12), failed to remediate the abnormalities in conidiation and sclerotial production of the ΔfluG strain (Fig. 5). At the region of contact, the ΔfluG strain still exhibited decreased conidiation and increased sclerotial production.
Fig 5.
Coculture of fluG-positive and ΔfluG strains. Cultures were grown on PDA at 30°C for 5 days in the dark. (A) Morphology before (left) and after (right) conidia were washed off the PDA plates. (B) Close-up of the boundary of the wild-type and ΔfluG strains.
DISCUSSION
Studies have shown that the A. nidulans fluG gene plays an important role in the initiation of conidiation and subsequent sterigmatocystin production (2, 27, 38). Unlike the A. nidulans fluffy ΔfluG strains, which are unable to conidiate (43), A. flavus ΔfluG strains, compared to the wild-type strain, show only delayed and decreased conidiation. The delayed conidiation in the A. flavus ΔfluG strains can be attributed to delayed expression of brlA. An approximately 2-day delay in brlA expression was observed when the ΔfluG strains were compared to the control and complemented strains (Table 1, 5 day versus 3 day). Fluffy mutants of A. nidulans exhibit altered brlA expression (2, 42); the level of BrlA reduction likely is more severe than that in A. flavus, resulting in an inability to form conidiophores. This halt in development in A. nidulans ΔfluG mutants also explains their inability to make secondary metabolites. For A. flavus, the delayed conidiation in the ΔfluG strains likely causes metabolic diversion, consequently shifting precursors, such as acetates, normally channeled to conidial pigment production to sclerotial formation (8, 41). The fluG complemented strains with the strong A. nidulans gpdA promoter conidiate earlier than those complemented strains under the control of the native fluG promoter, as evidenced by denser conidiation at and around the inoculation site on PDA (see Fig. S1 in the supplemental material). This earlier initiation of conidiation probably diverts the metabolic flux toward conidial formation, resulting in no sclerotia being produced, in contrast to the small amounts of sclerotia produced by the latter type of fluG complemented strains and the wild-type strain (Fig. 1B) (9). Conidiation and sclerotial production probably share common regulatory factors that enable the shift toward a developmental process dependent on either physiological or environmental conditions. A. flavus FluG may affect a signaling system that has opposite effects on conidial and sclerotial development.
Deletion of fluG in the A. flavus ΔlaeA strain decreases conidiation, which further confirms the positive effect of FluG on conidiation. LaeA is a global regulator of secondary metabolism in many fungi, and in A. nidulans, it forms a velvet complex with the developmental regulators VeA, VelB, and VosA (5). In A. nidulans and A. flavus, LaeA does not play a predominant role in the regulation of conidiation (11, 24). However, differences in function have been reported for the two LaeA regulators. A. nidulans LaeA inhibits formation of sexual cleistothecia in the light (37), while A. flavus LaeA is required for sclerotial formation in the dark (24) (Fig. 4). A. flavus FluG probably functions to fine tune the conidiation process by regulating the brlA expression level in response to certain physiological or developmental cues (signals). In the presence of LaeA, FluG may indirectly decrease or inhibit sclerotial formation, depending on the metabolic status of the fungus.
The A. flavus FluG amino acid sequence shows about 70% identity to those of A. nidulans and A. fumigatus (see Fig. S2 in the supplemental material). Aspergillus FluG is a fusion protein composed of an N-terminal amidohydrolase domain that shares homology with nodulins (Nod) and a C-terminal prokaryotic glutamine synthetase (GS) type I domain. Proteins similar to FluG are also found in plants (14, 30). FluG is predicted to be capable of carrying out condensation of glutamate with an amino-group-containing molecule and subsequent hydrolysis; the signaling molecule had been proposed to be aminobutyrate (22). However, no catalytic activity has been reported for FluG or FluG-like proteins (35). A. nidulans FluG does not directly synthesize but is associated with the synthesis of the diffusible signaling molecule dehydroaustinol (26, 36). The A. flavus coculture results suggest that, unlike A. nidulans, FluG is not associated with production of a diffusible effector metabolite (Fig. 5). An extensive search for potential dehydroaustinol biosynthesis clusters in the A. flavus genome did not find possible candidates. This suggests that the signaling pathways related to conidiation in the two related aspergilli must be different.
A. nidulans fluG fluffy strains have mutations primarily in the GS coding region (26), and the entire N-terminal half of A. nidulans FluG is dispensable without affecting conidiation (15). This suggests that FluG activity is mainly associated with the C-terminal GS domain. Since conidiation, sclerotial production, and secondary metabolism in A. flavus may be coordinated by the velvet complex and other interacting proteins, we performed tests to determine if FluG is able to interact with LaeA and VeA. Yeast two-hybrid assays showed that the GS domain of A. flavus FluG interacts with full-length LaeA and with the N-terminal half of LaeA, but not with VeA (see Fig. S3 in the supplemental material). However, we were unable to substantiate this specific interaction by bimolecular fluorescence complementation (BiFC) in fungal cells (data not shown). In A. nidulans, the fluG-dependent signal molecule is produced in the dark only in the strain with the veA1 mutation, and VeA appears to function as a negative regulator of conidiation in the dark at the posttranscriptional level (43). These observations suggest that a FluG-VeA interaction probably exists. Such an interaction may not occur in A. flavus, as suggested by the yeast two-hybrid results, because A. flavus normally conidiates in the dark. The Arabidopsis NodGS protein is part of a large heterogeneous protein complex (14). FluG-dependent conidiation in A. nidulans appears to be regulated by a suppressor, SfgA (38). FluG and other FluG-like proteins thus likely engage in protein-protein interactions. Whether FluG is an interacting partner of the Aspergillus velvet complex warrants further investigation.
While the downstream central pathway of conidiation regulated by brlA (i.e., abaA and wetA) is common to A. nidulans and A. fumigatus, the upstream pathways that activate brlA are distinct in the two species (28, 40, 44). In both A. nidulans and A. fumigatus, deletion of brlA eliminates conidiation completely, whereas deletion of fluG in A. fumigatus, as in our study, only reduces conidiation. In addition, in A. nidulans, deletion of fluG abolishes sterigmatocystin production, but deletion of fluG in A. flavus did not affect aflatoxin biosynthesis significantly (Fig. 3). Whole-genome comparison has concluded that A. fumigatus and A. oryzae/A. flavus are more closely related to each other than to A. nidulans (18). Our results suggest that the function of fluG in A. flavus is markedly different than in A. nidulans and that such potential variability should be taken into account when comparing the roles of developmental regulatory factors in different fungi.
Supplementary Material
Footnotes
Published ahead of print 17 August 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Adams TH, Boylan MT, Timberlake WE. 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54:353–362 [DOI] [PubMed] [Google Scholar]
- 2. Adams TH, Hide WA, Yager LN, Lee BN. 1992. Isolation of a gene required for programmed initiation of development by Aspergillus nidulans. Mol. Cell. Biol. 12:3827–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aguirre J. 1993. Spatial and temporal controls of the Aspergillus brlA developmental regulatory gene. Mol. Microbiol. 8:211–218 [DOI] [PubMed] [Google Scholar]
- 4. Andrianopoulos A, Timberlake WE. 1994. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol. Cell. Biol. 14:2503–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bayram O, et al. 2008. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320:1504–1506 [DOI] [PubMed] [Google Scholar]
- 6. Bok JW, Keller NP. 2004. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3:527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Breakspear A, Momany M. 2007. Aspergillus nidulans conidiation genes dewA, fluG, and stuA are differentially regulated in early vegetative growth. Eukaryot. Cell 6:1697–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Butler MJ, Gardiner RB, Day AW. 2009. Melanin synthesis by Sclerotinia sclerotiorum. Mycologia 101:296–304 [DOI] [PubMed] [Google Scholar]
- 9. Chang P-K, Bennett JW, Cotty PJ. 2002. Association of aflatoxin biosynthesis and sclerotial development in Aspergillus parasiticus. Mycopathologia 153:41–48 [DOI] [PubMed] [Google Scholar]
- 10. Chang P-K, Ehrlich KC, Hua SS. 2006. Cladal relatedness among Aspergillus oryzae isolates and Aspergillus flavus S and L morphotype isolates. Int. J. Food Microbiol. 108:172–177 [DOI] [PubMed] [Google Scholar]
- 11. Chang P-K, et al. 2012. Effects of laeA deletion on Aspergillus flavus conidial development and hydrophobicity may contribute to loss of aflatoxin production. Fungal Biol. 116:298–307 [DOI] [PubMed] [Google Scholar]
- 12. Chang P-K, Scharfenstein LL, Wei Q, Bhatnagar D. 2010. Development and refinement of a high-efficiency gene-targeting system for Aspergillus flavus. J. Microbiol. Methods 81:240–246 [DOI] [PubMed] [Google Scholar]
- 13. Cotty PJ. 1989. Virulence and cultural characteristics of two Aspergillus flavus strains pathogenic on cotton. Phytopathology 79:808–814 [Google Scholar]
- 14. Doskocilova A, et al. 2011. A nodulin/glutamine synthetase-like fusion protein is implicated in the regulation of root morphogenesis and in signalling triggered by flagellin. Planta 234:459–476 [DOI] [PubMed] [Google Scholar]
- 15. D'Souza CA, Lee BN, Adams TH. 2001. Characterization of the role of the FluG protein in asexual development of Aspergillus nidulans. Genetics 158:1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duran RM, Cary JW, Calvo AM. 2007. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 73:1158–1168 [DOI] [PubMed] [Google Scholar]
- 17. Etxebeste O, Garzia A, Espeso EA, Ugalde U. 2010. Aspergillus nidulans asexual development: making the most of cellular modules. Trends Microbiol. 18:569–576 [DOI] [PubMed] [Google Scholar]
- 18. Galagan JE, et al. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115 [DOI] [PubMed] [Google Scholar]
- 19. Geiser DM, Timberlake WE, Arnold ML. 1996. Loss of meiosis in Aspergillus. Mol. Biol. Evol. 13:809–817 [DOI] [PubMed] [Google Scholar]
- 20. Hicks JK, Yu JH, Keller NP, Adams TH. 1997. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA Gα protein-dependent signaling pathway. EMBO J. 16:4916–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Horn BW, Moore GG, Carbone I. 2009. Sexual reproduction in Aspergillus flavus. Mycologia 101:423–429 [DOI] [PubMed] [Google Scholar]
- 22. Iyer LM, Abhiman S, Maxwell Burroughs A, Aravind L. 2009. Amidoligases with ATP-grasp, glutamine synthetase-like and acetyltransferase-like domains: synthesis of novel metabolites and peptide modifications of proteins. Mol. Biosyst. 5:1636–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jin FJ, et al. 2011. SclR, a basic helix-loop-helix transcription factor, regulates hyphal morphology and promotes sclerotial formation in Aspergillus oryzae. Eukaryot. Cell 10:945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kale SP, et al. 2008. Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus. Fungal Genet. Biol. 45:1422–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim H, et al. 2002. The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet. Biol. 37:72–80 [DOI] [PubMed] [Google Scholar]
- 26. Lee BN, Adams TH. 1994. The Aspergillus nidulans fluG gene is required for production of an extracellular developmental signal and is related to prokaryotic glutamine synthetase I. Genes Dev. 8:641–651 [DOI] [PubMed] [Google Scholar]
- 27. Lee BN, Adams TH. 1996. FluG and FlbA function interdependently to initiate conidiophore development in Aspergillus nidulans through brlA beta activation. EMBO J. 15:299–309 [PMC free article] [PubMed] [Google Scholar]
- 28. Mah JH, Yu JH. 2006. Upstream and downstream regulation of asexual development in Aspergillus fumigatus. Eukaryot. Cell 5:1585–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marshall MA, Timberlake WE. 1991. Aspergillus nidulans wetA activates spore-specific gene expression. Mol. Cell. Biol. 11:55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mathis R, Gamas P, Meyer Y, Cullimore JV. 2000. The presence of GSI-like genes in higher plants: support for the paralogous evolution of GSI and GSII genes. J. Mol. Evol. 50:116–122 [DOI] [PubMed] [Google Scholar]
- 31. Mirabito PM, Adams TH, Timberlake WE. 1989. Interactions of three sequentially expressed genes control temporal and spatial specificity in Aspergillus development. Cell 57:859–868 [DOI] [PubMed] [Google Scholar]
- 32. Mooney JL, Hassett DE, Yager LN. 1990. Genetic analysis of suppressors of the veA1 mutation in Aspergillus nidulans. Genetics 126:869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ni M, Yu JH. 2007. A novel regulator couples sporogenesis and trehalose biogenesis in Aspergillus nidulans. PLoS One 2:e970 doi:10.1371/journal.pone.0000970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Raper KB, Thom CA. 1968. A manual of the penicilla. Williams & Wilkins, Baltimore, MD [Google Scholar]
- 35. Rexer HU, Schaberle T, Wohlleben W, Engels A. 2006. Investigation of the functional properties and regulation of three glutamine synthetase-like genes in Streptomyces coelicolor A3(2). Arch. Microbiol. 186:447–458 [DOI] [PubMed] [Google Scholar]
- 36. Rodriguez-Urra AB, et al. 2012. Signaling the induction of sporulation involves the interaction of two secondary metabolites in Aspergillus nidulans. ACS Chem. Biol. 7:599–606 [DOI] [PubMed] [Google Scholar]
- 37. Sarikaya Bayram O, et al. 2010. LaeA control of velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet. 6:e1001226 doi:10.1371/journal.pgen.1001226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seo JA, Guan Y, Yu JH. 2006. FluG-dependent asexual development in Aspergillus nidulans occurs via derepression. Genetics 172:1535–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sewall TC, Mims CW, Timberlake WE. 1990. abaA controls phialide differentiation in Aspergillus nidulans. Plant Cell 2:731–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tao L, Yu JH. 2011. AbaA and WetA govern distinct stages of Aspergillus fumigatus development. Microbiology 157:313–326 [DOI] [PubMed] [Google Scholar]
- 41. Tsai HF, Wheeler MH, Chang YC, Kwon-Chung KJ. 1999. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J. Bacteriol. 181:6469–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wieser J, Lee BN, Fondon J, III, Adams TH. 1994. Genetic requirements for initiating asexual development in Aspergillus nidulans. Curr. Genet. 27:62–69 [DOI] [PubMed] [Google Scholar]
- 43. Yager LN, Lee HO, Nagle DL, Zimmerman JE. 1998. Analysis of fluG mutations that affect light-dependent conidiation in Aspergillus nidulans. Genetics 149:1777–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu J-H. 2010. Regulation of development in Aspergillus nidulans and Aspergillus fumigatus. Mycobiology 38:229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.